Abstract
TOR is a serine-threonine kinase that was originally identified as a target of rapamycin in Saccharomyces cerevisiae and then found to be highly conserved among eukaryotes. In Drosophila melanogaster, inactivation of TOR or its substrate, S6 kinase, results in reduced cell size and embryonic lethality, indicating a critical role for the TOR pathway in cell growth control. However, the in vivo functions of mammalian TOR (mTOR) remain unclear. In this study, we disrupted the kinase domain of mouse mTOR by homologous recombination. While heterozygous mutant mice were normal and fertile, homozygous mutant embryos died shortly after implantation due to impaired cell proliferation in both embryonic and extraembryonic compartments. Homozygous blastocysts looked normal, but their inner cell mass and trophoblast failed to proliferate in vitro. Deletion of the C-terminal six amino acids of mTOR, which are essential for kinase activity, resulted in reduced cell size and proliferation arrest in embryonic stem cells. These data show that mTOR controls both cell size and proliferation in early mouse embryos and embryonic stem cells.
TOR (target of rapamycin) was originally identified in two mutant Saccharomyces cerevisiae strains, TOR1-1 and TOR2-1, that are resistant to the growth-inhibiting effect of the immunophilin-immunosuppressant complex FKBP (FK506 binding protein) and rapamycin (17). TOR1 and TOR2 are large proteins (≈280 kDa) and are ≈70% identical (26, 28). Single-copy TOR orthologs have been identified in Caenorhabditis elegans (31), Drosophila melanogaster (40, 55), mammals (2, 6, 46, 47), and Arabidopsis thaliana (35).
Mammalian TOR (mTOR; also known as FRAP [FKBP12-rapamycin-associated protein], RAFT [rapamycin and FKBP12 target], and RAPT [rapamycin target]) belongs to the phosphoinositide kinase-related kinase (PIKK) family (21, 48). mTOR and other members of this family, including ATM, ATR/FPR, and DNA-PKcs, contain C-terminal regions with high similarity to the catalytic domains of phosphoinositide (PI)-3 kinase and PI-4 kinase (26, 28). However, PIKK members are not lipid kinases but rather function as serine-threonine kinases (4, 20). The PIKK proteins contain a short segment at the extreme C terminus that is essential for protein kinase activity and is not present in PI-3 and PI-4 kinases (51).
Cell culture studies have demonstrated that mTOR controls protein synthesis, in part by phosphorylating downstream substrates, including p70 s6 kinase (p70S6K1) (3, 5, 20) and eukaryotic initiation factor 4E (eIF4E) binding protein 1 (4E-BP1) (4, 5, 13, 15). p70S6K phosphorylates the 40S ribosomal protein S6 and is proposed to play a crucial role in the translation of 5′-terminal oligopyrimidine tract mRNAs, which primarily encode ribosomal proteins and components of the translation apparatus (22, 23). Phosphorylation of 4E-BP1 disrupts its binding to eIF4E, a protein that binds the 5′ cap structure of mRNA. Released eIF4E then forms a functional translation initiation complex with eIF4G, eIF4A, and eIF3 ribosomes, enhancing translation (29, 45). Inactivation of 4E-BP1 and family proteins has a profound effect on translation of mRNAs with complex 5′ untranslated regions, which often encode regulatory proteins such as protooncogenes (45). The recent discoveries of a 150-kDa binding partner of mTOR, named raptor (regulatory-associated protein of mTOR) (14, 27), and its Saccharomyces cerevisiae ortholog, Kog1p (30), have provided new insights about how mTOR regulates p70S6K1 and 4E-BP1 phosphorylation; it has been proposed that raptor functions as a scaffolding protein for mTOR-catalyzed phosphorylation of p70S6K1 and 4E-BP1 (7, 14, 39).
TOR has been implicated in the regulation of cell growth (cell size control). In S. cerevisiae, TOR1 and TOR2 act as nutrient sensors and control cell size and proliferation (1, 8). In Drosophila melanogaster, inactivation of either the TOR homologue or the S6K homologue resulted in embryonic lethality accompanied by smaller cell sizes (36, 40, 55). Flies with partial loss of function in these molecules are significantly smaller in body size than their wild-type counterparts. In the mouse, disruption of p70S6K1 led to smaller sizes of the whole body (49) and of pancreatic β cells (42). Rapamycin treatment of mammalian cells resulted in smaller cell size, which was rescued by expression of rapamycin-resistant mTOR, rapamycin-resistant S6K, or wild-type eIF4E (11). In addition, rapamycin treatment resulted in impaired G1 progression, which was also rescued by rapamycin-resistant mTOR, rapamycin-resistant p70S6K1, or wild-type eIF4E (10). These data suggest that mTOR and the downstream p70S6K1 and 4EBP1 are important in cell size control and G1 progression.
However, the in vivo functions of mTOR remain elusive. The only mouse model in which mTOR is genetically altered is the “flat-top” mutant, generated by random chemical mutagenesis with ethylnitrosourea (19). Mutant embryos failed to upregulate cell proliferation in the telencephalon, the anteriormost region of the forebrain (18). Mutants also failed to rotate around the embryonic body axis and died at midgestation (12.5 days post) coitum. The gene responsible for this phenotype was subsequently shown to be mTOR (19); the mutant mice have a single nucleotide mutation in an intron, which resulted in aberrant splicing and a decrease in mTOR kinase activity to ≈20% of the normal level. Importantly, cell size was not altered in these mutants. The flat-top mutant was phenocopied by intraperitoneal administration of rapamycin into pregnant mice between 5.5 and 8.5 days postcoitum. These data argue that mTOR plays a crucial role in cell proliferation but not in cell growth (cell size control), in telencephalic vesicles, and that mTOR is dispensable at the peri-implantation stage of development in mice.
However, the flat-top is not a null mutation of mTOR; approximately 5% of mTOR transcripts are normally spliced (19). It is therefore still uncertain whether mTOR plays a role in peri-implantation mouse development and in cell size control. To answer these questions, we disrupted the mTOR gene by homologous recombination in mouse embryos and embryonic stem (ES) cells. Our results show that mTOR plays more important roles than had been anticipated from studies with rapamycin treatment and the flat-top mutants.
MATERIALS AND METHODS
Targeted disruption of the mouse mTOR gene.
To replace the kinase domain of mTOR with a cassette carrying the internal ribosome entry site (IRES) and a fusion of the β-galactosidase and neomycin resistance genes (βgeo) (38), a targeting vector with promoter trap selection was designed. The 5′ arm (a 4.4-kbp fragment containing exons 38 to 44) was amplified by the Expand long-template PCR system (Roche) with primers mTor-k-u9951-SacII (5′-CTC TGT GCC GCG GTT TCC TAG AGT-3′) and mTor-k-L14287-SpeI (5′-TCT CCC ACT AGT ACA AGC GAG ATG-3′). The 3′ arm (a 4.4-kbp fragment containing exons 48 to 50) was amplified with primers mTor-k-u20388 (5′-CAC ATG CCT GCA GTC CTG TAG CTT-3′) and mTor-k-L24742(X) (5′-CTC GAG TAG CCC AGG TCT ATC ATT-3′). The IRES-βgeo cassette was ligated between the two PCR fragments. The targeting vector was linearized with SacII and introduced into RF8 ES cells by electroporation (34). Genomic DNA from G418-resistant colonies was screened for homologous recombination by Southern blotting and PCR.
Southern blot screening for mTOR kinase domain knockout.
For 5′ Southern blot analysis, genomic DNA was digested with EcoRI, separated on a 0.8% agarose gel, and transferred to a nylon membrane as described previously (53). A 560-bp 5′ probe was amplified from intron 36 with primers TOR-kinase-probe-s2 (5′-GAC ACA CCA GAA GAG GGC ATC GGA-3′) and TOR-kinase-probe-as2 (5′-CCA ACT GGA GGC GGG CAC AGG AGT-3′). Hybridization with this probe revealed a 9.4-kbp band from the wild-type locus and a 5.3-kbp band from the targeted locus. For 3′ Southern blotting, genomic DNA was digested with KpnI. A 1,010-bp 3′ probe was amplified from exon 51 to intron 53 with primers TOR-kin-probe-3′-se (5′-TGA CCC TCC TGC TCT CGC CTG TCC-3′) and TOR-kin-probe-3′-as (5′-ATG CCA CTC CAG CTC TCA GAA CTG-3′). Hybridization with this probe revealed a 14-kbp band from the wild-type locus and a 7.9-kbp band from the targeted locus.
Genotyping of mTOR kinase domain knockout mice by PCR.
Mouse genomic DNA was extracted from tail tips as previously described (52). A sense primer, finTOR-k-tailu 6671 (5′-GCG GCA GGA TGA ACG AGT GAT GC-3′), was designed from exon 47 to amplify both the wild-type and targeted loci. An antisense primer, βgeo-screening 1 (5′-AAT GGG CTG ACC GCT TCC TCG TGC TT-3′), was designed from the βgeo cassette to amplify the targeted locus. Another antisense primer, TOR-kin-tail-L 20636 (5′-GTG ATC CGC CTG CCT CTG CCT CCT GT-3′), was designed from intron 47 to amplify the wild-type locus. Amplification with these three primers produced an 803-bp band from the wild-type locus and a 468-bp band from the targeted locus.
Conditional deletion of the extreme C terminus of mTOR.
A second targeting vector was designed to conditionally delete the extreme C-terminal six amino acids, which are essential for kinase activity (44, 51) The vector consists of a 5′ arm (a 3.3-kbp fragment from intron 56, exon 57, and intron 57), a loxP site-flanked (floxed) 515-bp fragment from intron 57 and exon 58, an IRES-βgeo cassette, and a 3′ arm (a 2.6-kbp fragment from exon 58 and the 3′-flanking region). The 5′ arm was amplified by PCR with primers CoTOR-u29338-s-SacII (5′-CCG CGG ATC CTC TTT CTC CTG TGG GGT A-3′) and coTOR-L32598-a-NotI (5′-GCG GCC GCC TGA GCT CCC AGA GAT CTC TC-3′). The floxed fragment was amplified with primers coTOR-u32622-s (5′-CGT TAG CCC TTG ACT AGT GAT ACT-3′) and coTOR-L33149-a (5′-CCC ATC CTG CAA ATG TCG CCA CTG-3′). The 3′ arm was amplified with primers coTOR-u33173-s-XhoI (5′-CTC GAG CCA ACA TGG CTA AGA GAG TCT GTC-3′) and coTOR-L35801-a-XhoI (5′-CTC GAG CTT CCC AAC CGC TGA GGT ATG TGA-3′). The vector was linearized with SacII and introduced into RF8 cells by electroporation. G418-resistant colonies were screened for homologous recombination by Southern blot analysis. We generated homozygous ES cell clones by selecting a heterozygous clone with a high concentration (6 mg/ml) of G418 (37).
Southern blot screening for conditional knockout.
For 5′ Southern blot analysis, genomic DNA was digested with HindIII. The probe was the same as that used for 3′ Southern blotting for the kinase domain knockout. Hybridization with this probe yielded a 12-kbp band from the wild-type locus and a 9.3-kbp band from the targeted locus. For 3′ Southern blot analysis, genomic DNA was digested with BamHI. A 550-bp 3′ probe was amplified from intron 58 with primers condTOR3′probe-S2 (5′-TAA CCC AGT GCC TGT GTC CGC CTA-3′) and condTOR3′probe-AS2 (5′-GAT CTT CGG GAG AAA TGG TGT GTT-3′). Hybridization with this probe yielded a 9.9-kbp band from the wild-type locus and a 6.2-kbp band from the targeted locus.
Cre-mediated recombination in ES cells.
A fusion protein consisting of a histidine tag, Tat peptide, simian virus 40 nuclear localization signal, and Cre (HTNC) was produced in Escherichia coli as previously described (41). The protein was dialyzed against a 1:1 mixture of Dulbecco's modified Eagle's medium and phosphate-buffered saline (PBS) and sterilized by filtration. mTORFlox/Flox cells were trypsinized with 0.25% trypsin, washed once in PBS, and resuspended in medium containing 2 μM HTNC. Cells were then plated on mitomycin C-treated STO cells (2.5 × 105 cells per well) in 24-well plates. After incubation with HTNC for 20 h, cells were washed once with PBS and cultured in ES medium for 24, 48, and 72 h. At each time point, we extracted genomic DNA and checked for deletion of the floxed fragment by PCR. Cell number was determined with a Coulter counter, and cell sizes and cell cycle patterns were checked by flow cytometry.
Genotyping HTNC-treated mTORFLox/Flox ES cells by PCR.
Deletion of the floxed fragment in mTORFLox/FLox cells was tested by PCR with primers Con-c-TOR_7660_RT-PCR (5′-CAA CCC AAG TGG AGC TGC TTA TCA-3′), designed from exon 57, and βgeo-screening-AS-6 (5′-GCG GAA TTC TCT AGA GTC CAG ATC-3′), designed from the βgeo cassette. A 1,200-bp band was amplified from the floxed locus, and a 640-bp band was amplified from the deleted locus.
Recloning of HTNC-treated ES cells.
mTORFlox/Flox ES cells (2.5 × 105 cells per well of 24-well plates) were treated with HTNC (41) as described above. After a 20-h incubation with HTNC, cells were washed with PBS, trypsinized, and replated at 1,000 cells per 10-cm dish on mitomycin C-treated STO cells. After 15 days, colonies were picked and genotyped by PCR.
Cell cycle analyses with flow cytometry.
ES cells were trypsinized, neutralized with ES cell medium, and washed twice with PBS. They were then resuspended in 1 ml of PBS containing 0.2% Triton X-100 for nuclear isolation. RNase A (0.5 mg/ml; Nacalai Tesque) and propidium iodide (25 μg/ml; Sigma) were added for RNA degradation and DNA staining. Flow cytometry analysis was performed with a FACSCalibur (Becton Dickinson). The cell cycle was analyzed with ModiFit software (Becton Dickinson).
Rapamycin treatment of ES cells and blastocysts.
Rapamycin (Calbiochem) was dissolved in ethanol (1 mg/ml). When treating ES cells or blastocysts, rapamycin was added daily to the culture medium to final concentrations of 200 nM for blastocysts and 20 nM for ES cells. Ethanol at the same final concentration was used in negative controls.
RESULTS
Ubiquitous expression of mTOR in mouse tissues and ES cells.
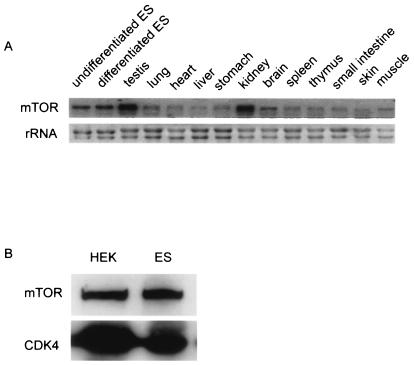
We first studied the expression patterns of mTOR in ES cells and somatic cells. Total RNA was isolated from undifferentiated ES cells, retinoic acid-treated ES cells, and 12 somatic organs of adult mice. Northern blot analysis with an mTOR probe detected transcripts in all the cells and organs (Fig. 1A), the highest expression being in undifferentiated ES cells, testis, and kidney. We also monitored mTOR expression at the protein level by Western blot analysis. Cell lysates were collected from human embryonic kidney (HEK) cells and ES cells. Immunoblotting with a monoclonal anti-mTOR antibody detected mTOR protein at comparable levels in both ES cells and HEK cells (Fig. 1B). These experiments showed that mTOR is expressed ubiquitously in most cells and tissues, including ES cells.
FIG. 1.
Expression profiles of mTOR. (A) Expression levels of mTOR transcripts in ES cells and in 12 adult mouse tissues were determined by Northern blot. (B) mTOR protein levels in human embryonic kidney (HEK) cells and ES cells were determined by Western blot analyses as previously described (16). CDK4 was used as a loading control.
Targeted disruption of the mouse mTOR gene in ES cells.
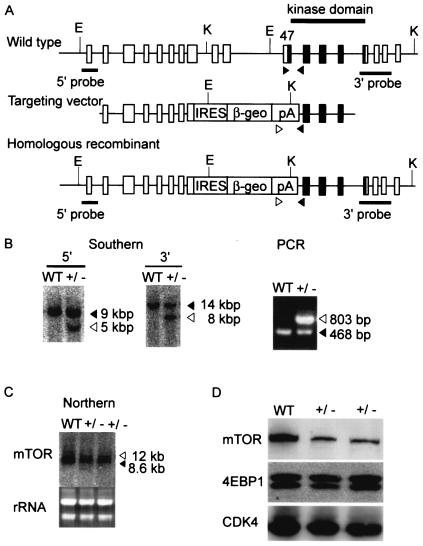
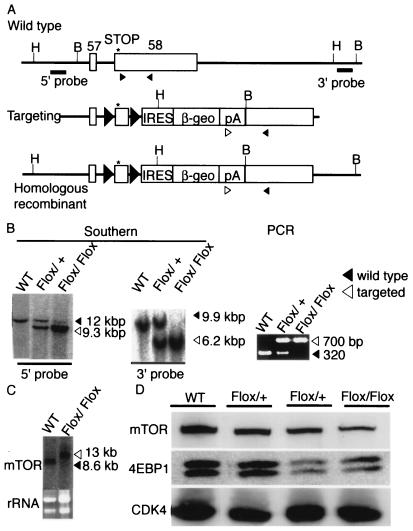
Based on the observation that ES cells express a high level of mTOR, we decided to use the promoter trap strategy (12) to achieve homologous recombination. Examination of sequences in the databases revealed that the mouse mTOR gene consists of 58 exons. We first constructed a targeting vector that replaced exons 2 and 3 with a fusion of the β-galactosidase and neomycin resistance (βgeo) genes. We expected that homologous recombination involving this vector would result in complete loss of mTOR function. However, we were unable to obtain any ES cell clones with homologous recombination from >600 colonies screened (data not shown). We therefore constructed another targeting vector that replaced exons 44 to 47 with the internal ribosome entry site (IRES)-βgeo cassette (Fig. 2A). Homologous recombination with this vector disrupts the C-terminal 20% of mTOR, including the conserved kinase domain.
FIG. 2.
Targeted disruption of the mouse mTOR gene. (A) We constructed a targeting vector that replaces exons 44 to 47 with an IRES-βgeo cassette. Shown are the locations of restriction enzyme sites (E, EcoRI; K, KpnI) and probes for Southern blot analyses. Exons corresponding to the kinase domain are shown by black boxes. Arrowheads indicate primers used for PCR. (B) Southern blot and PCR analyses. Solid arrows indicate bands corresponding to the wild-type locus, whereas open arrows indicate bands corresponding to the targeted locus. WT, wild type ES cells; +/−, heterozygous mutant ES cells. (C) Northern blot analysis showed that targeted cells expressed a larger transcript, corresponding to the fusion of mTOR and βgeo, in addition to the wild-type transcript. (D) Western blot analyses. The protein levels of mTOR, 4E-BP1, and CDK4 were determined as previously described (16).
The targeting vector was introduced into RF8 ES cells (34) by electroporation, and drug-resistant colonies were screened for homologous recombination. Two of 48 G418-resistant colonies were positive for homologous recombination by both Southern blot and PCR (Fig. 2B) analysis. Northern (Fig. 2C) and Western blot (Fig. 2D) analyses showed that mTOR expression was reduced, at both the RNA and protein levels, in these heterozygous clones. Northern blotting detected a larger band, corresponding to the fusion transcript of mTOR and IRES-βgeo, from which a truncated mTOR protein of ≈230 kDa was expected to be translated. However, no such band was detected, even after prolonged exposure of the Western blot (not shown), suggesting that the truncated protein was unstable or not translated. We did not observe differences in the phosphorylation status of 4E-BP1 (Fig. 2D). Cell proliferation, cell cycle, and cell size were also indistinguishable between the heterozygous and wild-type cells (not shown).
Generation of mTOR-deficient mice.
The two heterozygous ES clones were microinjected into C57BL/6 blastocysts to generate chimeric mice, and both were successfully transmitted through the germ line. The resulting mTOR heterozygous mutant mice were normal in gross appearance and body size and were fertile. To generate mTOR null mutant mice, heterozygous mutant mice were intercrossed, and the genotypes of 30-day-old offspring were determined by PCR. Of 111 offspring examined, 78 were heterozygous mutants and 33 were wild type. There were no homozygous mutant mice among the offspring. This result indicated that homozygous mTOR mutation resulted in embryonic lethality.
Early lethality in mTOR-deficient embryos.
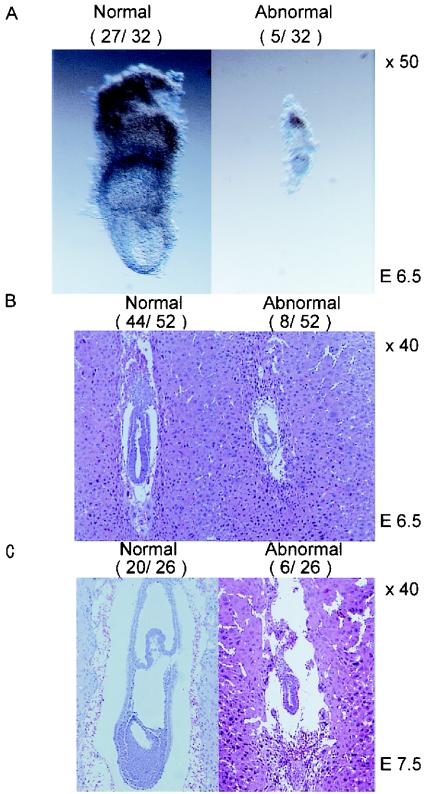
To determine the timing of lethality in the uterus, embryos from heterozygous intercrosses were analyzed and genotyped at 8.5, 9.5, 10.5, and 12.5 days postcoitum (Table 1). At all stages, 16.7 to 33.3% of deciduae either contained resorbed debris or were empty. At 6.5 days postcoitum, 5 of 32 embryos were much smaller than normal (Fig. 3A), but their genotypes could not be determined due to contamination with maternal tissue. Histological examination of 6.5-day-postcoitum embryos revealed that 44 of the 52 examined were normal and developed well-expanded epiblast as well as visceral and parietal endoderm. In the remaining eight embryos, these structures did exist but with significantly fewer cells (Fig. 3B). Among 26 embryos examined at 7.5 days postcoitum, 20 underwent normal gastrulation. In contrast, the other six embryos were similar in size and morphology to the 6.5-day-postcoitum mutant embryos (Fig. 3C). The high proportion of abnormal embryos indicated that these mutants were homozygous for mTOR deletion, and they died at around 6.5 to 7.5 days postcoitum due to impaired cell proliferation and gastrulation.
TABLE 1.
Genotypes of offspring from mTOR+/− intercrosses
| Day postcoitum | No. of embryos with genotype:
|
No. of embryos
|
|||
|---|---|---|---|---|---|
| +/+ | +/− | −/− | Resorbed | Empty | |
| 12.5 | 1 | 4 | 0 | 1 | |
| 10.5 | 6 | 12 | 0 | 8 | 1 |
| 9.5 | 10 | 13 | 0 | 3 | 4 |
| 8.5 | 19 | 29 | 0 | 7 | 7 |
| 3.5 | 3 | 19 | 10 | ||
| Total | 39 | 77 | 10 | 19 | 12 |
FIG. 3.
Embryonic lethality of mTOR mutants. (A) Embryos dissected at 6.5 days postcoitum. (B and C) Hematoxylin- and eosin-stained sections of paraffin-embedded embryos at 6.5 days postcoitum (B) and 7.5 days postcoitum (C). Numbers of embryos with normal and mutant (abnormal) morphologies are shown in parentheses.
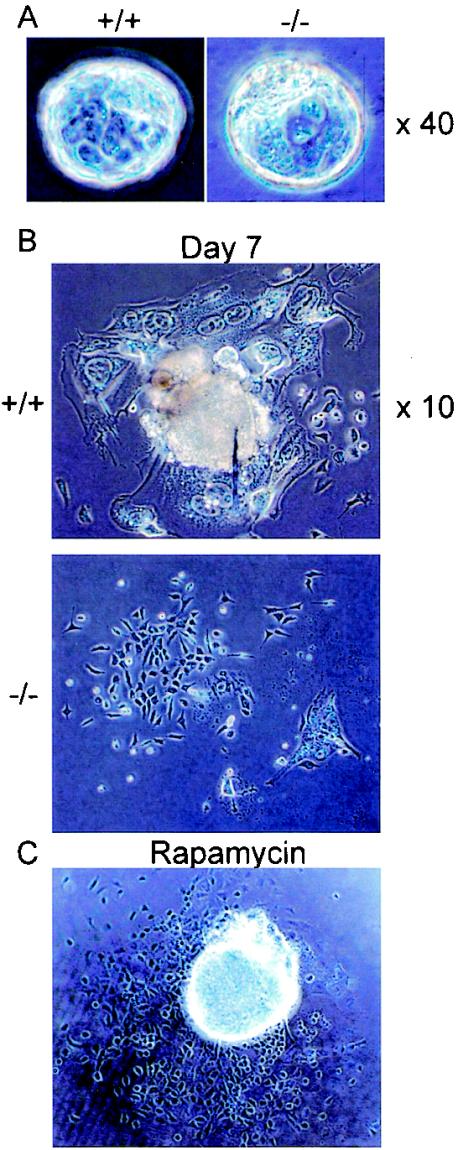
At the 3.5-day-postcoitum stage, 10 of 32 blastocysts were found to be homozygous by PCR (Table 1). They were indistinguishable from wild-type and heterozygous mutant embryos (Fig. 4A). However, the homozygous mutant blastocysts showed marked differences when cultured in vitro on gelatin-coated plates (Fig. 4B). Of 39 embryos cultured, 30 showed normal outgrowth of inner cell mass (ICM, pluripotent cells that produce all embryonic components) and trophoblasts (outer cells that produce placenta and extraembryonic tissues). In contrast, both ICM and trophoblasts failed to proliferate in nine blastocysts. Among the 30 embryos which proliferated properly, 20 were found to be heterozygous mutants and 10 were wild-type by PCR. Of the nine embryos that did not grow, five were homozygous mutants. The remaining four embryos could not be genotyped due to insufficient DNA recovery. These results indicate that neither ICM nor trophoblasts of mTOR homozygous mutant embryos are capable of proliferating in vitro.
FIG. 4.
Roles of mTOR in blastocyst proliferation. (A) Embryos were collected from heterozygous intercrosses at 3.5 days postcoitum and genotyped by PCR. Representative wild-type (+/+) and homozygous mutant (−/−) embryos are shown. (B) Wild-type and homozygous mutant blastocysts were cultured on gelatin-coated dishes for 7 days. (C) Wild-type blastocysts were cultured in vitro for 7 days with rapamycin (200 nM).
We then compared the effect of rapamycin with that of mTOR deletion. Blastocysts were cultured in ES medium with 200 nM rapamycin for 2 weeks. In all blastocysts, rapamycin effectively inhibited trophoblast outgrowth, mimicking the effect of mTOR deletion (Fig. 4C). In contrast, rapamycin did not impair ICM proliferation, suggesting that mTOR function in ICM is refractory to rapamycin.
Conditional inactivation of mTOR in ES cells.
To study the function of mTOR in ES cells, we first tried to generate homozygous cells from the heterozygous ES cells by selection with a higher concentration of G418 (37). However, no homozygous mutant clones were obtained (data not shown), suggesting that mTOR may be indispensable for ES cell proliferation and/or survival. We therefore undertook a conditional-knockout approach. It has been shown that several amino acids at the extreme C terminus of mTOR are essential for kinase activity (44, 51). We constructed a targeting vector in which the 5′ portion of the last exon, containing the C-terminal six amino acids, was flanked by two loxP sites (Fig. 5A).
FIG. 5.
Conditional deletion of the extreme C terminus of mTOR in ES cells. (A) The 3′ terminus of the mTOR gene, the targeting vector, and the correctly targeted mutant locus are shown. Solid triangles in the latter two sequences indicate loxP sites. Asterisks indicate translational termination codons of mTOR. The positions of restriction enzyme sites (B, BamH; H, HindIII) and probes for Southern blotting and the locations of PCR primers (arrowheads) are also shown. (B) Southern blot and PCR analyses. Solid arrowheads indicate bands corresponding to the wild-type locus, whereas open arrowheads indicate bands corresponding to the targeted locus. WT, wild-type ES cells; Flox/+, heterozygous targeted ES cells; Flox/Flox, homozygous targeted ES cells. (C) Northern blot analysis. (D) Western blot analyses showing the protein levels of mTOR, 4E-BP1, and CDK4.
The vector was introduced by electroporation into RF8 ES cells, and drug-resistant colonies were screened for homologous recombination. Two of 96 clones were positive by Southern blot and PCR analyses (Fig. 5B). Northern blotting showed the existence of chimeric transcripts consisting of mTOR and IRES-βgeo (Fig. 5C). To obtain ES cells in which both copies of the last exon were floxed (mTORFlox/Flox), mTORFlox/+ ES cells were cultured on gelatin-coated six-well plates in medium containing a high concentration of G418 (6 mg/ml). One ES clone was found to be mTORFlox/Flox by Southern blot (Fig. 5B), PCR (Fig. 5B) and Northern blot (Fig. 5C) analyses. This mTORFlox/Flox ES clone showed a decreased level of mTOR protein (Fig. 5D), suggesting that the modified 3′ untranslated region may cause decreased translation efficiency. In contrast, the mTORFlox/Flox ES clone was indistinguishable from wild-type cells in terms of the phosphorylation status of 4E-BP1 (Fig. 5D).
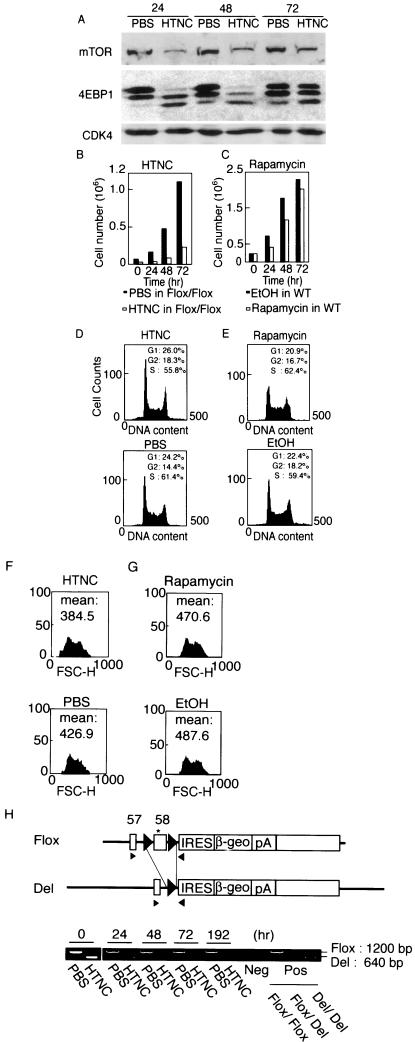
Addition of HTNC to the mTORFlox/Flox ES cells led to a further decrease in the mTOR protein level (Fig. 6A), suggesting that deletion of the six C-terminal amino acids impaired protein stability. HTNC treatment markedly decreased the phosphorylation status of 4E-BP1 in mTORFlox/Flox ES cells (Fig. 6A), confirming the importance of the extreme C terminus of mTOR for its kinase activity. mTORFlox/Flox ES cells barely proliferated after HTNC treatment (Fig. 6B). In contrast, HTNC had no such effect on wild-type ES cells (not shown). Flow cytometry analysis indicated impaired G1 progression in the HTNC-treated mTORFlox/Flox ES cells compared to control cells (Fig. 6D). HTNC also decreased cell sizes significantly, as judged from mean forward scatter height (Fig. 6F). In contrast, rapamycin treatment of ES cells decreased but did not block cell proliferation (Fig. 6C). The cell cycle was not significantly changed by rapamycin (Fig. 6E). Cell size was slightly decreased by rapamycin (Fig. 6G). These results showed that the effects of mTOR deletion were stronger than those induced by rapamycin.
FIG. 6.
Effect of mTOR inactivation in ES cells. (A) mTORFlox/Flox ES cells and wild-type ES cells were treated with a recombinant fusion protein consisting of a histidine tag, Tat peptide, simian virus 40 nuclear localization signal, and Cre recombinase (HTNC) or PBS. The proteins levels of mTOR, 4E-BP1, and CDK4 were determined by Western blot at 24, 48, and 72 h after HTNC treatment. (B) Cell numbers were counted every 24 h. (C) Wild-type ES cells were treated with rapamycin (20 nM) or solvent (ethanol) for the indicated time, and cell numbers were counted. (D and E) Cell cycle distribution was determined by flow cytometry analysis of the cells in panels B and C. (E and F) Cell size distribution (forward scatter, FSC-H) was determined by flow cytometry analysis of the cells in panels B and C. (H) Cre-mediated deletion of the floxed fragment was evaluated by PCR at the indicated time points after HTNC treatment. Arrowheads indicate primers used for PCR analyses. Del indicates the locus that underwent Cre-mediated deletion of the floxed region. The asterisk indicates the translation termination codon.
PCR analyses confirmed that the most of the floxed region was deleted by HTNC treatment (Fig. 6H), indicating that a large proportion of cells became homozygous for the C-terminal deletion (mTORDel/Del cells). However, the ratio of deleted band to floxed band intensity decreased during culture, confirming that mTORDel/Del cells grew significantly more slowly than either mTORDel/Flox cells or mTORFlox/Flox cells did. To obtain mTORDel/Del clones, we replated the HTNC-treated mTORFlox/Flox cells and isolated individual colonies. When HTNC-treated wild-type cells were plated, 240 colonies were obtained; in contrast, only 21 colonies were obtained from HTNC-treated mTORFlox/Flox cells. PCR analysis showed that 6 of the 21 clones remained mTORFlox/Flox and 12 were mTORDel/Flox. The other three clones did not grow, and no mTORDel/Del clones were obtained. These data, taken together, demonstrate that normal mTOR function is indispensable for ES cell proliferation and/or survival.
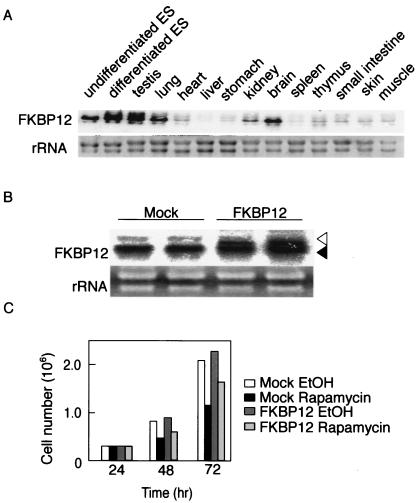
The observation that rapamycin showed weaker effects than did mTOR gene deletion in ES cells and ICM might have been due to the rapamycin receptor FKBP12 being expressed at an insufficient level in these cells. To test this possibility, we studied the expression levels of FKBP12 in ES cells and various adult tissues. Northern blot analysis showed ubiquitous expression of FKBP12 (Fig. 7A); notably, ES cells, testis, lung, and brain showed strong expression. To further confirm that low expression of FKBP12 was not the cause of the relative insensitivity of embryonic cells to rapamycin, we established ES cells expressing a higher level of FKBP12 (Fig. 7B). Despite a twofold increase in the expression of FKBP12, the effect of rapamycin on cell proliferation was not enhanced at all in these cells (Fig. 7C).
FIG. 7.
FKBP12 expression in ES cells. (A) Tissue distribution of FKBP12 transcripts was determined by Northern blot analysis. (B) Northern blot analysis confirming transgene expression of FKBP12 in MG1.19 ES cells. The open arrowhead indicates expression from the transgene, while the solid arrowhead indicates endogenous expression. As negative controls, cells were transfected with the parent plasmid (mock). (C) Effect of rapamycin (20 nM) on proliferation of cells transfected with the parent plasmid (mock) and the FKBP12-expressing plasmid.
DISCUSSION
In this study, we examined the functions of mTOR in vivo by gene targeting technology. Homozygous deletion of the kinase domain resulted in embryonic lethality shortly after implantation. Neither embryonic nor extraembryonic tissues proliferated properly in mutant embryos. Homozygous mutant blastocysts looked normal, but both ICM and trophoblasts failed to proliferate when cultured in vitro. Conditional deletion of the extreme C terminus, which is essential for mTOR kinase activity, decreased cell size and blocked proliferation in ES cells. These results demonstrate that in both early mouse embryos and ES cells, mTOR plays critical roles in cell proliferation and growth, as is the case in S. cerevisiae (1) and D. melanogaster (36, 55).
The phenotypes revealed by gene targeting were more severe than those induced by the mTOR inhibitor rapamycin. We showed that rapamycin treatment of blastocysts impaired proliferation of trophobasts but not of ICM. Hentges et al. (19) showed that intraperitoneal injection of rapamycin into pregnant female mice from 5.5 to 8.5 days postcoitum resulted in decreased proliferation in telencephalic primordium. However, most regions of these embryos developed normally despite the rapamycin treatment, and cell size was not affected. In ES cells, we found that rapamycin slowed but did not block proliferation, which is consistent with previous work by others (24, 25), who postulated that mTOR is not essential in peri-implantation mouse embryos and ES cells. However, our results contradict this notion.
The reason for the discrepancy is not clear. One possible mechanism is that FKBP12, the rapamycin receptor, is expressed at an insufficient level in early embryos and ES cells. However, we found that FKBP12 was highly expressed in undifferentiated and differentiated ES cells and that overexpression of FKBP12 in ES cells did not increase their sensitivity to rapamycin. Another possibility is the existence of a rapamycin-insensitive function for mTOR. In S. cerevisiae, there are two TOR-containing complexes, TORC1 and TORC2; only the former is rapamycin sensitive (30). Whether this is also the case in mammals is not known.
Another simpler and more likely possibility is that rapamycin does not completely inhibit mTOR activity and that the residual activity is sufficient to support cell proliferation and growth in early embryos and ES cells. This possibility is supported by the observation that the phenotypes of our mice were more severe than those observed in flat-top mice (19), which possess an ethylnitrosourea-induced nucleotide substitution in an intron of mTOR and show impaired cell proliferation of telencephalic primordium, a phenotype similar to that observed in rapamycin-treated embryos. The point mutation in the intron caused several types of aberrant splicing, all of which yielded proteins with impaired kinase activity. However, 5% of the transcripts were normally spliced and produced wild-type proteins, which may be sufficient to maintain cell proliferation in early embryos. It is likely that a small proportion of mTOR activity escapes inhibition by rapamycin and supports proliferation and growth in early embryos and ES cells.
The mTOR null mutant mice in our study displayed a significantly more severe phenotype than those with a homozygous mutation in the gene encoding its substrate, p70S6K1 (42, 49). Mice deficient in S6K1 are viable and have significantly smaller body sizes, indicating important roles for the kinase in cell size control. In contrast, S6K2-deficient mice tend to be slightly larger (43). Mice lacking both genes showed a marked reduction in viability due to perinatal lethality (43). However, those studies also showed that S6K differs from its upstream molecule mTOR in that it is dispensable for proliferation of embryonic cells. This is in marked contrast to D. melanogaster, in which inactivation of either TOR (40, 55) or S6K (36) alone resulted in embryonic lethality. The difference is at least partly attributable to S6 phosphorylation by a mitogen-activated protein kinase-dependent kinase in mammals (43). In addition, other targets may also contribute to proliferation control by mTOR. For example, mTOR has been shown to phosphorylate directly the transcription activator STAT3 (54); the authors showed that serine phosphorylation of STAT3 and STAT-dependent transcription were rapamycin sensitive. mTOR may thus regulate cell proliferation through STAT3 and/or other downstream targets.
It has been shown that PI-3 kinase plays important roles in ES cell proliferation. Treatment of ES cells with PI-3 kinase inhibitors markedly reduced cell proliferation (24), while inactivation of PTEN, a phosphatidylinositol 3,4,5-triphosphate phosphatase, resulted in enhancement of the PI-3 kinase pathway and cell proliferation (9). Recently, we showed that ES cells express a constitutively active RAS protein (ERas) that specifically activates the PI-3 kinase pathway and promotes ES cell proliferation (50). All of these results show that the PI-3 kinase pathway is important in the control of ES cell proliferation.
On the other hand, little is known about the molecules downstream of PI-3 kinase that are responsible for proliferation control in ES cells. Our data indicate that mTOR is a good candidate for such a downstream target. A number of recent studies have demonstrated that the tumor suppressors TSC1 and TSC2 form a protein complex that inhibits mTOR and that the inhibition is relieved by PI-3 kinase and its downstream target AKT (33). Moreover, the small GTPase Rheb (Ras ortholog enriched in brain) was recently identified as a molecular target of TSC (32). Rheb is a positive regulator of growth in D. melanogaster but requires TOR to achieve growth stimulation. Subsequent work established that Drosophila and mammalian TSC2, which contain a C-terminal GTPase-activator domain, directly stimulate Rheb GTPase in vitro. The effectors used by Rheb to control TOR function are as yet unknown. Thus, the mTOR/TSC/Rheb pathway is likely to be an important downstream effector of PI-3 kinase that enhances ES cell proliferation and growth.
In summary, we generated and analyzed mTOR gene knockout mice in this study. Our data reveal more essential and profound functions for mTOR in peri-implantation mouse development and ES cells than have been shown by either rapamycin treatment or studies with flat-top mutant mice. These findings, together with work by other groups, indicate that the PI-3 kinase and mTOR pathways may be the principal controllers of cell growth and proliferation in embryonic cells.
Acknowledgments
We thank Junko Iida, Yukiko Samitsu, Ikuyo Takeyama, Takashi Iwasaki, and Rie Kato for technical and administrative assistance. We also thank Robert Farese, Jr., for RF8 cells and Hitoshi Niwa for various plasmids and cell lines essential for this study. We are grateful to Kazutoshi Takahashi, Yoshimi Tokuzawa, Masayosi Maruyama, and Kiichiro Tomoda for useful discussion. We thank Ian Smith for critical reading of the manuscript.
This work was supported in part by research grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (S.Y. and K.Y.), Uehara Memorial Foundation (S.Y.), Naito Foundation (S.Y.), and Sumitomo Research Foundation (S.Y.). This work was also supported in part by a Grant-in-Aid for 21st Century COE Research from the Ministry of Education, Culture, Sports, Science and Technology.
REFERENCES
- 1.Barbet, N. C., U. Schneider, S. B. Helliwell, I. Stansfield, M. F. Tuite, and M. N. Hall. 1996. TOR controls translation initiation and early G1 progression in yeast. Mol. Biol. Cell 7:25-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, E. J., M. W. Albers, T. B. Shin, K. Ichikawa, C. T. Keith, W. S. Lane, and S. L. Schreiber. 1994. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 369:756-758. [DOI] [PubMed] [Google Scholar]
- 3.Brown, E. J., P. A. Beal, C. T. Keith, J. Chen, T. B. Shin, and S. L. Schreiber. 1995. Control of p70 s6 kinase by kinase activity of FRAP in vivo. Nature 377:441-446. [DOI] [PubMed] [Google Scholar]
- 4.Brunn, G. J., C. C. Hudson, A. Sekulic, J. M. Williams, H. Hosoi, P. J. Houghton, J. C. Lawrence, Jr., and R. T. Abraham. 1997. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science 277:99-101. [DOI] [PubMed] [Google Scholar]
- 5.Burnett, P. E., R. K. Barrow, N. A. Cohen, S. H. Snyder, and D. M. Sabatini. 1998. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc. Natl. Acad. Sci. USA 95:1432-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiu, M. I., H. Katz, and V. Berlin. 1994. RAPT1, a mammalian homolog of yeast Tor, interacts with the FKBP12/rapamycin complex. Proc. Natl. Acad. Sci. USA 91:12574-12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi, K. M., L. P. McMahon, and J. C. Lawrence, Jr. 2003. Two motifs in the translational repressor PHAS-I required for efficient phosphorylation by mammalian target of rapamycin and for recognition by raptor. J. Biol. Chem. 278:19667-19673. [DOI] [PubMed] [Google Scholar]
- 8.Di Como, C. J., and K. T. Arndt. 1996. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 10:1904-1916. [DOI] [PubMed] [Google Scholar]
- 9.Di Cristofano, A., B. Pesce, C. Cordon-Cardo, and P. P. Pandolfi. 1998. Pten is essential for embryonic development and tumour suppression. Nat. Genet. 19:348-355. [DOI] [PubMed] [Google Scholar]
- 10.Fingar, D. C., C. J. Richardson, A. R. Tee, L. Cheatham, C. Tsou, and J. Blenis. 2004. mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol. Cell. Biol. 24:200-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fingar, D. C., S. Salama, C. Tsou, E. Harlow, and J. Blenis. 2002. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 16:1472-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedrich, G., and P. Soriano. 1991. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 5:1513-1523. [DOI] [PubMed] [Google Scholar]
- 13.Gingras, A. C., S. P. Gygi, B. Raught, R. D. Polakiewicz, R. T. Abraham, M. F. Hoekstra, R. Aebersold, and N. Sonenberg. 1999. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 13:1422-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hara, K., Y. Maruki, X. Long, K. Yoshino, N. Oshiro, S. Hidayat, C. Tokunaga, J. Avruch, and K. Yonezawa. 2002. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110:177-189. [DOI] [PubMed] [Google Scholar]
- 15.Hara, K., K. Yonezawa, M. T. Kozlowski, T. Sugimoto, K. Andrabi, Q. P. Weng, M. Kasuga, I. Nishimoto, and J. Avruch. 1997. Regulation of eIF-4E BP1 phosphorylation by mTOR. J. Biol. Chem. 272:26457-26463. [DOI] [PubMed] [Google Scholar]
- 16.Hara, K., K. Yonezawa, Q. P. Weng, M. T. Kozlowski, C. Belham, and J. Avruch. 1998. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem. 273:14484-14494. [DOI] [PubMed] [Google Scholar]
- 17.Heitman, J., N. R. Movva, and M. N. Hall. 1991. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253:905-909. [DOI] [PubMed] [Google Scholar]
- 18.Hentges, K., K. Thompson, and A. Peterson. 1999. The flat-top gene is required for the expansion and regionalization of the telencephalic primordium. Development 126:1601-1609. [DOI] [PubMed] [Google Scholar]
- 19.Hentges, K. E., B. Sirry, A. C. Gingeras, D. Sarbassov, N. Sonenberg, D. Sabatini, and A. S. Peterson. 2001. FRAP/mTOR is required for proliferation and patterning during embryonic development in the mouse. Proc. Natl. Acad. Sci. USA 98:13796-13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isotani, S., K. Hara, C. Tokunaga, H. Inoue, J. Avruch, and K. Yonezawa. 1999. Immunopurified mammalian target of rapamycin phosphorylates and activates p70 S6 kinase alpha in vitro. J. Biol. Chem. 274:34493-34498. [DOI] [PubMed] [Google Scholar]
- 21.Jacinto, E., and M. N. Hall. 2003. Tor signalling in bugs, brain and brawn. Nat. Rev. Mol. Cell. Biol. 4:117-126. [DOI] [PubMed] [Google Scholar]
- 22.Jefferies, H. B., S. Fumagalli, P. B. Dennis, C. Reinhard, R. B. Pearson, and G. Thomas. 1997. Rapamycin suppresses 5′TOP mRNA translation through inhibition of p70s6k. EMBO J. 16:3693-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jefferies, H. B., C. Reinhard, S. C. Kozma, and G. Thomas. 1994. Rapamycin selectively represses translation of the “polypyrimidine tract” mRNA family. Proc. Natl. Acad. Sci. USA 91:4441-4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jirmanova, L., M. Afanassieff, S. Gobert-Gosse, S. Markossian, and P. Savatier. 2002. Differential contributions of ERK and PI3-kinase to the regulation of cyclin D1 expression and to the control of the G1/S transition in mouse embryonic stem cells. Oncogene 21:5515-5528. [DOI] [PubMed] [Google Scholar]
- 25.Kawasome, H., P. Papst, S. Webb, G. M. Keller, G. L. Johnson, E. W. Gelfand, and N. Terada. 1998. Targeted disruption of p70(s6k) defines its role in protein synthesis and rapamycin sensitivity. Proc. Natl. Acad. Sci. USA 95:5033-5038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keith, C. T., and S. L. Schreiber. 1995. PIK-related kinases: DNA repair, recombination, and cell cycle checkpoints. Science 270:50-51. [DOI] [PubMed] [Google Scholar]
- 27.Kim, D. H., D. D. Sarbassov, S. M. Ali, J. E. King, R. R. Latek, H. Erdjument-Bromage, P. Tempst, and D. M. Sabatini. 2002. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110:163-175. [DOI] [PubMed] [Google Scholar]
- 28.Kunz, J., R. Henriquez, U. Schneider, M. Deuter-Reinhard, N. R. Movva, and M. N. Hall. 1993. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell 73:585-596. [DOI] [PubMed] [Google Scholar]
- 29.Lawrence, J. C., Jr., and R. T. Abraham. 1997. PHAS/4E-BPs as regulators of mRNA translation and cell proliferation. Trends Biochem. Sci. 22:345-349. [DOI] [PubMed] [Google Scholar]
- 30.Loewith, R., E. Jacinto, S. Wullschleger, A. Lorberg, J. L. Crespo, D. Bonenfant, W. Oppliger, P. Jenoe, and M. N. Hall. 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 10:457-468. [DOI] [PubMed] [Google Scholar]
- 31.Long, X., C. Spycher, Z. Han, A. Rose, F. Muller, and J. Avruch. 2002. TOR Deficiency in C. elegans Causes Developmental Arrest and Intestinal Atrophy by Inhibition of mRNA Translation. Curr. Biol. 12:1448. [DOI] [PubMed] [Google Scholar]
- 32.Manning, B. D., and L. C. Cantley. 2003. Rheb fills a GAP between TSC and TOR. Trends Biochem. Sci. 28:573-576. [DOI] [PubMed] [Google Scholar]
- 33.Marygold, S. J., and S. J. Leevers. 2002. Growth signaling: TSC takes its place. Curr. Biol. 12:R785-R787. [DOI] [PubMed] [Google Scholar]
- 34.Meiner, V. L., S. Cases, H. M. Myers, E. R. Sande, S. Bellosta, M. Schambelan, R. E. Pitas, J. McGuire, J. Herz, and R. V. Farese, Jr. 1996. Disruption of the acyl-CoA:cholesterol acyltransferase gene in mice: evidence suggesting multiple cholesterol esterification enzymes in mammals. Proc. Natl. Acad. Sci. USA 93:14041-14046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Menand, B., T. Desnos, L. Nussaume, F. Berger, D. Bouchez, C. Meyer, and C. Robaglia. 2002. Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc. Natl. Acad. Sci. USA 99:6422-6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montagne, J., M. J. Stewart, H. Stocker, E. Hafen, S. C. Kozma, and G. Thomas. 1999. Drosophila S6 kinase: a regulator of cell size. Science 285:2126-2129. [DOI] [PubMed] [Google Scholar]
- 37.Mortensen, R. M., D. A. Conner, S. Chao, A. A. Geisterfer-Lowrance, and J. G. Seidman. 1992. Production of homozygous mutant ES cells with a single targeting construct. Mol. Cell. Biol. 12:2391-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mountford, P., B. Zevnik, A. Duwel, J. Nichols, M. Li, C. Dani, M. Robertson, I. Chambers, and A. Smith. 1994. Dicistronic targeting constructs: reporters and modifiers of mammalian gene expression. Proc. Natl. Acad. Sci. USA 91:4303-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nojima, H., C. Tokunaga, S. Eguchi, N. Oshiro, S. Hidayat, K. Yoshino, K. Hara, N. Tanaka, J. Avruch, and K. Yonezawa. 2003. The mammalian target of rapamycin (mTOR) partner, raptor, binds the mTOR substrates p70 S6 kinase and 4E-BP1 through their TOR signaling (TOS) motif. J. Biol. Chem. 278:15461-15464. [DOI] [PubMed] [Google Scholar]
- 40.Oldham, S., J. Montagne, T. Radimerski, G. Thomas, and E. Hafen. 2000. Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin. Genes Dev. 14:2689-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peitz, M., K. Pfannkuche, K. Rajewsky, and F. Edenhofer. 2002. Ability of the hydrophobic FGF and basic Tat peptides to promote cellular uptake of recombinant Cre recombinase: a tool for efficient genetic engineering of mammalian genomes. Proc. Natl. Acad. Sci. USA 99:4489-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pende, M., S. C. Kozma, M. Jaquet, V. Oorschot, R. Burcelin, Y. Le Marchand-Brustel, J. Klumperman, B. Thorens, and G. Thomas. 2000. Hypoinsulinaemia, glucose intolerance and diminished beta-cell size in S6K1-deficient mice. Nature 408:994-997. [DOI] [PubMed] [Google Scholar]
- 43.Pende, M., S. H. Um, V. Mieulet, M. Sticker, V. L. Goss, J. Mestan, M. Mueller, S. Fumagalli, S. C. Kozma, and G. Thomas. 2004. S6K1−/−/S6K2−/− mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol. Cell. Biol. 24:3112-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peterson, R. T., P. A. Beal, M. J. Comb, and S. L. Schreiber. 2000. FKBP12-rapamycin-associated protein (FRAP) autophosphorylates at serine 2481 under translationally repressive conditions. J. Biol. Chem. 275:7416-7423. [DOI] [PubMed] [Google Scholar]
- 45.Rousseau, D., A. C. Gingras, A. Pause, and N. Sonenberg. 1996. The eIF4E-binding proteins 1 and 2 are negative regulators of cell growth. Oncogene 13:2415-2420. [PubMed] [Google Scholar]
- 46.Sabatini, D. M., H. Erdjument-Bromage, M. Lui, P. Tempst, and S. H. Snyder. 1994. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 78:35-43. [DOI] [PubMed] [Google Scholar]
- 47.Sabers, C. J., M. M. Martin, G. J. Brunn, J. M. Williams, F. J. Dumont, G. Wiederrecht, and R. T. Abraham. 1995. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J. Biol. Chem. 270:815-822. [DOI] [PubMed] [Google Scholar]
- 48.Schmelzle, T., and M. N. Hall. 2000. TOR, a central controller of cell growth. Cell 103:253-262. [DOI] [PubMed] [Google Scholar]
- 49.Shima, H., M. Pende, Y. Chen, S. Fumagalli, G. Thomas, and S. C. Kozma. 1998. Disruption of the p70s6k/p85s6k gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J. 17:6649-6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takahashi, K., K. Mitsui, and S. Yamanaka. 2003. Role of ERas in promoting tumour-like properties in mouse embryonic stem cells. Nature 423:541-545. [DOI] [PubMed] [Google Scholar]
- 51.Takahashi, T., K. Hara, H. Inoue, Y. Kawa, C. Tokunaga, S. Hidayat, K. Yoshino, Y. Kuroda, and K. Yonezawa. 2000. Carboxyl-terminal region conserved among phosphoinositide-kinase-related kinases is indispensable for mTOR function in vivo and in vitro. Genes Cells 5:765-775. [DOI] [PubMed] [Google Scholar]
- 52.Tokuzawa, Y., E. Kaiho, M. Maruyama, K. Takahashi, K. Mitsui, M. Maeda, H. Niwa, and S. Yamanaka. 2003. Fbx15 is a novel target of Oct3/4 but is dispensable for embryonic stem cell self-renewal and mouse development. Mol. Cell. Biol. 23:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamanaka, S., X. Y. Zhang, M. Maeda, K. Miura, S. Wang, R. V. Farese, Jr., H. Iwao, and T. L. Innerarity. 2000. Essential role of NAT1/p97/DAP5 in embryonic differentiation and the retinoic acid pathway. EMBO J. 19:5533-5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yokogami, K., S. Wakisaka, J. Avruch, and S. A. Reeves. 2000. Serine phosphorylation and maximal activation of STAT3 during CNTF signaling is mediated by the rapamycin target mTOR. Curr. Biol. 10:47-50. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, H., J. P. Stallock, J. C. Ng, C. Reinhard, and T. P. Neufeld. 2000. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 14:2712-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]