Abstract
Common fragile sites are loci that form chromosome gaps or breaks when DNA synthesis is partially inhibited. Fragile sites are prone to deletions, translocations, and other rearrangements that can cause the inactivation of associated tumor suppressor genes in cancer cells. It was previously shown that ATR is critical to fragile-site stability and that ATR-deficient cells have greatly elevated fragile-site expression (A. M. Casper, P. Nghiem, M. F. Arlt, and T. W. Glover, Cell 111:779-789, 2002). Here we demonstrate that mouse and human cells deficient for BRCA1, due to mutation or knockdown by RNA interference, also have elevated fragile-site expression. We further show that BRCA1 functions in the induction of the G2/M checkpoint after aphidicolin-induced replication stalling and that this checkpoint function is involved in fragile-site stability. These data indicate that BRCA1 is important in fragile-site stability and that fragile sites are recognized by the G2/M checkpoint pathway, in which BRCA1 plays a key role. Furthermore, they suggest that mutations in BRCA1 or interacting proteins could lead to rearrangements at fragile sites in cancer cells.
Common fragile sites are loci that exhibit site-specific gaps and breaks on metaphase chromosomes when cells are grown under conditions that partially inhibit DNA synthesis, such as folate deficiency or treatment with aphidicolin (11). These fragile sites extend over hundreds of kilobases, with gaps and breaks occurring throughout the regions. Following aphidicolin treatment, 80% of all gaps and breaks are seen at just 20 fragile sites, with FRA3B (3p14.2) and FRA16D (16q23) being the most frequently broken, or expressed, fragile sites (11). Whereas rare fragile sites, such as FRAXA within the FMR1 gene, arise from mutation at di- or trinucleotide repeats, common fragile sites are found in all individuals and represent a normal component of chromosome structure.
Fragile sites are so-called hot spots for sister chromatid exchanges, translocations, deletions, and plasmid integration in cultured cells following replication stress (12, 14, 32, 40). Numerous studies have also shown that common fragile sites are prone to deletions and rearrangements in many cancers (1, 17, 21, 28, 33), and they may play a role in some gene amplification and viral integration events (6, 16, 26, 41). Some fragile sites lie within putative tumor suppressor genes, such as FHIT at FRA3B and WWOX at FRA16D (30, 33), leading to the model that fragile-site instability is a contributing factor in tumorigenesis.
Determining the mechanisms of fragile-site instability is important in understanding normal chromosome structure and DNA replication as well as the instability found at fragile sites in tumor cells. Sequence analysis of common fragile sites has not revealed why they are unstable. However, all sites studied to date are relatively AT rich and contain more areas of high flexibility than non-fragile-site regions (22, 24-27). Studies examining replication timing at common fragile sites have shown that they are late replicating (19, 39). Following addition of aphidicolin, an inhibitor of DNA polymerase α, the regions replicate even later, with indications that they are unreplicated as late as G2 in some cells (19). Such late or delayed replication likely contributes to instability at fragile sites, and the presence of unreplicated DNA in G2 implicates the G2/M checkpoint as being important in the process.
It has been shown that the replication checkpoint protein ATR is important in maintaining fragile site stability. Cells lacking ATR demonstrate an 8- to 10-fold increase in fragile-site expression after aphidicolin treatment and show measurable fragile-site expression without addition of replication inhibitors (3). ATR plays a central role in stabilizing stalled replication forks and in the induction of the intra-S and G2/M cell cycle checkpoints after replication inhibition (2, 4, 5, 7, 29), suggesting that either or both of these checkpoints are involved in fragile-site stability. It was proposed that fragile sites are single-stranded, unreplicated regions on metaphase chromosomes caused by stalled or collapsed replication forks that may give rise to double-strand breaks (DSBs) on some chromosomes (3). As such, common fragile sites provide a cytological assay for studying the pathways affecting stalled replication.
The BRCA1 protein and the CHK1 kinase are two primary downstream targets of ATR and ATM phosphorylation in response to DNA damage (8, 10, 36, 47). Following replication stress, both the S-phase and G2/M checkpoints appear to be activated via CHK1 (9, 20). BRCA1 functions upstream of CHK1 in this pathway and has been shown to activate CHK1 kinase activity in response to DNA DSBs formed by ionizing radiation (IR) in vitro (46). It is known that, following IR, BRCA1 is phosphorylated on multiple sites by ATM, thereby contributing to proper cell cycle arrest at the intra-S and G2/M checkpoints (43, 44, 46). Several studies have identified specific amino acid residues within BRCA1 that are important for particular checkpoints. Xu et al. (43) have shown that BRCA1 phosphorylation on serine 1423 is necessary for the G2/M checkpoint, but not for the intra-S-phase checkpoint, after induction of DSBs with IR. They also determined that phosphorylation of serine 1387 is necessary for proper induction of the intra-S-phase checkpoint but not the G2/M checkpoint (44).
There is evidence that BRCA1 is involved in these same checkpoints in response to stalled replication forks. Stalled replication induced by treatment of cells with UV or hydroxyurea results in an alteration in the pattern of BRCA1 nuclear foci as well as dose-dependent BRCA1 phosphorylation during S phase (31, 34, 36). This phosphorylation is ATR dependent after stalled replication (36). It has been demonstrated that ATM and ATR phosphorylate BRCA1 on the same residues, including serines 1387 and 1423 (8, 10, 36). These results suggest that ATR activates both the intra-S and G2/M checkpoints in response to stalled replication forks in a manner analogous to that of ATM-dependent induction of these checkpoints after exposure to IR and suggest that BRCA1 is involved in this response.
Based on these observations in ATR-deficient cells and the role of BRCA1 in the activation of cell cycle checkpoints, we hypothesized that BRCA1 plays a role in common-fragile-site stability via checkpoint activation and that cells lacking BRCA1 would show increased gaps and breaks at common fragile sites. We tested this hypothesis by using three independent methods. We first studied BRCA1-mutated HCC1937 breast cancer cells and compared the fragile-site expression level to that seen in cells transfected with a wild-type BRCA1 expression construct. Second, we examined fragile-site expression in a mouse embryo fibroblast cell line lacking Brca1. Third, we evaluated fragile site expression by using RNA interference (RNAi) to create BRCA1 deficiency in HCT116 and HeLa cells. In all three cases, we determined that cells lacking BRCA1 expression have a two- to threefold-increased level of fragile-site expression compared to that of isogenic controls.
To further determine if the checkpoint function of BRCA1 is involved in fragile-site stability, we stably transfected HCC1937 cells with BRCA1 expression constructs that have defined mutations at key residues involved in specific checkpoint functions. We found that HCC1937 cells expressing a BRCA1 mutant that lacks the G2/M checkpoint were characterized by the same elevated levels of fragile-site expression as untransfected cells. These data indicate that BRCA1 is an important factor in fragile-site stability, specifically through its role in the G2/M checkpoint. These results provide additional insight into the mechanisms of fragile-site instability, both in normal cells and cells with compromised checkpoint pathways, and further define the role of BRCA1 in the cellular response to replication stalling.
MATERIALS AND METHODS
Cell culture and fragile-site analysis.
HCC1937 cells were grown in RPMI medium supplemented with 15% fetal bovine serum (FBS), glucose, sodium pyruvate, and HEPES. Mouse cells were grown in minimum essential medium alpha containing 10% FBS. HCT116 and HeLa cells were grown in Dulbecco's modified essential medium supplemented with 10% FBS. All cell lines were grown at 37°C in a humidified atmosphere containing 5% CO2.
Fragile sites were induced by exposure of cells to 0.2 or 0.3 μM aphidicolin for 24 h prior to harvest. Cells were harvested for chromosome preparation by standard conditions of 45 min of Colcemid treatment (50 ng/ml) followed by an 18-min incubation in 0.075 M KCl at 37°C and multiple changes of Carnoy fixative (3:1 methanol:glacial acetic acid). Cells were dropped onto slides and baked overnight at 60°C before Giemsa staining or fluorescent in situ hybridization (FISH) protocols were carried out.
Yeast artificial chromosome (YAC) and bacterial artificial chromosome (BAC) probes that map to fragile-site regions were used for FISH analysis, following standard protocols (41). YAC 850A6 was used to detect human FRA3B, BAC 264L1 (RP-11) was used to detect human FRA16D, and YAC 158E12 was used to detect mouse Fra14A2. Probes were labeled with biotin-14-dATP or digoxigenin-11-dUTP with a BioNick Translation kit (Invitrogen, Carlsbad, Calif.). FISH signals were visualized by incubation with fluorescein isothiocyanate (FITC)-conjugated avidin-DCS (Vector Laboratories, Burlingame, Calif.) and fluorescein-conjugated anti-avidin immunoglobulin G. Chromosomes were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories). FISH results were analyzed with a Zeiss Axioscope epifluorescence microscope and Quips PathVysion imaging software (Vysis Inc., Downers Grove, Ill.).
Transfections.
HCC1937 cells were transfected with wild-type and mutant BRCA1 expression constructs (43, 44) by using Fugene transfection reagent (Roche Applied Science, Indianapolis, Ind.). Ten micrograms of linearized plasmid were used to transfect 1.5 × 105 cells in a 100-mm-diameter plate. After transfection, cells were grown in the presence of 300 μg of active G418 (Sigma, Saint Louis, Mo.)/ml. Stable, G418-resistant clones were picked and tested for BRCA1 expression by reverse transcription-PCR and Western blot.
RNA interference.
Small inhibitory RNA (siRNA) sequence pools directed against BRCA1 and pooled scrambled control siRNA sequences were obtained from Dharmacon Research, Inc. (Lafayette, Colo.). Transfection of siRNAs into HCT116 and HeLa cells was carried out with Oligofectamine (Invitrogen) based on protocols provided by Dharmacon. These two cell lines were chosen because they can be transfected with high efficiency. HCT116 was also chosen because it has a relatively normal karyotype. Fragile-site expression was induced by the addition of aphidicolin for 24 h 2 days posttransfection.
Immunoprecipitation and Western blots.
Cellular extracts were prepared by lysing cells in radioimmunoprecipitation assay buffer and incubating them on ice for 30 min. To detect BRCA1 expressed from the construct without detecting endogenous, mutant BRCA1 in HCC1937 cells, we performed immunoprecipitation with a C-terminal-specific antibody. This antibody does not recognize the endogenous, mutant form of BRCA1 found in HCC1937 cells. Ten micrograms of a C-terminal BRCA1 antibody, Ab-3 (Oncogene Research, San Diego, Calif.), was added to 400 μg of cell lysate and was incubated overnight at 4°C. Immune complexes were collected with 100 μl of protein G agarose beads (Invitrogen) and were washed three times with lysis buffer. Protein separation was performed by using 10% Tris-HCl polyacrylamide gels (Bio-Rad). Fifty micrograms of whole-cell lysate or the immunoprecipitated protein was loaded in each lane. Gels were transferred to polyvinylidene difluoride membranes (Millipore, Billerica, Mass.) by a Trans-Blot SD Semi-Dry Transfer gel (Bio-Rad Laboratories, Hercules, Calif.). Antibody hybridization and chemiluminescence detection were performed according to standard protocols. BRCA1 protein was detected with αBRCA1 antibodies Ab-3 and Ab-4 (Oncogene Research), Ab-2 (NeoMarkers, Fremont, Calif.), and MS110 (35). Horseradish peroxidase-conjugated anti-mouse antibodies were obtained from Amersham (Piscataway, N.J.).
G2/M checkpoint analysis.
Cells were harvested after 24 h of exposure to 0.5 μM aphidicolin and fixed in 70% ethanol at −20°C. The cells were suspended in 100 μl of phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA) and 0.75 μg of a polyclonal antibody that specifically recognizes the phosphorylated form of histone H3 (Upstate Biotechnology, Lake Placid, N.Y.), and they were incubated for 3 h at room temperature. The cells were then rinsed with PBS containing 1% BSA and were incubated with FITC-conjugated goat anti-rabbit immunoglobulin G antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, Pa.) diluted at a ratio of 1:30 in PBS containing 1% BSA. After a 30-min incubation at room temperature in the dark, the cells were stained with propidium iodide (Sigma) and cellular fluorescence was measured by a FACSCalibur flow cytometer. The proportion of cells in mitosis after aphidicolin treatment was compared to the proportion of cells in mitosis without aphidicolin treatment.
Statistical analysis.
Total gaps-and-breaks data were analyzed with the Student's t test for equal or unequal variance. Variance for each data set was determined with the sample variance F test. Fisher's exact test (two sided) was used for analysis of specific fragile-site expression data.
RESULTS
Cells lacking functional BRCA1 have increased expression of common fragile sites.
Three separate experiments were conducted to determine if wild-type BRCA1 is necessary for fragile-site stability. After low-dose aphidicolin treatment (0.2 to 0.3 μM), chromosomes from cells lacking wild-type BRCA1 were examined for total gaps and breaks, most of which occur at common fragile sites in normal cells under these conditions (11). Two of the most frequently expressed common fragile sites, FRA3B and FRA16D, were also specifically evaluated by FISH probes (Fig. 1).
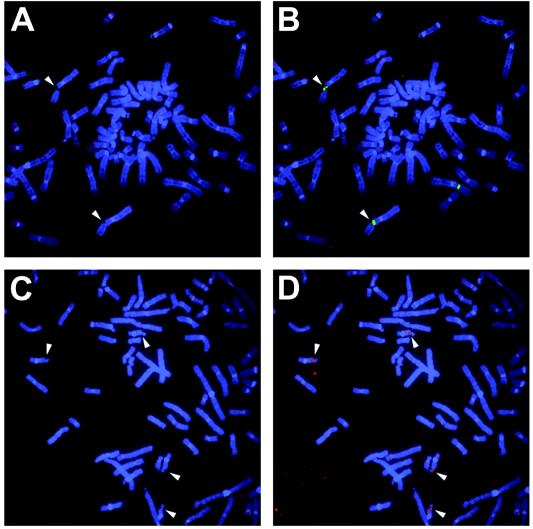
FIG. 1.
Examples of expressed common-fragile-site detection by FISH. (A) Partial metaphase of HCC1937 cells treated with 0.3 μM aphidicolin for 24 h. Chromosomes are stained with DAPI and demonstrate multiple fragile-site breaks. White arrows indicate positions of gaps and breaks at FRA3B. (B) Examples of FISH results on the same partial metaphase. FRA3B was probed with YAC 850A6 (green). White arrows indicate colocalization of FISH signal and broken FRA3B sites. (C) Partial metaphase of HCC1937 cells treated with 0.3 μM aphidicolin for 24 h. Chromosomes are stained with DAPI and demonstrate multiple fragile-site breaks. White arrows indicate positions of gaps and breaks at FRA16D. (D) Examples of FISH results on the same partial metaphase. FRA16D was probed with BAC 264L1 (red). White arrows indicate colocalization of FISH signal and broken FRA3B sites.
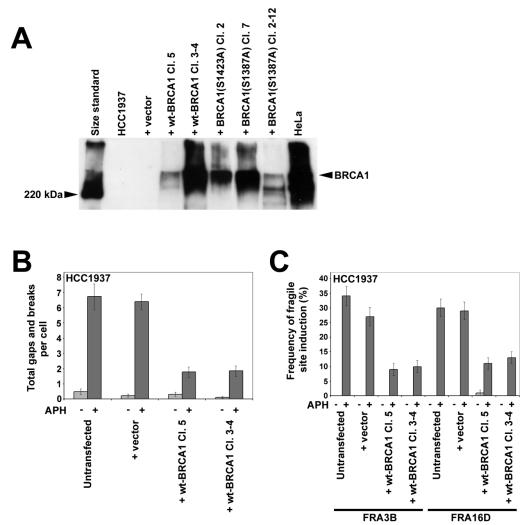
We first investigated fragile-site expression in HCC1937, a BRCA1-null breast cancer cell line (37). When treated with 0.3 μM aphidicolin for 24 h prior to harvesting, HCC1937 cells lacking functional BRCA1 showed an average of 6.7 gaps and breaks per cell. To determine if this breakage was enhanced by the lack of functional BRCA1, this experiment was conducted in parallel with isogenic control cells that had been stably transfected with a wild-type BRCA1 expression construct (Fig. 2A). Two independent HCC1937 clones expressing wild-type BRCA1 showed a fourfold decrease in total gaps and breaks compared to respective levels for untransfected HCC1937 and HCC1937 transfected with an empty vector (Fig. 2B). This difference is statistically significant (P < 0.001).
FIG. 2.
HCC1937 cells lacking BRCA1 have elevated fragile-site expression after aphidicolin treatment. (A) Western blot of BRCA1 immunoprecipitation from clones of HCC1937 stably transfected with wild-type BRCA1, BRCA1 (S1423A), or BRCA1 (S1387A) expression constructs or an empty vector. (B) Average total chromosomal gaps and breaks per cell in HCC1937 cells stably transfected with the indicated BRCA1 expression constructs after 24 h in the presence (dark gray) or absence (light gray) of 0.3 μM aphidicolin; n = 100 metaphases for each data set. Error bars indicate the 95% confidence interval. (C) Frequency (%) of gaps and breaks at specific fragile sites FRA3B and FRA16D in HCC1937 cells stably transfected with the indicated BRCA1 expression constructs after 24 h of treatment with 0 μM (light gray) or 0.3 μM (dark gray) aphidicolin; n = 88 to 108 sites examined. Fragile sites were identified by FISH with probes specific to these sites. Frequency of fragile-site induction is presented as the percentage of chromosome 3 or 16 homologs with breaks at FRA3B or FRA16D, respectively.
To verify that the increase in gaps and breaks in BRCA1−/− cells occurs at specific common fragile sites, the cell lines lacking functional BRCA1 were treated with aphidicolin and the expression (gaps and breaks) of two specific common fragile sites, FRA3B at 3p14 and FRA16D at 16q23, was examined by FISH analysis using YAC and BAC probes that map to these sites. Results are shown in Fig. 2C. Both of these sites were expressed at threefold lower frequencies in cells expressing wild-type BRCA1 compared to those of BRCA1−/− cells, a difference proportional to that seen with total gaps and breaks.
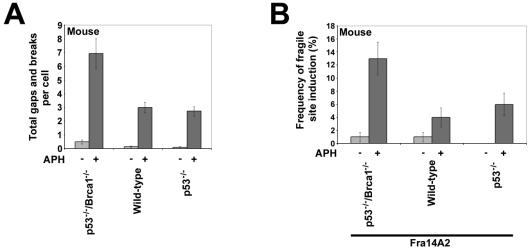
In the second set of experiments, we examined fragile-site expression in a mouse embryo fibroblast line with inactivating mutations in both p53 and Brca1 (48). A normal mouse embryo fibroblast line and a mouse embryo fibroblast line with biallelic inactivating mutations only in p53 were used as controls. Similar to HCC1937, cells expressing wild-type Brca1 showed more than a twofold decrease in aphidicolin-induced total gaps and breaks compared to levels of gaps and breaks for Brca1−/− cells (Fig. 3A). This difference is statistically significant (P < 0.001). No difference in total gaps and breaks was observed between wild-type and p53−/− cells (P = 0.152).
FIG. 3.
Mouse cells lacking BRCA1 have elevated fragile-site expression after aphidicolin treatment. (A) Average total chromosomal gaps and breaks per cell in mouse cells with the indicated genotypes after 24 h in the presence (dark gray) or absence (light gray) of 0.3 μM aphidicolin; n = 100 metaphases for each data set. Error bars indicate the 95% confidence interval. (B) Frequency (%) of gaps and breaks at specific fragile site Fra14A2 in mouse cells after 24 h of treatment with 0 μM (light gray) or 0.3 μM (dark gray) aphidicolin; n = 89 to 100 sites examined. The genotype of each cell line is indicated. Fragile sites were identified by FISH with a probe specific to Fra14A2. Fragile-site induction frequency is presented as the percentage of chromosome 14 homologs with breaks at Fra14A2.
The mouse ortholog of FRA3B, Fra14A2 (13), was examined by FISH with a YAC probe to this region. Wild-type and p53−/− cells had a two- to threefold decrease in aphidicolin-induced expression of Fra14A2 compared to expression in cells lacking Brca1 (Fig. 3B). This difference is proportional to that seen with total gaps and breaks and is significant (P = 0.035). In addition, there was no significant difference in Fra14A2 expression between wild-type and p53−/− cells (P = 0.747), suggesting that p53 has minimal involvement in fragile-site stability.
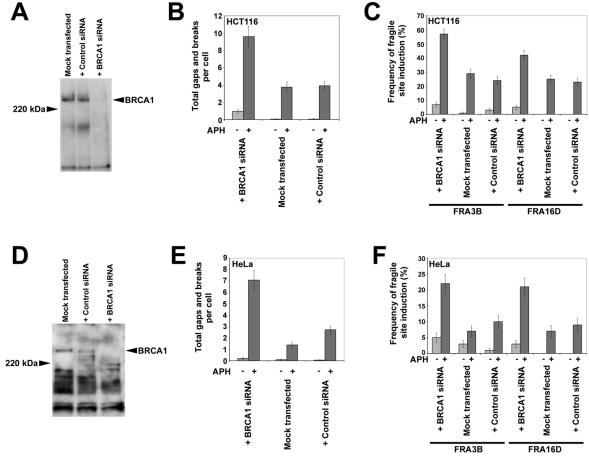
In the third set of experiments the effect of BRCA1 on fragile-site stability was evaluated in HCT116 and HeLa cells in which BRCA1 protein levels had been reduced through RNAi by using a siRNA pool specific to BRCA1. Reduction of BRCA1 protein levels was confirmed by Western blot (Fig. 4A and D). After aphidicolin treatment, HCT116 and HeLa cells transfected with BRCA1 siRNA were characterized by a threefold increase in total gaps and breaks compared to levels for control cells, a significant difference (P < 0.001 in both cases) (Fig. 4B and E). Induction of both FRA3B and FRA16D were also increased two- to threefold in cells targeted by BRCA1 siRNAs (Fig. 4C and F). As with total gaps and breaks, the differences in FRA3B and FRA16D expression were significant in both HCT116 (P < 0.001 and P = 0.016, respectively) and HeLa (P = 0.004 and P = 0.007, respectively).
FIG. 4.
Human cells with BRCA1 expression reduced via RNAi have elevated fragile-site expression after aphidicolin treatment. (A) Western blot probed with αBRCA1 antibodies showing reduced BRCA1 expression in HCT116 cells 48 h after transfection with BRCA1 SMARTpool siRNA. Untransfected cells and cells transfected with control siRNA show no reduction in BRCA1 expression. (B) Average total chromosomal gaps and breaks per cell in HCT116 cells after transfection with BRCA1 SMARTpool siRNA, no siRNA, or control siRNA; n = 100 metaphases for each data set. Error bars indicate the 95% confidence interval. RNAi reduction of BRCA1 levels was achieved 48 h before harvest. Fragile-site induction was achieved by addition of 0.2 μM aphidicolin 24 h before harvest. (C) Frequency (%) of gaps and breaks at specific fragile sites FRA3B and FRA16D in HCT116 cells after transfection with BRCA1 SMARTpool siRNA, no siRNA, or control siRNA. Fragile-site expression was measured after 24 h of treatment with 0 μM (light gray) or 0.3 μM (dark gray) aphidicolin; n = 80 to 101 sites examined. Frequency of fragile-site induction is presented as the percentage of chromosome 3 or 16 homologs with breaks at FRA3B or FRA16D, respectively. (D) Western blot probed with αBRCA1 antibodies showing reduced BRCA1 expression in HeLa cells 48 h after transfection with BRCA1 SMARTpool siRNA. Untransfected cells and cells transfected with control siRNA show no reduction in BRCA1 expression. (E) Average total chromosomal gaps and breaks per cell in HeLa cells after transfection with BRCA1 SMARTpool siRNA, no siRNA, or control siRNA; n = 100 metaphases for each data set. Error bars indicate the 95% confidence interval. RNAi reduction of BRCA1 levels was achieved 48 h before harvest. Fragile-site induction was achieved by addition of 0.2 μM aphidicolin 24 h before harvest. (F) Frequency (%) of gaps and breaks at specific fragile sites FRA3B and FRA16D in HeLa cells after transfection with BRCA1 SMARTpool siRNA, no siRNA, or control siRNA. Fragile-site expression was measured after 24 h of treatment with 0 μM (light gray) or 0.3 μM (dark gray) aphidicolin; n = 100 to 102 sites examined. Frequency of fragile-site induction is presented as the percentage of chromosome 3 or 16 homologs with breaks at FRA3B or FRA16D, respectively.
BRCA1 S1423 is necessary for the G2/M checkpoint in response to replication stalling via aphidicolin.
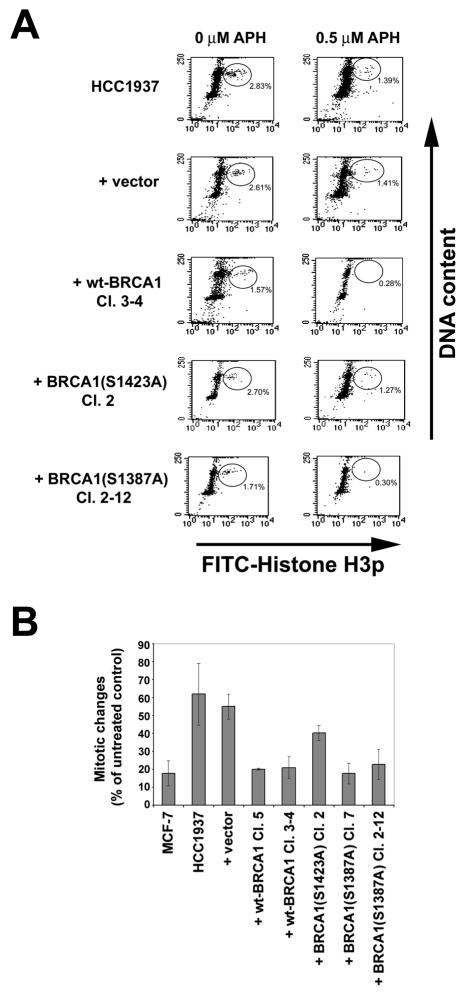
It has been demonstrated that BRCA1 induction of the G2/M checkpoint after IR is dependent on the phosphorylation of BRCA1 serine 1423 (43). In HCC1937 cells expressing a mutant BRCA1 that substitutes an alanine for this serine, the ability of BRCA1 to mediate the G2/M checkpoint is eliminated while leaving the intra-S checkpoint intact. In contrast, expressing a mutant BRCA1 that substitutes an alanine for serine 1387 eliminates the role of BRCA1 in the intra-S checkpoint while leaving the G2/M checkpoint intact (44). To determine if BRCA1 affects fragile-site stability via the G2/M checkpoint, it was first necessary to determine if serine 1423 is also required for induction of the G2/M checkpoint after cells undergo replication fork stalling via aphidicolin treatment.
HCC1937 cells lacking BRCA1 had reduced induction of the G2/M checkpoint after treatment with 0.5 μM aphidicolin as measured by flow cytometric analysis with staining for DNA content and histone H3 phosphorylation. In contrast, cells expressing wild-type BRCA1 induced the G2/M checkpoint after aphidicolin treatment to the same levels seen in a control cell line, MCF-7. The S1387A mutant was able to complement the aphidicolin-induced G2/M checkpoint defect in HCC1937 cells. However, the S1423A mutant failed to restore the aphidicolin-induced G2/M arrest (Fig. 5). It should be pointed out that cells lacking BRCA1 still showed a reduction in mitotic index after aphidicolin expression. This result likely indicates that cells lacking BRCA1 still retain some G2/M checkpoint function in response to aphidicolin. Alternatively, it is possible that aphidicolin treatment, which slows replication through toxic effect, may increase the proportion of cells in S phase, resulting in a reduction in mitotic cells. Regardless of the effect of other proteins, these results indicate that BRCA1 is involved in the G2/M checkpoint after aphidicolin treatment and that serine 1423 is necessary for this activity.
FIG. 5.
BRCA1 serine 1423, but not serine 1387, is necessary for G2/M checkpoint induction after aphidicolin treatment. (A) Flow cytometric profiles of cell cycle distribution of cells after 24 h in the presence of 0 and 0.5 μM aphidicolin (APH). Shown are HCC1937 cells stably transfected with constructs expressing wild-type BRCA1, BRCA1 containing serine-to-alanine mutations at serine 1423 or serine 1387, or vector alone. Cells were stained for DNA content with propidium iodide (y axis) and for histone H3 phosphorylation (x axis). The mitotic cell population is circled, and the percentage of total cells falling within that population is indicated. (B) Quantitation of flow cytometric profiles of cell cycle distribution following 24 h of exposure to 0.5 μM aphidicolin. Bars indicate the percentage of aphidicolin-treated cells that are in mitosis relative to untreated cells. Error bars indicate the standard deviation.
Cells expressing BRCA1 S1423A but not S1387A have an increased frequency of aphidicolin-induced common fragile sites.
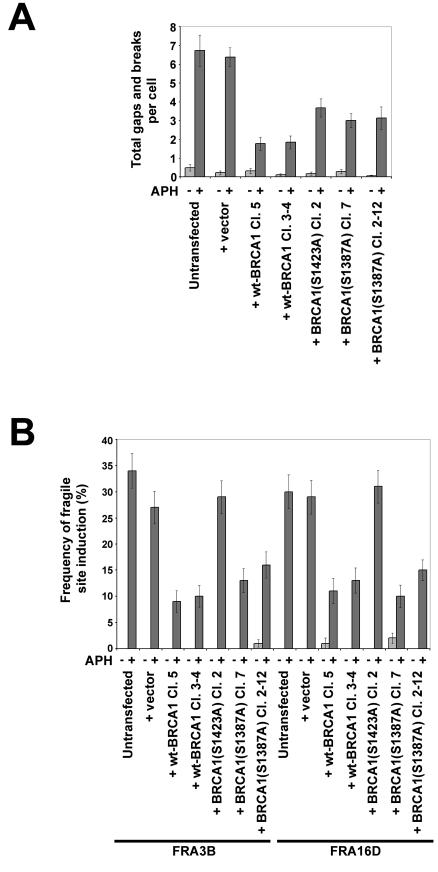
HCC1937 cells were stably transfected with constructs expressing each of two mutant forms of BRCA1, S1387A and S1423A (Fig. 2A). As described above, we have shown that the S1423A mutant, but not the S1387A mutant, reduces the ability of BRCA1 to activate the G2/M checkpoint after aphidicolin treatment. The frequency of chromosomal gaps and breaks after aphidicolin treatment was examined in independent clones stably transfected with each of these constructs.
When treated with 0.3 μM aphidicolin for 24 h prior to harvesting, cells expressing either mutant form of BRCA1 showed a frequency of chromosomal gaps and breaks that was midway between that observed for BRCA1−/− cells and that observed for cells expressing wild-type BRCA1 (Fig. 6A). One independent HCC1937 clone stably transfected with S1423A BRCA1 and two independent HCC1937 clones stably transfected with S1387A BRCA1 each had a significant, twofold decrease in total gaps and breaks, compared to total gaps and breaks of untransfected cells (P < 0.001 in each case). This reduction is approximately half the magnitude seen in HCC1937 cells expressing wild-type BRCA1.
FIG. 6.
G2/M checkpoint-deficient HCC1937 cells expressing a serine-to-alanine mutation at serine 1423 have increased fragile-site expression. (A) Average total chromosomal gaps and breaks per cell in HCC1937 cells stably transfected with the indicated BRCA1 expression constructs after 24 h in the presence (dark gray) or absence (light gray) of 0.3 μM aphidicolin; n = 100 metaphases for each data set. Error bars indicate the 95% confidence interval. (B) Frequency of fragile-site expression showing what percentage of specific fragile sites FRA3B and FRA16D were broken in HCC1937 cells stably transfected with the indicated BRCA1 expression constructs after 24 h of treatment with 0 μM (light gray) or 0.3 μM (dark gray) aphidicolin; n = 88 to 109 sites examined. Frequency of fragile-site induction is presented as the percentage of chromosome 3 or 16 homologs with breaks at FRA3B or FRA16D, respectively.
After treatment with 0.3 μM aphidicolin, the two HCC1937 clones expressing the S1387A mutant showed a significant, threefold decrease in expression of both FRA3B and FRA16D (P < 0.01 in all cases). The magnitude of the reduction in this G2/M checkpoint-proficient mutant is indistinguishable from that seen in BRCA1-positive cells (Fig. 6B). In contrast, expression of these two fragile sites in the S1423A (G2/M checkpoint-deficient) mutant cells was seen at the same levels as those in BRCA1−/− cells.
DISCUSSION
We have demonstrated that BRCA1 plays an important role in the stability of common fragile sites and that cells lacking BRCA1 show an increased expression of specific common fragile sites. In both mouse and human cells lacking functional BRCA1, aphidicolin-induced expression of specific fragile sites FRA3B, FRA16D (human), and Fra14A2 (mouse), as well as expression of total gaps and breaks, is increased two- to fourfold compared to levels for isogenic control cells that express BRCA1. While our results agree with those of previous studies that show increased aphidicolin-induced chromosome aberrations in BRCA1-deficient HCC1937 cells (38), we examined specific fragile sites to rule out random genomic instability and used isogenic controls to verify that BRCA1, as opposed to some other mutation in HCC1937 cells, is responsible for the effect. Furthermore, we have shown that the G2/M checkpoint function of BRCA1 is an important component of fragile-site stability. The experiments described herein expand upon our findings that ATR, which acts upstream of BRCA1 in cell cycle checkpoint pathways, plays a key role in fragile-site maintenance (3). In addition, these findings implicate the G2/M checkpoint as important in fragile-site stability and also help clarify the roles of BRCA1 and the G2/M checkpoint after partially inhibited DNA synthesis.
Of interest is the fact that BRCA1 is phosphorylated on serine 1423, in an ATR-dependent manner, after stalled replication (10, 31, 36). This residue has been shown to be important in the induction of the G2/M checkpoint after IR (31, 44). It is therefore likely that the ATR-dependent phosphorylation of this serine residue is involved in the activation of the G2/M checkpoint after aphidicolin-induced stalled replication. We examined the induction of the G2/M checkpoint by aphidicolin in cells expressing a BRCA1 construct that is mutated at the serine 1423 phosphorylation site. Cells expressing this mutant form of BRCA1 were unable to induce the G2/M checkpoint to the same extent as wild-type and BRCA1 S1387A-expressing cells. While other residues may also be important, these results indicate that serine 1423, but not serine 1387, of BRCA1 is involved in the G2/M checkpoint after stalled replication via aphidicolin treatment in a manner similar to its induction after IR.
HCC1937 cells expressing the G2/M checkpoint-proficient S1387A mutant had reduced breakage at FRA3B and FRA16D at the same low frequencies as cells expressing wild-type BRCA1. In contrast, HCC1937 cells stably transfected with the G2/M checkpoint-deficient S1423A mutant had breaks at these fragile sites at the same elevated levels seen in untransfected cells. While these experiments do not rule out other functions of BRCA1, such as DNA repair or the intra-S checkpoint, these results indicate that the G2/M checkpoint is important in the initial steps of common-fragile-site stability. The fact that both BRCA1 mutants were able to partially reduce the incidence of total gaps and breaks suggests that, in addition to FRA3B and FRA16D, BRCA1 also functions in maintaining stability at other, perhaps random, sites in the genome. It also illustrates the importance of examining specific fragile sites, in addition to total gaps and breaks, in such studies, because perturbing checkpoint and repair pathways may result in generalized instability that manifests itself as gaps and breaks at random sites in addition to or instead of at specific fragile sites.
Under similar treatment conditions, cells deficient in ATR activity show a substantially greater increase in common-fragile-site expression than do cells lacking BRCA1 (3). A likely explanation for these findings is that BRCA1 is only one of several downstream targets of ATR that include CHK1, which plays a major role in the G2/M checkpoint after stalled replication (20, 46). Thus, it is likely that inactivation of BRCA1 results in only partial inactivation of the G2/M checkpoint in response to aphidicolin, as has been shown following IR (43, 45). This hypothesis is consistent with our flow cytometry data, which showed a partial induction of the G2/M checkpoint by 0.5 μM aphidicolin (Fig. 5). On the other hand, when ATR expression is eliminated, the entire pathway is likely inactivated (15), resulting in a drastic reduction in the cell's ability to properly replicate common fragile sites.
The involvement of BRCA1 in the stability of common fragile sites after replication perturbations is of interest in light of the importance of BRCA1 in tumorigenesis. Loss of heterozygosity of BRCA1 has been associated with increased susceptibility to breast and ovarian cancer, and null mutations in BRCA1 have been found to persist in cancer cells (18, 42). Cells lacking BRCA1 will likely be prone to genomic alterations that lead to deletions of associated genes, including those at fragile sites, as has been seen in multiple cancers (1, 17, 21, 23, 28, 38).
Our finding that BRCA1 is involved in the stability of common fragile sites supports earlier work that the ATR checkpoint pathway is critical to the stability of these sites. It has previously been hypothesized that BRCA1, along with other DNA repair proteins, is involved in fragile-site maintenance through its DNA repair function (38). The data presented here demonstrate that the G2/M checkpoint function of BRCA1 is clearly important. Given the fact that fragile sites are likely to be late replicating and may even finish replication in G2, it is not surprising that the G2/M checkpoint is vital to fragile-site stability. It is significant that BRCA1, a protein mutated in breast and other cancers, plays a role in fragile-site stability. With some common fragile sites being linked to tumorigenesis (30, 33), these findings help elucidate the mechanisms of instability seen in tumor cells as well as the processes that maintain an integral feature of normal chromosome structure.
Acknowledgments
This work was supported by NIH grant CA43222 to T.W.G. Support for B.X. was provided by a grant from the Department of Defense (DAM17-03-1-0709) and by the Cancer Association of Greater New Orleans. M.B.K. was supported by grants from the NIH (CA86861, CA71387, and CA21765) and by the American Lebanese Syrian Associated Charities (ALSAC) of the St. Jude Children's Research Hospital.
We thank Shannon Callens for her technical support and Sara Hamon for her assistance with the statistical analysis. Mouse cell lines were kindly provided by Phang-Lang Chen and Wen-Hwa Lee. We thank J. Moran and M. Ljungman for helpful discussions.
REFERENCES
- 1.Arlt, M. F., D. E. Miller, D. G. Beer, and T. W. Glover. 2002. Molecular characterization of FRAXB and comparative common fragile site instability in cancer cells. Genes Chromosomes Cancer 33:82-92. [DOI] [PubMed] [Google Scholar]
- 2.Brown, E. J., and D. Baltimore. 2003. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev. 17:615-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casper, A. M., P. Nghiem, M. F. Arlt, and T. W. Glover. 2002. ATR regulates fragile site stability. Cell 111:779-789. [DOI] [PubMed] [Google Scholar]
- 4.Cha, R. S., and N. Kleckner. 2002. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297:602-606. [DOI] [PubMed] [Google Scholar]
- 5.Cliby, W. A., C. J. Roberts, K. A. Cimprich, C. M. Stringer, J. R. Lamb, S. L. Schreiber, and S. H. Friend. 1998. Overexpression of a kinase-inactive ATR protein causes sensitivity to DNA-damaging agents and defects in cell cycle checkpoints. EMBO J. 17:159-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coquelle, A., E. Pipiras, F. Toledo, G. Buttin, and M. Debatisse. 1997. Expression of fragile sites triggers intrachromosomal mammalian gene amplification and sets boundaries to early amplicons. Cell 89:215-225. [DOI] [PubMed] [Google Scholar]
- 7.Cortez, D., S. Guntuku, J. Qin, and S. J. Elledge. 2001. ATR and ATRIP: partners in checkpoint signaling. Science 294:1713-1716. [DOI] [PubMed] [Google Scholar]
- 8.Cortez, D., Y. Wang, J. Qin, and S. J. Elledge. 1999. Requirement of ATM-dependent phosphorylation of Brca1 in the DNA damage response to double-strand breaks. Science 286:1162-1166. [DOI] [PubMed] [Google Scholar]
- 9.Feijoo, C., C. Hall-Jackson, R. Wu, D. Jenkins, J. Leitch, D. M. Gilbert, and C. Smythe. 2001. Activation of mammalian Chk1 during DNA replication arrest: a role for Chk1 in the intra-S phase checkpoint monitoring replication origin firing. J. Cell Biol. 154:913-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gatei, M., B.-B. Zhou, K. Hobson, S. Scott, D. Young, and K. K. Khanna. 2001. Ataxia telangiectasia mutated (ATM) kinase and ATM and Rad3 related kinase mediate phosphorylation of Brca1 at distinct and overlapping sites. J. Biol. Chem. 276:17276-17280. [DOI] [PubMed] [Google Scholar]
- 11.Glover, T. W., C. Berger, J. Coyle, and B. Echo. 1984. DNA polymerase α inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Hum. Genet. 67:136-142. [DOI] [PubMed] [Google Scholar]
- 12.Glover, T. W., J. F. Coyle-Morris, F. P. Li, R. S. Brown, C. S. Berger, R. M. Gemmill, and F. Hecht. 1988. Translocation t(3;8)(p14.2;q24.1) in renal cell carcinoma affects expression of the common fragile site at 3p14(FRA3B) in lymphocytes. Cancer Genet. Cytogenet. 31:69-73. [DOI] [PubMed] [Google Scholar]
- 13.Glover, T. W., A. W. Hoge, D. E. Miller, J. E. Ascara-Wilke, A. N. Adam, S. L. Dagenais, C. M. Wilke, H. A. Dierick, and D. G. Beer. 1998. The murine Fhit gene is highly similar to its human orthologue and maps to a common fragile site region. Cancer Res. 58:3409-3414. [PubMed] [Google Scholar]
- 14.Glover, T. W., and C. K. Stein. 1987. Induction of sister chromatid exchanges at common fragile sites. Am. J. Hum. Genet. 41:882-890. [PMC free article] [PubMed] [Google Scholar]
- 15.Guo, Z., A. Kumagai, S. X. Wang, and W. G. Dunphy. 2000. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 14:2745-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, H., J. Qian, J. Proffit, K. Wilber, R. Jenkins, and D. I. Smith. 1998. FRA7G extends over a broad region: coincidence of human endogenous retroviral sequences (HERV-H) and small polydispersed circular DNAs (spcDNA) and fragile sites. Oncogene 16:2311-2319. [DOI] [PubMed] [Google Scholar]
- 17.Huang, H., C. P. Reed, A. Mordi, G. Lomberk, L. Wang, V. Shridhar, L. Hartmann, R. Jenkins, and D. I. Smith. 1999. Frequent deletions within FRA7G at 7q31.2 in invasive epithelial ovarian cancer. Genes Chromosomes Cancer 24:48-55. [PubMed] [Google Scholar]
- 18.Lallas, T. A., T. E. Buekers, and R. E. Buller. 1999. BRCA1 mutations in familial ovarian cancer. Mol. Gen. Metabol. 67:357-363. [DOI] [PubMed] [Google Scholar]
- 19.Le Beau, M. M., F. V. Rassool, M. E. Neilly, R. Espinosa III, T. W. Glover, D. I. Smith, and T. W. McKeithan. 1998. Replication of a common fragile site, FRA3B, occurs late in S phase and is delayed further upon induction: implications for the mechanism of fragile site induction. Hum. Mol. Genet. 7:755-761. [DOI] [PubMed] [Google Scholar]
- 20.Liu, Q., S. Guntuku, X.-S. Cui, S. Matsuoka, D. Cortez, K. Tamai, G. Luo, S. Carattini-Rivera, F. DeMayo, A. Bradley, L. A. Donehower, and S. J. Elledge. 2000. Chk1 is an essential kinase that is regulated by Atr and required for the G2/M DNA damage checkpoint. Genes Dev. 14:1448-1459. [PMC free article] [PubMed] [Google Scholar]
- 21.Mangelsdorf, M., K. Ried, E. Woollatt, S. Dayan, H. Eyre, M. Finnis, L. Hobson, J. Nancarrow, D. Venter, E. Baker, and R. I. Richards. 2000. Chromosomal fragile site FRA16D and DNA instability in cancer. Cancer Res. 60:1683-1689. [PubMed] [Google Scholar]
- 22.Matsuyama, A., T. Shiraishi, F. Trapasso, T. Kuroki, H. Alder, M. Mori, K. Huebner, and C. M. Croce. 2003. Fragile site orthologs FHIT/FRA3B and Fhit/Fra14A2: evolutionarily conserved but highly recombinogenic. Proc. Natl. Acad. Sci. USA 100:14988-14993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michael, D., D. G. Beer, C. W. Wilke, D. E. Miller, and T. W. Glover. 1997. Frequent deletions of FHIT and FRA3B in Barrett's metaplasia and esophageal adenocarcinomas. Oncogene 15:1653-1659. [DOI] [PubMed] [Google Scholar]
- 24.Mimori, K., T. Druck, H. Inoue, H. Alder, L. Berk, M. Mori, K. Huebner, and C. M. Croce. 1999. Cancer-specific chromosome alterations in the constitutive fragile region FRA3B. Proc. Natl. Acad. Sci. USA 96:7456-7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mishmar, D., Y. Mandel-Gutfreund, H. Margalit, A. Rahat, and B. Kerem. 1999. Common fragile sites: G-band characteristics within an R-band. Am. J. Hum. Genet. 64:908-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mishmar, D., A. Rahat, S. W. Scherer, G. Nyakatura, B. Hinzmann, Y. Kohwi, Y. Mandel-Gutfroint, J. R. Lee, B. Drescher, D. E. Sas, H. Margalit, M. Platzer, A. Weiss, L. C. Tsui, A. Rosenthal, and B. Kerem. 1998. Molecular characterization of a common fragile site (FRA7H) on human chromosome 7 by the cloning of a simian virus 40 integration site. Proc. Natl. Acad. Sci. USA 95:8141-8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morelli, C., E. Karayianni, C. Magnanini, A. J. Mungall, E. Thorland, M. Negrini, D. I. Smith, and G. Barbanti-Brodano. 2002. Cloning and characterization of the common fragile site FRA6F harboring a replicative senescence gene and frequently deleted in human tumors. Oncogene 21:7266-7276. [DOI] [PubMed] [Google Scholar]
- 28.Negrini, M., C. Monaco, I. Vorechovsky, M. Ohta, T. Druck, R. Baffa, K. Huebner, and C. M. Croce. 1996. The FHIT gene at 3p14.2 is abnormal in breast carcinomas. Cancer Res. 56:3173-3179. [PubMed] [Google Scholar]
- 29.Nghiem, P., P. K. Park, Y.-S. Kim, C. Vaziri, and S. L. Schreiber. 2001. ATR inhibition selectively sensitizes G1 checkpoint-deficient cells to lethal premature chromatin condensation. Proc. Natl. Acad. Sci. USA 98:9092-9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohta, M., H. Inoue, M. G. Cotticelli, K. Kastury, R. Baffa, J. Palazzo, Z. Siprashvili, M. Mori, P. McCue, T. Druck, C. M. Croce, and K. Huebner. 1996. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell 84:587-597. [DOI] [PubMed] [Google Scholar]
- 31.Okada, S., and T. Ouchi. 2003. Cell cycle differences in DNA damage-induced BRCA1 phosphorylation affect its subcellular localization. J. Biol. Chem. 278:2015-2020. [DOI] [PubMed] [Google Scholar]
- 32.Rassool, F. V., T. W. McKeithan, M. E. Neilly, E. van Melle, R. Espinosa III, and M. M. Le Beau. 1991. Preferential integration of marker DNA into the chromosomal fragile site at 3p14: an approach to cloning fragile sites. Proc. Natl. Acad. Sci. USA 88:6657-6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ried, K., M. Finnis, L. Hobson, M. Mangelsdorf, S. Sayan, J. K. Nancarrow, E. Woollatt, G. Kremmidiotis, A. Gardner, D. Venter, E. Baker, and R. I. Richards. 2000. Common chromosomal fragile site FRA16D sequence: identification of the FOR gene spanning FRA16D and homozygous deletions and translocation breakpoints in cancer cells. Hum. Mol. Genet. 9:1651-1663. [DOI] [PubMed] [Google Scholar]
- 34.Scully, R., J. Chen, R. L. Ochs, K. Keegan, M. Hoekstra, J. Feunteun, and D. M. Livingston. 1997. Dynamic changes of BRCA1 subnuclear location and phosphorylation state are initiated by DNA damage. Cell 90:425-435. [DOI] [PubMed] [Google Scholar]
- 35.Scully, R., S. Ganesan, M. Brown, J. A. De Caprio, S. A. Cannistra, J. Feunteun, S. Schnitt, and D. M. Livingston. 1996. Location of BRCA1 in human breast and ovarian cancer cells. Science 272:123-126. [DOI] [PubMed] [Google Scholar]
- 36.Tibbetts, R. S., D. Cortez, K. M. Brumbaugh, R. Scully, D. Livingston, S. J. Elledge, and R. T. Abraham. 2000. Functional interactions between BRCA1 and the checkpoint kinase ATR during genotoxic stress. Genes Dev. 14:2989-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomlinson, G. E., T. T.-L. Chen, V. A. Stastny, A. K. Virmani, M. A. Spillman, V. Tonk, J. L. Blum, N. R. Schneider, I. I. Wistuba, J. W. Shay, J. D. Minna, and A. F. Gazdar. 1998. Characterization of a breast cancer cell line derived from a germ-line BRCA1 mutation carrier. Cancer Res. 58:3237-3242. [PubMed] [Google Scholar]
- 38.Turner, B. C., M. Ottey, D. B. Zimonjic, M. Potoczek, W. W. Hauck, E. Pequignot, C. L. Keck-Waggoner, C. Devignani, C. M. Aldaz, P. A. McCue, J. Palazzo, K. Huebner, and N. C. Popescu. 2002. The fragile histidine triad/common chromosome fragile site 3B locus and repair-deficient cancers. Cancer Res. 62:4054-4060. [PubMed] [Google Scholar]
- 39.Wang, L., J. Darling, J.-S. Zhang, H. Huang, W. Liu, and D. I. Smith. 1999. Allele-specific late replication and fragility of the most active common fragile site, FRA3B. Hum. Mol. Genet. 8:431-437. [DOI] [PubMed] [Google Scholar]
- 40.Wang, L., W. Paradee, C. Mullins, R. Shridhar, R. Rosati, C. M. Wilke, T. W. Glover, and D. I. Smith. 1997. Aphidicolin-induced FRA3B breakpoints cluster in two distinct regions. Genomics 41:485-488. [DOI] [PubMed] [Google Scholar]
- 41.Wilke, C. M., B. K. Hall, A. Hoge, W. Paradee, D. I. Smith, and T. W. Glover. 1996. FRA3B extends over a broad region and contains a spontaneous HPV16 integration site: direct evidence for the coincidence of viral integration sites and fragile sites. Hum. Mol. Genet. 5:187-195. [DOI] [PubMed] [Google Scholar]
- 42.Wilson, C. A., L. Ramos, M. R. Villaseñor, K. H. Anders, M. F. Press, K. Clarke, B. Karlan, Chen-Jun-Jie, R. Scully, D. Livingston, R. H. Zuch, M. H. Kanter, S. Cohen, F. J. Calzone, and D. J. Slamon. 1999. Localization of human BRCA1 and its loss in high-grade, non-inherited breast carcinomas. Nat. Genet. 21:236-240. [DOI] [PubMed] [Google Scholar]
- 43.Xu, B., S.-T. Kim, and M. B. Kastan. 2001. Involvement of Brca1 in S-phase and G2-phase checkpoints after ionizing irradiation. Mol. Cell. Biol. 21:3445-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu, B., A. H. O'Donnell, S. T. Kim, and M. B. Kastan. 2002. Phosphorylation of serine 1387 in Brca1 is specifically required for the Atm-mediated S-phase checkpoint after ionizing irradiation. Cancer Res. 62:4588-4591. [PubMed] [Google Scholar]
- 45.Yamane, K., J. Chen, and T. J. Kinsella. 2003. Both DNA topoisomerase II-binding protein 1 and BRCA1 regulate the G2-M cell cycle checkpoint. Cancer Res. 63:3049-3053. [PubMed] [Google Scholar]
- 46.Yarden, R. I., S. Pardo-Reoyo, M. Sgagias, K. H. Cowan, and L. C. Brody. 2002. BRCA1 regulates the G2/M checkpoint by activating Chk1 kinase upon DNA damage. Nat. Genet. 30:285-289. [DOI] [PubMed] [Google Scholar]
- 47.Zhao, H., and H. Piwnica-Worms. 2001. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol. Cell. Biol. 21:4129-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhong, Q., T. G. Boyer, P.-L. Chen, and W.-H. Lee. 2002. Deficient nonhomologous end-joining activity in cell-free extracts from Brca1-null fibroblasts. Cancer Res. 62:3966-3970. [PubMed] [Google Scholar]