Abstract
We have employed a novel in vivo approach to study the structure and function of the eukaryotic kinetochore multiprotein complex. RNA interference (RNAi) was used to block the synthesis of centromere protein A (CENP-A) and Clip-170 in human cells. By coexpression, homologous kinetochore proteins from Saccharomyces cerevisiae were then tested for the ability to complement the RNAi-induced phenotypes. Cse4p, the budding yeast CENP-A homolog, was specifically incorporated into kinetochore nucleosomes and was able to complement RNAi-induced cell cycle arrest in CENP-A-depleted human cells. Thus, Cse4p can structurally and functionally substitute for CENP-A, strongly suggesting that the basic features of centromeric chromatin are conserved between yeast and mammals. Bik1p, the budding yeast homolog of human CLIP-170, also specifically localized to kinetochores during mitosis, but Bik1p did not rescue CLIP-170 depletion-induced cell cycle arrest. Generally, the newly developed in vivo complementation assay provides a powerful new tool for studying the function and evolutionary conservation of multiprotein complexes from yeast to humans.
Centromeres are eukaryotic cellular structures that are essential for faithful chromosomal segregation during mitotic and meiotic cell division. The kinetochore complex is a defined multiprotein structure on the mitotic chromosome that adheres to the centromere (18, 61). The kinetochore serves as the site of attachment for spindle microtubules, which facilitate the alignment and separation of chromosomes during mitosis (12, 13). Although the centromere's function is highly conserved among eukaryotes, centromeric morphology varies significantly, ranging from small, simple kinetochores in the budding yeast Saccharomyces cerevisiae to complex centromeres in multicellular eukaryotes (14). In mammalian cells, the centromere forms a visible primary constriction during metaphase and the kinetochore is a distinct structure that can be resolved into subregions (45, 47, 67). Finally, in holokinetic organisms such as the nematode Caenorhabditis elegans, multiple centromeres are dispersed throughout the chromosomes (1, 2). The primary nucleotide sequences of centromeric DNAs are also not conserved across phylogenies. The absolute size of the centromeric DNA can vary from ca. 125 bp in S. cerevisiae to tens of megabases in higher eukaryotes (11). Beyond the lack of size and sequence conservation between organisms, the centromere's function may be established not only at predefined sequences, but also at noncentromeric DNA elements, as illustrated by neocentromeres in human (11) and plant (93) cells. Finally, while in budding yeast the centromere DNA alone can nucleate centromere formation de novo, centromeres of metazoan cells strongly depend on epigenetic factors rather than DNA sequences for their activity (90). Thus, there is no primary sequence determinant in centromeric DNA that is conserved among eukaryotic species.
At the protein level, a series of kinetochore components show homology to proteins of other organisms and thus are evolutionarily conserved between eukaryotes (8, 13, 44, 85). The extent to which the molecular mechanisms of kinetochore function are conserved has been addressed by comparing centromere proteins from S. cerevisiae and humans (8, 36). More than 30 yeast kinetochore proteins have been identified. Based on their localization, function, or participation in distinct protein complexes, kinetochore proteins can be subgrouped into inner kinetochore, outer kinetochore, and spindle checkpoint factors (8, 36), although alternative classifications have also been suggested (48). Inner kinetochore proteins are directly associated with the centromeric DNA. In S. cerevisiae, these include the CBF3 complex, Cbf1p, Mif2p, and Cse4p (38, 50, 51). Outer kinetochore proteins connect chromosomes to spindle microtubules. Several additional multiprotein complexes (Ctf19p, Ndc80p, Dam1/Duo1p, Ipl/Sli15p, Mtw1p, and Spc105p) mediate the assembly of the outer kinetochore and represent the physical bridge between the core centromeres and the spindle components in budding yeast (5, 8, 32, 40, 55, 56, 91). The spindle checkpoint monitors the formation of bipolar kinetochore-microtubule attachments and is composed of members of the Mad and Bub protein families and the protein kinase Mps1 (3). Homology searches have revealed that components of each of these S. cerevisiae centromere or kinetochore elements show a different degree of sequence conservation with human proteins (36). While all of the spindle checkpoint components of budding yeast have highly conserved homologs in human cells, there is only limited similarity between the inner or outer kinetochore proteins from S. cerevisiae and the human centromere (36). Partial sequence homologies, for example, exist between the yeast centromere proteins Mif2p and Okp1p and the bona fide human centromere proteins C and F (CENP-C and CENP-F), respectively (52, 53, 57). Most strikingly, homologs of the CBF3 components Ndc10p, Cep3p, and Ctf13p, which constitute a fundamental and essential building unit of the yeast core centromere (8, 39), have not been found in human databases, and conversely, no homologs of the human constitutive centromere proteins CENP-B and CENP-H have been reported for S . cerevisiae (15, 42, 54, 56, 78, 79).
Despite this evidence of diversity, there appear to be at least some underlying common mechanisms for inner kinetochore structure and function. All centromeric DNAs studied so far bind a histone H3-related protein (CenH3), variously named CENP-A in vertebrates, Cid in Drosophila melanogaster, HCP-3 in C. elegans (7), and Cse4p in S. cerevisiae (for reviews, see references 27, 73, 74, and 81). CENP-A is a constitutive centromere component and localizes to the inner kinetochore plate of mitotic chromosomes (85, 86). Genetic and biochemical evidence suggests that CenH3 proteins replace histone H3 in centromere-specific nucleosomes (24, 58, 59, 72, 75, 80, 86, 88, 92). In CENP-A null mice, the centromeric chromatin organization is disrupted, suggesting that CENP-A is required for the assembly of a functional kinetochore (29). Human CENP-A and budding yeast Cse4p share extensive sequence homology in their histone cores, and this domain is required for centromeric localization (77, 82). Although CENP-A is not able to rescue either temperature-sensitive or null alleles of Cse4 in S. cerevisiae (53, 77), the two proteins may be regarded as true orthologs based on their similar properties within centromeric nucleosomes.
The microtubule-kinetochore connection represents another example of a strong structural-functional conservation between humans and budding yeast. The human homologs of Ndc80p, Nuf2p, and Bik1p, all of which functionally contribute to the kinetochore-mitotic spindle interface, were shown to specifically localize to kinetochores of HeLa cell mitotic chromosomes (19, 91). Human CLIP-170, the homolog of budding yeast Bik1p, is a microtubule binding protein that has been implicated in the attachment of endosomes to microtubules in interphase cells (60, 63) and the attachment of kinetochores to microtubules during mitosis (19). By analogy, Bik1p localizes to kinetochores (26) and has been found to contribute to the connection between kinetochores and microtubules in S. cerevisiae (41). CLIP-170 and Bik1p exhibit extensive sequence homology (60), appear to play similar roles at human and budding yeast kinetochores, respectively (41), and thus may also represent true orthologs, similar to the situation for CENP-A and Cse4p.
The structural and biochemical similarities between kinetochore proteins from different organisms raise the interesting question of whether they have identical kinetochore functions. We have developed a new approach to address this question. This approach uses RNA interference (RNAi) to specifically inhibit the synthesis of kinetochore proteins in human cells combined with the ectopic expression of homologous budding yeast proteins. Rescue of the RNAi-induced phenotype by expression of the yeast counterpart then serves as a readout for its ability to functionally substitute for the endogenous kinetochore protein. This assay was applied to the study of CENP-A and CLIP-170. The RNAi-induced depletion of CENP-A or CLIP-170 resulted in loss-of-proliferation phenotypes that were attributable to cell cycle arrest in interphase or mitosis, respectively. The coexpression of budding yeast Cse4p in CENP-A-depleted cells successfully complemented this phenotype, while attempts to rescue the CLIP-170 depletion phenotype by the use of Bik1p failed. Our results demonstrate that Cse4p can complement the CENP-A function across species.
MATERIALS AND METHODS
Expression constructs.
A plasmid encoding a green fluorescent protein (GFP)-Cse4p fusion protein was obtained by subcloning a KpnI-BamHI Cse4p PCR fragment (5′ primer GW21, 5′-GAC GGT ACC ATG TCA AGT AAA CAA CAA TGG GTT AG-3′; 3′ primer GW22, 5′-GAC GGA TCC CTA AAT AAA CTG TCC CCT GAT TCT TC-3′), obtained from plasmid pRB163 (33; kindly provided by Richard Baker, University of Massachusetts, Worcester) into a KpnI- and BamHI-linearized pEGFP-C1 vector (Clontech, Palo Alto, Calif.) (63). A Bik1p cDNA was obtained by PCR cloning from genomic DNA (a kind gift from T. Munder, Hans-Knöll-Institute, Jena, Germany), using 5′ primer GW37 (5′-GAC GGT ACC ATG GAT AGA TAT CAA AGA AA-3′), containing a KpnI restriction site, and 3′ primer GW38 (5′-GAC GGA TCC CTA GAA GAA CTG CTG GTT GTC AG-3′), containing a BamHI restriction site. PCRs were performed with these primers and yeast genomic chromatin as a template DNA. Plasmid pEGFP-AF8-CENP-A, encoding a GFP-CENP-A fusion protein (80), was a kind gift from K. Sugimoto (Osaka, Japan). A vector containing a CLIP-170-GFP fusion protein (pCB6-GFP-CLIP-170-full) (60) was a gracious gift of F. Perez (Paris, France). All fusion constructs, except for pCB6-GFP-CLIP-170-full, were verified by full-length sequencing.
Cell culture and transfection into HEp-2 cells.
HEp-2 (HeLa derivative) cells were obtained from the American Type Culture Collection (Manassas, Va.) and grown to subconfluence as recommended. The cells were cultured according to previously published methods (76) in Dulbecco's modified Eagle's medium (Sigma-Aldrich, Munich, Germany) supplemented with 10% fetal calf serum in a 7.5% CO2 atmosphere at 37°C. After aspiration of the medium, the cells were washed with magnesium- and calcium-containing phosphate-buffered saline (PBS) (Sigma-Aldrich), followed by detachment with trypsin-EDTA and reseeding on coverslips. The cells were transfected with pEGFP-C1 vectors containing the various budding yeast or human kinetochore cDNAs by use of the TransFast transfection reagent (Promega, Madison, Wis.) according to the protocols of the manufacturer. Cell lines stably expressing GFP fusion proteins were established by selection in a medium supplemented with 20 mM G-418 (Geniticin; Sigma-Aldrich) for several weeks, as described by Sugimoto et al. (80).
Antibodies and immunolocalization.
The following primary antibodies were used for indirect immunofluorescence or Western blot analyses: human CREST sera against centromere proteins CENP-A, -B, and -C (23) (see Fig. 2A); a mouse monoclonal anti-GFP antibody (sc-9996; Santa Cruz Biotechnology, Santa Cruz, Calif.); a mouse monoclonal antibody against lamin A/C (sc-7292; Santa Cruz Biotechnology); mouse monoclonal immunoglobulin M (IgM) antibodies against CLIP-170 (2D6 and 4D3; a kind gift of F. Perez, Paris, France); a mouse monoclonal antibody against HP1α (2HP-2G9; Euromedex, Mundolsheim, France); a rabbit antibody against acetylated histone H3 (06-599; Upstate Biotechnology, Lake Placid, N.Y.); and a rabbit antibody against the dimethylated lysine at position 9 of histone H3 (07-212; Upstate Biotechnology).
FIG. 2.
GFP fusion constructs are expressed as full-length proteins. (A) Total protein lysates from HEp-2 cells (lane 1) and HEp-2 cells expressing GFP (lane 2), GFP-CENP-A (lane 3), GFP-Cse4p (lane 4), GFP-CLIP-170 (lane 5), or GFP-Bik1p (lane 6) were separated by SDS-PAGE and transferred to nitrocellulose membranes. Single strips from the membranes were probed with anticentromere autoimmune serum (lane 1) or an anti-GFP antibody (lanes 2 to 6). The ACA serum detects CENP-A at ca. 20 kDa, CENP-B at ca. 80 kDa, and CENP-C at ca. 140 kDa. GFP fusions were detected as full-length proteins at their expected molecular masses. Numbers to the right indicate the positions of standard protein markers (in kilodaltons). (B) Chromatin fractionation. DNAs extracted from equivalent supernatant (S) and pellet (P) fractions after MNase digestion were electrophoresed in a 1.5% agarose gel, followed by ethidium bromide staining (chromatin). The migration positions of mononucleosomes (mn) and polynucleosomes (pn) are indicated. Whole-cell protein extracts (WCE) and aliquots of the chromatin fractionation procedure from different HEp-2 cell lines were subjected to SDS-17.5% PAGE and Western blotting with antibodies against N-terminally acetylated histone H3 (acH3), histone H3 methylated at lysine 9 (meH3), CENP-A, and GFP, as indicated. Nucleosomes containing acetylated histone H3 or histone H3 methylated at lysine 9 were released from pellet fractions into soluble supernatant fractions with increasing amounts of MNase. In contrast, CENP-A-, GFP-CENP-A-, and GFP-Cse4p-containing nucleosomes were not released into soluble fractions under these conditions. C, cytoplasmic fraction; N, nuclear fraction.
Cells grown on coverslips were fixed by incubation in 3.7% paraformaldehyde for 10 min at room temperature, followed by 5 min of permeabilization in 0.5% Triton X-100. Immunofluorescence was performed as described previously (35, 87). For immunofluorescence staining, primary antibodies from mouse and human sources were used and were detected with species-specific secondary antibodies linked to rhodamine (Jackson Immunoresearch, West Grove, Pa.). Cellular DNAs were stained with TO-PRO-3 and DAPI (4′,6′-diamidino-2-phenylindole) (Molecular Probes, Eugene, Oreg.) at individually established concentrations. The coverslips were then mounted onto microscope slides by using MobiGLOW mounting medium (MoBiTec, Goettingen, Germany). Microscopic images were collected with an Axiovert 200 M/LSM 510 META microscope (Carl Zeiss, Jena, Germany). The samples were scanned by using a 63×-1.40 Plan-Apochromat oil objective. GFP, rhodamine, and TO-PRO-3 dyes were excited by laser light at 488-, 543-, and 633-nm wavelengths, respectively. To avoid bleed-through effects in double or triple staining experiments, we scanned each dye independently (multitrack mode). Fluorescence signals were detected by using narrow-band-pass (±20-nm wavelengths) instead of long-pass filters. Thus, only the peak regions of the fluorescence signals were taken for data analysis. Single optical sections either were selected by eye-scanning the sample in the z axis for optimal fluorescence signals or were taken from stack projections. Images were electronically merged and stored as TIFF files. Figures were assembled from the TIFF files with Adobe Photoshop software.
Chromatin fractionation and Western blots.
Chromatin was fractionated according to previously described protocols (65). Briefly, HEp-2 cells were lysed in buffer N (0.3% NP-40, 15 mM Tris-HCl [pH 7.5], 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 1 mM CaCl2, 1 mM dithiothreitol, 2 mM sodium vanadate, 250 mM sucrose, complete protease inhibitor; Roche). Nuclear DNAs were digested by incubation with increasing amounts of micrococcal nuclease (MNase). After centrifugation, the supernatant fractions were removed and the pellets were resuspended in ice-cold 2 mM EDTA. Protein samples were incubated for 10 min at 100°C in a solution containing 2% sodium dodecyl sulfate (SDS), 0.1% bromophenol blue, 35 mM dithiothreitol, 25% glycerol, and 60 mM Tris-HCl (pH 6.8). The cell lysates were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and blotted onto Protran BA nitrocellulose (Schleicher & Schuell, Dassel, Germany). Bound antibodies were detected with horseradish peroxidase-conjugated anti-human and anti-mouse IgG antibodies (Jackson Immunoresearch Laboratories) at a dilution of 1:4,000 and by use of the ECL Advance system (Amersham Biosciences) according to the manufacturer's instructions. Chemiluminescence was detected by exposure on Biomax Light-1 film (Kodak).
RNAi.
Small interfering RNAs (20) specific for the N-terminal portion of the CENP-A and CLIP-170 mRNAs (IBA GmbH, Goettingen, Germany) were synthesized for RNAi against CENP-A (5′-GCCCGAGGCCCCGAGGAGGUU-3′) and CLIP-170 (5′-GCACAGCUCUGAAGACACCUU-3′).
As a positive control, the RNA sequence for lamin A/C (5′-CUGGACUUCCAGAAGAACAdTdT-3′) was used. The procedure for RNAi was adopted from published protocols (20). HEp-2 cells were grown to ca. 20% confluence in coverslip-containing dishes. The cells were transfected with ca. 1 ng of double-stranded small interfering RNA by the use of OligoFectamine (Invitrogen) according to the manufacturer's instructions. In 24-h time steps, the medium for the cells was replaced by fresh medium and the old medium was inspected for the presence of dead cells. In addition, a coverslip was removed to monitor cell viability properties. Seventy-two hours after transfection, the remaining cells were harvested by detachment with trypsin-EDTA. Aliquots of the cells were lysed (see above) and analyzed by Western blotting in order to verify the results of the RNAi treatment.
Mitotic index and cell viability assays.
DAPI staining of DNA was used to assess the loss-of-viability phenotype of CENP-A-depleted cells. Mitotic indices were determined by TO-PRO-3 and centromere staining and the quantitation of mitotic figures in confocal microscopic images (n > 400). In parallel, HEp-2 cells grown in the same petri dishes that contained the coverslips for microscopy were analyzed for frequencies of living cells. The supernatants and trypsin-EDTA-detached cells were combined, washed twice in PBS, and pelleted at 600 × g. The pellet was redissolved in 1 ml of PBS. Fifty microliters of this cell suspension was mixed with 50 μl of trypan blue and immediately loaded into a Neubauer chamber. Trypan blue exclusion served to discriminate and quantitate dead and living cells.
RESULTS
Yeast kinetochore proteins Cse4p and Bik1p specifically localize to centromeres in human cells.
The S. cerevisiae kinetochore components Cse4p and Bik1p are homologous to the human proteins CENP-A and CLIP-170, respectively (53, 63). In order to determine the functional properties of the budding yeast proteins in human cells and to compare them with their human counterparts, we established HEp-2 cell lines that stably expressed each of these proteins as GFP fusions (Fig. 1). The localization of the GFP fusion proteins in each cell line was analyzed by confocal microscopy after coimmunofluorescence staining of the centromeres with the human autoimmune serum ACA, which is specific for CENP-A, -B, and -C (23, 35) (Fig. 2A).
FIG. 1.
Budding yeast kinetochore proteins Cse4p and Bik1p are recruited to centromeres in human cells. HEp-2 cells stably expressing GFP-CENP-A (A and B), GFP-Cse4p (C and D), GFP-CLIP-170 (E to G), or GFP-Bik1p (H to J) were grown on coverslips and processed for indirect immunofluorescence for analyses of subcellular localization. Fixed cells were costained with a human anticentromere autoimmune serum to detect centromeres (ACA) and with ToPro-3 to visualize DNA. Single confocal sections were acquired from cells in interphase (i), telophase (t), or prometaphase (p). Fluorescence signals from each channel are displayed. To show the degree of colocalization, we also show overlay images of the GFP (green) and centromere signals (red). Bars, 5 μm.
As expected, the GFP-CENP-A construct was recruited to centromeres in both interphase cells (Fig. 1A) and mitotic cells (Fig. 1B). HEp-2 cells stably expressing GFP-Cse4p also showed a complete colocalization of the fusion protein with anticentromere antibodies in interphase and mitotic cells (Fig. 1C and D, respectively). This was consistent with the previous observation that H3-like proteins, including Cse4p, are specifically deposited at human centromeric heterochromatin upon transient transfection (28). We observed that the localization of GFP-CENP-A and GFP-Cse4 was dependent on the expression level of the fusion proteins: only low-level-expressing cells showed GFP signals that exclusively localized to kinetochores, while in high-level-expressing cells both GFP-CENP-A and GFP-Cse4 were also found distributed throughout the chromatin (data not shown). In our stably low-level-expressing cells, GFP-Cse4p was exclusively found at the centromeres and not in the peripheral centromeric heterochromatin during the cell cycle (Fig. 1C and D; also data not shown), suggesting a specific incorporation of GFP-Cse4p into human centromeric nucleosomes.
CLIP-170 is a cytoplasmic protein during interphase (66). It is recruited to prometaphase kinetochores during mitosis and disappears from the kinetochores in anaphase spindles (19). We observed an identical pattern of cell cycle-specific distribution for endogenous CLIP-170 in HEp-2 cells after antibody staining (not shown). Likewise, the GFP-CLIP-170 fusion protein was found to localize in the cytoplasm during interphase (Fig. 1E) and at kinetochores during prometaphase (Fig. 1F). During telophase, the CLIP-170 staining of centromeres was diminished (Fig. 1G). Surprisingly, an almost identical localization behavior was observed in mitotic HEp-2 cells stably expressing GFP-Bik1p (Fig. 1H to J). The only difference between GFP-CLIP-170 and GFP-Bik1p over the course of the cell cycle was a diffuse nuclear staining by GFP-Bik1p in addition to its cytoplasmic localization in interphase cells (compare Fig. 1E and H). These observations suggest that there is a specific recruiting mechanism of CLIP-170 to kinetochores during mitosis (19) which can also utilize Bik-1, a protein homolog of CLIP-170. The kinetochore localization of the fusion proteins cannot be attributed to the GFP moiety, since GFP alone distributed more or less evenly throughout all cell compartments (not shown).
A Western blot analysis demonstrated that all of the GFP fusion constructs were expressed as full-length proteins (Fig. 2A). In SDS-PAGE gels, GFP alone and the fusion proteins migrated at about the expected sizes of ca. 28 kDa (GFP), ca. 46 kDa (GFP-CENP-A; lane 3), ca. 52 kDa (GFP-Cse4p), ca. 190 kDa (GFP-CLIP-170), and ca. 85 kDa (GFP-Bik1p). This blot also shows the reactivity of the ACA serum against CENP-A, CENP-B, and CENP-C (Fig. 2A, lane 1).
The chromatin was fractionated in order to test whether Cse4p was incorporated into centromeric nucleosomes (Fig. 2B). Untransfected HEp-2 cells and cells that were transiently transfected with GFP-CENP-A or GFP-Cse4p vectors were fractionated to yield nuclear and cytoplasmic proteins. The nuclear fractions were further digested with increasing amounts of MNase. MNase-digested nuclei lead to the recovery of insoluble pellet fractions which mostly represent the transcriptionally inactive heterochromatin (68). Ethidium bromide staining showed classical nucleosomal ladders in the presence of increasing concentrations of MNase (Fig. 2B, chromatin). As the MNase concentration increased, the supernatant fractions were progressively enriched in mononucleosomes, while the pellet fractions contained polynucleosomes. Western blots of cytoplasmic, nuclear, and MNase fractions showed that CENP-A was, as expected, exclusively associated with the heterochromatin-containing pellet fractions (Fig. 2B, CENP-A). GFP-CENP-A and GFP-Cse4p fractionated in identical patterns, indicating the similar biochemical properties of the fusion proteins with those of CENP-A (Fig. 2B). Based on these observations and the exclusive localization of the fusion proteins to centromeres, we concluded that both GFP-CENP-A and GFP-Cse4p are correctly incorporated into nucleosomes of centromeric heterochromatin. Controls were used to analyze the MNase fractionation procedure. Acetylated and methylated histone H3 showed a fractionation pattern that was different from that of CENP-A (Fig. 2B, acH3 and meH3, respectively). With increasing amounts of MNase, these H3 isoforms (and heterochromatin protein HP1 [data not shown]) were released into the soluble supernatant, consistent with previous observations (65).
Depletion of CENP-A or CLIP-170 by RNAi causes block-of-proliferation phenotypes.
We next analyzed the phenotype of HEp-2 cells in which CENP-A or CLIP-170 was down-regulated by RNAi. As a control, we included the nuclear envelope protein lamin A/C, for which the RNAi phenotype is well described (20). Since protein levels after RNAi were maximally reduced after 72 h of treatment (Fig. 3 and unpublished data), we selected this time point for a detailed analysis of RNAi-induced phenotypes. Seventy-two hours after RNAi infection, the cells were analyzed by Western blotting, immunofluorescence, and bright-field microscopy. RNAi of CENP-A, CLIP-170, and lamin A/C resulted in the reduction of protein levels to <10% those of control cells, indicating the successful inhibition of protein synthesis (Fig. 3C). While CENP-A was down-regulated by CENP-A-specific RNAi, the amounts of CENP-B and CENP-C were not affected: the amounts of both proteins in Western blots remained constant (data not shown). During interphase, >50% of the CENP-A-depleted cells displayed malformed nuclei with local chromatin condensation (Fig. 3E), similar to cells in which the CENP-A function was inhibited by a microinjection of anti-CENP-A antibodies (22), while almost all of the control cells showed normal, round nuclei, as revealed by DAPI staining (Fig. 3D). Although the frequency of mitotic cells was elevated (18% versus 8% in control cells) (Fig. 4A), no accumulation of cells during mitosis was observed. While chromosome condensation and spindle formation appeared normal, we observed a high frequency of misaligned or lagging chromosomes (>60%) in mitotic cells (Fig. 3G). Mitotic control cells with no RNAi treatment of CENP-A showed normal behavior (Fig. 3F). CENP-A-depleted cells did not show any signs of apoptosis, such as condensed chromatin structures or blebbing, indicating that CENP-A depletion does not induce programmed cell death. Rather, these cells had reached a stage of nonviability that was consistent with similar findings for anti-CENP-A RNAi-treated HeLa cells after 68 h, as described by Goshima et al. (25). At 96 h, almost all of the cells were dead, as indicated by their detachment from the coverslips and by trypan blue exclusion (data not shown).
FIG.3.
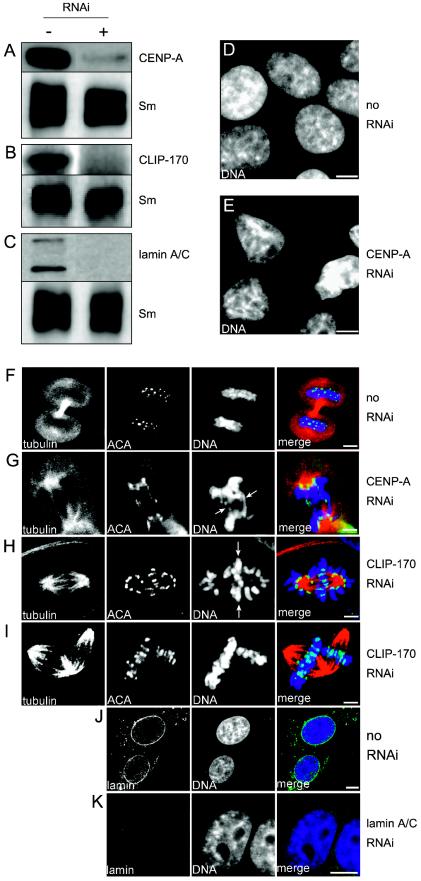
CENP-A and CLIP-170 reduction by RNAi causes severe cellular defects. RNAi was used to block the protein synthesis of CENP-A (A), CLIP-170 (B), and as a control, lamin A/C (C) in HEp-2 cells. The reduction in protein level was then analyzed by immunoblotting of protein lysates from cells treated without (−) or with (+) RNAi oligonucleotides for 72 h. An anti-splicing factor SmB/B′ antibody was used as a loading control (Sm). Control cells treated without RNAi (D) and cells treated for 72 h with anti-CENP-A RNAi oligonucleotides were fixed on coverslips and stained with a DNA dye to visualize the nuclei. While the control cells had normal, round nuclei and a typical chromatin distribution throughout the nucleus during interphase (D), CENP-A-depleted cells showed smaller nuclei displaying aberrant morphologies and partial chromatin condensation (E). (F to I) The mitotic phenotypes of cells treated with RNAi against CENP-A and CLIP-170 were assessed by triple fluorescence staining of tubulin (red), centromeres (ACA [green]), and DNA (ToPro-3 [blue]). Images show single confocal sections for each staining pattern as well as overlay views (merge). While control cells showed normal spindle morphology, centromere distribution, and DNA staining during telophase (F), CENP-A-depleted cells had lagging chromosomes (G, arrows). CLIP-170-depleted mitotic cells showed misaligned and/or lagging chromosomes (H, arrows) within anaphase spindles or multipolar spindles with misaligned chromosomes (I). (J) Lamin A/C antibody staining (green) in control cells showed a typical nuclear rim pattern, while in RNAi-treated cells, lamin A/C staining was strongly diminished (K). Note that the nuclear morphology was unaffected in lamin A/C-depleted cells. Bars, 5 μm.
FIG. 4.
Inhibition of proliferation by RNAi against CENP-A and CLIP-170. (A) The frequency of mitotic cells was assessed by immunofluorescence staining of centromeres and DNA in RNAi-untreated cells (mock) or cells incubated for 72 h with RNAi oligonucleotides specific for CENP-A, CLIP-170, or lamin A/C. The diagram shows mean values with standard deviations from four independent experiments (n > 400 for each RNAi experiment). (B) Quantitation of living cells after RNAi. Identical numbers of HEp-2 cells were incubated without (mock) or with the indicated RNAi oligonucleotides for 72 h. The frequency of living cells was then determined by trypan blue exclusion. Data represent mean values and standard deviations from at least five independent experiments (n > 200 for each RNAi experiment). The values were normalized to that for mock-treated cells (set to 100%) to directly compare the effects of RNAi treatments.
The RNAi experiment for CLIP-170 yielded a different phenotype. Seventy-two hours of RNAi exposure led to a massive accumulation of mitotic cells that displayed lagging chromosomes and/or multipolar spindles (Fig. 3H and I and 4A). None of these cells appeared to be able to complete anaphase, since telophase cells could not be observed. The RNAi kinetics of CLIP-170 revealed a gradual accumulation of mitotic cells, with a peak of abnormal mitotic cells at 48 h (not shown). At 72 h, the majority of cells were detached from coverslips due to their mitotic arrest, and the number of living cells was significantly reduced (Fig. 4B). The number of living cells further decreased after 96 h (data not shown). RNAi of lamin A/C diminished the nuclear rim staining of HEp-2 cells, indicating that the levels of lamin A/C had decreased (Fig. 3J and K). However, these cells divided almost normally, did not accumulate in mitosis, and did not show aberrant mitotic phenotypes (Fig. 3K), in accordance with previous observations of RNAi-mediated lamin A/C depletion in HeLa cells (20).
As a common functional readout for the RNAi effects, we also quantified the proportion of living cells after RNAi treatment (Fig. 4B). Identical numbers of cells were not treated or were treated with RNAi oligonucleotides to deplete CENP-A, CLIP-170, and lamin A/C for 72 h, and the numbers of living cells were determined by trypan blue exclusion. While the numbers of living cells after CENP-A and CLIP-170 reduction were reduced to 46% and 22%, respectively, RNAi of lamin A/C caused only a moderate decrease in the living cell number (83%) (Fig. 4B). This result is consistent with the phenotypes caused by the depletion of each of these proteins (Fig. 3B to H). We conclude that the reduction in the number of living cells after CENP-A or CLIP-170 RNAi is a consequence of the cell cycle arrest caused by the specific protein depletion. While CENP-A-depleted cells arrested in interphase, CLIP-170-depleted cells accumulated in mitosis. As a consequence, both phenotypes caused the cells to die and/or stop proliferation.
The S. cerevisiae protein Cse4p can substitute for the function of CENP-A at human centromeres.
The specific localization of yeast kinetochore proteins Cse4p and Bik1p to the centromeres of mitotic human cells suggested that they might fulfill functions similar to those of their human counterparts. To test this hypothesis, we developed a new approach that combines the RNAi-mediated depletion of human kinetochore proteins with the simultaneous ectopic expression of the respective S. cerevisiae protein homologs. The basic idea was to avoid RNAi-induced phenotypes of the endogenous kinetochore protein in HEp-2 cells by coexpressing the homologous yeast protein. Vectors encoding GFP fusion proteins were used in this assay (i) because the GFP fusion proteins had been shown to localize to kinetochores during interphase and mitosis (Fig. 1) and (ii) to monitor the fate of the yeast proteins in the kinetochore protein-depleted human cells. The initial analysis of different protocols in which the relative timing of RNAi oligonucleotide administration and transfection was tested systematically showed that the assay could be used successfully when expression of the exogenous protein occurred 12 h prior to RNAi depletion of the endogenous protein (Fig. 5A). All approaches in which transfection was done after the RNAi treatment did not result in complementation effects, suggesting that Cse4 must be present before CENP-A is diminished (data not shown). The assay was applied to test whether Cse4p and Bik1p are able to rescue the cell cycle arrest induced by RNAi of CENP-A and CLIP-170, respectively. As expected, RNAi of CENP-A caused a decrease in the number of living cells (Fig. 5B). However, when the cells where transfected with the vector encoding GFP-Cse4p before RNAi treatment, we found a relative amount of living cells of 88% ± 5%. The Cse4p-recovered cells showed an almost normal mitotic index (15% ± 2%), and the mitotic cells did not show chromosome misalignments, lagging chromosomes, or abnormal spindles (Fig. 5C). This demonstrated that the ectopically expressed Cse4p protein from S. cerevisiae is able to complement the phenotype induced by the depletion of its human homolog, CENP-A. Transfection of the GFP-CENP-A construct did not result in an increase in the number of living cells after RNAi depletion of endogenous CENP-A (Fig. 5B). This is most likely attributable to the ability of the RNAi oligonucleotides to inhibit protein synthesis from both endogenous CENP-A mRNA and vector-encoded GFP-CENP-A mRNA. The cotransfection of a vector encoding GFP without a fusion also showed no effect, indicating that the complementation effect is specific for Cse4p (data not shown).
FIG. 5.
Budding yeast Cse4p functionally substitutes for CENP-A in human cells. (A) Schematic representation of the assay developed to test the ability of S. cerevisiae kinetochore proteins to functionally complement centromere protein homologs in human cells. Twelve hours prior to RNAi treatment against the endogenous centromere protein, HEp-2 cells were transiently transfected with GFP expression plasmids encoding the homologous protein from budding yeast. The phenotypes of these cells were analyzed after 72 h of RNAi infection. (B) The procedure described in panel A was applied to cells that were not transfected with plasmids or RNAi treated (mock) and to CENP-A- and CLIP-170-depleted cells. The frequencies of living cells were determined at 72 h post-RNAi by trypan blue exclusion. The use of cotransfection plasmids encoding GFP fusion proteins of Cse4p, CENP-A, Bik1p, or CLIP-170 (as described in panel A) is indicated at the bottom. (C) CENP-A RNAi-treated cells cotransfected with GFP-Cse4p were fixed and immunostained to visualize DNA, centromeres, and GFP-Cse4p. The image shows individual channels of a single confocal section from a telophase cell. The Cse4p signals coincide with the positions of centromeres. (D) CLIP-170 RNAi-treated cells cotransfected with GFP-Bik1p were fixed and immunostained to visualize DNA, tubulin, and GFP-Bik1p. These cells show mitotic defects with misaligned chromosomes (upper cell) or multipolar spindles (lower cell). Bik1p is distributed throughout the whole cell, with preferred staining on chromatin. Bars, 5 μm. (E) Untreated HEp-2 cells (circles) and HEp-2 cells (open triangles) or stably Cse4-expressing HEp-2 cells (closed triangles) treated with anti-CENP-A RNAi were observed for 3 days post-RNAi oligonucleotide administration. The frequencies of living cells were determined for at least three independent experiments at 0, 24, 48, and 72 h. While the proliferation of HEp-2 cells was significantly reduced by CENP-A RNAi, the HEp-2/Cse4 cell line proliferated with kinetics similar to those of mock-treated HEp-2 cells.
The same set of experiments was performed with Bik1p and CLIP-170. The number of living cells for CLIP-170-depleted cells dropped to 22% ± 7% that for mock-treated cells (Fig. 5B). However, attempts to recover CLIP-170 function by coexpressing the GFP-Bik1p fusion protein failed. Coexpression even resulted in a smaller number of living cells (5% ± 3%), suggesting that Bik1p overexpression and CLIP-170 down-regulation exerted additive effects. Most of the remaining living cells displayed mitotic phenotypes with lagging chromosomes or multipolar spindles (Fig. 5D). In these cells, the coexpressed GFP-Bik1p fusion protein was diffusely distributed throughout the mitotic chromatin and the cytoplasm (Fig. 5D). Kinetochores were present, as indicated by staining with ACA serum, but GFP signals were never found to specifically colocalize to kinetochores in these cells (data not shown). The coexpression of GFP-CLIP-170 also did not result in a significant increase in the number of living cells (26% ± 5%) after RNAi against CLIP-170 (Fig. 5B).
The ability of Cse4 to rescue CENP-A-depleted cells was also analyzed in our stable cell lines (Fig. 5E). HEp-2 cells stably expressing GFP-Cse4 were treated with RNAi oligonucleotides against CENP-A, and the numbers of living cells were determined after 24, 48, and 72 h. While CENP-A RNAi caused an inhibition of proliferation of parental HEp-2 cells in a time-dependent fashion, the same treatment was not effective in cells that stably expressed Cse4. This cell line proliferated with the same kinetics as untreated HEp-2 cells (Fig. 5E). In contrast, the proliferation of HEp-2 cells that stably expressed GFP-Bik1 was strongly inhibited in the presence of RNAi oligonucleotides against CLIP-170. After 72 h of transient RNAi treatment, the relative amount of living HEp2/Bik1 cells decreased to 31% ± 11% (data not shown).
Taken together, these results show that Cse4p, but not Bik1p, from the budding yeast S. cerevisiae is capable of rescuing human cells in which the protein synthesis of its homologous counterpart, CENP-A, was inhibited by RNAi.
DISCUSSION
Although centromere function is conserved among eukaryotes, centromere morphology varies significantly. The kinetochore complex displays similarities between the budding yeast and human centromere functions, and a cross-species comparison of its components may be key to an understanding of the structure, function, and evolution of this multiprotein complex. We have established a new in vivo assay that utilizes RNAi in combination with ectopic protein expression in order to study the complementation ability of budding yeast kinetochore proteins in human cells. The underlying idea mirrors complementation assays in budding yeast in which the endogenous (yeast) protein is physically or functionally depleted and the depletion phenotype is then analyzed for its response to the expression of a protein homolog from related or unrelated organisms. With the new system, we have tested two proteins, the essential CENP-A protein and CLIP-170, which are components of the inner and outer kinetochore complex, respectively. The deleterious RNAi-induced CENP-A phenotype was rescued by coexpression of the budding yeast homolog Cse4p. To the best of our knowledge, this is the first demonstration of functional complementation of a human protein by its budding yeast homolog. The same strategy was applied for CLIP-170. In this case, the homologous protein Bik1p from S. cerevisiae, although present at the mitotic kinetochore complex in human cells after ectopic expression, was not able to complement the mitotic cell cycle arrest induced by the reduction of CLIP-170.
Functional complementation of CENP-A by Cse4p.
RNAi of CENP-A in HEp-2 cells resulted in misaligned or lagging chromosomes in mitotic cells and constricted nuclei in interphase cells (Fig. 3), similar to CENP-A RNAi phenotypes in HeLa cells (25). CENP-A-depleted HEp-2 cells did not accumulate during mitosis, although a slight increase in the number of mitotic cells was detected (18% versus 8% in control cells). This suggests a mitotic delay, probably due to chromosome missegregation during anaphase, but not a progression block of the cell cycle in mitosis. This interpretation is consistent with observations of mitotically blocked HeLa cells microinjected with anti-CENP-A antibodies. This treatment does not prevent the completion of mitosis, since these cells progress normally to the next interphase (22).
One may argue that the residual amount of CENP-A (10%) (Fig. 3A) in the RNAi-depleted cells might be sufficient to allow kinetochore localization by Cse4p and that residual CENP-A may be supported in its activity by the bound homologue. Such an interpretation is always a potential problem in any kind of knockouts of essential proteins, because 100% depletion of an essential gene renders the cells inaccessible for functional analysis of the protein. Although unlikely, it is hence not completely ruled out that the residual CENP-A may be sufficient to recruit ectopic Cse4p into centromeric nucleosomes and that Cse4p then helps to functionally activate the residual CENP-A population in RNAi-treated cells much beyond its original activity (at least twofold). However, for the protein activity of CENP-A, we consider this kind of “help” to be a functional complementation.
After 72 h of RNAi treatment, CENP-A-reduced interphase cells did not exhibit typical signs of apoptosis, such as extensive chromatin condensation or membrane blebbing (21), but rather they stayed in a cellular state of nonviability (Fig. 3E) and had lost the ability to divide (Fig. 4B), similar to G1-S-synchronized HeLa cells microinjected with an anti-CENP-A antibody (22). How can this phenotype be explained? While DNA replication and histone synthesis are tightly coupled during S phase (31), endogenous CENP-A expression and incorporation into centromeric nucleosomes occur in G2 (71, 72). This regulated expression of CENP-A is required for centromere maintenance (78) and may be a prerequisite for the maturation of centromeres in preparation of the cell for cell division (73, 74, 84). It is therefore conceivable that cells in G2 are most vulnerable to CENP-A synthesis inhibition by RNAi and that CENP-A deficiency-induced abnormalities in CENP-A nucleosome or centromere assembly cause the loss-of-viability and nonproliferation phenotype. However, this interpretation requires further cell cycle analysis of CENP-A-depleted interphase cells.
Experiments with human CENP-A and with Cse4 have shown that both proteins exhibit homotypic chromatin interactions in vivo (24, 34, 43, 53, 72, 82, 92). However, attempts to rescue either temperature-sensitive or null alleles of cse4 in S. cerevisiae by the high-level expression of CENP-A failed (77). In contrast, by the coexpression of Cse4p in HEp-2 cells, we were able to rescue the CENP-A depletion phenotype (Fig. 5), demonstrating that Cse4p can substitute functionally for CENP-A at the kinetochores of human cells. Specific centromere localization (Fig. 1) and the coelution of Cse4p with CENP-A nucleosomes during chromatin fractionation excluded the possibility that the yeast protein is only indirectly recruited to kinetochores rather than directly incorporated into kinetochore nucleosomes. These observations strongly support results from recent chromatin immunoprecipitation assays of budding yeast which indicate that there is an evolutionarily conserved molecular core of eukaryotic kinetochores (89). All of the centromere H3-like protein family members have a highly conserved C-terminal histone-fold domain (HFD) and distinctly different N termini (81). In contrast to the 44-amino-acid N-terminal region of human CENP-A, Cse4p has a unique 135-amino-acid N terminus extending from the HFD (9, 52, 53, 77). Unlike the N terminus of yeast H3, which can be deleted without a loss of cell viability (46), the Cse4p N terminus is essential (33). A thorough mutagenesis study has revealed an essential N-terminal domain (END) of 33 amino acids within the N-terminal region that is necessary and sufficient for the proper kinetochore function of Cse4p (9). The END is involved in the interaction between Cse4p and Ctf19p/Mcm21p/Okp1p (9), a protein complex that mediates protein-protein interactions at the budding yeast centromere, including the essential CFB3 complex (30, 49, 57). The END region is not present in CENP-A (data not shown). Based on these observations, it is likely that the failure of CENP-A to complement Cse4p function in S. cerevisiae is attributable to the lack of the END motif in CENP-A.
The HFDs of Cse4p and CENP-A are each sufficient to target these proteins to yeast and human kinetochores, respectively (9, 82). Transient expression experiments in HeLa cells have shown that GFP-tagged Cse4p localizes to centromeric regions, indicating that the protein sequence contains all of the information needed to deposit Cse4p in human centromeric nucleosomes (28). Further evidence for this conclusion is provided in the present study, firstly by the kinetochore localization of GFP-Cse4p in stably transfected HEp-2 cells (Fig. 1) and secondly by the identical biochemical behaviors of Cse4p during chromatin fractionation and of CENP-A (Fig. 2B). The HFD consists of a set of three α helices (helices I, II, and III) separated by two turn-β-sheet structures (strand A and strand B) (4). A mutational analysis revealed that a central portion consisting of strand A and helix II is primarily responsible for targeting CENP-A to centromeres, while substitutions or deletions in helix I, strand B, or the C-terminal part do not affect centromere targeting (72). Interestingly, the amino acid stretch of the adjacent strand A and helix II of Cse4p shows a higher degree of conservation (51% identity and 69% similarity) with CENP-A than does human histone H3 with CENP-A (46% identity and 67% similarity). In addition, a segment within helix II (LLTL in CENP-A) that is responsible for centromere targeting (72) is present in Cse4p (LLAL), but not in H3 (LCAI). The incorporation of Cse4p into centromeric nucleosomes of human cells may thus be mediated primarily by the conserved central region (strand A and helix II) of the HFD.
Besides proper localization and nucleosome incorporation, CENP-A complementation also requires the replacement of its functional properties by Cse4p. One main function of mammalian CENP-A nucleosomes may be to coordinate the binding of additional kinetochore components, such as CENP-C, CENP-H, CENP-F, and hZwintl-1, to the centromere (29). The precise nature of this function is not clear because direct protein interaction partners of the N-terminal tail of CENP-A have not been established yet. If critical interactions exist between CENP-A and adjacent centromere or kinetochore proteins, we have to assume that Cse4p is able to perform similar interactions. Although they are not conserved in sequence, the N-terminal tails of CENP-A and Cse4p show similarities in the distribution of posttranslationally modifiable lysine, arginine, and serine residues (73). Indeed, Ser7 phosphorylation of CENP-A is cell cycle dependent, highlighting the potential importance of CENP-A modifications in centromere assembly (37). Such epigenetic mechanisms of CENP-A functions may also act on Cse4p in human cells.
Bik1p does not complement CLIP-170 function in human cells.
The reduction of CLIP-170 protein levels by RNAi led to a massive accumulation (85%) of mitotic cells (Fig. 4A). The mitotic arrest was characterized by cells with multipolar spindles (in which all chromosomes were stalled in a metaphase configuration) or bipolar spindles (in which some chromosomes remained in the metaphase plate while others showed different localizations along the spindle). These observations confirmed that CLIP-170 is a kinetochore passenger protein (19) and established the fact that CLIP-170 plays an important role in the kinetochore function of mammalian cells during mitosis. Human CLIP-170 was originally identified as a microtubule (MT)-binding protein that mediates the association between MTs and endocytotic vesicles (63, 66). In interphase cells, CLIP-170 colocalizes with MT plus ends in the cytoplasm (17). During mitosis, CLIP-170 codistributes with dynein and dynactin at kinetochores (19). CLIP-170 may thus be a bifunctional capturing device in mammalian cells which connects MTs to peripheral organelles during interphase and to kinetochores during mitosis. Bik1p is the budding yeast homolog of human CLIP-170 (63). Like CLIP-170, Bik1p shows MT localization, with discrete dots located at the plus ends of MTs in interphase cells. During mitosis, Bik1p is a bona fide component of the budding yeast kinetochore (26, 41). Although Bik1p (51 kDa) is significantly smaller than CLIP-170, the three functional domains identified so far are conserved in both proteins (62). These include the NH2-terminal MT-binding domain (known as the CAP-Gly domain), the central coiled coil domain, and the carboxy-terminal metal-binding motif (cargo-binding domain), which is proposed to link MTs to various targets (19, 63, 69, 70). Furthermore, the MT-binding domains of Bik1p and CLIP-170 are functionally exchangeable in S. cerevisiae (41). The high level of sequence conservation, the overall similarity in the structural organization of Bik1p and CLIP-170, and the functional complementation of their MT-binding domains therefore suggested that these two proteins are true orthologs. Corroborating this conclusion, we showed here that the CLIP-170 deficiency phenotype is similar to that of S. cerevisiae cells from which Bik1p has been genetically deleted. Bik1pΔ mutant cells have highly abnormal spindles (41) and CLIP-170-diminished HEp-2 cells show a high frequency of multipolar spindles (Fig. 3I). Despite these striking similarities, Bik1p was unable to substitute for the CLIP-170 kinetochore function in HEp-2 cells, as the mitotic accumulation and cell cycle arrest of CLIP-170 RNAi cells were unaffected by Bik1p coexpression (Fig. 5). This may be explained by (i) as yet unidentified essential domains in CLIP-170 which are not present in Bik1p, (ii) the lack of key amino acid residues in conserved domains of Bik1p which mediate the proper kinetochore function of CLIP-170, or (iii) the lack of appropriate phosphorylation sites in Bik1p (10), since CLIP-170 functioning is regulated by phosphorylation (63). These differences between CLIP-170 and Bik1p may also disturb critical protein-protein interactions of Bik1p at the kinetochore, for example, with the dynein/dynactin complex (19) or with the kinetochore passenger LIS1 (16, 83).
Evolutionary considerations.
The point centromeres of S. cerevisiae likely represent the most basic iteration of centromeres, which expanded as organisms became more complex and evolved larger chromosomes. CenH3 proteins serve as heritable centromeric molecules which may have coevolved by positive selection driven by changing centromeric DNAs, as suggested by Henikoff et al. (27). Our finding that Cse4p can complement CENP-A across species was therefore unexpected, but in our view it does not contradict the Henikoff model of centromere evolution (27). The specific incorporation of Cse4 into human nucleosomes (Fig. 1 and 2) clearly demonstrates that although this protein may have diverged away from CENP-A along with a changing DNA specificity, the molecule (that is, the conserved core domain) still contains enough sequence information for nucleosome incorporation in human cells. The evolution of the N-terminal tails of CenH3 proteins, in contrast, may be driven by changing protein partners rather than by changing DNA specificities. The N terminus of Cse4p may have accumulated multiple substitutions and additions that are required for budding yeast-specific kinetochore functions, such as an interaction with CBF3 or Okp1p. Alternatively, CENP-A may have lost some of its (N-terminal) functions because they were not required anymore (there is no CBF3 complex homolog in mammalian cells) or because they may have been taken over by other components in the evolving mammalian centromere/kinetochore complex. Yet it remains to be elucidated how the N-terminal part of Cse4 precisely mediates CENP-A functions.
CLIP-170 and Bik1p share a similarly high degree of structural conservation as do CENP-A and Cse4p. The failure of Bik1p to rescue CLIP-170-depleted human cells suggests that Bik1p may lack some functional features which have evolved in the human kinetochore-MT interface (driven by changing protein partners). Thus, the extent to which yeast kinetochore proteins are able to functionally complement their human counterparts—as we determined here—yields information on the evolutionary conservation of both structure and function between human and yeast centromere proteins.
Studies to analyze further budding yeast kinetochore proteins in human cells are under way in our laboratory, and we are confident that this strategy will continue to reveal further common and divergent features of the two different but related kinetochore structures. The in vivo assay described here may also be useful for studying functional similarities and the evolutionary conservation of other multiprotein complexes in eukaryotes as well.
Acknowledgments
We thank M. Koch for technical assistance and F. Perez, T. Munder, and K. Sugimoto for plasmids and antibodies.
REFERENCES
- 1.Albertson, D. G., and J. N. Thomson. 1982. The kinetochores of Caenorhabditis elegans. Chromosoma 86:409-428. [DOI] [PubMed] [Google Scholar]
- 2.Albertson, D. G., and J. N. Thomson. 1993. Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans. Chromosome Res. 1:15-26. [DOI] [PubMed] [Google Scholar]
- 3.Amon, A. 1999. The spindle checkpoint. Curr. Opin. Genet. Dev. 9:69-75. [DOI] [PubMed] [Google Scholar]
- 4.Arents, G., R. W. Burlingame, B. C. Wang, W. E. Love, and E. N. Moudrianakis. 1991. The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix. Proc. Natl. Acad. Sci. USA 88:10148-10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biggins, S., and A. W. Murray. 1999. Sister chromatid cohesion in mitosis. Curr. Opin. Genet. Dev. 9:230-236. [DOI] [PubMed] [Google Scholar]
- 6.Brown, K. D., K. W. Wood, and D. W. Cleveland. 1996. The kinesin-like protein CENP-E is kinetochore-associated throughout poleward chromosome segregation during anaphase-A. J. Cell Sci. 109:961-969. [DOI] [PubMed] [Google Scholar]
- 7.Buchwitz, B. J., K. Ahmad, L. L. Moore, M. B. Roth, and S. Henikoff. 1999. A histone-H3-like protein in C. elegans. Nature 401:547-548. [DOI] [PubMed] [Google Scholar]
- 8.Cheeseman, I. M., D. G. Drubin, and G. Barnes. 2002. Simple centromere, complex kinetochore: linking spindle microtubules and centromeric DNA in budding yeast. J. Cell Biol. 157:199-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, Y., R. E. Baker, K. C. Keith, K. Harris, S. Stoler, and M. Fitzgerald-Hayes. 2000. The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol. Cell. Biol. 20:7037-7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi, J. H., P. G. Bertram, R. Drenan, J. Carvalho, H. H. Zhou, and X. F. Zheng. 2002. The FKBP12-rapamycin-associated protein (FRAP) is a CLIP-170 kinase. EMBO Rep. 3:988-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choo, K. H. 1997. Centromere DNA dynamics: latent centromeres and neocentromere formation. Am. J. Hum. Genet. 61:1225-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choo, K. H. 2000. Centromerization. Trends Cell Biol. 10:182-188. [DOI] [PubMed] [Google Scholar]
- 13.Choo, K. H. 2001. Domain organization at the centromere and neocentromere. Dev. Cell 1:165-177. [DOI] [PubMed] [Google Scholar]
- 14.Cleveland, D. W., Y. Mao, and K. F. Sullivan. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112:407-421. [DOI] [PubMed] [Google Scholar]
- 15.Cooke, C. A., R. L. Bernat, and W. C. Earnshaw. 1990. CENP-B: a major human centromere protein located beneath the kinetochore. J. Cell Biol. 110:1475-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coquelle, F. M., M. Caspi, F. P. Cordelieres, J. P. Dompierre, D. L. Dujardin, C. Koifman, P. Martin, C. C. Hoogenraad, A. Akhmanova, N. Galjart, J. R. De Mey, and O. Reiner. 2002. LIS1, CLIP-170's key to the dynein/dynactin pathway. Mol. Cell. Biol. 22:3089-3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diamantopoulos, G. S., F. Perez, H. V. Goodson, G. Batelier, R. Melki, T. E. Kreis, and J. E. Rickard. 1999. Dynamic localization of CLIP-170 to microtubule plus ends is coupled to microtubule assembly. J. Cell Biol. 144:99-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobie, K. W., K. L. Hari, K. A. Maggert, and G. H. Karpen. 1999. Centromere proteins and chromosome inheritance: a complex affair. Curr. Opin. Genet. Dev. 9:206-217. [DOI] [PubMed] [Google Scholar]
- 19.Dujardin, D., U. I. Wacker, A. Moreau, T. A. Schroer, J. E. Rickard, and J. R. De Mey. 1998. Evidence for a role of CLIP-170 in the establishment of metaphase chromosome alignment. J. Cell Biol. 141:849-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 21.El-Sheraby, A. M., and J. R. Hinchliffe. 1974. Apoptotic cells exhibit membrane phenotypes. J. Embryol. Exp. Morphol. 31:643-654. [PubMed]
- 22.Figueroa, J., R. Saffrich, W. Ansorge, and M. Valdivia. 1998. Microinjection of antibodies to centromere protein CENP-A arrests cells in interphase but does not prevent mitosis. Chromosoma 107:397-405. [DOI] [PubMed] [Google Scholar]
- 23.Fritzler, M. J., and T. D. Kinsella. 1980. The CREST syndrome: a distinct serologic entity with anticentromere antibodies. Am. J. Med. 69:520-526. [DOI] [PubMed] [Google Scholar]
- 24.Glowczewski, L., P. Yang, T. Kalashnikova, M. S. Santisteban, and M. M. Smith. 2000. Histone-histone interactions and centromere function. Mol. Cell. Biol. 20:5700-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goshima, G., T. Kiyomitsu, K. Yoda, and M. Yanagida. 2003. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 160:25-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He, X., D. R. Rines, C. W. Espelin, and P. K. Sorger. 2001. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell 106:195-206. [DOI] [PubMed] [Google Scholar]
- 27.Henikoff, S., K. Ahmad, and H. S. Malik. 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293:1098-1102. [DOI] [PubMed] [Google Scholar]
- 28.Henikoff, S., K. Ahmad, J. S. Platero, and B. van Steensel. 2000. Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl. Acad. Sci. USA 97:716-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howman, E. V., K. J. Fowler, A. J. Newson, S. Redward, A. C. MacDonald, P. Kalitsis, and K. H. Choo. 2000. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl. Acad. Sci. USA 97:1148-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hyland, K. M., J. Kingsbury, D. Koshland, and P. Hieter. 1999. Ctf19p: a novel kinetochore protein in Saccharomyces cerevisiae and a potential link between the kinetochore and mitotic spindle. J. Cell Biol. 145:15-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jackson, V. 1988. Deposition of newly synthesized histones: hybrid nucleosomes are not tandemly arranged on daughter DNA strands. Biochemistry 27:2109-2121. [DOI] [PubMed] [Google Scholar]
- 32.Janke, C., J. Ortiz, T. U. Tanaka, J. Lechner, and E. Schiebel. 2002. Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J. 21:181-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keith, K. C., R. E. Baker, Y. Chen, K. Harris, S. Stoler, and M. Fitzgerald-Hayes. 1999. Analysis of primary structural determinants that distinguish the centromere-specific function of histone variant Cse4p from histone H3. Mol. Cell. Biol. 19:6130-6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keith, K. C., and M. Fitzgerald-Hayes. 2000. CSE4 genetically interacts with the Saccharomyces cerevisiae centromere DNA elements CDE I and CDE II but not CDE III. Implications for the path of the centromere DNA around a Cse4p variant nucleosome. Genetics 156:973-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kiesslich, A., A. von Mikecz, and P. Hemmerich. 2002. Cell cycle-dependent association of PML bodies with sites of active transcription in nuclei of mammalian cells. J. Struct. Biol. 140:167-179. [DOI] [PubMed] [Google Scholar]
- 36.Kitagawa, K., and P. Hieter. 2001. Evolutionary conservation between budding yeast and human kinetochores. Nat. Rev. Mol. Cell. Biol. 2:678-687. [DOI] [PubMed] [Google Scholar]
- 37.Kunitoku, N., T. Sasayama, T. Marumoto, D. Zhang, S. Honda, O. Kobayashi, K. Hatakeyama, Y. Ushio, H. Saya, and T. Hirota. 2003. CENP-A phosphorylation by Aurora-A in prophase is required for enrichment of Aurora-B at inner centromeres and for kinetochore function. Dev. Cell 5:853-864. [DOI] [PubMed] [Google Scholar]
- 38.Lechner, J., and J. Carbon. 1991. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell 64:717-725. [DOI] [PubMed] [Google Scholar]
- 39.Lechner, J., and J. Ortiz. 1996. The Saccharomyces cerevisiae kinetochore. FEBS Lett. 389:70-74. [DOI] [PubMed] [Google Scholar]
- 40.Li, Y., J. Bachant, A. A. Alcasabas, Y. Wang, J. Qin, and S. J. Elledge. 2002. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev. 16:183-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin, H., P. de Carvalho, D. Kho, C. Y. Tai, P. Pierre, G. R. Fink, and D. Pellman. 2001. Polyploids require Bik1 for kinetochore-microtubule attachment. J. Cell Biol. 155:1173-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu, S. T., J. C. Hittle, S. A. Jablonski, M. S. Campbell, K. Yoda, and T. J. Yen. 2003. Human CENP-I specifies localization of CENP-F, MAD1 and MAD2 to kinetochores and is essential for mitosis. Nat. Cell Biol. 5:341-345. [DOI] [PubMed] [Google Scholar]
- 43.Lo, A. W., D. J. Magliano, M. C. Sibson, P. Kalitsis, J. M. Craig, and K. H. Choo. 2001. A novel chromatin immunoprecipitation and array (CIA) analysis identifies a 460-kb CENP-A-binding neocentromere DNA. Genome Res. 11:448-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malik, H. S., and S. Henikoff. 2001. Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics 157:1293-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maney, T., L. M. Ginkel, A. W. Hunter, and L. Wordeman. 2000. The kinetochore of higher eucaryotes: a molecular view. Int. Rev. Cytol. 194:67-131. [DOI] [PubMed] [Google Scholar]
- 46.Mann, R. K., and M. Grunstein. 1992. Histone H3 N-terminal mutations allow hyperactivation of the yeast GAL1 gene in vivo. EMBO J. 11:3297-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McEwen, B. F., Y. Ding, and A. B. Heagle. 1998. Relevance of kinetochore size and microtubule-binding capacity for stable chromosome attachment during mitosis in PtK1 cells. Chromosome Res. 6:123-132. [DOI] [PubMed] [Google Scholar]
- 48.McEwen, B. F., C. E. Hsieh, A. L. Mattheyses, and C. L. Rieder. 1998. A new look at kinetochore structure in vertebrate somatic cells using high-pressure freezing and freeze substitution. Chromosoma 107:366-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Measday, V., D. W. Hailey, I. Pot, S. A. Givan, K. M. Hyland, G. Cagney, S. Fields, T. N. Davis, and P. Hieter. 2002. Ctf3p, the Mis6 budding yeast homolog, interacts with Mcm22p and Mcm16p at the yeast outer kinetochore. Genes Dev. 16:101-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mellor, J., W. Jiang, M. Funk, J. Rathjen, C. A. Barnes, T. Hinz, J. H. Hegemann, and P. Philippsen. 1990. CPF1, a yeast protein which functions in centromeres and promoters. EMBO J. 9:4017-4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meluh, P. B., and D. Koshland. 1997. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 11:3401-3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meluh, P. B., and D. Koshland. 1995. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell 6:793-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meluh, P. B., P. Yang, L. Glowczewski, D. Koshland, and M. M. Smith. 1998. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94:607-613. [DOI] [PubMed] [Google Scholar]
- 54.Muro, Y., H. Masumoto, K. Yoda, N. Nozaki, M. Ohashi, and T. Okazaki. 1992. Centromere protein B assembles human centromeric alpha-satellite DNA at the 17-bp sequence, CENP-B box. J. Cell Biol. 116:585-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nekrasov, V. S., M. A. Smith, S. Peak-Chew, and J. V. Kilmartin. 2003. Interactions between centromere complexes in Saccharomyces cerevisiae. Mol. Biol. Cell 14:4931-4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nishihashi, A., T. Haraguchi, Y. Hiraoka, T. Ikemura, V. Regnier, H. Dodson, W. C. Earnshaw, and T. Fukagawa. 2002. CENP-I is essential for centromere function in vertebrate cells. Dev. Cell 2:463-476. [DOI] [PubMed] [Google Scholar]
- 57.Ortiz, J., O. Stemmann, S. Rank, and J. Lechner. 1999. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 13:1140-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Palmer, D. K., K. O'Day, H. L. Trong, H. Charbonneau, and R. L. Margolis. 1991. Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc. Natl. Acad. Sci. USA 88:3734-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Palmer, D. K., K. O'Day, M. H. Wener, B. S. Andrews, and R. L. Margolis. 1987. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 104:805-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Perez, F., G. S. Diamantopoulos, R. Stalder, and T. E. Kreis. 1999. CLIP-170 highlights growing microtubule ends in vivo. Cell 96:517-527. [DOI] [PubMed] [Google Scholar]
- 61.Pidoux, A. L., and R. C. Allshire. 2000. Centromeres: getting a grip of chromosomes. Curr. Opin. Cell Biol. 12:308-319. [DOI] [PubMed] [Google Scholar]
- 62.Pierre, P., R. Pepperkok, and T. E. Kreis. 1994. Molecular characterization of two functional domains of CLIP-170 in vivo. J. Cell Sci. 107:1909-1920. [DOI] [PubMed] [Google Scholar]
- 63.Pierre, P., J. Scheel, J. E. Rickard, and T. E. Kreis. 1992. CLIP-170 links endocytic vesicles to microtubules. Cell 70:887-900. [DOI] [PubMed] [Google Scholar]
- 64.Prasher, D. C., V. K. Eckenrode, W. W. Ward, F. G. Prendergast, and M. J. Cormier. 1992. Primary structure of the Aequorea victoria green-fluorescent protein. Gene 111:229-233. [DOI] [PubMed] [Google Scholar]
- 65.Remboutsika, E., Y. Lutz, A. Gansmuller, J. L. Vonesch, R. Losson, and P. Chambon. 1999. The putative nuclear receptor mediator TIF1alpha is tightly associated with euchromatin. J. Cell Sci. 112:1671-1683. [DOI] [PubMed] [Google Scholar]
- 66.Rickard, J. E., and T. E. Kreis. 1990. Identification of a novel nucleotide-sensitive microtubule-binding protein in HeLa cells. J. Cell Biol. 110:1623-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rieder, C. L., and E. D. Salmon. 1998. The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 8:310-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rose, S. M., and W. T. Garrard. 1984. Differentiation-dependent chromatin alterations precede and accompany transcription of immunoglobulin light chain genes. J. Biol. Chem. 259:8534-8544. [PubMed] [Google Scholar]
- 69.Schroer, T. A. 2000. Motors, clutches and brakes for membrane traffic: a commemorative review in honor of Thomas Kreis. Traffic 1:3-10. [DOI] [PubMed] [Google Scholar]
- 70.Schuyler, S. C., and D. Pellman. 2001. Microtubule “plus-end-tracking proteins”: the end is just the beginning. Cell 105:421-424. [DOI] [PubMed] [Google Scholar]
- 71.Shelby, R. D., K. Monier, and K. F. Sullivan. 2000. Chromatin assembly at kinetochores is uncoupled from DNA replication. J. Cell Biol. 151:1113-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shelby, R. D., O. Vafa, and K. F. Sullivan. 1997. Assembly of CENP-A into centromeric chromatin requires a cooperative array of nucleosomal DNA contact sites. J. Cell Biol. 136:501-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smith, M. M. 2002. Centromeres and variant histones: what, where, when and why? Curr. Opin. Cell Biol. 14:279-285. [DOI] [PubMed] [Google Scholar]
- 74.Smith, M. M. 2002. Histone variants and nucleosome deposition pathways. Mol. Cell 9:1158-1160. [DOI] [PubMed] [Google Scholar]
- 75.Smith, M. M., P. Yang, M. S. Santisteban, P. W. Boone, A. T. Goldstein, and P. C. Megee. 1996. A novel histone H4 mutant defective in nuclear division and mitotic chromosome transmission. Mol. Cell. Biol. 16:1017-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Spector, D. L., R. D. Goldman, and L. A. Leinwand. 1997. Cells, a laboratory manual, vol. 1 to 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 77.Stoler, S., K. C. Keith, K. E. Curnick, and M. Fitzgerald-Hayes. 1995. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 9:573-586. [DOI] [PubMed] [Google Scholar]
- 78.Sugata, N., S. Li, W. C. Earnshaw, T. J. Yen, K. Yoda, H. Masumoto, E. Munekata, P. E. Warburton, and K. Todokoro. 2000. Human CENP-H multimers colocalize with CENP-A and CENP-C at active centromere-kinetochore complexes. Hum. Mol. Genet. 9:2919-2926. [DOI] [PubMed] [Google Scholar]
- 79.Sugata, N., E. Munekata, and K. Todokoro. 1999. Characterization of a novel kinetochore protein, CENP-H. J. Biol. Chem. 274:27343-27346. [DOI] [PubMed] [Google Scholar]
- 80.Sugimoto, K., R. Fukuda, and M. Himeno. 2000. Centromere/kinetochore localization of human centromere protein A (CENP-A) exogenously expressed as a fusion to green fluorescent protein. Cell Struct. Funct. 25:253-261. [DOI] [PubMed] [Google Scholar]
- 81.Sullivan, K. F. 2001. A solid foundation: functional specialization of centromeric chromatin. Curr. Opin. Genet. Dev. 11:182-188. [DOI] [PubMed] [Google Scholar]
- 82.Sullivan, K. F., M. Hechenberger, and K. Masri. 1994. Human CENP-A contains a histone H3 related histone fold domain that is required for targeting to the centromere. J. Cell Biol. 127:581-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tai, C. Y., D. L. Dujardin, N. E. Faulkner, and R. B. Vallee. 2002. Role of dynein, dynactin, and CLIP-170 interactions in LIS1 kinetochore function. J. Cell Biol. 156:959-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Takahashi, K., E. S. Chen, and M. Yanagida. 2000. Requirement of Mis6 centromere connector for localizing a CENP-A-like protein in fission yeast. Science 288:2215-2219. [DOI] [PubMed] [Google Scholar]
- 85.Tyler-Smith, C., and G. Floridia. 2000. Many paths to the top of the mountain: diverse evolutionary solutions to centromere structure. Cell 102:5-8. [DOI] [PubMed] [Google Scholar]
- 86.Vafa, O., and K. F. Sullivan. 1997. Chromatin containing CENP-A and alpha-satellite DNA is a major component of the inner kinetochore plate. Curr. Biol. 7:897-900. [DOI] [PubMed] [Google Scholar]
- 87.von Mikecz, A., S. Zhang, M. Montminy, E. M. Tan, and P. Hemmerich. 2000. CREB-binding protein (CBP)/p300 and RNA polymerase II colocalize in transcriptionally active domains in the nucleus. J. Cell Biol. 150:265-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Warburton, P. E., C. A. Cooke, S. Bourassa, O. Vafa, B. A. Sullivan, G. Stetten, G. Gimelli, D. Warburton, C. Tyler-Smith, K. F. Sullivan, G. G. Poirier, and W. C. Earnshaw. 1997. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol. 7:901-904. [DOI] [PubMed] [Google Scholar]
- 89.Westermann, S., I. M. Cheeseman, S. Anderson, J. R. Yates III, D. G. Drubin, and G. Barnes. 2003. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J. Cell Biol. 163:215-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wiens, G. R., and P. K. Sorger. 1998. Centromeric chromatin and epigenetic effects in kinetochore assembly. Cell 93:313-316. [DOI] [PubMed] [Google Scholar]
- 91.Wigge, P. A., and J. V. Kilmartin. 2001. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 152:349-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yoda, K., S. Ando, S. Morishita, K. Houmura, K. Hashimoto, K. Takeyasu, and T. Okazaki. 2000. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc. Natl. Acad. Sci. USA 97:7266-7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yu, H. G., E. N. Hiatt, A. Chan, M. Sweeney, and R. K. Dawe. 1997. Neocentromere-mediated chromosome movement in maize. J. Cell Biol. 139:831-840. [DOI] [PMC free article] [PubMed] [Google Scholar]