Abstract
All-trans-retinoic acid (RA) stimulates differentiation of normal hematopoietic progenitors and acute myeloid leukemia cells. GATA-2 is a transcription factor expressed in early progenitor cells and implicated in the control of the fate of hematopoietic stem cells and progenitor cells. We have investigated the possibility that the GATA and nuclear hormone receptor pathways are functionally linked through direct protein-protein interaction. Here we demonstrate that in human myeloid KG1 cells, RA receptor alpha (RARα), the major RAR expressed in hematopoietic cells, associates with GATA-2. This association is mediated by the zinc fingers of GATA-2 and the DNA-binding domain of RARα. As a consequence of this interaction, RARα is tethered to the DNA sites that are recognized and bound by GATA-2, and the transcriptional activity of GATA-2 becomes RA responsive. The RA responsiveness of GATA-dependent transcription is eliminated by expression of either a dominant negative form of RARα or a GATA-2 mutant that fails to interact with RARα. Overexpression of RXRα inhibits RARα binding to the GATA-2-DNA complex, thus resulting in attenuation of the effects of RARα on GATA-2 activity. In addition, inhibition by RA of GATA-2-dependent hematopoietic colony formation in an embryonic stem cell model of hematopoietic differentiation provided biological evidence for functional cross talk between RA and GATA-2-dependent pathways.
Hematopoiesis is highly regulated in vertebrates and capable of numerous adaptive responses to changing conditions. Among the factors that modulate hematopoiesis are a number of nuclear receptor ligands such as the steroid and thyroid hormones, as well as vitamin A derivatives such as all-trans-retinoic acid (RA). Estrogens stimulate outgrowth of avian bone marrow-derived erythroid progenitor cells and delay their maturation. This delay is associated with reduced expression of many erythroid cell-specific genes (48). The role of thyroid hormones in erythropoiesis is reflected by the inhibitory effect of the dominant negative form of the thyroid hormone receptor (v-erbA) on erythropoiesis (47, 65).
Several lines of evidence support a role of RA receptor alpha (RARα) in regulating myeloid development, in particular along the granulocytic pathway. Acute promyelocytic leukemia (APL), which represents a block in granulocytic differentiation, is associated with chromosomal translocations involving RARα (66). The translocations give rise to fusion proteins that, at physiological concentrations of RA, act as dominant negative forms of wild-type RARα (30). Hematopoietic progenitor cells engineered to express dominant negative forms of RARα have been shown to be defective in granulocytic differentiation pathways (58-60). Similarly, antagonists of RARα inhibit myelopoiesis (34) and RARα agonists inhibit proliferation of primitive progenitor cells and stimulate a myeloid differentiation program (6, 52). Hematopoietic cells lacking RARs exhibit abnormalities in myeloid differentiation (25, 28), and expression of RARα is positively regulated by myelomonocytic growth factors (68).
The mechanisms by which steroid hormones can alter hematopoiesis are not fully understood, but one possible mode of their action may involve functional links with key transcriptional regulators of hematopoiesis. The GATA proteins may provide such an example. These comprise a family of transcriptional factors characterized by the ability to bind a common conserved DNA sequence (WGATAR) by virtue of evolutionarily conserved C4 zinc finger domains (42, 49). Of these, GATA-1, GATA-2, and GATA-3 are expressed in hematopoietic cells. GATA-1 is expressed at a high level in erythroid cells, mast cells, megakaryocytes, and eosinophils and at a low level in multipotent progenitors. GATA-2 is more broadly expressed among hematopoietic cells, with particularly prominent expression in early progenitor cells (42, 49). Loss-of-function experiments suggest that GATA-2 is critically involved in the survival and growth of multipotent progenitors (57). Forced-expression studies with factor-dependent cell lines and primary cells are also consistent with the involvement of GATA-2 in these processes (2, 15, 20, 26, 43). However, these experiments have revealed both positive and negative effects on progenitor cell proliferation and differentiation. These different results may reflect differences in cell context. They may also, in part, be attributable to the nature of the GATA-2 moieties involved, given that some of the studies made use of GATA-2-ER (estrogen receptor) fusion molecules that may not retain all of the properties of the native GATA-2 molecule (2, 15, 20, 26). Taken together these findings implicate GATA-2 as a key transcription factor controlling the fate of hematopoietic stem and progenitor cells.
Given the key roles played by GATA-2 and RARα in hematopoiesis, we postulated that there could be an important functional interaction between these factors. Here we present results indicating that GATA-2 interacts with RARα and suggesting that retinoids and GATA-2 cooperate to positively modulate myeloid differentiation programs.
MATERIALS AND METHODS
Expression plasmids.
An expression plasmid for human GATA-2 (GATA-2/pMT2) (13) was generously provided by S. H. Orkin (Harvard Medical School, Boston, Mass.). Flag-tagged GATA-2/pCMV has been described previously (62). The dominant negative form of RARα (RARα403) was constructed as previously described (58, 59). To construct expression vectors for Flag-tagged versions of RARα and RXRα, the coding regions of RARα and RXRα were produced by PCR with cDNAs for RARα and RXRα as templates and cloned into the pFLAG-CMV2 vector (Eastman Kodak, New Haven, Conn.). The encoding regions were fully sequenced to confirm the correct sequence.
The GATA-2 LW→AA mutant was made by PCR-mediated mutagenesis with primers G2SacII/S (ATCTTCCGCGGGGGGTA), G2BamHI/AS (CCGAGTCTGGATCCTT), NfLW/AA/S (GCAACCCCTGCCGCGCGGCGGGA),NfLW/AA/AS (TCCCGCCGCGCGGCAGGGGTTGC), CfLW/AA/S (ACCACCACCGCAGCGCGCCGAAAC), and CfLW/AA/AS (GTTTCGGCGCGCTGCGGTGGTGGT). Briefly, human GATA-2 cDNA was used as a template and two sets of PCRs were done with primer sets G2SacII/S-CfLW/AA/AS and G2BamHI/AS-CfLW/AA/S. The resulting PCR products were gel purified and mixed, and then a PCR was performed without added primers. Primers G2SacII/S and G2BamHI/AS were then added, and the mutant cDNA fragment from SacII to BamHI was amplified. The resulting PCR product was used as a template for the same procedure with primers NfLW/AA/S and NfLW/AA/AS in the place of primers CfLW/AA/S and CfLW/AA/AS. The resulting cDNA fragment encompassing SacII to BamHI with LW→AA substitutions in both the N and C fingers was digested with SacII and BamHI and inserted into the cDNA for human GATA-2, from which wild-type SacII-BamHI was cut out. The construct was validated by subsequent DNA sequencing.
Protein interaction assays.
293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum (FCS). Human leukemia KG1 cells were maintained in RPMI 1640 medium supplemented with 10% FCS. 293T cells (106) grown in 10-cm-diameter dishes were transfected with the indicated expression plasmids by a standard calcium phosphate coprecipitation method. The total amount of plasmids was equalized by the addition of corresponding empty vectors. Forty-eight hours later, nuclear extracts were prepared as described elsewhere (12) and immunoprecipitated with anti-Flag antibody (M2) in combination with protein G-agarose beads in the binding buffer (20 mM HEPES-KCl [pH 7.9], 140 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 5 mg of bovine albumin per ml, 5% protease inhibitor cocktail [Sigma, St. Louis, Mo.], 0.1% NP-40). After five washes with the binding buffer, immune complexes were analyzed by Western blotting with the indicated antibodies. For immunoprecipitations with KG1 cells, nuclear extracts from 107 cells and agarose-conjugated anti-GATA-2 antibody or agarose-conjugated mouse immunoglobulin (Ig; control) were used. Anti-Flag antibody M2 and protein G-agarose beads were purchased from Sigma. Agarose-conjugated anti-GATA-2 antibodies, agarose-conjugated mouse Ig, and polyclonal antibodies against GATA-2, RARα, and RXRα were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.).
One- and two-hybrid analyses were conducted essentially as detailed in the commercially obtained CheckMate Mammalian Two-Hybrid System (Promega). For the GAL4-GATA-2 fusion construct an NcoI fragment encompassing the entire cDNA for human GATA-2 except for the last amino acid was Klenow filled and inserted into the EcoRV site of the pBIND vector. For the VP16-RARα fusion construct the entire coding sequence of RARα was generated by PCR with primers with engineered BamHI sites and inserted into the BamHI site of pACT. Transient transfection of 293T cells was performed as described below.
Expression plasmids for glutathione S-transferase (GST) fusion proteins containing various parts of GATA-2 have been described previously (63). Fragments of cDNA for RARα were produced by PCR with the cDNA of RARα as a template in combinations with the following primers (restriction enzyme recognition sites are underlined): primer 1, CCAGAATTCATGGCCAGCAA-CAGCAGCT; primer 2, AGTACTCGAGCCCATAGTGGTAGCCTGAGGA; primer 3, CCAG-AATTCCCCTCGCC-ACCCCCTCTA; primer 4, AGTACTCGAGCTGGCAGAGGGCAG-GGAA; primer 5, ATAGAATTCAAAGCGCACCAGGAAACCTT; primer 6, ATACTCG-AGCGGTCACGGG-GAGTGGGT. Primers 1 and 2 were used for cloning of the A/B region of RARα, primers 3 and 4 were for the DNA-binding domain, primers 5 and 6 were for the ligand-binding domain, and primers 1 and 4 were for A/B plus the DNA-binding domain. The PCR products were digested with EcoRI and XhoI and inserted into the EcoRI/SalI site of GST fusion vector pGEX5x-1 (Pharmacia, Uppsala, Sweden). The bacterially expressed GST fusion proteins were purified in accordance with the manufacturer's instructions. Nuclear extracts of 293T cells transfected with an expression plasmid for Flag-RARα or Flag-GATA-2 were prepared as described previously and incubated with the indicated GST fusion proteins bound to the resin in the binding buffer (50 mM Tris-HCl [pH 7.5], 140 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 5 mg of bovine serum albumin per ml, 5% protease inhibitor cocktail, 0.1% NP-40) as described previously (63). The resin was washed five times with the binding buffer, and the bound protein was analyzed by Western blotting with anti-Flag antibody M2 (Eastman Kodak).
DNA-binding assays.
Nuclear extracts from the 293T cells transfected with the indicated expression plasmids or KG1 cells were incubated with 40 pmol of biotinylated double-stranded oligonucleotides containing the recognition sites for GATA-2 (TATTTTTATCTGATAGGAAGT [recognition site in boldface type]) in combination with streptavidin-agarose beads in the binding buffer described above, essentially as described previously (23, 32). After five washes with the binding buffer, proteins captured by the oligonucleotides were analyzed by Western blotting with the indicated antibodies. Biotinylated double-stranded oligonucleotides mutated in the core recognition sequence from GATA to TTTA were used as controls. For the analyses of protein binding to RA-responsive elements (RAREs), the following biotinylated oligonucleotides and their complementary antisense oligonucleotides were used after annealing: biotinylated DR2 (CGATCTAGGGTTCACCAGTTCACTCGGAT) and biotinylated DR5 (CGCACTAGGGTTCACCGAAAGTTCACTCGCTT). Biotinylated oligonucleotides mutated in the core recognition sequence from GGTTCA and AGTTCA to GGTAGT and AGTAGT (mutant DR2, mutant DR5), respectively, were used as controls. Electrophoretic mobility shift assays (EMSAs) with radiolabeled probes were performed as previously described (63). Dissociation assays were conducted as previously described (9).
Transactivation assays.
A luciferase reporter plasmid in which two copies of back-to-back double GATA sites in the mouse CD34 promoter were placed upstream of the β-globin minimal promoter driving a luciferase gene (designated CD34x2/Luc.) has already been described (63). A luciferase reporter plasmid in which a murine GATA-1 promoter (positions −798 to −574) containing a double GATA site was arrayed upstream of the β-globin minimal promoter (designated GATA-1/Luc.) was a gift from M. Yamamoto (Tsukuba University, Tsukuba, Japan) (24, 61). The mutant reporters in which core recognition sequences were changed from GATA to TTTA (designated mutant CD34x2/Luc. and mutant GATA-1/Luc.) were described previously (63). Luciferase reporter assays were conducted as previously described (63), with pRL-CMV-Renilla luciferase plasmids (Promega) used to monitor transfection efficiencies. All-trans-RA (Sigma) was added to the culture medium 24 h after transfection where indicated, and luciferase activities were measured after a further 24 h. The relative luciferase activities reflect duplicate values from a representation of no fewer than two independent experiments.
Semiquantitative RT-PCR.
HEL (human erythroleukemia) cells were cultivated in RPMI medium supplemented with 10% FCS and antibiotics. Untreated cells and those incubated with RA (final concentration, 10−6 M, 0.0005% ethanol; Sigma-Aldrich Company Ltd., Poole, United Kingdom) were harvested after 24 h. Total RNA was isolated with RNA-Bee (Biogenesis, Poole, England). Reverse transcription (RT) was carried out with 64 ng of RNA per μl, Moloney murine leukemia virus reverse transcriptase (Gibco Invitrogen, Paisley, United Kingdom), random hexamer primers, and reaction conditions suggested by the supplier.
Murine FCDPmix A4 cells were cultivated as described before (21, 68). All cells were maintained in a high concentration of interleukin-3 (IL-3; 10 ng/ml), except for induction of erythrocytic differentiation with Epo (1 U/ml) and hemin (2 × 10−4 M), where the IL-3 concentration was reduced to 0.05 ng/ml. RA (10−6 M) and the RARα antagonist Ro 41-5253 (10−5M) were used as previously described (68). Semiquantitative PCR was performed in the GeneAmp PCR system 9700 (Applied Biosystems, Warrington, United Kingdom) with the Expand High Fidelity PCR system (Roche Diagnostics, GmbH, Mannheim, Germany) and 500 nM each PCR primer. PCR primer pairs were derived from sequences present in different exons to avoid confounding results due to the possible presence of small amounts of genomic DNA in RNA samples. For detection of murine sequences, the following forward and reverse PCR primer pairs were used: mCD34, 5′-AAGCCACCAGAGCTATTCCC and 5′-GTTGTCTTGCTGAATGGCCG; mGATA1, 5′-TCACCATCAGATTCCACAGG and 5′-CCAAGAACGTGTTGTTGCTC; mGAPDH, 5′-GGGAAGCCCATCACCATCTT and 5′-GCCTTCTCCATGGTGGTGAA. The forward and reverse primers used to detect human sequences were as follows: GATA1, 5′-TGCTCTGGTGTCCTCCACAC and 5′-TGGGAGAGGAATAGGCTGCT; β2-microglobulin, 5′-TGACTTTGTCACAGCCCAAGATA and 5′-AATCCAAATGCGGCATCTTC. The GATA-2 primers used (5′-GACTATGGCAGCAGTCTCTTCC and 5′-GGTGGTTGTCGTCTGACAATT) detect both human and mouse GATA-2 transcripts. After an initial 2-min denaturation step at 94°C, the PCR amplification conditions were as follows: mCD34, 27 cycles of annealing (20 s), extension (30 s), and denaturation (20 s) at 60, 72, and 94°C, respectively; mGATA1, 27 cycles of annealing (30 s), extension (40 s), and denaturation (20 s) at 61, 72, and 94°C, respectively; mGAPDH, the same as for mCD34 but for 25 cycles; human GATA1 and β2-microglobulin, 25 cycles of annealing (20 s), extension (40 s), and denaturation (15 s) at 64, 72, and 95°C, respectively. Aliquots of each PCR mixture were analyzed by electrophoresis in 1.5% agarose gel and TAE buffer. The expected sizes of specific PCR product were as follows: mCD34, 290 bp; mGATA1, 325 bp; mGAPDH, 113 bp; human GATA1, 491 bp; human β2-microglobulin, 82 bp; mouse and human GATA-2, 297 bp.
Culture and differentiation of ES cells.
The various GATA-2-containing embryonic stem (ES) cell clones used in this study have been previously reported (26) and were maintained as previously described (39). Culture of OP9 stromal cells and in vitro differentiation induction to hematopoietic cells from ES cells on OP9 cells were performed as described previously (37, 38). In the OP9 system, primitive erythrocytes and definitive multipotent hematopoietic progenitors develop at day 5 of differentiation induction (36-38). GATA-2 expression was therefore induced by withdrawal of tetracycline (TET) after day 5 to allow examination of its function in hematopoiesis. Hematopoietic colonies were then counted 2 days after induction of GATA-2 expression.
RESULTS
Interaction of GATA-2 with RARα.
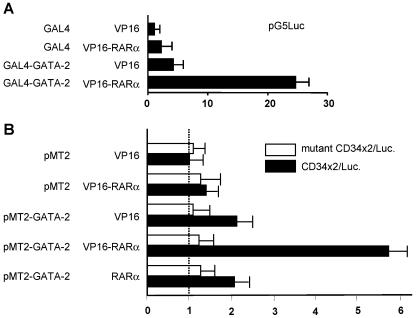
To examine a potential functional relationship between GATA-2 and RA in hematopoiesis, we first investigated whether GATA-2 can physically interact with RARα. Initial experiments were conducted with heterologous 293T cells and mammalian one- and two-hybrid assays. In the mammalian two-hybrid assay (Fig. 1A), significant activation of the pG5Luciferase reporter plasmid is only seen in the presence of the expression of both GAL4-GATA-2 and VP16-RARα. The one-hybrid data (Fig. 1B) are also indicative of an interaction between GATA-2 and RARα. Importantly, the one-hybrid data showed that the interaction of GATA-2 with VP16-RARα could stimulate the activity of a GATA-dependent reporter, suggesting that GATA-2 could recruit RARα to a GATA binding site.
FIG. 1.
Mammalian one- and two-hybrid analyses of GATA-2-RARα interaction. (A) Two-hybrid analysis conducted by transient transfection of 293T cells with the constructs indicated. The GAL4-GATA-2 fusion encompasses the entire human GATA-2 coding region save the last amino acid, which was mutagenized to facilitate subcloning into the pBIND expression vector (Promega). Similarly, the VP19-RARα fusion cloned into pACT (Promega) contains the entire RARα coding region. The relative activity of the pGL5 reporter plasmid (Promega) is plotted on the x axis. (B) One-hybrid analysis conducted with 293T cells and the constructs indicated by methods similar to those described above. Solid bars represent relative luciferase activities from the reporter designated CD34x2/Luc., which contains two copies of a double GATA site identified in the mouse CD34 promoter. Open bars represent activity from a version of this reporter in which these GATA sites have been mutated to abolish GATA binding.
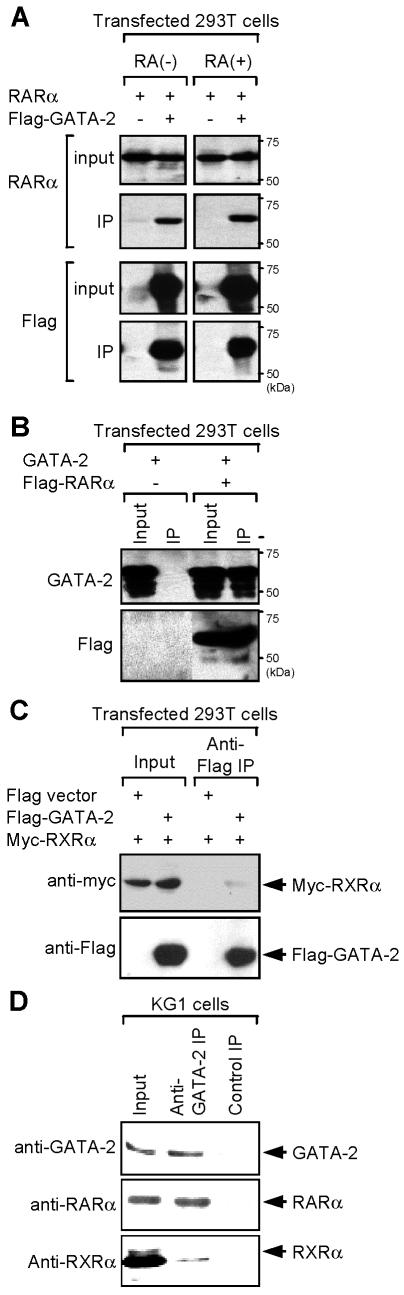
This interaction was next directly demonstrated by coimmunoprecipitation experiments performed with 293T cells transiently transfected with plasmids encoding RARα and Flag-tagged GATA-2 or GATA-2 and Flag-tagged RARα (Fig. 2A and B). The results of these experiments showed that RARα coimmunoprecipitated with GATA-2, further suggesting that GATA-2 and RARα could form a complex in vivo. The interaction of GATA-2 with RARα was not affected by the treatment with RA (Fig. 2A and B). Given that RXR is a well-established dimerization partner of RARs (5), we examined whether the GATA-2-RARα complex also contains the RXRα protein. The results of coimmunoprecipitation experiments showed that RXRα coprecipitated with GATA-2 (Fig. 2C), but at much lower levels than RARα.
FIG. 2.
Coimmunoprecipitation analysis of GATA-2-RARα interaction. (A) 293T cells were transfected with expression plasmids encoding the indicated proteins and treated with 1 μM RA [RA(+)] or diluent alone [RA(−)] 24 h after transfection. Cell lysates were prepared 24 h later, immunoprecipitated (IP) with anti-Flag antibody, and analyzed by Western blotting with anti-RARα (top) or anti-Flag (bottom) antibodies. Input (10%) nuclear extracts were analyzed as controls for the level of protein expression. Note that under these conditions GATA-2 binds RARα, irrespective of RA treatment. (B) Lysates of 293T cells transfected with the indicated expression plasmids were immunoprecipitated with anti-Flag antibody and analyzed by anti-GATA-2 (top) or anti-Flag (bottom) antibodies. (C) Cell lysates of 293T cells transfected with the expression plasmids for the indicated proteins were immunoprecipitated with anti-Flag antibody as described above. The precipitated proteins were analyzed by Western blotting with the indicated antibodies. Note that RXRα only weakly binds GATA-2. (D) Nuclear extracts of human myeloid KG1 cells were immunoprecipitated with anti-GATA-2 antibody. The precipitated materials were then analyzed by Western blotting with antibodies against GATA-2, RARα, and RXRα. Mouse IgG was used as a control. Input (10%) materials were used as controls. Molecular size markers are indicated on the right.
We next asked whether endogenous GATA-2 and RARα associate with each other in human immature myeloid KG1 cells. Nuclear extracts of KG1 cells were immunoprecipitated with anti-GATA-2 antibody, and the immunoprecipitated materials were analyzed by Western blotting with antibodies against GATA-2, RARα, and the RARα dimerization partner RXRα (Fig. 2D). Endogenous RARα readily coprecipitated with GATA-2. RXRα also coprecipitated with GATA-2, but to a much lesser extent than RARα, suggesting a much lower affinity for GATA-2.
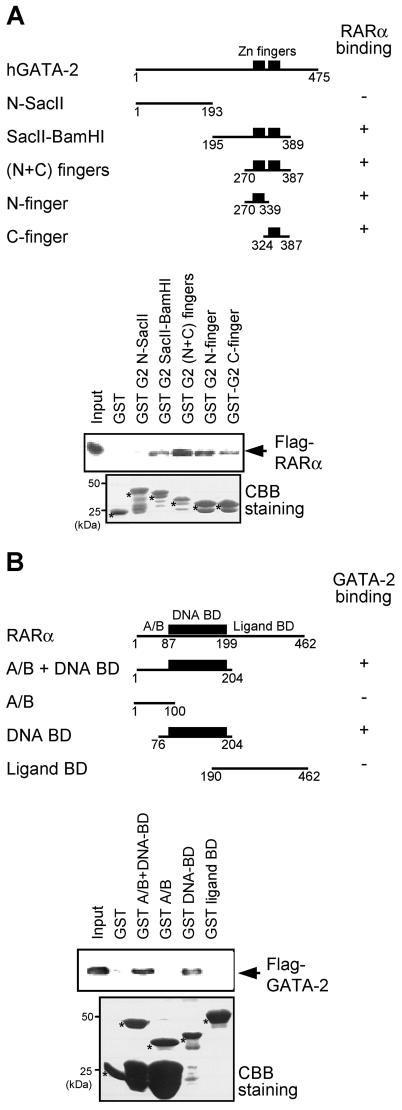
To delineate regions of interaction between GATA-2 and RARα, various parts of GATA-2 were expressed in bacteria as fusions to GST and the purified proteins were tested in vitro for interaction with FLAG-tagged RARα expressed in 293T cells (Fig. 3A). The results showed that the zinc finger domain of GATA-2 bound to RARα, with either the N or the C finger alone being sufficient for interaction. This interaction was specific to the zinc finger region, as the amino-terminal portion of GATA-2 (amino acids 1 to 193) did not bind RARα. Reciprocal experiments were conducted with GST fusion proteins encompassing various parts of RARα and Flag-tagged GATA-2 expressed in 293T cells (Fig. 3B). The DNA-binding domain of RARα was found to be responsible for the interaction with GATA-2. Neither the most N-terminal portion (designated A/B) nor the ligand-binding domain had affinity for GATA-2. Taken together, these results suggest that the zinc finger domain of GATA-2 and the DNA-binding domain of RARα are the regions that mediate the association between the two proteins.
FIG. 3.
GATA-2 and RARα interact through their zinc finger regions. (A) GST fusion proteins containing the indicated portions of GATA-2 were tested for the ability to bind Flag-tagged RARα present in 293T cell nuclear extract programmed with Flag-RARα expression plasmids. The first and last amino acids of the GATA-2 region present in the various GST fusions are indicated, and the abilities of the proteins to bind RARα are summarized schematically (top panel). Western blotting analysis of the pulldown materials with anti-Flag antibody is shown on the right (top), and Coomassie brilliant blue (CBB) staining is presented at the bottom to allow assessment of the quality and quantity of the various GST-GATA-2 proteins used. The values on the left indicate the positions of molecular size markers. (B) Reciprocal pulldown analyses in which various GST-RARα fusion proteins were analyzed for the ability to bind Flag-tagged GATA-2. BD, binding domain.
GATA-2 recruits RARα to its DNA target sites.
Given the association of GATA-2 with RARα, we next further explored the issue of recruitment of RARα by GATA-2 to a GATA binding motif in DNA (Fig. 4). Nuclear extracts of 293T cells programmed with expression plasmids for RARα and FLAG-tagged GATA-2 were incubated with biotinylated annealed oligonucleotides containing GATA binding sites. The biotinylated probes and the bound protein complexes were then captured by incubation with streptavidin-conjugated agarose beads. The results of this analysis showed that both GATA-2 and RARα were captured by the wild-type probe but not by the equivalent probe in which the GATA binding motif had been mutated (Fig. 4A). Since in the absence of GATA-2, RARα did not bind to the GATA probe, we concluded that the association of RARα with GATA motifs is mediated through interaction with GATA-2. Consistent with the immunoprecipitation results, only a small amount of RXRα was recruited to GATA motifs (Fig. 4B). As predicted by the data derived from transfection experiments, endogenous GATA-2 and RARα were pulled down together from KG-1 cells by the biotinylated GATA oligonucleotide DNAs (Fig. 4C). As expected, RXRα was not readily detected in the precipitated materials and the mutant GATA probe retained neither RARα nor RXRα (Fig. 4C). These results, which were consistent with the data derived from one-hybrid assays (Fig. 1), suggest that in hematopoietic cells GATA-2 can recruit RARα to its binding motifs in DNA.
FIG. 4.
RARα can be recruited to GATA motifs in DNA through interaction with GATA-2. 293T cells were transfected with the expression plasmids encoding RARα (A) or RXRα (B) and Flag-tagged GATA-2 as indicated. Nuclear extracts of the cells were then prepared and incubated with biotinylated oligonucleotides harboring GATA motifs (wild-type [wt] GATA oligonucleotides) or biotinylated mutant oligonucleotides in which GATA motifs were changed to TTTA (mutant [mt] GATA oligonucleotides). The oligonucleotides were then recovered by streptavidin-agarose beads, and the copurified proteins were analyzed by Western blotting with anti-RARα or anti-RXRα (top) and anti-Flag (bottom) antibodies. Input (10% input) was used as a control. (C) Nuclear extracts of human myeloid KG-1 cells were incubated with biotinylated oligonucleotides harboring GATA motifs and pulled down with streptavidin-agarose beads. The pulled-down materials were analyzed by Western blotting with antibodies against GATA-2, RARα, and RXRα.
RXRα inhibits recruitment of RARα to GATA-2-GATA DNA motif complex.
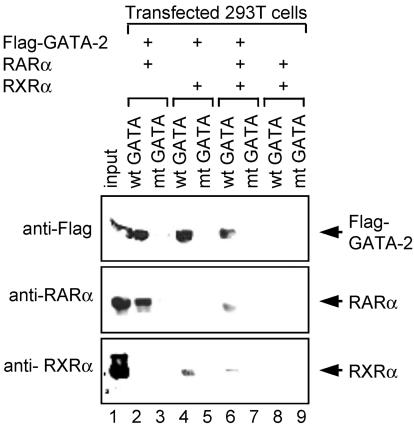
Given that some RXRα was coimmunoprecipitated with GATA-2, we next sought to determine whether the recruitment of RXRα to a GATA-2-DNA binding motif is due to the formation of a RXRα-GATA-2 complex or interaction of RXRα with RARα bound to GATA-2. To test these possibilities, we examined proteins copurified with the biotinylated GATA probe in combination with streptavidin-agarose beads (Fig. 5). After transfection of 293T cells with expression plasmids for either Flag-tagged GATA-2, RARα, or RXRα, the respective nuclear extracts were then mixed to obtain the combination of the desired proteins. A nuclear extract of 293T cells transfected with empty plasmid was used to make the total amount of the proteins in each mixture the same. This approach, in contrast to cotransfection of three expression plasmids, allows strict control of the protein levels used in a given experiment. When Flag-tagged GATA-2 and either RARα or RXRα were mixed, RARα or RXRα was copurified with Flag-GATA-2 bound to the GATA probe (Fig. 5, lanes 2 and 4). Consistent with results shown in Fig. 4B, the amount of copurified RXRα (lane 4) was less than that of RARα (lane 2). After addition of RARα with RXRα and GATA-2, the amount of RXRα copurified with GATA-2 decreased, albeit slightly (lane 6; compare lanes 4 and 6), suggesting that the recruitment of RXRα to the GATA-2-DNA complex is not due to the interaction of RXRα with RARα. It is noteworthy that when GATA-2 and RXRα are added with RARα, the amount of RARα bound to the GATA-2-DNA complex also decreased (lane 6) relative to that in experiments conducted in the absence of RXRα (lane 2). These results suggest that RXRα, which has a much lower affinity for GATA-2 than RARα, competes with GATA-2 for interaction with the RARα protein. Since RARα has a higher affinity for RXRα than GATA-2 (as revealed by mammalian two-hybrid assays [data not shown]), RARα may preferentially complex with RXRα rather than GATA-2. Given that cellular levels of RXR may be limiting (68), free RARα can be recruited to a GATA-2-DNA complex. One prediction that might emanate from these results is that signaling factors that increase RXRα expression would decrease levels of RARα associated with GATA-2.
FIG. 5.
RXRα inhibits recruitment of RARα to GATA-2-GATA motif DNA complex. 293T cells were transfected with an expression plasmid for either Flag-tagged GATA-2, RARα, or RXRα, separately. The resultant nuclear extracts were then mixed as indicated. Nuclear extract of the cells transfected with an empty vector was used to make the total amounts of nuclear proteins equal. The nuclear extracts containing the indicated proteins were then incubated with biotinylated oligonucleotides harboring GATA motifs (wild-type [wt] GATA), and the nucleotides were captured by streptavidin-agarose beads. The resultant copurified proteins were analyzed by Western blotting with anti-Flag, anti-RARα, and anti-RXRα antibodies. Nuclear extracts prior to mixing were analyzed as controls for appropriate expression of the proteins used (input; 10%). Biotinylated oligonucleotides in which GATA core recognition motifs were mutated to TTTA (mutant [mt] GATA) were used as controls.
Interaction of GATA-2 with RARα renders its activity RA regulated.
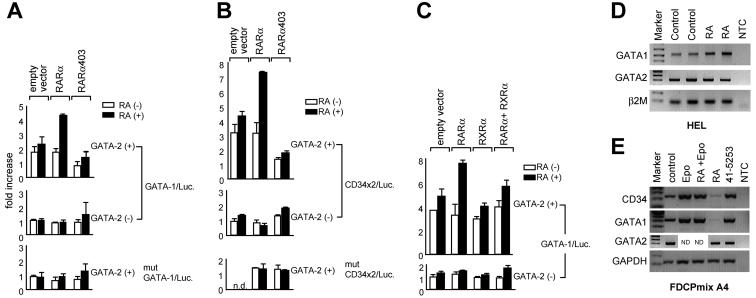
To examine effects of RARα on the transactivation activity of GATA-2, 293T cells were transfected with a luciferase reporter plasmid harboring GATA motifs from the mouse GATA-1 promoter (designated GATA-1-Luc.), or two GATA sites derived from the mouse CD34 promoter (CD34x2/Luc.), together with expression vectors for GATA-2 and RARα (Fig. 6A and B). In the absence of RA, GATA-2 alone induced luciferase activity to approximately 1.8-fold above the basal level and RA treatment increased the activity to ∼2.3-fold. This increase is likely to be due to the presence of endogenous RARα in 293T cells. RA treatment in the presence of cotransfected RARα increased this GATA-2 activity to ∼4.2-fold. Consistent with these results, an RARα mutant (RARα403) that retains the DNA-binding domain but lacks C-terminal activation function 2 (58-60) failed to activate GATA-2 transcriptional activity in the presence of RA; this was observed in the context of both of the reporters used (Fig. 6A and B). A decrease in reporter activity in the presence of RARα403 is likely due to ligand-insensitive recruitment of corepressor complexes by the mutated receptor (11) to GATA binding sites. It is noteworthy that hematopoietic progenitors engineered to express RARα403 are defective in myeloid differentiation programs (58, 59).
FIG. 6.
RARα renders GATA-2-dependent reporter activity RA responsive. 293T cells were transfected with a luciferase reporter plasmid containing a double GATA motif in the mouse GATA-1 promoter (designated GATA-1/Luc.; 0.5 μg) (A and C) or the reporter containing two copies of double GATA sites in the mouse CD34 promoter (designated CD34x2/Luc.; 0.5 μg) (B), together with expression plasmids for GATA-2 (GATA-2/pMT2, 100 ng) and RARα (0.5 μg), a C-terminally truncated form of RARα (RARα 403; 0.5 μg), RXRα (0.5 μg), or an empty vector (0.5 μg). Cells were then treated with RA (1 μM; solid bar) or diluent alone (open bar) 24 h after transfection, and the luciferase activities were measured another 24 h later. Luciferase activities are standardized against Renilla luciferase activity from cotransfected control reporter (pRL-CMV-Renilla luciferase) and expressed as fold increases over the activity of the reporter alone. The mutant reporters in which core recognition sites were changed from GATA to TTTA (mutant GATA-1/Luc. and mutant CD34x2/Luc.) were used as controls. Data are shown as means ± standard deviations of triplicate samples. (D) Expression of the endogenous GATA1 locus was measured by semiquantitative RT-PCR in HEL cells after 24 h of incubation with RA (10−6 M) and compared with that in untreated control samples. Analysis of GATA-2 expression is also shown, and parallel analysis of β2-microglobulin provided a control for normalization of RNA levels. NTC; no-template control. (E) Expression of endogenous CD34 and GATA-1 was measured in FDCPmix A4 cells by semiquantitative RT-PCR after 48 h of incubation with Epo (1 U/ml), RA (10−6 M), Epo plus RA, or the RARα antagonist Ro 41-5253 (10−5 M) and compared with that in an untreated control sample. Analysis of GATA-2 expression is also shown, and parallel analysis of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) provided a control for normalization of RNA levels. ND, not done.
To test effects of RXRα on RARα-dependent GATA-2 activity, 293T cells were transiently transfected with expression vectors for GATA-2, RARα, and RXRα and a luciferase reporter containing GATA binding sites from the GATA-1 promoter (Fig. 6C). When RARα was coexpressed with GATA-2, GATA-2 activity was potentiated by RA treatment, consistent with the results shown in Fig. 6A and B. In contrast, RXRα had little effect on GATA-2 activity in the presence of RA. Consistent with the interaction data, RXRα inhibited stimulation of GATA-2 activity in the presence of RARα and RA. Taken together, these results suggest that RXRα sequesters RARα from the GATA-2 complex, resulting in a reduced amount of RARα being recruited to the GATA-2-DNA complex.
These results raise the possibility that endogenous GATA-2-regulated genes may themselves exhibit RA responsiveness. Investigating this issue is complicated by the fact that bona fide GATA-2 target genes have not been identified. Also in vivo one might expect RA responsiveness to be critically dependent on the particular cell context in question and may or may not be a direct, rate-limiting, or assayable activity for any given gene and cell pair. Nevertheless, we examined whether the activity of the endogenous GATA-1 gene might be modulated by addition of RA with the human erythroleukemia progenitor cell line HEL as a model; HEL cells have been shown to express GATA-2 both as a transcript and as a DNA-binding activity (29, 67), and this was confirmed in the present study by RT-PCR. The results presented in Fig. 6D show a modest increase in GATA-1 expression in the presence of RA; this was paralleled by a decrease (0.6-fold) in GATA-1 expression in the presence of an RA antagonist (data not shown). Similar experiments were conducted with the murine progenitor cell line model FDCPmix A4 (Fig. 6E); these cells self-renew in IL-3 and exhibit myelomonocytic, as well as erythroid, differentiation in response to appropriate cytokines. GATA-2 expression has previously been documented in this cell line (10, 20) and was confirmed in this study by RT-PCR (Fig. 6E). Note the reduction in GATA-1 expression in cells that were treated with RA but maintained in IL-3 to prevent differentiation. A similar decrease in expression was noted for the CD34 gene. Treatment with an RARα-specific antagonist (lane 41-5253) resulted in the expected enhancement of expression of these genes over the nontreated control levels. Also note that in the presence of Epo, which in FDCPmix A4 cells induces erythroid differentiation, RA treatment had no effect on CD34 and GATA-1 expression. This is consistent with our previously published results showing that Epo rapidly down regulates expression of the RARα gene in these cells (68). These opposing effects on gene expression in the two different cell models examined are consistent with the variable differentiation and developmental stage-specific effects exhibited by RA in hematopoiesis, reinforcing the critical role of the cell context in determining the outcome of RA-mediated signaling (44, 45, 52). Taken together and within the constraints of the caveats alluded to above, these data are suggestive of a role for RA modulation of GATA-2-dependent target gene expression in vivo. As has already been alluded to above, the extent to which this may represent a direct or indirect effect remains unclear.
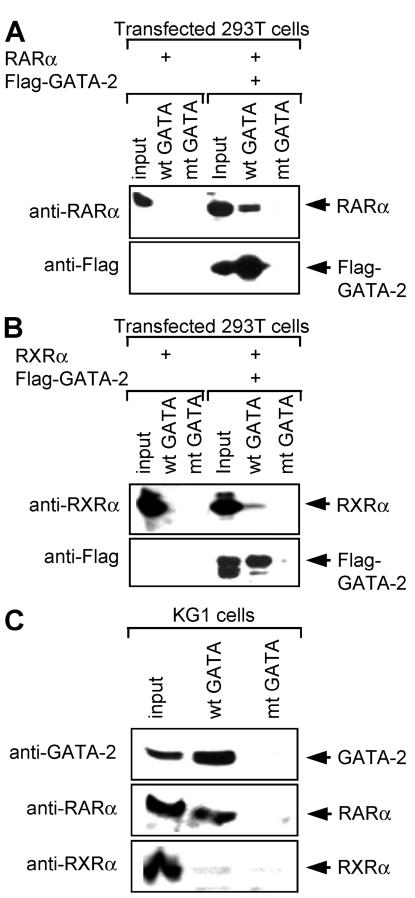
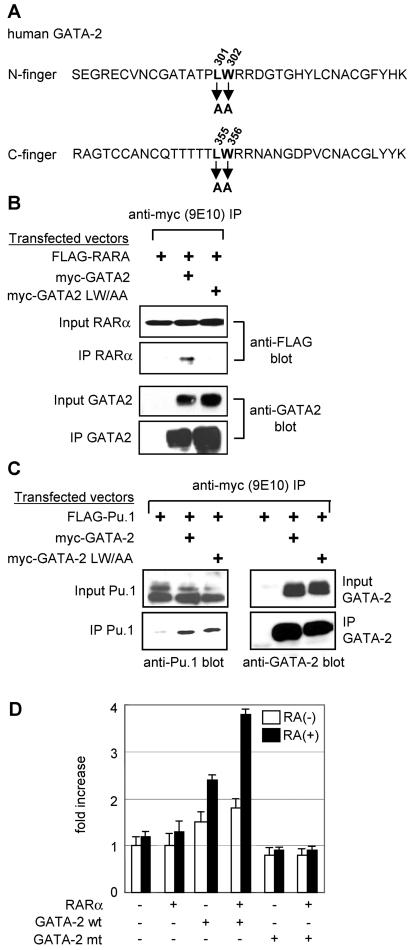
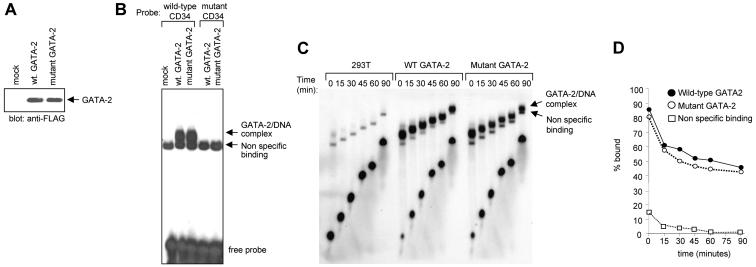
To further test the relevance of the interaction between GATA-2 and RARα we sought to generate a mutant form of GATA-2 that was unable to interact with RARα. We focused on the finger regions since this was where we had mapped the interaction by GST pulldown assay, as well as yeast and mammalian one- and two-hybrid assays. The nuclear magnetic resonance structure of the N finger of GATA-1 is available (27, 41) and predicts an exposed loop region in the vicinity of L214 that may therefore be particularly available for intermolecular interaction. Speculative mutations were made in the equivalent region of both fingers of GATA-2 (both the N and C fingers have the ability to interact with RARα). Mutant GATA-2 molecules were then tested for the ability to bind RARα. Figure 7 shows results obtained with a GATA-2 mutant form in which an LW pair has been mutated to AA at positions 301 and 302 and positions 355 and 356 in the N and C fingers, respectively (Fig. 7A). This mutant form does not interact with RARα, as judged by coimmunoprecipitation assays (Fig. 7B). This is unlikely to result from gross perturbation of the structure of the molecule since its ability to interact with Pu.1 is maintained (Fig. 7C). Similarly this mutant form of GATA-2 is unaffected in its ability to bind GATA-1, PML, FOG, or promyelocytic leukemia zinc finger (PLZF) (data not shown) although it is noteworthy that its ability to interact with myb is markedly reduced (data not shown). In contrast to wild-type GATA-2, and consistent with its inability to bind RARα, this mutant form of GATA-2 fails to exhibit RARα-dependent RA responsiveness in transactivation assays (Fig. 7D). To confirm that this failure to transactivate was not due to a reduced DNA-binding activity of the mutant GATA-2 molecule, we performed EMSAs with wild-type and mutant GATA-2. 293T cells were transfected with expression plasmids for Flag-tagged wild-type or mutant GATA-2. Nuclear extracts were prepared 48 h after transfection and assayed by Western blotting with anti-Flag antibody to confirm equivalent amounts of the respective proteins (Fig. 8A). Double-stranded oligonucleotide probes harboring either a wild-type GATA binding motif from the CD34 enhancer or the same GATA motif mutated to abolish GATA binding were radiolabeled and used in EMSAs as previously described (63). The results of this analysis are presented in Fig. 8B and indicate that the GATA-2 mutant, which is unable to interact with RARα, retains its ability to bind to GATA motifs in DNA. To further exclude the possibility that this mutant molecule may bind GATA motifs with less avidity than wild-type GATA-2, we conducted dissociation assays. For these experiments a 100-fold excess of unlabeled oligonucleotide probe was added to the binding reaction mixture after an initial 20-min incubation period had elapsed. Aliquots of the reaction mixture were sampled at subsequent time points and loaded onto polyacrylamide gels (Fig. 8C). The percentage of probe bound by GATA-2 was quantitated with imageanalyser and plotted on a log scale versus time (Fig. 8D). Results are also shown for a nonspecific binding activity that is variably seen in 293T cell extracts. The results indicate similar off rates for both wild-type and mutant GATA-2 molecules, indicating that they possess similar DNA-binding activities. Taken together these results suggest that GATA-2 activity can be rendered RA responsive in the context of an RARα-GATA-2 complex.
FIG. 7.
Analysis of a GATA-2 mutant that does not interact with RARα. (A) Amino acid sequences of GATA-2 N and C finger regions showing the positions of the mutagenized LW residues. (B) Coimmunoprecipitation analysis of mutant GATA-2 and RARα. 293T cells were transfected with the expression plasmids indicated. Cell lysates were prepared 24 h later, immunoprecipitated (IP) with anti-FLAG antibody, and analyzed by Western blotting with anti-FLAG and anti-GATA-2 antibodies. Input nuclear extracts were analyzed as a control for the levels of proteins expressed. (C) Coimmunoprecipitation analysis of mutant GATA-2 and Pu.1. Experiments were carried out essentially as described for panel B. (D) RA responsiveness of mutant (mt) GATA-2 activity. 293T cells were transfected with a GATA-dependent luciferase reporter plasmid, as well as the indicated combinations of expression vectors for RARα and mutant and wild-type (wt) GATA-2. Cells were then treated with RA (1 μM; solid bar) or diluent alone (open bar) 24 h after transfection, and the luciferase activities were measured another 24 h later. Luciferase activities are standardized against Renilla luciferase activity from a cotransfected control reporter (pRL-CMV-Renilla luciferase) and expressed as fold increases over the activity of the reporter alone. The data presented represent two independent experiments, each of which was performed in triplicate.
FIG. 8.
Comparison of wild-type (wt) and mutant GATA-2 DNA-binding activities. (A) Western blot analysis of 293T cell extracts programmed by transient expression of Flag-tagged versions of wild-type GATA-2 and the mutant GATA-2 defective in the ability to interact with RARα. (B) EMSA analysis with radiolabeled oligonucleotide probe harboring a GATA motif from the murine CD34 enhancer region. Note the presence of a nonspecific DNA-binding activity that is variably seen in 293T cell nuclear extracts. (C) Dissociation assays with control 293T cell extracts or extracts programmed by expression of wild-type or mutant GATA-2. Note the presence of a nonspecific DNA-binding activity. (D) Quantitative analysis of the results presented in panel C. The percentage of probe bound by GATA-2 is plotted on a log scale versus time.
GATA-2 is not a component of the RARα-RXRα-DNA complex.
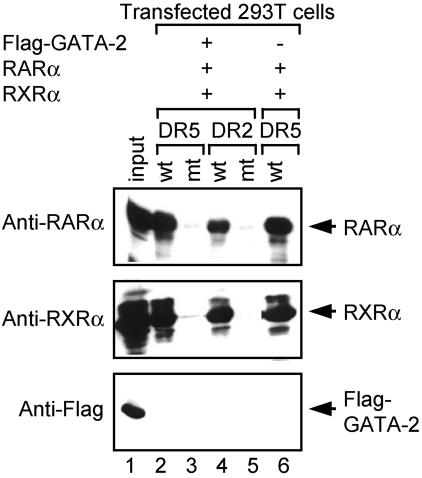
In reciprocal experiments we also examined whether GATA-2 could be recruited to RARα-RXRα complexes bound to RAREs in DNA (Fig. 9). 293T cells were transfected independently with either Flag-tagged GATA-2, RARα, RXRα, or an empty vector. The nuclear extracts were then mixed as before to obtain the combination of the desired proteins. The resultant protein solutions were incubated with biotinylated probes containing binding sites for the RAR-RXR heterodimer (DR5 and DR2) (33). The lysates were then incubated with streptavidin-agarose beads to capture the DNAs and the copurified proteins were analyzed by Western blotting with antibodies against RARα, RXRα, and the Flag epitope. Both RARα and RXRα were captured by the wild-type probe but not by mutant probes, indicating the specificity of the assay. However, GATA-2 was not copurified with the RARα-RXRα-RARE complexes. The amount of precipitated RARα or RXRα was not affected by the coexpression of GATA-2 (compare amounts of RARα and RXRα captured by DR5 in the presence of GATA-2 with those in the absence of GATA-2; lanes 2 and 6). These results suggest that GATA-2 does not bind to, nor have any effect on, RARα-RXRα complexes bound to RARE motifs in DNA. These results are consistent with data indicating that RXRα binds RARα more strongly than GATA-2 and its endogenous levels of expression are lower than those of RARα (68). In line with the interaction data, RXRα inhibited stimulation of GATA-2 activity in the presence of RARα and RA (Fig. 6C). Taken together, these results suggest that RXRα sequesters RARα from the GATA-2 complex, resulting in a reduced amount of RARα being recruited to the GATA-2-DNA complex. These data further implicate relative levels of GATA, RAR, and RXR as determining whether or not RA will stimulate GATA targets.
FIG. 9.
GATA-2 is not included in RARα-RXRα-DNA complexes. 293T cells were transfected with an expression plasmid for either RARα, RXRα, or Flag-tagged GATA-2, separately, and the resultant nuclear extracts were mixed as indicated. Nuclear extract of the cells transfected with the empty vector was used to make the total amount of nuclear extracts the same. The mixtures of the nuclear extracts were then incubated with biotinylated oligonucleotides harboring RARE of either the DR5 (wild-type [wt] DR5; lanes 2 and 6) or the DR2 (wt DR2; lane 4) type, and the nucleotides were captured with streptavidin-agarose beads. The copurified proteins were then analyzed by Western blotting with anti-RARα, anti-RXRα, and anti-Flag antibodies. Nuclear extracts prior to mixing were analyzed as controls for appropriate expression of the proteins used (10% input; lane 1). Biotinylated oligonucleotides in which core recognition sites of the RARα-RXRα complex were mutated from GGTTCA and AGTTCA to GGTAGT and AGTAGT, respectively, were used as controls (mutant [mt] DR5 and DR2, lanes 3 and 5). Note that in no combination was GATA-2 copurified with RARα-RXRα-RARE complexes. The amounts of RARα and RXRα bound to DR5 are similar whether or not GATA-2 is included (lanes 2 and 6).
Functional cross talk between RA and GATA-2.
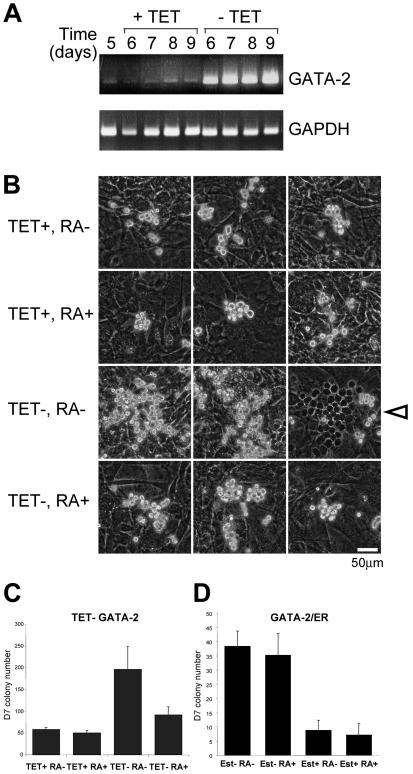
We next sought to gain some evidence for the functional relevance of a GATA-2-RARα interaction. Recently we have developed a TET-regulated system that affords conditional expression of GATA-2 in ES cells (26). An ES clone containing a TET-responsive GATA-2 expression construct was cultured on an OP9 stromal layer for 5 days in the presence of TET. The cells were then trypsinized and reseeded onto OP9 cells in the presence or absence of TET. Samples were collected over the following four days (days 6, 7, 8, and 9) and analyzed for GATA-2 expression by RT-PCR. The results of this analysis are presented in Fig. 10A and show the robust increase in GATA-2 expression that results from the withdrawal of TET. In this ES differentiation system, induction of exogenous GATA-2 expression by withdrawal of TET enhances the generation of immature, multipotent, definitive hematopoietic colonies when ES cells are plated on OP9 cell stroma under hematopoiesis-supportive culture conditions (26). We tested whether addition of RA might modulate the biological effects of exogenous GATA-2 expression seen in this system. Representative images of the hemopoietic colonies observed in our experiments are shown in Fig. 10B, and a summary of the number of colonies produced is shown in Fig. 10C. In the absence of induction of exogenous GATA-2 expression (TET+ samples), addition of RA had no effect on the frequency of hematopoietic colony formation (Fig. 10C) or the size of the colonies produced (Fig. 10B). Induction of exogenous GATA-2 activity (TET−) in the absence of RA resulted in the expected increase in the number of hematopoietic colonies (Fig. 10C). In addition, an increase in colony size was also observed (Fig. 10B). This was accompanied by an increase in the number of colonies that grew under the stromal layer with a cobblestone appearance. This phenomenon is known as pseudoemperipolesis and is thought to be an indicator of the relative immaturity of the colonies (26); an example of one such colony is indicated by the open arrowhead in Fig. 10B. Addition of RA clearly inhibited the GATA-2-dependent enhancement of both hematopoietic colony formation (Fig. 10C) and hematopoietic colony size (Fig. 10B); a reduction in the number of colonies exhibiting pseudoemperipolesis was also observed (not shown). Taken together these data provide evidence of functional cross talk between these two pathways.
FIG. 10.
RA modulates the frequency of GATA-2-dependent hematopoietic colony formation by ES cells. (A) RT-PCR analysis of GATA-2 expression in an ES cell clone containing a TET-regulatable GATA-2 expression vector. The first lane (day 5) is from an ES culture grown for 5 days on OP9 stroma in the presence of added TET. The remaining lanes are samples taken at further 1-day intervals (days 6 to 9) after the initial day 5 culture had been trypsinized and reseeded onto OP9 stroma in the presence or absence of TET. (B and C) When grown on OP9 stroma, ES cells give rise to immature, definitive, multipotent, hematopoietic progenitors which begin to emerge at day 5 of culture. The effects of (i) induction of exogenous GATA-2 expression, (ii) addition of RA, and (iii) both were assessed at this time point by withdrawal of TET and/or addition of 1 μM RA or control diluent. Colonies were examined 2 days later (day 7). Photomicrographs of representative colonies are shown in panel B, and the arrowhead indicates a colony that has undergone pseudoemperipolesis. A summary of the number of colonies produced is presented in panel C; the results presented represent the average of six cultures analyzed. Panel D shows colony frequency data obtained in a similar series of experiments (n = 6) conducted with an ES cell line expressing a GATA-2-ER chimera. In this case the activity of the exogenous GATA-2-ER was regulated by addition of 1 μM β-estradiol.
As an additional control we examined the effect of RA on the activity of a GATA-2-ER chimera that exhibits an altered activity to wild-type GATA-2 in this particular cell system; GATA-2-ER inhibits hematopoietic colony formation, and RA has no additional effect on the biological activity of this molecule (Fig. 10D).
DISCUSSION
In this report, we have presented evidence of functional cross talk between GATA-2 and RARα in hematopoietic cells. RARα is known to associate with RXR, and this interaction is required for the resulting complex to bind an RARE and activate transcription in response to RA. Our data suggest that RARα also has the ability to associate with GATA-2, thus allowing RA to regulate transcription from GATA binding motifs in DNA. This interaction and recruitment of RARα to GATA-2 binding sites is RXR independent. Our data also suggest that RXR competes with GATA-2 for RARα and attenuates the effects of RA on GATA-2 activity. Since GATA-2 expression does not affect RARα-RXRα interaction and its binding to RARE, it is not likely that GATA-2 interferes with authentic RARE-driven genes. Finally, we showed that RA influences the GATA-2-dependent emergence of hematopoietic colonies from ES cells.
Potentiation of GATA-2 activity through interaction with RARα.
The level of stimulation of GATA-2 activity by RA seen in these experiments is worthy of comment. Although only in the range of two- to threefold, it should be noted that GATA-2 itself is not a particularly potent transactivator (less than twofold) in these assays. Furthermore, relatively small changes in transcription factor level, and presumably therefore activity, have been demonstrated to have significant effects on cell fate (40). Haploinsufficiency of transcription factors such as AML-1 provides support for this notion (50, 51), and data from our own laboratory suggest that this may extend to GATA-2 (46; S. Delassus, K. Gale, and T. Enver, unpublished data).
On the basis of published studies demonstrating the binding of SCL-GATA complexes to bipartite E-box-GATA motifs (64), one possibility we have considered is that a GATA-2-RARα complex may be recruited to a subset of GATA-2 target genes that contain bipartite GATA-RARE motifs. However, in an extensive series of in vitro experiments with a range of oligomers containing variations of such motifs we have failed to find evidence for such a scenario (S. Tsuzuki and T. Enver, unpublished data). While these experiments do not conclusively exclude the existence or importance in vivo of such bipartite motifs, the ability of GATA-2 to recruit RARα to lone GATA motifs suggests that GATA-2-RARα complexes can function on single GATA sites.
The interaction region between GATA-2 and RARα has been mapped to structurally related cysteine-rich zinc finger regions. Importantly, specific point mutations made within the fingers of GATA-2 eliminate its ability to interact with RARα. The zinc fingers of the GATA factors, in addition to their roles in DNA binding, have also been implicated in protein-protein interaction. GATA factors have been shown to associate with other regulatory proteins (like Sp1, Lmo2, CBP, other GATA factors, and PML) by virtue of the C4C4 zinc fingers (4). Interactions between the N- and C-terminal fingers of GATA-1 have been postulated to modulate both DNA binding and transactivation (56). Since the zinc finger region has been evolutionarily conserved through the GATA family, it is not surprising that other members of GATA family, GATA-1 and GATA-3, also have the potential to bind RARα (Tsuzuki and Enver, unpublished data).
Potential effects on normal and leukemia transcriptional networks.
In prior studies we have demonstrated that GATA-2 could interact with the leukemia-associated proteins PML (63) and PLZF (62), as well as the PLZF homologous protein FAZF-ROG-TZFP (62), which has also been shown to interact with GATA-3 (35). In the case of PML, interaction is mediated by the zinc fingers of GATA-2 and the B-box region of PML. The GATA-2 zinc finger region is also involved in its interaction with the POZ and zinc finger domains of PLZF (62). Intriguingly, GATA-2 also interacts with t(15;17)- and t(11;17)-generated chimeric versions of PML (PML-RARα) and PLZF (PLZF-RARα) that include most of the native RAR molecule in the respective fusion proteins (62, 63). Our present results demonstrating that RAR in its native form can interact with GATA-2 expand the network of interactions that may be disrupted in t(15;17)- and t(11;17)-associated APL and add impetus to experiments aimed at identifying the spectrum of GATA-2 target genes normally regulated by RA in myeloid progenitor cells and potentially dysregulated in acute promyelocytic leukemia.
Since GATA-2 is the predominant GATA factor expressed in early myeloid cells, the role of RARα in myeloid differentiation may be functionally linked with GATA-2. Dominant negative forms of RARα (58, 60) and APL-associated RAR fusion oncoproteins have been shown to block myeloid differentiation at the promyelocytic stage (3, 7, 17-19). Consistent with these findings, antagonists of RARα inhibit (34) myelopoiesis while agonists of RARα stimulate myelopoiesis (6), suggesting that target genes regulated by RA are important for myeloid differentiation. Recent work from our own laboratories demonstrated that RA inhibits erythroid differentiation of multipotent progenitors (68); the extent to which these effects are mediated by GATA-2 or GATA-1, for that matter, are not understood. Unfortunately, bona fide target genes of GATA-2 in hematopoietic cells remain unidentified, but our results predict that some of the genes regulated by RA may overlap some of those regulated by GATA-2 as alluded to above. The transient transfection systems used in this report made use of isolated GATA motifs derived from the GATA-1 and CD34 promoters. However, analysis of the endogenous GATA-1 and CD34 loci in HEL and FDCPmix A4 cells revealed that their expression was indeed modulated by RA. Interestingly, different results were obtained with the two cell lines studied, emphasizing the critical importance of cell context in determining the output of RA signaling. In the same vein, our data predict that relative levels of GATA-2, RARα, and RXR will influence whether RA will result in activation of GATA-2 target genes as RXR has an inhibitory effect on RA-dependent GATA-2 potentiation by competing for RARα. An analysis of gene expression changes in RA-stimulated NB4 cells with a combination of cDNA microarray, suppression subtractive hybridization, and differential display PCR approaches has provided a number of candidate genes whose expression may be modulated by RA (31). Information regarding the cis-regulatory elements of these genes is quite limited, but a preliminary investigation has revealed that in many cases there is an abundance of GATA sites in the absence of any obvious RAREs (A. Zelent, unpublished data). Such genes are clearly candidates for GATA-dependent RA regulation, but considerable further work is required to confirm this possibility.
Biological effects of RA-GATA-2 cross talk in ES-derived hematopoietic development.
In terms of the biological effects of RA-potentiated GATA-2 activity, the nature of the output may vary according to cell type and differentiation stage, as well as which GATA factor or GATA factor combination predominates. This is additionally complicated by the fact that RA itself exhibits different activities at different stages of hematopoiesis. A clear example of this is provided by the work of Collins and colleagues, who demonstrated that RA promoted colony formation by primitive progenitors and delayed their differentiation but enhanced the differentiation of committed myeloid progenitors (44); more recently these workers have extended these studies to show that RA also enhances long-term repopulating activity (45). In the ES cell system we have studied in this report, conditional activation of GATA-2 leads to an increase in the production of hematopoietic colonies. Addition of RA inhibited this GATA-2-dependent effect. The simplest view consistent with current thoughts on the mechanisms underlying RAR action is that the association of GATA with RAR in the presence of ligand would be predicted to result in potentiation of GATA-2 activity. How this would result in inhibition of GATA-2-dependent colony formation is unclear, but one possibility is that expression of GATA-2 in the absence of RA may increase colony formation through repression of gene targets, with subsequent potentiation of GATA-2 activity by RA leading to derepression. In any event our results suggest that a combination of GATA-2 and RA produces a biological readout in this system that is similar to that achieved by expression of a GATA-2-ER chimera. This may in part reflect similarities in the ER and RAR moieties and suggests that a GATA-2-ER chimera may mimic the effect of a normal GATA-2-RARα complex. The comparison of RA effects seen in ES-derived hematopoiesis with those previously observed in primitive hemopoietic cells derived from adult bone marrow is intriguing. The generation of hematopoietic stem cells during ontogeny is, as a process, quite distinct from stem cell homeostasis in adulthood, and the roles of hematopoiesis-affiliated transcription factors in these two different processes may also be quite distinct (14). These caveats aside, our data provide evidence for functional cross talk between GATA and RA pathways. However, the extent to which the functional effects seen in ES cells mechanistically arise from direct interaction of GATA-2 and RARα remains an open question.
Signal-dependent regulation of GATA activity.
Perhaps surprisingly, given the importance of the GATA factor family, little is known about the signals that might impinge on its activity. We have previously demonstrated that the posttranslational modification of both GATA-2 and GATA-1 by phosphorylation is regulated by growth factor signaling in a mitogen-activated protein kinase (MAPK)-dependent manner (53, 54). The fact that GATA factors are known to be acetylated raises the possibility that these modifications could be similarly regulated (1, 22). Our present results demonstrating RA-dependent potentiation of GATA-2-RARα complexes provide a novel mechanism by which GATA activity could be rendered signal dependent. Furthermore, given that RA can stimulate MAPK signaling (16) and GATA proteins can be phosphorylated by MAPKs, the possibility exists that some of the effects we have observed in these studies may be mediated through a MAPK-dependent pathway.
Also interesting in this regard are the results of Trainor and colleagues (55), who identified a negatively acting hormone response-like element regulating element located in the first intron of the chicken GATA-1 gene (55). This negatively acting hormone response-like element binds a heterodimer of thyroid hormone receptor α and the chicken upstream promoter transcription factor. The inhibiting action of this complex can be overcome by GATA-1 itself or by v-erbA.
GATA factors and retinoid receptors represent families with important developmental functions and highly conserved zinc finger domains. The existence of a number of differentially expressed and functionally distinct GATA factors and RARs suggests that cross talk between GATA and RA signaling may not be restricted to hematopoiesis. Indeed, it has recently been shown that RXRα represses GATA-4-mediated transcription in cardiomyocytes (8).
Acknowledgments
This work was supported by Specialist Program grants from the Leukemia Research Fund of Great Britain (Tariq Enver and Arthur Zelent), the Medical Research Council (Tariq Enver), and a grant in aid for scientific research from the Japanese Society for the Promotion of Science (Shinobu Tsuzuki).
REFERENCES
- 1.Boyes, J., P. Byfield, Y. Nakatani, and V. Ogryzko. 1998. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature 396:594-598. [DOI] [PubMed]
- 2.Briegel, K., K. C. Lim, C. Plank, H. Beug, J. D. Engel, and M. Zenke. 1993. Ectopic expression of a conditional GATA-2/estrogen receptor chimera arrests erythroid differentiation in a hormone-dependent manner. Genes Dev. 7:1097-1099. [DOI] [PubMed] [Google Scholar]
- 3.Brown, D., S. Kogan, E. Lagasse, I. L. Weissman, M. Alcalay, P. G. Pelicci, S. Stwater, and J. M. Bishop. 1997. A PML-RARα transgene initiates murine acute promyelocytic leukemia. Proc. Natl. Acad. Sci. USA 94:2551-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cantor, A., and S. H. Orkin. 2002. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene 21:3368-3376. [DOI] [PubMed] [Google Scholar]
- 5.Chambon, P. 1996. A decade of molecular biology of retinoic acid receptors. FASEB J. 10:940-954. [PubMed] [Google Scholar]
- 6.Chen, J. W., J. Clifford, C. Zusi, J. Starrett, D. Tortolani, J. Ostrowski, P. R. Reczek, P. Chambon, and H. Gronemeyer. 1996. Two distinct actions of retinoid-receptor ligands. Nature 382:819-822. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, G. X., X. H. Zhu, X. Q. Men, L. Wang, Q. H. Huang, X. L. Jin, S. M. Xiong, J. Zhu, W. M. Guo, J. Q. Chen, S. F. Xu, E. So, L. C. Chan, S. Waxman, A. Zelent, G. Q. Chen, S. Dong, J. X. Liu, and S. J. Chen. 1999. Distinct leukemia phenotypes in transgenic mice and different corepressor interactions generated by promyelocytic leukemia variant fusion genes PLZF-RARα and NPM-RARα. Proc. Natl. Acad. Sci. USA 96:6318-6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clabby, M. L., T. A. Robison, H. F. Quigley, D. B. Wilson, and D. P. Kelly. 2003. Retinoid X receptor alpha represses GATA-4-mediated transcription via a retinoid-dependent interaction with the cardiac-enriched repressor FOG-2. J. Biol. Chem. 278:5760-5767. [DOI] [PubMed] [Google Scholar]
- 9.Crispino, J. D., M. B. Lodish, J. P. MacKay, and S. H. Orkin. 1999. Use of altered specificity mutants to probe a specific protein-protein interaction in differentiation: the GATA-1:FOG complex. Mol. Cell 3:219-228. [DOI] [PubMed] [Google Scholar]
- 10.Cross, M. A., C. M. Heyworth, A. M. Murrell, E. O. Bockamp, T. M. Dexter, and A. R. Green. 1994. Expression of lineage restricted transcription factors precedes lineage specific differentiation in a multipotent haemopoietic progenitor cell line. Oncogene 9:3013-3016. [PubMed] [Google Scholar]
- 11.Damm, K., R. A. Heyman, K. Umesono, and R. M. Evans. 1993. Functional inhibition of retinoic acid response by dominant negative retinoic acid receptor mutants. Proc. Natl. Acad. Sci. USA 90:2989-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorfman, D. M., D. B. Wilson, G. A. Bruns, and S. H. Orkin. 1992. Human transcription factor GATA-2. Evidence for regulation of preproendothelin-1 gene expression in endothelial cells. J. Biol. Chem. 267:1279-1285. [PubMed] [Google Scholar]
- 14.Enver, T., and M. Greaves. 1998. Loops, lineage, and leukemia. Cell 94:9-12. [DOI] [PubMed] [Google Scholar]
- 15.Ezoe, S., I. Matsumura, S. Nakata, K. Gale, K. Ishihara, N. Minegishi, T. Machii, T. Kitamura, M. Yamamoto, T. Enver, and Y. Kanakura. 2002. GATA-2/estrogen receptor chimera regulates cytokine-dependent growth of hematopoietic cells through accumulation of p21(WAF1) and p27(Kip1) proteins. Blood 100:3512-3520. [DOI] [PubMed] [Google Scholar]
- 16.Gianni, M., A. Bauer, E. Garattini, P. Chambon, and C. Rochette-Egly. 2002. Phosphorylation by p38MAPK and recruitment of SUG-1 are required for RA-induced RARγ degradation and transactivation. EMBO J. 21:3760-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grisolano, J. L., R. L. Wesselschmidt, P. G. Pelicci, and T. J. Ley. 1997. Altered myeloid development and acute leukemia in transgenic mice expressing PML-RARα under control of cathepsin G regulatory sequences. Blood 89:376-387. [PubMed] [Google Scholar]
- 18.He, L.-Z., A. Zelent, and P. P. Pandolfi. 1999. RARα-PLZF is a critical determinant of the leukemic phenotype in APL. Blood 94:368a.
- 19.He, L. Z., F. Guidez, C. Tribioli, D. Peruzzi, M. Ruthardt, A. Zelent, and P. P. Pandolfi. 1998. Distinct interactions of PML-RARα and PLZF-RARα with co-repressors determine differential responses to RA in APL. Nat. Genet. 18:126-135. [DOI] [PubMed] [Google Scholar]
- 20.Heyworth, C., K. Gale, M. Dexter, G. May, and T. Enver. 1999. A GATA-2/estrogen receptor chimera functions as a ligand-dependent negative regulator of self-renewal. Genes Dev. 13:1847-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heyworth, C. M., T. M. Dexter, O. Kan, and A. D. Whetton. 1990. The role of hemopoietic growth factors in self-renewal and differentiation of IL-3-dependent multipotent stem cells. Growth Factors 2:197-211. [DOI] [PubMed] [Google Scholar]
- 22.Hung, H. L., J. Lau, A. Y. Kim, M. J. Weiss, and G. A. Blobel. 1999. CREB-binding protein acetylates hematopoietic transcription factor GATA-1 at functionally important sites. Mol. Cell. Biol. 19:3496-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Igarashi, K., H. Hoshino, A. Muto, N. Suwabe, S. Nishikawa, H. Nakauchi, and M. Yamamoto. 1998. Multivalent DNA binding complex generated by small Maf and Bach1 as a possible biochemical basis for β-globin locus control region complex. J. Biol. Chem. 273:11783-11790. [DOI] [PubMed] [Google Scholar]
- 24.Igarashi, K., K. Kataoka, K. Itoh, N. Hayashi, M. Nishizawa, and M. Yamamoto. 1994. Regulation of transcription by dimerization of erythroid factor NF-E2 p45 with small Maf proteins. Nature 367:568-572. [DOI] [PubMed] [Google Scholar]
- 25.Kastner, P., H. J. Lawrence, C. Waltzinger, N. B. Ghyselinck, P. Chambon, and S. Chan. 2001. Positive and negative regulation of granulopoiesis by endogenous RARα. Blood 97:1314-1320. [DOI] [PubMed] [Google Scholar]
- 26.Kitajima, K., M. Masuhara, T. Era, T. Enver, and T. Nakano. 2002. GATA-2 and GATA-2/ER display opposing activities in the development and differentiation of blood progenitors. EMBO J. 21:3060-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kowalski, K., R. Czolij, G. F. King, M. Crossley, and J. P. Mackay. 1999. The solution structure of the N-terminal zinc finger of GATA-1 reveals a specific binding face for the transcriptional co-factor FOG. J. Biomol. NMR 13:249-262. [DOI] [PubMed] [Google Scholar]
- 28.Labrecque, J., D. Allan, P. Chambon, N. N. Iscove, D. Lohnes, and T. Hoang. 1998. Impaired granulocytic differentiation in vitro in hematopoietic cells lacking retinoic acid receptors α1 and γ. Blood 92:607-615. [PubMed] [Google Scholar]
- 29.Leonard, M., M. Brice, J. D. Engel, and T. Papayannopoulou. 1993. Dynamics of GATA transcription factor expression during erythroid differentiation. Blood 82:1071-1079. [PubMed] [Google Scholar]
- 30.Lin, R. J., D. A. Egan, and R. M. Evans. 1999. Molecular genetics of acute promyelocytic leukemia. Trends Genet. 15:179-184. [DOI] [PubMed] [Google Scholar]
- 31.Liu, T. X., J. W. Zhang, J. Tao, R. B. Zhang, Q. H. Zhang, C. J. Zhao, J. H. Tong, M. Lanotte, S. Waxman, S. J. Chen, M. Mao, G. X. Hu, L. Zhu, and Z. Chen. 2000. Gene expression networks underlying retinoic acid-induced differentiation of acute promyelocytic leukemia cells. Blood 96:1496-1504. [PubMed] [Google Scholar]
- 32.Liu, Y., H. Asch, and M. F. Kulesz-Martin. 2001. Functional quantification of DNA-binding proteins p53 and estrogen receptor in cells and tumor tissues by DNA affinity immunoblotting. Cancer Res. 61:5402-5406. [PubMed] [Google Scholar]
- 33.Mangelsdorf, D. J., and R. M. Evans. 1995. The RXR heterodimers and orphan receptors. Cell 83:841-850. [DOI] [PubMed] [Google Scholar]
- 34.Mehta, K., T. McQueen, T. Manshouri, M. Andreeff, S. Collins, and M. Albitar. 1997. Involvement of retinoic acid receptor-alpha-mediated signaling pathway in induction of CD38 cell-surface antigen. Blood 89:3607-3614. [PubMed] [Google Scholar]
- 35.Miaw, S. C., A. Choi, E. Yu, H. Kishikawa, and I. C. Ho. 2000. ROG, repressor of GATA, regulates the expression of cytokine genes. Immunity 12:323-333. [DOI] [PubMed] [Google Scholar]
- 36.Nakano, T. 1995. Lymphohematopoietic development from embryonic stem cells in vitro. Semin. Immunol. 7:197-203. [DOI] [PubMed] [Google Scholar]
- 37.Nakano, T., H. Kodama, and T. Honjo. 1994. Generation of lymphohematopoietic cells from embryonic stem cells in culture. Science 265:1098-1101. [DOI] [PubMed] [Google Scholar]
- 38.Nakano, T., H. Kodama, and T. Honjo. 1996. In vitro development of primitive and definitive erythrocytes from different precursors. Science 272:722-724. [DOI] [PubMed] [Google Scholar]
- 39.Niwa, H., T. Burdon, I. Chambers, and A. Smith. 1998. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 12:2048-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niwa, H., J. Miyazaki, and A. G. Smith. 2000. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24:372-376. [DOI] [PubMed] [Google Scholar]
- 41.Omichinski, J. G., G. M. Clore, O. Schaad, G. Felsenfeld, C. Trainor, E. Appella, S. J. Stahl, and A. M. Gronenborn. 1993. NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1. Science 261:438-446. [DOI] [PubMed] [Google Scholar]
- 42.Orkin, S. H. 1992. GATA binding transcription factors in hematopoietic cells. Blood 80:575-581. [PubMed] [Google Scholar]
- 43.Persons, D. A., J. A. Allay, E. R. Allay, R. A. Ashmun, D. Orlic, S. M. Jane, J. M. Cunningham, and A. W. Nienhuis. 1999. Enforced expression of the GATA-2 transcription factor blocks normal hematopoiesis. Blood 93:488-499. [PubMed] [Google Scholar]
- 44.Purton, L. E., I. D. Bernstein, and S. J. Collins. 1999. All-trans retinoic acid delays the differentiation of primitive hematopoietic precursors (lin-c-kit+Sca-1(+)) while enhancing the terminal maturation of committed granulocyte/monocyte progenitors. Blood 94:483-495. [PubMed] [Google Scholar]
- 45.Purton, L. E., I. D. Bernstein, and S. J. Collins. 2000. All-trans retinoic acid enhances the long-term repopulating activity of cultured hematopoietic stem cells. Blood 95:470-477. [PubMed] [Google Scholar]
- 46.Rodrigues, N. P., R. Forkert, D. M. Dombkowski, J. L. Zhang, K. W. Orford, S. H. Orkin, T. Enver, P. Vyas, and D. T. Scadden. 2003. Haploinsufficiency of GATA-2 effects adult stem cell homeostasis. Blood 102:565a. [DOI] [PubMed]
- 47.Sap, J., A. Munoz, K. Damm, Y. Goldberg, J. Ghysdael, A. Leutz, H. Beug, and B. Vennstrom. 1986. The c-erb-A protein is a high-affinity receptor for thyroid hormone. Nature 324:635-640. [DOI] [PubMed] [Google Scholar]
- 48.Schroeder, C., L. Gibson, C. Nordstrom, and H. Beug. 1993. The estrogen receptor cooperates with the TGFα receptor (c-erbB) in regulation of chicken erythroid progenitor self-renewal. EMBO J. 12:951-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shivdasani, R. A., and S. H. Orkin. 1996. The transcriptional control of hematopoiesis. Blood 87:4025-4039. [PubMed] [Google Scholar]
- 50.Song, W. J., M. G. Sullivan, R. D. Legare, S. Hutchings, X. Tan, D. Kufrin, J. Ratajczak, I. C. Resende, C. Haworth, R. Hock, M. Loh, C. Felix, D. C. Roy, L. Busque, D. Kurnit, C. Willman, A. M. Gewirtz, N. A. Speck, J. H. Bushweller, F. P. Li, K. Gardiner, M. Poncz, J. M. Maris, and D. G. Gilliland. 1999. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat. Genet. 23:166-175. [DOI] [PubMed] [Google Scholar]
- 51.Speck, N. A., and D. G. Gilliland. 2002. Core-binding factors in haematopoiesis and leukaemia. Nat. Rev. Cancer 2:502-513. [DOI] [PubMed] [Google Scholar]
- 52.Tocci, A., I. Parolini, M. Gabbianelli, U. Testa, L. Luchetti, P. Samoggia, B. Masella, G. Russo, M. Valtieri, and C. Peschle. 1996. Dual action of retinoic acid on human embryonic/fetal hematopoiesis: blockade of primitive progenitor proliferation and shift from multipotent/erythroid/monocytic to granulocytic differentiation program. Blood 88:2878-2888. [PubMed] [Google Scholar]
- 53.Towatari, M., M. Ciro, S. Ottolenghi, S. Tsuzuki, and T. Enver. 2004. Involvement of mitogen-activated protein kinase in the cytokine-regulated phosphorylation of transcription factor GATA-1. Hematol. J. 5:262-272. [DOI] [PubMed]
- 54.Towatari, M., G. E. May, R. Marais, G. R. Perkins, C. J. Marshall, S. Cowley, and T. Enver. 1995. Regulation of GATA-2 phosphorylation by mitogen-activated protein kinase and interleukin-3. J. Biol. Chem. 270:4101-4107. [DOI] [PubMed] [Google Scholar]
- 55.Trainor, C. D., T. Evans, and G. Felsenfeld. 1995. Negative regulation of chicken GATA-1 promoter activity mediated by a hormone response element. Mol. Endocrinol. 9:1135-1146. [DOI] [PubMed] [Google Scholar]
- 56.Trainor, C. D., R. Ghirlando, and M. A. Simpson. 2000. GATA zinc finger interactions modulate DNA binding and transactivation. J. Biol. Chem. 275:28157-28166. [DOI] [PubMed] [Google Scholar]
- 57.Tsai, F. Y., G. Keller, F. C. Kuo, M. Weiss, J. Chen, M. Rosenblatt, F. W. Alt, and S. H. Orkin. 1994. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature 371:221-226. [DOI] [PubMed] [Google Scholar]
- 58.Tsai, S., S. Bartelmez, R. Heyman, K. Damm, R. Evans, and S. J. Collins. 1992. A mutated retinoic acid receptor-alpha exhibiting dominant-negative activity alters the lineage development of a multipotent hematopoietic cell line. Genes Dev. 6:2258-2269. [DOI] [PubMed] [Google Scholar]
- 59.Tsai, S., S. Bartelmez, E. Sitnicka, and S. Collins. 1994. Lymphohematopoietic progenitors immortalized by a retroviral vector harboring a dominant-negative retinoic acid receptor can recapitulate lymphoid, myeloid, and erythroid development. Genes Dev. 8:2831-2841. [DOI] [PubMed] [Google Scholar]
- 60.Tsai, S., and S. J. Collins. 1993. A dominant negative retinoic acid receptor blocks neutrophil differentiation at the promyelocyte stage. Proc. Natl. Acad. Sci. USA 90:7153-7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsai, S. F., E. Strauss, and S. H. Orkin. 1991. Functional analysis and in vivo footprinting implicate the erythroid transcription factor GATA-1 as a positive regulator of its own promoter. Genes Dev. 5:919-931. [DOI] [PubMed] [Google Scholar]
- 62.Tsuzuki, S., and T. Enver. 2002. Interactions of GATA-2 with the promyelocytic leukemia zinc finger (PLZF) protein, its homologue FAZF, and the t(11;17)-generated PLZF-retinoic acid receptor alpha oncoprotein. Blood 99:3404-3410. [DOI] [PubMed] [Google Scholar]
- 63.Tsuzuki, S., M. Towatari, H. Saito, and T. Enver. 2000. Potentiation of GATA-2 activity through interactions with the promyelocytic leukemia protein (PML) and the t(15;17)-generated PML-retinoic acid receptor alpha oncoprotein. Mol. Cell. Biol. 20:6276-6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wadman, I. A., H. Osada, G. G. Grutz, A. D. Agulnick, H. Westphal, A. Forster, and T. H. Rabbitts. 1997. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 16:3145-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weinberger, C., C. C. Thompson, E. S. Ong, R. Lebo, D. J. Gruol, and R. M. Evans. 1986. The c-erb-A gene encodes a thyroid hormone receptor. Nature 324:641-646. [DOI] [PubMed] [Google Scholar]
- 66.Zelent, A., F. Guidez, A. Melnick, S. Waxman, and J. D. Licht. 2001. Translocations of the RARα gene in acute promyelocytic leukemia. Oncogene 20:7186-7203. [DOI] [PubMed] [Google Scholar]
- 67.Zhang, S. B., Q. Y. He, H. Zhao, C. Y. Gui, C. Jiang, and R. L. Qian. 2001. Function of GATA transcription factors in hydroxyurea-induced HEL cells. Cell Res. 11:301-310. [DOI] [PubMed] [Google Scholar]
- 68.Zhu, J., C. M. Heyworth, A. Glasow, Q. H. Huang, K. Petrie, M. Lanotte, G. Benoit, R. Gallagher, S. Waxman, T. Enver, and A. Zelent. 2001. Lineage restriction of the RARα gene expression in myeloid differentiation. Blood 98:2563-2567. [DOI] [PubMed] [Google Scholar]