Abstract
Exportin-5 is a nuclear export receptor for certain classes of double-stranded RNA (dsRNA), including pre-micro-RNAs, viral hairpin RNAs, and some tRNAs. It can also export the RNA binding proteins ILF3 and elongation factor EF1A. However, the rules that determine which RNA binding proteins are exportin-5 cargoes remain unclear. JAZ possesses an unusual dsRNA binding domain consisting of multiple C2H2 zinc fingers. We found that JAZ binds to exportin-5 in a Ran-GTP- and dsRNA-dependent manner. Exportin-5 stimulates JAZ shuttling, and gene silencing of exportin-5 reduces shuttling. Recombinant exportin-5 also stimulates nuclear export of JAZ in permeabilized cells. JAZ also binds to ILF3, and surprisingly, this interaction is RNA independent, even though it requires the dsRNA binding domains of ILF3. Exportin-5, JAZ, and ILF3 can form a heteromeric complex with Ran-GTP and dsRNA, and JAZ increases ILF3 binding to exportin-5. JAZ does not contain a classical nuclear localization signal, and in digitonin-permeabilized cells, nuclear accumulation of JAZ does not require energy or cytosol. Nonetheless, low temperatures prevent JAZ import, suggesting that nuclear entry does not occur via simple diffusion. Together, these data suggest that JAZ is exported by exportin-5 but translocates back into nuclei by a facilitated diffusion mechanism.
The movement of macromolecules into and out of the cell nucleus is usually mediated by soluble transport receptors that recognize specific sequences or structural characteristics of their cargoes (for reviews, see references 20, 23, and 25). These receptors can also interact with proteins in the nuclear pore complexes that span the double membrane of the nuclear envelope. A class of receptors called karyopherins is responsible for the nucleocytoplasmic transport of many proteins and of certain classes of RNA. Some karyopherins, called importins, are designed for nuclear import of cargo, while others, called exportins, remove their cargo from the nucleus. A small GTPase called Ran controls the assembly of the karyopherin-cargo complex. When Ran-GTP binds to an importin, it triggers the release of bound cargo; but when Ran-GTP binds to an exportin, it facilitates the assembly of the exportin-cargo complex. A steep gradient of Ran-GTP is maintained across the nuclear-cytoplasmic boundary by the asymmetric distribution of factors that regulate the guanine nucleotide-bound state of Ran. The exchange factor, Ran-GEF (also called RCC1), is exclusively nuclear, while the GTPase-activating protein, Ran-GAP, is cytoplasmic. This asymmetry ensures that import cargo is efficiently released only within the nucleus and that export cargo is released only in the cytoplasm
Most members of the karyopherin family carry protein cargoes, but two, exportin-t and exportin-5, are responsible for the transport of specific classes of small RNAs. Exportin-t provides the primary mechanism for the export of newly synthesized tRNAs (1, 16). Exportin-5 can also export some tRNAs, but its principal role is most likely the export of micro-RNA (miRNA) precursors (3, 8, 19, 36). Micro-RNAs are generated from large primary transcripts by a nuclear RNase III called Drosha. Drosha produces precursor miRNAs of ≈70 bp in length that form hairpins with short overhanging 3′ ends (17). Exportin-5 specifically recognizes double-stranded RNAs that possess this type of end structure (or similar structures that do not have a 5′ overhang) and forms a complex with the pre-miRNAs and Ran-GTP (13, 19, 36). In the cytoplasm, the Ran-GTP is hydrolyzed and the pre-miRNA is released so that it can be further processed into mature 21-bp miRNAs (17). Adenovirus produces a small RNA, VA1, with a similar hairpin structure that is also exported efficiently by exportin-5 (13). Moreover, hairpin RNAs expressed from vectors such as pSUPER are now widely used for gene silencing (7) and are exported from the nucleus by exportin-5 to the cytoplasm, where they are processed into 21-bp duplexes called short interfering RNAs (siRNAs) (4, 7).
Exportin-5 was initially identified as a transport factor for a dsRNA binding protein called ILF3 (also named NF90 and NFAR1) (6). The interaction with exportin-5 is RNA dependent, although it is inhibited by dsRNAs that possess a 5′ overhang (6, 14). These RNAs can bind ILF3 but not exportin-5. ILF3 is a nucleocytoplasmic shuttling protein that possesses a classical nuclear localization signal (NLS) (30). The function of ILF3 remains unclear, although it has been reported to regulate transcription and to stabilize certain mRNAs (15, 29, 31). However, it also facilitates the nuclear export of hairpin RNAs by exportin-5 (14). An interesting question is whether other dsRNA binding proteins also piggyback out of the nucleus via exportin-5 and whether these proteins function in miRNA processing.
As a first step towards addressing this issue, we have examined an unusual protein called JAZ, which does not contain classical dsRNA binding domains but instead binds RNA with high affinity through C2H2 zinc fingers (35). We show that JAZ is a nuclear protein at steady state but is highly dynamic within the nucleus and undergoes nucleocytoplasmic shuttling. JAZ associates with exportin-5 in the presence of Ran-GTP and a hairpin RNA, and nuclear export of JAZ requires exportin-5. However, JAZ also binds to ILF3 in an RNA-independent manner, and JAZ and ILF3 can form a heteromeric complex with exportin-5 and Ran-GTP. Unlike ILF3, JAZ does not contain a classical NLS. In principle, it could diffuse passively through the nuclear pores, and we show that import is indeed independent of soluble transport factors. Surprisingly, however, import is inhibited by wheat germ agglutinin and by low temperatures, which do not inhibit passive diffusion. This import pathway is reminiscent of those used by RanBP1, RCC1, and importin-α (22, 24, 26).
MATERIALS AND METHODS
Cloning, antibody, and recombinant protein expression.
Full-length cDNA encoding JAZ was generated by PCR amplification from a human brain cDNA library (Clontech Laboratories, Inc.) with primers to the 5′ and 3′ ends of the published sequence (accession number NM_012279). The entire JAZ cDNA was sequenced and then moved into the following vectors: pKH3, pKGFP, pKGFP3, pGEX-2T (Amersham Pharmacia Biotech), and pET-30a (Novagen). pKH3 is a mammalian vector that expresses fusion protein bearing an NH2-terminal triple hemagglutinin (HA) tag, and pKGFP and pKGFP3 are derivatives of this vector (5, 18). JAZ cDNA fragments were PCR amplified and ligated into pKGFP3. The pKmyc-ILF3, pKmyc-ILF3(F432/559A), and pKmyc-Exp-5 vectors have been described previously (6). Rabbit anti-human exportin-5, anti-JAZ, and anti-ILF3 antisera were produced against recombinant His6-tagged proteins. Antibodies were affinity purified with these proteins attached to cyanogen bromide beads and eluted at pH 2.5.
His6-S-JAZ and glutathione S-transferase (GST)-JAZ were expressed in Escherichia coli BL21(DE3) after growth at 37°C in Luria broth (LB) supplemented with 2% (vol/vol) ethanol to an optical density at 600 nm of ≈0.8, followed by induction with 400 μM isopropylthiogalactopyranoside (IPTG) at 18°C overnight. His6-S-JAZ was purified on nitrilotriacetic acid-Ni2+ beads (Qiagen). GST-JAZ was purified on glutathione-Sepharose beads (Amersham Pharmacia Biotech). Purification of GST-ILF3 fragments, exportin-5-His6, and Ran has been described previously (6, 18).
Yeast two-hybrid and conjugation assays.
Full-length wild-type ILF3 or ILF3(F432/559A) was used as the bait for a yeast two-hybrid screen of a random-primed murine 10-day embryo library. Mating assays were performed as described (27). ILF3 and ILF3(F432/559A) were expressed in the Saccharomyces cerevisiae HF7c (MATa) strain as COOH-terminal fusions to the DNA binding domain of the GAL3 protein. JAZ(178-276) was expressed in the W303 (MATα) strain as a COOH-terminal fusion to the transactivation domain of VP16.
Cell culture, transfection, and processing.
Cells for fusion and binding assays were usually transfected with calcium phosphate. At ≈18 h after transfection, the medium was replaced and the cells were incubated for an additional 24 h before use. For cotransfections of plasmids and siRNAs, FuGene 6 (Roche) was used to transfect plasmids according to the manufacturer's instructions, and 6 h later siRNAs were transfected with Oligofectamine (Invitrogen). Green fluorescent protein (GFP)3-JAZ fusions were transfected with Effectene (Qiagen), and the cells were imaged ≈18 h later.
Cells to be analyzed by fluorescence microscopy were fixed with 4% (wt/vol) paraformaldehyde in phosphate-buffered saline (PBS; 137 mM NaCl, 3 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4), permeabilized with −20°C methanol, and blocked in 10% (wt/vol) bovine serum albumin (BSA)-PBS at room temperature. Myc-tagged proteins were detected with 9E10 monoclonal antibody (2 μg/ml) and Texas red-conjugated goat anti-mouse immunoglobulin G (IgG) (1:500; Jackson ImmunoResearch Laboratories). HA3-tagged JAZ was detected with 12CA5 monoclonal antibody (1 μg/ml) and indodicarbocyanine-conjugated goat anti-rabbit IgG (1:500; Jackson ImmunoResearch Laboratories). Endogenous exportin-5 was detected with purified anti-exportin-5 antibodies (1:30), and GST-JAZ was detected with anti-GST monoclonal antibody (a gift from Tom Parsons, University of Virginia). DNA was stained with 4′,6′-diamidino-2-phenylindole (DAPI) (1 ng/ml), and then the slides were mounted with Antifade (Molecular Probes).
For live cell imaging, cells were washed with Ringer's solution (25 mM HEPES, pH 7.2, 110 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgSO4, 10 mM glucose, 1 mM KH2PO4, 1 mg of BSA per ml), incubated with Hoechst dye (5 μg/ml) for 15 min at 37°C, and washed and incubated in Ringer's solution. Images were captured with a 60× water immersion objective lens (n.a. 1.2) on a Nikon inverted microscope with a Hamamatsu charge-coupled device camera. All immunofluorescence data were obtained and processed with Openlab (Improvision) and Adobe Photoshop software as described previously (28).
RNA binding assays.
VARdm RNA (a gift from C. Dargemont, Centre National de la Recherche Scientifique, Paris, France) has been described elsewhere (12). [α-32P]ATP-labeled VARdm and unlabeled VARdm were synthesized by in vitro transcription with the Ampliscribe T7 transcription kit (Epicentre Technologies) as described by Gwizdek and Dargemont (12); 20-bp blunt-ended dsRNAs were obtained by annealing precursor single-stranded RNAs synthesized by Dharmacon. For the [α-32P]VARdm binding assays, 5 nM GST-JAZ and 7 nM [α-32P]VARdm were incubated in 200 μl of binding buffer [20 mM HEPES, pH 7.3, 150 mM potassium acetate, 2 mM magnesium acetate, 0.1% Tween 20, 7 mM β-mercaptoethanol, 1.5 μg of poly(dI[chemp]dC) per ml, 0.2% BSA, 10 μl of RNase inhibitor/40 ml of buffer] at 23°C for 1 h. At the indicated time points, 10 μl of the reaction mixture was removed and filtered through nitrocellulose. Protein-bound [α-32P]VARdm was quantified by scintillation counting.
Protein binding assays.
35S-labeled JAZ, wild-type ILF3, ILF3(F432/559A), and an unrelated protein called LGN were synthesized by in vitro transcription translation (Promega), with the pKH3-JAZ, pKmyc-ILF3, pKmyc-ILF3(F432/559A), and pKmyc-LGN vectors, respectively, as templates. Synthesized proteins were incubated in 100 μl of reaction mixture with 2 μg of 12CA5 monoclonal antibody and binding buffer (20 mM HEPES-KOH, pH 7.3, 150 mM potassium acetate, 2 mM magnesium acetate, 0.1% Tween 20, 14 mM β-mercaptoethanol, and 0.5% BSA) for 1 h at 4°C. Protein A beads were then added and incubated for 1 h at 4°C. Immunoprecipitates were washed three times in 1 ml of 0.05% NP-40-PBS, resuspended in Laemmli sample buffer, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and analyzed by fluorography.
To assay binding to GST fusion proteins, 35S-labeled proteins were incubated with the proteins attached to 10 μl of glutathione-Sepharose beads in binding buffer. Where indicated, 0.1 unit of RNase V1 was added. Reaction mixtures were incubated for 1 h at 4°C or room temperature when RNase V1 was included in the reaction, and bead-associated proteins were washed in 0.05% NP-40-PBS, dissolved in Laemmli sample buffer, separated by SDS-PAGE, and visualized by Coomassie brilliant blue staining or fluorography. GST fusion proteins were detected by horseradish peroxidase-conjugated anti-GST (1:500); His6-S-JAZ and exportin-5-His6 were detected by anti-His6 monoclonal antibody (1:1,000); and Ran was detected with anti-Ran monoclonal antibody (1:2,000; Transduction Labs), followed by horseradish peroxidase-conjugated goat anti-mouse IgG (1:20,000; Pierce Chemical Co.).
For coimmunoprecipitations from cells, HeLa cells were transfected with pKH3-JAZ or pKH3. The cells were washed with ice-cold PBS and lysed with lysis buffer (50 mM HEPES, pH 7.4, 300 mM sodium chloride, 5 mM EDTA, 15 mM magnesium chloride, 1% Tween 20, 4 mM β-mercaptoethanol, 20 μg each of aprotinin and leupeptin per ml, 1 mM phenylmethylsulfonyl fluoride) and then scraped off the plates, and the supernatant was cleared by centrifugation (14,000 rpm; 5 min). Protein A beads with and without 3 μg of 12CA5 monoclonal antibody were added to each cell lysate and incubated at 4°C for 1 h. Immunoprecipitates were washed three times in 1 ml of 0.05% NP-40/PBS, resuspended in Laemmli sample buffer, separated by SDS-PAGE, and analyzed by Western blotting. HA3-JAZ was detected by horseradish peroxidase-conjugated 12CA5 (1:500). ILF3 was detected by anti-ILF3 antibodies (1:500) and horseradish peroxidase-conjugated goat anti-rabbit IgG (1:20,000; Pierce Chemical Co.).
Microinjection.
Cells were cultured on Labtek coverglasses. The medium was changed to Ringer's solution before microinjection. GST-JAZ, GGNLS, and GST-GFP were injected at 1 mg/ml. Tetramethylrhodamine isothiocyanate- or fluorescein isothiocyanate (FITC)-labeled dextran (2 mg/ml; Sigma-Aldrich) was included as an injection site marker. After injections, samples were incubated for 30 to 45 min at 37°C before fixation.
Heterokaryon fusion assays.
HeLa and BHK21 cells were transfected with plasmids or siRNA as indicated. An acceptor cell line, GSN2, was a gift from Bryce Paschal (University of Virginia) (2). GSN2 with HeLa or GSN2 with BHK21 cells were coplated onto Labtek II slides overnight. Cells were treated with 50 μM cycloheximide for 30 min and then for 2 min with 50% (wt/vol) polyethylene glycol 8,000 warmed to 37°C. Fused cells were washed and incubated at 37°C for 1 h in the presence of cycloheximide and then fixed and processed as described above.
Permeabilized cell transport assays.
In vitro nuclear export assays were performed as described (6). HeLa cells were plated on Labtek II slides and transfected with Effectene. All export cocktails contained an energy-regenerating system (20 mM phosphocreatine, 1 mM GTP, 1 mM ATP, and 50 μg of creatine phosphokinase per ml) and wild-type Ran (3 μM) in transport buffer (20 mM HEPES-KOH, pH 7.4, 110 mM potassium acetate, 2 mM magnesium acetate, 2 mM dithiothreitol, 250 mM sucrose, 5% BSA). Exportin-5-His6 was added to a final concentration of 100 μg/ml, and where indicated, wheat germ agglutinin was added to 200 μg/ml. Export was performed for 30 min at 30°C, and then the cells were fixed, permeabilized, and processed as described above.
In vitro import assays were performed essentially as described (33). HeLa cells were permeabilized with digitonin (0.0025%) in transport buffer for 5 min on ice. Permeabilization was stopped by washing the samples in ice-cold import assay buffer and adding the energy-regenerating system in import assay buffer for 5 min at room temperature. Then cells were incubated with indicated factors at 30°C for 20 min. After washing, nuclei were fixed with 4% (wt/vol) paraformaldehyde-PBS, permeabilized with −20°C methanol, and stained as necessary.
RESULTS
JAZ is a nucleocytoplasmic shuttling protein.
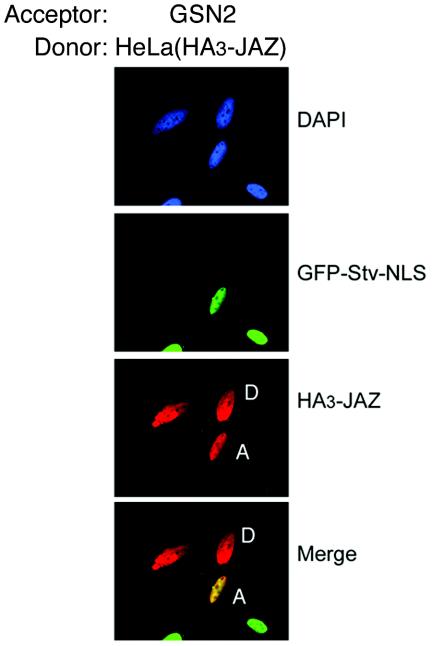
JAZ is predominantly nuclear at steady state, but like many other nuclear proteins, it is highly dynamic, as measured by fluorescence recovery after photobleaching of transfected GFP-JAZ (t1/2, ≈10 s; unpublished data). To determine whether JAZ shuttles between the nucleus and the cytoplasm, we used heterokaryon fusion assays. To construct heterokaryons, we fused HeLa cells expressing HA3-tagged JAZ with GSN2 cells. The latter cells stably express a GFP-streptavidin-NLS fusion protein that is constitutively nuclear and does not shuttle, and this cell line therefore both provides acceptor nuclei and functions as a negative control for shuttling (2). Shuttling of JAZ would lead to its equilibration into the nuclei of fused GSN2 cells. Figure 1A shows a representative heterokaryon consisting of two HeLa cell nuclei and one GSN2 nucleus. The merged image shows that the GSN2 nucleus contained HA3-JAZ, while the HeLa nuclei did not contain the GFP-streptavidin. Thus, although JAZ is predominantly nuclear at steady state, it is both dynamic within the nucleus and continuously traversing the nuclear envelope.
FIG. 1.
JAZ is a nucleocytoplasmic shuttling protein. GSN2 cells (labeled A for acceptor) and HeLa cells (labeled D for donor) that had been transiently transfected with pKH3-JAZ were coseeded onto coverslips. GSN2 cells constitutively express a nonshuttling GFP-streptavidin-NLS fusion protein as a marker of the acceptor cell nucleus (2). Cells were fused with polyethylene glycol and incubated for 1 h with cycloheximide. HA3-JAZ was detected with anti-HA antibody and Texas red-conjugated secondary antibody. Colocalization is shown as yellow.
Nuclear export of JAZ is dependent on exportin-5 in both digitonin-permeabilized cells and intact cells.
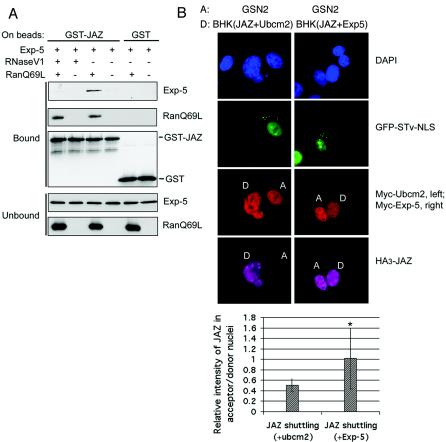
JAZ binds to dsRNA with high affinity (35). The transport factor exportin-5 can efficiently export certain classes of dsRNA from the nucleus, and we therefore asked if JAZ, a dsRNA binding protein, can bind to exportin-5 in the presence of Ran-GTP and dsRNA. Recombinant proteins were prepared from E. coli, which contains dsRNAs that remain associated with the JAZ and exportin-5 through purification. GST-JAZ was incubated with exportin-5-His6 with or without RanQ69L (a constitutively active mutant) and with and without RNase V1. RNase V1 efficiently degrades dsRNAs. The exportin-5 bound to recombinant GST-JAZ only in the presence of both RanQ69L and dsRNA (Fig. 2A). This result suggested that exportin-5 might behave as an export receptor for JAZ.
FIG. 2.
Exportin-5 binds to JAZ and stimulates JAZ shuttling in intact cells. (A) JAZ interacts with exportin-5 in a Ran-GTP-dependent manner that is mediated by dsRNA. GST-JAZ on beads was exposed to exportin-5-His6 with and without RanQ69L and with and without RNase V1. Bound proteins were detected with anti-His6 or anti-GST antibodies. (B) Overexpression of exportin-5 stimulates JAZ shuttling between nuclei of BHK and GSN2 cells. The heterotypic cell fusion assay was performed as described for Fig. 1. BHK21 cells cotransfected with pKH3-JAZ plus pKmyc-Ubcm2 or with pKH3-JAZ plus pKmyc-Exp-5 were used as donor cells. Quantification of shuttling is shown in arbitrary units ± standard deviation. Nuclei were stained with DAPI, and these images were used to define the nuclear boundaries. Ratios of the total pixel intensities in the donor and acceptor nuclei were measured. Data were analyzed by a two-tailed t test, P = 0.041; n = >10 fused cells/sample.
To test whether exportin-5 is involved in nuclear export of JAZ, we first used heterokaryon fusion assays. Our clone of BHK21 cells expresses only very low levels of exportin-5 and thus can be used as a “null” cell line (6). To construct heterokaryons, we fused GSN2 cells with BHK cells that coexpressed HA3-JAZ with either Myc-tagged exportin-5 or Myc-Ubcm2. Ubcm2 is a constitutively shuttling protein but not a transport receptor (27). Therefore, it was used here as an indicator of fused nuclei and also as a positive control for shuttling. As shown in Fig. 2B, both Ubcm2 and exportin-5 equilibrated between the donor and acceptor nuclei, but JAZ only equilibrated efficiently in fused nuclei that coexpressed exportin-5. Quantification of multiple fused cells showed that the JAZ equilibrated with significantly less efficiency in fused nuclei coexpressing Ubcm2 (Fig. 2B). This result suggests that the expression of exportin-5 can stimulate JAZ export in intact cells.
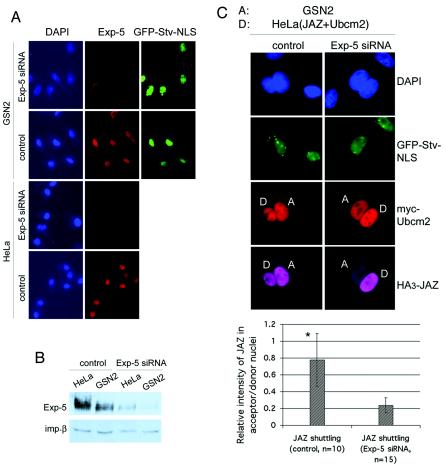
However, JAZ shuttling was not absent in the BHK-GSN2 fusions, probably because the GSN2 acceptor cells express exportin-5. Therefore, the heterokaryons will possess a variable amount of exportin-5, depending on the number of acceptor and donor cells that have fused to one another. This effect might account for the large variation in shuttling efficiency that we observed (see histogram, Fig. 2B). To overcome this problem, we used gene silencing by RNA interference. However, because the heterokaryon assay involves fusion of two cell types, we needed to silence exportin-5 expression in both the donor and acceptor cells. First, we transfected exportin-5 siRNA or nonspecific 20-bp dsRNA as a control into GSN2 cells and HeLa cells (9). As shown in Fig. 3A and B, the exportin-5 siRNA efficiently and uniformly decreased the expression level of exportin-5 in both cell types. The siRNA-transfected GSN2 cells were then fused with siRNA-transfected HeLa cells that expressed both HA3-JAZ and Myc-Ubcm2. Separately, control dsRNA-transfected GSN2 cells were fused with control dsRNA-transfected HeLa cells expressing HA3-JAZ and Myc-Ubcm2. Knockdown of exportin-5 in both donor and acceptor cells dramatically decreased the shuttling of JAZ, while transfection of control dsRNA did not affect the shuttling of JAZ (Fig. 3C). This result suggests that exportin-5 is responsible for export of JAZ in intact cells.
FIG. 3.
Exportin-5 is required for JAZ shuttling in intact cells. (A) Gene silencing of exportin-5 in HeLa cells and GSN2 cells. HeLa cells and GSN2 cells were transfected with exportin-5 siRNA or control dsRNA, and after 3 days the cells were fixed and stained for exportin-5 (red channel). The green channel shows the GFP in the nuclei of the GSN2 cells. (B) Cells were transfected as in panel A, but HeLa cells were also cotransfected with pKH3-JAZ plus pKmyc-Ubcm2. Cell lysates were analyzed by immunoblotting with anti-exportin-5 antibodies and anti-importin-β (as a loading control). (C) The cells were then fused as described for Fig. 1 and imaged to determine the extent of JAZ shuttling. Shuttling of JAZ was measured in fused cells as described for Fig. 2 and analyzed by an unpaired two-tailed t test, P < 0.0004.
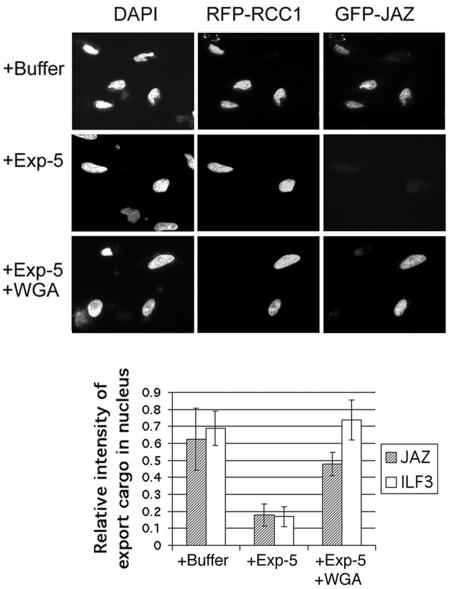
As a second approach to prove that exportin-5 is the export receptor of JAZ, we used a digitonin-permeabilized cell export assay (6). JAZ was expressed in HeLa cells as a GFP fusion protein together with a red fluorescent protein (RFP) fusion of RCC1. RCC1 is a constitutively nuclear, nonshuttling protein and therefore serves as a marker for transfected cells. The cell plasma membranes were then permeabilized with digitonin, and the cells were incubated with an energy-regenerating system in order to deplete their nuclei of endogenous transport factors. On addition of exportin-5 to these cells (together with energy and Ran), GFP-JAZ was exported rapidly from the nucleus, whereas no export was observed upon addition of buffer alone. Addition of wheat germ agglutinin, a protein that binds tightly to nuclear pores and inhibits receptor-mediated nucleocytoplasmic transport pathways (38), also blocked GFP-JAZ export (Fig. 4). As a positive control, we showed that exportin-5 was able to promote nuclear export of ILF3, as described previously (6). Together, these data demonstrate that exportin-5 is a nuclear export receptor for JAZ.
FIG. 4.
Exportin-5 mediates export of JAZ in digitonin-permeabilized cells. HeLa cells were cotransfected with pKGFP-JAZ plus pKRFP-RCC1 or pKGFP-ILF3 plus pKRFP-RCC1. pKRFP-RCC1 was used as a constitutive nuclear marker for transfected cells. The cells were permeabilized with digitonin, and export assays were performed as described previously in buffer alone or with buffer plus exportin-5-His6 (100 μg/ml) or plus exportin-5-His6 (100 μg/ml) plus wheat germ agglutinin (WGA, 200 μg/ml) (6). Export was quantified by measuring the nuclear intensity of GFP in transfected cells (red), using DAPI staining to mark the nuclear boundaries. Mean values for nuclear GFP-JAZ are shown in arbitrary units ± standard deviation (n = >30 cells/sample).
JAZ binds ILF3 independently of dsRNA.
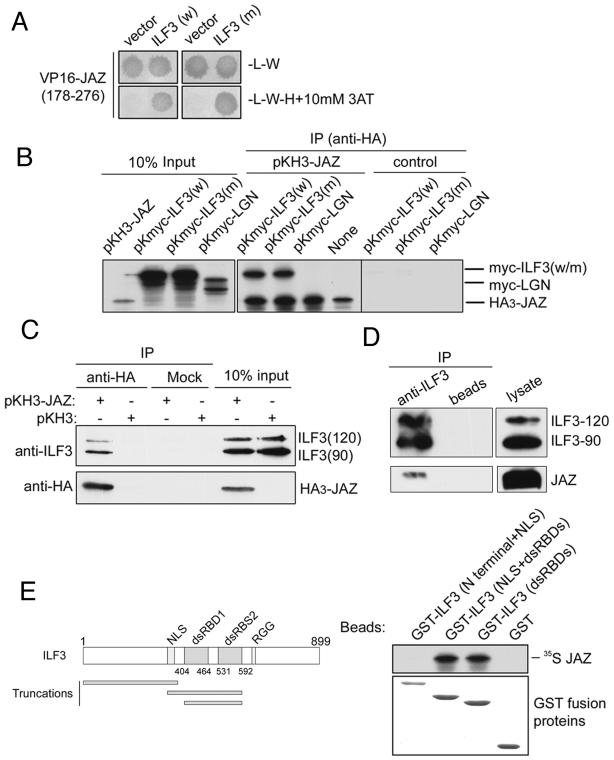
Exportin-5 also exports the dsRNA binding protein ILF3. During a yeast two-hybrid screen for ILF3 binding partners, we identified a fragment of JAZ. The identified JAZ clone included amino acid residues 178 to 276, which encode the third and fourth zinc finger domains of JAZ. These domains are essential for binding dsRNA, and we therefore assumed that the observed interaction was indirect and mediated by dsRNA, perhaps from proviruses present in the yeast genome. Surprisingly, however, a mutant of ILF3, ILF3(F432/559A), that is defective in dsRNA binding (6) also interacted with JAZ (Fig. 5A), suggesting that the interaction between ILF3 and JAZ does not involve dsRNA.
FIG. 5.
JAZ is a binding partner of ILF3. (A) Yeast two-hybrid conjugation assay. HF7c (MATa) yeast cells expressing the GAL4 DNA binding domain-ILF3 (w, as wild type) or ILF3(F432/559A) (m, as mutant) were mated with the W303 (MATα) strain expressing the VP16 fusion as indicated. Diploid yeast cells were selected on plates lacking leucine and tryptophan and replica-plated onto plates lacking leucine, tryptophan, histidine, and 3-aminotriazole. (B) Full-length JAZ interacts with ILF3 in vitro. 35S-labeled Myc-ILF3 (w), Myc-ILF3 (F432/559A) (m), or an unrelated protein Myc-LGN, was incubated with 35S-labeled HA3-JAZ on beads or beads alone. Bound proteins were visualized by fluorography. (C) JAZ interacts with ILF3 in vivo. HeLa cells were transfected with pKH3-JAZ or pKH3. Cytosol was precipitated with anti-HA monoclonal antibody (12CA5) or beads only (mock). Bound proteins were detected with anti-HA or anti-ILF3 antibodies. (D) Endogenous ILF3 binds endogenous JAZ. HeLa cell lysate was precleared by incubation with nonspecific rabbit IgG on protein A-Sepharose beads and then incubated with protein A beads alone or with beads to which anti-ILF3 antibodies were attached. After washing, bound JAZ was detected by immunoblotting with anti-JAZ antibodies. (E) JAZ interacts with the dsRNA binding domains of ILF3. 35S-labeled JAZ was incubated with GST-tagged ILF3(4-398), ILF3(366-592), ILF3(404-592), or GST on beads. Bead-bound proteins were visualized by Coomassie staining or fluorography.
To verify that full-length JAZ binds ILF3, HA3-tagged full-length JAZ was expressed by in vitro transcription and translation assays, immunoprecipitated with anti-HA monoclonal antibody, and incubated with Myc-tagged ILF3 or ILF3(F432/559A) expressed by TNT. Full-length JAZ interacted with both ILF3 and ILF3(F432/559A) but not the Myc-tagged control protein, LGN (Fig. 5B). This result suggested that JAZ interacts with ILF3 independently of dsRNA binding to ILF3. Additionally, when HA3-tagged JAZ was immunoprecipitated from transfected HeLa cells, we detected endogenous ILF3 (both the 90- and 120-kDa isoforms) associated with the protein (Fig. 5C). Moreover, a small amount of endogenous JAZ was immunoprecipitated with endogenous ILF3 (Fig. 5D). This result shows that JAZ interacts with endogenous ILF3 in vivo.
To refine the region of ILF3 required for JAZ binding, we tested the ability of JAZ to bind to several ILF3 fragments. JAZ only interacted with ILF3(366-592) and ILF3(404-592) (Fig. 5E). The overlapping regions of these two fragments are the dsRNA binding domains (dsRBDs) of ILF3. This result suggests that the dsRNA binding domains of ILF3 are the region required for binding to JAZ.
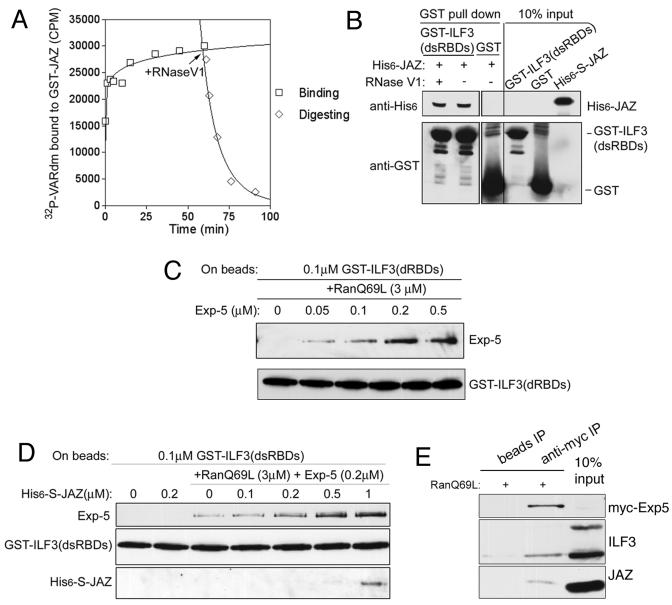
Since both JAZ and ILF3 are dsRNA binding proteins, the experiments described above do not completely exclude the possibility that the interaction is bridged by dsRNA. To test this more rigorously, we added RNase V1 to the GST-ILF3(dsRBDs) and His6-S-JAZ. RNase V1 is an endonuclease specific for dsRNA (34). First, to validate that RNase V1 can efficiently destroy protein-bound RNA, we added RNase V1 to a complex of [32P]VARdm RNA and GST-JAZ and measured the amount of remaining RNA bound to the GST-JAZ filtering through nitrocellulose at intervals (Fig. 6A). VARdm is derived from VA1 RNA (12). It contains a ≈24-bp imperfect base-paired dsRNA and 3′ protrusion and can bind both exportin-5 and ILF3 (14). We used VARdm here as a substrate dsRNA for JAZ. JAZ has a high affinity for VARdm. However, addition of RNase V1 rapidly destroyed the JAZ-bound RNA (Fig. 6A). Nonetheless, RNase V1 treatment did not affect the interaction between JAZ and ILF3 (Fig. 6B). Nor did the addition of the VARdm RNA alter the binding of JAZ to ILF3. These results confirm that JAZ interacts directly with ILF3, independently of RNA binding to either protein.
FIG. 6.
JAZ binds directly to ILF3, independently of dsRNA, and can interact with ILF3 and exportin-5 simultaneously. (A) RNase V1 can digest dsRNA bound to JAZ. GST-JAZ (5 nM) was incubated with 7 nM [α-32P]VARdm. After 1 h, 0.1 unit of RNase V1 was added to the reaction mixture, and aliquots of reaction mixture were taken at the indicated times for analysis. The amount of [α-32P]VARdm bound to GST-JAZ was detected by filter binding to nitrocellulose and scintillation counting. (B) JAZ interacts directly with ILF3. GST-ILF3(dsRBDs) on beads was exposed to His6-S-JAZ with and without RNase V1 for 1 h. Bound proteins were detected with anti-His6 or anti-GST antibodies. (C) Relative binding affinity between ILF3 and exportin-5; 0.1 μM GST-ILF3(dsRBDs) on beads was exposed to 3 μM RanQ69L and increasing concentrations of exportin-5-His6. Bound proteins were detected by anti-His6 or anti-GST antibodies. (D) JAZ increases the association of ILF3 and exportin-5. GST-ILF3(dsRBDs) at 0.1 μM on glutathione-beads was exposed to 3 μM RanQ69L, 0.2 μM exportin-5-His6, and increasing concentrations of His6-S-JAZ. Bound proteins were detected by anti-His6 or anti-GST antibodies. (E) Immunoprecipitation of a heteromeric complex of exportin-5, ILF3, Ran-GTP, and JAZ. HeLa cells were transfected with pKmyc-Exp-5. Cytosol was precipitated with anti-Myc monoclonal antibody or beads only in the presence of RanQ69L. Bound proteins were detected with anti-Myc, anti-ILF3, or anti-JAZ antibodies.
To further refine the region in JAZ required for ILF3 binding, we tested the ability of GST-ILF3(dsRBDs) to bind JAZ fragments. GST-ILF3(dsRBDs) interacts strongly with full-length JAZ but very weakly with JAZ(1-186) and JAZ(178-276) and not with JAZ(211-294) (data not shown), suggesting that zinc fingers (ZFs) I and II, ZFs III and IV, or ZF IV alone is not sufficient for ILF3 binding.
JAZ, ILF3, and exportin-5 can form a heteromeric complex.
Previously we have shown that the interactions between ILF3 and exportin-5 and between JAZ and exportin-5 are each bridged by dsRNA (14). Since JAZ binds the dsRNA binding domains of ILF3 when they interact, dsRNA and JAZ binding to ILF3 might involve overlapping residues. Therefore, bound JAZ might conceivably promote, prevent, or have no affect on the interaction of ILF3 with exportin-5. To estimate the affinity of the ILF3 for exportin-5, GST-ILF3(dsRBDs) was incubated with RanQ69L and increasing concentrations of exportin-5. The Kapp was determined to be ≈0.1 μM (Fig. 6C). GST-ILF3(dsRBDs) was then incubated with RanQ69L, exportin-5, and increasing concentrations of His6-S-JAZ. JAZ joined the complex and increased the binding of ILF3 to exportin-5 (Fig. 6D). This result suggests that JAZ, ILF3, Ran-GTP, and dsRNA can form a heteromeric complex with exportin-5 in vitro and that JAZ promotes ILF3 binding. However, the coexpression of ILF3 with GFP-JAZ did not discernibly increase the export of the JAZ from permeabilized cells (data not shown).
Additionally, when Myc-exportin-5 was immunoprecipitated from transfected HeLa cell lysate, we detected endogenous ILF3 (predominantly the 90-kDa isoform) and endogenous JAZ associated with the protein in the presence of RanQ69L (Fig. 6E). This result indicates that a heteromeric complex of exportin-5, Ran-GTP, ILF3, and JAZ can form in vivo. However, the expression of exportin-5 was too low, as detected by our antibody, and the complex was probably too transient to enable detection with endogenous exportin-5.
JAZ is imported into the nucleus by a nonclassical mechanism.
The size of JAZ is below the diffusion limit for the nuclear pore complex, which is about 60 kDa for a globular protein (32). In principle, therefore, JAZ could accumulate in the nucleus either by an active mechanism or by passive diffusion followed by nuclear retention. Previously, it was reported that the zinc finger domains of JAZ are required for its nuclear localization, but this study did not distinguish sequestration in the nucleus from nuclear import (35). There is one cluster of basic residues (207 to 210) in the protein that might be recognized as a classical NLS, but this sequence is within the third zinc finger domain, so it is unlikely to be recognized by an import receptor.
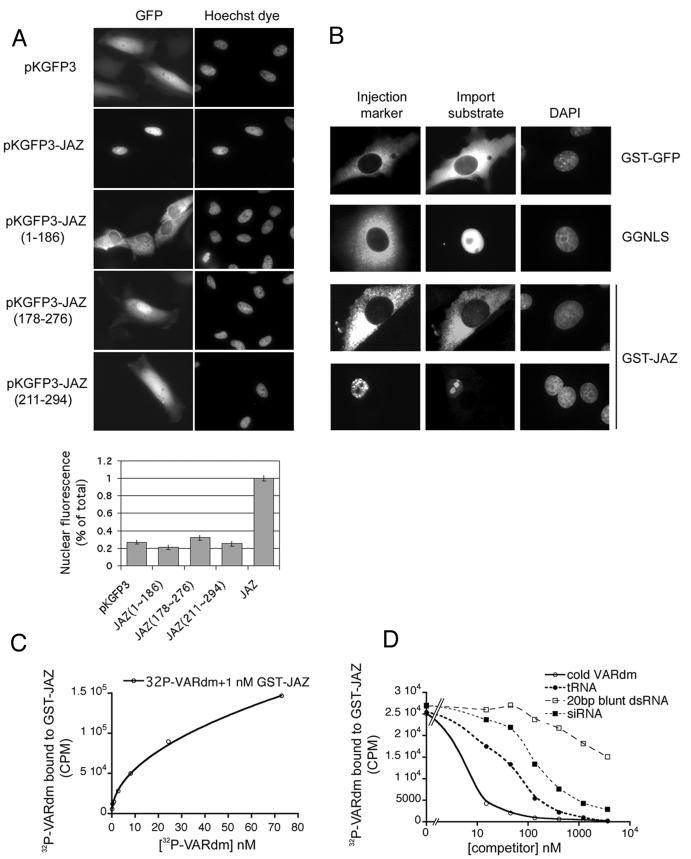
To test whether an NLS is present in JAZ, we first expressed residues 1 to 186, 178 to 276, 211 to 284, and full-length JAZ as amino-terminal fusions to GFP3 in mammalian cells. The triple GFP construct (≈80 kDa) was used to decrease passive diffusion through the nuclear pore complex. As shown in Fig. 7A, none of the truncations accumulated efficiently in the nucleus. Since the three truncations overlap and cover the entire length of JAZ, we conclude that there is no classical NLS sequence in JAZ. However, it is possible that the protein contains some type of multipartite NLS that extends beyond the overlapping regions used in this experiment.
FIG. 7.
Nuclear import of JAZ. (A) pKGFP3, pKGFP3-JAZ, pKGFP3-JAZ(1-186), pKGFP3-JAZ(178-276), and pKGFP3-JAZ(211-294) were transfected into HeLa cells. At 18 h posttransfection, the DNA was stained with Hoechst dye, and the relative nuclear and cytoplasmic fluorescence levels of the constructs were analyzed by live cell imaging. Quantification of import is shown in arbitrary units ± standard deviation (n = >15 cells/sample). (B) GST-JAZ does not accumulate rapidly in the nuclei of intact cells. GST-JAZ, GST-GFP-NLS (GGNLS), and GST-GFP were injected into BHK21 cells in either the nuclei or cytoplasm. Cells were fixed after 30 to 45 min of incubation. The localization of GST-JAZ was analyzed with anti-GST and a Texas red-conjugated secondary antibody. (C and D) GST-JAZ is properly folded and binds specifically to certain types of dsRNA; 1 nM GST-JAZ was exposed to increasing concentrations of [α-32P]VARdm in panel C, and 1 nM GST-JAZ plus 15 nM [α-32P]VARdm were incubated with increasing concentrations of VARdm, tRNA, 20-bp dsRNA, or siRNA in panel D. [α-32P]VARdm bound to GST-JAZ was analyzed by filter binding to nitrocellulose and scintillation counting.
To further examine the mechanism by which JAZ enters the nucleus, we asked if recombinant GST-JAZ could enter the nucleus after microinjection into cells (Fig. 7B). Surprisingly, however, the GST-JAZ remained mostly cytoplasmic when injected into the cytoplasm of BHK cells. One interpretation of this result is that JAZ enters the nucleus by diffusion rather than active import, and GST-JAZ (≈122 kDa as a dimer) is too large to cross the nuclear pore complex. Alternatively, GST-JAZ might not be not functional, or the GST tag might interfere with the JAZ transport mechanism. We have some evidence, however, that the GST-JAZ fusion is properly folded and functional, since it can bind ILF3, exportin-5, and dsRNA (Fig. 7C and D). Moreover, the GST-JAZ has a high affinity for VARdm (Fig. 7C). Its binding affinities with other dsRNAs tested were VARdm > tRNA > blunt-ended 20-bp dsRNA > siRNA (Fig. 7D). Therefore, we thought it more likely that either JAZ diffuses passively through the nuclear pore complex or GST interferes with the JAZ transport mechanism.
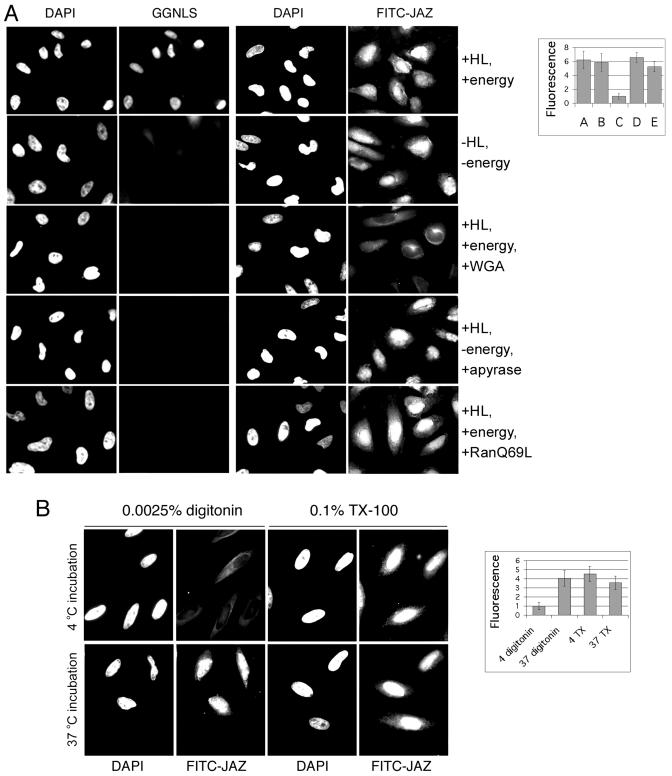
To address this issue, we used digitonin-permeabilized cells that were competent for nuclear protein transport. As a substrate, we used FITC-labeled His6-S-JAZ. GST-GFP-NLS (GGNLS), which is imported by a classical temperature-dependent and importin-α/β-dependent import pathway, was used as a control (33). As expected, the import of GGNLS was inhibited by removal of soluble factors (HeLa cell lysate), addition of the pore-binding protein wheat germ agglutinin, removal of energy, and addition of RanQ69L (Fig. 8A). In contrast, however, JAZ accumulated in the nucleus in the absence of either soluble factors or energy. Moreover, JAZ import was not inhibited by addition of RanQ69L. Treatment of the JAZ with RNase VI also did not affect nuclear import (data not shown). These results are consistent with passive diffusion.
FIG. 8.
JAZ is imported into nuclei by a nonclassical pathway. (A) Nuclear accumulation of JAZ does not require energy or soluble factors. HeLa cells were permeabilized with 0.0025% digitonin and overlaid with either 3 μM FITC-labeled His6-S-JAZ or 3 μM GGNLS in the presence of an energy-regenerating system, HeLa cell lysate, 100 U of apyrase/ml, 100 mg of RanQ69L/ml, or 200 mg of wheat germ agglutinin (WGA)/ml. Cells were incubated at 30°C for 20 min and then fixed. DNA was stained by DAPI. Import was measured from the nuclear pixel intensities for >30 cells/sample, using the DAPI staining to define the nuclear boundaries. Quantification of import is shown in arbitrary units ± standard deviation (n = >40 cells/sample). (B) Nuclear accumulation of JAZ is inhibited by low temperature. HeLa cells were permeabilized with 0.0025% digitonin or 0.1% Triton X-100 and overlaid with 3 μM FITC-labeled His6-S-JAZ, with HeLa cell lysate (HL) and energy-regenerating system. The cells were incubated on ice or at 30°C for 20 min and then fixed. DNA was stained by DAPI. Nuclear FITC-JAZ was measured as described for panel A.
Surprisingly, however, FITC-JAZ import was efficiently inhibited either by addition of wheat germ agglutinin or by reducing the temperature to 4°C (Fig. 8A and B). The passive diffusion of fluorescent dextrans and of small proteins such as GFP is not blocked by wheat germ agglutinin, which interacts with glycosylated proteins in the nuclear pore complex (10). Moreover, passive diffusion is proportional to the absolute temperature, so a drop from 23 to 4°C would have a negligible effect, strongly arguing that JAZ import cannot be a passive diffusive process. An alternative explanation, however, is that cold temperatures disrupt the retention of JAZ in the nucleus, perhaps by altering nucleolar structure. To address this point, HeLa cells were permeabilized with Triton X-100 to remove the nuclear envelope and incubated with FITC-JAZ. As shown in Fig. 8B, the binding of JAZ to nucleoli was independent of temperature, confirming that the effect of cooling is on import rather than on nuclear retention.
DISCUSSION
JAZ is an unusual dsRNA-binding protein that binds its target RNAs with high affinity via C2H2 zinc fingers (35). Although GFP-JAZ is predominantly associated with nucleoli, we found that it is highly dynamic within the nucleus and also shuttles between the nuclear and cytoplasmic compartments. Several lines of evidence identify exportin-5 as the export factor for JAZ, but import is independent of classical transport factors. Exportin-5 recognizes a class of small, double-stranded RNAs that have a “minihelix” structure lacking a 5′ overhang. These RNAs can simultaneously bind to certain dsRNA binding proteins, such as ILF3. The function of ILF3 is not known, although it has been reported to be a transcriptional regulator and to stabilize certain mRNAs. In addition, it appears to facilitate RNA export via exportin-5, although the mechanism for this effect remains unclear. ILF3 also binds directly to JAZ, and surprisingly this interaction is independent of RNA. Thus, ILF3 and JAZ can cobind to exportin-5 in the presence of Ran-GTP and dsRNA and be exported in a “piggyback” fashion from the nucleus. We have been unable to detect any increase in JAZ export when ILF3 was coexpressed, however, suggesting that ILF3 is not a limiting cofactor for JAZ export.
The import mechanism for JAZ is puzzling. By certain criteria it behaves as though translocation through the nuclear pore complex is a passive diffusive process. For instance, no soluble factors are required, GTP is not required, and import is not inhibited by dominant mutants of Ran. Moreover, a GST-JAZ fusion (which is above the diffusion limit) is not imported when injected into the cytoplasm. In contrast to passive diffusion, however, JAZ import was clearly inhibited by low temperatures and by wheat germ agglutinin. These properties are reminiscent of those found previously for RanBP1, β-catenin, and importin-α (22, 26, 37). Conceivably, JAZ binds directly to nucleoporins, although we have been unable to detect such an interaction by pulldown assays (data not shown). It is also possible that an unidentified transporter is retained within the nuclear pore complex during permeabilization, to which JAZ binds, but presumably this transporter would not be coupled to Ran-GTP hydrolysis.
The biological function of JAZ, like that of most dsRNA binding proteins, remains unclear. However, the recent discovery that exportin-5 is required for the export of micro-RNA precursors, coupled with our present results showing that JAZ piggybacks out of the nucleus bound to exportin-5-RNA complexes, suggest that these dsRNA binding proteins might participate in some way in the processing of micro-RNAs or might protect them from inappropriate destruction by nuclear RNases.
Acknowledgments
This work was supported by grant GM50526 from the National Institutes of Health, DHHS.
REFERENCES
- 1.Arts, G. J., M. Fornerod, and I. W. Mattaj. 1998. Identification of a nuclear export receptor for tRNA. Curr. Biol. 8:305-314. [DOI] [PubMed] [Google Scholar]
- 2.Black, B. E., L. Levesque, J. M. Holaska, T. C. Wood, and B. M. Paschal. 1999. Identification of an NTF2-related factor that binds Ran-GTP and regulates nuclear protein export. Mol. Cell. Biol. 19:8616-8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bohnsack, M. T., K. Regener, B. Schwappach, R. Saffrich, E. Paraskeva, E. Hartmann, and D. Gorlich. 2002. Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J. 21:6205-6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bridge, A. J., S. Pebernard, A. Ducraux, A. L. Nicoulaz, and R. Iggo. 2003. Induction of an interferon response by RNAi vectors in mammalian cells. Nat. Genet. 34:263-264. [DOI] [PubMed] [Google Scholar]
- 5.Brownawell, A. M., G. J. Kops, I. G. Macara, and B. M. Burgering. 2001. Inhibition of nuclear import by protein kinase B (Akt) regulates the subcellular distribution and activity of the forkhead transcription factor AFX. Mol. Cell. Biol. 21:3534-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brownawell, A. M., and I. G. Macara. 2002. Exportin-5, a novel karyopherin, mediates nuclear export of double-stranded RNA binding proteins. J. Cell Biol. 156:53-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 8.Calado, A., N. Treichel, E. Muller, A. Otto, and U. Kutay. 2002. Exportin-5-mediated nuclear export of eukaryotic elongation factor 1A and tRNA. EMBO J. 21:6216-6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 10.Finlay, D., D. Newmeyer, T. Price, and D. Forbes. 1987. Inhibition of in vitro nuclear transport by a lectin that binds to nuclear pores. J. Cell Biol. 104:189-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607-660. [DOI] [PubMed] [Google Scholar]
- 12.Gwizdek, C., E. Bertrand, C. Dargemont, J. C. Lefebvre, J. M. Blanchard, R. H. Singer, and A. Doglio. 2001. Terminal minihelix, a novel RNA motif that directs polymerase III transcripts to the cell cytoplasm. Terminal minihelix and RNA export. J. Biol. Chem. 276:25910-25918. [DOI] [PubMed] [Google Scholar]
- 13.Gwizdek, C., B. Ossareh-Nazari, A. M. Brownawell, A. Doglio, E. Bertrand, I. G. Macara, and C. Dargemont. 2003. Exportin-5 mediates nuclear export of minihelix-containing RNAs. J. Biol. Chem. 278:5505-5508. [DOI] [PubMed] [Google Scholar]
- 14.Gwizdek, C., B. Ossareh-Nazari, A. M. Brownawell, S. Evers, I. G. Macara, and C. Dargemont. 2003. Minihelix-containing RNAs mediate exportin-5-dependent nuclear export of the double-stranded RNA binding protein ILF3. J. Biol. Chem. 279:884-891. [DOI] [PubMed] [Google Scholar]
- 15.Isken, O., C. Grassmann, R. Sarisky, M. Kann, S. Zhang, F. Grosse, P. N. Kao, and S. Behrens. 2003. Members of the NF90/NFAR protein group are involved in the life cycle of a positive-strand RNA virus. EMBO J. 22:5655-5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kutay, U., G. Lipowsky, E. Izaurralde, F. R. Bischoff, P. Schwarzmaier, E. Hartmann, and D. Gorlich. 1998. Identification of a tRNA-specific nuclear export receptor. Mol. Cell 1:359-369. [DOI] [PubMed] [Google Scholar]
- 17.Lee, Y., C. Ahn, J. Han, H. Choi, J. Kim, J. Yim, J. Lee, P. Provost, O. Radmark, S. Kim, and V. Kim. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425:415-419. [DOI] [PubMed] [Google Scholar]
- 18.Lounsbury, K. M., S. A. Richards, K. L. Carey, and I. G. Macara. 1996. Mutations within the Ran/Tc4 Gtpas—effects on regulatory factor interactions and subcellular localization. J. Biol. Chem. 271:32834-32841. [DOI] [PubMed] [Google Scholar]
- 19.Lund, E., S. Guttinger, A. Calado, J. Dahlberg, and U. Kutay. 2004. Nuclear export of micro-RNA precursors. Science 303:95-98. [DOI] [PubMed] [Google Scholar]
- 20.Macara, I. G. 2001. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65:570-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melchior, F., and L. Gerace. 1998. Two-way trafficking with ran. Trends Cell Biol. 8:175-179. [DOI] [PubMed] [Google Scholar]
- 22.Miyamoto, Y., M. Hieda, M. T. Harreman, M. Fukumoto, T. Saiwaki, A. E. Hodel, A. H. Corbett, and Y. Yoneda. 2002. Importin alpha can migrate into the nucleus in an importin beta- and Ran-independent manner. EMBO J. 21:5833-5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakielny, S., and G. Dreyfuss. 1999. Transport of proteins and RNAs in and out of the nucleus. Cell 99:677-690. [DOI] [PubMed] [Google Scholar]
- 24.Nemergut, M. E., and I. G. Macara. 2000. Nuclear import of the Ran exchange factor, RCC1, is mediated by at least two distinct mechanisms. J. Cell Biol. 149:835-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pemberton, L. F., G. Blobel, and J. S. Rosenblum. 1998. Transport routes through the nuclear pore complex. Curr. Opin. Cell Biol. 10:392-399. [DOI] [PubMed] [Google Scholar]
- 26.Plafker, K., and I. G. Macara. 2000. Facilitated nucleocytoplasmic shuttling of the Ran binding protein RanBP1. Mol. Cell. Biol. 20:3510-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plafker, S. M., and I. G. Macara. 2000. Importin-11, a nuclear import receptor for the ubiquitin-conjugating enzyme, UbcM2. EMBO J. 19:5502-5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plafker, S. M., and I. G. Macara. 2002. Ribosomal protein L12 uses a distinct nuclear import pathway mediated by importin 11. Mol. Cell. Biol. 22:1266-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reichman, T., L. Muniz, and M. B. Mathews. 2002. The RNA binding protein nuclear factor 90 functions as both a positive and negative regulator of gene expression in mammalian cells. Mol. Cell. Biol. 22:343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saunders, L. R., V. Jurecic, and G. N. Barber. 2001. The 90- and 110-kDa human NFAR proteins are translated from two differentially spliced mRNAs encoded on chromosome 19p13. Genomics 71:256-259. [DOI] [PubMed] [Google Scholar]
- 31.Shim, J., H. Lim, R. Y. J., and M. Karin. 2002. Nuclear export of NF90 is required for interleukin-2 mRNA stabilization. Mol. Cell 10:1331-1344. [DOI] [PubMed] [Google Scholar]
- 32.Stoffler, D., B. Fahrenkrog, and U. Aebi. 1999. The nuclear pore complex: from molecular architecture to functional dynamics. Curr. Opin. Cell Biol. 11:391-401. [DOI] [PubMed] [Google Scholar]
- 33.Welch, K., J. Franke, M. Kohler, and I. G. Macara. 1999. RanBP3 contains an unusual nuclear localization signal that is imported preferentially by importin-alpha3. Mol. Cell. Biol. 19:8400-8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wyatt, J., and G. Walker. 1989. Deoxynucleotide-containing oligoribonucleotide duplexes: stability and susceptibility to RNase V1 and RNase H. Nucleic Acids Res. 17:7833-7842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang, M., W. S. May, and T. Ito. 1999. JAZ requires the double-stranded RNA-binding zinc finger motifs for nuclear localization. J. Biol. Chem. 274:27399-27406. [DOI] [PubMed] [Google Scholar]
- 36.Yi, R., Y. Qin, I. Macara, and B. Cullen. 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17:3011-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yokoya, F., N. Imamoto, T. Tachibana, and Y. Yoneda. 1999. β-catenin can be transported into the nucleus in a Ran-unassisted manner. Mol. Biol. Cell 10:1119-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoneda, Y., N. Imamoto-Sonobe, M. Yamaizumi, and T. Uchida. 1987. Reversible inhibition of protein import into the nucleus by wheat germ agglutinin injected into cultured cells. Exp. Cell Res. 173:586-595. [DOI] [PubMed] [Google Scholar]