Abstract
The stability of mRNAs undergoing translation has long been a controversial question. Here, we systematically investigate links between mRNA turnover and translation during the endoplasmic reticulum (ER) stress response, a process during which protein synthesis is potently regulated. cDNA array-based approaches to assess the stability and translational status of each mRNA were devised. First, ER stress-triggered changes in mRNA stability were studied by comparing differences in steady-state mRNA levels with differences in gene transcription. Second, changes in translational status were monitored by studying ER stress-induced shifts in the relative distribution of each mRNA along sucrose gradients. Together, the array-derived data reveal complex links between mRNA stability and translation, with all regulatory groups represented: both stabilized and destabilized mRNAs were found among translationally induced as well as translationally suppressed mRNA collections. Remarkably, however, the subset of stabilized mRNAs was prominently enriched in translationally suppressed transcripts, suggesting that ER stress was capable of causing the stabilization of mRNAs associated with a global reduction in protein synthesis. The cDNA array-based approach described here can be applied to global analyses of mRNA turnover and translation and can serve to investigate subsets of mRNAs subject to joint posttranscriptional control.
Mammalian cells respond to stressful stimuli of both environmental and endogenous origins by effecting a series of adaptive responses that are collectively termed the stress response. Central to the mammalian stress response is the implementation of changes in gene expression patterns. In addition to the extensively studied transcriptional regulatory events, knowledge of stress-triggered posttranscriptional gene regulatory mechanisms has escalated in recent years. Such posttranscriptional control is exerted through processes such as splicing, mRNA turnover (stabilization and destabilization), mRNA transport (nuclear export, cytoplasmic compartmentalization, and polysomal localization), and mRNA translation. According to emerging views, gene regulation comprises an integrated set of events whereby carefully orchestrated transcriptional and posttranscriptional processes allow precise and rapid changes in the production of stress response genes. However, the relative contribution of transcriptional and posttranscriptional events is likely to differ greatly, depending on the stress in question. For instance, in response to DNA-damaging treatments, posttranscriptional regulatory mechanisms are particularly critical, given that global transcriptional events can be strongly inhibited (12, 40), while oxidants, developmental cues, immune response factors, and heat shock have been shown to potently induce the transcription of specific sets of genes (14, 34, 35).
Stresses affecting endoplasmic reticulum (ER) function can elicit a global inhibition of translation while selectively enhancing the transcription and translation of ER stress response gene products (reviewed in reference 36). ER stress is triggered by a number of pathophysiological conditions such as hypoxia and hypoglycemia and by agents that perturb ER function and cause an accumulation of proteins requiring posttranslational modification and folding (22, 37). The rapid repression of protein biosynthesis is mediated by phosphorylation of the α subunit of the eukaryotic initiation factor 2 (eIF2α) complex by the ER resident kinase PERK, which is kept in an inactive state through binding to the ER chaperone GRP78 (BiP) in unstimulated cells. Under conditions of ER stress, PERK dissociates from GRP78, becomes activated, and phosphorylates eIF2α, a modification that in turn inhibits eIF2B activity and thereby prevents the formation of the GTP-bound (active) form of the eIF2 complex. Since active eIF2 is necessary for recruiting the charged initiator methionyl-tRNA to the small ribosomal subunit, eIF2α phosphorylation can efficiently prevent the assembly of the 80S ribosome and thus attenuate translational initiation and protein synthesis (9).
In addition to core translation factors that carry out the various steps of protein biosynthesis, a growing number of ancillary proteins that modulate translation have been described. For example, the iron response proteins associate with the iron-responsive element in the 5′ untranslated region (UTR) of specific mRNAs and repress translation in an iron concentration-dependent manner, while the poly(A)-binding protein associates with the poly(A) tract on the 3′ end of the mRNA and can enhance or repress translation (reviewed in references 43 and 44). Other accessory proteins (such as TIA-1, TIAR, and members of the ELAV protein family) can bind to specific AU-rich elements (AREs) present in the 3′ UTRs of many posttranscriptionally regulated mRNAs and modulate their translation (3, 13, 18, 20, 21, 29, 32, 33, 47). Importantly, AREs have also been extensively implicated in modulating the stability of ARE-bearing mRNAs such as those encoding cytokines, growth factors, protooncogenes, cell cycle proteins, and many other transcripts. Together, these observations lend support to a long-standing hypothesis that mRNA stability and translation might be functionally linked.
However, the interplay between translation and stability is likely to be fairly complex, based on a few reports available to-date. Some translationally engaged mRNAs have been reported to exhibit both enhanced and reduced stability, while similar disparities in mRNA turnover regarding mRNAs that are not bound to polysomes have been described (3, 5, 6, 21, 46); it is important, however, that these reports describe primarily single-gene studies. In the present investigation, we sought to systematically investigate the relationship between mRNA turnover and translation on a global level. We devised a cDNA array-based strategy to comprehensively investigate links between the stability of a given mRNA and its translational status. We assessed mRNA stability by comparing the information obtained from cDNA arrays measuring gene transcription (nuclear run-on [NRO] arrays, as previously described [16]) with that obtained from cDNA arrays in which steady-state mRNA levels were studied. In parallel, translational status was monitored by cDNA array-based assessment of the relative distribution of mRNAs on polysomes that had been fractionated through sucrose gradients. From these comparisons, we were able to identify the contribution of mRNA turnover to the steady-state levels of sets of mRNAs while simultaneously gaining information on their translational condition. Using ER stress as the study system, we present evidence that altered mRNA turnover prominently influences steady-state mRNA levels, and stabilized mRNAs are preferentially subject to translational repression.
MATERIALS AND METHODS
Cell culture and treatment of cells.
Human cervical carcinoma HeLa cells were cultured in Dulbecco's modified essential medium containing 10% fetal bovine serum (HyClone, South Logan, Utah). ER stress was elicited by treatment with either 2.4 μM tunicamycin (Tn) or 2 μM thapsigargin (Tg) for the times indicated.
Polysome analysis.
HeLa cells (5 × 106 cells per sample) that were plated 16 h before treatment were used at ∼80% confluence. After treatment, cells were incubated for 3 min in 0.1 mg of cycloheximide/ml and then lifted into 1 ml of PEB lysis buffer (0.3 M NaCl, 15 mM MgCl2, 15 mM Tris-HCl [pH 7.6], 1% Triton X-100, 1 mg of heparin/ml, 0.1 mg of cycloheximide/ml) and lysed on ice for 10 min. Nuclei were pelleted at 10,000 × g for 10 min, and the resulting supernatant was layered onto a 10 to 50% linear sucrose gradient; gradients were centrifuged at 35,000 × g for 3 h at 4°C with a swinging bucket rotor. The eluted fractions were collected at a rate of 1 ml per min by using a syringe pump and a needle piercing system (Brandel, Gaithersburg, Md.) and were monitored at 254 nm with a UV-6 detector (ISCO, Lincoln, Nebr.). RNA in each fraction was extracted with 8 M guanidine-HCl. Equal volumes from each fraction were used for cDNA array analysis, Northern blotting, and Western blotting.
Northern and Western blotting.
Northern blot analysis was performed by using RNA isolated either from whole cells (using standard methodologies) or from each fraction in the sucrose gradients (extracted with 8 M guanidine-HCl) and used for Northern blotting. Equal volumes from each fraction were used for Northern blot analysis. To detect mRNAs encoding GRP78, ATF4 or β-actin, cDNA probes were labeled by using random primers in the presence of [α-32P]dATP and Klenow enzyme. Oligonucleotides CCCGTGAACCCTTCTCTTTCCGTGGTGTGTCCTTTTTAGGT, GGGAGAGCGCAGAGAGAAGTCGATGTGCTTCTGGGAATCCAG, TCCAGGTAGATGTGGTGGTCACTCAGAGCCTTGTAGACAG, GGAGCTCTAGGGCCTCATAGATACCAGTTGAAGCACCACT, TCAGGTCCTAGTCCCATCTCTTAGGGTGCTGATCTTCCA, GACAAGCACAGAGTCACTATTGCGGTTAGAAGTTGGCAGCATGGG, CCTCATATCTCTTCTGGCTGTAGGGTGGCTCAGTGGAATC, GCCTTTGTTCTTATTGGCGGCTGTGTAAGAGTATCCAGGGGCC, CACCCATCCCTTCTGCATATTAGCAACTTGTCACTCCTGAGC, GTCTTGTCAGTCTTGGTGACAGTGACATTGAAGGTGGGGGCC, and ACGGTATCTGATCGTCTTCGAACC, which were end labeled with [α-32P]dATP and terminal transferase, were used for the detection of eEF1γ, RPS9, aldolase A, enolase 1, BAK1, cofilin 1, eEF1α1, cytochrome c, H3 histone 3B, profilin 1, and 18S, respectively. For mRNA half-life assessments, three independent experiments were performed. Actinomycin D (2 μg/ml) was added and total RNA was prepared at the times indicated; mRNA half-lives were calculated after measuring mRNA signals on Northern blots, normalizing them to 18S rRNA signals, plotting them on logarithmic scales, and calculating the time period required for a given transcript to undergo a reduction to one-half of its initial abundance (at time zero, before adding actinomycin D). The Northern data showing the abundance of mRNAs in each polysome fraction are representative of three independently performed experiments, each yielding highly reproducible patterns of mRNA distribution. RNA patterns and relative abundance in each fraction and each treatment group were routinely monitored by staining with ethidium bromide.
For Western blot analysis, whole-cell (10 μg), cytoplasmic (20 μg), and nuclear (10 μg) lysates were prepared as described previously (42) 9 h after treatment with either Tn or Tg. Protein samples were size fractionated by electrophoresis through Tris-HCl gels (Bio-Rad, Life Science, Hercules, Calif.) and transferred onto polyvinylidene difluoride membranes. Monoclonal antibodies recognizing β-tubulin (control cytoplasmic and cytoskeletal protein) as well as a polyclonal antibodies recognizing hnRNP C1/C2 (control nuclear protein), GRP78 (control protein of the ER lumen), GADD153, aldolase A, enolase 1, ATF4, BAK1, cofilin 1, histone H3, and profilin 1 were from Santa Cruz Biotech (Santa Cruz, Calif.). A monoclonal antibody against β-actin was obtained from Abcam (Cambridge, Mass.), a polyclonal antibody recognizing phosphorylated eIF2α was obtained from BioSource International, Inc. (Camarillo, Calif.), and a polyclonal antibody recognizing the IP3 receptor 1 (control protein present on ER membranes) was obtained from Chemicon International, Inc. (Temecula, Calif.). Following secondary antibody incubations, signals were detected by enhanced chemiluminescence.
Analysis of newly translated protein.
New synthesis of β-actin was measured by incubating 106 cells with 1 mCi of l-[35S]methionine and l-[35S]cysteine (Easy TagEXPRESS; NEN/Perkin Elmer, Boston, Mass.) per 60-mm plate for 20 min, whereupon cells were lysed with TSD lysis buffer (50 mM Tris-HCl [pH 7.5], 1% sodium dodecyl sulfate [SDS], and 5 mM dithiothreitol). Immunoprecipitations were carried out for 1 h at 4°C by using either a monoclonal antibody recognizing β-actin or immunoglobulin G1. Following extensive washes in TNN buffer (50 mM Tris-HCl [pH 7.5], 250 mM NaCl, 5 mM EDTA, 0.5% NP-40), immunoprecipitated material was resolved by electrophoresis through SDS-containing 12% polyacrylamide gels, transferred onto polyvinylidene difluoride filters, and visualized with a PhosphorImager (Molecular Dynamics).
cDNA array analysis.
Whole-cell RNA, RNA in unbound fractions (fractions 1 and 2, designated as U), and RNA from each fraction associated with either ribosome subunits or an increasing number of ribosomes (fractions 3 through 11), were reverse transcribed in the presence of [α-33P]dCTP. Newly transcribed RNA was prepared from nuclei of HeLa cells in a NRO reaction in the presence of [α-33P]UTP, as described previously (16). Radiolabeled cDNA and RNA were then used for the hybridization of cDNA array membranes (http://www.grc.nia.nih.gov/branches/rrb/dna/array.htm; Mammalian Genome Collection arrays, 9,600 genes [6,385 unique]). All of the data were analyzed with Array Pro software (Media Cybernetics, Inc., Carlsbad, Calif.) and then normalized by Z score transformation (11). Based on Z scores, Z ratios were calculated to ascertain changes in gene expression (for the Z ratio, average Z score in control cells is subtracted from the average Z score in ER stress-treated cells, which is divided by the standard deviation of all of the differences in signal intensity in the array). Z scores of steady-state RNA signals, newly transcribed RNA signals, and RNA in each sucrose gradient fraction from Tn- or Tg-treated cells were clustered and visualized with Cluster and TreeView software, respectively (Eisen's Laboratory, University of California at Berkeley). Positive Z ratios (>0) in both fractions 9 and 10 for both treatments was the criterion used to deem a gene translationally activated in ER stress-treated cells; negative Z ratios (<0) in both fractions 9 and 10 in both the Tn and Tg groups was the criterion for translational repression. Z ratio values of >0.5 and <−0.5 were used to determine significant changes in either steady-state abundance (Total RNA arrays) or transcription (NRO arrays). Complete listings of all of the genes in each regulatory group are available at the National Institute on Aging website (http://www.grc.nia.nih.gov/branches/rrb/dna/index/dnapubs.htm#2).
RESULTS
Response of HeLa cells to ER stress agents.
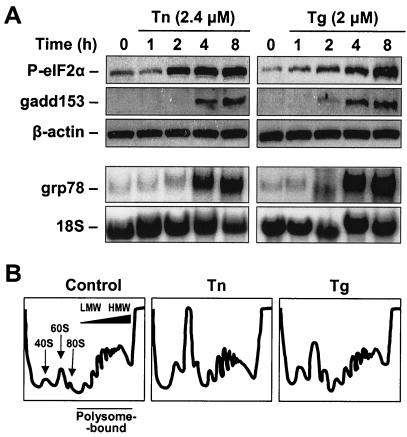
ER stress was triggered in human cervical carcinoma HeLa cells through treatment with either Tn (2.4 μM), an inhibitor of protein glycosylation that prevents maturation of secretory proteins, or Tg (2 μM), which depletes ER calcium and thereby impairs protein folding. Treatment with Tn or Tg caused increased phosphorylation of eIF2α, a hallmark of ER stress, and elevated the expression levels of the ER stress response genes gadd153 and grp78 (Fig. 1A). Also characteristic of ER stress were the overall reduction in high-molecular-weight (HMW) polysomes and the increase in ribosomal subunits (40S and 60S) and free ribosomes (80S) seen on sucrose gradient profiles following treatment with either Tn or Tg (Fig. 1B).
FIG. 1.
Treatment of HeLa cells with Tn and Tg triggers the ER stress response. (A) Markers of ER stress were monitored at the times shown after the addition of either 2.4 μM Tn or 2 μM Tg to asynchronous, actively growing HeLa cells. The levels of phosphorylated eIF2 (P-eIF2α), GADD153, and loading control β-actin were monitored by Western blotting (top), while the abundance of GRP78 mRNA and loading control 18S rRNA was assessed by Northern blotting (bottom). (B) Cytoplasmic extracts from HeLa cells that were either left untreated (control) or treated with Tn or Tg were centrifuged through continuous sucrose gradients to obtain fractions of increasing molecular weight. Absorbance at 254 nm served to monitor the sample preparation and to identify the fractions containing ribosomal subunits (40S and 60S), ribosomes (80S), and polysomes of increasing weight (polysome-bound). LMW, low-molecular-weight polysomes. HMW, high-molecular-weight polysomes.
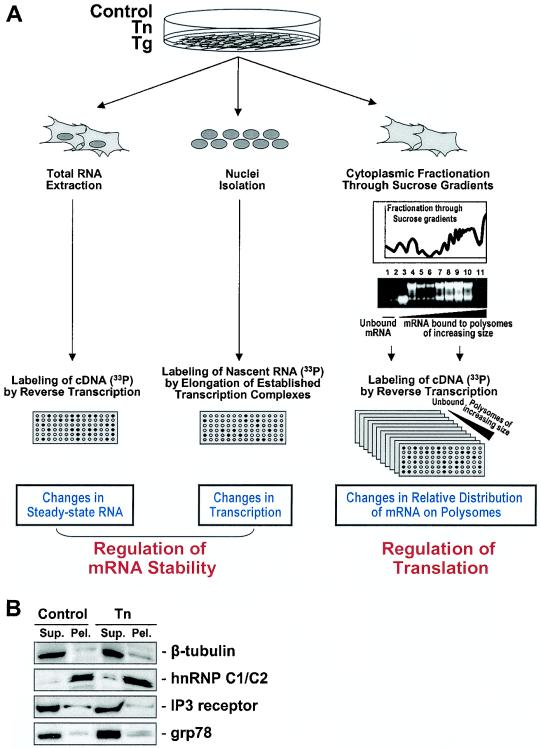
HeLa cells subjected to ER stress were used to systematically investigate links between mRNA turnover and translation. The strategy for the analysis, largely based on the use of cDNA arrays (see Materials and Methods), is depicted in Fig. 2A. First, ER stress-regulated mRNA turnover was studied on a global level by comparing changes in steady-state mRNA levels with changes in gene transcription rates by using a previously described NRO assay adapted to microarray analysis (16). In this comparison, genes whose steady-state mRNA levels were increased but their transcription was not correspondingly elevated were deemed to be subject to mRNA stabilization; conversely, genes whose decreased steady-state mRNA abundance was not accompanied by reduced gene transcription were considered to be subject to mRNA destabilization. Second, the translational status was monitored by hybridization of cDNA arrays with RNA isolated from each of the 11 fractions that were obtained after centrifugation through sucrose gradients. Here, mRNAs shifting their association towards heavier polysomal fractions indicate a greater engagement in translation, while mRNAs shifting their association to lighter polysomal fractions, monosomes or unbound pools, revealed a decrease in their translational status. Preliminary experiments indicated that analysis of fewer than 11 fractions failed to detect subtle changes in the relative distribution of certain mRNAs, while assessment of more than 11 fractions did not provide additional information (data not shown). Fractions 1 and 2 were combined, given that they jointly represent the unbound (U) collection of mRNAs. Together, these cDNA array-based comparisons allowed us to simultaneously identify transcripts whose stability was altered in response to ER stress agents and study changes in their translational profiles by monitoring the mRNA's relative distribution among polysomes of differing molecular weights.
FIG. 2.
Strategy to comparatively study changes in mRNA stability and translation by using cDNA arrays. (A) Cells were either left untreated or treated with Tn or Tg, whereupon three sets of radiolabeled probes were prepared. Total RNA was isolated from whole cells 6 h after treatment and then used to prepare radiolabeled cDNA through reverse transcription in the presence of [α-33P]dCTP (left). Nuclei were isolated 3 h after treatment and used to obtain newly transcribed RNA that was radiolabeled in an NRO reaction in the presence of [α-33P]UTP (center). Cytoplasmic fractions were prepared 6 h after treatment, and RNA isolated from each of the 11 resulting fractions was processed as explained above for total RNA (right). Fractions 1 and 2 (typically pooled and designated U) were completely devoid of ribosomes and ribosome components, fractions 3 and 4 routinely included the “free” small and large ribosomal subunits, respectively, and fractions 5 to 11 contained polysomes of increasing weight. Probes were used for hybridization of cDNA arrays (described in Materials and Methods). Comparison of changes in steady-state mRNA levels after ER stress with changes in transcription was used to ascertain to what extent the regulation of mRNA stability contributed to the implementation of altered steady-state mRNA profiles; monitoring of the relative abundance of specific genes along the polysome fractions served to study the regulation of translation. Cell fractions and cDNA array analyses were performed three times, each as a completely independent experiment. (B) Western blot analysis of the cytoplasmic (supernatant [Sup.]) and nuclear (pellet [Pel.]) fractions to detect β-tubulin (a cytoskeletal protein), hnRNP C1 and C2 (nuclear proteins), the IP3 receptor (a component of the ER membrane), and GRP78 (a protein present in the ER lumen).
The quality of the cytoplasmic (supernatant) and nuclear (pellet) fractionation procedure was assessed by monitoring the abundance of proteins specifically present in the different cellular compartments (Fig. 2B). The presence of β-tubulin in the cytoplasmic fraction revealed that cytoskeletal components were not spun down during the fractionation process and that cytoplasmic components did not contaminate nuclear preparations. By contrast, nuclear proteins hnRNP C1 and C2 were found essentially in the pellet, revealing that the nuclear fraction had not “leaked” significantly into the cytoplasmic fraction. IP3 receptor, a component of the ER membrane, was found almost in its entirety in the supernatant fraction, indicating that the ER membrane had not been pelleted down, and GRP78, a protein present in the ER lumen, was also found almost exclusively in the supernatant fraction, further indicating that ER components had not been spun down during the fractionation procedure (Fig. 2B).
ER stress-triggered changes in gene expression.
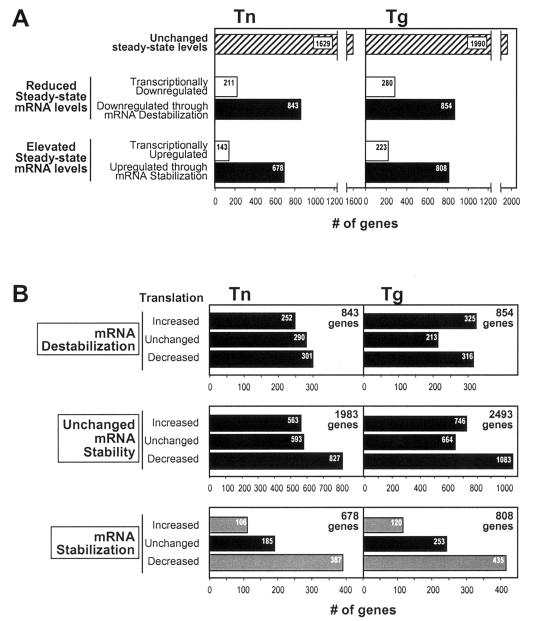
The analysis of cDNA arrays was performed according to the aforementioned methodology and employing the conditions described above in Materials and Methods. Various significance criteria were tested, and Z ratios (11) that were either >0.5 or <−0.5 (comparing Tn or Tg groups with the control) were finally set as significant. These limits, applied to comparisons of steady-state mRNA levels and to comparisons of gene transcription, allowed us to include adequate numbers of genes in the study and thus be able to investigate associations between mRNA stability and translation. Of the 6,385 genes on the array, the steady-state levels of 1,054 and 1,134 mRNAs were lower after 6 h of treatment with Tn and Tg, respectively (Fig. 3A). Strikingly, transcriptional decreases were observed for only 211 genes (for Tn) and 280 genes (for Tg), indicating that for a high proportion of the reduced mRNAs (843 and 854, respectively), transcription was not significantly suppressed, and the mRNAs were therefore subject to ER stress-triggered destabilization (i.e., ∼39 and ∼44% of all steady-state mRNA changes were significantly influenced by enhanced mRNA degradation in the Tn and Tg treatment groups, respectively). Increased steady-state levels were seen for 821 and 1,031 mRNAs in Tn- and Tg-treated cells, respectively, but a transcriptional enhancement was measurable in only 143 and 223 genes, respectively, revealing a comparably large subset of increased mRNAs (678 and 808 genes) that were subject to ER stress-triggered stabilization (i.e., ∼36% of all steady-state mRNA changes after Tn or Tg treatment were attributed to enhanced mRNA stability). Limitations of the methodology employed here in order to assess transcriptional and mRNA stability are discussed below (see Discussion).
FIG. 3.
Global changes in mRNA turnover and translational status of ER stress-regulated mRNAs. (A) Classification of mRNA populations according to their steady-state levels following ER stress. Groups undergoing changes in steady-state levels were further divided according to their transcriptional profiles: when decreases or elevations in mRNA abundance were accompanied by corresponding changes in transcription, genes were considered to be transcriptionally downregulated or transcriptionally upregulated, respectively; when no significant changes in gene transcription were found, genes were considered to be downregulated through mRNA destabilization or upregulated through mRNA stabilization, respectively. (B) Classification of mRNA populations in each turnover category according to their translational profiles. The number of genes in each turnover subcategory (increased, unchanged, and decreased) are indicated inside the corresponding bars. The most notable differences in translational status within each turnover category were seen in the mRNA stabilization group (gray bars).
mRNAs in each turnover category (i.e., “mRNA destabilization,” “unchanged mRNA stability,” and “mRNA stabilization”) were then subdivided according to their translational status, following the criteria described above in Materials and Methods. As shown in Fig. 3B, most genes were found to be in the “decreased translation” subcategories, in agreement with the well-documented translational repression by ER stress (36); a total of 1,515 (301 + 827 + 387) and 1,834 (316 + 1,083 + 435) mRNAs (the sum of genes in the mRNA destabilization, unchanged mRNA stability, and mRNA stabilization groups, respectively) were found to be translationally decreased in the Tn and Tg treatment groups, respectively. By contrast, 921 (252 + 563 + 106) and 1,191 (325 + 746 + 120) mRNAs were found in the “increased translation” subcategories and 1,068 (290 + 593 + 185) and 1,130 (213 + 664 + 253) mRNAs were found in the “unchanged translation” subcategories in the Tn and Tg groups, respectively.
The most striking difference among these categories was seen in the mRNA stabilization group (Fig. 3B); here, a distinctly low number of mRNAs were found in the increased translation subcategory (106 for Tn and 120 for Tg) compared with the decreased translation subcategory (387 for Tn and 435 for Tg) (Fig. 3). This >3.5-fold enrichment in translationally decreased mRNAs in the mRNA stabilization group was substantially higher than that seen among either of the other stability groups (unchanged mRNA stability and mRNA destabilization, where differences were <1.5- and <1.2-fold, respectively). These findings support the existence of links between inhibition of mRNA translation and mRNA stabilization.
Array data in each category of posttranscriptional gene regulation.
Examples of gene clusters in each stability and translation category are depicted in Fig. 4, 5, and 6 with Cluster and TreeView software packages. Comparisons between treated and untreated cells are shown as follows: the NRO column represents transcriptional differences, and the “total” column depicts steady-state mRNA differences; the columns U through 11 reflect differences in relative mRNA abundance in gradient fractions of increasing molecular weight, starting from unbound mRNA (cytosolic) and gradually increasing to monosome-bound mRNA and mRNA bound to low-molecular-weight and HMW polysome fractions. For each stability group, representative examples of translationally induced and translationally repressed mRNAs are provided; examples of unchanged translation are not shown. Complete listings of all of the genes in each regulatory group are available at the National Institute on Aging website (http://www.grc.nia.nih.gov/branches/rrb/dna/index/dnapubs.htm#2). Various criteria were tested for consideration of genes in the translationally regulated categories (data not shown); among them, significantly increased abundance in fractions 9 and 10 was found to provide the most reproducible differences and was thus adopted for this classification.
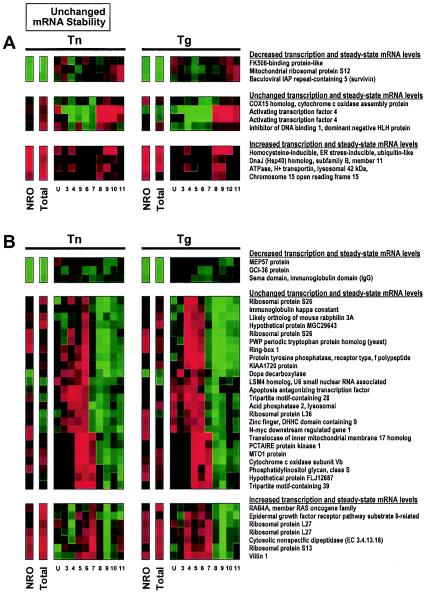
FIG. 4.
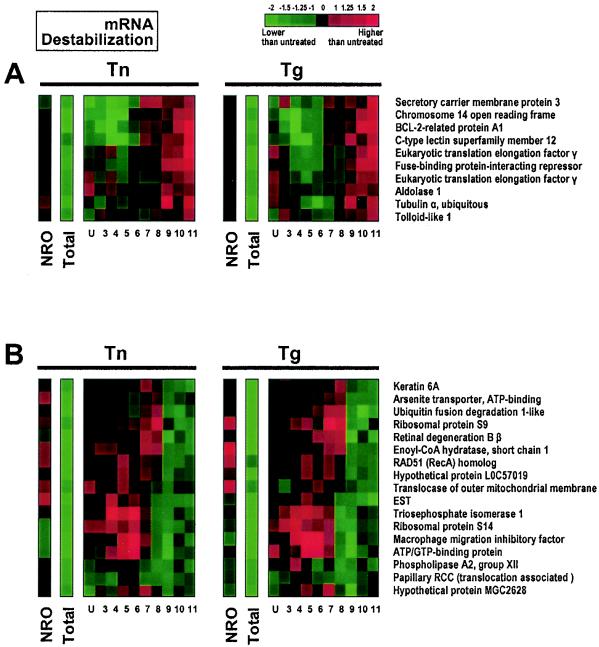
Examples of translational regulation of destabilized mRNAs. Genes encoding mRNAs whose stability decreased with ER stress are shown. In each case, steady-state mRNA levels (total) are markedly lower in both the Tn and Tg treatment groups than in the control groups (green signals), while transcription rates for the same genes, as measured by NRO analysis, did not change significantly (dark signals). (A) Destabilized mRNAs subject to translational increase. (B) Destabilized mRNAs subject to translational decrease. Lanes U through 11 represent RNA from sucrose fractions of increasing molecular weight, spanning unbound (U) RNA, RNA bound to ribosome components, and RNA bound to single ribosomes and to polysomes of low and increasingly higher molecular weight.
FIG. 5.
Examples of translational regulation of mRNAs exhibiting unchanged stability. Shown are genes encoding mRNAs whose stability was unaltered by ER stress which fall into three categories: both transcription and steady-state mRNA levels are reduced (top), both transcription and steady-state mRNA levels remain unchanged (middle), and both transcription and steady-state mRNA levels increase (bottom). (A) mRNAs undergoing ER stress-induced translational increase. (B) mRNAs subject to ER stress-induced translational decrease. Data labels are the same as those described in the legend of Fig. 4.
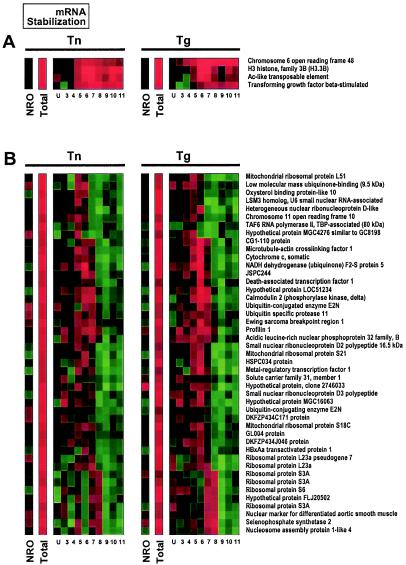
FIG. 6.
Examples of translational regulation of stabilized mRNAs. Shown are genes encoding mRNAs whose stability was increased by ER stress. In each case, steady-state mRNA levels (total) are markedly higher in both the Tn and Tg treatment groups than in the control groups (red signals), while transcription rates for the same genes, as measured by NRO analysis, did not change significantly (dark signals). (A) Stabilized mRNAs undergoing ER stress-induced translational increase. (B) Stabilized mRNAs undergoing ER stress-induced translational decrease. Data labels are the same as those described in the legend of Fig. 4.
Figure 4 illustrates the collection of destabilized mRNAs that are subject to either increased translation (Fig. 4A) or decreased translation (Fig. 4B) in response to Tn or Tg. According to the criteria for inclusion in this group, NRO signals typically show an absence of change (black), while total signals are markedly lower in the Tn or Tg group than in the control group (bright green). In Fig. 4A, the increased mRNA signals in the HMW polysome-bound fractions (red, specifically seen in fractions 9 and 10) indicate that the mRNAs in question, though less abundant overall after Tn or Tg treatment (note the “total” columns), associate more actively with HMW polysomes for translation, while mRNA abundance typically decreased (green) in lower-molecular-weight fractions (up to approximately fraction 8). In Fig. 4B, by contrast, mRNA abundance increases in the lower-molecular-weight fractions, while it is strongly reduced in higher-molecular-weight fractions (specifically fractions 9 and 10), in keeping with an overall reduction in translation rates.
As shown in Fig. 5, mRNAs that are not subject to regulated turnover fall into three categories: transcription and steady-state mRNA levels increase (both NRO and total signals are bright red), neither transcription nor steady-state mRNA levels change (NRO and total signals are black), and both transcription and steady-state mRNA levels decrease (NRO and total signals are bright green). Examples of translationally elevated (Fig. 5A) and translationally reduced (Fig. 5B) mRNAs are shown. The interpretation of these data is similar to that of Fig. 4.
Finally, stabilized mRNAs (black NRO signals and red total signals) are also divided into translationally induced and translationally repressed subgroups (Fig. 6A and B, respectively). Illustrated by the representative genes shown (Fig. 6B), the latter group was of particular interest to this investigation, given that it was significantly larger than stabilized mRNAs either undergoing translational induction (Fig. 6A) or showing no change in translation (data not shown).
Regarding Fig. 4, 5, and 6, it is important that in the case of genes that were spotted several times on the arrays, replicate samples typically clustered in close proximity, providing an initial indication of the internal reproducibility of the data. In instances in which signals in fractions U through 11 do not appear to “add up” to the differences in total signals, perhaps changes in the nuclear mRNA pool (not represented in the polysome fractions) may help to explain the discrepancy; moreover, the signal intensity in the fractions is a reflection of the difference in abundance between the control and ER stress-treated groups (even if the absolute amount of mRNA in particular fractions is very low). It is also worth remarking that changes in NRO, total, and fraction-associated RNA signals were closely mirrored by the two stresses despite their different modes of action.
Validation of mRNA turnover and translational status of genes identified by array analysis.
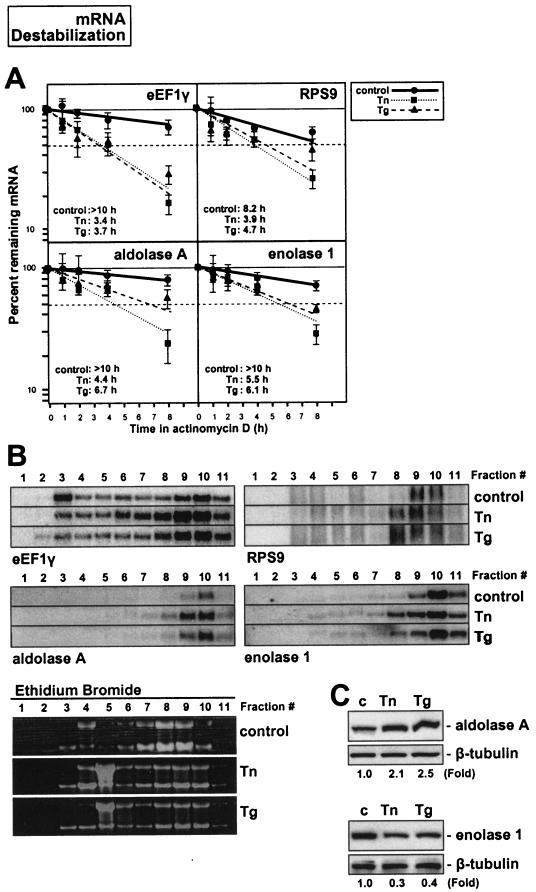
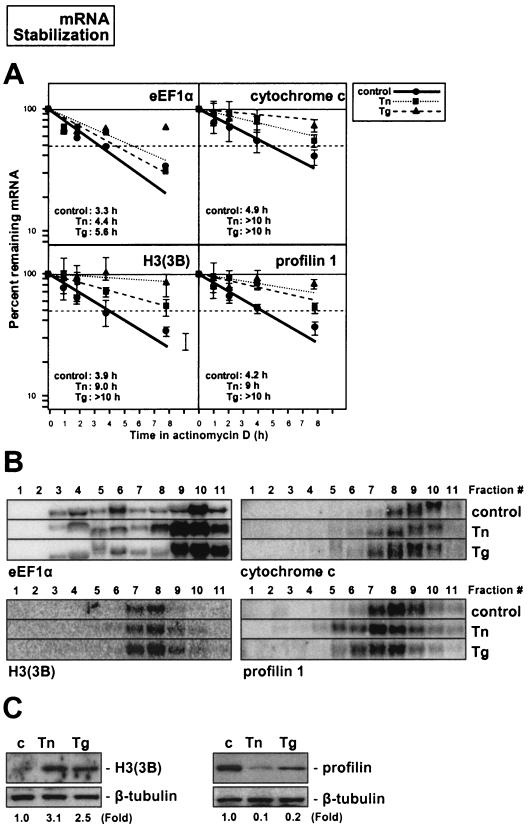
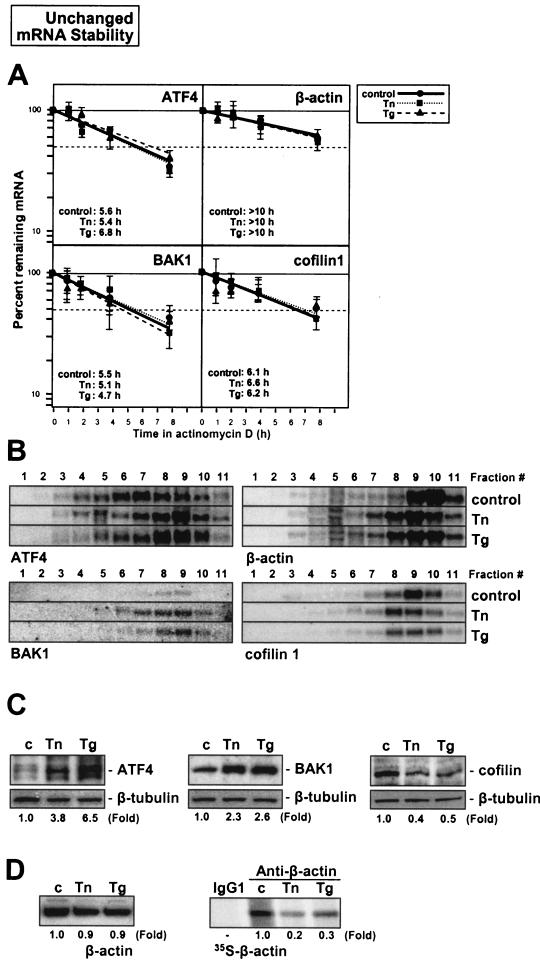
Confirmation of the validity of the array results was obtained through three experimental approaches, as illustrated in Fig. 7, 8, and 9. First, actinomycin D-based estimations of mRNA half-life, carried out in triplicate, corroborated the suitability of comparing array-derived changes in steady-state mRNA signals with NRO data to infer changes in mRNA half-life after ER stress (Fig. 7 to 9, panels A). As shown, mRNAs encoding eEF1γ, RPS9, aldolase A, and enolase 1, which were predicted to be rendered less stable by ER stress agents, indeed exhibited reduced half-lives in both the Tn and Tg treatment groups (e.g., for eEF1γ mRNA, the half-life was reduced from >10 h to 3.4 and 3.7 h after Tn and Tg treatments, respectively; comparable reductions in stability were seen for the other mRNAs tested). Similarly, mRNAs encoding ATF4, β-actin, BAK1, and cofilin 1, predicted to show unchanged stability by ER stress treatments, indeed showed little variability in half-life following Tn or Tg treatment (Fig. 8A). By contrast, mRNAs encoding eEF1α, cytochrome c, histone H3, and profilin 1 were found to have increased half-lives, as predicted (Fig. 9A).
FIG. 7.
Validation of translational regulation of ER stress-stabilized mRNAs. Assessment of the half-life, translational status, and resulting protein synthesis for representative mRNAs undergoing ER stress-triggered destabilization. (A) Six hours after treatment with either Tn or Tg, mRNA half-lives were measured for the genes shown; RNA was prepared at the times indicated, mRNA signals were quantified and normalized to 18S rRNA signals, the results were plotted on logarithmic scales, and the time periods required for each mRNA to undergo a reduction to one-half of the initial abundance (at time zero, before adding actinomycin D) were calculated. Horizontal dashed lines, 50% of untreated cells. Data represent the mean ± standard deviations from three experiments. An ethidium bromide-stained gel showing representative RNA abundance in each of the fractions tested is shown (bottom). (B) Relative distribution of mRNAs along polysome gradients (left, translationally upregulated mRNAs; right, translationally repressed mRNAs); data are representative of three experiments performed independently. (C) Western blot analysis to measure the abundance of representative proteins in whole-cell lysates (10 μg) after 9 h of treatment with either Tn or Tg. β-Tubulin levels were assessed to monitor differences in sample loading and transfer.
FIG. 9.
Validation of translational regulation of ER stress-stabilized mRNAs. Assessments of the half-life (A), translational status (B), and resulting protein products (C) for representative mRNAs displaying increased stability after ER stress were performed as explained in the legend of Fig. 7.
FIG. 8.
Validation of translational regulation of mRNA displaying unchanged mRNA stability. Assessments of the half-life (A), translational status (B), and resulting protein products (C) for representative mRNAs displaying unchanged stability after ER stress were performed as explained in the legend of Fig. 7. (D) Western blot analysis of β-actin expression 9 h after treatment with ER stress agents (left); after 6 h of treatment with either Tn or Tg, cells were incubated for 20 min in the presence of l-[35S]methionine and l-[35S]cysteine, whereupon nascent β-actin was visualized by immunoprecipitation as described in Materials and Methods (right). Samples were resolved by electrophoresis in SDS-containing 12% polyacrylamide gels. Radiolabeled β-actin signal is shown.
Second, Northern blotting confirmed the adequacy of employing cDNA arrays for monitoring shifts in transcript abundance along the sucrose gradients. For each stability group, transcripts that were subject to increased translation are shown on the left, and those subject to decreased translation are shown on the right (Fig. 7 to 9, panels B). In general, fractions 9 and 10 showed the predicted differences in transcript abundance after treatment with either Tn or Tg (lower in translationally repressed and higher in translationally induced subgroups), although changes in transcript abundance were also seen in other fractions for some of the mRNAs tested (Fig. 7 to 9, panels B). The Northern data showing mRNA abundance in each polysome fraction (Fig. 7 to 9, panels B) are representative of three independently performed experiments, each yielding highly reproducible patterns of transcript distribution. Figure 7B shows representative images of RNA patterns and relative abundance in each fraction and each treatment group after staining with ethidium bromide.
Third, Western blotting served to ascertain the appropriateness of studying shifts in mRNA abundance among polysome fractions in order to predict alterations in translation of the corresponding polypeptides (Fig. 7 to 9, panels C). While this is not an ideal method for monitoring protein synthesis, as other factors like preexisting protein levels and ER stress-regulated proteolysis cannot be excluded, it did reveal the expected trends in protein abundance: by 9 h after adding Tn or Tg treatment, proteins that were predicted to be translationally upregulated indeed showed higher expression levels (aldolase A [Fig. 7C], ATF4 and BAK [Fig. 8C], and H3 histone 3B [Fig. 9C]), while those predicted to be translationally suppressed were found in lower abundance (enolase 1 [Fig. 7C], cofilin 1 [Fig. 8C], and profilin 1 [Fig. 9C]). For this analysis, the preferred method involved the monitoring of nascent protein synthesis through a brief (20-min) incubation in l-[35S]methionine and l-[35S]cysteine followed by immunoprecipitation with specific antibodies, as was previously reported (17, 32); this approach allows the direct measurement of newly synthesized protein without the confounding effects of the aforementioned regulatory processes. Unfortunately, the signal incorporation is very inefficient due to the short labeling period, so the usefulness of this method depends heavily on the availability of very good antibodies. For the panel of proteins tested, no such antibodies were available except for β-actin (Fig. 8D). Indeed, β-actin showed virtually no change in steady-state abundance (Fig. 8D, left), but the incorporation of radiolabeled amino acids was markedly reduced, demonstrating that its translation was suppressed by ER stress, as anticipated from the polysome profiles (Fig. 8B). In summary, the array-derived information accurately reflected changes in ER stress-triggered mRNA turnover and was a good indicator of changes in protein abundance dictated by ER stress.
Together, the methodology presented here provides a systematic approach for the analysis of global gene regulatory events associated with transcription, mRNA stability, and translation. Our findings underscore the extensive contribution of mRNA turnover towards the implementation of steady-state mRNA profiles following ER stress and further reveal that stabilized mRNAs are almost four times more likely to be translationally repressed.
DISCUSSION
By focusing on ER stress as a study system and the outlined cDNA array-based strategy, we have investigated links between mRNA turnover and translation. The discovery that global changes in mRNA stability strongly influenced steady-state mRNA profiles was not unexpected (Fig. 3A), given that regulated mRNA turnover was responsible for effecting changes in steady-state mRNA abundance in response to other stress agents such as genotoxins and heat shock (16), and also considering studies in which specific mRNAs were reported to be stabilized during ER stress (8, 45). Moreover, abrupt fluctuations in cytosolic calcium concentrations, such as those caused by treatment with Tg, have been shown to stabilize collections of labile mRNAs (27). The finding that ER stress caused the translational repression of large groups of transcripts (Fig. 3B) is also in keeping with an extensive body of literature implicating eIF2α phosphorylation in the transient attenuation of protein synthesis (19, 39).
A recent cDNA array-based study of Saccharomyces cerevisiae revealed no apparent correlations between polysomal distributions of mRNAs and mRNA stabilities (4). In mammalian cells, however, with the exception of nonsense-mediated decay, a mechanism whereby cells identify and degrade aberrant mRNAs that prematurely terminate translation (31), the association between mRNA turnover and translation has not been formally investigated on a genome-wide basis. A reduced number of single-gene studies have addressed this question directly or indirectly, yielding inconsistent conclusions. In some cases, as described for the cationic amino acid transporter cat-1, the glucose transporter glut-1, and neurofilament M, enhanced mRNA stability was found associated with enhanced translation (3, 6, 21, 46); conversely, mRNA degradation coupled with translational repression has also been reported (5). In the reverse scenario, it has also been reported that mRNA stability and translation are regulated in opposite directions. The ability of active translation to trigger mRNA decay was first postulated based on the transcript-stabilizing effects of treatments with translation inhibitors (such as cycloheximide). Aharon and Schneider and Curatola et al. studied this process by focusing on the ARE instability determinant of the granulocyte-macrophage colony-stimulating factor mRNA. Their studies demonstrated that rapid mRNA degradation by the granulocyte-macrophage colony-stimulating factor ARE was activated by translation and that strong secondary structures blocking ribosome transit inhibited ARE-mediated decay (1, 15). However, the universality of such translation-coupled ARE-mediated decay remains unclear, since other studies have shown that modulation of the translational state of an RNA containing the c-fos ARE (another potent instability determinant) had no influence on mRNA turnover (28). The most plausible explanation for the complex links between mRNA stability and translation is that many factors contribute to these multistep processes, including the metabolic conditions of the cell, the nature of the stimulus, the participating RNA-binding factors, and the sequence of the target mRNA of interest.
It is important that the en mass analysis of transcription employed here, based on the use of the NRO methodology, has significant limitations. First, NRO data obtained either by using arrays or by using conventional methods are only semiquantitative. Second, the data can lack in sensitivity, specificity, or both due to background hybridization among genes of related sequence, artifacts derived from the “pausing” of RNA polymerase II (as described previously for the c-myc gene [7]), and the possibility that some endogenous antisense transcripts could also hybridize to the double-stranded cDNAs spotted on the arrays. Nevertheless, several genes previously reported to be transcriptionally induced by ER stress were found among those in the transcriptionally upregulated subset (143 and 223 genes for Tn and Tg, respectively) (Fig. 3A), including GADD45α, asparagine synthetase, and the homocysteine-inducible, ER stress-inducible, ubiquitin-like domain member 1 (http://www.grc.nia.nih.gov/branches/rrb/dna/index/dnapubs.htm#2). Third, the approach undertaken in this investigation cannot comprehensively reflect important distinctions between changes that are due exclusively to alterations in transcription or mRNA turnover and changes that are due to a combination of both processes. Indeed, a growing number of genes are found to be subject to gene regulation as described in the latter category, wherein transcriptional and posttranscriptional events are orchestrated to achieve maximal and rapid induction or repression of a particular gene (10, 30, 38). Given the limitations of the approach employed here, some of these gene regulatory categories have been grouped together and are not represented individually. All of these issues should be taken into consideration when interpreting the transcription and mRNA stability data.
Recently described in mammalian cells (2), stress granules (SG) are cytoplasmic foci that may provide a partial explanation for this multifaceted response, particularly in the case of translationally suppressed mRNAs. As proposed by Kedersha and Anderson, SGs forming transiently in response to various stress stimuli including heat shock, oxidative stress, metabolic poisons, and UV light irradiation (23) represent dynamic sites of abortive or stalled translational initiation where mRNAs, 40S ribosomal subunits, and possibly also eIF2 complexes assemble (26). The discovery that SGs also recruit a number of RNA-binding proteins that promote both translational suppression and activation (TIA-1, TIAR, and HuR), as well as both mRNA stabilization and decay (HuR and TTP) (23), led Kedersha and Anderson to hypothesize that SGs represent sites of “mRNA triage,” where mRNAs are targeted for either translation, degradation, or sequestration into translationally silent mRNA-protein complexes. Indeed, the accumulation of unfolded proteins was also shown to trigger the formation of SGs (26). In the present study, treatment with ER stress agents was also found to induce the formation of SGs earlier in the process, but all SGs had disappeared by 3 h following the addition of the ER stress agents (data not shown). Initial efforts to biochemically isolate SGs from HeLa cells for the systematic analysis of participating mRNAs and proteins were unfruitful. Therefore, whether the decisions made in SGs several hours earlier in the ER stress response are still reflected in the fate of one or several of the mRNA subsets studied here remains to be formally investigated.
The data gathered in this investigation support the existence of all of the aforementioned stability and translation groups and provide examples of mRNAs whose stability and translation are regulated in similar and in opposite directions. While all three stability groups (increased, unchanged, and decreased mRNA stability) were represented among all three translational groups (induced, unchanged, and decreased translation), the most notable differences were found among genes whose mRNAs were stabilized, where prominent >3.5-fold mRNAs were translationally repressed. The latter set of data supports the model put forth by Aharon and Schneider (1); however, further experiments will be needed to formally discern whether a block in the translation of these mRNAs contributed to rendering them stable or, instead, mRNA association with a putative stabilizing factor caused their translational suppression. In this regard, it is becoming increasingly appreciated that cellular mRNAs are dynamically associated with many RNA-binding proteins that function as chaperones and regulators of their posttranscriptional fate (24, 25). A scenario could thus be envisioned whereby transit of a given mRNA through the polysome might lead to the displacement of RNA-binding proteins entrusted with protecting the integrity of the mRNA. Alterations in the composition of the ribonucleoprotein complex could, in turn, affect the interaction of the poly(A)-binding protein with poly(A) and thus influence the deadenylation process, the rate-limiting step in mRNA degradation (41).
In summary, we have developed a systematic approach for investigating links between mRNA turnover and translation using cDNA arrays. Following exposure to ER stress agents, a comparison of the changes in steady-state mRNA levels and in gene transcription rates allowed us to elucidate the extent to which mRNA stabilization and destabilization influenced mRNA expression profiles. The ER stress-elicited changes in translation were monitored by studying shifts in the relative abundance of mRNAs along polysome gradients. The conclusions derived from this investigation contribute to a growing body of evidence that critical gene regulatory events are implemented at a posttranscriptional level. The methodology described provides a robust approach for studying the coordination of such molecular mechanisms as we seek more comprehensive insight into posttranscriptional gene regulatory networks.
Acknowledgments
We are grateful to our colleagues K. G. Becker and D. Teichberg for assistance with the preparation and analysis of the array data.
REFERENCES
- 1.Aharon, T., and R. J. Schneider. 1993. Selective destabilization of short-lived mRNAs with the granulocyte-macrophage colony-stimulating factor AU-rich 3′ noncoding region is mediated by a cotranslational mechanism. Mol. Cell. Biol. 13:1971-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, P., and N. Kedersha. 2002. Stressful initiations. J. Cell Sci. 115:3227-3234. [DOI] [PubMed] [Google Scholar]
- 3.Antic, D., N. Lu, and J. D. Keene. 1999. ELAV tumor antigen, Hel-N1, increases translation of neurofilament M mRNA and induces formation of neurites in human teratocarcinoma cells. Genes Dev. 13:449-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arava, Y., Y. Wang, J. D. Storey, C. L. Liu, P. O. Brown, and D. Herschlag. 2003. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 100:3889-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Audic, Y., M. Garbrecht, B. Fritz, M. D. Sheets, and R. S. Hartley. 2002. Zygotic control of maternal cyclin A1 translation and mRNA stability. Dev. Dyn. 225:511-521. [DOI] [PubMed] [Google Scholar]
- 6.Aulak, K. S., R. Mishra, L. Zhou, S. L. Hyatt, W. de Jonge, W. Lamers, M. Snider, and M. Hatzoglou. 1999. Post-transcriptional regulation of the arginine transporter Cat-1 by amino acid availability. J. Biol. Chem. 274:30424-30432. [DOI] [PubMed] [Google Scholar]
- 7.Bentley, D. L., and M. Groudine. 1986. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature 321:702-706. [DOI] [PubMed] [Google Scholar]
- 8.Berger, B. J., T. S. Muller, I. R. Buschmann, K. Peters, M. Kirsch, B. Christ, and F. Prols. 2003. High levels of the molecular chaperone Mdg1/ERdj4 reflect the activation state of endothelial cells. Exp. Cell Res. 290:82-92. [DOI] [PubMed] [Google Scholar]
- 9.Bertolotti, A., Y. Zhang, L. M. Hendershot, H. P. Harding, and D. Ron. 2000. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2:326-332. [DOI] [PubMed] [Google Scholar]
- 10.Briata, P., C. Ilengo, G. Corte, C. Moroni, M. G. Rosenfeld, C. Y. Chen, and R. Gherzi. 2003. The Wnt/β-catenin→Pitx2 pathway controls the turnover of Pitx2 and other unstable mRNAs. Mol. Cell 12:1201-1211. [DOI] [PubMed] [Google Scholar]
- 11.Cheadle, C., M. P. Vawter, W. J. Freed, and K. G. Becker. 2003. Analysis of microarray data using Z score transformation. J. Mol. Diagn. 5:73-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cline, S. D., and P. C. Hanawalt. 2003. Who's on first in the cellular response to DNA damage? Nat. Rev. Mol. Cell Biol. 4:361-372. [DOI] [PubMed] [Google Scholar]
- 13.Cok, S. J., S. J. Acton, and A. R. Morrison. 2003. The proximal region of the 3′-untranslated region of cyclooxygenase-2 is recognized by a multimeric protein complex containing HuR, TIA-1, TIAR, and the heterogeneous nuclear ribonucleoprotein U. J. Biol. Chem. 278:36157-36162. [DOI] [PubMed] [Google Scholar]
- 14.Costa, R. H., V. V. Kalinichenko, A. X. Holterman, and X. Wang. 2003. Transcription factors in liver development, differentiation, and regeneration. Hepatology 38:1331-1347. [DOI] [PubMed] [Google Scholar]
- 15.Curatola, A. M., M. S. Nadal, and R. J. Schneider. 1995. Rapid degradation of AU-rich element (ARE) mRNAs is activated by ribosome transit and blocked by secondary structure at any position 5′ to the ARE. Mol. Cell. Biol. 15:6331-6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan, J., X. Yang, W. Wang, W. H. Wood III, K. G. Becker, and M. Gorospe. 2002. Global analysis of stress-regulated mRNA turnover using cDNA arrays. Proc. Natl. Acad. Sci. USA 99:10611-10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galbán, S., J. L. Martindale, K. Mazan-Mamczarz, I. López de Silanes, J. Fan, W. Wang, J. Decker, and M. Gorospe. 2003. Influence of the RNA-binding protein HuR in pVHL-regulated p53 expression in renal carcinoma cells. Mol. Cell. Biol. 23:7083-7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gueydan, C., L. Droogmans, P. Chalon, G. Huez, D. Caput, and V. Kruys. 1999. Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor mRNA. J. Biol. Chem. 274:2322-2326. [DOI] [PubMed] [Google Scholar]
- 19.Harding, H. P., Y. Zhang, and D. Ron. 1999. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397:271-274. [DOI] [PubMed] [Google Scholar]
- 20.Jacobson, A. 1996. Poly(A) metabolism and translation: the closed-loop model, p.451-480. In J. Hershey, et al., Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Jain, R. G., L. G. Andrews, K. M. McGowan, P. H. Pekala, and J. D. Keene. 1997. Ectopic expression of Hel-N1, an RNA-binding protein, increases glucose transporter (GLUT1) expression in 3T3-L1 adipocytes. Mol. Cell. Biol. 17:954-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaufman, R. J. 1999. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 13:1211-1233. [DOI] [PubMed] [Google Scholar]
- 23.Kedersha, N., and P. Anderson. 2002. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30:963-969. [DOI] [PubMed] [Google Scholar]
- 24.Keene, J. D. 2001. Ribonucleoprotein infrastructure regulating the flow of genetic information between the genome and the proteome. Proc. Natl. Acad. Sci. USA 98:7018-7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keene, J. D., and S. A. Tenenbaum. 2002. Eukaryotic mRNPs may represent posttranscriptional operons. Mol. Cell 9:1161-1167. [DOI] [PubMed] [Google Scholar]
- 26.Kimball, S. R., R. L. Horetsky, D. Ron, L. S. Jefferson, and H. P. Harding. 2003. Mammalian stress granules represent sites of accumulation of stalled translation initiation complexes. Am. J. Physiol. Cell Physiol. 284:C273-C284. [DOI] [PubMed] [Google Scholar]
- 27.Klein, N., A. M. Curatola, and R. J. Schneider. 1999. Calcium-induced stabilization of AU-rich short-lived mRNAs is a common default response. Gene Expr. 7:357-365. [PMC free article] [PubMed] [Google Scholar]
- 28.Koeller, D. M., J. A. Horowitz, J. L. Casey, R. D. Klausner, and J. B. Harford. 1991. Translation and the stability of mRNAs encoding the transferrin receptor and c-fos. Proc. Natl. Acad. Sci. USA 88:7778-7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kullmann, M., U. Gopfert, B. Siewe, and L. Hengst. 2002. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′UTR. Genes Dev. 16:3087-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindstein, T., C. H. June, J. A. Ledbetter, G. Stella, and C. T. Thompson. 1989. Regulation of lymphokine messenger RNA stability by a surface-mediated T cell activation pathway. Science 244:339-343. [DOI] [PubMed] [Google Scholar]
- 31.Maquat, L. E. 2002. Nonsense-mediated mRNA decay. Curr. Biol. 12:R196-R197. [DOI] [PubMed] [Google Scholar]
- 32.Mazan-Mamczarz, K., S. Galbán, I. López de Silanes, J. L. Martindale, U. Atasoy, J. D. Keene, and M. Gorospe. 2003. RNA-binding HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl. Acad. Sci. USA 100:8354-8359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piecyk, M., S. Wax, A. R. P. Beck, N. Kedersha, M. Gupta, B. Maritim, S. Chen, C. Gueydan, V. Kruys, M. Streuli, and P. Anderson. 2000. TIA-1 is a translational silencer that selectively regulates the expression of TNF-α. EMBO J. 19:4154-4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pirkkala, L., P. Nykanen, and L. Sistonen. 2001. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 15:1118-1131. [DOI] [PubMed] [Google Scholar]
- 35.Refojo, D., A. C. Liberman, D. Giacomini, A. Carbia Nagashima, M. Graciarena, C. Echenique, M. Paez Pereda, G. Stalla, F. Holsboer, and E. Arzt. 2003. Integrating systemic information at the molecular level: cross-talk between steroid receptors and cytokine signaling on different target cells. Ann. N. Y. Acad. Sci. 992:196-204. [DOI] [PubMed] [Google Scholar]
- 36.Ron, D. 2002. Translational control in the endoplasmic reticulum stress response. J. Clin. Investig. 110:1383-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rutkowski, D. T., and R. J. Kaufman. 2003. All roads lead to ATF4. Dev. Cell 4:442-444. [DOI] [PubMed] [Google Scholar]
- 38.Shaw, J., K. Meerovitch, R. C. Bleackley, and V. Paetkau. 1988. Mechanisms regulating the level of IL-2 mRNA in T lymphocytes. J. Immunol. 140:2243-2248. [PubMed] [Google Scholar]
- 39.Shi, Y., K. M. Vattem, R. Sood, J. An, J. Liang, L. Stramm, and R. C. Wek. 1998. Identification and characterization of pancreatic eukaryotic initiation factor 2 α-subunit kinase, PEK, involved in translational control. Mol. Cell. Biol. 18:7499-7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Svejstrup, J. Q. 2002. Mechanisms of transcription-coupled DNA repair. Nat. Rev. Mol. Cell. Biol. 3:21-29. [DOI] [PubMed] [Google Scholar]
- 41.Tucker, M., and R. Parker. 2000. Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae. Annu. Rev. Biochem. 69:571-595. [DOI] [PubMed] [Google Scholar]
- 42.Wang, W., H. Furneaux, H. Cheng, M. C. Caldwell, D. Hutter, Y. Liu, N. J. Holbrook, and M. Gorospe. 2000. HuR regulates p21 mRNA stabilization by UV light. Mol. Cell. Biol. 20:760-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wickens, M., P. Anderson, and R. J. Jackson. 1997. Life and death in the cytoplasm: messages from the 3′ end. Curr. Opin. Genet. Dev. 7:220-232. [DOI] [PubMed] [Google Scholar]
- 44.Wilkie, G. S., K. S. Dickson, and N. K. Gray. 2003. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem. Sci. 28:182-188. [DOI] [PubMed] [Google Scholar]
- 45.Yaman, I., J. Fernandez, B. Sarkar, R. J. Schneider, M. D. Snider, L. E. Nagy, and M. Hatzoglou. 2002. Nutritional control of mRNA stability is mediated by a conserved AU-rich element that binds the cytoplasmic shuttling protein HuR. J. Biol. Chem. 277:41539-41546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yaman, I., J. Fernandez, H. Liu, M. Caprara, A. A. Komar, A. E. Koromilas, L. Zhou, M. D. Snider, D. Scheuner, R. J. Kaufman, and M. Hatzoglou. 2003. The zipper model of translational control: a small upstream ORF is the switch that controls structural remodeling of an mRNA leader. Cell 113:519-531. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, T., V. Kruys, G. Huez, and C. Gueydan. 2002. AU-rich element-mediated translational control: complexity and multiple activities of trans-activating factors. Biochem. Soc. Trans. 30:952-958. [DOI] [PubMed] [Google Scholar]