Abstract
The ADAMTS (A Disintegrin and Metalloproteinase with Thrombospondin motifs) enzymes are secreted, multi-domain matrix-associated zinc metalloendopeptidases that have diverse roles in tissue morphogenesis and patho-physiological remodeling, in inflammation and in vascular biology. The human family includes 19 members that can be sub-grouped on the basis of their known substrates, namely the aggrecanases or proteoglycanases (ADAMTS1, 4, 5, 8, 9, 15 and 20), the procollagen N-propeptidases (ADAMTS2, 3 and 14), the cartilage oligomeric matrix protein-cleaving enzymes (ADAMTS7 and 12), the von-Willebrand Factor proteinase (ADAMTS13) and a group of orphan enzymes (ADAMTS6, 10, 16, 17, 18 and 19). Control of the structure and function of the extracellular matrix (ECM) is a central theme of the biology of the ADAMTS, as exemplified by the actions of the procollagen-N-propeptidases in collagen fibril assembly and of the aggrecanases in the cleavage or modification of ECM proteoglycans. Defects in certain family members give rise to inherited genetic disorders, while the aberrant expression or function of others is associated with arthritis, cancer and cardiovascular disease. In particular, ADAMTS4 and 5 have emerged as therapeutic targets in arthritis. Multiple ADAMTSs from different sub-groupings exert either positive or negative effects on tumorigenesis and metastasis, with both metalloproteinase-dependent and -independent actions known to occur. The basic ADAMTS structure comprises a metalloproteinase catalytic domain and a carboxy-terminal ancillary domain, the latter determining substrate specificity and the localization of the protease and its interaction partners; ancillary domains probably also have independent biological functions. Focusing primarily on the aggrecanases and proteoglycanases, this review provides a perspective on the evolution of the ADAMTS family, their links with developmental and disease mechanisms, and key questions for the future.
Gene organization and evolutionary history
Mammalian genomes contain 19 ADAMTS genes numbered 1 to 20, the designation ADAMTS11 not being employed because it was assigned to a gene previously identified as ADAMTS5 [1, 2]. Like their relatives, the matrix metalloproteinases (MMPs) and the ADAMs (A Disintegrin And Metalloproteinases), the ADAMTSs belong to the metzincin protease superfamily, named for the conserved methionine residue close to the zinc ion-dependent metalloproteinase active site [3]. Representatives of the ADAMTS family are found in all metazoans, although to date they have not been identified in single-cell organisms or in plants.
All ADAMTSs are secreted, extracellular enzymes that have a compound domain organization (Fig. 1), comprising, from the amino-terminus: a signal peptide followed by a pro-region of variable length; a metalloproteinase domain; a disintegrin-like domain; a central thrombospondin type 1 sequence repeat (TSR) motif; and a cysteine-rich domain followed by a spacer region. This basic organization is manifest by ADAMTS4, and built upon in other family members with a variety of further carboxy-terminal modules, including one or more additional TSRs. The entire carboxy-terminal region downstream of the central TSR is termed the ancillary domain, and this is where the greatest differences between ADAMTS family members occur. Unlike their ADAM relatives, the ADAMTSs lack epidermal growth factor (EGF)-like, transmembrane and cytoplasmic modules. Separate from the ADAMTSs, another family of seven ADAMTS-like genes (ADAMTSL) encode proteins that resemble the ancillary domains of ADAMTS but lack their catalytic domains. These ADAMTSL proteins, which include ADAMTSL 1 to 6 and papilin, may function to modulate the activities of the ADAMTSs [2, 4].
Fig. 1.
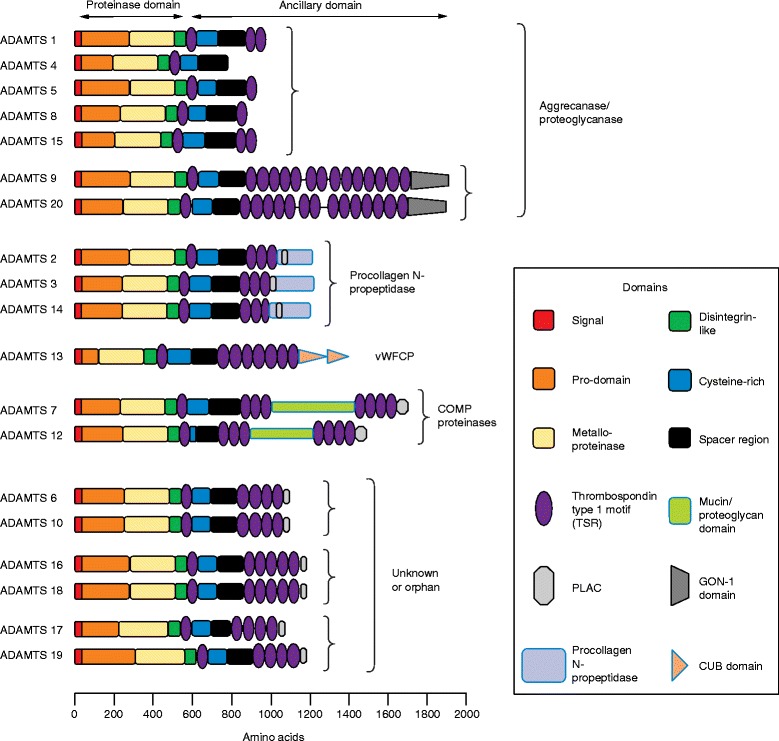
The ADAMTS (A Disintegrin And Metalloproteinase with ThromboSpondin motifs) family. The basic domain organization of the 19 ADAMTS family members and their major functional groups. Structurally the ADAMTS members are broadly organized into a proteinase domain and an ancillary domain. The proteinase domain comprises the signal, pro, metalloproteinase and disintegrin-like domains. The greatest variability between ADAMTS members is found in the ancillary domain, which is composed of one or more thrombospondin type 1 sequence repeats (TSRs), a cysteine-rich domain and a spacer domain. Some family members also have one or more specialist domains as part of their ancillary domain, as listed in the key on the right. The diagram is drawn to scale
The 19 human ADAMTS proteins can be assembled into eight ‘clades’ on the basis of their domain organization and their known functions. The aggrecanase and proteoglycanase clades (ADAMTS1, 4, 5, 8, 15, and ADAMTS9 and 20) can cleave hyaluronan-binding chondroitin sulfate proteoglycan (CSPG) extracellular proteins, including aggrecan, versican, brevican and neurocan [5]. This sub-group has also been labeled ‘angioinhibitory’ on the basis of the original identification of ADAMTS1 and 8 as anti-angiogenic factors [6]; nevertheless, ADAMTSs in other clades also have effects on angiogenesis. Another group (ADAMTS2, 3 and 14) are pro-collagen N-propeptidases that are essential for the maturation of triple helical collagen fibrils [7]. A lone family member, ADAMTS13, is the von-Willebrand factor (vWF)-cleaving protease (vWFCP). This protease processes large multimeric vWF precursor proteins under fluid shear stress conditions to generate vWF proteins of optimal size for proper blood coagulation [8]. Another clade (ADAMTS7 and 12) has been recognized as cleaving cartilage oligomeric matrix protein (also known as thrombospondin-5) [9, 10]. This clade is unique in the ADAMTS family in possessing a mucin domain to which chondroitin sulfate chains are attached, conferring proteoglycan status upon these two enzymes [11]. The remaining three subgroups, which are defined on the basis of their domain organizations, each contain a pair of enzymes (ADAMTS6 and 10; ADAMTS16 and 18; ADAMTS17 and 19) whose physiological substrates have yet to be identified, and thus are currently called the ‘orphan’ sub-groups. Like those ADAMTSs whose functions are better understood, several orphan enzymes have important physiological roles that are emerging from their associations with inherited human genetic disorders and acquired diseases.
The pairs of ADAMTS proteins that share similar domain structures suggest the occurrence of gene duplication during evolution [12–16]. Comparison of the genomes of deuterostomes, namely those of the vertebrates Homo, Mus and Xenopus and that of the chordate Ciona, with those of protostome invertebrates (Drosophila and Caenorhabditis) has provided the view of the evolutionary history of the ADAMTS family summarized in the schematic and the phylogenetic tree in Fig. 2 [13, 16]. Caenorhabditis elegans and Drosophila melanogaster have four and three ADAMTS orthologs, respectively [12, 13]. Both have a single gene - Gon-1 in the nematode and CG6107 in the fly - representing the right-hand branch of the human family shown in Fig. 2a; these genes are related to ADAMTS9/20. The left-hand branch is also represented by a single gene in Drosophila (CG4096) but it is missing entirely in C. elegans, which likely reflects loss of the founder gene during nematode evolution. Nevertheless, the remaining Drosophila gene (named stall) and the three C. elegans ADAMTS-related genes (adt-1, adt-2 and T19D2.1) cluster as a protostome-specific sub-family that have no counterparts in deuterostomes [12].
Fig. 2.
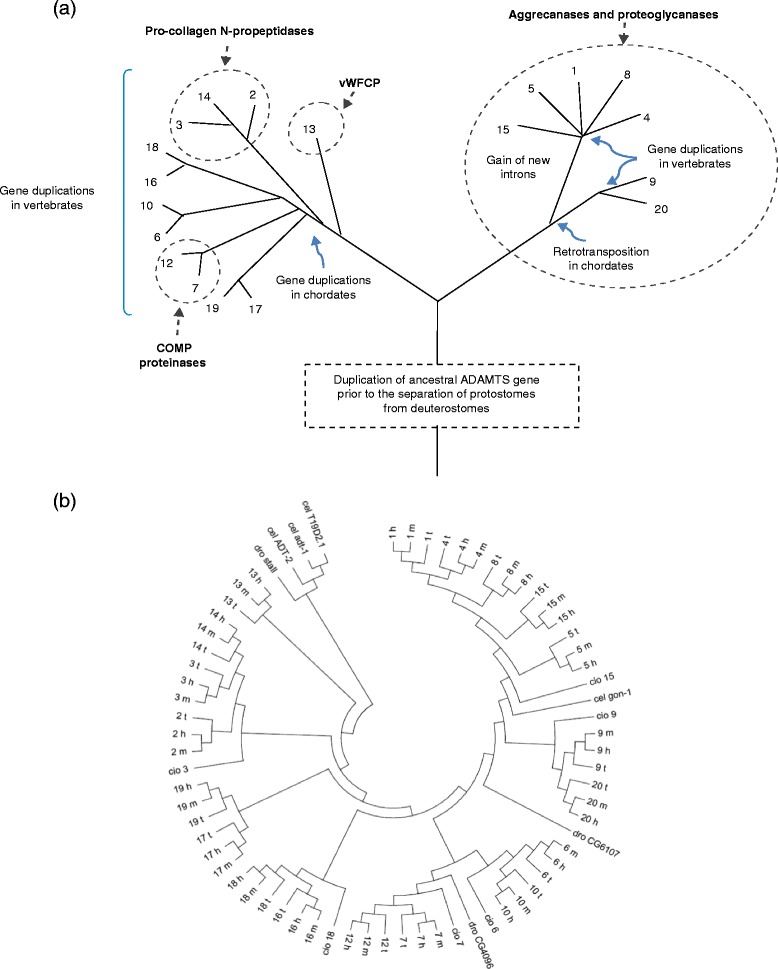
Evolution of the ADAMTS family. a A schematic representation of the relationships of the eight vertebrate ADAMTS clades and the probable events (gene duplications and a retrotransposition) that have contributed to the expansion of the family. The figure is not to scale in terms of evolutionary distance. COMP, cartilage oligomeric protein; vWFCP, von-Willebrand-factor-cleaving protease. b Phylogenetic tree of the ADAMTS genes inferred by the maximum likelihood method based on the JTT matrix-based model [144]. The bootstrap consensus tree inferred from 1,000 replicates was taken to represent the evolutionary history of the taxa analyzed [145]. Branches corresponding to partitions reproduced in less than 50 % bootstrap replicates were collapsed. Initial tree(s) for the heuristic search were obtained by applying the neighbor-joining method to a matrix of pairwise distances estimated using a JTT model. The analysis involved 70 amino acid sequences. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA6 [146]. The vertebrate ADAMTS genes are indicated by a number followed by a single letter code indicating the species: for example, 1 t represents 1_t ADAMTS1 from Xenopus tropicalis; 15 h is 15_h ADAMTS15 from Homo sapiens; 8 m is ADAMTS8 from Mus musculus. For Drosophila melanogaster (dro), Caenorhabditis elegans (cel) and Ciona intestinalis (cio), the annotation is species followed by the gene number; for example, cio 6 is cio_6 ADAMTS6 from Ciona
Six ADAMTS genes are present in the basal chordate Ciona intestinalis, one of the closest invertebrate relatives of the vertebrates, each of which is the root of one of the eight mammalian clades (ADAMTS3, 6, 7, 9, 15 and 18), the two mammalian clades not represented in Ciona being ADAMTS13 and the ADAMTS17/19 pair [12]. This evidence argues that the gene pairs in vertebrates arose by duplication during their evolution from their chordate ancestors. The likely sequence of events is therefore that an ancestral ADAMTS gene duplicated approximately 650 million years ago, prior to the divergence of the protostomes (that is, the insects, crustaceans and nematodes) from the deuterostomes (chordates and vertebrates). One of these early duplicated genes gave rise ultimately to the aggrecanase/proteoglycanase sub-group (the right-hand branch in Fig. 2), while the other duplicate was the founder of the remaining family members. Subsequently, three or four gene duplications occurred during chordate evolution, with further duplications during vertebrate evolution.
The ADAMTS9 and 20 gene pair in the aggrecanase/proteoglycanase sub-group share a high degree of similarity with the C. elegans Gon-1 gene, suggesting their close relationship with the ancestral gene that founded the aggrecanases/proteoglycanases. On the basis of their intron and exon structure, however, the other related genes (ADAMTS1, 4, 5, 8, 15) seem likely to be derived from a retrotransposition event that occurred early in deuterostome evolution involving the same founder gene that also gave rise to ADAMTS9/20. This event seems to have produced an intron-less gene that subsequently acquired new introns at different positions. This evolutionary pathway is supported by the lack of orthologs of the ADAMTS1, 4, 5, 8, 15 clade in protostomes [12–14]. Subsequently, this founder gene underwent a duplicative chromosome inversion to generate two adjacent ADAMTS genes in head-to-head orientation. A later duplication of this pair of genes resulted in two sets of paired genes, ADAMTS1 and ADAMTS5 on human 21q21, and the ADAMTS8/15 pair on human11q24.
The deuterostome sea urchin Strongylocentrus purpuratus has eight ADAMTS orthologs that correspond to five of the eight ADAMTS clades present in vertebrates, but it lacks any representative for the ADAMTS1/4/5/8/15 and ADAMTS9/20 aggrecanase/proteoglycanase clades, suggesting that the progenitor of these genes has been lost [14]. This sea urchin has two genes with similarity to ADAMTS13, which had previously been thought to be a vertebrate-specific gene [12]. The presence of 16 ADAMTS genes in pufferfish (Fugu rubripes, which like Ciona lacks ADAMTS13 and 17/19), 17 genes in zebrafish (Danio rerio, which lacks ADAMTS4 and 19) and all 19 ADAMTS genes in Xenopus (X. tropicalis and X. laevis) argues that most of these gene duplications occurred prior to the divergence of fish and mammals. Among the zebrafish gene complement there are two copies each of ADAMTS2, 8 and 15, which result from a teleost-specific whole-genome duplication event [16]. However, only 17 ADAMTS genes have been found in several species of birds: chicken (Gallus gallus), duck (Anas platyrhynchos) and zebra finch (Taeniopygia guttata) lack ADAMTS4 and 16, suggesting that these two genes were lost during the divergence of birds. The expansion of the ADAMTS family during vertebrate evolution goes hand-in-hand with the increased complexity of the ECM, which has also arisen through the duplication, retention and modification of ancestral genes [17].
Characteristic structural features and mechanism
Pro-domain
The pro-domains of the metzincins generally maintain latency and direct proper folding of the enzymes; but in the ADAMTSs they have additional functions. In general, ADAMTSs lack the ‘cysteine switch’ that controls activation of the MMPs, though curiously there is evidence for this in ADAMTS15. All ADAMTSs contain at least one site (R/KXnR/K↓R) for furin-like pro-protein convertases (PPCs) and some (such as pro-ADAMTS1 and −4) have been shown to be activated by PPCs in the trans-Golgi network, leading to secretion of active enzyme [18, 19]. Other ADAMTS precursors (such as pro-ADAMTS5) are activated by furin not in the Golgi but extracellularly [20], whereas pro-ADAMTS9 is activated on the cell surface when in a complex with the chaperone heat shock protein gp96/GRP94 [21]. Loss of the pro-domain actually reduces the activity of ADAMTS9 to cleave its substrate, versican; likewise pro-ADAMTS13 does not require pro-domain removal for catalytic activity [21–23]. The ADAMTS pro-domain may act to chaperone proper folding and secretion rather than in maintaining latency, although this is not the case for ADAMTS13. This enzyme has an unusually short pro-domain that probably acts by influencing binding to other proteins or by regulating catalytic activity in some other way [22].
Catalytic domain: metalloproteinase and disintegrin-like modules
The ADAMTSs contain a consensus HEXXHXBG(/N/S)BXHD catalytic motif, in which the three histidines coordinate a Zn2+ ion [3]; B represents a large non-polar residue. A methionine, which in ADAMTSs lies 14 to 20 residues downstream of the third histidine, defines the ‘Met-turn’ common to the catalytic domains of all metzincin metalloproteinases [3]. In contrast to the ADAMs, in which 8 of the 21 human family members have lost a functional Zn-binding motif and are therefore proteolytically inactive [24], all ADAMTSs are predicted to be catalytically functional.
The ADAMTS metalloproteinase domains are predicted to adopt the typical metzincin architecture: a globular structure with an amino-terminal sub-domain comprising a five-stranded β-sheet on the top; and on the bottom, a carboxy-terminal sub-domain composed of α-helices in which the Met-turn is positioned, forming a hydrophobic pillow underneath the catalytic Zn2+. This creates an active site cleft into which substrates bind in essentially an extended, linear configuration [25]. A distinctive feature of ADAMTS metalloproteinase domains, when compared to those of the MMPs, is the presence of four disulphide bonds that stabilize the structure (MMPs have none) [26].
Unlike their ADAM relatives, no ADAMTS have been reported to interact with integrins via their disintegrin-like domains, and it has been suggested that this domain is misnamed in the ADAMTSs. Crystal structure data for ADAMTS1 [26] and for ADAMTS4 and 5 [27] reveal that the disintegrin-like domain is a cysteine-rich region that stacks against the metalloproteinase active-site cleft, so it is appropriate that we consider it part of the catalytic domain. A surprising finding from the crystal structures of the catalytic domains of ADAMTS4 and 5 is the identification of two alternative conformations of the active sites that may exist in equilibrium: an ‘open’ structure with an additional Ca2+ ion bound and a ‘closed’, inaccessible structure in which the Ca2+ ion is released [27]. The existence of two distinct conformational states has not been seen for MMPs or ADAMs (though crystal data are still sparse) and it may be an attribute of the ADAMTSs that is used to regulate their catalytic actions via binding accessory proteins, substrates or other ADAMTS domains.
Ancillary domain
The ADAMTS enzymes rely upon their carboxy-terminal ancillary domains for their association with the ECM, for regulation of their activity, and for specification of their substrate-binding preferences. The ancillary domain in all ADAMTS enzymes contains an approximately 50-amino-acid thrombospondin-like repeat (TSR) that is similar to the type I repeats of thrombospondins 1 and 2 [28], followed by a cysteine-rich region of slightly more than 100 amino acid residues (in all except ADAMTS12) that contains 10 conserved cysteine residues, and finally, a more variable cysteine-free spacer region, which ranges from 103 to 160 amino acids in length. With the sole exception of ADAMTS4, the spacer domain is followed by 1 to 14 further TSR modules and additional motifs that are characteristic of particular subgroups. The ADAMTS9/20 pair has the largest number of TSRs and each concludes with a GON-1 module (first described in C. elegans Gon-1), which contains 10 conserved cysteine residues [23]. ADAMTS13 is unique in having two CUB modules. Several ADAMTSs (ADAMTS2, 3, 6, 7, 10, 12, 14, 16, 17, 18 and 19) possess a PLAC (protease and lacunin) module that contains six conserved cysteine residues. In ADAMTS7 and 12, a mucin/proteoglycan domain is interposed in the middle of the seven carboxy-terminal TSRs.
Post-translational modifications
ADAMTS enzymes are glycosylated and their ancillary domains can be proteolytically processed, with both types of modification affecting the enzymes’ secretion, localization, activation or catalytic functions. All ADAMTSs with the exception of ADAMTS4 are N-glycosylated at NxS/T sites, with N-glycosylation within the pro-domain of ADAMTS9 being necessary for its secretion [21]. The TSRs are sites for C-mannosylation at tryptophans in conserved WxxW motifs, and O-fucosylation of serine or threonine residues in CxxS/TCG motifs, as demonstrated for the type I repeats in thrombospondins [29]. The O-fucosylation of ADAMTS13 regulates its secretion [30] and is probably a quality-control mechanism that ensures proper protein folding [2]. The mucin domain of ADAMTS7 is modified by the addition of CS GAG chains, which together with N-glycosylation of the pro-domain may regulate the enzyme’s cell-surface association and sequential processing by furin [11]. Proteolytic processing within the ancillary domains has been reported for many ADAMTS enzymes, and in some cases the cleavages are autolytic [18, 31, 32].
Inhibitors of the ADAMTSs
Like the ADAMs, ADAMTSs show restricted susceptibility to inhibition by the four tissue inhibitors of metalloproteinases (TIMPs) [33]. Where multiple TIMPs have been tested, as is the case for ADAMTS2 [34] and ADAMTS4 [35], TIMP-3 emerges as the most effective inhibitor. The aggrecanase activity of ADAMTS1 is inhibited by both TIMP-2 and −3, but not by TIMP-1 and −4 [36]. The ancillary domains of ADAMTS4 and 5 promote interactions with TIMP-3 [37]. Inhibition of ADAMTS4 aggrecanase activity by TIMP-3 is enhanced by aggrecan, through a mechanism that involves the interaction of aggrecan GAG chains with the TSR and spacer regions of ADAMTS4 [38].
Another key difference between the ADAMTSs and the MMPs relates to their mode of inhibition by TIMPs. For MMP inhibition, the amino-terminal cysteine residue of a mature TIMP molecule is essential to coordinate with the active site Zn2+ [33]. Extension of TIMP-3 at the amino terminus by an alanine residue abrogates MMP inhibitory activity, but potent inhibition of ADAMTS4 and 5 is retained [39]. This may be significant in the design of selective ADAMTS inhibitors, such as cis-1(S)2(R)-amino-2-indanol-based compounds. These compounds are potent inhibitors of ADAMTS4 and 5, with Ki values in the low nM range and selectivity two orders of magnitude greater than that of the MMPs, suggesting that they are good platforms for the development of highly selective aggrecanase inhibitors [40]. Interference with substrate interactions by binding to exosites in the ancillary domains of ADAMTS is also a viable strategy, as shown by the use of calcium pentosan polysulfate for ADAMTS4 aggrecanase inhibition [41]. Also, granulin-epithelin precursor binds to the carboxy-terminal TSR motifs of ADAMTS7 and 12, blocking the abilities of these enzymes to cleave cartilage oligomeric protein (COMP) [42].
The activity of the ADAMTSs is also controlled by their internalization and degradation. For ADAMTS4 and 5 these processes have been shown to involve interaction with low-density lipoprotein-related protein-1 (LRP-1) [43, 44]. Differential affinity for LRP-1 results in different half-lives for extracellular ADAMTS4 and 5 [44]. As LRP-1 also binds and internalizes TIMP-3, and as this interaction is blocked by heparan sulfate GAGs, which also potentiates TIMP-3 activity, there is potential for modulating the tissue activities of ADAMTS using sulfated glycan mimics [45].
Localization and function
The ADAMTS proteases have important roles in tissue development and maintenance, and their dysregulation or mutation is associated with a number of diseases. In the sections that follow we provide an overview of the current knowledge of the functions and regulation of ADAMTSs that has emerged from work on human pathologies and gene-knockout mice, concentrating on the aggrecanase/proteoglycanases. Other recent reviews have focused on the roles of ADAMTSs in arthritis [5, 46], cancer [47–51], atherosclerosis [52] and central nervous system injury and disorders [53].
Mutations in several ADAMTS genes are associated with human autosomal recessive Mendelian inherited disorders; this topic has been the focus of an excellent recent review [4]. The extreme skin fragility displayed by sufferers of Ehlers-Danlos syndrome type VIIC, which corresponds to dermatosparaxis in cattle and sheep, arises due to inactivation of ADAMTS2 [41]. Recessive mutations in ADAMTS13 are responsible for a condition called thrombotic thrombocytopenic purpura (TTP), which is caused by a failure to cleave the otherwise pro-thrombogenic ultra-large von Willebrand Factor multimers in the circulation, resulting in platelet aggregation and vessel occlusion [54]. Autoantibodies to ADAMTS13 also give rise to acquired TTP [55]. The connective tissue disorder Weill-Marchesani syndrome (WMS) manifests by short stature, brachydactyly, joint stiffness, cardiac valve stenosis and ectopia lentis (lens dislocation), and can be inherited in both autosomal dominant and recessive modes. Autosomal dominant WMS is attributable to mutations in the ECM protein fibrillin-1, which is required for formation of tissue microfibrils [56]. By contrast, autosomal recessive WMS is attributable to mutations in ADAMTS10 [57]. ADAMTS10 binds to fibrillin-1 and −2 and promotes microfibril formation in the ECM [58], but it has been suggested that its function may be independent of its protease activity [4]. Significantly, mutations in two members of the ADAMTSL gene family (ADAMTSL2 and 4) result in syndromes whose phenotypes overlap the ocular, skeletal and cardiac features of WMS, and are also probably linked to proper microfibril formation in the affected tissues [4]. Likewise, mutations in ADAMTS17 cause a recessive WMS-like phenotype [59]. Another inherited ocular syndrome, microcornea, myopic chorioretinal atrophy and telechanthus (MMCAT) is caused by mutations in the orphan gene ADAMTS18 [60, 61]. Although not yet detected in human genetic disorders, inactivation of the Adamts16 gene in rodents has been shown to lead to hypertension, cryptorchidism and male infertility, and aberrant renal development [62–64].
Table 1 summarizes the major tissue locations in which ADAMTS genes are expressed (see also the BioGPS database - http://biogps.org) and known ADAMTS substrates. Many ADAMTS genes are transcriptionally regulated by cytokines, growth factors, hormones and inflammatory mediators: major inducing and repressing stimuli or regulators are shown, though it must be emphasized that this list is not exhaustive. In Table 2 we have also listed the phenotypes of Adamts knockout animals, which are covered more extensively in another recent review [4].
Table 1.
ADAMTS genes: their chromosomal positions, major tissue expression locations, expression-inducing factors, substrates, and associations with pathologies
| Gene | Location | Expression | Factors inducing or (repressing) expression | Substrates | Pathology associations |
|---|---|---|---|---|---|
| ADAMTS1 | 21q21 | Ovary, bronchial epithelial cells, fetal lung, placenta, smooth muscle, uterus, adrenal cortex, adipocyte, ciliary ganglion, prostate, olfactory bulb, breast stromal fibroblasts and myoepithelial cells | Progesterone, Brg1, IL-1, S100A8, S100A9, TNFα | Aggrecan, versican, syndecan 4, TFPI-2, semaphorin 3C, nidogen-1, −2, desmocollin-3, dystroglycan, mac-2, gelatin (denatured collagen type I), amphiregulin, TGF-α, heparin-binding EGF | Cancer (both pro- and anti-tumorigenic/metastatic), anti-angiogenic |
| ADAMTS2 | 5q35 | Adipocyte, skeletal muscle, superior cervical ganglion, uterus, placenta, heart, liver, lung, tongue, smooth muscle, breast stromal fibroblasts | Glucocorticoids (in monocytes), IL-6 | Fibrillar procollagens types I-III and V | Ehlers-Danlos syndrome type VIIc, dermatosparaxis (in sheep and cattle) |
| ADAMTS3 | 4q21 | CD105+ endothelial cells, CD34+ cells, pineal gland, cartilage, bone, skeletal muscle, tendon, breast myoepithelial cells | Fibrillar procollagen type II, biglycan | ||
| ADAMTS4 | 1q23 | Ovary, spinal cord, adrenal cortex, ciliary ganglion, trigeminal ganglion, brain, retina, pancreas (islets), fetal lung, breast myoepithelial cells | IL-1 + oncostatin M, TNFα, S100A8, S100A9, leptin, IL-6 | Aggrecan, versican, reelin, biglycan, brevican, matrilin-3, α2-macroglobulin, COMP | Arthritis |
| (HDAC inhibitors, pentosan polysulfate) | |||||
| ADAMTS5 | 21q21 | Adipocyte, uterus, breast myoepithelial cells | IL-1, TNFα, S100A8, S100A9, leptin, IL-6 | Aggrecan, versican, reelin, biglycan, matrilin-4, brevican, α2-macroglobulin | Arthritis, cancer (anti-tumorigenic, anti-angiogenic) |
| (HDAC inhibitors) | |||||
| ADAMTS6 | 5q12 | Superior cervical ganglion, trigeminal ganglion, appendix, heart, breast myoepithelial cells | TNFα, | - | |
| ADAMTS7 | 5q24 | Trigeminal ganglion, adrenal cortex, liver, heart, skeletal muscle, intervertebral disc, breast stromal fibroblasts | PTHrP | COMP | Coronary artery disease (smooth muscle cell migration) |
| (miR-29a/b) | |||||
| ADAMTS8 | 11q24 | Skeletal muscle, heart, liver, superior cervical ganglion, adrenal cortex, breast stromal fibroblasts and luminal epithelial cells | Aggrecan | ||
| ADAMTS9 | 3p14 | Dorsal root ganglion, breast myoepithelial cells | TNFα, IL1 + oncostatin M, leptin | Aggrecan, versican | Cancer (anti-angiogenic) |
| (HDAC inhibitors) | |||||
| ADAMTS10 | 19p13 | CD8+ T-cells, brain, uterus, breast stromal fibroblasts | Fibrillin-1 | Weill-Marchesani syndrome | |
| ADAMTS12 | 5p13 | Liver, bone marrow, atrioventricular node, intervertebral disc, breast stromal fibroblasts and myoepithelial cells | COMP | Cancer (pro- and anti-tumorigenic) | |
| ADAMTS13 | 9q34 | Liver, CD71+ early erythroid cells, lung, thyroid, breast myoepithelial cells | (IL-1) | vWF | Thrombotic thrombocytopenic purpura |
| ADAMTS14 | 10q22 | Thalamus, bone marrow, fetal thyroid, adipocyte, cerebellum, bone, skin, fibroblasts, breast myoepithelial and luminal epithelial cells | Fibrillar procollagen type I (pNα1 and pNα2 chains) | ||
| ADAMTS15 | 11q24 | Colon, brain, heart, uterus, musculoskeletal system, breast myoepithelial cells | Aggrecan, versican | Cancer (anti-tumorigenic/metastatic, anti-angiogenic) | |
| ADAMTS16 | 5p15 | Breast myoepithelial cells | Follicle stimulating hormone; forskolin (cAMP); | - | Hypertension |
| Transcription factors: Wilm’s tumor-1; Egr-1, Sp1 | |||||
| ADAMTS17 | 15q26 | Breast myoepithelial cells | - | Weill-Marchesani-like syndrome | |
| ADAMTS18 | 16q23 | Ciliary ganglion, heart, skin, brain, breast myoepithelial cells | - | ||
| ADAMTS19 | 5q23 | Dorsal root ganglion, breast myoepithelial cells | - | ||
| ADAMTS20 | 2q12 | Brain, appendix, heart, liver, skeletal muscle, pituitary, trigeminal ganglion, breast myoepithelial cells | Versican |
Table 2.
ADAMTS knockout and mutant mouse phenotypes
| Gene | Phenotype of gene knockout or mutant mice | Reference(s) |
|---|---|---|
| Adamts1 | Growth retardation, adipose tissue malformation | [91] |
| Impaired fertility with defective ovulation | [153] | |
| Severe kidney abnormalities: enlarged renal calices with fibrosis leading to obstruction of uteropelvic junction; abnormal adrenal medullary architecture with no formation of capillaries | [92] [148] | |
| [150] | ||
| Defective follicular development during ovulation, delay in development of ovarian lymphatic vessels | [94, 95] | |
| Impaired skin wound healing; effects on keratinocyte and fibroblast migration | [98] | |
| No defects in aggrecan turnover in vivo or in vitro | [145] | |
| Reduced tumorigenesis and metastasis in PyMT mammary cancer, with increased apoptosis | [117] | |
| Defective myocardial morphogenesis | [96] | |
| Selective decline in synaptic protein levels in frontal cortex of female Adamts1−/− mice | [58] | |
| Adamts2 | Fragile skin at 1–2 months postnatal; male sterility | [144] |
| Widespread defects in procollagen III processing; abnormal lungs | [142] | |
| Reduced extent and stability of carbon tetrachloride-induced hepatic fibrosis | [141] | |
| Adamts4 | No phenotype unchallenged | [69] |
| Perinatal lethality, exacerbation of renal phenotype in Adamts1−/−;Adamts4−/− double knockout mice | [138] | |
| Adamts5 | No phenotype unchallenged. Protection in surgery-induced osteoarthritis and antigen-induced arthritis models | [69] |
| [68] | ||
| Adamts4−/−;Adamts5−/− double knockout mice phenotypically normal; osteoarthritis phenotype same as Adamts5−/− mice | [146] | |
| Blockade of fibrosis and accumulation of aggrecan in joints in the DMM and TTR models of osteoarthritis | [143] | |
| [139] | ||
| Reduced changes in subchondral bone in DMM model of osteoarthritis | [152] | |
| Altered biomechanical properties of tendon | [81] | |
| Cardiac valve defects resembling myxomatous valve disease; rescued in versican (Vcan) heterozygotic animals | [147] | |
| Partial reduction of interdigital web regression | [151] | |
| Impaired dermal repair in excisional skin wound healing; aggrecan accumulation, altered transforming growth factor β (TGFβ) signaling | [140] | |
| Dermal fibroblasts have myofibroblastic phenotype showing increased contractility in three-dimensional collagen gels, rescued in versican (Vcan) heterozygotic animals | ||
| Adamts9 | Embryonic lethal at E7.5 days post coitum | [83] |
| Partial reduction of interdigital web regression, enhanced in Adamts5−/−;Adamts9−/+; bt/bt mice | [82] | |
| Abnormal cardiac development in Adamts9+/− mice | [80] | |
| Adamts12 | No phenotype unchallenged; Elevated tumor growth and angiogenesis | [104] |
| Exacerbated inflammation and airway dysfunction in allergen-induced airways disease | [149] | |
| More severe inflammation and delayed recovery following colitis, endotoxic sepsis and pancreatitis induction | [130] | |
| Adamts13 | Little phenotype unchallenged; loss of ADAMTS13 is pro-thrombotic but insufficient to generate thrombotic thromboscytopenic purpura | [137] |
| Adamts20 | Mutations in Adamts20 are found in belted (bt) mice, causing white spotting in the torso due to defective melanoblast survival | [83] |
| Partial reduction of interdigital web regression, enhanced in Adamts5−/−;Adamts9−/+; bt/bt mice | [82] | |
| Adamts9+/−;bt/bt mice have cleft palate | [86] |
Development
A principal developmental role for the aggrecanases/proteoglycanases is in the cleavage of the CSPG versican. Versican is an essential ECM component during embryogenesis as it gives rise to a loose, hydrated hyaluronan-rich matrix that provides structural support while allowing dynamic remodeling during morphogenesis. It influences the adhesion, migration and proliferation of many cell types; versican-null mice die around E10 because of cardiac defects [65]. Versican cleavage is regulated during development to dismantle transitional structures, but versican cleavage fragments are themselves bioactive factors that modulate diverse cell signaling pathways [66]. Cleavage in vivo liberates the amino-terminal 70-kDa G1 hyaluronan-binding domain with a DPEAAE neo-epitope at its carboxyl terminus, which has been termed ‘versikine’ [67]. Antibodies against DPEAAE have shown that ADAMTS1, 4, 5, 9, 15 and 20 are versicanases in various contexts [68–70], including cardiac development [71, 72], limb morphogenesis [73], palate formation [74], skin pigmentation [75], and myogenesis [76, 77].
Developmental defects in versican cleavage underpin the phenotypes displayed by mice that are deficient in Adamts1, 5, 9 and 20 (Table 2). During heart formation, the initially immature versican-rich ECM is replaced by a collagen, proteoglycan and elastin-containing matrix. Adamts9-null mice die prior to gastrulation, but hemizygous Adamts9+/− mice have heart malformations that reflect decreased detection of the DPEAAE neo-epitope and resulting accumulation of intact versican [71]. Adamts5−/− mice have enlarged heart valves by late fetal stages, correlating with reduced versican cleavage [72]. This phenotype was rescued by in vivo reduction of the level of versican by intercrossing with Vcan heterozygous mice. Thus, ADAMTS5 is required during heart development for the clearance of the early versican-rich matrix.
A similar requirement for removal of versican by ADAMTS5 is seen in skin development [78]. In this process, it is unnecessary to invoke a role for a neo-active versikine as partial depletion of versican restored normality. A dramatically different outcome is seen, however, in other morphogenetic events, including inter-digital web regression during autopod development, failure of which results in syndactyly, or webbing of the fingers and toes. Mice carrying combinations of null alleles for Adamts5, Adamts9 and Adamts20 (the latter gene is also known as the ‘belted’ locus, bt) show a failure of web regression, along with reduced versican cleavage and apoptosis [73]. Thus, the combined proteolytic activities of ADAMTS5, 9 and 20 are required to keep versican proteolysis above a threshold required for web regression. The strategy of reducing in vivo versican levels by heterozygosity resulted in 100 % penetrant syndactyly when combined with the absence of a single protease, ADAMTS20. In this context, therefore, the bioactivity of a versican cleavage fragment is probably required to promote web regression, a hypothesis that was confirmed by administration of recombinant G1-DPEAAE versikine. A similar outcome was observed with regard to palate formation [74]. Cleft palate occurs as a result of the failure of the two lateral palatal epithelial shelves to fuse and undergo epithelial-mesenchymal transition. Adamts9+/−;bt/bt mice show fully penetrant cleft palate, which is associated with decreased versican cleavage. Once again, versican heterozygosity exacerbated the phenotype seen with mice deficient in ADAMTS20 alone, suggesting that ADAMTS9 and 20 co-operate in the fusing palate. Functional co-operation of ADAMTS9 and 20 is also seen in their roles in melanoblast survival linked to coat pigmentation [75].
Versican cleavage by ADAMTS1 also underlies the roles of this enzyme in ovulation and cardiac development. Adamts1-null mice have extensive perinatal lethality, with surviving animals showing decreased growth and abnormalities in ureteral, adrenal and adipose tissues and infertility in female mice, indicating the importance of this enzyme in organogenesis and ovulation [79, 80]. ADAMTS1 expression by the granulosa cells of ovarian follicles is induced by progesterone, leading to follicle rupture by cleavage of the surrounding versican-rich matrix, which allows release of the oocyte [81, 82]. In Adamts1−/− mice, both ovulation and subsequent fertilization were severely impaired as a result of the persistence of versican. In cardiac development in mice, Adamts1 expression is repressed by a chromatin remodeling protein, Brg1, leading to an extracellular environment within the cardiac jelly that supports the formation of the myocardial trabeculae [83]. Subsequently, ADAMTS1 production is switched on to terminate trabeculation, which coincides with the disappearance of versican. The participation of several ADAMTS enzymes in cardiac development suggests that they may also be involved in cardiomyopathies and heart failure in humans.
Cardiovascular disease
In addition to the roles of versican in development, there is growing - and conflicting - evidence that this proteoglycan and its cleavage by ADAMTS enzymes may be associated with vascular pathologies [52]. It is present in atherosclerotic intimal thickenings and in advanced lesions, where it contributes to lipid retention and may influence the adhesion and recruitment of macrophages. In atherosclerotic lesions, ADAMTS1 is expressed by smooth muscle cells, whereas ADAMTS4 is a product of macrophages and its levels increase during lesion development [84]. Versican cleavage by ADAMTS1 could promote atherosclerosis through the activity of versikine in stimulating the migration of vascular smooth muscle cells (VSMCs) [85]. By contrast, ADAMTS5 has been suggested to be protective as it is depleted in atherosclerotic aortas, leading to accumulation of versican and biglycan [86]. ADAMTS7 is also linked to vascular disease because it promotes VSMC migration mediated by cleavage of COMP [87, 88]. Genome-wide association studies have identified ADAMTS16 as a candidate locus involved in inherited hypertension [89], and this candidacy is supported by targeted disruption of Adamts16 in a rat model [90].
Arthritis
The ‘aggrecanase’ moniker for the aggrecanase/proteoglycanase group of enzymes originated in the arthritis field. Osteoarthritis (OA) emerges as a result of a progressive loss of aggrecan from cartilage, leading to exposure of the collagen matrix and its breakdown by MMP13 [91]. The most significant aggrecan cleavage site for OA pathogenesis is located at a highly conserved sequence TEGE373↓374ARGS, at which MMPs do not cut. Antibodies that recognize the 374ARGS neo-epitope led to the discovery of aggrecanase-1, which proved to be ADAMTS4 [92] and aggrecanase-2, which is ADAMTS5 [93]. Subsequently, other related ADAMTS enzymes, including ADAMTS1, 8, 9 and 15, were shown to have aggrecanase activity [28, 36, 94–96]. ADAMTS16 and 18 are also weak aggrecanases [97].
Various lines of evidence indicate that ADAMTS4 and 5 are the principal enzymes involved in the pathogenesis of the arthritides [91, 98]. They are the major aggrecanases present in cartilage, though in vitro ADAMTS5 is about 1,000 times more potent than ADAMTS4 [99]. In human cartilage explants and chondrocytes, knockdown of either ADAMTS4 or 5 (but not ADAMTS1) attenuated aggrecan breakdown [100], suggesting that both enzymes may be involved in human tissues. Expression of these two enzymes is augmented by cytokines such as interleukin-1 and oncostatin-M, which provoke aggrecan breakdown in tissues [101]. In mice, however, ADAMTS5 alone is the critical enzyme, as Adamts5−/− mice show significantly reduced joint destruction when compared to wild-type or Adamts4−/− mice in surgical and allergen-induced models of arthritis (Table 2) [102, 103]. Other ADAMTSs may be physiologically relevant aggrecanases in tissues other than cartilage: for example, aggrecan cleavage co-localized with ADAMTS1 in the developing kidney and was reduced in Adamts1−/− embryos when compared to wild-type animals. Local cofactors such as Fibulin-1, which binds to ADAMTS1 and increases its aggrecanase activity, may be important in determining which enzyme has the principal activity in a particular tissue context [104]. Also, ancillary domain cleavages change enzyme activities: autolytic and MMP17-mediated processing of ADAMTS4 in its ancillary domain enhance its ability to cleave aggrecan [105, 106]. Thus, ADAMTS5 (and in humans ADAMTS4) is a target for therapeutic inhibition. Selective inhibitors are yielding promise, as shown by the protective effects of the aggrecanase-selective inhibitor AGG-523 in a rat joint-instability-induced model of arthritis [107].
Cancer
Numerous ADAMTS genes have been linked to cancer development and progression, with both promoting and antagonistic actions apparent. These dual roles probably reflect the effects of ADAMTS enzymes on the tumor microenvironment, affecting the interplay between malignant cells, the local stroma and the immune system. For many family members, the dominant theme is tumor suppression as they show epigenetic silencing (ADAMTS1, 8, 9, 12 and 18) or mutational inactivation (ADAMTS15) in several cancer types [48, 51, 108–114]. ADAMTS1, 3, 5, 8, 9, 10 and 18 are down-regulated in human breast carcinomas compared to normal tissue, while only ADAMTS4, 6, 14 and 20 are up-regulated [115].
Though originally reported for ADAMTS1 and 8, many ADAMTSs have proved to be anti-angiogenic, a property that may contribute to their tumor-suppressive actions [6, 116–121]. Metalloproteinase-dependent and -independent activities have been observed, with ADAMTS1 displaying both types of mechanism [122]. This enzyme cleaves matrix-bound thrombospondins-1 and −2, generating bioactive anti-angiogenic fragments [123], and also sequesters angiogenic factors such as the vascular endothelial growth factor VEGF165 [124] and basic fibroblast growth factor [125]. Metalloproteinase-independent inhibition of neovascularization has also been seen for ADAMTS2 [116], ADAMTS4 [118], ADAMTS5 [120] and ADAMTS12 [117]. The TSR domain may be responsible for these protease-independent activities since peptides (termed ‘Adamtsostatins’) corresponding to the TSRs of several family members are anti-angiogenic, indicating that this may be a general activity of the ADAMTSs [121, 126]. Metalloproteinase-dependent anti-angiogenic activity is displayed by ADAMTS9, but the mechanism is as yet unknown as thrombospondin cleavage has been ruled out [119]. The identities of the cryptic angio-inhibitory substrates of ADAMTS9 and ADAMTS15 [96] will be important to elucidate.
That ADAMTSs can have both pro- and anti-tumorigenic functions, depending on the cancer and its context, is best illustrated by ADAMTS1, which is down-regulated in breast, primary head and neck carcinomas, and epigenetically silenced in around 85 % of colorectal cancer cell lines [115, 127, 128]. Nevertheless, it appears to promote metastasis where its expression is activated later in tumor progression [122, 127, 129]. Its tumor-promoting action dominates in the highly aggressive MMTV-PyMT mouse mammary cancer model: Adamts1−/−;PyMT mice show reduced tumor growth and metastasis as a result of increased tumor cell apoptosis, while the cell proliferation and vascularity of their tumors were unaffected [130]. Tumors in mice displayed reduced cleaved versican and increased presence of cytotoxic immune cells, suggesting that ADAMTS1 creates a tumor-promoting microenvironment in the mammary gland. This may involve the shedding of epidermal growth factor receptor (EGFR) ligands, which has been linked to promotion of metastasis in other syngeneic mouse models [131] and to the collaboration of ADAMTS1 with MMP-1 to promote bone metastasis of human xenografts [132]. Other mechanisms that potentially contribute to the metastasis-enhancing effects of ADAMTS1 include promotion of cell migration by cleavage of Syndecan 4 and Semaphorin 3C [133, 134], and induction of endothelial mimicry by melanoma and sarcoma cells [135].
Tumor-suppressive effects of the ADAMTSs have been linked to the deactivation of key proliferation or survival signaling pathways, including suppression of Erk signaling by ADAMTS8 [136], ADAMTS12 [137] and ADAMTS15 [113], and of Akt/mTOR activity by ADAMTS9 [138]. Recently, ADAMTS15 was shown to inhibit breast cancer cell migration independently of its catalytic activity by increasing cell surface syndecan-4 levels [96].
Frontiers
We are still short of understanding the functions of the ADAMTS family in development and disease. This truth is patent for the orphan enzymes for which substrates have yet to be discovered. Even for those ADAMTS (such as the aggrecanases) for which we have some knowledge of (some of) their substrates and the importance of their cleavage, it is likely that we will find new specialized roles in particular tissue contexts linked to the proteolysis of novel targets. There is a need, therefore, to apply proteomics technologies to unravel the ADAMTS substrate degradomes, working at the levels of both cells and intact tissues [139]. Building on this base, the identification of extracellular bioactive fragments that are generated by ADAMTSs’ action and investigation of the functions of these fragments will be priorities, along with determining the ways in which cofactors such as the fibulins [104, 140] influence binding and cleavage preferences.
Gene knockout mice have determined the developmental roles of certain ADAMTS family members, though coverage is incomplete [4]. The early lethality in Adamts9−/− mice indicates an essential developmental function for ADAMTS9 but restricts investigations to the study of heterozygous animals. Now a new conditional Adamts9 model has been generated and a limb-specific knockout has confirmed an essential role for ADAMTS9 in interdigital web regression [141]. The generation of conditional knockout strains for other Adamts genes to chart their involvement in morphogenesis and disease will be essential.
At the protein level, there are many gaps in our knowledge of ADAMTS structure. In particular, we need to know how features within the pro- and ancillary domains of these enzymes conspire with the distinctive attributes of the ADAMTS catalytic domains to control enzyme localization, activation, trafficking, substrate binding, cleavage, inhibition and turnover. This knowledge will also prove useful in the design of novel therapeutics. Active-site-directed inhibitors that are selective for ADAMTSs over other metalloproteinases are a reality, but it may be difficult to build in an ability to discriminate between ADAMTS family members. Thus, although ADAMTS4 and ADAMTS5 inhibitors could be useful anti-arthritics, they may have unwanted toxicities if they also hit the pro-collagen-N-proteinases, for example. We would also need to know whether such agents have longer-term consequences, for instance in vascular function and angiogenesis. There is, however, potential to build on knowledge of exosite interactions to develop novel inhibitors that may block cleavage of specific substrates, while leaving other catalytic actions of the targeted enzyme unaltered.
Finally, an area that has not been covered extensively in this review is the regulation of ADAMTS expression, which is important for understanding disease pathogenesis. There is also the possibility of prevention of disease, potentially through dietary consumption of protective phytochemicals that down-tune ADAMTS expression [142, 143]. At the post-transcriptional level, our understanding of the functions of small RNAs as master regulators of gene systems that include the ADAMTSs is now advancing rapidly. Finally, given the roles of ADAMTSs in human diseases, it is likely that genetic polymorphisms that affect either their transcriptional or post-transcriptional control will be revealed.
Acknowledgements
We wish to thank the Breast Cancer Campaign, Cancer Research-UK and Big C for their support over many years. This work was also facilitated by funding from the European Union Framework Programmes 6 (Cancerdegradome) and FP7 (SaveMe) to DRE and DevCom (a Marie Curie Initial Training Networks action, 607142) to GNW and ID. We also thank Norfolk Fundraisers and Mrs Margaret Doggett for their help, and members of the Cellular Protease Group at the University of East Anglia for their ongoing encouragement.
Abbreviations
- ADAM
A Disintegrin and Metalloproteinases
- ADAMTS
A Disintegrin and Metalloproteinase with Thrombospondin motifs
- ADAMTSL
ADAMTS-like gene
- COMP
Cartilage oligomeric protein
- CSPG
Chondroitin sulfate proteoglycan
- ECM
Extracellular matrix
- EGF
Epidermal growth factor
- LRP-1
Low-density lipoprotein-related protein-1
- MMP
Matrix metalloproteinase
- OA
Osteoarthritis
- PPC
Pro-protein convertase
- TIMP
Tissue inhibitors of metalloproteinases
- TSR
Thrombospondin type 1 sequence repeat
- TTP
Thrombotic thrombocytopenic purpura
- VSMC
Vascular smooth muscle cell
- vWF
von-Willebrand factor
- vWFCP
von-Willebrand-factor-cleaving protease
- WMS
Weill-Marchesani syndrome
Footnotes
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Richard Kelwick, Email: r.kelwick@imperial.ac.uk.
Ines Desanlis, Email: i.desanlis@uea.ac.uk.
Grant N Wheeler, Email: grant.wheeler@uea.ac.uk.
Dylan R Edwards, Email: dylan.edwards@uea.ac.uk.
References
- 1.Porter S, Clark IM, Kevorkian L, Edwards DR. The ADAMTS metalloproteinases. Biochem J. 2005;386:15–27. doi: 10.1042/BJ20040424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Apte SS. A disintegrin-like and metalloprotease (reprolysin-type) with thrombospondin type 1 motif (ADAMTS) superfamily: functions and mechanisms. J Biol Chem. 2009;284:31493–7. doi: 10.1074/jbc.R109.052340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gomis-Ruth FX. Catalytic domain architecture of metzincin metalloproteases. J Biol Chem. 2009;284:15353–7. doi: 10.1074/jbc.R800069200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubail J, Apte SS. Insights on ADAMTS proteases and ADAMTS-like proteins from mammalian genetics. Matrix Biol. 2015. doi: 10.1016/j.matbio.2015.03.001. [DOI] [PubMed]
- 5.Stanton H, Melrose J, Little CB, Fosang AJ. Proteoglycan degradation by the ADAMTS family of proteinases. Biochim Biophys Acta. 1812;2011:1616–29. doi: 10.1016/j.bbadis.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 6.Vazquez F, Hastings G, Ortega MA, Lane TF, Oikemus S, Lombardo M, et al. METH-1, a human ortholog of ADAMTS-1, and METH-2 are members of a new family of proteins with angio-inhibitory activity. J Biol Chem. 1999;274:23349–57. doi: 10.1074/jbc.274.33.23349. [DOI] [PubMed] [Google Scholar]
- 7.Colige A, Vandenberghe I, Thiry M, Lambert CA, Van Beeumen J, Li SW, et al. Cloning and characterization of ADAMTS-14, a novel ADAMTS displaying high homology with ADAMTS-2 and ADAMTS-3. J Biol Chem. 2002;277:5756–66. doi: 10.1074/jbc.M105601200. [DOI] [PubMed] [Google Scholar]
- 8.Fujikawa K, Suzuki H, McMullen B, Chung D. Purification of human von Willebrand factor-cleaving protease and its identification as a new member of the metalloproteinase family. Blood. 2001;98:1662–6. doi: 10.1182/blood.v98.6.1662. [DOI] [PubMed] [Google Scholar]
- 9.Liu CJ, Kong W, Ilalov K, Yu S, Xu K, Prazak L, et al. ADAMTS-7: a metalloproteinase that directly binds to and degrades cartilage oligomeric matrix protein. FASEB J. 2006;20:988–90. doi: 10.1096/fj.05-3877fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu CJ, Kong W, Xu K, Luan Y, Ilalov K, Sehgal B, et al. ADAMTS-12 associates with and degrades cartilage oligomeric matrix protein. J Biol Chem. 2006;281:15800–8. doi: 10.1074/jbc.M513433200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Somerville RP, Longpre JM, Apel ED, Lewis RM, Wang LW, Sanes JR, et al. ADAMTS7B, the full-length product of the ADAMTS7 gene, is a chondroitin sulfate proteoglycan containing a mucin domain. J Biol Chem. 2004;279:35159–75. doi: 10.1074/jbc.M402380200. [DOI] [PubMed] [Google Scholar]
- 12.Huxley-Jones J, Apte SS, Robertson DL, Boot-Handford RP. The characterisation of six ADAMTS proteases in the basal chordate Ciona intestinalis provides new insights into the vertebrate ADAMTS family. Int J Biochem Cell Biol. 2005;37:1838–45. doi: 10.1016/j.biocel.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 13.Nicholson AC, Malik SB, Logsdon JM, Jr, Van Meir EG. Functional evolution of ADAMTS genes. evidence from analyses of phylogeny and gene organization. BMC Evol Biol. 2005;5:11. doi: 10.1186/1471-2148-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Angerer L, Hussain S, Wei Z, Livingston BT. Sea urchin metalloproteases: a genomic survey of the BMP-1/tolloid-like. MMP and ADAM families. Dev Biol. 2006;300:267–81. doi: 10.1016/j.ydbio.2006.07.046. [DOI] [PubMed] [Google Scholar]
- 15.Brocker CN, Vasiliou V, Nebert DW. Evolutionary divergence and functions of the ADAM and ADAMTS gene families. Hum Genomics. 2009;4:43–55. doi: 10.1186/1479-7364-4-1-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brunet FG, Fraser FW, Binder MJ, Smith AD, Kintakas C, Dancevic CM, et al. The evolutionary conservation of the A Disintegrin-like and Metalloproteinase domain with Thrombospondin-1 motif metzincins across vertebrate species and their expression in teleost zebrafish. BMC Evol Biol. 2015;15:281. doi: 10.1186/s12862-015-0281-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huxley-Jones J, Robertson DL, Boot-Handford RP. On the origins of the extracellular matrix in vertebrates. Matrix Biol. 2007;26:2–11. doi: 10.1016/j.matbio.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez-Manzaneque JC, Milchanowski AB, Dufour EK, Leduc R, Iruela-Arispe ML. Characterization of METH-1/ADAMTS1 processing reveals two distinct active forms. J Biol Chem. 2000;275:33471–9. doi: 10.1074/jbc.M002599200. [DOI] [PubMed] [Google Scholar]
- 19.Wang P, Tortorella M, England K, Malfait AM, Thomas G, Arner EC, et al. Proprotein convertase furin interacts with and cleaves pro-ADAMTS4 (Aggrecanase-1) in the trans-Golgi network. J Biol Chem. 2004;279:15434–40. doi: 10.1074/jbc.M312797200. [DOI] [PubMed] [Google Scholar]
- 20.Longpre JM, McCulloch DR, Koo BH, Alexander JP, Apte SS, Leduc R. Characterization of proADAMTS5 processing by proprotein convertases. Int J Biochem Cell Biol. 2009;41:1116–26. doi: 10.1016/j.biocel.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 21.Koo BH, Longpre JM, Somerville RP, Alexander JP, Leduc R, Apte SS. Regulation of ADAMTS9 secretion and enzymatic activity by its propeptide. J Biol Chem. 2007;282:16146–54. doi: 10.1074/jbc.M610161200. [DOI] [PubMed] [Google Scholar]
- 22.Majerus EM, Zheng X, Tuley EA, Sadler JE. Cleavage of the ADAMTS13 propeptide is not required for protease activity. J Biol Chem. 2003;278:46643–8. doi: 10.1074/jbc.M309872200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Somerville RP, Longpre JM, Jungers KA, Engle JM, Ross M, Evanko S, et al. Characterization of ADAMTS-9 and ADAMTS-20 as a distinct ADAMTS subfamily related to Caenorhabditis elegans GON-1. J Biol Chem. 2003;278:9503–13. doi: 10.1074/jbc.M211009200. [DOI] [PubMed] [Google Scholar]
- 24.Edwards DR, Handsley MM, Pennington CJ. The ADAM metalloproteinases. Mol Aspects Med. 2008;29:258–89. doi: 10.1016/j.mam.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomis-Ruth FX. Structural aspects of the metzincin clan of metalloendopeptidases. Mol Biotechnol. 2003;24:157–202. doi: 10.1385/MB:24:2:157. [DOI] [PubMed] [Google Scholar]
- 26.Gerhardt S, Hassall G, Hawtin P, McCall E, Flavell L, Minshull C, et al. Crystal structures of human ADAMTS-1 reveal a conserved catalytic domain and a disintegrin-like domain with a fold homologous to cysteine-rich domains. J Mol Biol. 2007;373:891–902. doi: 10.1016/j.jmb.2007.07.047. [DOI] [PubMed] [Google Scholar]
- 27.Mosyak L, Georgiadis K, Shane T, Svenson K, Hebert T, McDonagh T, et al. Crystal structures of the two major aggrecan degrading enzymes, ADAMTS4 and ADAMTS5. Protein Sci. 2008;17:16–21. doi: 10.1110/ps.073287008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adams JC, Lawler J. The thrombospondins. Cold Spring Harb Perspect Biol. 2011;3:a009712. doi: 10.1101/cshperspect.a009712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hofsteenge J, Huwiler KG, Macek B, Hess D, Lawler J, Mosher DF, et al. C-mannosylation and O-fucosylation of the thrombospondin type 1 module. J Biol Chem. 2001;276:6485–98. doi: 10.1074/jbc.M008073200. [DOI] [PubMed] [Google Scholar]
- 30.Ricketts LM, Dlugosz M, Luther KB, Haltiwanger RS, Majerus EM. O-fucosylation is required for ADAMTS13 secretion. J Biol Chem. 2007;282:17014–23. doi: 10.1074/jbc.M700317200. [DOI] [PubMed] [Google Scholar]
- 31.Gao G, Westling J, Thompson VP, Howell TD, Gottschall PE, Sandy JD. Activation of the proteolytic activity of ADAMTS4 (aggrecanase-1) by C-terminal truncation. J Biol Chem. 2002;277:11034–41. doi: 10.1074/jbc.M107443200. [DOI] [PubMed] [Google Scholar]
- 32.Kashiwagi M, Enghild JJ, Gendron C, Hughes C, Caterson B, Itoh Y, et al. Altered proteolytic activities of ADAMTS-4 expressed by C-terminal processing. J Biol Chem. 2004;279:10109–19. doi: 10.1074/jbc.M312123200. [DOI] [PubMed] [Google Scholar]
- 33.Murphy G. Tissue inhibitors of metalloproteinases. Genome Biol. 2011;12:233. doi: 10.1186/gb-2011-12-11-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang WM, Ge G, Lim NH, Nagase H, Greenspan DS. TIMP-3 inhibits the procollagen N-proteinase ADAMTS-2. Biochem J. 2006;398:515–9. doi: 10.1042/BJ20060630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hashimoto G, Aoki T, Nakamura H, Tanzawa K, Okada Y. Inhibition of ADAMTS4 (aggrecanase-1) by tissue inhibitors of metalloproteinases (TIMP-1, 2, 3 and 4) FEBS Lett. 2001;494:192–5. doi: 10.1016/s0014-5793(01)02323-7. [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez-Manzaneque JC, Westling J, Thai SN, Luque A, Knauper V, Murphy G, et al. ADAMTS1 cleaves aggrecan at multiple sites and is differentially inhibited by metalloproteinase inhibitors. Biochem Biophys Res Commun. 2002;293:501–8. doi: 10.1016/S0006-291X(02)00254-1. [DOI] [PubMed] [Google Scholar]
- 37.Troeberg L, Fushimi K, Scilabra SD, Nakamura H, Dive V, Thogersen IB, et al. The C-terminal domains of ADAMTS-4 and ADAMTS-5 promote association with N-TIMP-3. Matrix Biol. 2009;28:463–9. doi: 10.1016/j.matbio.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wayne GJ, Deng SJ, Amour A, Borman S, Matico R, Carter HL, et al. TIMP-3 inhibition of ADAMTS-4 (Aggrecanase-1) is modulated by interactions between aggrecan and the C-terminal domain of ADAMTS-4. J Biol Chem. 2007;282:20991–8. doi: 10.1074/jbc.M610721200. [DOI] [PubMed] [Google Scholar]
- 39.Lim NH, Kashiwagi M, Visse R, Jones J, Enghild JJ, Brew K, et al. Reactive-site mutants of N-TIMP-3 that selectively inhibit ADAMTS-4 and ADAMTS-5: biological and structural implications. Biochem J. 2010;431:113–22. doi: 10.1042/BJ20100725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tortorella MD, Tomasselli AG, Mathis KJ, Schnute ME, Woodard SS, Munie G, et al. Structural and inhibition analysis reveals the mechanism of selectivity of a series of aggrecanase inhibitors. J Biol Chem. 2009;284:24185–91. doi: 10.1074/jbc.M109.029116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nusgens BV, Verellen-Dumoulin C, Hermanns-Le T, De Paepe A, Nuytinck L, Pierard GE, et al. Evidence for a relationship between Ehlers-Danlos type VII C in humans and bovine dermatosparaxis. Nat Genet. 1992;1:214–7. doi: 10.1038/ng0692-214. [DOI] [PubMed] [Google Scholar]
- 42.Guo F, Lai Y, Tian Q, Lin EA, Kong L, Liu C. Granulin-epithelin precursor binds directly to ADAMTS-7 and ADAMTS-12 and inhibits their degradation of cartilage oligomeric matrix protein. Arthritis Rheum. 2010;62:2023–36. doi: 10.1002/art.27491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamamoto K, Troeberg L, Scilabra SD, Pelosi M, Murphy CL, Strickland DK, et al. LRP-1-mediated endocytosis regulates extracellular activity of ADAMTS-5 in articular cartilage. FASEB J. 2013;27:511–21. doi: 10.1096/fj.12-216671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamamoto K, Owen K, Parker AE, Scilabra SD, Dudhia J, Strickland DK, et al. Low density lipoprotein receptor-related protein 1 (LRP1)-mediated endocytic clearance of a disintegrin and metalloproteinase with thrombospondin motifs-4 (ADAMTS-4): functional differences of non-catalytic domains of ADAMTS-4 and ADAMTS-5 in LRP1 binding. J Biol Chem. 2014;289:6462–74. doi: 10.1074/jbc.M113.545376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Troeberg L, Lazenbatt C, Anower EKMF, Freeman C, Federov O, Habuchi H, et al. Sulfated glycosaminoglycans control the extracellular trafficking and the activity of the metalloprotease inhibitor TIMP-3. Chem Biol. 2014;21:1300–9. doi: 10.1016/j.chembiol.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin EA, Liu CJ. The role of ADAMTSs in arthritis. Protein Cell. 2010;1:33–47. doi: 10.1007/s13238-010-0002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rocks N, Paulissen G, El Hour M, Quesada F, Crahay C, Gueders M, et al. Emerging roles of ADAM and ADAMTS metalloproteinases in cancer. Biochimie. 2008;90:369–79. doi: 10.1016/j.biochi.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 48.Wagstaff L, Kelwick R, Decock J, Edwards DR. The roles of ADAMTS metalloproteinases in tumorigenesis and metastasis. Front Biosci (Landmark Ed) 2011;16:1861–72. doi: 10.2741/3827. [DOI] [PubMed] [Google Scholar]
- 49.Kumar S, Rao N, Ge R. Emerging roles of ADAMTSs in angiogenesis and cancer. Cancers (Basel) 2012;4:1252–99. doi: 10.3390/cancers4041252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Noel A, Gutierrez-Fernandez A, Sounni NE, Behrendt N, Maquoi E, Lund IK, et al. New and paradoxical roles of matrix metalloproteinases in the tumor microenvironment. Front Pharmacol. 2012;3:140. doi: 10.3389/fphar.2012.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cal S, Lopez-Otin C. ADAMTS proteases and cancer. Matrix Biol. 2015. doi: 10.1016/j.matbio.2015.01.013. [DOI] [PubMed]
- 52.Salter RC, Ashlin TG, Kwan AP, Ramji DP. ADAMTS proteases: key roles in atherosclerosis? J Mol Med (Berl) 2010;88:1203–11. doi: 10.1007/s00109-010-0654-x. [DOI] [PubMed] [Google Scholar]
- 53.Gottschall PE, Howell MD. ADAMTS expression and function in central nervous system injury and disorders. Matrix Biol. 2015. doi:10.1016/j.matbio.2015.01.014. [DOI] [PMC free article] [PubMed]
- 54.Levy GG, Nichols WC, Lian EC, Foroud T, McClintick JN, McGee BM, et al. Mutations in a member of the ADAMTS gene family cause thrombotic thrombocytopenic purpura. Nature. 2001;413:488–94. doi: 10.1038/35097008. [DOI] [PubMed] [Google Scholar]
- 55.Zheng XL. ADAMTS13 and von Willebrand factor in thrombotic thrombocytopenic purpura. Annu Rev Med. 2015;66:211–25. doi: 10.1146/annurev-med-061813-013241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Faivre L, Gorlin RJ, Wirtz MK, Godfrey M, Dagoneau N, Samples JR, et al. In frame fibrillin-1 gene deletion in autosomal dominant Weill-Marchesani syndrome. J Med Genet. 2003;40:34–6. doi: 10.1136/jmg.40.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dagoneau N, Benoist-Lasselin C, Huber C, Faivre L, Megarbane A, Alswaid A, et al. ADAMTS10 mutations in autosomal recessive Weill-Marchesani syndrome. Am J Hum Genet. 2004;75:801–6. doi: 10.1086/425231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kutz WE, Wang LW, Bader HL, Majors AK, Iwata K, Traboulsi EI, et al. ADAMTS10 protein interacts with fibrillin-1 and promotes its deposition in extracellular matrix of cultured fibroblasts. J Biol Chem. 2011;286:17156–67. doi: 10.1074/jbc.M111.231571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morales J, Al-Sharif L, Khalil DS, Shinwari JM, Bavi P, Al-Mahrouqi RA, et al. Homozygous mutations in ADAMTS10 and ADAMTS17 cause lenticular myopia, ectopia lentis, glaucoma, spherophakia, and short stature. Am J Hum Genet. 2009;85:558–68. doi: 10.1016/j.ajhg.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aldahmesh MA, Khan AO, Mohamed JY, Alkuraya H, Ahmed H, Bobis S, et al. Identification of ADAMTS18 as a gene mutated in Knobloch syndrome. J Med Genet. 2011;48:597–601. doi: 10.1136/jmedgenet-2011-100306. [DOI] [PubMed] [Google Scholar]
- 61.Peluso I, Conte I, Testa F, Dharmalingam G, Pizzo M, Collin RW, et al. The ADAMTS18 gene is responsible for autosomal recessive early onset severe retinal dystrophy. Orphanet J Rare Dis. 2013;8:16. doi: 10.1186/1750-1172-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jacobi CL, Rudigier LJ, Scholz H, Kirschner KM. Transcriptional regulation by the Wilms tumor protein, Wt1, suggests a role of the metalloproteinase Adamts16 in murine genitourinary development. J Biol Chem. 2013;288:18811–24. doi: 10.1074/jbc.M113.464644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abdul-Majeed S, Mell B, Nauli SM, Joe B. Cryptorchidism and infertility in rats with targeted disruption of the adamts16 locus. PLoS One. 2014;9:e100967. doi: 10.1371/journal.pone.0100967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pyun JA, Kim S, Kwack K. Interaction between thyroglobulin and ADAMTS16 in premature ovarian failure. Clin Exp Reprod Med. 2014;41:120–4. doi: 10.5653/cerm.2014.41.3.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mjaatvedt CH, Yamamura H, Capehart AA, Turner D, Markwald RR. The Cspg2 gene, disrupted in the hdf mutant, is required for right cardiac chamber and endocardial cushion formation. Dev Biol. 1998;202:56–66. doi: 10.1006/dbio.1998.9001. [DOI] [PubMed] [Google Scholar]
- 66.Wight TN. Versican - a versatile extracellular matrix proteoglycan in cell biology. Curr Opin Cell Biol. 2002;14:617–23. doi: 10.1016/s0955-0674(02)00375-7. [DOI] [PubMed] [Google Scholar]
- 67.Nandadasa S, Foulcer S, Apte SS. The multiple, complex roles of versican and its proteolytic turnover by ADAMTS proteases during embryogenesis. Matrix Biol. 2014;35:34–41. doi: 10.1016/j.matbio.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sandy JD, Westling J, Kenagy RD, Iruela-Arispe ML, Verscharen C, Rodriguez-Mazaneque JC, et al. Versican V1 proteolysis in human aorta in vivo occurs at the Glu441-Ala442 bond, a site that is cleaved by recombinant ADAMTS-1 and ADAMTS-4. J Biol Chem. 2001;276:13372–8. doi: 10.1074/jbc.M009737200. [DOI] [PubMed] [Google Scholar]
- 69.Russell DL, Doyle KM, Ochsner SA, Sandy JD, Richards JS. Processing and localization of ADAMTS-1 and proteolytic cleavage of versican during cumulus matrix expansion and ovulation. J Biol Chem. 2003;278:42330–9. doi: 10.1074/jbc.M300519200. [DOI] [PubMed] [Google Scholar]
- 70.Kintakas C, McCulloch DR. Emerging roles for ADAMTS5 during development and disease. Matrix Biol. 2011;30:311–7. doi: 10.1016/j.matbio.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 71.Kern CB, Wessels A, McGarity J, Dixon LJ, Alston E, Argraves WS, et al. Reduced versican cleavage due to Adamts9 haploinsufficiency is associated with cardiac and aortic anomalies. Matrix Biol. 2010;29:304–16. doi: 10.1016/j.matbio.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dupuis LE, McCulloch DR, McGarity JD, Bahan A, Wessels A, Weber D, et al. Altered versican cleavage in ADAMTS5 deficient mice; a novel etiology of myxomatous valve disease. Dev Biol. 2011;357:152–64. doi: 10.1016/j.ydbio.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McCulloch DR, Nelson CM, Dixon LJ, Silver DL, Wylie JD, Lindner V, et al. ADAMTS metalloproteases generate active versican fragments that regulate interdigital web regression. Dev Cell. 2009;17:687–98. doi: 10.1016/j.devcel.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Enomoto H, Nelson CM, Somerville RP, Mielke K, Dixon LJ, Powell K, et al. Cooperation of two ADAMTS metalloproteases in closure of the mouse palate identifies a requirement for versican proteolysis in regulating palatal mesenchyme proliferation. Development. 2010;137:4029–38. doi: 10.1242/dev.050591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Silver DL, Hou L, Somerville R, Young ME, Apte SS, Pavan WJ. The secreted metalloprotease ADAMTS20 is required for melanoblast survival. PLoS Genet. 2008;4:e1000003. doi: 10.1371/journal.pgen.1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stupka N, Kintakas C, White JD, Fraser FW, Hanciu M, Aramaki-Hattori N, et al. Versican processing by a disintegrin-like and metalloproteinase domain with thrombospondin-1 repeats proteinases-5 and −15 facilitates myoblast fusion. J Biol Chem. 2013;288:1907–17. doi: 10.1074/jbc.M112.429647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dancevic CM, Fraser FW, Smith AD, Stupka N, Ward AC, McCulloch DR. Biosynthesis and expression of a disintegrin-like and metalloproteinase domain with thrombospondin-1 repeats-15: a novel versican-cleaving proteoglycanase. J Biol Chem. 2013;288:37267–76. doi: 10.1074/jbc.M112.418624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hattori N, Carrino DA, Lauer ME, Vasanji A, Wylie JD, Nelson CM, et al. Pericellular versican regulates the fibroblast-myofibroblast transition: a role for ADAMTS5 protease-mediated proteolysis. J Biol Chem. 2011;286:34298–310. doi: 10.1074/jbc.M111.254938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shindo T, Kurihara H, Kuno K, Yokoyama H, Wada T, Kurihara Y, et al. ADAMTS-1: a metalloproteinase-disintegrin essential for normal growth, fertility, and organ morphology and function. J Clin Invest. 2000;105:1345–52. doi: 10.1172/JCI8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mittaz L, Russell DL, Wilson T, Brasted M, Tkalcevic J, Salamonsen LA, et al. Adamts-1 is essential for the development and function of the urogenital system. Biol Reprod. 2004;70:1096–105. doi: 10.1095/biolreprod.103.023911. [DOI] [PubMed] [Google Scholar]
- 81.Robker RL, Russell DL, Espey LL, Lydon JP, O’Malley BW, Richards JS. Progesterone-regulated genes in the ovulation process: ADAMTS-1 and cathepsin L proteases. Proc Natl Acad Sci U S A. 2000;97:4689–94. doi: 10.1073/pnas.080073497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Brown HM, Dunning KR, Robker RL, Boerboom D, Pritchard M, Lane M, et al. ADAMTS1 cleavage of versican mediates essential structural remodeling of the ovarian follicle and cumulus-oocyte matrix during ovulation in mice. Biol Reprod. 2010;83:549–57. doi: 10.1095/biolreprod.110.084434. [DOI] [PubMed] [Google Scholar]
- 83.Stankunas K, Hang CT, Tsun ZY, Chen H, Lee NV, Wu JI, et al. Endocardial Brg1 represses ADAMTS1 to maintain the microenvironment for myocardial morphogenesis. Dev Cell. 2008;14:298–311. doi: 10.1016/j.devcel.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wagsater D, Bjork H, Zhu C, Bjorkegren J, Valen G, Hamsten A, et al. ADAMTS-4 and −8 are inflammatory regulated enzymes expressed in macrophage-rich areas of human atherosclerotic plaques. Atherosclerosis. 2008;196:514–22. doi: 10.1016/j.atherosclerosis.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 85.Jonsson-Rylander AC, Nilsson T, Fritsche-Danielson R, Hammarstrom A, Behrendt M, Andersson JO, et al. Role of ADAMTS-1 in atherosclerosis: remodeling of carotid artery, immunohistochemistry, and proteolysis of versican. Arterioscler Thromb Vasc Biol. 2005;25:180–5. doi: 10.1161/01.ATV.0000150045.27127.37. [DOI] [PubMed] [Google Scholar]
- 86.Didangelos A, Mayr U, Monaco C, Mayr M. Novel role of ADAMTS-5 protein in proteoglycan turnover and lipoprotein retention in atherosclerosis. J Biol Chem. 2012;287:19341–5. doi: 10.1074/jbc.C112.350785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Patel RS, Ye S. ADAMTS7: a promising new therapeutic target in coronary heart disease. Expert Opin Ther Targets. 2013;17:863–7. doi: 10.1517/14728222.2013.816287. [DOI] [PubMed] [Google Scholar]
- 88.Pu X, Xiao Q, Kiechl S, Chan K, Ng FL, Gor S, et al. ADAMTS7 cleavage and vascular smooth muscle cell migration is affected by a coronary-artery-disease-associated variant. Am J Hum Genet. 2013;92:366–74. doi: 10.1016/j.ajhg.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Joe B, Saad Y, Dhindaw S, Lee NH, Frank BC, Achinike OH, et al. Positional identification of variants of Adamts16 linked to inherited hypertension. Hum Mol Genet. 2009;18:2825–38. doi: 10.1093/hmg/ddp218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gopalakrishnan K, Kumarasamy S, Abdul-Majeed S, Kalinoski AL, Morgan EE, Gohara AF, et al. Targeted disruption of Adamts16 gene in a rat genetic model of hypertension. Proc Natl Acad Sci U S A. 2012;109:20555–9. doi: 10.1073/pnas.1211290109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Troeberg L, Nagase H. Proteases involved in cartilage matrix degradation in osteoarthritis. Biochim Biophys Acta. 1824;2012:133–45. doi: 10.1016/j.bbapap.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Arner EC, Pratta MA, Trzaskos JM, Decicco CP, Tortorella MD. Generation and characterization of aggrecanase. A soluble, cartilage-derived aggrecan-degrading activity. J Biol Chem. 1999;274:6594–601. doi: 10.1074/jbc.274.10.6594. [DOI] [PubMed] [Google Scholar]
- 93.Abbaszade I, Liu RQ, Yang F, Rosenfeld SA, Ross OH, Link JR, et al. Cloning and characterization of ADAMTS11, an aggrecanase from the ADAMTS family. J Biol Chem. 1999;274:23443–50. doi: 10.1074/jbc.274.33.23443. [DOI] [PubMed] [Google Scholar]
- 94.Kuno K, Okada Y, Kawashima H, Nakamura H, Miyasaka M, Ohno H, et al. ADAMTS-1 cleaves a cartilage proteoglycan, aggrecan. FEBS Lett. 2000;478:241–5. doi: 10.1016/s0014-5793(00)01854-8. [DOI] [PubMed] [Google Scholar]
- 95.Collins-Racie LA, Flannery CR, Zeng W, Corcoran C, Annis-Freeman B, Agostino MJ, et al. ADAMTS-8 exhibits aggrecanase activity and is expressed in human articular cartilage. Matrix Biol. 2004;23:219–30. doi: 10.1016/j.matbio.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 96.Kelwick R, Wagstaff L, Decock J, Roghi C, Cooley LS, Robinson SD, et al. Metalloproteinase-dependent and -independent processes contribute to inhibition of breast cancer cell migration, angiogenesis and liver metastasis by a disintegrin and metalloproteinase with thrombospondin motifs-15. Int J Cancer. 2015;136:E14–26. doi: 10.1002/ijc.29129. [DOI] [PubMed] [Google Scholar]
- 97.Zeng W, Corcoran C, Collins-Racie LA, Lavallie ER, Morris EA, Flannery CR. Glycosaminoglycan-binding properties and aggrecanase activities of truncated ADAMTSs: comparative analyses with ADAMTS-5, −9, −16 and −18. Biochim Biophys Acta. 2006;1760:517–24. doi: 10.1016/j.bbagen.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 98.Fosang AJ, Little CB. Drug insight: aggrecanases as therapeutic targets for osteoarthritis. Nat Clin Pract Rheumatol. 2008;4:420–7. doi: 10.1038/ncprheum0841. [DOI] [PubMed] [Google Scholar]
- 99.Gendron C, Kashiwagi M, Lim NH, Enghild JJ, Thogersen IB, Hughes C, et al. Proteolytic activities of human ADAMTS-5: comparative studies with ADAMTS-4. J Biol Chem. 2007;282:18294–306. doi: 10.1074/jbc.M701523200. [DOI] [PubMed] [Google Scholar]
- 100.Song RH, Tortorella MD, Malfait AM, Alston JT, Yang Z, Arner EC, et al. Aggrecan degradation in human articular cartilage explants is mediated by both ADAMTS-4 and ADAMTS-5. Arthritis Rheum. 2007;56:575–85. doi: 10.1002/art.22334. [DOI] [PubMed] [Google Scholar]
- 101.Koshy PJ, Lundy CJ, Rowan AD, Porter S, Edwards DR, Hogan A, et al. The modulation of matrix metalloproteinase and ADAM gene expression in human chondrocytes by interleukin-1 and oncostatin M: a time-course study using real-time quantitative reverse transcription-polymerase chain reaction. Arthritis Rheum. 2002;46:961–7. doi: 10.1002/art.10212. [DOI] [PubMed] [Google Scholar]
- 102.Glasson SS, Askew R, Sheppard B, Carito B, Blanchet T, Ma HL, et al. Deletion of active ADAMTS5 prevents cartilage degradation in a murine model of osteoarthritis. Nature. 2005;434:644–8. doi: 10.1038/nature03369. [DOI] [PubMed] [Google Scholar]
- 103.Stanton H, Rogerson FM, East CJ, Golub SB, Lawlor KE, Meeker CT, et al. ADAMTS5 is the major aggrecanase in mouse cartilage in vivo and in vitro. Nature. 2005;434:648–52. doi: 10.1038/nature03417. [DOI] [PubMed] [Google Scholar]
- 104.Lee NV, Rodriguez-Manzaneque JC, Thai SN, Twal WO, Luque A, Lyons KM, et al. Fibulin-1 acts as a cofactor for the matrix metalloprotease ADAMTS-1. J Biol Chem. 2005;280:34796–804. doi: 10.1074/jbc.M506980200. [DOI] [PubMed] [Google Scholar]
- 105.Gao G, Plaas A, Thompson VP, Jin S, Zuo F, Sandy JD. ADAMTS4 (aggrecanase-1) activation on the cell surface involves C-terminal cleavage by glycosylphosphatidyl inositol-anchored membrane type 4-matrix metalloproteinase and binding of the activated proteinase to chondroitin sulfate and heparan sulfate on syndecan-1. J Biol Chem. 2004;279:10042–51. doi: 10.1074/jbc.M312100200. [DOI] [PubMed] [Google Scholar]
- 106.Flannery CR, Zeng W, Corcoran C, Collins-Racie LA, Chockalingam PS, Hebert T, et al. Autocatalytic cleavage of ADAMTS-4 (Aggrecanase-1) reveals multiple glycosaminoglycan-binding sites. J Biol Chem. 2002;277:42775–80. doi: 10.1074/jbc.M205309200. [DOI] [PubMed] [Google Scholar]
- 107.Chockalingam PS, Sun W, Rivera-Bermudez MA, Zeng W, Dufield DR, Larsson S, et al. Elevated aggrecanase activity in a rat model of joint injury is attenuated by an aggrecanase specific inhibitor. Osteoarthritis Cartilage. 2011;19:315–23. doi: 10.1016/j.joca.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 108.Lopez-Otin C, Matrisian LM. Emerging roles of proteases in tumour suppression. Nat Rev Cancer. 2007;7:800–8. doi: 10.1038/nrc2228. [DOI] [PubMed] [Google Scholar]
- 109.Kuno K, Bannai K, Hakozaki M, Matsushima K, Hirose K. The carboxyl-terminal half region of ADAMTS-1 suppresses both tumorigenicity and experimental tumor metastatic potential. Biochem Biophys Res Commun. 2004;319:1327–33. doi: 10.1016/j.bbrc.2004.05.105. [DOI] [PubMed] [Google Scholar]
- 110.Moncada-Pazos A, Obaya AJ, Fraga MF, Viloria CG, Capella G, Gausachs M, et al. The ADAMTS12 metalloprotease gene is epigenetically silenced in tumor cells and transcriptionally activated in the stroma during progression of colon cancer. J Cell Sci. 2009;122:2906–13. doi: 10.1242/jcs.050468. [DOI] [PubMed] [Google Scholar]
- 111.Lo PH, Leung AC, Kwok CY, Cheung WS, Ko JM, Yang LC, et al. Identification of a tumor suppressive critical region mapping to 3p14.2 in esophageal squamous cell carcinoma and studies of a candidate tumor suppressor gene, ADAMTS9. Oncogene. 2007;26:148–57. doi: 10.1038/sj.onc.1209767. [DOI] [PubMed] [Google Scholar]
- 112.Lung HL, Lo PH, Xie D, Apte SS, Cheung AK, Cheng Y, et al. Characterization of a novel epigenetically-silenced, growth-suppressive gene, ADAMTS9, and its association with lymph node metastases in nasopharyngeal carcinoma. Int J Cancer. 2008;123:401–8. doi: 10.1002/ijc.23528. [DOI] [PubMed] [Google Scholar]
- 113.Viloria CG, Obaya AJ, Moncada-Pazos A, Llamazares M, Astudillo A, Capella G, et al. Genetic inactivation of ADAMTS15 metalloprotease in human colorectal cancer. Cancer Res. 2009;69:4926–34. doi: 10.1158/0008-5472.CAN-08-4155. [DOI] [PubMed] [Google Scholar]
- 114.Jin H, Wang X, Ying J, Wong AH, Li H, Lee KY, et al. Epigenetic identification of ADAMTS18 as a novel 16q23.1 tumor suppressor frequently silenced in esophageal, nasopharyngeal and multiple other carcinomas. Oncogene. 2007;26:7490–8. doi: 10.1038/sj.onc.1210559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Porter S, Scott SD, Sassoon EM, Williams MR, Jones JL, Girling AC, et al. Dysregulated expression of adamalysin-thrombospondin genes in human breast carcinoma. Clin Cancer Res. 2004;10:2429–40. doi: 10.1158/1078-0432.ccr-0398-3. [DOI] [PubMed] [Google Scholar]
- 116.Dubail J, Kesteloot F, Deroanne C, Motte P, Lambert V, Rakic JM, et al. ADAMTS-2 functions as anti-angiogenic and anti-tumoral molecule independently of its catalytic activity. Cell Mol Life Sci. 2010;67:4213–32. doi: 10.1007/s00018-010-0431-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.El Hour M, Moncada-Pazos A, Blacher S, Masset A, Cal S, Berndt S, et al. Higher sensitivity of Adamts12-deficient mice to tumor growth and angiogenesis. Oncogene. 2010;29:3025–32. doi: 10.1038/onc.2010.49. [DOI] [PubMed] [Google Scholar]
- 118.Hsu YP, Staton CA, Cross N, Buttle DJ. Anti-angiogenic properties of ADAMTS-4 in vitro. Int J Exp Pathol. 2012;93:70–7. doi: 10.1111/j.1365-2613.2011.00802.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Koo BH, Coe DM, Dixon LJ, Somerville RP, Nelson CM, Wang LW, et al. ADAMTS9 is a cell-autonomously acting, anti-angiogenic metalloprotease expressed by microvascular endothelial cells. Am J Pathol. 2010;176:1494–504. doi: 10.2353/ajpath.2010.090655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kumar S, Sharghi-Namini S, Rao N, Ge R. ADAMTS5 functions as an anti-angiogenic and anti-tumorigenic protein independent of its proteoglycanase activity. Am J Pathol. 2012;181:1056–68. doi: 10.1016/j.ajpath.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 121.Rodriguez-Manzaneque JC, Fernandez-Rodriguez R, Rodriguez-Baena FJ, Iruela-Arispe ML. ADAMTS proteases in vascular biology. Matrix Biol. 2015. doi: 10.1016/j.matbio.2015.02.004. [DOI] [PMC free article] [PubMed]
- 122.Tan Ide A, Ricciardelli C, Russell DL. The metalloproteinase ADAMTS1: a comprehensive review of its role in tumorigenic and metastatic pathways. Int J Cancer. 2013;133:2263–76. doi: 10.1002/ijc.28127. [DOI] [PubMed] [Google Scholar]
- 123.Lee NV, Sato M, Annis DS, Loo JA, Wu L, Mosher DF, et al. ADAMTS1 mediates the release of antiangiogenic polypeptides from TSP1 and 2. EMBO J. 2006;25:5270–83. doi: 10.1038/sj.emboj.7601400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Luque A, Carpizo DR, Iruela-Arispe ML. ADAMTS1/METH1 inhibits endothelial cell proliferation by direct binding and sequestration of VEGF165. J Biol Chem. 2003;278:23656–65. doi: 10.1074/jbc.M212964200. [DOI] [PubMed] [Google Scholar]
- 125.Krampert M, Kuenzle S, Thai SN, Lee N, Iruela-Arispe ML, Werner S. ADAMTS1 proteinase is up-regulated in wounded skin and regulates migration of fibroblasts and endothelial cells. J Biol Chem. 2005;280:23844–52. doi: 10.1074/jbc.M412212200. [DOI] [PubMed] [Google Scholar]
- 126.Karagiannis ED, Popel AS. Anti-angiogenic peptides identified in thrombospondin type I domains. Biochem Biophys Res Commun. 2007;359:63–9. doi: 10.1016/j.bbrc.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Demircan K, Gunduz E, Gunduz M, Beder LB, Hirohata S, Nagatsuka H, et al. Increased mRNA expression of ADAMTS metalloproteinases in metastatic foci of head and neck cancer. Head Neck. 2009;31:793–801. doi: 10.1002/hed.21045. [DOI] [PubMed] [Google Scholar]
- 128.Lind GE, Kleivi K, Meling GI, Teixeira MR, Thiis-Evensen E, Rognum TO, et al. ADAMTS1, CRABP1, and NR3C1 identified as epigenetically deregulated genes in colorectal tumorigenesis. Cell Oncol. 2006;28:259–72. doi: 10.1155/2006/949506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rocks N, Paulissen G, Quesada Calvo F, Polette M, Gueders M, Munaut C, et al. Expression of a disintegrin and metalloprotease (ADAM and ADAMTS) enzymes in human non-small-cell lung carcinomas (NSCLC) Br J Cancer. 2006;94:724–30. doi: 10.1038/sj.bjc.6602990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ricciardelli C, Frewin KM, Tan Ide A, Williams ED, Opeskin K, Pritchard MA, et al. The ADAMTS1 protease gene is required for mammary tumor growth and metastasis. Am J Pathol. 2011;179:3075–85. doi: 10.1016/j.ajpath.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liu YJ, Xu Y, Yu Q. Full-length ADAMTS-1 and the ADAMTS-1 fragments display pro- and antimetastatic activity, respectively. Oncogene. 2006;25:2452–67. doi: 10.1038/sj.onc.1209287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lu X, Wang Q, Hu G, Van Poznak C, Fleisher M, Reiss M, et al. ADAMTS1 and MMP1 proteolytically engage EGF-like ligands in an osteolytic signaling cascade for bone metastasis. Genes Dev. 2009;23:1882–94. doi: 10.1101/gad.1824809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rodriguez-Manzaneque JC, Carpizo D, Plaza-Calonge Mdel C, Torres-Collado AX, Thai SN, Simons M, et al. Cleavage of syndecan-4 by ADAMTS1 provokes defects in adhesion. Int J Biochem Cell Biol. 2009;41:800–10. doi: 10.1016/j.biocel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Esselens C, Malapeira J, Colome N, Casal C, Rodriguez-Manzaneque JC, Canals F, et al. The cleavage of semaphorin 3C induced by ADAMTS1 promotes cell migration. J Biol Chem. 2010;285:2463–73. doi: 10.1074/jbc.M109.055129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Casal C, Torres-Collado AX, Plaza-Calonge Mdel C, Martino-Echarri E, Ramon YCS, Rojo F, et al. ADAMTS1 contributes to the acquisition of an endothelial-like phenotype in plastic tumor cells. Cancer Res. 2010;70:4676–86. doi: 10.1158/0008-5472.CAN-09-4197. [DOI] [PubMed] [Google Scholar]
- 136.Choi GC, Li J, Wang Y, Li L, Zhong L, Ma B, et al. The metalloprotease ADAMTS8 displays antitumor properties through antagonizing EGFR-MEK-ERK signaling and is silenced in carcinomas by CpG methylation. Mol Cancer Res. 2014;12:228–38. doi: 10.1158/1541-7786.MCR-13-0195. [DOI] [PubMed] [Google Scholar]
- 137.Llamazares M, Obaya AJ, Moncada-Pazos A, Heljasvaara R, Espada J, Lopez-Otin C, et al. The ADAMTS12 metalloproteinase exhibits anti-tumorigenic properties through modulation of the Ras-dependent ERK signalling pathway. J Cell Sci. 2007;120:3544–52. doi: 10.1242/jcs.005751. [DOI] [PubMed] [Google Scholar]
- 138.Du W, Wang S, Zhou Q, Li X, Chu J, Chang Z, et al. ADAMTS9 is a functional tumor suppressor through inhibiting AKT/mTOR pathway and associated with poor survival in gastric cancer. Oncogene. 2013;32:3319–28. doi: 10.1038/onc.2012.359. [DOI] [PubMed] [Google Scholar]
- 139.Auf Dem Keller U, Prudova A, Eckhard U, Fingleton B, Overall CM. Systems-level analysis of proteolytic events in increased vascular permeability and complement activation in skin inflammation. Sci Signal. 2013;6:rs2. doi: 10.1126/scisignal.2003512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Fontanil T, Rua S, Llamazares M, Moncada-Pazos A, Quiros PM, Garcia-Suarez O, et al. Interaction between the ADAMTS-12 metalloprotease and fibulin-2 induces tumor-suppressive effects in breast cancer cells. Oncotarget. 2014;5:1253–64. doi: 10.18632/oncotarget.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Dubail J, Aramaki-Hattori N, Bader HL, Nelson CM, Katebi N, Matuska B, et al. A new Adamts9 conditional mouse allele identifies its non-redundant role in interdigital web regression. Genesis. 2014;52:702–12. doi: 10.1002/dvg.22784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Vankemmelbeke MN, Jones GC, Fowles C, Ilic MZ, Handley CJ, Day AJ, et al. Selective inhibition of ADAMTS-1, −4 and −5 by catechin gallate esters. Eur J Biochem. 2003;270:2394–403. doi: 10.1046/j.1432-1033.2003.03607.x. [DOI] [PubMed] [Google Scholar]
- 143.Cudic M, Burstein GD, Fields GB, Lauer-Fields J. Analysis of flavonoid-based pharmacophores that inhibit aggrecanases (ADAMTS-4 and ADAMTS-5) and matrix metalloproteinases through the use of topologically constrained peptide substrates. Chem Biol Drug Des. 2009;74:473–82. doi: 10.1111/j.1747-0285.2009.00885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–82. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 145.Felsenstein J. Confidence-limits on phylogenies - an approach using the bootstrap. Evolution. 1985;39:783–91. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 146.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]


