Abstract
Background
Members of fastidious Granulicatella and Aggregatibacter genera belong to normal oral flora bacteria that can cause serious infections, such as infective endocarditis. Aggregatibacter actinomycetemcomitans has long been implicated in aggressive periodontitis, whereas DNA-based methods only recently showed an association between Granulicatella spp. and dental diseases. As bacterial coaggregation is a key phenomenon in the development of oral and nonoral multispecies bacterial communities it would be of interest knowing coaggregation pattern of Granulicatella species with A. actinomycetemcomitans in comparison with the multipotent coaggregator Fusobacterium nucleatum.
The aim was to investigate coaggregation and biofilm formation of Granulicatella elegans and Granulicatella adiacens with A. actinomycetemcomitans and F. nucleatum strains.
Results
F. nucleatum exhibited significantly (p < 0.05) higher autoaggregation than all other test species, followed by A. actinomycetemcomitans SA269 and G. elegans. A. actinomycetemcomitans CU1060 and G. adiacens did not autoaggregate. G. elegans with F. nucleatum exhibited significantly (p < 0.05) higher coaggregation than most others, but failed to grow as biofilm together or separately. With F. nucleatum as partner, A. actinomycetemcomitans strains SA269, a rough-colony wild-type strain, and CU1060, a spontaneous smooth-colony laboratory variant, and G. adiacens were the next in coaggregation efficiency. These dual species combinations also were able to grow as biofilms. While both G. elegans and G. adiacens coaggregated with A. actinomycetemcomitans strain SA269, but not with CU1060, they grew as biofilms with both A. actinomycetemcomitans strains.
Conclusions
G. elegans failed to form biofilm with F. nucleatum despite the strongest coaggregation with it. The ability of Granulicatella spp. to coaggregate and/or form biofilms with F. nucleatum and A. actinomycetemcomitans strains suggests that Granulicatella spp. have the potential to integrate into dental plaque biofilms.
Keywords: Coaggregation, Biofilm, Granulicatella, Aggregatibacter actinomycetemcomitans, Fusobacterium nucleatum, Autoaggregation
Background
Granulicatella adiacens and Granulicatella elegans were previously known as “nutritionally variant streptococci (NVS)” [1]. The NVS group of bacteria were assigned an independent genus “Abiotrophia” [2] and later on, based on 16S rRNA sequence phylogeny, they were further divided into genera Abiotrophia and Granulicatella [3]. Granulicatella spp. are nonmotile, nonspore-forming, facultatively anaerobic Gram-positive cocci requiring complex nutrient-rich media for their growth. They are part of the normal oral flora [4,5], but similar to other oral species such as viridans streptococci and HACEK (Haemophilus, Aggregatibacter, Cardiobacterium, Eikenella, Kingella)-group of bacteria Granulicatella spp. can cause severe infections including infective endocarditis. Furthermore, most likely due to advancements in molecular biological methods, recent DNA-based studies have reported increased detection rates of Granulicatella spp. in periodontitis [6], caries [7] and endodontic infections [8,9].
Bacterial adhesion to surfaces is an important step in colonization and biofilm formation [10,11]. In the case of oral multispecies plaque biofilm development, early colonizer species adhere to nascent hard or soft tissues and provide a substratum for subsequent colonizers of the plaque biofilm. Essential for the development of multispecies biofilm communities is bacterial coaggregation, the adhesion of different bacterial species to each other. Furthermore, coaggregation is a key phenomenon that facilitates interaction among different bacterial species in the biofilm [12,13]. The interactions may occur between protein adhesins and polysaccharide receptors [14,15] or between proteinaceous adhesin-receptors [16].
Fusobacterium nucleatum, a Gram-negative obligate anaerobe in the oral cavity, plays a crucial role in the development and maturation of dental plaque biofilm due to its strong ability to coaggregate with early plaque colonizers, such as streptococci, and with the late colonizing Gram-negative anaerobes [15,17,18]. Apart this central role played by F. nucleatum, P. gingivalis, a late colonizer, also directly coaggregates with Streptococcus mitis [19], Streptococcus oralis [20], and Actinomyces viscosus [21]. Similarly, coaggregation between Prevotella spp. and P. gingivalis [22], and between Tannerella forsythia, streptococci and P. gingivalis [23] have been reported. Thus, it seems that in addition to using coaggregation as a mechanism of resistance to bacterial clearance, bacteria coaggregate with different oral species for specific additional benefits. For example, S. gordonii benefits from its coaggregation with A. naeslundii in arginine-deficient conditions through increase in the expression of arginine biosynthesis genes [24] and A. naeslundii also protects S. gordonii from H2O2-mediated oxidative damage [25]. Recently, it was suggested that by binding to streptococci, F. nucleatum overcomes resistance by oral microbiota and thus gets integrated into oral microbial community [26]. Further, adhesive capacity of P. gingivalis was reported to increase upon binding to Treponema denticola [27].
G. adiacens, G. elegans and Aggregatibacter actinomycetemcomitans are normal oral flora bacteria. While Granulicatella spp. belong to oral streptococci that bind to salivary pellicle on tooth surface, A. actinomycetemcomitans is a Gram-negative bacterium regarded as a late colonizer in dental plaque biofilm. Studies on A. actinomycetemcomitans interaction with oral streptococci have shown that H2O2 produced by S. gordonii enhances expression of the autotransporter ApiA, which leads to greater resistance against host’s immune response [28]. However, coaggregation between Granulicatella spp. and A. actinomycetemcomitans has not been studied. Investigating specific bacterial interactions may shed light on cooperation among these species of interest in biofilms. Therefore, our aim was to study coaggregation and biofilm formation of Granulicatella spp. with A. actinomycetemcomitans, a member of the HACEK group [29] in comparison with the “multipotent” coaggregator F. nucleatum.
Methods
Bacteria and culture conditions
Granulicatella elegans CCUG 38949 and Granulicatella adiacens CCUG 27809-T were cultured on chocolate blood agar with or without 0.001 % pyridoxal HCl [30] for 2 days at 37 °C and 5 % CO2 in air. A. actinomycetemcomitans strains were cultured on tryptic soy agar and incubated for 3 days at 37 °C and 5 % CO2 in air. A. actinomycetemcomitans SA269, a serotype d strain isolated from a 14-year-old female patient with localized aggressive periodontitis [31], was chosen to represent wild-type rough-colony strains which are fimbriated and strongly adhere to surfaces and form tenacious clumps in suspension [32]. A. actinomycetemcomitans CU1060 (a gift from D. Fine, Rutgers University, New Jersey, USA), a serotype f strain, is a spontaneous smooth-colony laboratory variant of the strain CU1000 isolated from a 13-year-old patient with localized aggressive periodontitis [33]. Fusobacterium nucleatum ssp. polymorphum NCTC 10562 (ATCC 10953) (isolated by H. Hoffman 1951; source “inflamed gingiva”) [34], was cultured on brucella blood agar containing 5 % defibrinated sheep blood and incubated at 37 °C for 2 days in anaerobic atmosphere (10 % H2, 5 % CO2, 85 % N2) using Anoxomat™ MarkII anaerobic gas filling system (Mart Microbiology, The Netherlands). Identities of the reference strains were confirmed by 16S rDNA sequencing as described earlier [35].
Scanning electron microscopy
Agar blocks containing colonies of Granulicatella spp. were cut from the plates using sterile scalpels. For fixation, the blocks were immersed in PBS containing 3 % glutaraldehyde for 2 h on a rotator and then kept in a refrigerator overnight. After washing in PBS 3×, the agar blocks were incubated in 1 % osmium tetroxide for 2 h. The blocks were rinsed again as above and dehydrated in increasing concentrations of acetone from 30–100 %, 10 min in each on a rotator. The samples were then placed in a critical point dryer for complete drying, mounted on stubs by carbon double adhesive tape and finally coated with gold and stored in desiccator until observation. The samples were observed on Zeiss Leo SUPRA® 50VP field emission scanning electron microscope.
Coaggregation assay
Previously published protocol was used with some modifications [36]. After the incubation, the bacterial colonies were suspended in sterile coaggregation buffer [10 mM Tris (pH 8.0), 1 mM CaCl2, 1 mM MgCl2, 150 mM NaCl]. Absorbance of the suspension was adjusted to an OD600 = 1. Five hundred microliters of cell suspensions from each of the test species and the partner species were added into a 1-ml cuvette. For autoaggregation, 1000-μl aliquots of each species were added into separate cuvettes. The cuvettes were incubated at room temperature and absorbance was read at 600 nm every 15 min for 2 h. Decline in optical density of the bacterial suspensions with time was used as a measure of autoaggregation or coaggregation. Percent coaggregation or autoaggregation [37] was calculated as follows:
Phase contrast microscopy
Auto- and coaggregation of the test bacterial species were also studied by phase contrast microscopy. After preparing bacterial suspensions with or without partner species in coaggregation buffer as described above, 10 μl from each preparation was mounted on a microscopic glass slide with a coverslip and observed at 1000× magnification using phase contrast optics on Leica DMLM microscope.
Biofilm growth
Static biofilm cultures were setup in 24-well cell culture plates using a previously established method [38]. Briefly, G. elegans CCUG 38949, G. adiacens CCUG 27809-T, F. nucleatum NCTC 10562 and A. actinomycetemcomitans strains SA269 and CU1060 were grown on their respective culture media as described above. Colonies were harvested, suspended in brucella broth and the cell suspensions adjusted to OD600 = 1 in brucella broth. A 100-μl aliquot from each strain separately or 50 μl from each of two partner strains was added into wells of a 24-well plate containing 900 μl brucella broth with 0.001 % pyridoxal. In parallel, a 20-μl aliquot from each bacterial stock suspension was streaked on respective growth media to ensure that the inocula used for biofilm culture were viable. Wells containing only broth served as negative control. After incubating for 3 days in anaerobic atmosphere at 37 °C, broth supernatant was removed and biofilms were washed once gently with sterile PBS. The plates were air-dried for 10 min at room temperature. One ml of 2 % crystal violet stain was added to each well and the plate was allowed to stay at room temperature for 10 min. The crystal violet stain was removed and the wells were washed 7–8 times with distilled water. To each well, 250 μl 95 % ethanol was added and the plate was incubated at room temperature on a shaker for 10 min. One hundred μl from each well was added into wells of a 96-well plate in duplicates and the absorbance was read at 590 nm using a microplate reader (Eon, BioTek® Instruments Inc., USA).
Statistics
Mann-Whitney U test was used to compare groups. A p value of <0.05 was considered statistically significant. All experiments were performed in duplicates and 3 independent experiments were run.
Results
Pleomorphism of Granulicatella spp. cells
As seen in scanning electron micrographs (Fig. 1), G. elegans and G. adiacens showed pleomorphic cell morphology when grown in the absence of pyridoxal supplement. The cells were elongated and often bulged in the middle. However, when the culture medium was supplemented with pyridoxal, both species assumed coccal shape. Further, Gram variability, which was evident when grown in the absence of pyridoxal, was no longer observed and the majority of the cells were Gram positive (data not shown).
Fig. 1.
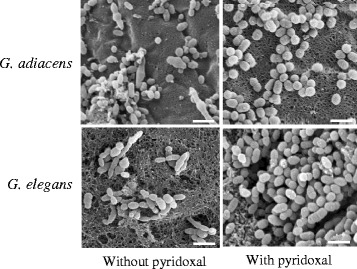
Pleomorphic cell morphology of Granulicatella spp. G. elegans CCUG 38949 and G. adiacens CCUG 27809-T were grown on chocolate blood agar supplemented with or without pyridoxal as described in the methods section. Scanning electron microscopy was performed as described in methods. The images were acquired at 10,000× magnification using Zeiss Leo SUPRA® 50VP scanning electron microscope. Bar represents 2 μm
Autoaggregation
F. nucleatum was the strongest autoaggregating strain of all test species (Fig. 2A and Fig. 3A). It showed significantly (p < 0.05) higher percent of autoaggregation than all other test species at time points 30–120 min except A. actinomycetemcomitans SA269 at 15 and 30 min. Mean (SD) autoaggregation of F. nucleatum increased from 21.3 % (15 %) at 30 min to 48 % (14 %) at 45 min, 64.4 % (7.6 %) at 60 min and finally to 80 % (5 %) at 120 min. The next in autoaggregation efficiency was A. actinomycetemcomitans SA269, a rough-colony clinical isolate, which showed a mean (SD) autoaggregation of 19 % (7.5 %) at 30 min, 32 % (11.5 %) at 45 min and reached 53.6 % (10.4 %) at the end of the follow up. G. elegans began to autoaggregate after 45 min with a mean (SD) autoaggregation of 11.6 % (2.3 %), 18 % (4.9 %) at 75 min, 31 % (4 %) at 90 min finally reaching 49 % (1.4 %) at 120 min. G. elegans autoaggregation was significantly (p < 0.05) lower than that of F. nucleatum and A. actinomycetemcomitans SA269 at 45 min. However, at the end of the follow up, only F. nucleatum, but not A. actinomycetemcomitans SA269, showed significantly higher (p < 0.05) percent of autoaggregation than G. elegans. G. adiacens and A. actinomycetemcomitans CU1060, a smooth-colony variant, did not exhibit autoaggregation.
Fig. 2.
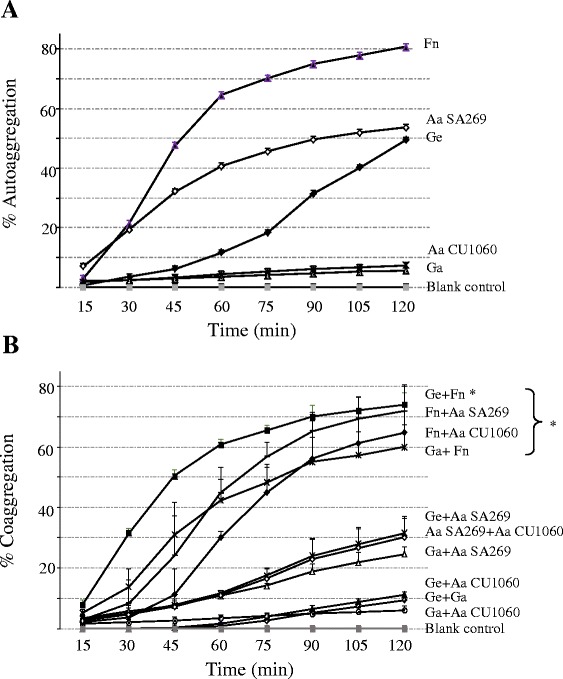
Autoaggregation and coaggregation of Granulicatella spp. Cell suspensions of each test bacterial strain separately or in combination with a partner species were prepared as described in the methods section. OD600 was measured every 15 min and the OD600 values were converted into percent autoaggregation (panel A) or coaggregation (panel B) using the formula described in the methods part. Abbreviations: Aa A. actinomycetemcomitans; Ge G. elegans; Ga G. adiacens and Fn F. nucleatum. The results are means (SD) from 3 independent experiments. *p < 0.05
Fig. 3.
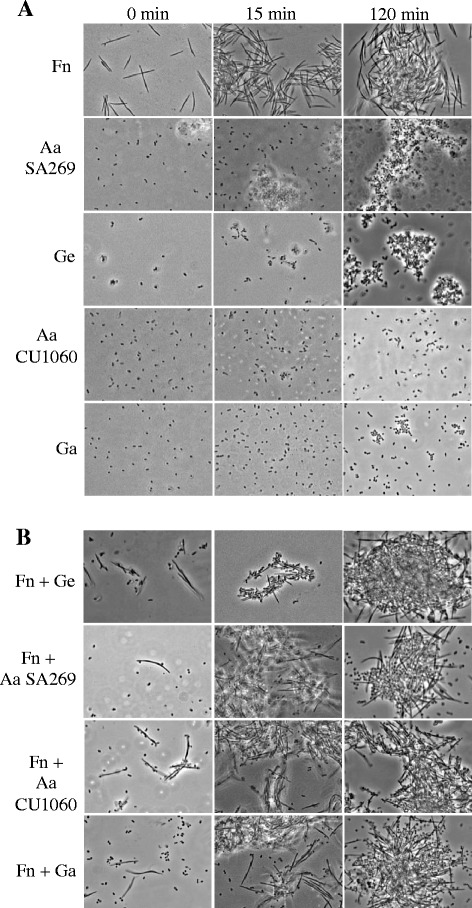
Auto- and coaggregation of Granulicatella spp. Bacterial cell suspensions of test species alone or in combination of the partner species were prepared as described in the methods section. A 10-μl aliquot from each suspension was applied on a glass slide with a cover slip and autoaggregation (panel A) and coaggregation (panel B) were assessed visually by observing at 1000× using phase contrast optics on a Leica DMLM microscope. Abbreviations: Fn F. nucleatum, Ge G. elegans, Ga G. adiacens, Aa A. actinomycetemcomitans
Coaggregation
Overall, 3 distinct groups were observed based on coaggregation efficiency (Fig. 2B and Fig. 3B): 1) Strong coaggregators: G. elegans with F. nucleatum, F. nucleatum with A. actinomycetemcomitans SA269, G. adiacens with F. nucleatum, F. nucleatum with A. actinomycetemcomitans CU1060. 2) Moderate coaggregators: G. elegans with A. actinomycetemcomitans SA269, A. actinomycetemcomitans strains SA269 with CU1060 and G. adiacens with A. actinomycetemcomitans SA269. 3) Weak to Non-coaggregators: G. elegans with A. actinomycetemcomitans CU1060, G. adiacens with A. actinomycetemcomitans CU1060 and G. elegans with G. adiacens.
G. elegans together with F. nucleatum exhibited strongest (p < 0.05) coaggregation compared to all other species combinations. Mean (SD) coaggregation of G. elegans with F. nucleatum nearly doubled from 31.5 % (3.6 %) at 30 min to 60.6 % (2 %) at 60 min, finally reaching 74 % (3.5 %) at 120 min. Mean (SD) percent coaggregation of G. elegans with F. nucleatum was significantly (p < 0.05) higher [(31.5 % (3.6 %)] than that of G. adiacens with F. nucleatum [(13.7 % (5.6 %)] at 30 min. The difference was also significant (p < 0.05) at all time points starting from 30 min.
F. nucleatum in combination with A. actinomycetemcomitans SA269 showed mean (SD) percent coaggregation of 24 % (15.8 %) at 45 min, 44.7 % (7.6 %) at 60 min and finally increased to 71.8 % (7.2 %) at 120 min. The combination of G. adiacens and F. nucleatum showed mean (SD) percent coaggregation of 31 % (5.5 %) at 45 min, which increased to 60 % (6.6 %) at 120 min. G. elegans or G. adiacens exhibited moderate coaggregation with A. actinomycetemcomitans SA269 at later time points from 60 min to 120 min. However, neither the combinations of G. elegans or G. adiacens with A. actinomycetemcomitans CU1060 nor the combination of G. elegans and G. adiacens showed coaggregation within first 60 min, while a weak coaggregation of 6–10 % was evident at the end of the follow-up period.
Phase-contrast microscopy (Fig. 3) revealed that by 15 min, large numbers of cells of all partner species were bound to F. nucleatum cells. Additionally, numerous cells of partner species were also seen in close proximity to F. nucleatum cells. At the end of the follow-up period, i.e., 120 min, large clumps of F. nucleatum together with each of the partner species were abundantly present (Fig. 3B). Unlike G. elegans which was found to be binding to F. nucleatum cells already at 0 min, G. adiacens showed binding only at later time points, but not at 0 min. On the other hand, both A. actinomycetemcomitans strains, SA269 and CU1060, were seen bound to F. nucleatum at 0 min. Species combinations that included a partner species other than F. nucleatum are not shown as species were not distinguishable based on cell morphology.
Biofilm formation
In the case of monospecies biofilm cultures, A. actinomycetemcomitans SA269 showed highest biofilm mass with a mean (SD) OD590 of 1.9 (0.5), followed by A. actinomycetemcomitans CU1060 and G. adiacens, which showed OD590 of 1.8 (0.3) and 1.04 (0.5), respectively, in the decreasing order (Fig. 4). OD590 values for F. nucleatum 0.17 (0.04) and G. elegans 0.16 (0.06) consistently showed poor biofilm formation ability in all three experiments. When G. elegans and F. nucleatum were grown as dual species biofilms, the biofilm mass did not increase compared to their respective monospecies cultures. In contrast, G. adiacens and F. nucleatum dual species biofilm exhibited a significant (p < 0.05) increase in biofilm mass compared to their monospecies cultures. Both G. elegans and G. adiacens showed significantly (p < 0.05) higher biofilm mass when grown together with either of the two A. actinomycetemcomitans strains. Dual species culture of G. elegans and G. adiacens showed significantly (p < 0.05) higher biofilm mass than their respective monospecies biofilm cultures did. In dual species biofilms, highest biofilm formation with a mean (SD) OD590 of 3 (1.5) was observed when F. nucleatum and A. actinomycetemcomitans SA269 were grown together. However, the biofilm mass was not significantly (p > 0.05) higher compared to A. actinomycetemcomitans SA269 cultured alone. Similarly, dual species culture of F. nucleatum and A. actinomycetemcomitans CU1060 did not show significantly (p > 0.05) higher biofilm mass compared to A. actinomycetemcomitans CU1060 cultured alone. When the A. actinomycetemcomitans strains SA269 and CU1060 were grown together, the biofilm mass was significantly (p < 0.05) higher than CU1060, but not SA269, cultured alone.
Fig 4.
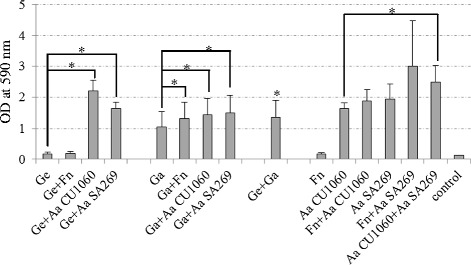
Biofilm formation by Granulicatella spp. in the presence or absence of F. nucleatum and A. actinomycetemcomitans strains. Bacterial strains were cultured separately or together with a partner species in 24-well plates in brucella broth in anaerobic atmosphere for 3 days as described in the methods section. After the incubation period, biofilms were washed gently to remove unattached cells and stained with 2 % crystal violet for quantifying biofilm mass. Abbreviations: Fn F. nucleatum, Ge G elegans, Ga G. adiacens, Aa A. actinomycetemcomitans. The results are means (SD) from 3 independent experiments. *p < 0.05
Discussion
This study demonstrates that both G. elegans and G. adiacens have the ability to coaggregate with F. nucleatum and A. actinomycetemcomitans. These Granulicatella species also grew together with A. actinomycetemcomitans and F. nucleatum as dual species biofilms, except that G. elegans, despite being the strongest coaggregator with F. nucleatum, failed to grow as biofilm with this species.
Coaggregation efficiency was evaluated by a quantitative spectrophotometric method similar to several previous studies [39,40]. In this method, coaggregation is assessed by the decrease in the optical density of the bacterial suspension by time. The method allows definitive quantification of coaggregation in contrast to the visual scoring method used by several other studies [17,41,42]. Furthermore, we measured coaggregation for 120 min with 15 min time interval, which enabled us to investigate whether there were slow or late coaggregators among our test species. Varying incubation times from 1 to 30 min have been used for visual scoring of coaggregation [36,43]. Other methods of studying coaggregation among oral bacteria have been the use of radiolabelled [44,45] or fluorescently labeled bacteria [46]. Although these labeling based methods were claimed to be more sensitive, procedures like labeling of bacteria with fluorescent dyes and the need to pretreat cells in the case of certain dyes made them less simple compared to the spectrophotometric method used in this study. For biofilm studies, we used a standard static biofilm model, which was sufficient for direct quantification of biofilms by crystal violet staining.
Our interest towards Granulicatella species derived from recent findings that, besides systemic infections, granulicatellas also occur relatively frequently in dental infections [6-9]. The strains of G. elegans and G. adiacens we chose for this study were originally isolated from patients with endocarditis [47,48] and have been commonly used by medical researchers. On the other hand, the rationale for including F. nucleatum ssp. polymorphum (NCTC 10562) in this study was that it has been studied extensively for its strong coaggregation property and therefore known as a “bridge” organism between early- and late colonizing species during the process of plaque biofilm development. The A. actinomycetemcomitans strains selected for the present study were considered interesting, since both originated from young patients with aggressive periodontitis. However, the strains were of different serotypes and had dissimilar surface structures and autoaggregation behavior, as previously shown [32,33]. The strain SA269 (serotype d) is a representative of rough-colony, wild-type strains and strain CU1060 (serotype f) is a smooth-colony, non-fimbriated, spontaneous laboratory variant of the clinical strain CU1000 [33]. Using these A. actinomycetemcomitans strains we hoped to find differences in their behavior in the present experiments, which could be hypothesis generating in our further studies. As long known, A. actinomycetemcomitans is regarded as a late colonizer in dental plaque and coaggregates with other bacteria, e.g., F. nucleatum [43] and P. gingivalis [49] via serotype-specific polysaccharide antigen.
In our autoaggregation studies, F. nucleatum was the strongest autoaggregator followed by A. actinomycetemcomitans SA269 and G. elegans. Although, F. nucleatum has been extensively studied in the context of coaggregation, little is known about its autoaggregation [39,50,51]. Using a similar optical density method as in our study, F. nucleatum ATCC 25586, but not the strain Fusobacterium nucleatum ssp. nucleatum (ATCC 23726), was found to begin autoaggregating within 30 min, similar to our results; however, only in the presence of saliva [39], in contrast to our study where saliva was not required for autoaggregation. Thus, it seems that autoaggregation of F. nucleatum is strain-dependent and occurs via both saliva-dependent and -independent mechanisms. Autoaggregation of A. actinomycetemcomitans rough-colony isolates is well known and is mainly attributed to their long bundled fimbriae, while the smooth-colony strains are known to be non-autoaggregating [52,53].
In our study, the most efficient coaggregation, i.e., 45–65 %, was seen in the first 75 min in the case of “strong coaggregators” group in which F. nucleatum exhibited highest coaggregation with G. elegans. Majority of the earlier reports using either visual scoring or optical density method have demonstrated that F. nucleatum coaggregates strongly and rapidly (usually within 5–30 min) with most bacterial species in the oral cavity belonging to both early and late colonizer groups [45,54].
G. elegans and F. nucleatum exhibited strongest coaggregation, but failed to form biofilm together. This was unpredicted since coaggregation efficiency is thought to directly enhance biofilm formation in mixed species cultures [13]. It seems plausible that coaggregation alone was not adequate for a dual-species biofilm development by G. elegans and F. nucleatum. These species probably require presence of a third species for multispecies cooperation for successful biofilm formation. This has been demonstrated in a study where F. nucleatum did show some coaggregation but failed to grow as dual species biofilm with Streptococcus oralis, but when Actinomyces naeslundii was added, abundant biofilm growth was observed [55]. In that study, the authors concluded that F. nucleatum required A. naeslundii in the consortium to produce catalase to counter hydrogen peroxide produced by Streptococcus oralis. It should be remembered that in multispecies biofilms bacterial interactions are enormously complex and certain species exert antagonistic effect on others. For example, arginine deiminase produced by Streptococcus cristatus inhibited fimA expression in P. gingivalis [56]. Also, Candida albicans biofilm formation was inhibited by A. actinomycetemcomitans [57]. It seems that A. actinomycetemcomitans down-regulates expression of several proteins of other species in multispecies biofilm [58]. Thus, if the expression of a bacterial component essential for biofilm growth, e.g., quorum sensing signal molecules, such as autoinducer-2 [59], in G. elegans is down-regulated by the partner species, G. elegans might not be able to grow and persist in biofilm. Moreover, G. elegans might use coaggregation as a mechanism of cell-cell interaction to simply evade washout from saliva. It is likely that G. elegans requires presence of and interaction with other bacterial species in addition to F. nucleatum to successfully integrate itself into a dual- or multispecies biofilm.
In strong contrast to G. elegans, G. adiacens did not only coaggregate, but also grew together in biofilms with F. nucleatum. Both G. elegans and G. adiacens coaggregated with and grew as biofilms in the presence of either of the A. actinomycetemcomitans strains with the same efficiency. Our results confirm previous reports that A. actinomycetemcomitans coaggregates with F. nucleatum [36,43]. Importantly, both the fimbriated strain SA269 and the non-fimbriated strain CU1060 coaggregated with F. nucleatum with near-equal efficiency, suggesting that fimbriae did not contribute to coaggregation. In fact, previous studies have found that serotype-specific polysaccharide is responsible for coaggregation of A. actinomycetemcomitans with F. nucleatum [36,60] and P. gingivalis [49]. This is unlike autoaggregation of A. actinomycetemcomitans, which is solely attributed to fimbriae [32,53]. Furthermore, contradicting results have been reported regarding coaggregation of A. actinomycetemcomitans with F. nucleatum. While in one study F. nucleatum ATCC 10953 (equivalent of NCTC 10562 used in this study) did not coaggregate with any of the six A. actinomycetemcomitans serotypes [36], it did exhibit coaggregation with A. actinomycetemcomitans serotype b in several other studies [43,60]. Thus, no clear correlation has been established between A. actinomycetemcomitans serotypes and ability to coaggregate with other oral bacteria.
Since F. nucleatum and A. actinomycetemcomitans SA269 possess autoaggregation property, one might argue that coaggregation observed with their partner species was due to their autoaggregation. However, G. elegans, which did not show any autoaggregation during first 60 min, but not the strong autoaggregator A. actinomycetemcomitans SA269, showed strongest coaggregation with F. nucleatum. Further, G. elegans and G. adiacens showed only a weak coaggregation with the autoaggregating A. actinomycetemcomitans SA269 (Fig. 2B). Microscopic examination (Fig. 3B) of the coaggregates provided further evidence to specific coaggregation since partner species were seen avidly bound to F. nucleatum. Thus, our results clearly demonstrate that the observed coaggregation efficiencies were due to specific interaction between the partner species and not due to the autoaggregation property.
The ability of G. elegans and G. adiacens to coaggregate with and grow as biofilms together with F. nucleatum and A. actinomycetemcomitans is remarkable and suggests that they may form an important part of dental plaque biofilm. Further, since Granulicatella spp. belong to streptococci group of bacteria that are known to be the early colonizers of dental plaque, it was not surprising that Granulicatella spp. coaggregated with F. nucleatum, the so-called “bridge organism”. However, Granulicatella spp. may have additional benefits of this partnership. For example, if Granulicatella spp. lack β-lactamase, similar to some other oral streptococci [61], F. nucleatum producing the enzyme, may protect granulicatellas against β-lactam antibiotics.
Conclusions
The ability of G. adiacens and G. elegans to coaggregate and form biofilms with F. nucleatum and A. actinomycetemcomitans probably offers granulicatellas additional benefits besides evasion from clearance in the oral cavity.
Acknowledgements
This study was funded by the Kuwait University General Facility Grant GD01/11 and by the Kuwait Foundation for the Advancement of Sciences under project code: 2011–5502–01. We thank Nanoscopy Center of Kuwait University for help with scanning electron microscopy. We also thank the Research Core Facility at the Health Science Center of Kuwait University, Grant SRUL02/13.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Author contributions
MK: Design and conception of the experiments, data analysis, and manuscript preparation. RB: Execution of the experiments. SA: Design and conception of the experiments, data analysis, and manuscript preparation. All authors have read and approved the manuscript.
Contributor Information
Maribasappa Karched, Email: mkarched@hsc.edu.kw.
Radhika G. Bhardwaj, Email: radhika@hsc.edu.kw
Sirkka E. Asikainen, Email: sirkka.asikainen@hsc.edu.kw
References
- 1.Frenkel A, Hirsch W. Spontaneous development of L forms of streptococci requiring secretions of other bacteria or sulphydryl compounds for normal growth. Nature. 1961;191:728–30. doi: 10.1038/191728a0. [DOI] [PubMed] [Google Scholar]
- 2.Kawamura Y, Hou XG, Sultana F, Liu S, Yamamoto H, Ezaki T. Transfer of Streptococcus adjacens and Streptococcus defectivus to Abiotrophia gen. nov. as Abiotrophia adiacens comb. nov. and Abiotrophia defectiva comb. nov., respectively. Int J Syst Bacteriol. 1995;45(4):798–803. doi: 10.1099/00207713-45-4-798. [DOI] [PubMed] [Google Scholar]
- 3.Collins MD, Lawson PA. The genus Abiotrophia (Kawamura et al.) is not monophyletic: proposal of Granulicatella gen. nov., Granulicatella adiacens comb. nov., Granulicatella elegans comb. nov. and Granulicatella balaenopterae comb. nov. Int J Syst Evol Microbiol. 2000;50 Pt 1:365–9. doi: 10.1099/00207713-50-1-365. [DOI] [PubMed] [Google Scholar]
- 4.Mikkelsen L, Theilade E, Poulsen K. Abiotrophia species in early dental plaque. Oral Microbiol Immunol. 2000;15(4):263–8. doi: 10.1034/j.1399-302x.2000.150409.x. [DOI] [PubMed] [Google Scholar]
- 5.Sato S, Kanamoto T, Inoue M. Abiotrophia elegans strains comprise 8% of the nutritionally variant streptococci isolated from the human mouth. J Clin Microbiol. 1999;37(8):2553–6. doi: 10.1128/jcm.37.8.2553-2556.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belstrom D, Fiehn NE, Nielsen CH, Kirkby N, Twetman S, Klepac-Ceraj V, et al. Differences in bacterial saliva profile between periodontitis patients and a control cohort. J Clin Periodontol. 2014;41(2):104–12. doi: 10.1111/jcpe.12190. [DOI] [PubMed] [Google Scholar]
- 7.Kanasi E, Dewhirst FE, Chalmers NI, Kent R, Jr, Moore A, Hughes CV, et al. Clonal analysis of the microbiota of severe early childhood caries. Caries Res. 2010;44(5):485–97. doi: 10.1159/000320158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsiao WW, Li KL, Liu Z, Jones C, Fraser-Liggett CM, Fouad AF. Microbial transformation from normal oral microbiota to acute endodontic infections. BMC Genomics. 2012;13:345. doi: 10.1186/1471-2164-13-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siqueira JF, Jr, Rocas IN. Catonella morbi and Granulicatella adiacens: new species in endodontic infections. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2006;102(2):259–64. doi: 10.1016/j.tripleo.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 10.Dunne WM., Jr Bacterial adhesion: seen any good biofilms lately? Clin Microbiol Rev. 2002;15(2):155–66. doi: 10.1128/CMR.15.2.155-166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolenbrander PE, Palmer RJ, Jr, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol. 2010;8(7):471–80. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- 12.Foster JS, Kolenbrander PE. Development of a multispecies oral bacterial community in a saliva-conditioned flow cell. Appl Environ Microbiol. 2004;70(7):4340–8. doi: 10.1128/AEM.70.7.4340-4348.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rickard AH, Gilbert P, High NJ, Kolenbrander PE, Handley PS. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 2003;11(2):94–100. doi: 10.1016/S0966-842X(02)00034-3. [DOI] [PubMed] [Google Scholar]
- 14.Kline KA, Falker S, Dahlberg S, Normark S, Henriques-Normark B. Bacterial adhesins in host-microbe interactions. Cell Host Microbe. 2009;5(6):580–92. doi: 10.1016/j.chom.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 15.Kolenbrander PE, Palmer RJ, Jr, Rickard AH, Jakubovics NS, Chalmers NI, Diaz PI. Bacterial interactions and successions during plaque development. Periodontol 2000. 2006;42:47–79. doi: 10.1111/j.1600-0757.2006.00187.x. [DOI] [PubMed] [Google Scholar]
- 16.Daep CA, Lamont RJ, Demuth DR. Interaction of Porphyromonas gingivalis with oral streptococci requires a motif that resembles the eukaryotic nuclear receptor box protein-protein interaction domain. Infect Immun. 2008;76(7):3273–80. doi: 10.1128/IAI.00366-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolenbrander PE, Andersen RN. Inhibition of coaggregation between Fusobacterium nucleatum and Porphyromonas (Bacteroides) gingivalis by lactose and related sugars. Infect Immun. 1989;57(10):3204–9. doi: 10.1128/iai.57.10.3204-3209.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lancy P, Jr, Dirienzo JM, Appelbaum B, Rosan B, Holt SC. Corncob formation between Fusobacterium nucleatum and Streptococcus sanguis. Infect Immun. 1983;40(1):303–9. doi: 10.1128/iai.40.1.303-309.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murakami Y, Nagata H, Amano A, Takagaki M, Shizukuishi S, Tsunemitsu A, et al. Inhibitory effects of human salivary histatins and lysozyme on coaggregation between Porphyromonas gingivalis and Streptococcus mitis. Infect Immun. 1991;59(9):3284–6. doi: 10.1128/iai.59.9.3284-3286.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagata H, Amano A, Ojima M, Tanaka M, Kataoka K, Shizukuishi S. Effect of binding of fibrinogen to each bacterium on coaggregation between Porphyromonas gingivalis and Streptococcus oralis. Oral Microbiol Immunol. 1994;9(6):359–63. doi: 10.1111/j.1399-302X.1994.tb00286.x. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Ellen RP. Relative adherence of Bacteroides species and strains to Actinomyces viscosus on saliva-coated hydroxyapatite. J Dent Res. 1989;68(9):1308–12. doi: 10.1177/00220345890680090301. [DOI] [PubMed] [Google Scholar]
- 22.Sato T, Nakazawa F. Coaggregation between Prevotella oris and Porphyromonas gingivalis. J Microbiol Immunol Infect. 2014;47(3):182–6. doi: 10.1016/j.jmii.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 23.Shimotahira N, Oogai Y, Kawada-Matsuo M, Yamada S, Fukutsuji K, Nagano K, et al. The surface layer of Tannerella forsythia contributes to serum resistance and oral bacterial coaggregation. Infect Immun. 2013;81(4):1198–206. doi: 10.1128/IAI.00983-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jakubovics NS, Gill SR, Iobst SE, Vickerman MM, Kolenbrander PE. Regulation of gene expression in a mixed-genus community: stabilized arginine biosynthesis in Streptococcus gordonii by coaggregation with Actinomyces naeslundii. J Bacteriol. 2008;190(10):3646–57. doi: 10.1128/JB.00088-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jakubovics NS, Gill SR, Vickerman MM, Kolenbrander PE. Role of hydrogen peroxide in competition and cooperation between Streptococcus gordonii and Actinomyces naeslundii. FEMS Microbiol Ecol. 2008;66(3):637–44. doi: 10.1111/j.1574-6941.2008.00585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He X, Hu W, Kaplan CW, Guo L, Shi W, Lux R. Adherence to streptococci facilitates Fusobacterium nucleatum integration into an oral microbial community. Microb Ecol. 2012;63(3):532–42. doi: 10.1007/s00248-011-9989-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meuric V, Martin B, Guyodo H, Rouillon A, Tamanai-Shacoori Z, Barloy-Hubler F, et al. Treponema denticola improves adhesive capacities of Porphyromonas gingivalis. Mol Oral Microbiol. 2013;28(1):40–53. doi: 10.1111/omi.12004. [DOI] [PubMed] [Google Scholar]
- 28.Ramsey MM, Whiteley M. Polymicrobial interactions stimulate resistance to host innate immunity through metabolite perception. Proc Natl Acad Sci U S A. 2009;106(5):1578–83. doi: 10.1073/pnas.0809533106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Das M, Badley AD, Cockerill FR, Steckelberg JM, Wilson WR. Infective endocarditis caused by HACEK microorganisms. Annu Rev Med. 1997;48:25–33. doi: 10.1146/annurev.med.48.1.25. [DOI] [PubMed] [Google Scholar]
- 30.Christensen JJ, Facklam RR. Granulicatella and Abiotrophia species from human clinical specimens. J Clin Microbiol. 2001;39(10):3520–3. doi: 10.1128/JCM.39.10.3520-3523.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paul-Satyaseela M, Karched M, Bian Z, Ihalin R, Boren T, Arnqvist A, et al. Immunoproteomics of Actinobacillus actinomycetemcomitans outer-membrane proteins reveal a highly immunoreactive peptidoglycan-associated lipoprotein. J Med Microbiol. 2006;55(Pt 7):931–42. doi: 10.1099/jmm.0.46470-0. [DOI] [PubMed] [Google Scholar]
- 32.Karched M, Paul-Satyaseela M, Asikainen S. A simple viability-maintaining method produces homogenic cell suspensions of autoaggregating wild-type Actinobacillus actinomycetemcomitans. J Microbiol Methods. 2007;68(1):46–51. doi: 10.1016/j.mimet.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 33.Fine DH, Furgang D, Schreiner HC, Goncharoff P, Charlesworth J, Ghazwan G, et al. Phenotypic variation in Actinobacillus actinomycetemcomitans during laboratory growth: implications for virulence. Microbiology. 1999;145(Pt 6):1335–47. doi: 10.1099/13500872-145-6-1335. [DOI] [PubMed] [Google Scholar]
- 34.Hoffman H. Gaseous requirements for the cultivation of fusobacteria. J Bacteriol. 1951;61(2):241–2. doi: 10.1128/jb.61.2.241-242.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karched M, Bhardwaj RG, Inbamani A, Asikainen SE: Quantitation of biofilm and planktonic life forms of coexisting periodontal species. Anaerobe 2015, In press. [DOI] [PubMed]
- 36.Rupani D, Izano EA, Schreiner HC, Fine DH, Kaplan JB. Aggregatibacter actinomycetemcomitans serotype f O-polysaccharide mediates coaggregation with Fusobacterium nucleatum. Oral Microbiol Immunol. 2008;23(2):127–30. doi: 10.1111/j.1399-302X.2007.00399.x. [DOI] [PubMed] [Google Scholar]
- 37.Nagaoka S, Hojo K, Murata S, Mori T, Ohshima T, Maeda N. Interactions between salivary Bifidobacterium adolescentis and other oral bacteria: in vitro coaggregation and coadhesion assays. FEMS Microbiol Lett. 2008;281(2):183–9. doi: 10.1111/j.1574-6968.2008.01092.x. [DOI] [PubMed] [Google Scholar]
- 38.Oscarsson J, Karched M, Thay B, Chen C, Asikainen S. Proinflammatory effect in whole blood by free soluble bacterial components released from planktonic and biofilm cells. BMC Microbiol. 2008;8:206. doi: 10.1186/1471-2180-8-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Merritt J, Niu G, Okinaga T, Qi F. Autoaggregation response of Fusobacterium nucleatum. Appl Environ Microbiol. 2009;75(24):7725–33. doi: 10.1128/AEM.00916-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma A, Inagaki S, Sigurdson W, Kuramitsu HK. Synergy between Tannerella forsythia and Fusobacterium nucleatum in biofilm formation. Oral Microbiol Immunol. 2005;20(1):39–42. doi: 10.1111/j.1399-302X.2004.00175.x. [DOI] [PubMed] [Google Scholar]
- 41.Bradshaw DJ, Marsh PD, Watson GK, Allison C. Role of Fusobacterium nucleatum and coaggregation in anaerobe survival in planktonic and biofilm oral microbial communities during aeration. Infect Immun. 1998;66(10):4729–32. doi: 10.1128/iai.66.10.4729-4732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okuda T, Kokubu E, Kawana T, Saito A, Okuda K, Ishihara K. Synergy in biofilm formation between Fusobacterium nucleatum and Prevotella species. Anaerobe. 2012;18(1):110–6. doi: 10.1016/j.anaerobe.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 43.Periasamy S, Kolenbrander PE. Aggregatibacter actinomycetemcomitans builds mutualistic biofilm communities with Fusobacterium nucleatum and Veillonella species in saliva. Infect Immun. 2009;77(9):3542–51. doi: 10.1128/IAI.00345-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kinder SA, Holt SC. Localization of the Fusobacterium nucleatum T18 adhesin activity mediating coaggregation with Porphyromonas gingivalis T22. J Bacteriol. 1993;175(3):840–50. doi: 10.1128/jb.175.3.840-850.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kolenbrander PE, Andersen RN, Moore LV. Coaggregation of Fusobacterium nucleatum, Selenomonas flueggei, Selenomonas infelix, Selenomonas noxia, and Selenomonas sputigena with strains from 11 genera of oral bacteria. Infect Immun. 1989;57(10):3194–203. doi: 10.1128/iai.57.10.3194-3203.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bachrach G, Ianculovici C, Naor R, Weiss EI. Fluorescence based measurements of Fusobacterium nucleatum coaggregation and of fusobacterial attachment to mammalian cells. FEMS Microbiol Lett. 2005;248(2):235–40. doi: 10.1016/j.femsle.2005.05.055. [DOI] [PubMed] [Google Scholar]
- 47.Bouvet A, Grimont F, Grimont PAD. Streptococcus defectivus sp. nov. and Streptococcus adjacens Human Clinical Specimens sp. nov. Nutritionally Variant Streptococci. Int J Syst Bacteriol. 1989;39:290–4. doi: 10.1099/00207713-39-3-290. [DOI] [PubMed] [Google Scholar]
- 48.Roggenkamp A, Abele-Horn M, Trebesius KH, Tretter U, Autenrieth IB, Heesemann J. Abiotrophia elegans sp. nov., a possible pathogen in patients with culture-negative endocarditis. J Clin Microbiol. 1998;36(1):100–4. doi: 10.1128/jcm.36.1.100-104.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suzuki N, Nakano Y, Kiyoura Y. Characterizing the specific coaggregation between Actinobacillus actinomycetemcomitans serotype c strains and Porphyromonas gingivalis ATCC 33277. Oral Microbiol Immunol. 2006;21(6):385–91. doi: 10.1111/j.1399-302X.2006.00309.x. [DOI] [PubMed] [Google Scholar]
- 50.Khemaleelakul S, Baumgartner JC, Pruksakom S. Autoaggregation and coaggregation of bacteria associated with acute endodontic infections. J Endod. 2006;32(4):312–8. doi: 10.1016/j.joen.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 51.Shen S, Samaranayake LP, Yip HK. Coaggregation profiles of the microflora from root surface caries lesions. Arch Oral Biol. 2005;50(1):23–32. doi: 10.1016/j.archoralbio.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 52.Inouye T, Ohta H, Kokeguchi S, Fukui K, Kato K. Colonial variation and fimbriation of Actinobacillus actinomycetemcomitans. FEMS Microbiol Lett. 1990;57(1-2):13–7. doi: 10.1111/j.1574-6968.1990.tb04167.x. [DOI] [PubMed] [Google Scholar]
- 53.Kachlany SC, Planet PJ, Bhattacharjee MK, Kollia E, DeSalle R, Fine DH, et al. Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in bacteria and archaea. J Bacteriol. 2000;182(21):6169–76. doi: 10.1128/JB.182.21.6169-6176.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagayama M, Sato M, Yamaguchi R, Tokuda C, Takeuchi H. Evaluation of co-aggregation among Streptococcus mitis, Fusobacterium nucleatum and Porphyromonas gingivalis. Lett Appl Microbiol. 2001;33(2):122–5. doi: 10.1046/j.1472-765x.2001.00964.x. [DOI] [PubMed] [Google Scholar]
- 55.Periasamy S, Chalmers NI, Du-Thumm L, Kolenbrander PE. Fusobacterium nucleatum ATCC 10953 requires Actinomyces naeslundii ATCC 43146 for growth on saliva in a three-species community that includes Streptococcus oralis 34. Appl Environ Microbiol. 2009;75(10):3250–7. doi: 10.1128/AEM.02901-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin X, Lamont RJ, Wu J, Xie H. Role of differential expression of streptococcal arginine deiminase in inhibition of fimA expression in Porphyromonas gingivalis. J Bacteriol. 2008;190(12):4367–71. doi: 10.1128/JB.01898-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bachtiar EW, Bachtiar BM, Jarosz LM, Amir LR, Sunarto H, Ganin H, et al. AI-2 of Aggregatibacter actinomycetemcomitans inhibits Candida albicans biofilm formation. Front Cell Infect Microbiol. 2014;4:94. doi: 10.3389/fcimb.2014.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bao K, Bostanci N, Selevsek N, Thurnheer T, Belibasakis GN. Quantitative Proteomics Reveal Distinct Protein Regulations Caused by Aggregatibacter actinomycetemcomitans within Subgingival Biofilms. PLoS One. 2015;10(3) doi: 10.1371/journal.pone.0119222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xavier KB, Bassler BL. Interference with AI-2-mediated bacterial cell-cell communication. Nature. 2005;437(7059):750–3. doi: 10.1038/nature03960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rosen G, Nisimov I, Helcer M, Sela MN. Actinobacillus actinomycetemcomitans serotype b lipopolysaccharide mediates coaggregation with Fusobacterium nucleatum. Infect Immun. 2003;71(6):3652–6. doi: 10.1128/IAI.71.6.3652-3656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kuriyama T, Karasawa T, Nakagawa K, Nakamura S, Yamamoto E. Antimicrobial susceptibility of major pathogens of orofacial odontogenic infections to 11 beta-lactam antibiotics. Oral Microbiol Immunol. 2002;17(5):285–9. doi: 10.1034/j.1399-302X.2002.170504.x. [DOI] [PubMed] [Google Scholar]


