Abstract
At least 10 hepatitis B virus (HBV) genotypes (A to J) with distinct geographic distributions and several HBV mutants, including precore/core promoter mutations and pre-S/S deletion mutations, have been recognized to be not only predictive of liver disease progression but also associated with response to antiviral therapy. HBV genotype–specific pathogenesis may contribute to heterogeneous clinical outcomes in chronic hepatitis B patients across the world. For example, patients with HBV genotypes C and D infection have a lower rate of spontaneous HBeAg seroconversion. In addition, HBV genotypes C and D have a higher frequency of core promoter and pre-S mutations than genotypes A and B. Genotypes C and D also carry a higher lifetime risk of cirrhosis and HCC development than genotypes A and B. Core promoter and pre-S mutations also correlate with an increased risk of hepatocellular carcinoma (HCC). Therapeutically, genotypes A and B patients have a better response to interferon-based therapy than genotypes C and D patients, but the response to nucleos(t)ide analogs is comparable across different HBV genotypes. In conclusion, HBV genotypes and variants may serve as viral genetic markers to predict disease progression as well as help practicing physicians optimize individualized antiviral therapy in clinical practice.
At least 10 hepatitis B virus genotypes exist globally. Some of these genotypes are associated with increased risk of liver disease, and others are predictive of therapeutic response.
Hepatitis B virus (HBV) is one of the most common viral infections in humans, which is endemic in Asia, Pacific Islands, Africa, Southern Europe and Latin America. The prevalence of chronic HBV infection in the general population of different countries ranges from 2% to 20% (Kao and Chen 2002). Persistent HBV infection has a wide spectrum of clinical manifestations, including inactive carrier state, chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (HCC) (Kao and Chen 2002). Eventually, 15%–40% of HBV carriers have a lifetime risk to develop cirrhosis, liver failure, or HCC (Fattovich et al. 2008).
HBV is the smallest human DNA virus with a genome of 3200 bp (Lau and Wright 1993). Through reverse transcription, pregenomic RNA can be transcribed from covalently closed circular DNA (cccDNA) to serve as the template of negative-strand DNA and then fully double-stranded DNA through DNA polymerase within the nucleocapsid, finally with the assembly of envelope protein to form mature HBV virions (Beck and Nassal 2007). Because of the spontaneous error rate of viral reverse transcriptase, HBV genome evolves with an estimated rate of nucleotide substitution at 1.4–3.2 × 10−5/sites/yr (Lau and Wright 1993). This unique replication strategy leads to the occurrence of various genotypes, subtypes, mutants, recombinants, and even quasispecies in the context of the long-term evolution of HBV (Hunt et al. 2000; Kao 2002, 2003; Kao and Chen 2006).
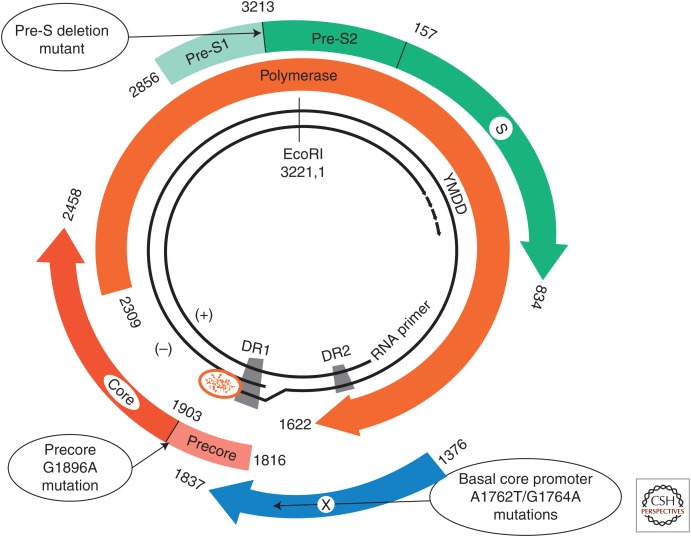
On the basis of recent advances in molecular biology techniques, genotypic classifications of HBV and their geographic/ethnic distributions have been increasingly recognized. Ample evidence reveals that HBV genotype is associated with HBV endemicity, transmission mode, as well as clinical outcomes (Kao 2002, 2003; Kao and Chen 2006; Lin and Kao 2011, 2013c). In addition, naturally occurring variations in the HBV genome also have clinical and epidemiological implications. Several HBV mutants, including precore/core promoter mutations and pre-S/S deletion mutations, have been reported to be associated with progressive liver disease and risk of HCC development (Fig. 1) (Chotiyaputta and Lok 2009; Kao et al. 2010). In this article, recent advances regarding the impact of HBV genotypes and variants on the disease progression, risk of HCC development, and responses to antiviral treatments in chronic hepatitis B (CHB) patients will be reviewed and discussed.
Figure 1.
The partially double-stranded circular DNA of hepatitis B virus encodes four overlapping open reading frames: S for the surface or envelope gene; C for the core gene; P for the polymerase gene; and X for the X gene. Naturally occurring mutant strains including mutations in precore, core promoter, and deletion mutation in pre-S genes have been reported to be associated with the pathogenesis of progressive liver disease and risk of hepatocellular carcinoma (HCC) development. (From Kao et al. 2010; adapted, with permission, Elsevier © 2010.)
GLOBAL DISTRIBUTION OF HBV GENOTYPES AND SUBTYPES
The hepatitis B surface antigen (HBsAg) derived from different HBV strains carries serologically defined group-specific antigenic determinants. Based on different serological reactivities of HBsAg, HBV can be classified into four serotypes, adw, adr, ayw, and ayr (Le Bouvier et al. 1972; Tiollais et al.1985), with specific geographical distributions (Nishioka et al. 1975; Sung and Chen 1977). In addition, according to the homogeneity of the virus sequence, >8% or 4–8% genetic divergence, at least 10 HBV genotypes (A to J) and several subtypes have been identified, also with specific geographical distributions (Datta 2008; Cao 2009; McMahon 2009; Kurbanov et al. 2010). For example, genotype A is highly prevalent in Africa, Europe, India, and America. Genotypes B and C are common in the Asia-Pacific region. Genotype D is prevalent in Africa, Europe, the Mediterranean region, and India. Genotype E is restricted to West Africa. Genotype F is found in Central and South America. Genotype G has been reported in France, Germany, and the Americas. Genotype H is found in Central America and Mexico. Genotype I was isolated in Vietnam and Laos (Tran et al. 2008; Phung et al. 2010). The newest HBV genotype, J, was identified in Japan (Tatematsu et al. 2009). The correlation and geographical distributions of HBV serotypes, genotypes, and subtypes are shown in Table 1. A molecular epidemiologic study showed that, in Taiwan, adw and adr account for 70% and 30% of HBV strains, respectively. All adr strains are genotype C, whereas 81% and 12% of the adw strains are genotypes B and C, respectively (Liu et al. 2002). Another study from Korea revealed extraordinary predominance of HBV genotype C2 in CHB patients; 92.3% and 5.7% were adr and adw serotypes, respectively (Kim et al. 2007). Similar to the specific global distribution of HBV genotypes, different transmission modes of HBV are shown (Kao and Chen 2002). For example, genotypes B and C are prevalent in highly endemic areas, such as Asian countries, where perinatal or mother-to-infant transmission plays an important role in spreading HBV; whereas the remaining genotypes are frequently found in areas in which horizontal transmission (close personal contact between young children, blood, or sexual interactions between adults) is the most common route of HBV infection. Thus, distinct modes of HBV transmission have been associated with a specific geographical distribution of the HBV genotypes. In Taiwan, HBV genotyping has been applied to investigate the modes of intrafamilial HBV transmission (Lin et al. 2005); the prevalence of HBsAg carriage in children from families clustering with HBV carriers was significantly higher than that in the general population (77.8% vs. 15%). Possible intrafamilial modes of transmission were determined by identifying the concordant HBV genotype between carrier children and their parents (Lin et al. 2005). The modes of transmission may also influence the distribution of HBV in a given country in which universal hepatitis B vaccination has not yet been launched. For example, through promiscuous sexual contacts, HBV genotype A is prevalent in patients with acute hepatitis B in Japan (Yotsuyanagi et al. 2005). In a nationwide survey, Matsuura et al. further found that the prevalence of HBV genotype A in chronic hepatitis B patients in Japan increased from 1.7% in year 2000 to 3.5% in 2006 (Matsuura et al. 2009). Therefore, HBV genotyping can serve as an epidemiologic marker to determine the correlation of HBV genotype distribution with modes of transmission, linked to important social behavioral changes.
Table 1.
Geographic distribution of hepatitis B virus genotypes and subtypes
| Genotypes | Serotypes | Subtypes | Geographic location |
|---|---|---|---|
| A | adw | A1 | Sub-Saharan Africa and India |
| A2 | Northern Europe and India | ||
| A3 | Western Africa | ||
| B | adw, ayw | B1 | Japan |
| B2–5 | East Asia, Taiwan, China, Indonesia, Vietnam, and the Philippines | ||
| B6 | Alaska, Northern Canada, and Greenland | ||
| C | adw, ayr, adr | C1–3 | Taiwan, China, Korea, and Southeast Asia |
| C4 | Australia | ||
| C5 | The Philippines and Vietnam | ||
| C6–11 | Indonesia | ||
| D | ayw | D1–6 | Africa, Europe, Mediterranean countries, India, and Indonesia |
| E | ayw | Restricted to West Africa | |
| F | adw | F1–4 | Central and South America |
| G | adw | France, Germany, and the United States | |
| H | adw | Central America | |
| I | adw | Vietnam and Laos | |
| J | Japan |
Intergenotypic Recombination
The quasi-species nature of HBV infection has been documented (Ngui and Teo 1997). In addition, high prevalence of more than one dominant genotype in a certain region is common, especially for genotypes B and C in Asian-Pacific regions, or genotypes A and D in Western countries. Therefore, mixed infection with different HBV genotypes is not uncommon and of great virological and clinical interest. For example, previous large cohort studies in Taiwan showed that the prevalence of mixed HBV genotype infections was 16.3% and 34.4%, respectively, in HBsAg-positive and occult HBV–infected intravenous drug users (Chen et al. 2004a; Lin et al. 2007a). Coinfection with different HBV genotypes further leads to recombination of different viral strains (Locarnini et al. 2013). Intergenotypic recombination of HBV strains has been widely reported. Most of the recombinants were genotypes B/C or A/D hybrids. Genotypes A and C showed a higher recombination tendency than did other genotypes (Yang et al. 2006). For example, HBV genotype B2 is a recombinant, in which the majority of the genetic framework is HBV genotype B, and the precore/core region is from genotype C (Sugauchi et al. 2002). HBV genotypes C/D recombinant is dominant in the Qinghai-Tibet plateau in Southwest China (Wang et al. 2005; Zhou et al. 2011; Shen et al. 2014). HBV genotype I, a novel intergenotypic recombination among genotypes A, C, and G was isolated in Vietnam and Laos (Tran et al. 2008; Phung et al. 2010). The newest HBV genotype, J, was identified in the Ryukyu Islands in Japan, and this genotype has a close relationship with gibbon genotypes and human genotype C (Tatematsu et al. 2009). Recombination is important for HBV evolution. Further studies are required to clarify the mechanisms of intergenotypic recombination and the clinical significance of these HBV recombinants.
THE IMPACT OF HBV GENOTYPES AND VARIANTS ON THE NATURAL COURSE OF CHB
As a noncytopathic virus, the immunopathogenesis of HBV infection is mainly mediated by cellular responses to epitopes of HBV proteins expressed on the surface of hepatocytes with consequent liver injury (Chisari and Ferrari 1995; Ganem and Prince 2004; Rehermann 2013). Persistent HBV replication may trigger strong and continued immune responses against the virus and result in severe liver damage (Perrillo 2001). The relationship of HBV genotypes and the tendency of chronic infection has been elucidated. Ito et al. reported 212 adult patients with acute hepatitis B recruited from 2005 to 2010 in Japan. The persistence of HBV infection was higher in patients with genotype A infection (23.4%) than those with non-A genotype (8.6%) infection (P = 0.003). In addition, multivariate logistic regression analysis revealed that only genotype A was independently associated with viral persistence following acute hepatitis B (Ito et al. 2014). These data suggest that infection with genotype A prevails in patients with acute hepatitis B in Japan where genotypes B and C are common, and tends to persist. In a European study of 65 patients, genotype D was more prevalent in patients with acute, self-limited hepatitis B as compared with genotype A (80% vs. 10%, P < 0.01). In contrast, genotype A predominated over genotype D in patients with chronic HBV infection (80% vs. 11%, P < 0.01) (Mayerat et al. 1999). In general, the rate of chronicity following acute genotypes A and D infection were reported to be high compared with genotypes B and C (Heijtink et al. 1999; Mayerat et al. 1999; Kobayashi et al. 2004; Leblebicioglu and Eroglu 2004; Suzuki et al. 2005; Ito et al. 2014). Taken together, the persistence of HBV infection after acute infection may be attributable to the variable strength of host–viral interactions, the modes of transmission as well as the varying distribution of genotypes.
In the natural history of chronic HBV infection, seroconversion of hepatitis B e antigen (HBeAg) and seroclearance of HBsAg are landmark events in the control of HBV infection. Earlier HBeAg seroconversion with subsequent low viral load (<2000 IU/mL) usually confers a favorable clinical outcome (Liu et al. 2013a), whereas late or absent HBeAg seroconversion after multiple hepatitis flares may accelerate the progression of chronic hepatitis leading to cirrhosis; it, therefore, has a poor clinical outcome (Chen et al. 2002; Hsu et al. 2002; Yang et al. 2002; Chu and Liaw 2007; Liu et al. 2013a). The pathogenic differences among various HBV genotypes have been partially clarified and influence the clinical outcomes of HBV infection (Table 2). In our cohort study on 272 Taiwanese patients with chronic HBV infection, genotype C patients were more likely to have HBeAg-positive chronic hepatitis B despite multiple hepatitis flares (Kao et al. 2002a). In addition, genotype C infection was associated with lower rates of spontaneous HBeAg seroconversion than genotype B (27% vs. 47%, P < 0.025) during the follow-up. The estimated annual rates of HBeAg seroconversion in genotypes B and C infections were 15.5% and 7.9%, respectively (Kao et al. 2004). Furthermore, a long-term follow-up study with 460 Taiwanese HBV chronically infected children indicated that the positive rate of HBeAg after 20 years of follow-up was 70% in genotype C and 40% in genotype B carriers (Ni et al. 2004). Accordingly, genotype C patients experience delayed HBeAg seroconversion and, thus, have a longer duration of high HBV replication than genotype B patients.
Table 2.
HBV genotype–specific implications in patients with chronic hepatitis B
| Genotypes compareda | B versus C | A versus D | ||
|---|---|---|---|---|
| B | C | A | D | |
| Virological implications | ||||
| Serum HBV DNA level | Lower | Higher | NDb | ND |
| Frequency of precore A1896 mutation | Higher | Lower | Lower | Higher |
| Frequency of basal core promoter A1762T/G1764A mutation | Lower | Higher | Lower | Higher |
| Frequency of pre-S deletion mutation | Lower | Higher | ND | ND |
| Intracellular expression of HBV DNA | Lower | Higher | Lower | Higher |
| Secretion of HBeAg | Lower | Higher | ND | ND |
| Immunologic implications | ||||
| Tendency of chronic infection | Higher | Lower | Lower | Lower |
| HBeAg seroconversion | Earlier | Later | Earlier | Later |
| HBsAg seroclearance | More | Less | More | Less |
| Histologic activity | Lower | Higher | Lower | Higher |
| Clinical implications | ||||
| Modes of transmission | Perinatal/vertical | Perinatal/vertical | Horizontal | Horizontal |
| Incidence of progression to cirrhosis and hepatocellular carcinoma (HCC) | Lower | Higher | Lower | Higher |
| Response to interferon-based therapy | Higher | Lower | Higher | Lower |
| Response to nucleos(t)ide analogs | c | c | c | c |
aBecause of the unique distribution of HBV genotypes in Asian and Western countries, sufficient data for meaningful comparisons are available only for comparisons between genotypes B and C or between genotypes A and D.
bND, no available data.
cNo significant difference between genotypes A to D.
Regarding genotypes A and D, one prospective study of Spanish patients with chronic HBV infection did not reveal any differences in the probability of HBeAg seroconversion between patients infected with genotypes A and D. However, the rate of sustained remission after HBeAg seroconversion was higher in genotype A patients (55% vs. 32%, P < 0.01) (Sanchez-Tapias et al. 2002). In addition, compared with genotypes C and D patients, genotypes A and B patients had a higher rate of spontaneous HBsAg seroclearance (Sanchez-Tapias et al. 2002; Yuen et al. 2004a). Taking these lines of evidence together, genotypes C and D patients, compared with genotypes A and B patients, have late or absent HBeAg seroconversion after multiple hepatitis flares that may accelerate the progression of chronic hepatitis, thereby conferring a worse clinical outcome.
The possible influence of other genotypes on the natural history of HBV infection remains limited. A previous study on vaccinated children in the Gambia showed that breakthrough HBV infection in these children was mainly caused by the wild-type genoytype E strain and that immune escape mutants were uncommon (Mendy et al. 2008). On the contrary, among vaccinated Taiwanese children (genotypes B and C infection are common) with breakthrough HBV infection, S gene mutations have become prominent (Chang 2010). Further investigations are awaited to determine the relationship between HBV genotype and vaccine response.
A recent retrospective study from Argentina revealed that 62% of acute hepatitis B and 37% of chronic hepatitis B were caused by genotype F HBV. Among chronic cases, genotype F tended to display higher histological activity indexes (Pezzano et al. 2011). Roman et al. (2010) further reported a high prevalence (14%) of occult HBV infection in Mexico natives, a low endemic area of HBV infection. In addition, HBV genotype H was predominant in those with occult HBV infection in Mexico (Panduro et al. 2013). The low incidence of cirrhosis and HCC in Mexico may be reasoned by the virulence of genotype H is different from other HBV genotypes in high endemic areas (Roman et al. 2009).
Naturally occurring HBV mutants can display important variation in epitopes that are important in host immune recognition, enhance virulence with increased levels of HBV replication, or facilitate cell attachment/penetration, thereby having basic and clinical significance (Fig. 1) (Hunt et al. 2000; Kao et al. 2010). These mutant strains, including the HBV precore nucleotide 1896 stop codon mutation and basal core promoter (BCP) A1762T/G1764A variants, are known to be responsible for ongoing HBV DNA replication even after HBeAg seroconversion and lead to prolongation of chronic hepatic inflammation (Lin and Kao 2008; Mansourian et al. 2013). These findings suggest that hepatitis B viral characteristics may play an important role in HBV genotype–specific pathogenesis. For example, it has been shown that different HBV genotypes have different HBV replication capacities, both in vitro and in vivo. In an in vitro study, intracellular expression of HBV DNA was higher for genotypes C than B and genotypes D than A (Sugiyama et al. 2006). Furthermore, the secretion of HBeAg in genotype B was lower than that in genotype C (Liu et al. 2011). The intracellular accumulation of HBV DNA may, thus, induce liver cell damage. In addition, the higher replication capacity of genotype C explains why this genotype is associated with more severe liver disease than others. Further investigation revealed an increase of intracellular core protein expression when BCP A1762T/G1764A variants were introduced in the genotype C strains (Liu et al. 2011). Our in vivo study also revealed that HBV BCP A1762T/G1764A variants were significantly associated with cytoplasmic localization of intracellular HBcAg, which were closely related to active necroinflammation of hepatocytes (Liu et al. 2009a). Similarly, genotype D–infected patients who had more progressive liver disease showed a higher prevalence of BCP A1762T/G1764A variants than those with genotype A infection (Sharma et al. 2010).
In summary, the specific virological manifestations of HBV genotypes and variants include (1) genotype C shows a higher frequency of BCP A1762T/G1764A variants and pre-S deletion mutations than genotype B; (2) serum HBV viral load is higher in genotype C than genotype B; (3) the expression of intracellular HBV DNA increases in genotype C; (4) the expression of intracellular core protein increases in genotype C with BCP A1762T/G1764A variants; and (5) more HBeAg is secreted by genotype C than by genotype B. These findings may partly explain why genotype C is associated with more severe liver disease than other genotypes (Kao 2011; Vutien et al. 2013).
HBV GENOTYPES AND RISK OF HCC DEVELOPMENT
As already highlighted, most retrospective or case-control studies indicated that patients with genotype C infection have more severe liver disease, including cirrhosis and HCC, than those with genotype B (Kao et al. 2000a, 2003; Chan et al. 2004; Yuen et al. 2004b). A community-based prospective cohort study on 2762 Taiwanese HBV carriers showed that HBV genotype C was associated with an increased risk of HCC than genotype B; the adjusted hazard ratio (HR) was 2.35 (95% confidence interval [CI]: 1.68–3.30; P < 0.001) (Yang et al. 2008). In a prospective study with 4841 Taiwanese, male, and HBV-infected patients without HCC at enrollment, Yu et al. found that HBV viral load was higher in genotype C than genotype B patients, whereas genotype C–infected patients who also had very high viral load had a 26-fold higher risk of HCC than those with other genotypes and low or undetectable viral loads (Yu et al. 2005). Our recent hospital-based cohort study on 2688 Taiwanese HBsAg-positive patients without evidence of cirrhosis for a mean time period of 14.7 years (elucidation of risk factors for disease control or advancement in Taiwanese hepatitis B carriers [ERADICATE-B] cohort) also showed that genotype C patients have a higher annual incidence rate of HCC than genotype B patients by univariate analysis (Tseng et al. 2012a). These findings confirm that genotype C correlates with a higher risk of HCC development. Of interest, several reports showed HBV genotype B is associated with risk of HCC development at a young age, whereas genotype C is associated with HCC development at an older age (Kao et al. 2000a; Ni et al. 2004; Yin et al. 2008).
HBV genotype also influences the clinicopathological features of patients with resectable HCC. In Taiwan, among 193 HBV-related HCC patients receiving surgery, genotype B patients had a higher rate of solitary tumor (94% vs. 86%, P = 0.048) but more satellite nodules (22% vs. 12%, P = 0.05) than genotype C patients (Chen et al. 2004b; Lin et al. 2007b). Wu et al. (2009) also reported that in HCC patients, liver inflammation activity was higher in HBV genotype C patients than in genotype B patients (P = 0.023), and more genotype C patients tended to have a higher viral load (>106 copies/ml) than genotype B patients (52.3% vs. 37.6%, P = 0.067). These characteristics may contribute to the recurrence patterns and prognosis of HBV-related HCC patients with genotypes B or C infection.
Compared with HBV genotypes, the clinical significance of HBV subtypes remains less clear. A study analyzed the distribution of HBV subtypes in 296 HBV-related HCC patients collected from all across Japan (Orito et al. 2005). They found HBV subtype B2 in 4.4%, B1 in 7.4%, and genotype C in 86.5%. Interestingly, in the Tohoku district and Okinawa, subtypes B2, B1, and genotype C were found in 6.7%, 40.0%, and 48.9%, respectively, compared with 4.0%, 1.6%, and 93.2% in the other districts in Japan. These data suggest that HBV subtype B1 may run a more indolent course than subtype B2. A study of 242 Taiwanese HBsAg carriers revealed that there was no significant difference in the distribution of the HBV genotype C subtypes among patients with different stages of liver disease, suggesting that subtypes of genotype C may have minimal impact on liver disease progression of chronic hepatitis B in Taiwan (Tseng et al. 2007). However, another prospective study on 1006 CHB patients with a median follow-up of 7.7 years from Hong Kong showed that subtype C2 has the highest risk of HCC (HR: 2.75; 95% CI: 1.66–4.56; P < 0.0001) and subtype C1 has intermediate risk (HR: 1.70; 95% CI: 1.09–2.64; P = 0.020) compared with genotype B (Chan et al. 2008). In addition, multiple mutations in subtype C4 were associated with more rapid liver disease progression and an increased risk of HCC in indigenous Northern Australian populations (Littlejohn et al. 2014). Genotypes B2 and C4 have been shown to be recombinants with other genotypes, which may play an important role in natural history and pathogenesis. Further studies from different parts of world with detailed characterization of the strains, are needed to confirm the association between HBV subtypes and risk of HCC development.
Although limited studies have examined the relationship between other HBV genotypes and the risk of HCC development, HCC is more frequent in patients with HBV genotypes D and F infection than those with genotype A infection (Thakur et al. 2002; Livingston et al. 2007). A prospective study with Alaska native HBV carriers revealed that the risk of HCC development was significantly higher for genotype F than genotypes A–D (P < .001; OR: 7.73; 95% CI: 3.69–16.4; P < 0.001) (Livingston et al. 2007).
HBV VARIANTS AND RISK OF HCC DEVELOPMENT
Of the various naturally occurring HBV mutants, several mutations in the X gene of the HBV genome are frequently found in patients with HCC (Yeh et al. 2000; Chen et al. 2005; Guo et al. 2008; Ma et al. 2008), suggesting that these mutants may play a significant role in the malignant potential of HBV. Several studies showed that 3′-end X gene is frequently deleted in HCC cells, leading to a carboxy-terminal truncated HBx protein (Iavarone et al. 2003; Wang et al. 2004). Subsequent studies showed carboxy-terminal truncated HBx in ∼80% of HCC tissues, and may contribute to hepatocarcinogenesis via the activation of cell proliferation and loss of proapoptotic ability (Ma et al. 2008).
In a cohort study of genotypes B– or C–infected HBV carriers, genotype C had a higher prevalence of BCP A1762T/G1764A variants than genotype B (odds ratio [OR]: 5.18; 95% CI: 2.59–10.37; P < 0.001). Patients with BCP A1762T/G1764A variants were more significantly associated with the development of HCC than those without (HR: 10.6; 95% CI: 4.92–22.86; P < 0.001) (Kao et al. 2003). These findings were further confirmed by a long-term follow-up study involving 1526 HBV carriers (risk evaluation of viral load elevation and associated liver disease/cancer-hepatitis B virus [REVEAL-HBV] study), revealing the presence of BCP A1762T/G1764A variants as an independent predictor for progression to HCC (HR: 1.73; 95% CI: 1.13–2.67; P = 0.013) (Yang et al. 2008). In addition, a meta-analysis produced a summary odds ratio of HCC for BCP A1762T/G1764A variants that was 3.79 (95% CI: 2.71–5.29) (Liu et al. 2009b). Recently, BCP A1762T/G1764A variants were determined both qualitatively and quantitatively to correlate with cirrhosis in Taiwanese HBV carriers with genotypes B or C infection. BCP A1762T/G1764A variants served as an independent risk factor for cirrhosis development (HR: 4.26; 95% CI: 1.32–13.77). Quantitative analysis using pyrosequencing revealed that risk of cirrhosis was higher in patients with BCP A1762T/G1764A variants ≥45% compared with <45% (adjusted OR: 2.81; 95% CI: 1.40–5.67; P = 0.004) (Tseng et al. 2014). Finally, the BCP A1762T/G1764A variants also affect the carboxy-terminal codons 130 and 131 of the X protein (K130M and V131I) because of the overlapping nature of the HBV genome. Taking these lines of evidence together, BCP A1762T/G1764A variants may select out amino acid changes in the X protein, which increase the fibrogenic activity and promote hepatocarcinogenesis, regardless of HBV genotypes.
Mutations in enhancer II (C1653T) and elsewhere in the basal core promoter (T1753V) have also been found to be associated with HCC development. A case-control study from Hong Kong revealed that patients with C1653T mutation had a significantly higher risk of HCC than those without (OR: 2.43; 95% CI: 1.08–5.54; P = 0.028) (Yuen et al. 2008). Another cohort study from Taiwan indicated that patients with the T1753V mutation had a significantly higher risk for developing HCC than those without this change (OR: 2.43; 95% CI: 1.33–4.44; P = 0.028) (Chou et al. 2008).
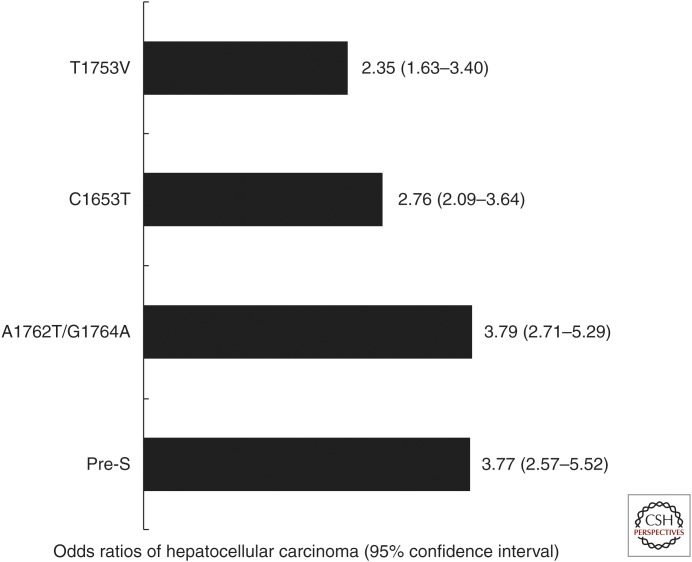
Previous reports also showed that the deletion mutations within the pre-S gene were significantly associated with the development of cirrhosis and HCC (Chen et al. 2007; Lin et al. 2007c; Fang et al. 2008). The proposed mechanism is that the endoplasmic reticulum stress associated with pre-S accumulation induced oxidative DNA damage and so, pre-S gene deletion mutation variants may lead to mutagenesis of the host genome, thereby contributing to hepatocarcinogenesis (Hsieh et al. 2004). In our case-control study, the presence of pre-S deletion was an independent risk factor associated with HCC development (OR: 3.72; 95% CI: 1.44–9.65; P = 0.007). In addition, the frequency of pre-S deletion was significantly higher in genotype C patients than genotype B patients (Lin et al. 2007c). A meta-analysis further confirmed that the OR of HCC for pre-S deletion was 3.77 (95% CI: 2.57–5.52). Of particular note, the summary OR for pre-S deletion was higher in genotype C patients than genotype B patients (Liu et al. 2009b). Our previous mapping study of pre-S regions suggested that all the deletion regions encompassed T- and B-cell epitopes, and most of them lost one or more functional sites, including the polymerized human serum albumin-binding site and nucleocapsid-binding site. Therefore, HBV pre-S deletion mutations may also lead to defective immunity against HBV and contribute directly to progressive liver cell damage and finally hepatocarcinogenesis (Chen et al. 2006). The odds ratios of HCC for HBV carriers by mutations in core promoter and pre-S regions are shown in Figure 2. Furthermore, subsequent investigations have shown that the combination of pre-S deletion and core promoter mutations rather than either single mutation profile was strongly associated with disease progression and development of HCC (Chen et al. 2006, 2007; Yuen et al. 2008; Liu et al. 2009b).
Figure 2.
Odds ratios of hepatocellular carcinoma for HBV carriers by mutations in core promoter and pre-S regions in a meta-analysis. (Imaged created from data in Liu et al. 2009b.)
In addition to the potential hepatocarcinogenesis, HBV variants also influence the postoperative prognosis of patients with HCC. In a study of 185 liver samples obtained from the noncancerous part of surgically removed HBV-associated HCC tissues, the presence of the BCP mutation independently predicted disease-free survival (adjusted HR: 2.075; 95% CI: 1.203–3.579). In addition, short stretch (<100 bp) pre-S deletions were significantly associated with poorer disease-free survival (P = 0.005) (Yeh et al. 2010).
CLINICAL SIGNIFICANCE OF QUANTITATIVE HBsAg AND ITS CORRELATION WITH HBV GENOTYPES AND VARIANTS
Recently, quantitative hepatitis B surface antigen (qHBsAg) has been increasingly recognized to be a promising biomarker to predict both favorable and adverse outcomes of HBV carriers (Tseng et al. 2011a, 2012a,b, 2013; Lin and Kao 2013a). An Italian study showed that the combination of qHBsAg <1000 IU/ml and serum HBV DNA level <2000 IU/ml can predict the inactive HBV carrier state in HBV genotype D patients (Brunetto et al. 2010). Our recent study further showed that low serum levels of HBsAg (<100 IU/ml) at 1 year after HBeAg seroconversion could predict HBsAg loss in patients with HBV genotypes B or C infection (Tseng et al. 2011a). In addition, qHBsAg was better than serum HBV DNA level for the prediction of spontaneous HBsAg loss in HBeAg-negative carriers with a low viral load (<2000 IU/mL). Low serum HBsAg level was the strongest predictor of spontaneous HBsAg seroclearance in patients with a low viral load (Tseng et al. 2012b; Liu et al. 2013b). In the recent update of REVEAL-HBV study with more than 3000 HBV carriers, qHBsAg, HBV DNA levels, and HBV genotype C were all independent predictors of HCC development. Of particular note, qHBsAg was significantly associated with cirrhosis and HCC in a dose-response manner (P for trend <0.001) in the HBeAg-negative patients with low serum HBV DNA levels (<200,000 IU/mL) (Lee et al. 2013). Our hospital-based ERADICATE-B cohort study also showed similar findings. A total of 2688 noncirrhotic Taiwanese chronic hepatitis B patients were followed for a mean of 14.7 years HCC risk increased when patients had increased HBV DNA levels (HR: 4.7; 95% CI: 2.2–10.0), increased qHBsAg (HR: 7.2; 95% CI: 1.8–28.6), and elevated ALT levels (HR: 6.6; 95% CI: 2.2–19.8). In addition, for patients with low serum HBV DNA levels (<2000 IU/mL), qHBsAg ≥ 1000 IU/mL was an independent risk factor for HCC development (HR: 13.7; 95% CI: 4.8–39.3) (Tseng et al. 2012a). A recent study evaluated the correlation between HBV DNA and HBsAg levels according to HBV genotype. These investigators found that serum HBsAg level tended to correlate with HBV DNA level for genotype A, but did not reach significant for genotype D (Tuaillon et al. 2012). We recently followed 187 patients with chronic HBV infection for a median of 8 years. We found that inactive carriers had a significantly lower qHBsAg at baseline and during follow-up compared with patients with elevated serum HBV DNA levels. However, the longitudinal qHBsAg change was independent of genotypes B or C, the most common genotypes in Taiwan (Su et al. 2013). More studies are definitely warranted to clarify the relationship between HBV genotype and qHBsAg level in different clinical situations as well as other geographical locations.
THE IMPACT OF HBV GENOTYPES ON RESPONSE TO ANTIVIRAL THERAPY
The therapeutic endpoints for chronic hepatitis B treatment include sustained suppression of HBV replication to below the detection limit of real-time PCR assays, biochemical remission, histological improvement, HBeAg loss, or HBeAg seroconversion for HBeAg-positive patients and, ideally, HBsAg loss or even HBsAg seroconversion (Lok and McMahon 2009; EASL 2012; Liaw et al. 2012). Currently, two types of therapy are recommended: standard interferon (IFN) or pegylated interferon (PEG-IFN) and nucleos(t)ide analogs (NUCs), including lamivudine, telbivudine, entecavir, adefovir dipivoxil, and tenofovir disoproxil (Lok and McMahon 2009; EASL 2012; Liaw et al. 2012). The impact of HBV genotype on therapeutic responses to both IFN-based and NUCs has been increasingly recognized (Liu et al. 2005; Liu and Kao 2008; Lin and Kao 2013b,c). Because patients infected with genotypes E–J are less common, their responses to antiviral therapy remain largely unknown. Therefore, the influences of HBV genotypes on response to antiviral therapy can only be reliably shown for genotypes A, B, C, and D (Table 2).
Interferon-Based Therapy
In HBeAg-positive patients treated with standard IFN, patients with genotypes A and B had a significantly higher rate of HBeAg seroconversion posttreatment than those with genotypes C and D (Kao et al. 2000b; Hou et al. 2001; Wai et al. 2002; Erhardt et al. 2005). For the HBeAg-positive Asian population, genotype B HBV–infected patients show higher response rates with IFN-based therapies, regardless of pegylated or standard type IFN products, whereas genotype C–infected patients have a higher likelihood of response to PEG-IFN compared with standard IFN (Cooksley et al. 2003; Lau et al. 2005). Furthermore, HBV genotype B infection and younger age were independent factors associated with sustained response of low-dose, 24-wk IFN regimen in HBeAg-positive Chinese patients (Zhao et al. 2007). Another multicenter study on PEG-IFN for HBeAg-positive patients revealed that the rate of HBeAg clearance also differed according to the infecting HBV genotype: genotype A, 47%; genotype B, 44%; genotype C, 28%; and genotype D, 25% (Janssen et al. 2005). Subsequent analysis consistently showed a higher rate of HBsAg clearance in genotype A compared with other genotypes in both HBeAg-positive and HBeAg-negative CHB patients (Flink et al. 2006). In addition, compared with genotypes C and D patients, durable loss of HBeAg at 3 yr after PEG-IFN treatment was higher in genotypes A and B patients (Buster et al. 2008). Among HBeAg-negative patients treated with PEG-IFN, HBsAg clearance was significantly higher in genotype A (20%) than genotype B (6%), genotype C (9%), and genotype D (6%) (Marcellin et al. 2009). On the basis of existing data, a meta-analysis further confirmed that HBV genotypes are informative concerning responses to IFN-based therapy. HBV genotype A has a better response to IFN treatment than genotype D patients, regardless of HBeAg status. HBV genotype B has a higher response rate to IFN treatment than genotype C in HBeAg-positive patients (Wiegand et al. 2008). Recent pooled data from two largest global trials of HBeAg-positive patients with PEG-IFN treatment showed that higher levels of ALT and lower levels of HBV DNA predicted a sustained response to PEG-IFN therapy for genotypes A–, B–, and C–infected patients. On the contrary, genotype D–infected patients had the lowest chance of sustained response, irrespective of ALT or HBVDNA levels (Buster et al. 2009).
More on-treatment decline of qHBsAg has been shown in patients with IFN-based therapy than in those with NUC therapy (Su et al. 2010). Recent data further revealed the correlation between the declines of qHBsAg and HBV genotype in patients receiving IFN-based therapy (Tseng and Kao 2013). The on-treatment decline of qHBsAg level has proven a useful predictor of response for IFN-based therapy. For HBeAg-positive genotypes B and C patients, HBeAg seroconversion was significantly associated with serum HBsAg level <1500 IU/mL at wk 12 of PEG-IFN treatment, whereas patients with serum HBsAg level >20,000 IU/mL at wk 12 of PEG-IFN treatment did not respond (Liaw et al. 2011; Sonneveld et al. 2013).
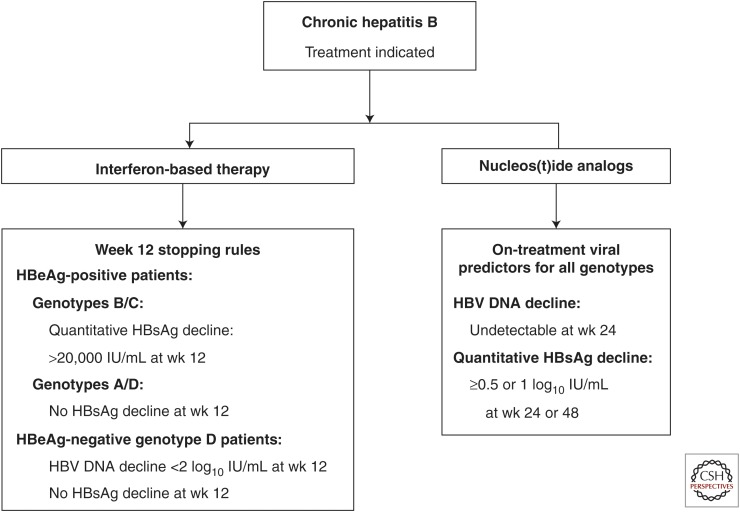
Similarly, for HBeAg-positive genotypes A and D patients, no change in the serum HBsAg level at wk 12 of PEG-IFN predicted a poor response of HBeAg loss at 26 wk after treatment (Sonneveld et al. 2010). Recently, a futility rule has been developed in which no HBsAg decline and <2 log copies/ml decline in HBV DNA at wk 12 of therapy with PEG-IFN has been proposed. This has been validated in genotype D HBeAg–negative patients (Rijckborst et al. 2010, 2012). The HBsAg kinetics during PEG-IFN treatment also varied among different HBV genotypes. For example, at the end of treatment, the mean decrease of HBsAg level was highest with genotype A infection, intermediate in genotypes B and D, and lowest in genotypes C and E. During follow-up, serum HBsAg levels continued to decrease in genotypes A and D, whereas rebound was observed in genotypes B, C, and E (Moucari et al. 2009). A recent study further revealed that high positive predictive values for long-term virological response at 5-yr post-PEG-IFN treatment could be obtained by applying end-of-treatment genotype-specific qHBsAg cutoff levels: 75%, 47%, 71%, and 75% for genotypes A (<400 IU/ml), B (<50 IU/ml), C (<75 IU/ml), and D (<1000 IU/ml), respectively (Brunetto et al. 2013). Therefore, monitoring HBV genotype–specific qHBsAg may improve response-guided treatment of HBeAg-negative chronic hepatitis B (Fig. 3).
Figure 3.
Hypothetical algorithm for HBV genotype–specific antiviral treatment in patients with chronic hepatitis B.
NUCLEOSIDE AND NUCLEOTIDE ANALOGS
In contrast to IFN-based therapy, the therapeutic responses to NUCs as well as the development of resistance were comparable among patients with different genotypes (Kao et al. 2002b; Chan et al. 2003; Westland et al. 2003; Yuen et al. 2003; Hou et al. 2008; Wiegand et al. 2008; Chen et al. 2013). Although HBV genotypes seem not to have an impact on the response and resistance to NUC treatment, our retrospective study found that HBV genotype B was independently associated with an earlier detection of lamivudine-resistant strains. In addition, genotype B was significantly associated with development of lamivudine resistance within the first 12 mo of lamivudine therapy compared with genotype C (OR: 8.27; P = 0.004) (Hsieh et al. 2009). Therefore, more frequent monitoring of genotypic resistance might be needed for particular HBV genotypes during NUC therapy.
Clearance of HBsAg has been reported in patients treated with NUCs. HBsAg loss was achieved in 8% of HBeAg-positive patients after 3 yr of tenofovir therapy (Heathcote et al. 2011). Similarly, the HBsAg loss rates for HBeAg-positive patients treated with entecavir or lamivudine for 2 yr were 5% and 3%, respectively (Gish et al. 2010). More specifically, most patients with NUC-associated HBsAg loss had HBV genotypes A or D infection (Gish et al. 2010; Wursthorn et al. 2010; Heathcote et al. 2011), and the clearance of HBsAg by NUCs seems very rare in Asian patients with genotypes B or C infection. Although the proportion of patients with HBsAg loss is too small to reach any conclusion, the association between HBV genotype and NUC-induced HBsAg loss deserve further study.
THE IMPACT OF HBV VARIANTS ON RESPONSE TO ANTIVIRAL THERAPY
The impact of HBV variants on HBV treatment remains largely unknown. Previous studies have shown that the precore G1896A mutation or BCP A1762T/G1764A variants have been associated with response to IFN-α or NUC therapy (Tseng et al. 2010). Our previous study on 116 HBeAg-positive CHB patients receiving short-term lamivudine therapy revealed that precore G1896A mutation, compared with wild-type (75% vs. 52%, P = 0.045), correlated with the loss of HBeAg at the end of therapy (Tseng et al. 2009). Similarly, a recent study on 137 HBeAg-positive NUC-treated CHB patients revealed that the chance of HBeAg seroconversion was higher in patients with mixed populations of precore and/or BCP mutants (P = 0.01) (Zoutendijk et al. 2013). Another study on 115 HBeAg-positive patients receiving PEG-IFN-α-2a for 6 mo indicated that the presence of BCP A1762T/G1764A variants was associated with a combined response defined as HBeAg seroconversion, HBV DNA levels less than 20,000 IU/mL, as well as ALT normalization at 6 mo off therapy (OR: 8.04; 95% CI: 2.00–32.28) (Tseng et al. 2011b). On the contrary, recent analysis on 214 HBeAg-positive patients receiving PEG-IFN-α with or without lamivudine for 1 yr revealed that patients without presence of precore and BCP mutants at baseline were more likely to achieve HBeAg loss with HBV DNA <10,000 copies/mL and HBsAg clearance at 6 mo off therapy than patients with presence of precore or BCP mutants. The wild type of precore and BCP region of virus at baseline was an independent predictor of response (OR: 2.90; 95% CI: 1.15–7.31; P = 0.023) and HBsAg clearance (OR: 5.58; 95% CI: 1.26–24.63; P = 0.013) (Sonneveld et al. 2012). Using a new assay employing pyrosequencing to quantify the precore G1896A and BCP A1762T/G1764A variants percentages, a correlation between dynamic changes of these mutants and IFN-associated HBeAg seroconversion was determined in 203 HBeAg-positive CHB patients (Yang et al. 2013). The chance of HBeAg seroconversion increased by 2.2% (OR: 1.022; 95% CI: 1.009–1.034; P = 0.001) and 2.3% (OR: 1.023; 95% CI: 1.010–1.037; P = 0.001) per 1% increase of the pretreatment precore G1896A and BCP A1762T/G1764A variants percentages, respectively (Yang et al. 2013). Confirmatory data are limited and, therefore, further large-scale studies are required to explore the association of common HBV variants with treatment response to currently available antiviral agents.
CONCLUSIONS AND PERSPECTIVES
Over the past decade, ample evidence in molecular studies has clarified the clinical implications of HBV genotypes and variants. In brief, compared with genotypes A and B patients, genotypes C and D patients have a higher risk of cirrhosis and HCC, leading to a poorer clinical outcome. Mutations in core promoter and the pre-S regions are also associated with an increased risk of HCC. In addition, genotypes A and B patients have a better response to IFN-based therapy than genotypes C and D patients. However, the association between HBV genotypes/variants and therapeutic response to NUCs seems minimal. Despite numerous lines of evidence connecting HBV genotypes/variants and the disease progression as well as responses to antiviral therapy (Chen 2011), HBV genotyping is still not recommended as a part of the management of chronic hepatitis B in the recent update guidelines for the management of HBV infection (Lok and McMahon 2009; EASL 2012; Liaw et al. 2012). Nevertheless, HBV genotypes and variants have shown potential to be useful viral biomarkers for the prediction of disease progression as well as help practicing clinicians identify patients who can benefit most from IFN-based therapy. In the foreseeable future, clinical trials stratified by different genotypes/variants and treatment regimens are mandatory to implement individualized therapies for chronic hepatitis B patients.
Footnotes
Editors: Christoph Seeger and Stephen Locarnini
Additional Perspectives on Hepatitis B and Delta Viruses available at www.perspectivesinmedicine.org
REFERENCES
- Beck J, Nassal M. 2007. Hepatitis B virus replication. World J Gastroenterol 13: 48–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetto MR, Oliveri F, Colombatto P, Moriconi F, Ciccorossi P, Coco B, Romagnoli V, Cherubini B, Moscato G, Maina AM, et al. 2010. Hepatitis B surface antigen serum levels help to distinguish active from inactive hepatitis B virus genotype D carriers. Gastroenterology 139: 483–490. [DOI] [PubMed] [Google Scholar]
- Brunetto MR, Marcellin P, Cherubini B, Yurdaydin C, Farci P, Hadziyannis SJ, Rothe V, Regep L, Bonino F. 2013. Response to peginterferon α-2a (40 KD) in HBeAg-negative CHB: On-treatment kinetics of HBsAg serum levels vary by HBV genotype. J Hepatol 59: 1153–1159. [DOI] [PubMed] [Google Scholar]
- Buster EH, Flink HJ, Cakaloglu Y, Simon K, Trojan J, Tabak F, So TM, Feinman SV, Mach T, Akarca US, et al. 2008. Sustained HBeAg and HBsAg loss after long-term follow-up of HBeAg-positive patients treated with peginterferon α-2b. Gastroenterology 135: 459–467. [DOI] [PubMed] [Google Scholar]
- Buster EH, Hansen BE, Lau GK, Piratvisuth T, Zeuzem S, Steyerberg EW, Janssen HL. 2009. Factors that predict response of patients with hepatitis B e antigen-positive chronic hepatitis B to peginterferon-α. Gastroenterology 137: 2002–2009. [DOI] [PubMed] [Google Scholar]
- Cao GW. 2009. Clinical relevance and public health significance of hepatitis B virus genomic variations. World J Gastroenterol 15: 5671–5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan HL, Wong ML, Hui AY, Chim AM, Tse AM, Hung LC, Chan FK, Sung JJ. 2003. Hepatitis B virus genotype has no impact on hepatitis B e antigen seroconversion after lamivudine treatment. World J Gastroenterol 9: 2695–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan HL, Hui AY, Wong ML, Tse AM, Hung LC, Wong VW, Sung JJ. 2004. Genotype C hepatitis B virus infection is associated with an increased risk of hepatocellular carcinoma. Gut 53: 1494–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan HL, Tse CH, Mo F, Koh J, Wong VW, Wong GL, Lam Chan S, Yeo W, Sung JJ, Mok TS. 2008. High viral load and hepatitis B virus subgenotype Ce are associated with increased risk of hepatocellular carcinoma. J Clin Oncol 26: 177–182. [DOI] [PubMed] [Google Scholar]
- Chang MH. 2010. Breakthrough HBV infection in vaccinated children in Taiwan: Surveillance for HBV mutants. Antivir Ther 15: 463–469. [DOI] [PubMed] [Google Scholar]
- Chen DS. 2011. Fighting against viral hepatitis: Lessons from Taiwan. Hepatology 54: 381–392. [DOI] [PubMed] [Google Scholar]
- Chen YC, Sheen IS, Chu CM, Liaw YF. 2002. Prognosis following spontaneous HBsAg seroclearance in chronic hepatitis B patients with or without concurrent infection. Gastroenterology 123: 1084–1089. [DOI] [PubMed] [Google Scholar]
- Chen JD, Liu CJ, Lee PH, Chen PJ, Lai MY, Kao JH, Chen DS. 2004a. Hepatitis B genotypes correlate with tumor recurrence after curative resection of hepatocellular carcinoma. Clin Gastroenterol Hepatol 2: 64–71. [DOI] [PubMed] [Google Scholar]
- Chen BF, Chen PJ, Jow GM, Liu CJ, Kao JH, Chen DS. 2004b. High prevalence of mixed genotype infections in hepatitis B virus infected intravenous drug users. J Med Virol 74: 536–542. [DOI] [PubMed] [Google Scholar]
- Chen GG, Li MY, Ho RL, Chak EC, Lau WY, Lai PB. 2005. Identification of hepatitis B virus X gene mutation in Hong Kong patients with hepatocellular carcinoma. J Clin Virol 34: 7–12. [DOI] [PubMed] [Google Scholar]
- Chen BF, Liu CJ, Jow GM, Chen PJ, Kao JH, Chen DS. 2006. High prevalence and mapping of pre-S deletion in hepatitis B virus carriers with progressive liver diseases. Gastroenterology 130: 1153–1168. [DOI] [PubMed] [Google Scholar]
- Chen CH, Hung CH, Lee CM, Hu TH, Wang JH, Wang JC, Lu SN, Changchien CS. 2007. Pre-S deletion and complex mutations of hepatitis B virus related to advanced liver disease in HBeAg-negative patients. Gastroenterology 133: 1466–1474. [DOI] [PubMed] [Google Scholar]
- Chen XL, Li M, Zhang XL. 2013. HBVgenotypeB/C and response to Lamivudine therapy: A systematic review. Biomed Res Int 2013: 672614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisari FV, Ferrari C. 1995. Hepatitis B immunopathogenesis. Annu Rev Immunol 13: 29–60. [DOI] [PubMed] [Google Scholar]
- Chotiyaputta W, Lok AS. 2009. Hepatitis B virus variants. Nat Rev Gastroenterol Hepatol 6: 453–462. [DOI] [PubMed] [Google Scholar]
- Chou YC, Yu MW, Wu CF, Yang SY, Lin CL, Liu CJ, Shih WL, Chen PJ, Liaw YF, Chen CJ. 2008. Temporal relationship between hepatitis B virus enhancer II/basal core promoter sequence variation and risk of hepatocellular carcinoma. Gut 57: 91–97. [DOI] [PubMed] [Google Scholar]
- Chu CM, Liaw YF. 2007. Chronic hepatitis B virus infection acquired in childhood: Special emphasis on prognostic and therapeutic implication of delayed HBeAg seroconversion. J Viral Hepat 14: 147–152. [DOI] [PubMed] [Google Scholar]
- Cooksley WG, Piratvisuth T, Lee SD, Mahachai V, Chao YC, Tanwandee T, Chutaputti A, Chang WY, Zahm FE, Pluck N. 2003. Peginterferon α-2a (40 kDa): An advance in the treatment of hepatitis B e antigen-positive chronic hepatitis B. J Viral Hepat 10: 298–305. [DOI] [PubMed] [Google Scholar]
- Datta S. 2008. An overview of molecular epidemiology of hepatitis B virus (HBV) in India. Virol J 5: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt A, Blondin D, Hauck K, Sagir A, Kohnle T, Heintges T, Häussinger D. 2005. Response to interferon α is hepatitis B virus genotype dependent: Genotype A is more sensitive to interferon than genotype D. Gut 54: 1009–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Association for the Study of the Liver. 2012. EASL Clinical Practice Guidelines: Management of chronic hepatitis B virus infection. J Hepatol 57: 167–185. [DOI] [PubMed] [Google Scholar]
- Fang ZL, Sabin CA, Dong BQ, Wei SC, Chen QY, Fang KX, Yang JY, Huang J, Wang XY, Harrison TJ. 2008. Hepatitis B virus pre-S deletion mutations are a risk factor for hepatocellular carcinoma: A matched nested case-control study. J Gen Virol 89: 2882–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattovich G, Bortolotti F, Donato F. 2008. Natural history of chronic hepatitis B: Special emphasis on disease progression and prognostic factors. J Hepatol 48: 335–352. [DOI] [PubMed] [Google Scholar]
- Flink HJ, van Zonneveld M, Hansen BE, de Man RA, Schalm SW, Janssen HL HBV 99-01 Study Group. 2006. Treatment with Peg-interferon α-2b for HBeAg-positive chronic hepatitis B: HBsAg loss is associated with HBV genotype. Am J Gastroenterol 101: 297–303. [DOI] [PubMed] [Google Scholar]
- Ganem D, Prince AM. 2004. Hepatitis B virus infection—Natural history and clinical consequences. N Engl J Med 350: 1118–1129. [DOI] [PubMed] [Google Scholar]
- Gish RG, Chang TT, Lai CL, de Man R, Gadano A, Poordad F, Yang J, Brett-Smith H, Tamez R. 2010. Loss of HBsAg antigen during treatment with entecavir or lamivudine in nucleoside-naïve HBeAg-positive patients with chronic hepatitis B. J Viral Hepat 17: 16–22. [DOI] [PubMed] [Google Scholar]
- Guo X, Jin Y, Qian G, Tu H. 2008. Sequential accumulation of the mutations in core promoter of hepatitis B virus is associated with the development of hepatocellular carcinoma in Qidong, China. J Hepatol 49: 718–725. [DOI] [PubMed] [Google Scholar]
- Heathcote EJ, Marcellin P, Buti M, Gane E, De Man RA, Krastev Z, Germanidis G, Lee SS, Flisiak R, Kaita K, et al. 2011. Three-year efficacy and safety of tenofovir disoproxil fumarate treatment for chronic hepatitis B. Gastroenterology 140: 132–143. [DOI] [PubMed] [Google Scholar]
- Heijtink RA, Paulij W, van Roosmalen M, Hellings JA, Niesters HG, Schalm SW, Osterhaus AD. 1999. Characteristics of the early phase of chronicity in acute hepatitis B infection. J Med Virol 57: 331–336. [DOI] [PubMed] [Google Scholar]
- Hou J, Schilling R, Janssen HLA. 2001. Molecular characteristics of hepatitis B virus genotype A confer a higher response to interferon treatment. J Hepatol 34: 15–16. [Google Scholar]
- Hou J, Yin YK, Xu D, Tan D, Niu J, Zhou X, Wang Y, Zhu L, He Y, Ren H, et al. 2008. Telbivudine versus lamivudine in Chinese patients with chronic hepatitis B: Results at 1 year of a randomized, double-blind trial. Hepatology 47: 447–454. [DOI] [PubMed] [Google Scholar]
- Hsieh YH, Su IJ, Wang HC, Chang WW, Lei HY, Lai MD, Chang WT, Huang W. 2004. Pre-S mutant surface antigens in chronic hepatitis B virus infection induce oxidative stress and DNA damage. Carcinogenesis 25: 2023–2032. [DOI] [PubMed] [Google Scholar]
- Hsieh TH, Tseng TC, Liu CJ, Lai MY, Chen PJ, Hsieh HL, Chen DS, Kao JH. 2009. Hepatitis B virus genotype B has an earlier emergence of lamivudine resistance than genotype C. Antivir Ther 14: 1157–1163. [DOI] [PubMed] [Google Scholar]
- Hsu YS, Chien RN, Yeh CT, Sheen IS, Chiou HY, Chu CM, Liaw YF. 2002. Long-term outcome after spontaneous HBeAg seroconversion in patients with chronic hepatitis B. Hepatology 35: 1522–1527. [DOI] [PubMed] [Google Scholar]
- Hunt CM, McGill JM, Allen MI, Condreay LD. 2000. Clinical relevance of hepatitis B viral mutations. Hepatology 31: 1037–1044. [DOI] [PubMed] [Google Scholar]
- Iavarone M, Trabut JB, Delpuech O, Carnot F, Colombo M, Kremsdorf D, Bréchot C, Thiers V. 2003. Characterisation of hepatitis B virus X protein mutants in tumour and non-tumour liver cells using laser capture microdissection. J Hepatol 39: 253–261. [DOI] [PubMed] [Google Scholar]
- Ito K, Yotsuyanagi H, Yatsuhashi H, Karino Y, Takikawa Y, Saito T, Arase Y, Imazeki F, Kurosaki M, Umemura T, et al. 2014. Risk factors for long-term persistence of serum hepatitis B surface antigen following acute hepatitis B virus infection in Japanese adults. Hepatology 59: 89–97. [DOI] [PubMed] [Google Scholar]
- Janssen HL, van Zonneveld M, Senturk H, Zeuzem S, Akarca US, Cakaloglu Y, Simon C, So TM, Gerken G, de Man RA, et al. 2005. Pegylated interferon α-2b alone or in combination with lamivudine for HBeAg-positive chronic hepatitis B: A randomised trial. Lancet 365: 123–129. [DOI] [PubMed] [Google Scholar]
- Kao JH. 2002. Hepatitis B viral genotypes: Clinical relevance and molecular characteristics. J Gastroenterol Hepatol 17: 643–650. [DOI] [PubMed] [Google Scholar]
- Kao JH. 2003. Hepatitis B virus genotypes and hepatocellular carcinoma in Taiwan. Intervirology 46: 400–407. [DOI] [PubMed] [Google Scholar]
- Kao JH. 2011. Molecular epidemiology of Hepatitis B Virus. Korean J Intern Med 26: 255–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao JH, Chen DS. 2002. Global control of hepatitis B virus infection. Lancet Infect Dis 2: 395–403. [DOI] [PubMed] [Google Scholar]
- Kao JH, Chen DS. 2006. HBV Genotypes: Epidemiology and implications regarding natural history. Current Hepatitis Reports 5: 5–13. [Google Scholar]
- Kao JH, Chen PJ, Lai MY, Chen DS. 2000a. Hepatitis B genotypes correlate with clinical outcomes in patients with chronic hepatitis B. Gastroenterology 118: 554–559. [DOI] [PubMed] [Google Scholar]
- Kao JH, Wu NH, Chen PJ, Lai MY, Chen DS. 2000b. Hepatitis B genotypes and the response to interferon therapy. J Hepatol 33: 998–1002. [DOI] [PubMed] [Google Scholar]
- Kao JH, Chen PJ, Lai MY, Chen DS. 2002a. Genotypes and clinical phenotypes of hepatitis B virus in patients with chronic hepatitis B virus infection. J Clin Microbiol 40: 1207–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao JH, Liu CJ, Chen DS. 2002b. Hepatitis B viral genotypes and lamivudine resistance. J Hepatol 36: 303–304. [DOI] [PubMed] [Google Scholar]
- Kao JH, Chen PJ, Lai MY, Chen DS. 2003. Basal core promoter mutations of hepatitis B virus increase the risk of hepatocellular carcinoma in hepatitis B carriers. Gastroenterology 124: 327–334. [DOI] [PubMed] [Google Scholar]
- Kao JH, Chen PJ, Lai MY, Chen DS. 2004. Hepatitis B virus genotypes and spontaneous hepatitis B e antigen seroconversion in Taiwanese hepatitis B carriers. J Med Virol 72: 363–369. [DOI] [PubMed] [Google Scholar]
- Kao JH, Chen PJ, Chen DS. 2010. Recent advances in the research of hepatitis B virus-related hepatocellular carcinoma: Epidemiologic and molecular biological aspects. Adv Cancer Res 108: 21–72. [DOI] [PubMed] [Google Scholar]
- Kim H, Jee YM, Song BC, Shin JW, Yang SH, Mun HS, Kim HJ, Oh EJ, Yoon JH, Kim YJ, et al. 2007. Molecular epidemiology of hepatitis B virus (HBV) genotypes and serotypes in patients with chronic HBV infection in Korea. Intervirology 50: 52–57. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Suzuki F, Arase Y, Akuta N, Suzuki Y, Hosaka T, Saitoh S, Kobayashi M, Tsubota A, Someya T, et al. 2004. Infection with hepatitis B virus genotype A in Tokyo, Japan during 1976 through 2001. J Gastroenterol 39: 844–850. [DOI] [PubMed] [Google Scholar]
- Kurbanov F, Tanaka Y, Mizokami M. 2010. Geographical and genetic diversity of the human hepatitis B virus. Hepatol Res 40: 14–30. [DOI] [PubMed] [Google Scholar]
- Lau JY, Wright TL. 1993. Molecular virology and pathogenesis of hepatitis B. Lancet 342: 1335–1340. [DOI] [PubMed] [Google Scholar]
- Lau GK, Piratvisuth T, Luo KX, Marcellin P, Thongsawat S, Cooksley G, Gane E, Fried MW, Chow WC, Paik SW, et al. 2005. Peginterferon α-2a, lamivudine, and the combination for HBeAg-positive chronic hepatitis B. N Engl J Med 352: 2682–2695. [DOI] [PubMed] [Google Scholar]
- Le Bouvier GL, McCollum RW, Hierholzer WJ Jr, Irwin GR, Krugman S, Giles JP. 1972. Subtypes of Australia antigen and hepatitis-B virus. JAMA 222: 928–930. [DOI] [PubMed] [Google Scholar]
- Leblebicioglu H, Eroglu C, Members of the Hepatitis Study Group. 2004. Acute hepatitis B virus infection in Turkey: Epidemiology and genotype distribution. Clin Microbiol Infect 10: 537–541. [DOI] [PubMed] [Google Scholar]
- Lee MH, Yang HI, Liu J, Batrla-Utermann R, Jen CL, Iloeje UH, Lu SN, You SL, Wang LY, Chen CJ, et al. 2013. Prediction models of long-term cirrhosis and hepatocellular carcinomarisk in chronic hepatitis B patients: Risk scores integrating host and virus profiles. Hepatology 58: 546–554. [DOI] [PubMed] [Google Scholar]
- Liaw YF, Jia JD, Chan HL, Han KH, Tanwandee T, Chuang WL, Tan DM, Chen XY, Gane E, Piratvisuth T, et al. 2011. Shorter durations and lower doses of peginterferon α-2a are associated with inferior hepatitis B e antigen seroconversion rates in hepatitis B virus genotypes B or C. Hepatology 54: 1591–1599. [DOI] [PubMed] [Google Scholar]
- Liaw YF, Kao JH, Piratvisuth T, Chan HL, Chien RN, Liu CJ, Gane E, Locarnini S, Lim SG, Han KH, et al. 2012. Asian-Pacific consensus statement on the management of chronic hepatitis B: A 2012 update. Hepatol Int 6: 531–561. [DOI] [PubMed] [Google Scholar]
- Lin CL, Kao JH. 2008. Hepatitis B viral factors and clinical outcomes of chronic hepatitis B. J Biomed Sci 15: 137–145. [DOI] [PubMed] [Google Scholar]
- Lin CL, Kao JH. 2011. The clinical implications of hepatitis B virus genotype: Recent advances. J Gastroenterol Hepatol 26: 123–130. [DOI] [PubMed] [Google Scholar]
- Lin CL, Kao JH. 2013a. Risk stratification for hepatitis B virus related hepatocellular carcinoma. J Gastroenterol Hepatol 28: 10–17. [DOI] [PubMed] [Google Scholar]
- Lin CL, Kao JH. 2013b. Hepatitis B virus genotypes: Clinical relevance and therapeutic implications. Curr Hepat Rep 12: 124–132. [Google Scholar]
- Lin CL, Kao JH. 2013c. Hepatitis B viral factors and treatment responses in chronic hepatitis B. J Formos Med Assoc 112: 302–311. [DOI] [PubMed] [Google Scholar]
- Lin CL, Kao JH, Chen BF, Chen PJ, Lai MY, Chen DS. 2005. Application of hepatitis B virus genotyping and phylogenetic analysis in intrafamilial transmission of hepatitis B virus. Clin Infect Dis 41: 1576–1581. [DOI] [PubMed] [Google Scholar]
- Lin CL, Liu CJ, Chen PJ, Lai MY, Chen DS, Kao JH. 2007a. High prevalence of occult hepatitis B virus infection in Taiwanese intravenous drug users. J Med Virol 79: 1674–1678. [DOI] [PubMed] [Google Scholar]
- Lin CL, Chen JD, Liu CJ, Lee PH, Chen PJ, Lai MY, Kao JH, Chen DS. 2007b. Clinicopathological differences between hepatitis B viral genotype B- and C-related resectable hepatocellular carcinoma. J Viral Hepat 14: 64–69. [DOI] [PubMed] [Google Scholar]
- Lin CL, Liu CH, Chen W, Huang WL, Chen PJ, Lai MY, Chen DS, Kao JH. 2007c. Association of pre-S deletion mutant of hepatitis B virus with risk of hepatocellular carcinoma. J Gastroenterol Hepatol 22: 1098–1103. [DOI] [PubMed] [Google Scholar]
- Littlejohn M, Davies J, Yuen L, Edwards R, Sozzi T, Jackson K, Cowie B, Tong S, Davis J, Locarnini S. 2014. Molecular virology of hepatitis B virus, sub-genotype C4 in northern Australian indigenous populations. J Med Virol 86: 695–706. [DOI] [PubMed] [Google Scholar]
- Liu CJ, Kao JH. 2008. Genetic variability of hepatitis B virus and response to antiviral therapy. Antivir Ther 13: 613–624. [PubMed] [Google Scholar]
- Liu CJ, Kao JH, Chen PJ, Lai MY, Chen DS. 2002. Molecular epidemiology of hepatitis B viral serotypes and genotypes in Taiwan. J Biomed Sci 9: 166–170. [DOI] [PubMed] [Google Scholar]
- Liu CJ, Kao JH, Chen DS. 2005. Therapeutic implications of hepatitis B virus genotypes. Liver Int 25: 1097–1107. [DOI] [PubMed] [Google Scholar]
- Liu CJ, Jeng YM, Chen CL, Cheng HR, Chen PJ, Chen TC, Liu CH, Lai MY, Chen DS, Kao JH. 2009a. Hepatitis B virus basal core promoter mutation and DNA load correlate with expression of hepatitis B core antigen in patients with chronic hepatitis B. J Infect Dis 199: 742–749. [DOI] [PubMed] [Google Scholar]
- Liu S, Zhang H, Gu C, Yin J, He Y, Xie J, Cao G. 2009b. Associations between hepatitis B virus mutations and the risk of hepatocellular carcinoma: A meta-analysis. J Natl Cancer Inst 101: 1066–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CJ, Cheng HR, Chen CL, Chen TC, Tseng TC, Wang ZL, Chen PJ, Liu CH, Chen DS, Kao JH. 2011. Effects of hepatitis B virus precore and basal core promoter mutations on the expression of viral antigens: Genotype B vs C. J Viral Hepat 18: e482–490. [DOI] [PubMed] [Google Scholar]
- Liu J, Yang HI, Lee MH, Lu SN, Jen CL, Batrla-Utermann R, Wang LY, You SL, Hsiao CK, Chen PJ, et al. 2013a. Spontaneous seroclearance of hepatitis B seromarkers and subsequent risk of hepatocellularcarcinoma. Gut 63: 1648–1657. [DOI] [PubMed] [Google Scholar]
- Liu J, Lee MH, Batrla-Utermann R, Jen CL, Iloeje UH, Lu SN, Wang LY, You SL, Hsiao CK, Yang HI, et al. 2013b. A predictive scoring system for the seroclearance of HBsAg in HBeAg-seronegative chronic hepatitis B patients with genotype B or C infection. J Hepatol 58: 853–860. [DOI] [PubMed] [Google Scholar]
- Livingston SE, Simonetti J, McMahon B, Bulkow LR, Hurlburt KJ, Homan CE, Snowball MM, Cagle HH, Williams JL, Chulanov VP. 2007. Hepatitis B virus genotypes in Alaska native people with hepatocellular carcinoma: Preponderance of genotype F. J Infect Dis 195: 5–11. [DOI] [PubMed] [Google Scholar]
- Locarnini S, Littlejohn M, Aziz MN, Yuen L. 2013. Possible origins and evolution of the hepatitis B virus (HBV). Semin Cancer Biol 23: 561–575. [DOI] [PubMed] [Google Scholar]
- Lok AS, McMahon BJ. 2009. Chronic hepatitis B: update 2009. Hepatology 50: 661–662. [DOI] [PubMed] [Google Scholar]
- Ma NF, Lau SH, Hu L, Xie D, Wu J, Yang J, Wang Y, Wu MC, Fung J, Bai X, et al. 2008. COOH terminal truncated HBV X protein plays key role in hepatocarcinogenesis. Clin Cancer Res 14: 5061–5068. [DOI] [PubMed] [Google Scholar]
- Mansourian PG, Ghany MG, Thomas E. 2013. Spontaneous mutations in the HBV genome and their clinical implications. Curr Hepat Rep 12: 79–87. [Google Scholar]
- Marcellin P, Bonino F, Lau GK, Farci P, Yurdaydin C, Piratvisuth T, Jin R, Gurel S, Lu ZM, Wu J, et al. ; Peginterferon α-2a in HBeAg-Negative Chronic Hepatitis B Study Group. 2009. Sustained response of hepatitis B e antigen-negative patients 3 years after treatment with peginterferon α-2a. Gastroenterology 136: 2169–2179. [DOI] [PubMed] [Google Scholar]
- Matsuura K, Tanaka Y, Hige S, Yamada G, Murawaki Y, Komatsu M, Kuramitsu T, Kawata S, Tanaka E, Izumi N, et al. 2009. Distribution of hepatitis B virus genotypes among patients with chronic infection in Japan shifting toward an increase of genotype A. J Clin Microbiol 47: 1476–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayerat C, Mantegani A, Frei PC. 1999. Does hepatitis B virus (HBV) genotype influence the clinical outcome of HBV infection? J Viral Hepat 6: 299–304. [DOI] [PubMed] [Google Scholar]
- McMahon BJ. 2009. The influence of hepatitis B virus genotype and subgenotype on the natural history of chronic hepatitis B. Hepatol Int 3: 334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendy M, D'Mello F, Kanellos T, Oliver S, Whittle H, Howard CR. 2008. Envelope protein variability among HBV-Infected asymptomatic carriers and immunized children with breakthrough infections. J Med Virol 80: 1537–1546. [DOI] [PubMed] [Google Scholar]
- Moucari R, Martinot-Peignoux M, Mackiewicz V, Boyer N, Ripault MP, Castelnau C, Leclere L, Dauvergne A, Valla D, Vidaud M, et al. 2009. Influence of genotype on hepatitis B surface antigen kinetics in hepatitis B e antigen-negative patients treated with pegylated interferon-α2a. Antivir Ther 14: 1183–1188. [DOI] [PubMed] [Google Scholar]
- Ngui SL, Teo CG. 1997. Hepatitis B virus genomic heterogeneity: Variation between quasispecies may confound molecular epidemiological analyses of transmission incidents. J Viral Hepat 4: 309–315. [DOI] [PubMed] [Google Scholar]
- Ni YH, Chang MH, Wang KJ, Hsu HY, Chen HL, Kao JH, Yeh SH, Jeng YM, Tsai KS, Chen DS. 2004. Clinical relevance of hepatitis B virus genotype in children with chronic infection and hepatocellular carcinoma. Gastroenterology 127: 1733–1738. [DOI] [PubMed] [Google Scholar]
- Nishioka K, Levin AG, Simons MJ. 1975. Hepatitis B antigen, antigen subtypes, and hepatitis B antibody in normal subjects and patients with liver disease. Bull World Health Organ 52: 293–300. [PMC free article] [PubMed] [Google Scholar]
- Orito E, Sugauchi F, Tanaka Y, Ichida T, Sata M, Tanaka E, Okanoue T, Sakugawa H, Watanabe H, Miyakawa H, et al. 2005. Differences of hepatocellular carcinoma patients with hepatitis B virus genotypes of Ba, Bj or C in Japan. Intervirology 48: 239–245. [DOI] [PubMed] [Google Scholar]
- Panduro A, Maldonado-Gonzalez M, Fierro NA, Roman S. 2013. Distribution of HBV genotypes F and H in Mexico and Central America. Antivir Ther 18: 475–484. [DOI] [PubMed] [Google Scholar]
- Perrillo RP. 2001. Acute flares in chronic hepatitis B: the natural and unnatural history of an immunologically mediated liver disease. Gastroenterology 120: 1009–1022. [DOI] [PubMed] [Google Scholar]
- Pezzano SC, Torres C, Fainboim HA, Bouzas MB, Schroder T, Giuliano SF, Paz S, Alvarez E, Campos RH, Mbayed VA. 2011. Hepatitis B virus in Buenos Aires, Argentina: Genotypes, virological characteristics and clinical outcomes. Clin Microbiol Infect 17: 223–231. [DOI] [PubMed] [Google Scholar]
- Phung TB, Alestig E, Nguyen TL, Hannoun C, Lindh M. 2010. Genotype X/C recombinant (putative genotype I) of hepatitis B virus is rare in Hanoi, Vietnam—Genotypes B4 and C1 predominate. J Med Virol 82: 1327–1333. [DOI] [PubMed] [Google Scholar]
- Rehermann B. 2013. Pathogenesis of chronic viral hepatitis: Differential roles of T cells and NK cells. Nat Med 19: 859–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijckborst V, Hansen BE, Cakaloglu Y, Ferenci P, Tabak F, Akdogan M, Simon K, Akarca US, Flisiak R, Verhey E, et al. 2010. Early on treatment prediction of response to peginterferon α-2a for HBeAg-negative chronic hepatitis B using HBsAg and HBV DNA levels. Hepatology 52: 454–461. [DOI] [PubMed] [Google Scholar]
- Rijckborst V, Hansen B, Ferenci P, Brunetto MR, Tabak F, Cakaloglu Y, Lanza AG, Messina V, Iannacone C, Massetto B, et al. 2012. Validation of a stopping rule at week 12 using HBsAg and HBV DNA for HBeAg-negative patients treated with peginterferon α-2a. J Hepatol 56: 1006–1011. [DOI] [PubMed] [Google Scholar]
- Roman S, Panduro A, Aguilar-Gutierrez Y, Maldonado M, Vazquez-Vandyck M, Martinez-Lopez E, Ruiz-Madrigal B, Hernandez-Nazara Z. 2009. A low steady HBsAg seroprevalence is associated with a low incidence of HBV-related liver cirrhosis and hepatocellular carcinoma in Mexico: A systematic review. Hepatol Int 3: 343–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman S, Tanaka Y, Khan A, Kurbanov F, Kato H, Mizokami M, Panduro A. 2010. Occult hepatitis B in the genotype H-infected Nahuas and Huichol native Mexican population. J Med Virol 82: 1527–1536. [DOI] [PubMed] [Google Scholar]
- Sanchez-Tapias JM, Costa J, Mas A, Bruguera M, Rodés J. 2002. Influence of hepatitis B virus genotype on the long-term outcome of chronic hepatitis B in western patients. Gastroenterology 123: 1848–1856. [DOI] [PubMed] [Google Scholar]
- Sharma S, Sharma B, Singla B, Chawla YK, Chakraborti A, Saini N, Duseja A, Das A, Dhiman RK. 2010. Clinical significance of genotypes and precore/basal core promoter mutations in HBV related chronic liver disease patients in North India. Dig Dis Sci 55: 794–802. [DOI] [PubMed] [Google Scholar]
- Shen L, Yin W, Zheng H, Cui F, Zhang S, Wang F, Wang F, Zhang Y, Liang X, Bi S. 2014. Molecular epidemiological study of hepatitis B virus genotypes in Southwest, China. J Med Virol 86: 1307–1313. [DOI] [PubMed] [Google Scholar]
- Sonneveld MJ, Rijckborst V, Boucher CA, Hansen BE, Janssen HL. 2010. Prediction of sustained response to peginterferon α-2b for hepatitis B e antigen-positive chronic hepatitis B using on-treatment hepatitis B surface antigen decline. Hepatology 52: 1251–1257. [DOI] [PubMed] [Google Scholar]
- Sonneveld MJ, Rijckborst V, Zeuzem S, Heathcote EJ, Simon K, Senturk H, Pas SD, Hansen BE, Janssen HL. 2012. Presence of precore and core promoter mutants limits the probability of response to peginterferon in hepatitis B e antigen-positive chronic hepatitis B. Hepatology 56: 67–75. [DOI] [PubMed] [Google Scholar]
- Sonneveld MJ, Hansen BE, Piratvisuth T, Jia JD, Zeuzem S, Gane E, Liaw YF, Xie Q, Heathcote EJ, Chan HL, et al. 2013. Response-guided peginterferon therapy in hepatitis B e antigen-positive chronic hepatitis B using serum hepatitis B surface antigen levels. Hepatology 58: 872–880. [DOI] [PubMed] [Google Scholar]
- Su TH, Hsu CS, Chen CL, Liu CH, Huang YW, Tseng TC, Liu CJ, Chen PJ, Lai MY, Chen DS, et al. 2010. Serum hepatitis B surface antigen concentration correlates with HBV DNA level in patients with chronic hepatitis B. Antivir Ther 15: 1133–1139. [DOI] [PubMed] [Google Scholar]
- Su TH, Liu CJ, Tseng TC, Liu CH, Yang HC, Chen CL, Chen PJ, Kao JH, Chen DS. 2013. Longitudinal change of HBsAg in HBeAg-negative patients with genotype B or C infection. PLoS ONE 8: e55916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugauchi F, Orito E, Ichida T, Kato H, Sakugawa H, Kakumu S, Ishida T, Chutaputti A, Lai CL, Ueda R, et al. 2002. Hepatitis B virus of genotype B with or without recombination with genotype C over the precore region plus the core gene. J Virol 76: 5985–5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama M, Tanaka Y, Kato T, Orito E, Ito K, Acharya SK, Gish RG, Kramvis A, Shimada T, Izumi N. 2006. Influence of hepatitis B virus genotypes on the intra- and extracellular expression of viral DNA and antigens. Hepatology 44: 915–924. [DOI] [PubMed] [Google Scholar]
- Sung JL, Chen DS. 1977. Geographical distribution of the subtype of hepatitis B surface antigen in Chinese. Gastroenterol Jpn 12: 58–63. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Kobayashi M, Ikeda K, Suzuki F, Arfase Y, Akuta N, Hosaka T, Saitoh S, Kobayashi M, Someya T, et al. 2005. Persistence of acute infection with hepatitis B virus genotype A and treatment in Japan. J Med Virol 76: 33–39. [DOI] [PubMed] [Google Scholar]
- Tatematsu K, Tanaka Y, Kurbanov F, Sugauchi F, Mano S, Maeshiro T, Nakayoshi T, Wakuta M, Miyakawa Y, Mizokami M. 2009. A genetic variant of hepatitis B virus divergent from known human and ape genotypes isolated from a Japanese patient and provisionally assigned to new genotype J. J Virol 83: 10538–10547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur V, Guptan RC, Kazim SN, Malhotra V, Sarin SK. 2002. Profile, spectrum and significance of HBV genotypes in chronic liver disease patients in the Indian subcontinent. J Gastroenterol Hepatol 17: 165–170. [DOI] [PubMed] [Google Scholar]
- Tiollais P, Pourcel C, Dejean A. 1985. The hepatitis B virus. Nature 317: 489–495. [DOI] [PubMed] [Google Scholar]
- Tran TT, Trinh TN, Abe K. 2008. New complex recombinant genotype of hepatitis B virus identified in Vietnam. J Virol 82: 5657–5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng TC, Kao JH. 2013. Clinical utility of quantitative HBsAg in natural history and nucleos(t)ide analogue treatment of chronic hepatitis B: New trick of old dog. J Gastroenterol 48: 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng TC, Liu CJ, Chen PJ, Lai MY, Lin CL, Kao JH, Chen DS. 2007. Subgenotypes of hepatitis B virus genotype C do not correlate with disease progression of chronic hepatitis B in Taiwan. Liver Int 27: 983–988. [DOI] [PubMed] [Google Scholar]
- Tseng TC, Liu CJ, Wang CC, Chen PJ, Lai MY, Chen DS, Kao JH. 2009. Association of baseline viral factors with response to lamivudine therapy in chronic hepatitis B patients with high serum alanine aminotransferase levels. Antivir Ther 14: 203–210. [PubMed] [Google Scholar]
- Tseng TC, Liu CJ, Kao JH. 2010. Implications of hepatitis B virus genomic variations on treatment outcomes. Curr Pharmacogenomics Person Med 8: 280–288. [Google Scholar]
- Tseng TC, Liu CJ, Su TH, Wang CC, Chen CL, Chen PJ, Chen DS, Kao JH. 2011a. Serum hepatitis B surface antigen levels predict surface antigen loss in hepatitis B e antigen seroconverters. Gastroenterology 141: 517–525. [DOI] [PubMed] [Google Scholar]
- Tseng TC, Yu ML, Liu CJ, Lin CL, Huang YW, Hsu CS, Liu CH, Kuo SF, Pan CJ, Yang SS, et al. 2011b. Impact of host and viral factors on HBeAg-positive chronic hepatitis B patients receiving peginterferon α-2a therapy. Antivir Ther 16: 629–637. [DOI] [PubMed] [Google Scholar]
- Tseng TC, Liu CJ, Yang HC, Su TH, Wang CC, Chen CL, Kuo SF, Liu CH, Chen PJ, Chen DS, et al. 2012a. High levels of hepatitis B surface antigen increase risk of hepatocellular carcinoma in patients with low HBV load. Gastroenterology 142: 1140–1149. [DOI] [PubMed] [Google Scholar]
- Tseng TC, Liu CJ, Yang HC, Su TH, Wang CC, Chen CL, Kuo SF, Liu CH, Chen PJ, Chen DS, et al. 2012b. Determinants of spontaneous surface antigen loss in hepatitis B e antigen-negative patients with a low viral load. Hepatology 55: 68–76. [DOI] [PubMed] [Google Scholar]
- Tseng TC, Liu CJ, Yang HC, Su TH, Wang CC, Chen CL, Hsu CA, Kuo SF, Liu CH, Chen PJ, et al. 2013. Serum hepatitis B surface antigen levels help predict disease progression in patients with low hepatitis B virus loads. Hepatology 57: 441–450. [DOI] [PubMed] [Google Scholar]
- Tseng TC, Liu CJ, Yang HC, Chen CL, Yang WT, Tsai CS, Kuo SF, Verbree FC, Su TH, Wang CC, et al. 2014. Higher proportion of viral basal core promoter mutant increases the risk of liver cirrhosis in hepatitis B carriers. Gut 10.1136/gutjnl-2014-306977. [DOI] [PubMed] [Google Scholar]
- Tuaillon E, Mondain AM, Nagot N, Ottomani L, Kania D, Nogue E, Rubbo PA, Pageaux GP, Van de Perre P, Ducos J. 2012. Comparison of serum HBsAg quantitation by four immunoassays, and relationships of HBsAg level with HBV replication and HBV genotypes. PLoS ONE 7: e32143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vutien P, Trinh HN, Nguyen K, Garcia RT, Nguyen HA, Levitt BS, Nguyen L, Ha NB, Ahmed A, Daugherty T. 2013. Precore and basal core promoter mutations in Asian American patients with hepatitis B e antigen-positive chronic hepatitis B. Aliment Pharmacol Ther 37: 464–472. [DOI] [PubMed] [Google Scholar]
- Wai CT, Chu CJ, Hussain M, Lok AS. 2002. HBV genotype B is associated with better response to interferon therapy in HBeAg+ chronic hepatitis than genotype C. Hepatology 36: 1425–1430. [DOI] [PubMed] [Google Scholar]
- Wang Y, Lau SH, Sham JST, Wu MC, Wang T, Guan XY. 2004. Characterization of HBV integrants in 14 hepatocellular carcinomas: Association of truncated X gene and hepatocellular carcinogenesis. Oncogene 23: 142–148. [DOI] [PubMed] [Google Scholar]
- Wang Z, Liu Z, Zeng G, Wen S, Qi Y, Ma S, Naoumov NV, Hou J. 2005. A new intertype recombinant between genotypes C and D of hepatitis B virus identified in China. J Gen Virol 86: 985–990. [DOI] [PubMed] [Google Scholar]
- Westland C, Delaney W 4th, Yang H, Chen SS, Marcellin P, Hadziyannis S, Gish R, Fry J, Brosgart C, Gibbs C. 2003. Hepatitis B virus genotypes and virologic response in 694 patients in phase III studies of adefovir dipivoxil. Gastroenterology 125: 107–116. [DOI] [PubMed] [Google Scholar]
- Wiegand J, Hasenclever D, Tillmann HL. 2008. Should treatment of hepatitis B depend on hepatitis B virus genotypes? A hypothesis generated from an explorative analysis of published evidence. Antivir Ther 13: 211–220. [PubMed] [Google Scholar]
- Wu JC, Huang YH, Chau GY, Su CW, Lai CR, Lee PC, Huo TI, Sheen IJ, Lee SD, Lui WY. 2009. Risk factors for early and late recurrence in hepatitis B-related hepatocellular carcinoma. J Hepatol 51: 890–897. [DOI] [PubMed] [Google Scholar]
- Wursthorn K, Jung M, Riva A, Goodman ZD, Lopez P, Bao W, Manns MP, Wedemeyer H, Naoumov NV. 2010. Kinetics of hepatitis B surface antigen decline during 3 years of telbivudine treatment in hepatitis B e antigen-positive patients. Hepatology 52: 1611–1620. [DOI] [PubMed] [Google Scholar]
- Yang HI, Lu SN, Liaw YF, You SL, Sun CA, Wang LY, Hsiao CK, Chen PJ, Chen DS, Chen CJ, et al. 2002. Hepatitis B e antigen and the risk of hepatocellular carcinoma. N Engl J Med 347: 168–174. [DOI] [PubMed] [Google Scholar]
- Yang J, Xing K, Deng R, Wang J, Wang X. 2006. Identification of Hepatitis B virus putative intergenotype recombinants by using fragment typing. J Gen Virol 87: 2203–2215. [DOI] [PubMed] [Google Scholar]
- Yang HI, Yeh SH, Chen PJ, Iloeje UH, Jen CL, Su J, Wang LY, Lu SN, You SL, Chen DS, et al. 2008. Associations between hepatitis B virus genotype and mutants and the risk of hepatocellular carcinoma. J Natl Cancer Inst 100: 1134–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HC, Chen CL, Shen YC, Peng CY, Liu CJ, Tseng TC, Su TH, Chuang WL, Yu ML, Dai CY, et al. 2013. Distinct evolution and predictive value of hepatitis B virus precore and basal core promoter mutations in interferon-induced HBeAg seroconversion. Hepatology 57: 934–943. [DOI] [PubMed] [Google Scholar]
- Yeh CT, Shen CH, Tai DI, Chu CM, Liaw YF. 2000. Identification and characterization of a prevalent hepatitis B virus X protein mutant in Taiwanese patients with hepatocellular carcinoma. Oncogene 19: 5213–5220. [DOI] [PubMed] [Google Scholar]
- Yeh CT, So M, Ng J, Yang HW, Chang ML, Lai MW, Chen TC, Lin CY, Yeh TS, Lee WC. 2010. Hepatitis B virus-DNA level and basal core promoter A1762T/G1764A mutation in liver tissue independently predict postoperative survival in hepatocellular carcinoma. Hepatology 52: 1922–1933. [DOI] [PubMed] [Google Scholar]
- Yin J, Zhang H, Li C, Gao C, He Y, Zhai Y, Zhang P, Xu L, Tan X, Chen J. 2008. Role of hepatitis B virus genotype mixture, subgenotypes C2 and B2 on hepatocellular carcinoma: Compared with chronic hepatitis B and asymptomatic carrier state in the same area. Carcinogenesis 29: 1685–1691. [DOI] [PubMed] [Google Scholar]
- Yotsuyanagi H, Okuse C, Yasuda K, Orito E, Nishiguchi S, Toyoda J, Tomita E, Hino K, Okita K, Murashima S. 2005. Japanese Acute Hepatitis B Group. Distinct geographic distributions of hepatitis B virus genotypes in patients with acute infection in Japan. J Med Virol 77: 39–46. [DOI] [PubMed] [Google Scholar]
- Yu MW, Yeh SH, Chen PJ, Liaw YF, Lin CL, Liu CJ, Shih WL, Kao JH, Chen DS, Chen CJ. 2005. Hepatitis B virus genotype and DNA level and hepatocellular carcinoma: A prospective study in men. J Natl Cancer Inst 97: 265–272. [DOI] [PubMed] [Google Scholar]
- Yuen MF, Wong DK, Sablon E, Yuan HJ, Sum SM, Hui CK, Chan AO, Wang BC, Lai CL. 2003. Hepatitis B virus genotypes B and C do not affect the antiviral response to lamivudine. Antivir Ther 8: 531–534. [PubMed] [Google Scholar]
- Yuen MF, Wong DK, Sablon E, Tse E, Ng IO, Yuan HJ, Siu CW, Sander TJ, Bourne EJ, Hall JG. 2004a. HBsAg seroclearance in chronic hepatitis B in the Chinese: Virological, histological, and clinical aspects. Hepatology 39: 1694–1701. [DOI] [PubMed] [Google Scholar]
- Yuen MF, Tanaka Y, Mizokami M, Yuen JC, Wong DK, Yuan HJ, Sum SM, Chan AO, Wong BC, Lai CL. 2004b. Role of hepatitis B virus genotypes Ba and C, core promoter and precore mutations on hepatocellular carcinoma: A case control study. Carcinogenesis 25: 1593–1598. [DOI] [PubMed] [Google Scholar]
- Yuen MF, Tanaka Y, Shinkai N, Poon RT, But DY, Fong DY, Fung J, Wong DK, Yuen JC, Mizokami M. 2008. Risk for hepatocellular carcinoma with respect to hepatitis B virus genotypes B/C, specific mutations of enhancer II/core promoter/precore regions and HBV DNA levels. Gut 57: 98–102. [DOI] [PubMed] [Google Scholar]
- Zhao H, Kurbanov F, Wan MB, Yin YK, Niu JQ, Hou JL, Wei L, Wang GQ, Tanaka Y, Mizokami M. 2007. Genotype B and younger patient age associated with better response to low-dose therapy: A trial with pegylated/nonpegylated interferon-α-2b for hepatitis B e antigen-positive patients with chronic hepatitis B in China. Clin Infect Dis 44: 541–548. [DOI] [PubMed] [Google Scholar]
- Zhou B, Xiao L, Wang Z, Chang ET, Chen J, Hou J. 2011. Geographical and ethnic distribution of the HBV C/D recombinant on the Qinghai-Tibet plateau. PLoS ONE 6: e18708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoutendijk R, Sonneveld MJ, Reijnders JG, van Vuuren AJ, Biesta P, Hansen BE, Boonstra A, Janssen HL. 2013. Precore and core promoter mutants are associated with higher HBeAg seroconversion but low disease remission rates in HBV patients treated with nucleos(t)ide analogues. J Viral Hepat 20: 322–327. [DOI] [PubMed] [Google Scholar]