Abstract
This review provides an overview of the unique features of DNA methylation in the human placenta. We discuss the importance of understanding placental development, structure, and function in the interpretation of DNA methylation data. Examples are given of how DNA methylation is important in regulating placental-specific gene expression, including monoallelic expression and X-chromosome inactivation in the placenta. We also discuss studies of global DNA methylation changes in the context of placental pathology and environmental exposures.
Placental DNA has a unique methylation profile (e.g., global hypomethylation and monoallelic methylation of specific genes). This profile changes throughout gestation and in response to in utero conditions.
Placental DNA is uniquely methylated as compared with that from the embryo or even other extraembryonic membranes, reflecting this organ's early developmental origin and distinct function. This DNA methylation profile changes throughout gestation, due both to changes in cell composition and changes in gene expression within individual cell types. Furthermore, the placenta shows considerable developmental plasticity, which is important for responding to fetal and maternal signals and to compensate for defects that may occur in the placenta. Thus, the placental methylome may be a rich source of information about a pregnancy, relevant to understanding development, assessing health, and providing clues about intrauterine exposures. We hereby review some of the unique features of DNA methylation in the human placenta and their relevance to clinical applications.
DNA METHYLATION
Epigenetic marks are important for the regulation of tissue- and developmental age-specific gene expression. Such marks include DNA methylation and histone modifications, which affect the accessibility of DNA to the binding of transcription factors and other regulatory proteins. DNA methylation typically involves the methylation of a cytosine residue located in a CpG dinucleotide. There are ∼28 million CpGs in the genome, most of which are methylated. However, those clustered within “CpG islands” (regions with more concentrated CpG dinucleotides commonly found at gene promoters) tend to be unmethylated (Weber et al. 2007). Some CpG islands become methylated during early differentiation and play a role in programmed repression of cell-specific or tissue-specific genes (Ching et al. 2005; Shen et al. 2007). The less CpG-dense edges of the CpG islands are referred to as “island shores.” DNA methylation at island shores may be reduced during periods of active gene transcription, perhaps owing to relaxation of the chromatin structure (Irizarry et al. 2009) or the physical presence of transcription factors. DNA methylation can also regulate the use of alternative gene promoters (Maunakea et al. 2010) or regulatory regions (enhancers, insulators, or suppressors) altogether outside of gene promoter(s) (Weber et al. 2007). Hence, the DNA methylation code contains information about many different functional regions of the genome that work together to influence (or respond to) gene transcription.
In most tissues, DNA methylation is relatively stable within a cell type once established and can yield information about normal developmental processes. However, the permanency of epigenetic marks is not absolute. There are extensive alterations to DNA methylation in the placenta throughout gestation (Novakovic et al. 2011) and in response to changes to in utero conditions (Hogg et al. 2012). Studying DNA methylation can therefore yield information about pathological changes and in utero exposures (Hogg et al. 2014a).
UNIQUE FEATURES OF DNA METHYLATION WITHIN THE PLACENTA
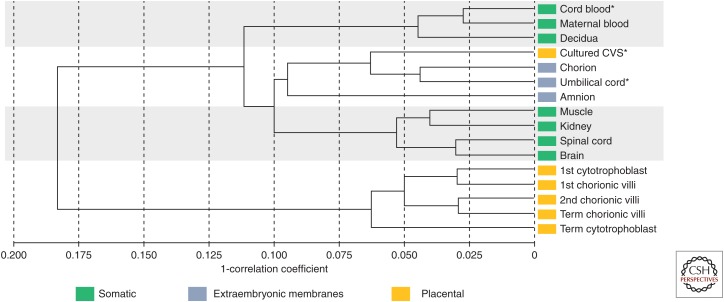
Studies of the placenta generally sample chorionic villi, which are embryonically derived and comprise the bulk of the placenta. The chorionic villi have a distinctive DNA methylation profile compared with embryonic tissues, maternal decidua, or the extraembryonic membranes (amnion and chorion) (Fig. 1). The chorionic villi are composed of a mix of trophectoderm (outer layer of the blastocyst) and inner cell mass (ICM)-derived cells, whereas amnion and chorion are most likely derived primarily from the epiblast (derived from the ICM) just before primitive streak formation (Peñaherrera et al. 2012). Correspondingly, the DNA methylation profile of these latter two tissues is more similar to fetal tissues than placenta (chorionic villi), based on Illumina Infinium microarray profiling (Fig. 1) and X-chromosome inactivation (XCI) studies (Peñaherrera et al. 2012). We summarize below and in Table 1 the global DNA methylation features of the placenta, which are unique among healthy tissues, but many of which are also observed in cancer (Novakovic and Saffery 2013). Cancer cells may exploit pathways normal in placental development for invasion of surrounding tissue and avoidance/modulation of the host/maternal immune system.
Figure 1.
Correlation dendrogram of DNA methylation at 473,369 CpG sites. Correlation dendogram using Illumina HumanMethylation450 BeadChip data filtered for X and Y chromosome probes, probes cross-hybridizing to the X or Y, probes with SNPs in the target CpG site and probes that could not be distinguished from the background signal (detection p value >0.05). Sample sizes for each tissue group are: 3 female cord blood, 10 maternal blood, 3 decidua, 6 female cultured CVS (chorionic villus sampling), 3 female and 3 male umbilical cord, 3 male amnion, 7 female and 6 male muscle, 8 female and 7 male kidney, 4 female and 5 male spinal cord, 6 female and 5 male brain, 4 FACS-sorted cytotrophoblast, 2 female and 3 male first trimester chorionic villi (<13 wk gestation), 8 female and 6 male second trimester chorionic villi (≥13 to <24 wk gestation), 10 female and 11 male term chorionic villi (≥37 wk gestation), 5 density-gradient separated term cytotrophoblast. *Indicates a pooled sample of several individuals. Colored labels indicate the origin of each tissue.
Table 1.
Characteristic features of DNA methylation in the placenta
| Feature | Cause and consequence |
|---|---|
| Global hypomethylation | Hypomethylation of the placenta may reflect (1) the major contribution of trophectoderm-derived cells; (2) reduced DNMT1 expression; and/or (3) less maintenance of methylation during rapid growth phase early in development. Methylation at gene regulatory regions is not, on average, reduced and this does not appear to reflect unimportance of methylation in regulating gene expression. |
| Presence of partially methylated domains | The origin of this unique organization of chromatin into hypomethylated domains in the placenta is unclear. Disruption of this organization could potentially have widespread effects. |
| Reduced L1 and HERV methylation | Reduced retrotransposon methylation may reflect a general reduction in methylation. L1/HERV promoters may be used more often for placental-specific genes. |
| X-chromosome hypomethylation of gene promoters (female) | X-chromosome inactivation (XCI) could be less stable in the placenta or more genes might escape XCI. However, there is no clear evidence for this as yet. |
| Increased methylation of strong CpG island promoters; reduced methylation at nonisland promoters | Embryonic “housekeeping genes” are turned down in placenta. This may reflect different features in regulation of transcription in placenta. |
| Monoallelic methylation | In addition to imprinted loci, which are well established to be important in placental development, a number of other genes have shown variable or polymorphic monoallelic methylation. This may allow gene expression to be more fine-tuned in placenta as part of its developmental plasticity. |
See text for citations and further explanation.
Global Hypomethylation
A key feature of epigenetic reprogramming in the zygote and preimplantation embryo is the loss of DNA methylation, such that the late morula/early blastocyst shows less DNA methylation than at any other developmental time point (Smith et al. 2012). Although in the ICM, this is followed by a rapid wave of de novo DNA methylation, the trophectoderm remains hypomethylated (Santos et al. 2010). In fact, chorionic villi show ∼10%–25% less cytosine methylation than fetal tissues (Ehrlich et al. 1982). As the villous trophoblast (derived from trophectoderm) is the predominant cell type present in the placenta throughout gestation (Grigoriu et al. 2011; Peñaherrera et al. 2012), its distinct DNA methylation profile may in part reflect the exemption of the trophoblast from this early reprogramming (Fig. 1). In addition, DNMT1, a gene encoding a maintenance DNA methyltransferase, is specifically down-regulated by monoallelic promoter DNA methylation in trophoblast cells and cultured and uncultured chorionic villi. This may contribute to, but seems not to be essential for, placental DNA hypomethylation (Novakovic et al. 2010).
Partially Methylated Domains (PMDs)
Placental DNA hypomethylation is not uniform across the genome, but occurs largely in “partially methylated domains” (PMDs). These are large (>100 kb) regions of reduced DNA methylation interspersed with regions of higher DNA methylation (Schroeder et al. 2013). The occurrence of PMDs is unique to placenta, some cultured cells, such as fetal lung fibroblasts, foreskin fibroblasts and adipocytes, and cancer (Lister et al. 2011; Schroeder and LaSalle 2013; Schroeder et al. 2013). Placental PMDs are estimated to cover nearly 40% of the genome and are relatively gene-poor. Placental genes within PMDs tend to be tissue-specific, and show higher promoter DNA methylation and reduced expression as compared with somatic tissues (Schroeder et al. 2013). The function of PMDs is unclear, and they have been hypothesized to arise as a consequence of the rapid cell divisions occurring in early placental development or as a programmed mechanism for regulating placental-specific gene expression (Schroeder and LaSalle 2013). If occurring as a result of epigenetic programming, then disruption of such organization could theoretically lead to significant placental dysfunction, including perhaps implantation failure or miscarriage.
Retrotransposable Element Hypomethylation
The placenta also shows reduced DNA methylation at some types of repetitive DNA (Price et al. 2012). Retrotransposable elements (RE) (e.g., L1, Alu, human endogenous retrovirus [HERV]) constitute nearly 50% of the human genome and are generally highly methylated as a silencing mechanism of the host. L1 DNA methylation (based on a consensus sequence present in a subset of L1s) is substantially reduced and more variable across individuals with ∼60% DNA methylation in the placenta as compared with 80%–90% in fetal tissues (Cotton et al. 2009; Price et al. 2012). In contrast, average Alu DNA methylation is similar in placenta and fetal tissues using the same technique (Gama-Sosa et al. 1983; Price et al. 2012). DNA methylation of any individual Alu or L1 element depends on multiple factors such as its location in the genome and age of the element (Szpakowski et al. 2009; Xie et al. 2009; Price et al. 2012). Similarly, DNA methylation of HERV families in placenta show on average reduced, but widely variable, levels across different HERVs (Reiss et al. 2007). Reduced DNA methylation at a subset of such elements may result in the increased usage of RE-derived promoters for placental-specific expression of genes or gene-isoforms (Rawn and Cross 2008; Cohen et al. 2009). Some examples of placental-specific genes using HERV-derived promoters include: (1) syncytin 1 and syncytin 2, involved in the fusion of trophoblast cells to form a multinucleate syncytium and in the formation of exosomes (Vargas et al. 2009); (2) the placental-specific promoter of IL2RB (interleukin-2 receptor b subunit), which plays a role in NK cell differentiation at the maternal–fetal interface (Cohen et al. 2011); and (3) PEG10, a maternally imprinted gene essential for placental development (Clark et al. 2007).
Hypomethylation of X-Chromosome Gene Promoters in Females
To achieve dosage compensation between males (XY) and females (XX), one of the two X-chromosome is normally inactivated in all female cells. In somatic cells, the inactive X is characterized by high gene promoter DNA methylation and hypomethylation of gene bodies. Although autosomal gene promoters are roughly similarly methylated in placenta and embryonic tissues, there is a striking hypomethylation of X-linked CpG island promoters in female placentas (Cotton et al. 2009). DNA methylation at X-linked CpG island promoters is on average 27% less in the female placenta as compared with female blood, but this is still higher than X-linked CpG island promoter methylation observed in the male placenta (Cotton et al. 2009). The reason for reduced methylation on the X is not known. Given this pattern, one might expect greater expression of genes from the inactive X-chromosome in female placenta as compared with blood, but this does not generally appear to be the case (Nguyen and Disteche 2006). Although there is no evidence for X-chromosome reactivation in healthy female placental tissues, this was observed in cultures of villous stroma from first trimester miscarriages (Migeon et al. 2005).
While the reduction in DNA methylation on the X in female placentas might not have functional consequences for XCI, it does affect the interpretation of DNA methylation-based assays of XCI in placental tissues. The human androgen receptor (HUMARA) assay commonly used clinically to assess XCI in human tissues is based on differential DNA methylation at an HpaII site within exon1 of the androgen receptor. In fetuses carrying X mutations or chromosomal rearrangements, assessment of nonrandom or skewed XCI may be relevant to interpreting potential phenotype. However, application of HUMARA or similar assays to placental tissues is limited for several reasons. First, this site is incompletely methylated on the inactive X in trophoblast and mesenchyme of the placenta as well as in amnion and chorion (Peñaherrera et al. 2012). It was also reported that this site was unmethylated in microdissected stromal tissue from placenta and umbilical cord (Looijenga et al. 1999). Second, the villous trees of the placenta grow in a highly clonal manner (see below), creating extensive site-to-site differences in XCI status (Moriera de Mello et al. 2010; Peñaherrera et al. 2012). Last, the average level of skewing over multiple sampled sites shows little correlation with skewing observed in fetal somatic tissues (Peñaherrera et al. 2012). In one X-autosome translocation female, skewed XCI was observed in cord blood, umbilical cord, and amnion, but was largely random in the placenta (Peñaherrera et al. 2003). Hence, placental tissue should not be used to predict skewing status in the somatic tissues of carriers of X-linked mutations.
Gene Promoter Methylation
Despite the global hypomethylation of placenta, DNA methylation is clearly an important mechanism for gene regulation in the placenta. On average, autosomal gene promoters are methylated similarly in placenta as in somatic tissues (Cotton et al. 2009; Price et al. 2012), with high density CpG island promoters tending to be more methylated and nonisland promoters slightly less methylated in the placenta as compared with fetal tissues (Price et al. 2012). However, this is very much gene specific. Some examples of genes with placental-specific expression that are also specifically unmethylated at their gene promoters include chorionic gonadotropin-beta subunit (CGB) (Campain et al. 1993) and placental lactogen (CSH1) (Cho et al. 2001). Likewise, many genes are methylated at their promoter and down-regulated specifically in the placenta. For example, methylation and down-regulation of OCT4 is associated with normal trophoblast differentiation early in development (Zhang et al. 2008) and methylation of tumor suppressor genes, such as RASSF1 or APC, is associated with trophoblast invasiveness (Novakovic et al. 2008). Further evidence for the importance of DNA methylation for gene expression in placental trophoblast comes from in vitro studies using 5-Aza-2′-deoxycytidine to inhibit DNA methylation in choriocarcinoma cells (Hogg et al. 2014b).
Monoallelic Methylation and Genomic Imprinting
Monoallelic DNA methylation leading to monoallellic repression/expression of one of the two gene copies can occur in an imprinted (parent-of-origin dependent) manner or can be seemingly random. The importance of genomic imprinting in placental development has been widely discussed in the literature (Coan et al. 2005; Frost and Moore 2010; Fowden et al. 2011). Many imprinted genes are associated with an “imprinting control region” (ICR), the methylation of which regulates monoallelic gene repression. For example, two large microRNA clusters (C14MC and C19MC) on chromosome 14 and 19 are each regulated by an ICR such that both alleles are methylated and repressed in somatic tissues, but there is monoallelic DNA methylation of the ICR and expression of these microRNAs in placental tissues (Noguer-Dance et al. 2010; Mouillet et al. 2011). As previously mentioned, the DNA methyltransferase gene DNMT1 shows placental-specific imprinting with maternal-allele DNA methylation and paternal-allele expression in the placenta (Das et al. 2013). Some imprinted genes, such as CDKN1C and IGF2, have placental-specific promoters (Monk et al. 2006; Yuen et al. 2011a), illustrating that placental imprinted differentially methylated regions (DMRs) may differ from somatic ones for a given imprinted gene. Polymorphic or variable imprinting across cells has also been reported in placenta (Lambertini et al. 2008), for example, in the genes IGF2R (Xu et al. 1993) and WT1 (Jinno et al. 1994).
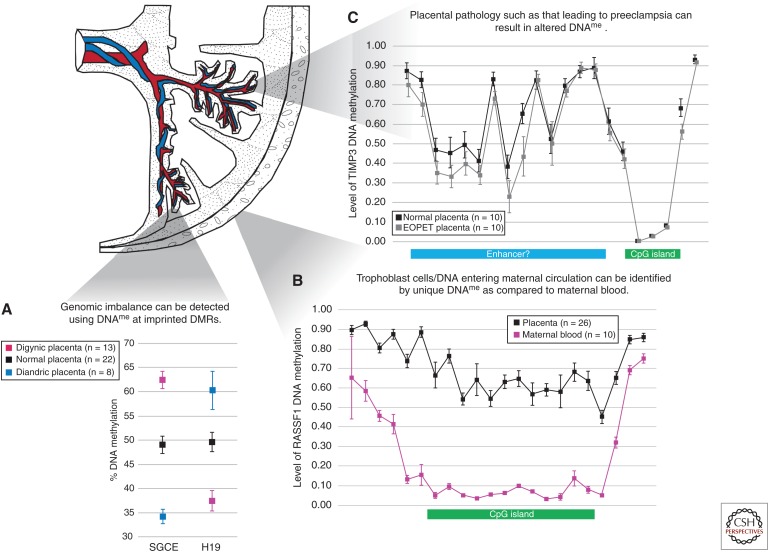
As many imprinted DMRs/ICRs are tightly maintained in placenta, DNA methylation testing can be used for diagnosis of chromosomal imbalance in the placenta (Bourque et al. 2011). Therefore, the parental origin of triploidy or the level of androgenetic cells in samples from placentas with placental mesenchymal dysplasia can be diagnosed from DNA methylation ratios at imprinted ICRs (Fig. 2). Comparing the DNA methylation profile of placentas with unbalanced parental contributions (i.e., triploidy, hydatidiform moles) has also allowed for the identification of novel imprinted genes (Choufani et al. 2011; Yuen et al. 2011a). Abnormal DNA methylation values at imprinted DMRs have been associated with in vitro fertilization, miscarriage, and intrauterine growth restriction (Table 2).
Figure 2.
Applications of placental DNA methylation. (A) DNA methylation at imprinting control regions (ICRs) such as those associated with the genes SGCE and H19 can be used to identify genomic imbalance in the placenta. Mean and standard deviation of DNA methylation levels for digynic and diandric triploidy and controls are indicated. (Adapted from Bourque et al. 2010.) (B) Regions of the genome with distinct patterns of placental DNA methylation as compared with that in maternal blood, for example at the CpG island of RASSF1 depicted here, can be used to distinguish fetal DNA in maternal circulation. Each data point represents the mean level of DNA methylation at a single CpG site obtained from the Illumina HumanMethylation450 BeadChip array; error bars represent standard deviation of the mean (WP Robinson and EM Price, unpubl.). (C) Altered DNA methylation may be detected in the placentas of complicated pregnancies, such as in early onset preeclampsia (EOPET). These may not be restricted to CpG islands; in TIMP3 (chr22: 33195342–33257640), for example, lower DNA methylation is observed in the putative enhancer region. Each data point represents the mean level of DNA methylation at a single CpG site obtained from the Illumina HumanMethylation450 BeadChip array; error bars represent standard deviation of the mean. (Adapted from Blair et al. 2013.) DNA methylation, DNAme.
Table 2.
Studies of genome-wide placental DNA methylation with pregnancy complications
| Condition/criteria | Main findings | Reference(s) |
|---|---|---|
| Miscarriage (normal chromosomes) | More variable DNA methylation at imprinted genes and reduced global DNA methylation were reported in chromosomally normal miscarriages compared with elective terminations. | Pliushch et al. 2010; Yin et al. 2012; Hanna et al. 2013 |
| Placental mesenchymal dysplasia (PMD)/ triploidy/ complete hydatidiform mole (CHM) | DNA methylation changes at imprinted DMRs consistent with excess paternal alleles (PMD, CHM, diandric triploidy) or maternal alleles (digynic triploidy) and at affected regions of the placenta. | Bourque et al. 2011; Yuen et al. 2011a |
| Trisomy | Global hypermethylation in trisomy 21 placentas. Widespread changes in trisomy 16 miscarriage and confined placental trisomy 16. The latter overlap changes with preeclampsia. | Jin et al. 2013; Tolmacheva et al. 2013; Blair et al. 2014 |
| Preeclampsia | Widespread DNA methylation changes in early onset preeclampsia, much fewer changes in late-onset (less severe) preeclampsia. Changes in genes associated with syncytial trophoblast and/or cell adhesion (an important process in trophoblast differentiation and function) among many others. | Yuen et al. 2010; Jia et al. 2012; Blair et al. 2013; Anton et al. 2014 |
| Intrauterine growth restriction (IUGR)/small for gestational age | Inconsistent results, largely focused on imprinted genes. Criteria for IUGR vary by study. Correlation between L1 methylation and birth weight reported. | Yuen et al. 2010; Banister et al. 2011; Ferreira et al. 2011; Koukoura et al. 2012; Wilhelm-Benartzi et al. 2012 |
| Assisted reproductive technology | One study identified multiple CpG sites with lower DNA methylation in placentas of ART babies and another, no difference in DNA methylation at repetitive Alu, L1 or α-satellite repeat elements; no loss of imprinting identified at 25 imprinted genes in ART placentae versus spontaneously conceived. | Katari et al. 2009; Camprubi et al. 2013 |
As in other tissues (Gimelbrant et al. 2007; Chess 2012), monoallelic expression can occur in the absence of imprinting. Variable degrees of nonimprinted monoallelic DNA methylation have been reported for the leptin gene (LEP) (Hogg et al. 2013b). Interestingly, placentas associated with early-onset preeclampsia show hypomethylation of LEP, which is in turn associated with more biallelic LEP expression compared with controls (Hogg et al. 2013b). Methylation allelic polymorphism (MAP), or an “on−off” type of DNA methylation (0% or 50% corresponding to biallelic/monoallelic expression patterns) that cannot be explained by a nearby genetic polymorphism, has been reported in placenta for WNT2, TUSC3, EPHB4, and CGB5 (Yuen et al. 2009; Uuskula et al. 2011). There was suggestive evidence for an association of the TUSC3 MAP with preeclampsia (Yuen et al. 2009) and the CGB5 MAP to early miscarriage (Uuskula et al. 2011). Overall, the possibility of variable monoallelic DNA methylation as a way to fine tune gene expression or increase diversity is intriguing.
INFLUENCE OF PLACENTAL ARCHITECTURE
Villous Tree Structure
In a human placenta, the fetal vessels enter through the chorionic plate and branch into a network covered by two main cell layers, an outer trophoblast layer and an inner mesenchyme layer (a mix of immune, endothelial, and fibroblast cells) (Castellucci et al. 2000). There are an estimated 60–70 of these chorionic villus trees that grow throughout development in a clonal manner (Peñaherrera et al. 2012). Each tree may show a varying degree of maturation or pathology, which may be reflected in localized differences in DNA methylation. Variation in maternal and fetal blood supplies may also contribute to spatial differences in gene expression and DNA methylation (Wyatt et al. 2005). As the placenta is a dynamic structure, showing considerable ability to adapt to a variety pathological influences (Redline 2008), sampling strategy (in relation to organ anatomy) needs to be considered carefully in the planning and interpretation of placental DNA methylation studies (Burton et al. 2014; Hogg et al. 2014a).
Measured DNA methylation level is an average across all cells present in a sample. Cellular composition may vary by sampling site, placental pathology, or gestational age. The DNA methylation profiles of individual cell types derived from the mesenchyme core have not been well-studied and are expected to be quite distinct from each other. For example, long-term cultures of chorionic villi, typically used for prenatal cytogenetic analysis, show a DNA methylation profile more similar to amnion and chorion than the chorionic villi as a whole (Fig. 1). Understanding the DNA methylation profiles of individual cells will be an important step to understanding DNA methylation in healthy placentas and the changes that occur in pathological ones.
Changes with Gestational Age
Numerous changes in placental DNA methylation occur throughout gestation, with term placentas showing on average higher and more variable DNA methylation than those from earlier in gestation (Fuke et al. 2004; Novakovic et al. 2011). For example, the level of HERV expression has been shown to correspondingly decline with gestational age (Fuke et al. 2004). Changes to DNA methylation likely reflect a combination of developmentally programmed changes in cell composition and gene expression as well as environmentally influenced changes.
In early gestation, primary functions of the placenta include invasion into the maternal endometrium, remodeling of maternal vasculature, and secretion of hormones important to maintain pregnancy. Trophoblast cells predominate within the chorionic villi as the early hypoxic conditions favor their proliferation. However, once there is maternal blood flow to the placenta (10–12 wk gestation), trophoblast growth slows and embryo/fetal growth along with fetal vasculature increases. Hence, in the first trimester, the DNA methylation profile of whole chorionic villi is more similar to that of isolated trophoblast than it is later in gestation (Fig. 1).
While a dominant syncytiotrophoblast layer remains throughout pregnancy, there is a relative decline in cytotrophoblast and mesoderm with an increase in placental vasculature and Hofbauer cells (placental macrophages) (Wang and Zhao 2010). Interestingly, the ratio of syncytiotrophoblast to cytotrophoblast nuclei remains constant through gestation (10 wk to term) at 8:1, but cellular volume is altered through gestation (Mayhew et al. 1999). The origin of specific cell types may also change with gestation, which could affect the placental DNA methylation profile. For example, Hofbauer cells derive from villous mesenchymal stem cells in the first trimester, but later in gestation likely come from fetal monocytes (Tang et al. 2011).
An increase in hematopoietic cells through gestation may explain why the pathways most enriched amongst genes differentially methylated between first, second, or third trimester placentas were those involved in “communication between innate and adaptive immune cells” and other immune-related pathways (Novakovic et al. 2011). Altered T-cell pathways include the NFAT pathway (an important regulator of parturition [Tabata et al. 2009]) and pathways associated with autoimmune disorders (presumably related to immune tolerance). This agrees with gene expression studies, which also show substantial enrichment of immune regulators amongst the most differentially expressed genes in placenta at different gestational ages (Winn et al. 2007; Mikheev et al. 2008).
In the last half of pregnancy, the placenta also shows changes in gene expression in response to the increasing demands of the fetus for nutrients (Constancia et al. 2005). These include, for example, up-regulation of System A transporters, which are responsible for transporting amino acids to the fetus (Coan et al. 2010; Burton and Fowden 2012), iron transport proteins (Bradley et al. 2004), which absorb iron from maternal blood, and placental corticotropin releasing hormone (CRH), which may stimulate maternal glucose production needed to support the growing brain (Gangestad et al. 2012). CRH gene promoter DNA methylation has been correlated with its expression in the placenta (Jiang et al. 2012); increased gestational age was associated with decreased DNA methylation at CpG sites associated with the CRH gene as well as other cortisol signaling and steroidogenic genes in the placenta (Hogg et al. 2013a). Increased DNA methylation at the promoter region of SLC2A3, the gene encoding the glucose transporter GLUT3, was associated with decreased GLUT3 expression through gestation (Novakovic et al. 2013).
ALTERED DNA METHYLATION IN CLINICAL CONDITIONS
Pregnancy Complication
Altered placental DNA methylation has been studied in the context of pregnancy complications including miscarriage, preeclampsia, intrauterine growth restriction (IUGR), and trisomy (see Table 2). Generally speaking, many changes in DNA methylation are observed with gross placental pathology (i.e., hydatidiform mole, triploidy, early-onset/severe preeclampsia), although more subtle changes occur when there is not a distinct pathology. For example, early-onset/severe preeclampsia is associated with decreased placental perfusion (characterized by increased syncytial knots, aggregates of intervillous fibrin, vascular lesions) and smaller placentas (Redline 2008; Nelson et al. 2014). There are correspondingly widespread and large changes in DNA methylation associated with early-onset preeclampsia (Yuen et al. 2010; Jia et al. 2012; Blair et al. 2013; Anton et al. 2014). Although a subset of these changes overlap sites altered in syncytial trophoblast differentiation and hypoxia exposure (Yuen et al. 2013), the overall relationship of the DNA methylation changes to the observed pathology is unknown and likely complex (Blair et al. 2013). Regardless, DNA methylation profiling should prove clinically useful to group placentas that show similar underlying pathologies as well as to identify DNA methylation marks that could be used to quantify or characterize placental DNA circulating in maternal blood during pregnancy (Yuen et al. 2011b).
DNA methylation changes that have been reported to be associated with IUGR tend to be of small magnitude and have not always been reproduced in other studies. An example is altered DNA methylation associated with the H19/IGF2 ICR in IUGR, which has been reported in some but not all studies (Guo et al. 2008; Bourque et al. 2010; Tabano et al. 2010; Cordeiro et al. 2014). This may be because of both technical aspects and different clinical criteria used in these studies. Many studies have used small-for-gestational age (SGA) (<10th percentile) as a surrogate for IUGR, although only a subset of SGA cases are associated with placental-mediated IUGR. Some studies may also be complicated by inclusion of cases that co-occur with preeclampsia. Larger samples sizes with stricter case criteria may help clarify consistent placental DNA methylation changes associated with IUGR. Genome-wide DNA methylation profiling may prove useful as an approach to classification of cases by etiology in a manner unbiased by clinical presentation.
Chromosomally normal first trimester miscarriages show relatively few DNA methylation changes compared with placentas obtained from elective terminations (Hanna et al. 2013), possibly because of the heterogeneous etiology of miscarriage in such samples. Nonetheless, a number of significant differences in DNA methylation have been noted, including an increased number of extreme values at imprinted DMRs (Pliushch et al. 2010; Hanna et al. 2013). It remains to be determined if such changes are causative or a consequence of associated placental changes in these pregnancies (e.g., delayed or arrested development).
Even when a distinct etiology for abnormal placental function is apparent, it is important to consider gestational age effects. This can be illustrated by our studies of placentas with trisomy 16 (Blair et al. 2014). Although widespread changes in placental DNA methylation were found in both first trimester trisomy 16 (ascertained from miscarriages) and third trimester trisomy 16 (in the context of confined placental mosaicism), there was relatively little overlap between the major changes at these different gestational ages. Overall there were a greater number of changes in the third trimester, which may reflect the greater time for changes to accumulate in response to such a genetic insult.
DNA Methylation Changes in Response to Exposure
The placenta has received attention in the field of developmental origins of health and disease (DOHaD) because of its role in moderating the fetal microenvironment. In addition to its immunologic and synthetic roles, placental tissue transfers molecules in the maternal blood to the fetus as well as consuming some itself. By changing structure, cell composition or gene expression, the placenta may respond to, buffer against, or adapt to the contents of maternal blood. For example, smaller and/or lighter placentas have been documented in offspring of mothers who smoke (Anblagan et al. 2013), were pregnant during Ramadan fasting (Alwasel et al. 2013), or were exposed to higher levels of air pollution (van den Hooven et al. 2012). Larger and/or heavier placentas have been documented in offspring of mothers who are obese or endure psychosocial stress during pregnancy (Tegethoff et al. 2010). Furthermore, structural and functional changes in the placenta might correlate with changes in the fetus. Raised blood pressure in offspring was found to be associated with smaller placenta (weight and size) in children born to mothers of short stature and low socioeconomic status (Barker et al. 2010).
Analyzing site-specific and/or genome-wide placental DNA methylation may thus afford a resource for assessing maternal environmental conditions that shape fetal development and postnatal disease. A measured change in placental DNA methylation may reflect one of several effects of an exposure: (1) altered placental morphology; (2) changes in gene expression; or (3) changes in the establishment/maintenance of DNA methylation. The placenta shows a remarkable degree of developmental plasticity (Yuen and Robinson 2011). Therefore, it is important to consider that evidence of an exposure in the placenta will not necessarily be associated with an effect in the fetus. But while numerous studies have examined maternal exposures by targeting individual genes likely to be involved in placental adaptation, far fewer have examined the placental methylome in association with environmental exposures (Tables 2 and 3). With the exception of chromosomal abnormalities, preeclampsia, and perhaps maternal smoking, little difference in genome-wide placental DNA methylation has been consistently observed. Some of the difficulties facing DOHaD epigenome-wide association studies (EWAS) in placenta include the following.
Table 3.
Studies of genome-wide placental DNA methylation with environmental exposures
| Condition/criteria | Main findings | Reference(s) |
|---|---|---|
| Gestational diabetes mellitus (GDM) | One study found reduced genome-wide DNA methylation in GDM-exposed placentas, and another no differential DNA methylation after correction for multiple comparisons. | Ruchat et al. 2013; Nomura et al. 2014 |
| Maternal prepregnancy BMI | No differential DNA methylation at repetitive L1 elements or on the global scale. | Michels et al. 2011; Janssen et al. 2013; Nomura et al. 2014 |
| Tobacco | Moderate alteration of genome-wide DNA methylation and Alu repetitive elements, but not L1 repetitive elements with maternal tobacco use. | Suter et al. 2011; Wilhelm-Benartzi et al. 2012 |
| Alcohol | No change in global DNA methylation with maternal alcohol use during pregnancy; a trend toward lower L1 DNA methylation was identified in a study with small sample size. | Wilhelm-Benartzi et al. 2012; Janssen et al. 2013 |
| Acetaminophen | Increased global DNA methylation with maternal acetaminophen use during pregnancy. | Janssen et al. 2013 |
| Pollution | Reduced global DNA methylation with increased exposure to ambient particulate matter, with the strongest association with first trimester exposure. | Janssen et al. 2013 |
Study design: Amassing a large group of human samples with a homogeneous exposure is difficult. Heterogeneous genetic background and environmental modifiers further complicate phenotype and necessitate an even larger sample size.
Statistical power and biological significance: Investigators should first consider if there is reason to expect alterations in DNA methylation on the site-specific or genome-wide level, if at all. The power of EWAS studies is heavily burdened by the essential correction for multiple testing. Furthermore, the magnitude of change in DNA methylation that might be consequent to an exposure and/or result in functional changes is unknown and likely to vary across the genome.
Exposure conditions: Study of the blood of individuals born to mothers exposed to the Dutch famine suggests that differential DNA methylation at some imprinted genes may be dependent on the timing of exposure (i.e., periconceptional vs. late gestation) as well as sex of the fetus (Heijmans et al. 2008; Tobi et al. 2009). Variables such as the type, timing, and duration/dose of exposure may result in differential adaptation and in turn differential patterns of DNA methylation.
With attention to these factors, the placenta should yield valuable information as to the in utero exposures that may affect neonatal and postnatal health. Deciphering the epigenetic record of the placenta is an exciting and growing area of research, and focusing on changes in specific cell populations within the placenta may help distinguish changes in placental growth/structure from changes caused by the epigenome.
CONCLUDING REMARKS
Currently our understanding of the human placental methylome is in the exploratory stage. Its unique features make it attractive to study to understand the role of epigenetic processes in development. It also can hold extensive clues to adaptive and pathological processes that occur in development. We are hopeful that, as our understanding of variation in DNA methylation across the placenta and across individuals improves, it may be used as a source of clinical biomarkers during pregnancy and possibly for prediction of future well-being of the neonate.
ACKNOWLEDGMENTS
We would like to thank Maria Peñaherrera for editorial comments. W.P.R. is funded by a salary award from the Child & Family Research Institute, Vancouver, British Columbia; E.M.P. is funded through a Doctoral award from the Canadian Institutes for Health Research.
Footnotes
Editors: Diana W. Bianchi and Errol R. Norwitz
Additional Perspectives on Molecular Approaches to Reproductive and Newborn Medicine available at www.perspectivesinmedicine.org
REFERENCES
- Alwasel SH, Harrath A, Aljarallah JS, Abotalib Z, Osmond C, Al Omar SY, Khaled I, Barker DJ. 2013. Intergenerational effects of in utero exposure to Ramadan in Tunisia. Am J Hum Biol 25: 341–343. [DOI] [PubMed] [Google Scholar]
- Anblagan D, Jones NW, Costigan C, Parker AJ, Allcock K, Aleong R, Coyne LH, Deshpande R, Raine-Fenning N, Bugg G, et al. 2013. Maternal smoking during pregnancy and fetal organ growth: A magnetic resonance imaging study. PLoS ONE 8: e67223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton L, Brown AG, Bartolomei MS, Elovitz MA. 2014. Differential methylation of genes associated with cell adhesion in preeclamptic placentas. PLoS ONE 9: e100148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banister CE, Koestler DC, Maccani MA, Padbury JF, Houseman EA, Marsit CJ. 2011. Infant growth restriction is associated with distinct patterns of DNA methylation in human placentas. Epigenetics 6: 920–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker DJ, Thornburg KL, Osmond C, Kajantie E, Eriksson JG. 2010. The surface area of the placenta and hypertension in the offspring in later life. Int J Dev Biol 54: 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair JD, Yuen RK, Lim BK, McFadden DE, von Dadelszen P, Robinson WP. 2013. Widespread DNA hypomethylation at gene enhancer regions in placentas associated with early-onset pre-eclampsia. Mol Hum Reprod 19: 697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair JD, Langlois S, McFadden DE, Robinson WP. 2014. Overlapping DNA methylation profile between placentas with trisomy 16 and early-onset preeclampsia. Placenta 35: 216–222. [DOI] [PubMed] [Google Scholar]
- Bourque DK, Avila L, Peñaherrera MS, von Dadelszen P, Robinson WP. 2010. Decreased placental methylation at the H19/IGF2 imprinting control region is associated with normotensive intrauterine growth restriction but not preeclampsia. Placenta 31: 197–202. [DOI] [PubMed] [Google Scholar]
- Bourque D, Peñaherrera M, Yuen R, Van Allen M, McFadden D, Robinson W. 2011. The utility of quantitative methylation assays at imprinted genes for the diagnosis of fetal and placental disorders. Clin Genet 79: 169–175. [DOI] [PubMed] [Google Scholar]
- Bradley J, Leibold EA, Harris ZL, Wobken JD, Clarke S, Zumbrennen KB, Eisenstein RS, Georgieff MK. 2004. Influence of gestational age and fetal iron status on IRP activity and iron transporter protein expression in third-trimester human placenta. Am J Physiol Regul Integr Comp Physiol 287: R894–R901. [DOI] [PubMed] [Google Scholar]
- Burton GJ, Fowden AL. 2012. Review: The placenta and developmental programming: Balancing fetal nutrient demands with maternal resource allocation. Placenta 33: S23–S27. [DOI] [PubMed] [Google Scholar]
- Burton GJ, Sebire NJ, Myatt L, Tannetta D, Wang YL, Sadovsky Y, Staff AC, Redman CW. 2014. Optimising sample collection for placental research. Placenta 35: 9–22. [DOI] [PubMed] [Google Scholar]
- Campain JA, Gutkin DW, Cox GS. 1993. Differential DNA methylation of the chorionic gonadotropin β-subunit multigene family. Mol Endocrinol 7: 1331–1346. [DOI] [PubMed] [Google Scholar]
- Camprubi C, Iglesias-Platas I, Martin-Trujillo A, Salvador-Alarcon C, Rodriguez MA, Barredo DR, Court F, Monk D. 2013. Stability of genomic imprinting and gestational-age dynamic methylation in complicated pregnancies conceived following assisted reproductive technologies. Biol Reprod 89: 50. [DOI] [PubMed] [Google Scholar]
- Castellucci M, Kosanke G, Verdenelli F, Huppertz B, Kaufmann P. 2000. Villous sprouting: Fundamental mechanisms of human placental development. Hum Reprod Update 6: 485–494. [DOI] [PubMed] [Google Scholar]
- Chess A. 2012. Mechanisms and consequences of widespread random monoallelic expression. Nat Rev Genet 13: 421–428. [DOI] [PubMed] [Google Scholar]
- Ching TT, Maunakea AK, Jun P, Hong C, Zardo G, Pinkel D, Albertson DG, Fridlyand J, Mao JH, Shchors K, et al. 2005. Epigenome analyses using BAC microarrays identify evolutionary conservation of tissue-specific methylation of SHANK3. Nat Genet 37: 645–651. [DOI] [PubMed] [Google Scholar]
- Cho JH, Kimura H, Minami T, Ohgane J, Hattori N, Tanaka S, Shiota K. 2001. DNA methylation regulates placental lactogen I gene expression. Endocrinology 142: 3389–3396. [DOI] [PubMed] [Google Scholar]
- Choufani S, Shapiro JS, Susiarjo M, Butcher DT, Grafodatskaya D, Lou Y, Ferreira JC, Pinto D, Scherer SW, Shaffer LG, et al. 2011. A novel approach identifies new differentially methylated regions (DMRs) associated with imprinted genes. Genome Res 21: 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MB, Janicke M, Gottesbuhren U, Kleffmann T, Legge M, Poole ES, Tate WP. 2007. Mammalian gene PEG10 expresses two reading frames by high efficiency-1 frameshifting in embryonic-associated tissues. J Biol Chem 282: 37359–37369. [DOI] [PubMed] [Google Scholar]
- Coan PM, Burton GJ, Ferguson-Smith AC. 2005. Imprinted genes in the placenta—A review. Placenta 26: S10–S20. [DOI] [PubMed] [Google Scholar]
- Coan PM, Vaughan OR, Sekita Y, Finn SL, Burton GJ, Constancia M, Fowden AL. 2010. Adaptations in placental phenotype support fetal growth during undernutrition of pregnant mice. J Physiol 588: 527–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen CJ, Lock WM, Mager DL. 2009. Endogenous retroviral LTRs as promoters for human genes: A critical assessment. Gene 448: 105–114. [DOI] [PubMed] [Google Scholar]
- Cohen CJ, Rebollo R, Babovic S, Dai EL, Robinson WP, Mager DL. 2011. Placenta-specific expression of the interleukin-2 (IL-2) receptor β subunit from an endogenous retroviral promoter. J Biol Chem 286: 35543–35552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constancia M, Angiolini E, Sandovici I, Smith P, Smith R, Kelsey G, Dean W, Ferguson-Smith A, Sibley CP, Reik W, et al. 2005. Adaptation of nutrient supply to fetal demand in the mouse involves interaction between the Igf2 gene and placental transporter systems. Proc Natl Acad Sci 102: 19219–19224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordeiro A, Neto AP, Carvalho F, Ramalho C, Doria S. 2014. Relevance of genomic imprinting in intrauterine human growth expression of CDKN1C, H19, IGF2, KCNQ1 and PHLDA2 imprinted genes. J Assist Reprod Genet 31: 1361–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton AM, Avila L, Peñaherrera MS, Affleck JG, Robinson WP, Brown CJ. 2009. Inactive X chromosome-specific reduction in placental DNA methylation. Hum Mol Genet 18: 3544–3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R, Lee YK, Strogantsev R, Jin S, Lim YC, Ng PY, Lin XM, Chng K, Yeo G, Ferguson-Smith AC, et al. 2013. DNMT1 and AIM1 imprinting in human placenta revealed through a genome-wide screen for allele-specific DNA methylation. BMC Genomics 14: 685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, Gehrke C. 1982. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res 10: 2709–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira JC, Choufani S, Grafodatskaya D, Butcher DT, Zhao C, Chitayat D, Shuman C, Kingdom J, Keating S, Weksberg R. 2011. WNT2 promoter methylation in human placenta is associated with low birthweight percentile in the neonate. Epigenetics 6: 440–449. [DOI] [PubMed] [Google Scholar]
- Fowden AL, Coan PM, Angiolini E, Burton GJ, Constancia M. 2011. Imprinted genes and the epigenetic regulation of placental phenotype. Prog Biophys Mol Biol 106: 281–288. [DOI] [PubMed] [Google Scholar]
- Frost JM, Moore GE. 2010. The importance of imprinting in the human placenta. PLoS Genet 6: e1001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuke C, Shimabukuro M, Petronis A, Sugimoto J, Oda T, Miura K, Miyazaki T, Ogura C, Okazaki Y, Jinno Y. 2004. Age related changes in 5-methylcytosine content in human peripheral leukocytes and placentas: An HPLC-based study. Ann Hum Genet 68: 196–204. [DOI] [PubMed] [Google Scholar]
- Gama-Sosa MA, Midgett RM, Slagel VA, Githens S, Kuo KC, Gehrke CW, Ehrlich M. 1983. Tissue-specific differences in DNA methylation in various mammals. Biochim Biophys Acta 740: 212–219. [DOI] [PubMed] [Google Scholar]
- Gangestad SW, Caldwell Hooper AE, Eaton MA. 2012. On the function of placental corticotropin-releasing hormone: A role in maternal-fetal conflicts over blood glucose concentrations. Biol Rev Camb Philos Soc 87: 856–873. [DOI] [PubMed] [Google Scholar]
- Gimelbrant A, Hutchinson JN, Thompson BR, Chess A. 2007. Widespread monoallelic expression on human autosomes. Science 318: 1136–1140. [DOI] [PubMed] [Google Scholar]
- Grigoriu A, Ferreira JC, Choufani S, Baczyk D, Kingdom J, Weksberg R. 2011. Cell specific patterns of methylation in the human placenta. Epigenetics 6: 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Choufani S, Ferreira J, Smith A, Chitayat D, Shuman C, Uxa R, Keating S, Kingdom J, Weksberg R. 2008. Altered gene expression and methylation of the human chromosome 11 imprinted region in small for gestational age (SGA) placentae. Dev Biol 320: 79–91. [DOI] [PubMed] [Google Scholar]
- Hanna CW, McFadden DE, Robinson WP. 2013. DNA methylation profiling of placental villi from karyotypically normal miscarriage and recurrent miscarriage. Am J Pathol 182: 2276–2284. [DOI] [PubMed] [Google Scholar]
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. 2008. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci 105: 17046–17049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg K, Price EM, Hanna CW, Robinson WP. 2012. Prenatal and perinatal environmental influences on the human fetal and placental epigenome. Clin Pharmacol Ther 92: 716–726. [DOI] [PubMed] [Google Scholar]
- Hogg K, Blair JD, McFadden DE, von Dadelszen P, Robinson WP. 2013a. Early onset pre-eclampsia is associated with altered DNA methylation of cortisol-signalling and steroidogenic genes in the placenta. PLoS ONE 8: e62969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg K, Blair JD, von Dadelszen P, Robinson WP. 2013b. Hypomethylation of the LEP gene in placenta and elevated maternal leptin concentration in early onset pre-eclampsia. Mol Cell Endocrinol 367: 64–73. [DOI] [PubMed] [Google Scholar]
- Hogg K, Price EM, Robinson WP. 2014a. Improved reporting of DNA methylation data derived from studies of the human placenta. Epigenetics 9: 333–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg K, Robinson WP, Beristain AG. 2014b. Activation of endocrine-related gene expression in placental choriocarcinoma cell lines following DNA methylation knock-down. Mol Hum Reprod 20: 677–689. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. 2009. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 41: 178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen BG, Godderis L, Pieters N, Poels K, Kicinski M, Cuypers A, Fierens F, Penders J, Plusquin M, Gyselaers W, et al. 2013. Placental DNA hypomethylation in association with particulate air pollution in early life. Part Fibre Toxicol 10: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia RZ, Zhang X, Hu P, Liu XM, Hua XD, Wang X, Ding HJ. 2012. Screening for differential methylation status in human placenta in preeclampsia using a CpG island plus promoter microarray. Int J Mol Med 30: 133–141. [DOI] [PubMed] [Google Scholar]
- Jiang X, Yan J, West AA, Perry CA, Malysheva OV, Devapatla S, Pressman E, Vermeylen F, Caudill MA. 2012. Maternal choline intake alters the epigenetic state of fetal cortisol-regulating genes in humans. FASEB J 26: 3563–3574. [DOI] [PubMed] [Google Scholar]
- Jin S, Lee YK, Lim YC, Zheng Z, Lin XM, Ng DP, Holbrook JD, Law HY, Kwek KY, Yeo GS, et al. 2013. Global DNA hypermethylation in down syndrome placenta. PLoS Genet 9: e1003515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinno Y, Yun K, Nishiwaki K, Kubota T, Ogawa O, Reeve AE, Niikawa N. 1994. Mosaic and polymorphic imprinting of the WT1 gene in humans. Nat Genet 6: 305–309. [DOI] [PubMed] [Google Scholar]
- Katari S, Turan N, Bibikova M, Erinle O, Chalian R, Foster M, Gaughan JP, Coutifaris C, Sapienza C. 2009. DNA methylation and gene expression differences in children conceived in vitro or in vivo. Hum Mol Genet 18: 3769–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koukoura O, Sifakis S, Spandidos DA. 2012. DNA methylation in the human placenta and fetal growth (review). Mol Med Rep 5: 883–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertini L, Diplas AI, Lee MJ, Sperling R, Chen J, Wetmur J. 2008. A sensitive functional assay reveals frequent loss of genomic imprinting in human placenta. Epigenetics 3: 261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O’Malley R, Castanon R, Klugman S, et al. 2011. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471: 68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looijenga LH, Gillis AJ, Verkerk AJ, van Putten WL, Oosterhuis JW. 1999. Heterogeneous X inactivation in trophoblastic cells of human full-term female placentas. Am J Hum Genet 64: 1445–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, et al. 2010. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466: 253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayhew TM, Leach L, McGee R, Ismail WW, Myklebust R, Lammiman MJ. 1999. Proliferation, differentiation and apoptosis in villous trophoblast at 13–41 weeks of gestation (including observations on annulate lamellae and nuclear pore complexes). Placenta 20: 407–422. [DOI] [PubMed] [Google Scholar]
- Michels KB, Harris HR, Barault L. 2011. Birthweight, maternal weight trajectories and global DNA methylation of LINE-1 repetitive elements. PLoS ONE 6: e25254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migeon BR, Axelman J, Jeppesen P. 2005. Differential X reactivation in human placental cells: Implications for reversal of X inactivation. Am J Hum Genet 77: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikheev AM, Nabekura T, Kaddoumi A, Bammler TK, Govindarajan R, Hebert MF, Unadkat JD. 2008. Profiling gene expression in human placentae of different gestational ages: An OPRU Network and UW SCOR Study. Reprod Sci 15: 866–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk D, Sanches R, Arnaud P, Apostolidou S, Hills FA, Abu-Amero S, Murrell A, Friess H, Reik W, Stanier P, et al. 2006. Imprinting of IGF2 P0 transcript and novel alternatively spliced INS-IGF2 isoforms show differences between mouse and human. Hum Mol Genet 15: 1259–1269. [DOI] [PubMed] [Google Scholar]
- Moriera de Mello JC, Souza de Aroujo ES, Stabellini R, Fraga AM, Santana de Souza JE, Sumita DR, Camargo AA, Periera LV. 2010. Random X inactivation and extensive mosaicism in human placenta revealed by analysis of allele-specific gene expression along the X chromosome. PLoS ONE 5: e10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouillet JF, Chu T, Sadovsky Y. 2011. Expression patterns of placental microRNAs. Birth Defects Res A Clin Mol Teratol 91: 737–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DB, Ziadie MS, McIntire DD, Rogers BB, Leveno KJ. 2014. Placental pathology suggesting that preeclampsia is more than one disease. Am J Obstet Gynecol 210: 66.e1–66.e7. [DOI] [PubMed] [Google Scholar]
- Nguyen DK, Disteche CM. 2006. Dosage compensation of the active X chromosome in mammals. Nat Genet 38: 47–53. [DOI] [PubMed] [Google Scholar]
- Noguer-Dance M, Abu-Amero S, Al-Khtib M, Lefevre A, Coullin P, Moore GE, Cavaille J. 2010. The primate-specific microRNA gene cluster (C19MC) is imprinted in the placenta. Hum Mol Genet 19: 3566–3582. [DOI] [PubMed] [Google Scholar]
- Nomura Y, Lambertini L, Rialdi A, Lee M, Mystal EY, Grabie M, Manaster I, Huynh N, Finik J, Davey M, et al. 2014. Global methylation in the placenta and umbilical cord blood from pregnancies with maternal gestational diabetes, preeclampsia, and obesity. Reprod Sci 21: 131–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novakovic B, Saffery R. 2013. Placental pseudo-malignancy from a DNA methylation perspective: Unanswered questions and future directions. Front Genet 4: 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novakovic B, Rakyan V, Ng HK, Manuelpillai U, Dewi C, Wong NC, Morley R, Down T, Beck S, Craig JM, et al. 2008. Specific tumour-associated methylation in normal human term placenta and first-trimester cytotrophoblasts. Mol Hum Reprod 14: 547–554. [DOI] [PubMed] [Google Scholar]
- Novakovic B, Wong NC, Sibson M, Ng HK, Morley R, Manuelpillai U, Down T, Rakyan VK, Beck S, Hiendleder S, et al. 2010. DNA methylation-mediated down-regulation of DNA methyltransferase-1 (DNMT1) is coincident with, but not essential for, global hypomethylation in human placenta. J Biol Chem 285: 9583–9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novakovic B, Yuen R, Gordon L, Peñaherrera M, Sharkey A, Moffett A, Craig J, Robinsonn W, Saffery R. 2011. Evidence for widespread changes in promoter methylation profile in human placenta in response to increasing gestational age and environmental/stochastic factors. BMC Genomics 12: 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novakovic B, Gordon L, Robinson WP, Desoye G, Saffery R. 2013. Glucose as a fetal nutrient: Dynamic regulation of several glucose transporter genes by DNA methylation in the human placenta across gestation. J Nutr Biochem 24: 282–288. [DOI] [PubMed] [Google Scholar]
- Peñaherrera MS, Ma S, Ho Yuen B, Brown CJ, Robinson WP. 2003. X-chromosome inactivation (XCI) patterns in placental tissues of a paternally derived bal t(X;20) case. Am J Med Genet A 118A: 29–34. [DOI] [PubMed] [Google Scholar]
- Peñaherrera MS, Jiang R, Avila L, Yuen RK, Brown CJ, Robinson WP. 2012. Patterns of placental development evaluated by X chromosome inactivation profiling provide a basis to evaluate the origin of epigenetic variation. Hum Reprod 27: 1745–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pliushch G, Schneider E, Weise D, El Hajj N, Tresch A, Seidmann L, Coerdt W, Muller AM, Zechner U, Haaf T. 2010. Extreme methylation values of imprinted genes in human abortions and stillbirths. Am J Pathol 176: 1084–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price EM, Cotton AM, Peñaherrera MS, McFadden DE, Kobor MS, Robinson W. 2012. Different measures of “genome-wide” DNA methylation exhibit unique properties in placental and somatic tissues. Epigenetics 7: 652–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawn SM, Cross JC. 2008. The evolution, regulation, and function of placenta-specific genes. Annu Rev Cell Dev Biol 24: 159–181. [DOI] [PubMed] [Google Scholar]
- Redline RW. 2008. Placental pathology: A systematic approach with clinical correlations. Placenta 29: S86–S91. [DOI] [PubMed] [Google Scholar]
- Reiss D, Zhang Y, Mager DL. 2007. Widely variable endogenous retroviral methylation levels in human placenta. Nucleic Acids Res 35: 4743–4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchat SM, Houde AA, Voisin G, St-Pierre J, Perron P, Baillargeon JP, Gaudet D, Hivert MF, Brisson D, Bouchard L. 2013. Gestational diabetes mellitus epigenetically affects genes predominantly involved in metabolic diseases. Epigenetics 8: 935–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F, Hyslop L, Stojkovic P, Leary C, Murdoch A, Reik W, Stojkovic M, Herbert M, Dean W. 2010. Evaluation of epigenetic marks in human embryos derived from IVF and ICSI. Hum Reprod 25: 2387–2395. [DOI] [PubMed] [Google Scholar]
- Schroeder DI, LaSalle JM. 2013. How has the study of the human placenta aided our understanding of partially methylated genes? Epigenomics 5: 645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder DI, Blair JD, Lott P, Yu HO, Hong D, Crary F, Ashwood P, Walker C, Korf I, Robinson WP, et al. 2013. The human placenta methylome. Proc Natl Acad Sci 110: 6037–6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Kondo Y, Guo Y, Zhang J, Zhang L, Ahmed S, Shu J, Chen X, Waterland RA, Issa JP. 2007. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet 3: 2023–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A. 2012. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484: 339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter M, Ma J, Harris A, Patterson L, Brown KA, Shope C, Showalter L, Abramovici A, Aagaard-Tillery KM. 2011. Maternal tobacco use modestly alters correlated epigenome-wide placental DNA methylation and gene expression. Epigenetics 6: 1284–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpakowski S, Sun X, Lage JM, Dyer A, Rubinstein J, Kowalski D, Sasaki C, Costa J, Lizardi PM. 2009. Loss of epigenetic silencing in tumors preferentially affects primate-specific retroelements. Gene 448: 151–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabano S, Colapietro P, Cetin I, Grati FR, Zanutto S, Mando C, Antonazzo P, Pileri P, Rossella F, Larizza L, et al. 2010. Epigenetic modulation of the IGF2/H19 imprinted domain in human embryonic and extra-embryonic compartments and its possible role in fetal growth restriction. Epigenetics 5: 313–324. [DOI] [PubMed] [Google Scholar]
- Tabata C, Ogita K, Sato K, Nakamura H, Qing Z, Negoro H, Kumasawa K, Temma-Asano K, Tsutsui T, Nishimori K, et al. 2009. Calcineurin/NFAT pathway: A novel regulator of parturition. Am J Reprod Immunol 62: 44–50. [DOI] [PubMed] [Google Scholar]
- Tang Z, Abrahams VM, Mor G, Guller S. 2011. Placental Hofbauer cells and complications of pregnancy. Ann NY Acad Sci 1221: 103–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegethoff M, Greene N, Olsen J, Meyer AH, Meinlschmidt G. 2010. Maternal psychosocial stress during pregnancy and placenta weight: Evidence from a national cohort study. PLoS ONE 5: e14478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobi EW, Lumey LH, Talens RP, Kremer D, Putter H, Stein AD, Slagboom PE, Heijmans BT. 2009. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet 18: 4046–4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolmacheva EN, Kashevarova AA, Skriabin NA, Lebedev IN. 2013. Epigenetic effects of trisomy 16 in the human placenta. Mol Biol (Mosk) 47: 423–432. [DOI] [PubMed] [Google Scholar]
- Uuskula L, Rull K, Nagirnaja L, Laan M. 2011. Methylation allelic polymorphism (MAP) in chorionic gonadotropin β5 (CGB5) and its association with pregnancy success. J Clin Endocrinol Metab 96: E199–E207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hooven EH, Pierik FH, de Kluizenaar Y, Hofman A, van Ratingen SW, Zandveld PY, Russcher H, Lindemans J, Miedema HM, Steegers EA, et al. 2012. Air pollution exposure and markers of placental growth and function: The generation R study. Environ Health Perspect 120: 1753–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas A, Moreau J, Landry S, LeBellego F, Toufaily C, Rassart E, Lafond J, Barbeau B. 2009. Syncytin-2 plays an important role in the fusion of human trophoblast cells. J Mol Biol 392: 301–318. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhao S. 2010. Vascular biology of the placenta. Morgan & Claypool Life Sciences, San Rafael, CA. [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. 2007. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 39: 457–466. [DOI] [PubMed] [Google Scholar]
- Wilhelm-Benartzi CS, Houseman EA, Maccani MA, Poage GM, Koestler DC, Langevin SM, Gagne LA, Banister CE, Padbury JF, Marsit CJ. 2012. In utero exposures, infant growth, and DNA methylation of repetitive elements and developmentally related genes in human placenta. Environ Health Perspect 120: 296–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn VD, Haimov-Kochman R, Paquet AC, Yang YJ, Madhusudhan MS, Gormley M, Feng KT, Bernlohr DA, McDonagh S, Pereira L, et al. 2007. Gene expression profiling of the human maternal-fetal interface reveals dramatic changes between midgestation and term. Endocrinology 148: 1059–1079. [DOI] [PubMed] [Google Scholar]
- Wyatt SM, Kraus FT, Roh CR, Elchalal U, Nelson DM, Sadovsky Y. 2005. The correlation between sampling site and gene expression in the term human placenta. Placenta 26: 372–379. [DOI] [PubMed] [Google Scholar]
- Xie H, Wang M, Bonaldo Mde F, Smith C, Rajaram V, Goldman S, Tomita T, Soares MB. 2009. High-throughput sequence-based epigenomic analysis of Alu repeats in human cerebellum. Nucleic Acids Res 37: 4331–4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Goodyer CG, Deal C, Polychronakos C. 1993. Functional polymorphism in the parental imprinting of the IGF2R gene. Biochem Biophys Res Comm 197: 747–754. [DOI] [PubMed] [Google Scholar]
- Yin LJ, Zhang Y, Lv PP, He WH, Wu YT, Liu AX, Ding GL, Dong MY, Qu F, Xu CM, et al. 2012. Insufficient maintenance DNA methylation is associated with abnormal embryonic development. BMC Med 10: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen R, Robinson W. 2011. Review: A high capacity of the human placenta for genetic and epigenetic variation: Implications for assessing pregnancy outcome. Placenta 32: S136–S141. [DOI] [PubMed] [Google Scholar]
- Yuen RK, Avila L, Peñaherrera MS, von Dadelszen P, Lefebvre L, Kobor MS, Robinson WP. 2009. Human placental-specific epipolymorphism and its association with adverse pregnancy outcomes. PLoS ONE 4: e7389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen RK, Peñaherrera MS, von Dadelszen P, McFadden DE, Robinson WP. 2010. DNA methylation profiling of human placentas reveals promoter hypomethylation of multiple genes in early-onset preeclampsia. Eur J Hum Genet 18: 1006–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen RK, Jiang R, Peñaherrera MS, McFadden DE, Robinson WP. 2011a. Genome-wide mapping of imprinted differentially methylated regions by DNA methylation profiling of human placentas from triploidies. Epigenetics Chromatin 4: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen RK, Manokhina I, Robinson WP. 2011b. Are we ready for DNA methylation-based prenatal testing? Epigenomics 3: 387–390. [DOI] [PubMed] [Google Scholar]
- Yuen RK, Chen B, Blair JD, Robinson WP, Nelson DM. 2013. Hypoxia alters the epigenetic profile in cultured human placental trophoblasts. Epigenetics 8: 192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HJ, Siu MK, Wong ES, Wong KY, Li AS, Chan KY, Ngan HY, Cheung AN. 2008. Oct4 is epigenetically regulated by methylation in normal placenta and gestational trophoblastic disease. Placenta 29: 549–554. [DOI] [PubMed] [Google Scholar]