Abstract
Members of the homeobox family of transcription factors are major regulators of hematopoiesis. Overexpression of either HOXB4 or HOXA9 in primitive marrow cells enhances the expansion of hematopoietic stem cells (HSCs). However, little is known of how expression or function of these proteins is regulated during hematopoiesis under physiological conditions. In our previous studies we demonstrated that thrombopoietin (TPO) enhances levels of HOXB4 mRNA in primitive hematopoietic cells (K. Kirito, N. Fox, and K. Kaushansky, Blood 102:3172-3178, 2003). To extend our studies, we investigated the effects of TPO on HOXA9 in this same cell population. Although overall levels of the transcription factor were not affected, we found that TPO induced the nuclear import of HOXA9 both in UT-7/TPO cells and in primitive Sca-1+/c-kit+/Gr-1− hematopoietic cells in a mitogen-activated protein kinase-dependent fashion. TPO also controlled MEIS1 expression at mRNA levels, at least in part due to phosphatidylinositol 3-kinase activation. Collectively, TPO modulates the function of HOXA9 by leading to its nuclear translocation, likely mediated by effects on its partner protein MEIS1, and potentially due to two newly identified nuclear localization signals. Our data suggest that TPO controls HSC development through the regulation of multiple members of the Hox family of transcription factors through multiple mechanisms.
Hematopoiesis is regulated by interactions between intracellular molecules, including multiple transcription factors, and environmental signals from cytokines and adhesion molecules. Of the small number of cytokines found to nonredundantly control the self-renewal, proliferation, and differentiation of hematopoietic stem cells (HSCs), thrombopoietin (TPO), the primary regulator of platelet production (16), is perhaps the most surprising; nevertheless, data establishing its critical role in HSC biology are impressive. Primitive hematopoietic cells that display repopulating ability all express c-Mpl, the receptor for TPO (41). Analysis by cDNA expression profiling also revealed that c-Mpl is highly expressed in marrow cell populations enriched for HSCs (42). Several in vitro studies demonstrated that TPO alone or in combination with other early-acting cytokines, such as stem cell factor, interleukin-3, or Flt-3 ligand, enhances the proliferation of primitive hematopoietic cells (24, 40). In vivo studies also revealed that TPO is crucial for HSC biology; both the number of committed progenitors of all hematopoietic lineage and CFU-S, a more primitive hematopoietic progenitor, are reduced in c-Mpl−/− mice (19). More recently, we demonstrated that following bone marrow transplantation, expansion of HSCs in adult bone marrow is 10 to 20 times less robust in Tpo−/− mice than in normal animals (12). Although it is obvious that TPO is important for the growth and expansion of HSCs, the mechanisms by which the hormone regulates HSC development remains poorly understood.
Homeodomain proteins are transcription factors which display a conserved DNA-binding domain, known as the homeobox. Recently, many studies have provided strong evidence that homeodomain proteins play key roles in HSC biology (reviewed in reference 31). Among the Homeobox family proteins, HOXB4 has been shown to enhance the expansion of HSC when overexpressed in bone marrow cells (3) or cord blood cells (37). These observations prompted us to test whether TPO might affect HOXB4 expression. In a previously reported study we found that TPO induced a two- to threefold increase in HOXB4 mRNA in the primitive hematopoietic cell lines EML and UT-7/TPO (20). More importantly, Sca-1+/c-kit+/Gr-1− cells from tpo-deficient mice have two- to fivefold-lower levels of HOXB4 mRNA than do wild-type cells (20). These results suggested that TPO-induced HOXB4 expression is one of the mechanisms by which TPO controls HSC expansion. However, in contrast to the finding that overexpression of HOXB4 can enhance HSC expansion, in vivo studies with null mice revealed that the loss of HOXB4 function results in only very modest effects on HSC number and repopulating capacity (6); there were no significant differences in the numbers of bone marrow-derived primitive hematopoietic progenitor cells or colony-forming abilities between control and Hoxb4−/− mice. And although competitive repopulation studies revealed that Hoxb4−/− cells have reduced repopulating capacity, the level was only twofold lower (6), even when combined with deletion of Hoxb3 (5). Because the magnitude of the stem cell defect in Tpo−/− mice is much greater than those in Hoxb4−/− or Hoxb3/b4−/− animals, we hypothesized that other downstream molecules, including members of the Homeobox-containing family, might contribute to TPO-dependent HSC self-renewal and expansion.
HOXA9, another member of the Homeodomain-containing family of proteins, might be one such candidate. Like HOXB4, HOXA9 is also selectively expressed in primitive hematopoietic cells (34), and HOXA9-deficient mice display a 5- to 12-fold reduction in HSC numbers (H. J. Lawrence, S. T. Fong, Y. H. Hsiang, G. Sauvageau, R. K. Humphries, and C. Largman, Abstr. 40th Annu. Meet. Am. Soc. Hematol., Blood 92(Suppl. 1):55a, 1998). However, in contrast to HOXB4, TPO did not enhance HOXA9 mRNA levels either in UT-7/TPO or primary marrow cells. Moreover, HOXA9 levels in Sca-1+/c-kit+ cells from Tpo−/− mice were equal to those in cells from wild-type mice (20).
Many cytokines regulate the function of transcription factors through posttranslational mechanisms, including covalent modifications, by affecting their interaction with cofactors or by altering nuclear transport or DNA binding ability. In this study, we investigated whether TPO affects HOXA9 function at posttranslational levels. We report here that TPO enhances HOXA9 nuclear localization and its interaction with the cofactor, MEIS1 (myeloid ecotropic viral integration site), and utilizes two newly identified nuclear localization signals (NLS). In addition, MEIS1 expression levels are affected by TPO, suggesting that homeodomain-containing proteins may be major downstream mediators of the hormone.
MATERIALS AND METHODS
Cell culture.
UT-7/TPO cells (22) were a gift of Norio Komatsu (Jichi Medical School, Tochigi, Japan) and were maintained in liquid culture with Iscove’s modified Dulbecco’s medium containing 10% fetal calf serum and 10 ng of human TPO/ml, a gift of Don Foster (Zymogenetics, Seattle, Wash.). UT-7/TPO is a primitive human hematopoietic cell line and responds to multiple cytokines including granulocyte-macrophage colony-stimulating factor (GM-CSF), stem cell factor, and TPO. HeLa cells were cultured with Dulbecco's modified Eagle's medium containing 10% fetal calf serum. U-937 cells were obtained from American Type Culture Collection (CRL-1593.2) and maintained in RPMI 1640 with 10% fetal calf serum. GM-CSF was purchased from PeproTech (Rocky Hill, N.J.).
Reagents.
G418 was purchased from Sigma (St. Louis, Mo.). LY294002 and PD98059 were purchased from CALBIOCHEM (La Jolla, Calif.). A rabbit anti-HOXA9 antibody was purchased from Upstate Biotechnology (Lake Placid, N.Y.). Antibody to the goat N-terminal domain of HOXA9 (N-20), goat antibody to MEIS1 (C-17), and rabbit antibody to green fluorescent protein (GFP) and TFIIH were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, Calif.). Antibodies to pre-B-cell homeobox (PBX) were as follows: a rabbit antibody to PBX from Santa Cruz (C-20) and a mouse anti-human PBX monoclonal antibody from BD Biosciences (San Diego, Calif.). Alexa-Fluor 488-goat anti-rabbit immunoglobulin G (IgG) antibodies were obtained from Molecular Probes (Eugene, Ore.). Fluorescein isothiocyanate-conjugated anti-Gr-1 and phycoerythrin-conjugated anti-c-kit antibodies were obtained from BD Biosciences/Pharmingen (San Diego, Calif.).
RNA preparation and real-time RT-PCR.
Total cellular RNA was extracted from cells using RNeasy Midi kit (QIAGEN, Valencia, Calif.). Quantitative-real time reverse transcription-PCR (RT-PCR) assays for MEIS1 and PBX2 transcripts were developed using the iCycler (Bio-Rad Laboratories, Hercules, Calif.) with SYBR green PCR core reagent (Applied Biosystems, Foster City, Calif.). The primer pairs for MEIS1 and PBX2 were previously reported (10). The primer pair for murine Mesi1 is as follows: Forward primer, 5′-CCTCGGTCAATGACGCTTTA-3′; reverse primer, 5′-GAATCTGTTTGGCGAACACC-3′. All primers were synthesized by Invitrogen Life Technologies (Carlsbad, Calif.). For analysis of Meis1 expression, 50 ng of total RNA was amplified and quantified in 40 μl of reaction mixture containing 2.5 mM MgCl2, 300 μM deoxynucleoside triphosphates, 0.2 μl AmpliTaq Gold (Applied Biosystems), 0.05 μl of Moloney murine leukemia virus reverse transcriptase (Invitrogen Life Technologies), 0.4 μl of RNasin RNase inhibitor (Promega, Madison, Wis.), 100 nM forward primer, and 100 nM reverse primer. For analysis of PBX2 expression, forward and reverse primers were used at 300 nM concentrations. All reactions were performed in triplicate. For quantification of gene expression, we used a relative standard curve method as described previously (20). As an internal control we used glyceraldehyde-3-phosphate dehydrogenase (GAPDH) forward and reverse primers included in the GAPDH control reagents kit (Applied Biosystems).
Preparation of cell lysates and Western blotting.
Nuclear and cytoplasmic extracts were prepared from UT-7/TPO cells according to methods previously described (21). Briefly, 107 cells were washed with ice-cold phosphate-buffered saline (PBS) containing 2 mM Na3VO4, resuspended in a hypotonic buffer (20 mM HEPES [pH 7.5], 10 mM KCl, 1 mM MgCl2, 10% glycerol, 0.5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 10 μg of aprotinin/ml, 10 μg of leupeptin/ml, 2 mM benzamide, 10 mM NaF, and 2 mM Na3VO4, with 0.2% Nonidet P-40), and homogenized. After centrifugation at 1,000 × g for 5 min, the supernatant was separated from the nuclear pellet and then centrifuged at 14,000 × g for 20 min at 4°C. The debris was removed, and the supernatants were collected as cytoplasmic extracts. The nuclear pellets were resuspended in hypotonic buffer with 300 mM NaCl; debris was removed by centrifugation (14,000 × g for 20 min), and the supernatants were collected as nuclear extracts. Cell lysate proteins were size fractioned by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then electroblotted onto polyvinylidene difluoride membranes. The blots were incubated with primary antibodies and visualized with a chemiluminescence detection kit (LumiGLO; Cell Signaling, Beverly, Mass.). For immunoprecipitation studies, cell lysates were precleared with normal IgG and protein A/G-agarose (Santa Cruz Biotechnology Inc.) and incubated with primary antibody over-night and with protein A/G agarose for 6 h, and the agarose beads were then washed five times and eluted with sodium dodecyl sulfate sample buffer.
Immunofluorescence microscopy.
Cytospin preparations were fixed in PBS containing 4.0% paraformaldehyde for 10 min and then permeabilized with 0.2% Triton X-100 for 5 min at room temperature. After a wash with Tris-buffered saline, the slides were blocked for 1 h with 10% goat serum (Sigma) and stained with anti-HOXA9 antibody overnight. After a second wash with Tris-buffered saline, the slides were incubated for 60 min with Alexa Fluor 488 goat anti-rabbit IgG (H+L) conjugate (Molecular Probes) and then mounted in VECTASHIELD mounting medium with 4′,6′-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, Calif.). Immunofluorescent images were obtained using SPOT RT software (DIAGNOSTIC instruments, Sterling Heights, Mich.). After the images were made, they were blindly scored into nuclear, cytoplasmic, or mixed nuclear/cytoplasmic staining patterns by a single observer.
Construction of GFP-HOXA9 fusion plasmids.
A HOXA9 cDNA was a gift from Takuro Nakamura (The Cancer Institute, Japanese Foundation for Cancer Research, Tokyo, Japan). All HOXA9 mutant cDNAs were generated using the QuikChange mutagenesis kit (Stratagene), and all sequences were verified. To add a GFP tag, wild-type and mutant HOXA9 cDNA were cloned into pEGFP-C1 vector (Clonetech, Palo Alto, Calif.). The identities of the fusion constructs were verified by Western blotting with both anti-GFP and anti-HOXA9 antibodies.
Establishment of GFP-HOXA9-expressing UT-7/TPO cells.
The pEGFP-C1 vectors containing GFP-tagged wild-type and mutant forms of HOXA9 were introduced into UT-7/TPO cells with Tfx-20 (Promega) according to the manufacture's instructions, and overexpressing cells were selected by culturing in 800 μg of G418/ml.
Transient-transfection and fluorescence studies.
HeLa cells were seeded onto Lab-TekII Chamber slides (Nalgen Nunc International, Naperville, Ill.), and the next day they were transfected with GFP-HOXA9 constructs using Tfx-20 reagents. After 48 h of culture, the cells were fixed in PBS containing 4.0% paraformaldehyde for 10 min, permeabilized 0.2% Triton X-100 for 5 min, and then mounted in VECTASHIELD mounting medium with DAPI (Vector Laboratories). Fluorescent images were obtained using SPOT RT software (DIAGNOSTIC instruments).
Isolation of immature hematopoietic cells.
Sca-1+/c-kit+/Gr-1− fraction cells were prepared from C57BL/6, BDF-1, and Tpo−/− mice as previously described (20). Mice were housed in a specific-pathogen-free environment, and the Animal Care Program of the University of California, San Diego, approved all protocols involving mice.
Statistical analysis.
Statistical analysis was performed by using the Student t test. A P value of <0.05 was considered significant.
RESULTS
TPO increases nuclear HOXA9 protein in UT-7/TPO cells.
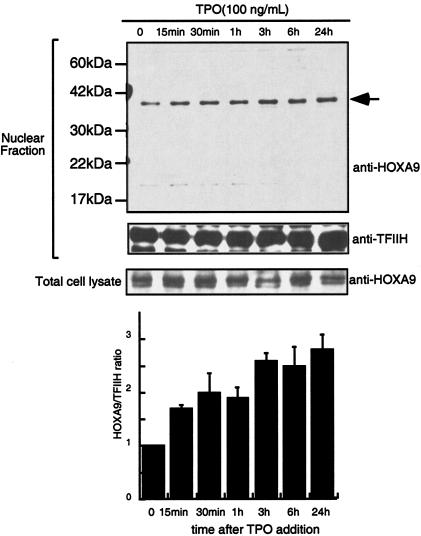
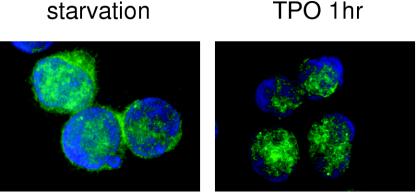
To address whether TPO controls HOXA9 function, we initially analyzed the effects of TPO on levels of the transcription factor using UT-7/TPO cells, which display characteristics of immature hematopoietic cells. To examine HOXA9 protein levels, we performed Western blotting with a rabbit polyclonal anti-HOXA9 antibody. The HOXA9 proteins were detected on gels with an apparent molecular mass of approximately 35 kDa (Fig. 1). To confirm the identity of the detected bands, the cell lysates were immunoprecipitated with a goat anti-HOXA9 antibody, and the immunoprecipitates were analyzed by Western blotting using rabbit anti-HOXA9 antibody. The immunoprecipitated HOXA9 band was detected at the same migration position (data not shown). In our previous studies we found that TPO did not alter HOXA9 mRNA levels in UT-7/TPO cells (20). In accord with this result, TPO did not change total HOXA9 protein levels in the present studies (Fig. 1). Even after a 24-h TPO starvation, HOXA9 protein levels were not decreased in UT-7/TPO cells and the addition of TPO did not enhance HOXA9 expression. In contrast, HOXA9 levels in the nuclear fraction of UT-7/TPO cells were enhanced 2.5-fold by TPO (Fig. 1). Nuclear HOXA9 levels increased rapidly (15 min) in response to TPO and reached a maximum 24 h after addition of the hormone. To confirm these results, we employed immunofluorescence in UT-7/TPO cells using a specific antibody for HOXA9. After TPO starvation, the majority of the HOXA9 was detected in the cytoplasm (Fig. 2, left panel). In contrast, after a 1-h incubation with TPO, we found HOXA9 signals almost exclusively within nuclear speckles (Fig. 2, right panel).
FIG. 1.
TPO increases nuclear HOXA9 levels. After a 24-h starvation, UT-7/TPO cells were treated with 100 ng of TPO/ml for the indicated time periods and total cell and nuclear extracts were prepared. HOXA9 levels were analyzed by Western blotting. For an internal control, the membranes were reprobed with anti-TFIIH antibody. (B) The film was subjected to densitometric analysis, and the HOXA9/TFIIH ratio was calculated. Each column represents the average ± standard deviation for two independent experiments.
FIG. 2.
TPO induces nuclear localization of HOXA9 in UT-7/TPO cells. UT-7/TPO cells were starved for 24 h and then stimulated with 100 ng of TPO/ml for 1 h. Cells were fixed and stained with anti-HOXA9 primary antibody and Alexa488-conjugated secondary antibody (green). Cells were counterstained with DAPI to visualize nuclear DNA (blue). An example of the predominant staining pattern under each condition is shown.
TPO regulates HOXA9 subcellular localization in primitive hematopoietic cells.
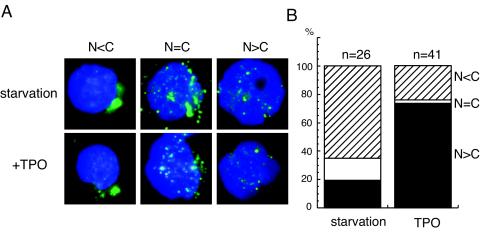
In an attempt to assess whether TPO has an effect on HOXA9 nuclear localization in a more physiological setting, we isolated Sca-1+/c-kit+/Gr-1− cells, a population highly enriched in hematopoietic stem and primitive progenitor cells, and analyzed HOXA9 distribution in response to TPO. After 6 h of culture without cytokines, 65% of the cells showed predominant cytoplasmic expression of HOXA9, and only 19% showed a predominant nuclear pattern (N>C) (Fig. 3B). TPO treatment dramatically increased the proportion of cells that displayed predominant nuclear HOXA9 staining (Fig. 3B); after 1 h of incubation with TPO, 70% of the cells showed a N>C pattern.
FIG. 3.
TPO stimulates HOXA9 nuclear localization in Sca-1+/c-kit+/Gr-1− fraction cells. (A) Bone marrow cells were prepared from BDF-1 mice, and Sca-1+/c-kit+/Gr-1− cells were selected by cell sorting. The cells were cultured without cytokines for 6 h and then stimulated with 100 ng of human TPO/ml, equipotent to 10 ng of murine TPO/ml, for 1 h. Nuclear localization of HOXA9 was analyzed by immunofluorescence microscopy. According to the distribution pattern of the HOXA9, the cells were divided into three groups: N<C represents cells in which nuclear staining is less than cytoplasmic staining (shown on the left two panels), N=C represents cells which have roughly equal levels of HOXA9 in the nucleus and cytoplasm (the middle two panels), and N>C denotes cells displaying stronger nuclear staining than cytoplasmic staining (shown in the right two panels). (B) Under starvation or TPO-stimulated conditions, the indicated number of cells were blindly scored and categorized into the three groups. The columns show the ratio of each group.
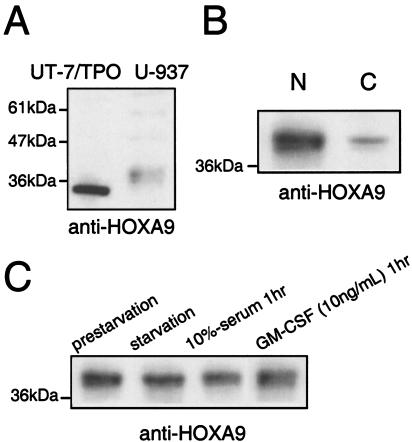
Subcellular distribution of HOXA9 in myeloid leukemia cell lines.
In addition to immature hematopoietic stem cells, myeloid lineage cells also express HOXA9. Thus, in order to assess whether mitogenic stimulation of myeloid cells also results in the same translocation effect, we investigated whether nuclear localization of HOXA9 is also controlled in the human myelomonocytic leukemia cell line U-937, known to express HOXA9. As shown in Fig. 4, U-937 cells express the transcription factor mainly in the nucleus, a pattern unaffected by serum starvation, serum refeeding, or the presence of 10-ng/ml GM-CSF.
FIG. 4.
Subcellular distribution of HOXA9 in myeloid leukemia cell lines. (A) Nuclear extracts were prepared from UT-7/TPO and U-937 cells, and equal amounts of extract were probed for HOXA9 expression with a rabbit polyclonal antibody. (B) Nuclear and cytoplasmic extracts were prepared from serum-replete cultures of U-937 cells, and the HOXA9 protein in each fraction was analyzed by Western blotting. (C) U-937 cells were starved of serum for 24 h and then stimulated with 10% serum or 10 ng of GM-CSF/ml for 1 h, and nuclear extracts were prepared and analyzed by Western blotting for HOXA9 expression.
TPO stimulates MEIS1 expression.
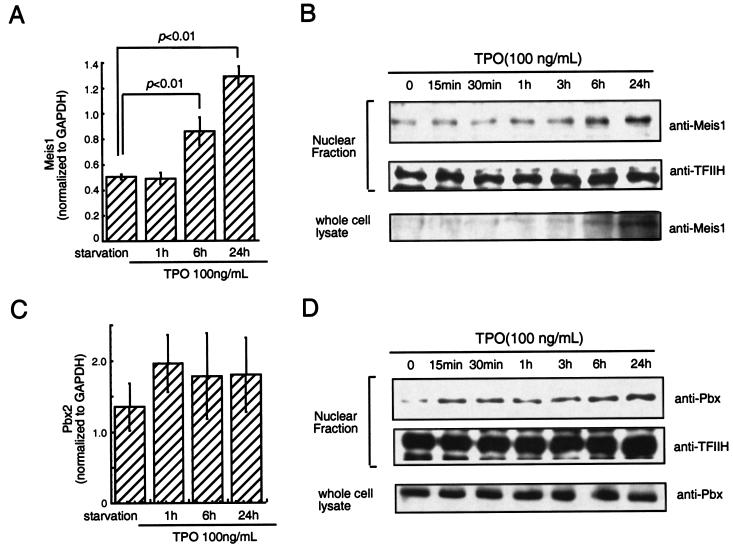
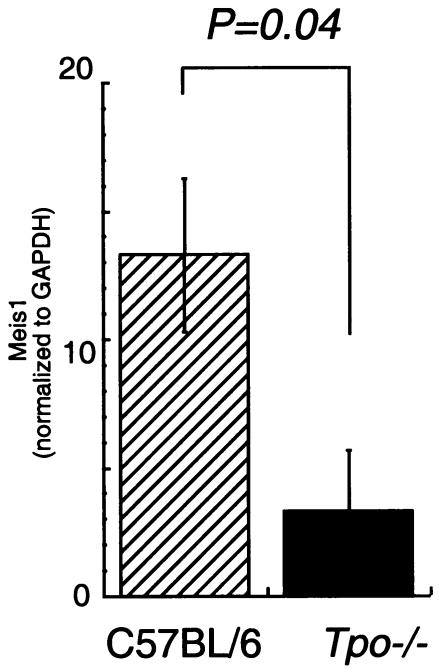
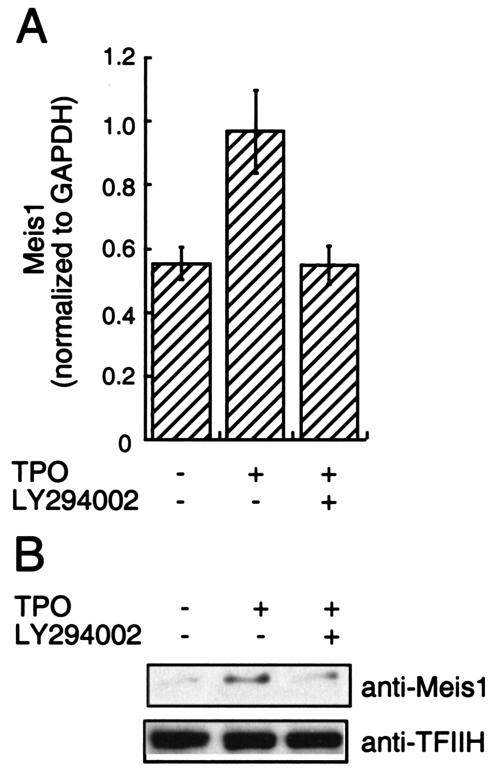
It has been suggested that HOX protein function requires binding to members of the TALE (three-amino-acid loop extension) subclass of homeodomain-containing proteins, including MEIS1 and PBX, proteins that contain NLS (36). Moreover, trimolecular complexes composed of HOXA9, PBX, and MEIS1 have been reported to form in some hematopoietic cell lines (39). These findings prompted us to analyze whether TPO regulates the subcellular localization of HOXA9 through induction of HOXA9-MEIS1 and/or HOXA9-PBX complexes. To address this question we first investigated whether TPO affects expression of MEIS1 and PBX. As shown in Fig. 5A, TPO induced two- to threefold increases of MEIS1 mRNA in UT-7/TPO cells. Consistent with this result, we found increases of MEIS1 protein levels not only in the nuclear fraction of UT-7/TPO cells but also in total cell lysates. In contrast to MEIS1, TPO did not affect the PBX2 mRNA level or total PBX protein levels. However, we found a rapid increase of PBX protein in the nuclear fraction of TPO-stimulated UT-7/TPO cells. These results indicate that TPO controls MEIS1 levels at a transcriptional level and that it leads to the nuclear localization of both HOXA9-interacting proteins. To confirm the effects of TPO on MEIS1 expression under more physiological conditions, mRNA levels were also analyzed using Sca-1+/c-kit+/Gr1− fraction cells from Tpo−/− and control mice. As demonstrated in Fig. 6, Meis1 mRNA levels were five times lower in primitive hematopoietic cells from Tpo−/− mice than in controls. Next, we investigated which signal transduction molecules were involved in TPO-induced MEIS1 expression by using several well-characterized chemical inhibitors of known TPO-responsive signal transduction pathways. We found that LY294002, an inhibitor of phosphatidylinositol 3-kinase (PI3K), inhibited MEIS1 expression at both the mRNA and protein levels (Fig. 7) but that neither the ERK1/2 inhibitor PD98059 nor the p38 mitogen-activated protein kinase (MAPK) inhibitor SB203580 affected MEIS1 expression (data not shown).
FIG. 5.
TPO stimulates MEIS1 transcription but not that of PBX. (A and C) After 24 h of starvation, UT-7/TPO cells were treated with 100 ng of TPO/ml for the indicated time periods and RNA was prepared for real-time PCR analysis for MEIS1 (A) or PBX2 (C). Each column represents the average MEIS1 or PBX2 level normalized to a GAPDH internal control ± the standard deviation for three independent experiments. Statistically significant differences from starvation conditions are noted. (B and D) UT-7/TPO cells were starved for 24 h and then stimulated with 100 ng of TPO/ml for the indicated times. After each culture period, total cellular and nuclear lysates were prepared, and the protein levels of MEIS1 (B) or PBX (D) were analyzed by Western blotting. As a loading control for nuclear proteins, the membranes were reprobed with an anti-TFIIH antibody.
FIG. 6.
MEIS1 levels in Tpo−/− mice. Whole bone marrow cells were prepared from control C57BL/6 and Tpo−/− mice. The cells were pooled (for experiment 1, n = 4 C57BL/6 mice and n = 4 [Tpo−/− mice]; for experiment 2, n = 4 C57BL/6 mice and n = 5 Tpo−/− mice), and Sca-1+/c-kit+/Gr-1− cells were collected for real-time PCR analysis. Each column represents an average of MEIS1 mRNA levels normalized to a GAPDH internal control ± standard deviation for two experiments (hatched column, C57BL/6; black column, Tpo−/−).
FIG. 7.
Blockade of PI3K inhibits TPO-induced MEIS1 expression. After 24 h of starvation, UT-7/TPO cells were treated with 20 μM LY294002 for 1 h and then incubated with TPO for 24 h, and nuclear protein and total cellular mRNA were prepared. Expression levels of MEIS1 were analyzed by real-time RT-PCR (A) and by Western blotting (B). (A) Each column represents the average MEIS1 level normalized to the GAPDH internal control ± standard deviation for three independent experiments. (B) Western blotting analysis of MEIS1 in the nuclear fraction. For an internal control the membrane was reprobed with an anti-TFIIH antibody.
TPO induces the association of HOXA9 with MEIS1.
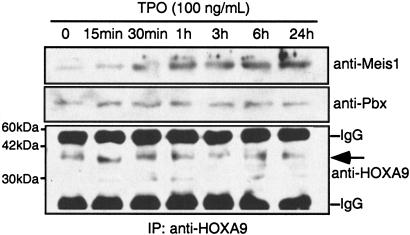
Next, using an immunoprecipitation assay, we analyzed whether the association of HOXA9 and either MEIS1 or PBX was enhanced by TPO. As shown in Fig. 8, TPO increased the association of HOXA9 with MEIS1 in a time-dependent fashion. In contrast, HOXA9 consistently associated with PBX independently of the presence of TPO.
FIG. 8.
Interaction of HOXA9 with MEIS1 is enhanced by TPO. UT-7/TPO cells were starved for 24 h, and the cells were then stimulated with 100 ng of TPO/ml for the times indicated. Total cell lysates were prepared and immunoprecipitated with a rabbit polyclonal anti-HOXA9 antibody. The immunoprecipitates were analyzed by Western blotting with a goat polyclonal anti-MEIS1 antibody and a mouse monoclonal anti-PBX antibody. The membrane was also reprobed with a rabbit polyclonal anti-HOXA9 antibody. The predicted molecular mass of HOXA9 is indicated by an arrow.
The MEIS1 interaction domain is crucial for nuclear localization of HOXA9.
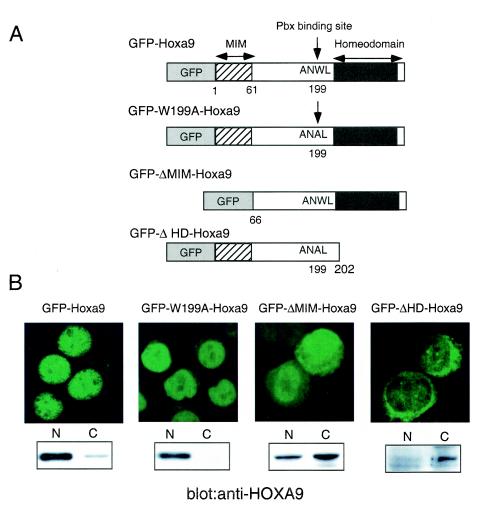
To explore the roles of MEIS1 and PBX in TPO-induced HOXA9 nuclear localization, we constructed cDNA for mutant forms of HOXA9; ΔMIM-HOXA9 lacks the N-terminal 65 amino acids of the protein required for MEIS1 interaction (38), W199A-HOXA9 carries a mutation in the PBX binding site (8), and ΔHD-HOXA9 lacks the entire homeodomain (Fig. 9A). In previously reported studies, interaction of Homeodomain-containing protein with MEIS1 or PBX was disrupted in the respective mutants (8, 38). cDNA for wild-type HOXA9 and these mutant forms of the transcription factor were fused to GFP cDNA and introduced into UT-7/TPO cells, and stable transfectant clones were established. We analyzed the subcellular localization of the proteins under normal TPO-stimulated culture conditions by fluorescence microscopy and Western blotting. GFP-tagged wild-type HOXA9 was distributed mainly in nucleus (Fig. 9B) under these conditions, as was GFP-W199A-HOXA9. In contrast, most GFP-ΔMIM-HOXA9 was distributed both in the cytoplasm and in the nucleus. Most ΔHD-HOXA9 was distributed in the cytoplasm. These results indicate that the MEIS-interacting domain of HOXA9 is important for TPO-induced nuclear localization of the protein.
FIG. 9.
The MEIS interaction motif is required for effective nuclear localization of HOXA9. (A) A schematic illustration of the wild-type and mutant HOXA9 cDNA fused to GFP used in this study. MIM, MEIS interaction motif; ANWL represents the amino acid sequence required for interaction with PBX. (B) UT-7/TPO clones stably expressing each fusion were established, and the intracellular distribution of the fusion protein in each clone was examined by fluorescence microscopy (upper panels). Nuclear and cytoplasmic (N and C) lysates were prepared from each clone, and expression of each fusion protein was analyzed by Western blotting using a rabbit polyclonal anti-HOXA9 antibody (lower panels). To detect ΔHD-HOXA9, a goat polyclonal anti-HOXA9 antibody was used.
Activation of ERK is required for TPO-induced HOXA9-MEIS1 complex formation and subsequent nuclear localization of HOXA9.
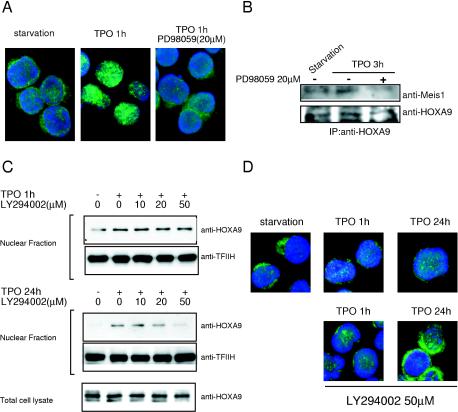
Next, we sought to identify the signal transduction pathway(s) required for TPO-induced HOXA9 nuclear localization. UT-7/TPO cells were pretreated with chemical inhibitors that block several well-characterized signal transduction pathways stimulated by TPO in megakaryocytes, and TPO-induced HOXA9 nuclear accumulation was analyzed by immunofluorescence microscopy. We found that PD98059, a potent inhibitor of MEK1 and hence ERK1/2, diminished TPO-induced HOXA9 transfer to the nuclear fraction of UT-7/TPO cells soon after stimulation (Fig. 10A). Inhibition of the ERK pathway also disrupted the TPO-induced interaction between HOXA9 and MEIS1 (Fig. 10B). In contrast, inhibitors for PI3K, p38 MAPK, and protein kinase A pathways did not affect TPO-induced HOXA9 nuclear accumulation at early time points (Fig. 10C and D; also data not shown). However, at 24 h, inhibition of PI3K had a profound affect on TPO-induced nuclear localization of HOXA9 (Fig. 10C and D). These results suggest that TPO influences nuclear localization of HOXA9 through two distinct pathways: an ERK-dependent rapid relocalization and a PI3K-dependent delayed mechanism.
FIG. 10.
ERK activation is required for TPO-enhanced nuclear localization of HOXA9. (A) After 24 h of starvation, UT-7/TPO cells were treated with 20 μM PD98059 for 30 min and then stimulated with 100 ng of TPO/ml for 1 h. Cells were then fixed and stained with an anti-HOXA9 primary antibody and an Alexa488-conjugated secondary antibody (green). Cells were counterstained with DAPI to visualize nuclear DNA (blue). (B) After 24 h of starvation, UT-7/TPO cells were treated with 20 μM PD98059 for 30 min and then stimulated with 100 ng of TPO/ml for 3 h. Whole-cell lysates were then prepared and immunoprecipitated with an anti-HOXA9 antibody. The immunoprecipitates were analyzed by Western blotting for MEIS1 interaction. The membrane was also probed with an anti-HOXA9 antibody to confirm that equal amounts of HOXA9 were precipitated. (C) After 24 h of starvation, UT-7/TPO cells were pretreated with the indicated concentrations of LY294002 for 30 min and then stimulated with 100 ng of TPO/ml for 1 h (upper panel) or 24 h (lower panel). Nuclear extracts were prepared, and HOXA9 levels were analyzed by Western blotting. For an internal control, the membranes were reprobed with anti-TFIIH antibody. To analyze HOXA9 expression, total cell lysates were also prepared after 24 h of culture with LY294002 and subjected to Western blotting. (D) After 24 h of starvation, UT-7/TPO cells were treated with 50 μM LY294002 for 30 min and then stimulated with 100 ng of TPO/ml for the indicated time periods. The cells were then fixed and stained with an anti-HOXA9 primary antibody and Alexa488-conjugated secondary antibody (green); the cells were counterstained with DAPI to visualize nuclear DNA (blue).
Identification of two NLS in HOXA9.
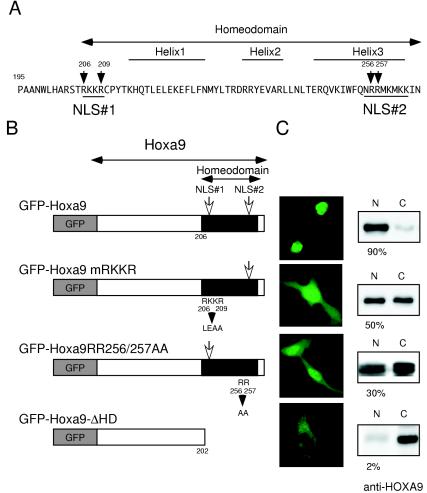
Our results revealed that the interaction with MEIS1 is a crucial step for HOXA9 nuclear localization in response to TPO. The nuclear import of the many proteins (>40 kDa) requires specific amino acid sequences known as NLS. Thus, we next investigated whether HOXA9 bears an NLS. First, we searched for putative NLS in HOXA9 using the PSORTII program and The Predict Protein Server. These programs predicted that HOXA9 contains two putative NLS sites, one at amino acids 206 to 209 (RKKR), located in the N-terminal arm of the homeodomain, and one at amino acids 256 to 262 (RRMKMKK), found in the third helix of the homeodomain (Fig. 11A). To verify whether these sequences actually function as NLS, we introduced mutations in each site of HOXA9 and fused the mutants to GFP (Fig. 11B); subcellular localization of these fusion proteins was examined in HeLa cells. We found that GFP alone was uniformly distributed between the nucleus and cytoplasm, as previously reported (1). In contrast, wild-type HOXA9 fused to GFP was found almost exclusively in the nuclei of transduced HeLa cells. Western blotting also revealed that more than 90% of the overexpressed GFP-HOXA9 proteins localized to the nuclear fraction (Fig. 11C). Replacement of the amino acid sequence RKKR (amino acid residues 206 to 209) in predicted NLS 1 to LEAA blunted the nuclear localization of the fusion protein (Fig. 11C). A mutant HOXA9 protein carrying two Arg-to-Ala mutations (R256 and R257) in the second putative NLS also led to a uniform nuclear/cytoplasmic distribution in HeLa cells. Deletion of the homeodomain completely resulted in exclusion of the fusion protein from the nucleus. A similar result was found in UT-7/TPO cells transduced with HOXA9ΔHD (Fig. 9). These results indicate that both predicted NLS sequences are required for nuclear distribution of HOXA9. Furthermore, our results clearly indicate that the homeodomain is indispensable for nuclear localization of HOXA9. Together with the above observation, our results suggest that both the MEIS1 interaction motif and the homeodomain including the two NLS are responsible for TPO-induced nuclear accumulation of HOXA9.
FIG. 11.
HOXA9 displays two nuclear localization signals. (A) A schematic representation of the amino acid sequence of the HOXA9 homeodomain is shown, with the predicted NLS sites underlined. The numbers represent the amino acid sequence. (B) A schematic structure of the wild-type and mutant HOXA9-GFP fusion cDNA used in this study. In the mRKKR mutant, amino acids 206 to 209, corresponding to NLS 1, were changed to LEAA. The RR256/257AA construct carries mutations in NLS 2. In the ΔHD mutant, the entire homeodomain was deleted. (C) Each construct was transfected into HeLa cells, and their subcellular localization was examined by fluorescence microscopy. Nuclear and cytoplasmic lysates were also prepared from the transfected HeLa cells, and the distribution of the fusion proteins was analyzed by Western blotting with an anti-HOXA9 antibody. For analysis of ΔHD-HOXA9 distribution, we used an anti-HOXA9 antibody that recognizes the N-terminal region of HOXA9.
DISCUSSION
Although initially characterized as the primary regulator of platelet production (16), numerous studies have now established that TPO plays an important and nonredundant role in the self-renewal and expansion of HSCs. Unfortunately, the molecular mechanism(s) of these effects is poorly understood, although it is attractive to postulate that alterations in the transcription factors critical for stem cell biology are at least partially responsible. Recent studies using both gain-of-function and loss-of-function strategies have determined that the transcription factor HOXA9 also plays an important role in stem cell physiology (25, 43), mediated by alterations in a wide variety of gene families (9). In the present study we demonstrate that although TPO fails to affect total cellular levels of HOXA9, the hormone affects nuclear localization of the transcription factor in a primitive hematopoietic cell line and in primary Sca-1+/c-kit+/Gr-1− hematopoietic cells, a population highly enriched with stem cells. We also found that TPO influences HOXA9 nuclear import by enhancing HOXA9-MEIS1 complex formation in an ERK1/2-dependent fashion and by enhancing MEIS1 mRNA expression, a response dependent on activation of PI3K.
Regulation of nuclear localization by cytokine stimulation is an important mechanism for controlling the function of transcription factors. Several studies have revealed that certain classes of homeodomain-containing proteins are so regulated. For example, insulin (11) or high concentrations of glucose (27, 35) induce nuclear translocation of pancreatic/duodenal homeobox-1, while oxidative stress promoted its nuclear export (17). Mechanistically, a number of signaling pathways have been identified to support such changes; for example, nuclear localization of PBX1 is PKA dependent (18). In the present study we focused on HOXA9, a member of the Abd-B-like Hox paralog group, and found its nuclear localization to be regulated by TPO in primitive hematopoietic cells. To our knowledge, this is the first report for nuclear localization control of class I Hox proteins.
The studies reported here also establish that MEIS1 plays a central role in TPO-induced HOXA9 nuclear localization and begin to reveal its molecular mechanism. We found that TPO induced the rapid association of HOXA9 and MEIS1 and that deletion of the MEIS interaction domain of HOXA9 disrupted its nuclear localization in UT-7/TPO cells stimulated with TPO. In accord with our results, several reports have established that MEIS1 supports the nuclear localization of another homeodomain-containing protein, PBX1 (36). Moreover, nuclear import of Extradenticle (EXD), a Drosophila homolog of PBX1, is regulated by Homothorax, a MEIS family member (2, 4, 32). In these cases, MEIS binding blocks the nuclear export signal (NES) on EXD or PBX, making the NLS signal the predominant influence on the proteins. As discussed below, we found that HOXA9 contains two NLS. Furthermore, Maizel and colleagues found that a NES present in Engrailed (which is also present in other homeodomain-containing proteins, including HOXA9 (28), must be overcome by phosphorylation for nuclear localization. Thus, our data are quite consistent with these findings and indicate that the TPO-induced HOXA9/MEIS1 association overcomes a NES on HOXA9, allowing the NLS we identified to drive the protein into the nucleus.
Based on inhibitor studies, we found that within an hour of exposure to TPO, activation of the ERK pathway was responsible for the association of HOXA9 and MEIS1 and hence for nuclear accumulation of the transcription factor complex. At present, we do not have an explanation for how ERK activation controls the association of HOXA9 and MEIS1 and increases nuclear import of the complex. One possibility is that ERK phosphorylates HOXA9 and/or MEIS1 to enhance formation of the HOXA9-MEIS1 complex. Some precedent for this process exists; nuclear localization of PDX1 requires its phosphorylation (11). Both HOXA9 and MEIS1 contain several consensus ERK phosphorylation sequences, and further investigation of this hypothesis is under way in our laboratory.
In addition to the direct ERK-mediated TPO effect on induction of HOXA9-MEIS1 complex formation, our observations also establish that TPO influences HOXA9-MEIS1 complex levels by increasing MEIS1 expression. In UT-7/TPO cells, HOXA9 nuclear levels gradually increased in step with MEIS1 expression levels (Fig. 8), a response dependent on the PI3K pathway, at least at 24 h following stimulation by TPO (Fig. 7). In contrast, this inhibitor did not affect early-phase (within 1 h) elevation of HOXA9. Saleh and colleagues reported that MEIS1 expression is required for PBX1 nuclear localization and activation during mouse forelimb development (36). Likewise, in Drosophila, EXD is nuclear only in the presence of Homothorax (2). Together with these observations, it is possible that HOXA9 may work only in MEIS1-expressing cells. Consistent with this notion, both HOXA9 and MEIS1 are selectively expressed in Sca-1+/Lin− primitive hematopoietic cells (34). Coexpression of HOXA9 and MEIS1 is also frequently found in primary leukemia cells (26), and in that setting the latter augments the leukemogenicity of the former (23, 33). Thus, it is becoming clear that MEIS1 plays important roles in HOXA9-dependent normal and pathological hematopoiesis by regulating HOXA9 nuclear localization.
Our observations that MEIS1 is required for effective nuclear translocation of HOXA9 differ from those recently reported by Calvo and colleagues (7). These investigators introduced a MEIS1 cDNA into HOXA9-transformed murine bone marrow cells and analyzed intracellular localization of HOXA9. They found that the exogenous HOXA9 was constitutively located in the nucleus and that MEIS1 overexpression did not alter its subcellular localization. One possible explanation of this discrepancy is that endogenous MEIS1 in those cells was sufficient to induce nuclear import of the exogenous HOXA9, or that the transformed nature of the cells affected HOXA9 localization. Furthermore, they analyzed HOXA9 localization in the presence of GM-CSF, a cytokine that supports the growth of these cells; it is thus possible that as with our experience with TPO, GM-CSF induces the association of HOXA9 and endogenous MEIS1. Alternately, the high levels of HOXA9 in the transformed marrow cells reported by Calvo could have been responsible for the predominant nuclear localization; consistent with this postulate we found that 5- to 10-fold overexpression of GFP-HOXA9 led to its predominant nuclear localization in HeLa cells (Fig. 11).
In addition to its interaction with MEIS1, our results indicate that HOXA9 requires its own NLS to shift into the nucleus. In this work we identified two NLS in the homeobox domain of HOXA9. Kasahara and Izumo demonstrated that the N-terminal region of the homeodomain contains a highly conserved Arg/Lys-rich sequence and also demonstrated that this region of the cardiac tissue specifying Csx/Nkx5.2 homeoprotein functions as an NLS (15), a region that corresponds to the first NLS that we identified in the amino terminus of HOXA9. The second NLS we identified was located in helix 3 of the HOXA9 homeodomain, a region that also shares substantial homology with other homeodomain-containing proteins. Two groups reported that this region of PDX1 functions as an NLS (13, 29). Together with our observations it appears that the homeodomain of HOXA9 not only works as a DNA binding motif but also is important for the nuclear localization of the protein.
In the present study we also found that MEIS1 expression is induced by TPO, and more importantly, MEIS1 mRNA levels were decreased substantially in Sca-1+/c-kit+/Gr-1− cells from Tpo−/− mice compared to levels for controls. In addition to class I homeodomain proteins, including HOXB4 and HOXA9, MEIS1 has been shown to play a crucial role in hematopoietic cell development. MEIS1 expression is highly restricted in primitive hematopoietic cells (34), and primitive cell populations from MEIS1-deficient mice display reduced repopulating activity and reduced numbers of committed progenitor cells of multiple lineages (14). Thus, together with our data it is suggested that the hematopoietic defects seen in Tpo−/− and MEIS−/− mice lie in the same mechanistic pathway.
Recent studies have also demonstrated that MEIS1 plays a crucial role in megakaryopoiesis. Okada and colleagues reported that MEIS1 is expressed in megakaryocytes and regulates expression of the megakaryocyte-specific gene platelet factor 4 (30). Meis1−/− mice display a severe abnormality of megakaryopoiesis; the fetal liver is lacking in CD41+ cells, a specific marker of the megakaryocyte lineage (14). Together with our present observation that TPO controls its expression, these results raise the possibility that MEIS1 plays important roles in megakaryopoiesis downstream of TPO.
We have previously reported that TPO controls HOXB4 mRNA expression in a p38 MAPK-dependent fashion and that the kinase activates upstream stimulating factor 1 to affect the homeodomain-containing protein (20). In this study we demonstrate that HOXA9 function is regulated by TPO by affecting the nuclear localization of the transcription factor, an effect mediated by its interaction with, and levels of, MEIS1. It should also be noted that HOXA9 is present in the nucleus of some starved UT-7/TPO cells and in a proportion of unstimulated primitive murine marrow cells. Whether this residual level of nuclear HOXA9 is due to other stimuli present in the cultures or during preparation of the marrow cells is currently unclear, nor is it known if these levels are sufficient to foster the full repertoire of HOXA9 transcriptional effects. Nevertheless, our demonstration that TPO can influence the intracellular pattern of HOXA9 is novel and provides a new insight into the interaction of cytokines and transcription factors. Collectively, these data indicate that homeodomain-containing proteins are major effectors of TPO, and potentially other growth factors, and serve as an excellent example of how cytokines can determine the developmental fate of marrow cells through influencing the level and/or function of transcription factors critical for hematopoiesis.
Acknowledgments
We thank Takuro Nakamura (The Cancer Institute, Japanese Foundation for Cancer Research, Tokyo, Japan) for kindly providing a HOXA9 cDNA and Don Foster (Zymogenetics, Seattle, Wash.) for the kind gift of TPO.
This work was supported by grants R01 CA31615 and R01 DK49855 from the National Institutes of Health.
REFERENCES
- 1.Abramovich, C., E. A. Chavez, P. M. Lansdorp, and R. K. Humphries. 2002. Functional characterization of multiple domains involved in the subcellular localization of the hematopoietic Pbx interacting protein (HPIP). Oncogene 21:6766-6771. [DOI] [PubMed]
- 2.Abu-Shaar, M., H. D. Ryoo, and R. S. Mann. 1999. Control of the nuclear localization of Extradenticle by competing nuclear import and export signals. Genes Dev. 13:935-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antonchuk, J., G. Sauvageau, and R. K. Humphries. 2002. HOXB4-induced expansion of adult hematopoietic stem cells ex vivo. Cell 109:39-45. [DOI] [PubMed] [Google Scholar]
- 4.Berthelsen, J., C. Kilstrup-Nielsen, F. Blasi, F. Mavilio, and V. Zappavigna. 1999. The subcellular localization of PBX1 and EXD proteins depends on nuclear import and export signals and is modulated by association with PREP1 and HTH. Genes Dev. 13:946-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bjornsson, J. M., N. Larsson, A. C. Brun, M. Magnusson, E. Andersson, P. Lundstrom, J. Larsson, E. Repetowska, M. Ehinger, R. K. Humphries, and S. Karlsson. 2003. Reduced proliferative capacity of hematopoietic stem cells deficient in Hoxb3 and Hoxb4. Mol. Cell. Biol. 23:3872-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brun, A. C., J. M. Bjornsson, M. Magnusson, N. Larsson, P. Leveen, M. Ehinger, E. Nilsson, and S. Karlsson. 2004. Hoxb4 deficient mice have normal hematopoietic development but exhibit a mild proliferation defect in hematopoietic stem cells. Blood 103:4126-4133. [DOI] [PubMed]
- 7.Calvo, K. R., P. S. Knoepfler, D. B. Sykes, M. P. Pasillas, and M. P. Kamps. 2001. Meis1a suppresses differentiation by G-CSF and promotes proliferation by SCF: potential mechanisms of cooperativity with Hoxa9 in myeloid leukemia. Proc. Natl. Acad. Sci. USA 98:13120-13125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calvo, K. R., D. B. Sykes, M. Pasillas, and M. P. Kamps. 2000. Hoxa9 immortalizes a granulocyte-macrophage colony-stimulating factor-dependent promyelocyte capable of biphenotypic differentiation to neutrophils or macrophages, independent of enforced Meis expression. Mol. Cell. Biol. 20:3274-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorsam, S. T., C. M. Ferrell, G. P. Dorsam, M. K. Derynck, U. Vijapurkar, D. Khodabakhsh, B. Pau, H. Bernstein, C. M. Haqq, C. Largman, and H. J. Lawrence. 2004. The transcriptome of the leukemogenic homeoprotein HOXA9 in human hematopoietic cells. Blood 103:1676-1684. [DOI] [PubMed] [Google Scholar]
- 10.Drabkin, H. A., C. Parsy, K. Ferguson, F. Guilhot, L. Lacotte, L. Roy, C. Zeng, A. Baron, S. P. Hunger, M. Varella-Garcia, R. Gemmill, F. Brizard, A. Brizard, and J. Roche. 2002. Quantitative HOX expression in chromosomally defined subsets of acute myelogenous leukemia. Leukemia 16:186-195. [DOI] [PubMed] [Google Scholar]
- 11.Elrick, L. J., and K. Docherty. 2001. Phosphorylation-dependent nucleocytoplasmic shuttling of pancreatic duodenal homeobox-1. Diabetes 50:2244-2252. [DOI] [PubMed] [Google Scholar]
- 12.Fox, N., G. Priestley, T. Papayannopoulou, and K. Kaushansky. 2002. Thrombopoietin expands hematopoietic stem cells after transplantation. J. Clin. Investig. 110:389-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hessabi, B., P. Ziegler, I. Schmidt, C. Hessabi, and R. Walther. 1999. The nuclear localization signal (NLS) of PDX-1 is part of the homeodomain and represents a novel type of NLS. Eur. J. Biochem. 263:170-177. [DOI] [PubMed] [Google Scholar]
- 14.Hisa, T., S. E. Spence, R. A. Rachel, M. Fujita, T. Nakamura, J. M. Ward, D. E. Devor-Henneman, Y. Saiki, H. Kutsuna, L. Tessarollo, N. A. Jenkins, and N. G. Copeland. 2004. Hematopoietic, angiogenic and eye defects in Meis1 mutant animals. EMBO J. 23:450-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasahara, H., and S. Izumo. 1999. Identification of the in vivo casein kinase II phosphorylation site within the homeodomain of the cardiac tissue-specifying homeobox gene product Csx/Nkx2.5. Mol. Cell. Biol. 19:526-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaushansky, K., S. Lok, R. D. Holly, V. C. Broudy, N. Lin, M. C. Bailey, J. W. Forstrom, M. M. Buddle, P. J. Oort, F. S. Hagen, et al. 1994. Promotion of megakaryocyte progenitor expansion and differentiation by the c-Mpl ligand thrombopoietin. Nature 369:568-571. [DOI] [PubMed] [Google Scholar]
- 17.Kawamori, D., Y. Kajimoto, H. Kaneto, Y. Umayahara, Y. Fujitani, T. Miyatsuka, H. Watada, I. B. Leibiger, Y. Yamasaki, and M. Hori. 2003. Oxidative stress induces nucleo-cytoplasmic translocation of pancreatic transcription factor PDX-1 through activation of c-Jun NH(2)-terminal kinase. Diabetes 52:2896-2904. [DOI] [PubMed] [Google Scholar]
- 18.Kilstrup-Nielsen, C., M. Alessio, and V. Zappavigna. 2003. PBX1 nuclear export is regulated independently of PBX-MEINOX interaction by PKA phosphorylation of the PBC-B domain. EMBO J. 22:89-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura, S., A. W. Roberts, D. Metcalf, and W. S. Alexander. 1998. Hematopoietic stem cell deficiencies in mice lacking c-Mpl, the receptor for thrombopoietin. Proc. Natl. Acad. Sci. USA 95:1195-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirito, K., N. Fox, and K. Kaushansky. 2003. Thrombopoietin stimulates Hoxb4 expression: an explanation for the favorable effects of TPO on hematopoietic stem cells. Blood 102:3172-3178. [DOI] [PubMed] [Google Scholar]
- 21.Kirito, K., M. Uchida, M. Yamada, Y. Miura, and N. Komatsu. 1997. A distinct function of STAT proteins in erythropoietin signal transduction. J. Biol. Chem. 272:16507-16513. [DOI] [PubMed] [Google Scholar]
- 22.Komatsu, N., M. Kunitama, M. Yamada, T. Hagiwara, T. Kato, H. Miyazaki, M. Eguchi, M. Yamamoto, and Y. Miura. 1996. Establishment and characterization of the thrombopoietin-dependent megakaryocytic cell line, UT-7/TPO. Blood 87:4552-4560. [PubMed] [Google Scholar]
- 23.Kroon, E., J. Krosl, U. Thorsteinsdottir, S. Baban, A. M. Buchberg, and G. Sauvageau. 1998. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 17:3714-3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ku, H., Y. Yonemura, K. Kaushansky, and M. Ogawa. 1996. Thrombopoietin, the ligand for the Mpl receptor, synergizes with steel factor and other early cytokines in supporting proliferation of primitive hematopoietic progenitors of mice. Blood 87:4544-4551. [PubMed] [Google Scholar]
- 25.Lawrence, H. J., C. D. Helgason, G. Sauvageau, S. Fong, D. J. Izon, R. K. Humphries, and C. Largman. 1997. Mice bearing a targeted interruption of the homeobox gene HOXA9 have defects in myeloid, erythroid, and lymphoid hematopoiesis. Blood 89:1922-1930. [PubMed] [Google Scholar]
- 26.Lawrence, H. J., S. Rozenfeld, C. Cruz, K. Matsukuma, A. Kwong, L. Komuves, A. M. Buchberg, and C. Largman. 1999. Frequent co-expression of the HOXA9 and MEIS1 homeobox genes in human myeloid leukemias. Leukemia 13:1993-1999. [DOI] [PubMed] [Google Scholar]
- 27.Macfarlane, W. M., C. M. McKinnon, Z. A. Felton-Edkins, H. Cragg, R. F. James, and K. Docherty. 1999. Glucose stimulates translocation of the homeodomain transcription factor PDX1 from the cytoplasm to the nucleus in pancreatic beta-cells. J. Biol. Chem. 274:1011-1016. [DOI] [PubMed] [Google Scholar]
- 28.Maizel, A., O. Bensaude, A. Prochiantz, and A. Joliot. 1999. A short region of its homeodomain is necessary for engrailed nuclear export and secretion. Development 126:3183-3190. [DOI] [PubMed] [Google Scholar]
- 29.Moede, T., B. Leibiger, H. G. Pour, P. Berggren, and I. B. Leibiger. 1999. Identification of a nuclear localization signal, RRMKWKK, in the homeodomain transcription factor PDX-1. FEBS Lett. 461:229-234. [DOI] [PubMed] [Google Scholar]
- 30.Okada, Y., R. Nagai, T. Sato, E. Matsuura, T. Minami, I. Morita, and T. Doi. 2003. Homeodomain proteins MEIS1 and PBXs regulate the lineage-specific transcription of the platelet factor 4 gene. Blood 101:4748-4756. [DOI] [PubMed] [Google Scholar]
- 31.Owens, B. M., and R. G. Hawley. 2002. HOX and non-HOX homeobox genes in leukemic hematopoiesis. Stem Cells 20:364-379. [DOI] [PubMed] [Google Scholar]
- 32.Pai, C. Y., T. S. Kuo, T. J. Jaw, E. Kurant, C. T. Chen, D. A. Bessarab, A. Salzberg, and Y. H. Sun. 1998. The Homothorax homeoprotein activates the nuclear localization of another homeoprotein, extradenticle, and suppresses eye development in Drosophila. Genes Dev. 12:435-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pineault, N., C. Abramovich, H. Ohta, and R. K. Humphries. 2004. Differential and common leukemogenic potentials of multiple NUP98-Hox fusion proteins alone or with Meis1. Mol. Cell. Biol. 24:1907-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pineault, N., C. D. Helgason, H. J. Lawrence, and R. K. Humphries. 2002. Differential expression of Hox, Meis1, and Pbx1 genes in primitive cells throughout murine hematopoietic ontogeny. Exp. Hematol. 30:49-57. [DOI] [PubMed] [Google Scholar]
- 35.Rafiq, I., G. da Silva Xavier, S. Hooper, and G. A. Rutter. 2000. Glucose-stimulated preproinsulin gene expression and nuclear trans-location of pancreatic duodenum homeobox-1 require activation of phosphatidylinositol 3-kinase but not p38 MAPK/SAPK2. J. Biol. Chem. 275:15977-15984. [DOI] [PubMed] [Google Scholar]
- 36.Saleh, M., H. Huang, N. C. Green, and M. S. Featherstone. 2000. A conformational change in PBX1A is necessary for its nuclear localization. Exp. Cell Res. 260:105-115. [DOI] [PubMed] [Google Scholar]
- 37.Schiedlmeier, B., H. Klump, E. Will, G. Arman-Kalcek, Z. Li, Z. Wang, A. Rimek, J. Friel, C. Baum, and W. Ostertag. 2003. High-level ectopic HOXB4 expression confers a profound in vivo competitive growth advantage on human cord blood CD34+ cells, but impairs lymphomyeloid differentiation. Blood 101:1759-1768. [DOI] [PubMed] [Google Scholar]
- 38.Shen, W. F., J. C. Montgomery, S. Rozenfeld, J. J. Moskow, H. J. Lawrence, A. M. Buchberg, and C. Largman. 1997. AbdB-like Hox proteins stabilize DNA binding by the Meis1 homeodomain proteins. Mol. Cell. Biol. 17:6448-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen, W. F., S. Rozenfeld, A. Kwong, L. G. Kom ves, H. J. Lawrence, and C. Largman. 1999. HOXA9 forms triple complexes with PBX2 and MEIS1 in myeloid cells. Mol. Cell. Biol. 19:3051-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sitnicka, E., N. Lin, G. V. Priestley, N. Fox, V. C. Broudy, N. S. Wolf, and K. Kaushansky. 1996. The effect of thrombopoietin on the proliferation of murine hematopoietic stem cells. Blood 87:4998-5005. [PubMed] [Google Scholar]
- 41.Solar, G. P., W. G. Kerr, F. C. Zeigler, D. Hess, C. Donahue, F. J. de Sauvage, and D. L. Eaton. 1998. Role of c-mpl in early hematopoiesis. Blood 92:4-10. [PubMed] [Google Scholar]
- 42.Terskikh, A. V., T. Miyamoto, C. Chang, L. Diatchenko, and I. L. Weissman. 2003. Gene expression analysis of purified hematopoietic stem cells and committed progenitors. Blood 102:94-101. [DOI] [PubMed] [Google Scholar]
- 43.Thorsteinsdottir, U., E. Kroon, L. Jerome, F. Blasi, and G. Sauvageau. 2001. Defining roles for HOX and MEIS1 genes in induction of acute myeloid leukemia. Mol. Cell. Biol. 21:224-234. [DOI] [PMC free article] [PubMed] [Google Scholar]