Abstract
RAG1 and RAG2 catalyze the first DNA cleavage steps in V(D)J recombination. We demonstrate that the isolated central domain of RAG1 has inherent single-stranded (ss) DNA cleavage activity, which does not require, but is enhanced by, RAG2. The central domain, therefore, contains the active-site residues necessary to perform hydrolysis of the DNA phosphodiester backbone. Furthermore, the catalytic activity of this domain on ss DNA is abolished by addition of the C-terminal domain of RAG1. The inhibitory effects of this latter domain are suppressed on substrates containing double-stranded (ds) DNA. Together, the activities of the reconstituted domains on ss versus mixed ds-ss DNA approximate the activity of intact RAG1 in the presence of RAG2. We propose how the combined actions of the RAG1 domains may function in V(D)J recombination and also in aberrant cleavage reactions that may lead to genomic instability in B and T lymphocytes.
RAG1 and RAG2 mediate the generation of functional immunoglobulin and T-cell receptor genes by catalyzing the initial site-specific DNA cleavage steps in the V(D)J recombination mechanism (5, 11, 14). The sites of DNA cleavage are specified by the recombination signal sequence (RSS) that flanks each of the V, D, and J gene segments available for rearrangement. The RSS consists of conserved heptamer and nonamer sequence elements separated by a spacer of either 12 ± 1 or 23 ± 1 base pairs (referred to as a 12- or 23-RSS). Recombination is restricted by the 12/23 rule, which requires that gene segments must be flanked by signals of different spacer lengths in order to be joined together. The RAG proteins catalyze double-stranded (ds) DNA cleavage between the coding segment and the flanking RSS in two enzymatic steps (29). First, a DNA nick is generated between the coding gene segment and the heptamer of the RSS. Second, the resulting 3′OH on the coding end is coupled to the phosphate group between the RSS and the gene segment on the opposite strand. At each RSS, the resulting products are a 5′-phosphorylated ds break at the signal end and a covalently closed hairpin at the coding end (29). Appropriate joining of both the coding and signal ends in V(D)J recombination requires factors that mediate nonhomologous DNA end joining, including DNA-PKcs, Ku70/80, Artemis, XRCC4, and DNA ligase IV (11, 28).
The core regions of the RAG proteins retain all DNA cleavage activity, are more soluble than their full-length counterparts, and have thus been used in the majority of in vitro characterizations (14). The murine forms of the core proteins include residues 384 to 1008 (of 1,040 total residues) in full-length RAG1 and residues 1 to 387 (of 527 total residues) in full-length RAG2 (14). Core RAG1 was previously reported to contain the active site for both nicking and hairpin formation, formed by two essential aspartate residues at positions 600 and 708 (13, 23, 24) and a glutamate at position 962 (23, 24). Together these residues comprise a DDE motif, an active-site arrangement utilized by bacterial transposases and retroviral integrases (17). In all such enzymes the triad of active-site residues functions to coordinate one or more divalent cations in the active site (38). The divalent cations in turn activate a water molecule for the hydrolytic DNA nicking reaction or activate the 3′-OH group for the transesterification reaction in hairpin formation.
In addition to the active site, core RAG1 has been shown to bind to the RSS, independently of RAG2, with specificity for both the nonamer and heptamer of the RSS (3, 7, 34, 40, 45). Conversely, core RAG2 does not bind to DNA in the absence of RAG1, but it does enhance both the binding affinity and specificity of RAG1 to the RSS (15, 18, 32). The core RAG proteins have been shown to bind a 12-RSS or 23-RSS independently (single RSS complex) or to both simultaneously to form the paired complex (18, 33, 47). In vivo, DNA nicking occurs in either the single RSS complex or the paired complex, while hairpin formation occurs nearly simultaneously at both signals bound in the paired complex (8, 52). In vitro, this coupled cleavage reaction is most closely reproduced in Mg2+-containing buffers, whereas in the presence of Mn2+, the reaction is uncoupled, with nicking and hairpin formation readily occurring on a single RSS (29).
The regions of RAG1 that bind with site specificity to the conserved RSS heptamer and nonamer sites have been identified. The RSS nonamer is recognized by the N-terminal region of core RAG1 (7, 45), referred to as the nonamer-binding region (NBR). On a single RSS, the NBR is contributed from a separate subunit of the dimeric RAG1 protein than that containing the active-site residues (46). While the NBR is essential for in vivo recombination activity, it is dispensable in vitro for nicking and hairpin formation (42) and also for DNA cleavage at the ds-ss junction of 3′ overhang substrates (43) in the presence of Mn2+. This latter activity is DNA sequence independent and is proposed to contribute to coding end processing in V(D)J recombination (43).
Arbuckle et al. found previously that the RSS heptamer-binding site lies in the central domain (residues 528 to 760), which was identified through domain analysis of core RAG1 (4). Moreover, while the central domain of RAG1 interacts poorly with ds RSS heptamer, it binds with substantially enhanced affinity and specificity to the single-stranded (ss) form compared to the ds form (35). This latter finding is of particular interest since there is evidence that DNA helical distortions, including base unpairing, are induced at the border between the RSS heptamer and the coding flank upon RAG binding to the ds RSS (6, 37). As has recently been proposed, the introduction of ss conformation in the RSS heptamer would allow specific association of the RAG1 central domain and thus close proximity of D600 and D708 active-site residues to the DNA cleavage site (35).
Besides recognizing the RSS, the RAG proteins also interact with the coding segment that flanks the heptamer. Protein-DNA cross-linking studies have demonstrated that both RAG proteins are in close proximity to the coding flank (9, 32, 48). However, RAG1 formed the strongest contacts (48), which were found to be mediated at least in part by a region near the C-terminal end of core RAG1 (31). Consistent with this latter report, we have identified a topologically independent C-terminal domain (residues 761 to 979) within core RAG1 that binds efficiently to non-sequence-specific ds DNA (4), a necessary property for interaction with the nonconserved V, D, and J gene segments.
As outlined above, insights into the DNA-binding capabilities of RAG1 were gained by a domain analysis study (4). Investigations of the isolated central and C-terminal domains have yielded information into RAG1 function that would have been difficult to obtain by studying the intact protein. However, it is not clear how the RAG1 domains function together to carry out the DNA cleavage activities of the intact protein. Furthermore, there are many issues concerning the intact RAG proteins that remain unresolved, including the role of RAG2 in V(D)J recombination. Several possibilities exist for the function of RAG2 in RSS cleavage activity, none of which are mutually exclusive. RAG2 may be required to activate RAG1 through a conformational change, as has been proposed previously (15, 16, 48). It is also possible that RAG2 is required for an essential DNA-binding interaction that is not yet characterized. For example, RAG2 has been predicted to form specific contacts with the RSS based on mutagenesis studies (12). Finally, RAG2 may contribute essential residues to the active site in RAG1, although no evidence for this has been provided.
To determine how the domains function together in the intact protein, we expanded our domain studies of core RAG1. In turn, our results demonstrate that RAG2 functions to activate RAG1 for DNA cleavage. Specifically, we show that the isolated central domain of RAG1, as well as a larger fragment that also contains the NBR, can endonucleolytically cleave ss DNA. This DNA cleavage activity requires the two aspartate active-site residues located in the central domain. Furthermore, while the isolated central domain is active in the absence of RAG2, a substantial enhancement in ss DNA cleavage activity occurs with the addition of the latter protein. Conversely, the C-terminal domain suppresses the activity of the central domain on ss DNA, regardless of the presence of RAG2. However, the inhibitory effect of the C-terminal domain is much less pronounced on substrates containing some ds regions. Together, the combined domains recapitulate the activity of core RAG1 and core RAG2 on ss and mixed ds-ss substrates. Overall, our results indicate distinct roles for the RAG1 domains in catalysis and in autoregulation and support a role for RAG2 as an activator of RAG1.
MATERIALS AND METHODS
Construction of plasmids and site-directed mutagenesis.
Fusion proteins joining maltose-binding protein (MBP) to RAG1 residues 384 to 1008, 528 to 760, 377 to 760, 761 to 979, and 528 to 721 were encoded by plasmids pCJM233, pRS3, pRS5, pJLA1, and pAEC5, respectively. The constructions of plasmids pCJM233 (40), pRS3 and pJLA1 (4), and pAEC5 (35) have been reported previously.
Plasmid pRS5 was constructed as follows. The RAG1 gene fragment encoding residues 377 to 760 was created by amplification of the appropriate region from the full-length murine RAG1 gene. The primers introduced a BamHI site at the 5′ end of the product and two stop codons and a SalI site at the 3′ end of the product. The fragment was inserted into the BamHI and SalI sites of the multiple cloning site of pMAL-c2 (New England Biolabs).
The active-site mutant of the RAG1 central domain [D600,708A R1(CD)] was derived from pRS3 by using a QuikChange site-directed mutagenesis kit (Stratagene) following the manufacturer's protocols.
Protein expression and purification.
The MBP-RAG1 fusion proteins were expressed in Escherichia coli and purified as described previously (4). Core RAG2 (fused to glutathione S-transferase [GST]) was expressed by transfection in 293T cells and was purified as described previously (45). Purified MBP was obtained from New England Biolabs, Inc.
Oligonucleotide substrates for cleavage assays.
Oligonucleotides were commercially synthesized and purified by polyacrylamide gel electrophoresis (PAGE) (Integrated DNA Technologies). The 59-base top-strand sequences of the wild-type (WT) 12-RSS and the mutant heptamer, mutant nonamer (MHMN) 12-RSS have been reported previously (4). The sequence of the 53-base bottom-strand sequence of the WT 12-RSS is 5′-TCGATTGGAGGTTTTTGTTAAGGTCTATATCACTGTGTAAGACGAGCCATATC-3′. The sequences for oligonucleotides O1, O2, and O2-m are 5′-AATTACTTTGCCAAAACTTTGGCAGCTGTCCCTGA-3′, 5′-GAAGCCTCTGGCGCAGTCTACATCTGTAC-3′, and 5′-GAAGCCTCTGGCGCTACATGGGTCTGTAC-3′respectively.
5′-Labeled oligonucleotides used in the DNA cleavage assays were labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs). 3′-Labeled oligonucleotides used in the DNA cleavage assays were labeled with [α-32P]ATP by using terminal deoxynucleotidyl transferase (New England Biolabs). For ds DNA substrates, the 32P-labeled top strand was annealed to its complement. The 3′ overhang substrate was constructed by annealing the 32P-labeled top strand with a 16-base unlabeled complementary strand (5′-TAAGACGAGCCATATC-3′).
Fe-induced RAG1 central domain cleavage.
MBP alone and MBP fused to the WT and D600,708A R1(CD) were mixed with Fe(NH4)SO4 (final concentration, 0.02 mM) and ascorbic acid (final concentration, 20 mM) and incubated at 25°C for 30 min. Hydrogen peroxide (final concentration, 0.02 or 0.0275%) was added to each sample for 10 s, and the reactions were stopped by adding sodium dodecyl sulfate (SDS) gel-loading solution and heated to 95°C. The proteins were immediately resolved by SDS-12% PAGE and electrotransferred to a polyvinylidene difluoride membrane. Subsequently, RAG1 peptides that remained fused to MBP were identified by Western blotting using rabbit polyclonal anti-MBP antibody (Santa Cruz Biotechnology).
Cleavage assays for ss substrate.
The ss DNA cleavage assays were performed in a total volume of 10 μl with a buffer composed of 10 mM Tris (pH 8.0), 2 mM dithiothreitol, 6% glycerol, and 100 mM NaCl (buffer A), plus 5 mM MgCl2. The indicated proteins (at the amounts given) were incubated with 1 nM 32P-labeled ss WT or MHMN 12-RSS substrate at 25°C for 30 min, unless stated otherwise. If separate proteins were combined, the proteins were first incubated together at 4°C for 30 min prior to incubation with the oligonucleotide substrate. Proteins were diluted at least 10-fold in the cleavage assay buffer prior to addition of DNA to ensure that the final buffer conditions in each sample were the same. Reactions were stopped by adding an equal volume of formamide gel-loading buffer and heating at 95°C for 5 min. DNA products were resolved on 10% denaturing gels, and 32P-labeled DNA was visualized by autoradiography and phosphorimaging on a Molecular Dynamics STORM phosphorimager.
Cleavage assays for ds substrate.
The ds DNA cleavage assays were performed in a total volume of 10 μl in buffer A and either 5 mM MgCl2 or 5 mM MnCl2. Core RAG1 and a fragment that contained both the central domain and the NBR of core RAG1 fused to MBP [R1(N+CD)] were incubated with core RAG2 for 30 min at 4°C, followed by addition of 1 nM 32P-labeled ds WT 12-RSS and incubation for 2 h at 37°C. The remaining procedure was the same as that described above.
Cleavage assays on 3′ overhang substrates.
The 3′ overhang DNA cleavage assays were performed in a total volume of 10 μl in buffer A and 5 mM MnCl2. The indicated proteins (at the amounts given) were incubated with 1 nM 32P-labeled 3′ overhang DNA at 37°C for 2 h. The remaining procedure was the same as that described above.
Electrophoretic mobility shift assays.
Electrophoretic mobility shift assays were accomplished using the 32P-labeled ss WT 12-RSS oligonucleotide substrate. The assays were conducted as previously described (35).
RESULTS
The carboxylate active-site residues in the isolated central domain bind divalent metal ions.
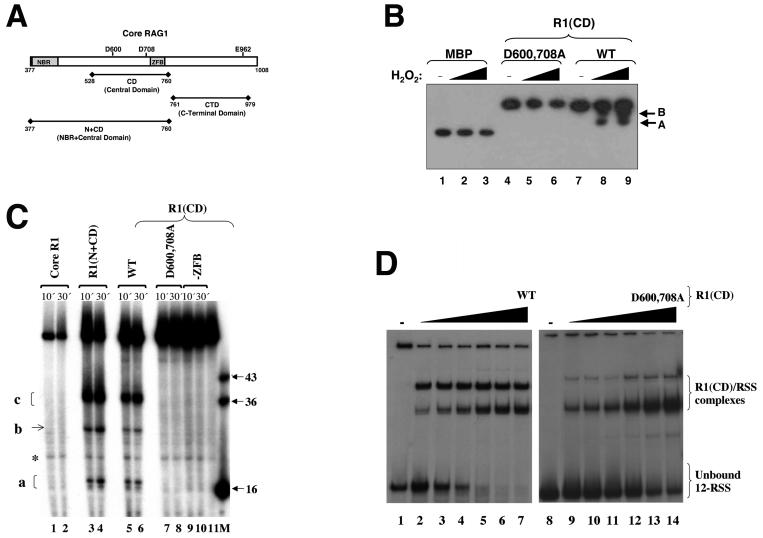
The DDE active-site residues in core RAG1 include D600 and D708 in the central domain and E962 located in the C-terminal domain (Fig. 1A). A previous study demonstrated that the active-site residues D600 and D708 in core RAG1 bound directly to divalent metal ions, specifically Fe2+, as determined using an iron-induced hydroxyl radical protein cleavage assay (23). The core RAG proteins demonstrated DNA cleavage activity in Fe2+-containing buffers, indicating that one or more ferrous ions bound within the RAG1 active site (23). However, efforts to demonstrate E962-coordinated ferrous ions were unsuccessful, bringing into question its role in the active site (23). We investigated whether D600 and D708 in the isolated central domain could coordinate divalent metal ions. To determine this, we used a hydroxyl radical cleavage assay similar to that mentioned above, but with the central domain in place of intact core RAG1. In this case, the central domain, referred to as R1(CD), was fused to MBP, and the fusion protein was incubated with Fe2+, ascorbic acid, and hydrogen peroxide. The resulting peptide products were separated by SDS-PAGE and analyzed by Western blotting against MBP (Fig. 1B). WT R1(CD) demonstrated two prominent cleavage products (Fig. 1B, lanes 7 to 9). Both cleavage products had a higher molecular mass than MBP alone (Fig. 1B, lane 1), indicating that the cleavage of the fusion protein was due exclusively to Fe2+ bound within the central domain. This latter point is further supported by the fact that peptide cleavage did not occur in MBP alone (Fig. 1B, lanes 1 to 3). Moreover, the D600,708A R1(CD) double mutant was completely resistant to hydroxyl radical cleavage, demonstrating that D600 and D708 coordinate divalent metal ions in the isolated central domain (Fig. 1B, lanes 4 to 6). Thus, the central domain requires the same carboxylate residues as intact core RAG1 to coordinate at least one metal ion in the active site.
FIG. 1.
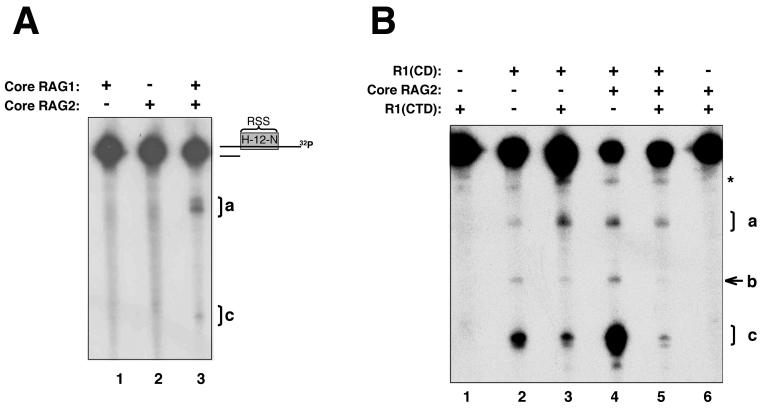
Metal-binding and ss DNA cleavage activities of the central domain in core RAG1. (A) Schematic of core RAG1. The positions of the three DDE active-site residues (D600, D708, and E962) are shown. The relative locations of the central domain, the C-terminal domain, and the NBR plus central domain are shown below the schematic of core RAG1. (B) Iron-induced hydroxyl radical peptide cleavage of the core RAG1 central domain. Lanes 1 to 3 contain MBP, lanes 4 to 6 contain the D600,708A R1(CD) double mutant, and lanes 7 to 9 contain the WT R1(CD). Both WT and mutant R1(CD) were fused to MBP. The reactions were initiated with the addition of either 0.02 or 0.0275% H2O2. The absence (−) of H2O2 from lanes 1, 4, and 7 is indicated. Western blotting with anti-MBP antibody revealed cleavage products (labeled A and B) in the reactions containing WT R1(CD). (C) ss DNA cleavage activity of the RAG1 domains. Cleavage reactions were performed using the ss WT 12-RSS (top) oligonucleotide substrate. The 5′-labeled substrate was incubated with core RAG1 (at 30 μg/ml) and the respective domains [the R1(CD) proteins at 30 μg/ml each and R1(N+CD) at 4 μg/ml] for the times indicated prior to electrophoresis. The major cleavage products are labeled a, b, and c. A band that is present with the substrate only is labeled with an asterisk. A RAG1 fragment that includes residues 528 to 721 is denoted as -ZFB. M indicates the marker lane with the lengths (in bases) of each marker indicated. Lane 11 contained only substrate. (D) Electrophoretic mobility shift assay demonstrating the interaction of WT and D600,708A R1(CD) with ss 12-RSS. The radiolabeled ss 12-RSS substrate was titrated with increasing concentrations of either WT or D600,708A R1(CD). The protein concentrations ranged from 0.2 to 0.8 μM [lanes 2 to 7 for WT and lanes 9 to 14 for D600,708A R1(CD)]. The samples were subjected to electrophoresis on a 6% nondenaturing polyacrylamide gel.
The central domain of core RAG1 is sufficient to catalyze cleavage of ss DNA.
As the central domain contains two metal-coordinating active-site residues, and also interacts preferentially with ss over ds RSS (35), we examined whether the isolated domain was capable of catalytic activity on an ss RSS substrate. To address this question, R1(CD) was incubated, in the presence of Mg2+, with an ss oligonucleotide containing the 12-RSS [the strand normally nicked in V(D)J recombination, referred to here as the top strand]. Subsequent to incubation, the DNA products were separated on a denaturing polyacrylamide gel. The central domain demonstrated significant cleavage of the ss oligonucleotide substrate within a 10-min time period at 25°C (Fig. 1C, lanes 5 to 6). These results were reproducible up to an incubation temperature of 37°C (data not shown). Notably, the DNA cleavage activity of the central domain occurred in the absence of RAG2, which reduces the likelihood that RAG2 provides essential active-site residues to RAG1, at least in the DNA nicking reaction. Moreover, mutation of D600 and D708 to alanine (D600,708A) completely abolished DNA cleavage (Fig. 1C, lanes 7 to 8). Compared with the WT domain, the mutant bound to ss 12-RSS with only slightly less affinity (Fig. 1D). Interestingly, however, significantly smaller amounts of the dimeric form of the mutant appeared to form a complex with the ss 12-RSS. Nevertheless, the complete absence of DNA cleavage was not due to the inability of the mutant to bind to the DNA substrate, thereby demonstrating that the observed DNA cleavage activity of the central domain on ss 12-RSS required the active site utilized in V(D)J recombination. Additionally, these results reveal that the active site in the central domain can function in the absence of additional residues from RAG1, particularly the putative glutamate residue of the DDE motif, E962.
A C2H2 zinc finger within the central domain, termed ZFB (39) (Fig. 1A), was previously found to contribute to the interaction between the central domain and ss RSS heptamer (35) and also to that between a RAG1-RAG2 complex and ds RSS (20). To determine whether the ZFB had any significant effect on ss DNA cleavage activity by the central domain, a truncated fusion protein of MBP fused to the central domain minus the ZFB was used. This protein did not exhibit catalytic activity on the ss substrate (Fig. 1C, lanes 9 to 10), indicating that the ZFB plays an important role in the cleavage activity of the central domain, perhaps by providing stability to the formation of the active site. Further studies of the ZFB will be useful in elucidating the function of this motif in the ss DNA cleavage activity of the central domain and possibly in the overall catalytic activities of the intact RAG proteins.
R1(N+CD) (Fig. 1A) produced identical DNA products as the central domain when ss substrate labeled at either the 5′ end (Fig. 1C, lanes 3 to 4) or the 3′ end (data not shown) was used. In addition, R1(N+CD) demonstrated more-efficient cleavage activity than the central domain, as lower protein concentrations were sufficient to yield the same cleavage efficiency. This latter result may be due to the presence of the additional DNA-binding site, NBR, contained in R1(N+CD). In contrast to the observed DNA cleavage activities of R1(CD) and R1(N+CD), intact core RAG1 failed to cleave the ss DNA substrate (Fig. 1C, lanes 1 to 2). Inclusion of the C-terminal region (residues 761 to 1008) in core RAG1 may inhibit the ss DNA cleavage activity of the intact core protein, perhaps by preventing access of the DNA substrate to the active-site residues in the central domain.
The ss DNA cleavage activities discussed above were reproducible for three separate protein preparations of each RAG1 protein. Moreover, the purification scheme for each protein consisted of three chromatographic separations with size exclusion chromatography as the final step (4). The size exclusion chromatography elution profiles for R1(N+CD) and core RAG1, which are active and inactive in ss DNA cleavage, respectively, overlapped considerably, demonstrating that the cleavage activity was not due to coelution of a contaminating nuclease. In addition, the WT and D600,708A mutant of the central domain showed identical chromatographic properties, and importantly, the absence of cleavage activity by the mutant definitively showed that the observed cleavage of the ss DNA is catalyzed by the RAG1 active-site residues in the central domain.
Core RAG1 central domain cleaves preferentially at A-C-A sequences.
In a highly precise manner, the RAG proteins nick the DNA immediately 5′ to the ds RSS heptamer in normal V(D)J recombination reactions (11). Conversely, the ss DNA cleavage catalyzed by the RAG1 central domain did not exhibit such precise sequence specificity. Rather, R1(CD) and R1(N+CD) yielded cleavage products in three areas of the oligonucleotide (producing 16- to 18-nt, 28-nt, and 36- to 37-nt fragments) from the 59-nt substrate (Fig. 1C, lanes 3 to 6). The 16- to 18-nt product (labeled a in Fig. 1C) resulted from cleavage at or near the heptamer-coding flank border, with the 16-nt product corresponding to the nicked product in a normal V(D)J recombination reaction. The 28-nt and 36- to 37-nt products (labeled b and c, respectively, in Fig. 1C) resulted from cleavage within the 12-base spacer and near the 5′ end of the nonamer, respectively. The preferred site of cleavage was typically at an A-C-A sequence, with cleavage in the 12-base spacer occurring at an A-G-A sequence (Fig. 2A). The observed ss DNA cleavage activity was due to endonucleolytic cleavage as corresponding products of the correct lengths were observed using ss substrate labeled on the 3′ end (Fig. 2B). Substantial increases in the protein concentration (up to 10-fold) of either R1(CD) or R1(N+CD) resulted in nonspecific DNA cleavage activity, which ultimately degraded the oligonucleotide substrate (data not shown). However, this occurred under nonphysiological conditions in which the binding of the central domain to the ss substrate had reached saturation (35), and therefore in the present work we utilized the appropriate protein concentrations to examine the endonucleolytic cleavage events.
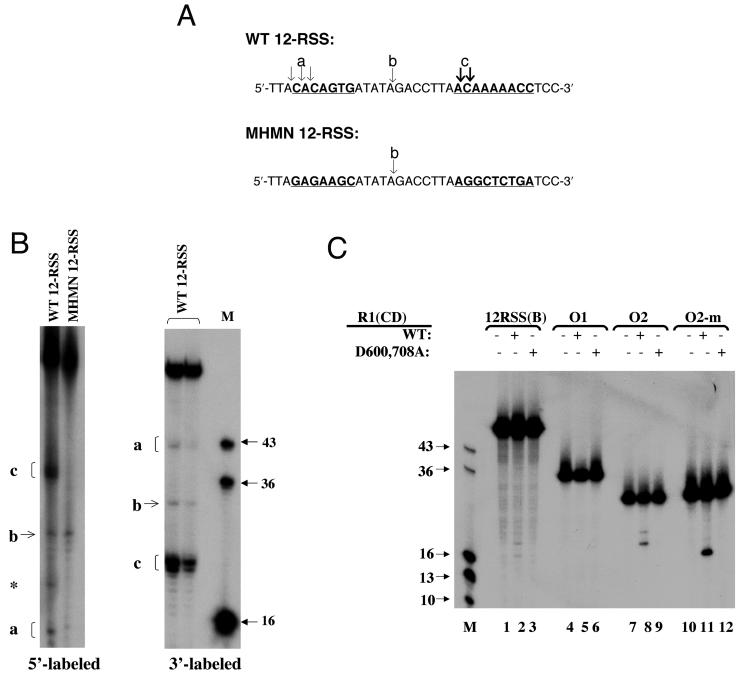
FIG. 2.
Pattern of ss DNA cleavage by R1(N+CD) and R1(CD). (A) Sites of cleavage on the top strands of WT 12-RSS (5′ and 3′ labeled) and MHMN 12-RSS (5′ labeled). The conserved heptamer and nonamer sequences in WT 12-RSS and the corresponding sequences in MHMN 12-RSS are underlined and indicated in bold letters. Arrows indicate the sites of cleavage. The bold arrows indicate enhanced cleavage. The labels a, b, and c correspond to the DNA products in panel B. (B) The left autoradiogram illustrates cleavage of 5′-labeled ss WT 12-RSS (top) and ss MHMN 12-RSS (top) by 30 μg of R1(CD)/ml. A band in the WT substrate that is independent of protein is marked with an asterisk. The right autoradiogram illustrates cleavage of 3′-labeled ss WT 12-RSS (top) by 30 and 15 μg of R1(CD)/ml in the first and second lanes, respectively. (C) Cleavage of the following 5′-labeled substrates: ss WT 12-RSS(B), O1, O2, and O2-m by 30 μg of WT and D600,708A R1(CD)/ml. In panels B and C, M indicates the marker lane with the lengths (in bases) of each marker indicated.
The cleavage pattern with the 12-RSS substrate (top strand) was changed upon use of an ss substrate (termed MHMN) in which the heptamer and nonamer sequences of the RSS were altered to remove A-C-A sequences (Fig. 2). The 28-nt fragment (product b) was observed with the MHMN substrate due to cleavage in the spacer (Fig. 2B), which was identical to what was seen with the WT substrate. However, the 36- to 37-nt fragments (product c) did not form with the MHMN substrate, demonstrating a disruption of the interaction with the mutated substrate. Similarly, cleavage 5′ to the heptamer was disrupted; however, minor amounts of the 16- to 18-nt fragments were observed (Fig. 2B), likely due to weak cleavage at the overlapping A-G-A sites in the mutant heptamer sequence.
The most efficient cleavage event within the WT substrate occurred at the 5′ end of the nonamer (product c), which yielded the 36- to 37-nt and 23- to 24-nt fragments with the 5′- and 3′-labeled WT substrates, respectively (Fig. 2B). The basis for a preferred cleavage site near the 5′ end of the nonamer is not clear and will require further study. Moreover, cleavage in the spacer and 5′ to the nonamer sequence shows that even though the central domain binds to the ss RSS heptamer with 10-fold-higher affinity than nonspecific DNA sequences (35), the domain apparently can still sample other sequences in the oligonucleotide substrate. Furthermore, it might be expected that R1(N+CD), which contains the NBR, may alter the sequence specificity of DNA binding and cleavage. However, the similar cleavage pattern suggests that the NBR may show less specificity for the RSS nonamer in the ss than in the ds form.
To further demonstrate the sequence preference for R1(CD) cleavage at ss A-C-A sequences, we utilized additional oligonucleotide substrates (Fig. 2C). First, we examined whether R1(CD) could cleave the complementary (bottom) strand of the ss 12-RSS [12-RSS(B)]. However, as 12-RSS(B) did not contain an A-C-A sequence, only minor cleavage products were formed (Fig. 2C, lane 2). Second, we tested the activity of R1(CD) on non-RSS-containing oligonucleotides. When an oligonucleotide that did not contain A-C-A sequences (O1) was used, no cleavage products were formed upon addition of R1(CD) (Fig. 2C, lane 5). However, with an A-C-A sequence in the center of a separate oligonucleotide (O2), a major product (of 20 bases) resulting from cleavage 5′ of the C and a minor product resulting from cleavage 3′ of the same base were evident after incubation with R1(CD) (Fig. 2C, lane 8). Furthermore, a variant of O2, O2-m, was created in which the position of the A-C-A within the sequence was moved 5′ by 4 bases. The resulting cleavage product of O2-m corresponded to the position of the A-C-A, yielding a major product of 16 bases (Fig. 2C, lane 11). As a control in the above cleavage reactions, we also tested the D600,708A mutant of the central domain, which did not produce cleavage products for any of the oligonucleotide substrates, as expected (Fig. 2C, lanes 3, 6, 9, and 12). In sum, we conclude that the major cleavage site in ss DNA for the RAG1 central domain is at an A-C-A sequence, with minor cleavage sites occurring at other A-containing sequences.
RAG2 enhances the ss DNA cleavage activity of the central domain.
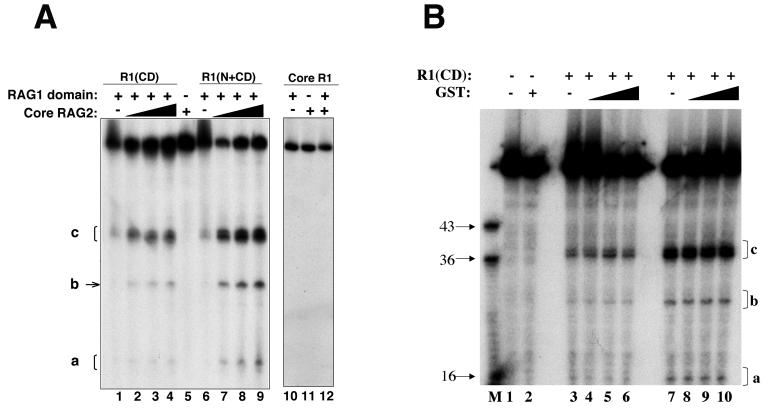
We investigated how RAG2 may affect the ss DNA cleavage activity of R1(CD) and R1(N+CD), since the central domain contains the ZFB, which is the predominant binding site for RAG2 (2). Addition of core RAG2 fused to GST led to dramatically enhanced cleavage of ss 12-RSS by both R1(CD) (Fig. 3A, lanes 1 to 4) and R1(N+CD) (Fig. 3A, lanes 6 to 9). As expected, RAG2 alone did not catalyze cleavage of the ss DNA substrate (Fig. 3A, lane 5). Moreover, the enhancement in cleavage activity is specific for RAG2, as addition of GST alone (Fig. 3B) or bovine serum albumin (data not shown) did not alter the levels of cleavage products formed, even with molar ratios of GST to R1(CD) of 2:1 and regardless of the input concentration of R1(CD). These results are consistent with RAG2 inducing conformational changes in the RAG1 central domain, perhaps resulting in greater access of the ss DNA substrate to the active site. Although it was recently reported that core RAG2 inhibited the binding of the central domain to ss DNA (35), it is likely that this result was due to increased DNA cleavage upon the addition of RAG2, resulting in less-efficient complex formation of the central domain with the cleaved products.
FIG. 3.
Effect of RAG2 on ss DNA cleavage by R1(N+CD), R1(CD), and core RAG1. (A) Cleavage of a 5′-labeled ss WT 12-RSS substrate is shown. Each lane contains the indicated proteins, with R1(CD), R1(N+CD), and core RAG1 at 15, 2, and 10 μg/ml, respectively. Lanes 2 to 4 and 7 to 9 contain increasing concentrations of core RAG2 from 1 to 10 μg/ml. The concentration of core RAG2 in lanes 5, 11, and 12 was 10 μg/ml. The cleavage products are labeled a, b, and c. (B) GST has no effect on the cleavage activity of R1(CD) on ss DNA. Increasing concentrations of GST were added to R1(CD) and ss 12-RSS. Lanes 3 to 6 contain 15 μg of R1(CD)/ml, with lanes 4 to 6 containing increasing concentrations of GST at 0.5-, 1-, and 2-fold molar ratios of GST to R1(CD), respectively. Lanes 7 to 10 contain 30 μg of R1(CD)/ml, with lanes 8 to 10 containing increasing concentrations of GST at 0.5-, 1-, and 2-fold molar ratios of GST to R1(CD), respectively. Lane 2 contains GST alone at the same concentrations as in lanes 6 and 9. M indicates the marker lane with the lengths (in bases) of each marker indicated.
Next, we examined whether core RAG2 would be sufficient to activate the ss DNA cleavage activity of the intact core RAG1 protein. However, even though core RAG2 enhanced cleavage activity of the central domain, core RAG2 combined with intact core RAG1 demonstrated no cleavage activity on the ss substrate (Fig. 3A, lanes 10 to 12). Therefore, in the absence of ds DNA, core RAG1 remains inactive regardless of the addition of RAG2, consistent with previous results obtained by using an entirely ss substrate (43). It is possible that the C-terminal region continues its autoinhibitory function on ss DNA cleavage activity even in the presence of RAG2.
Addition of the C-terminal domain suppresses ss DNA cleavage activity of the central domain.
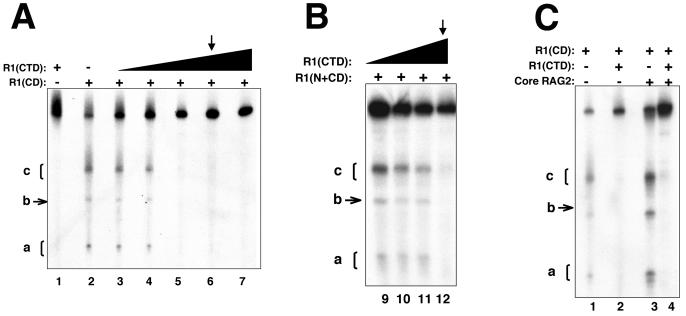
To test for an autoinhibitory function in the C-terminal region of core RAG1, we utilized a topologically independent domain, referred to as the C-terminal domain, which consists of residues 761 to 979 (Fig. 1A), a substantial portion of the C-terminal region. The addition of increasing concentrations of the C-terminal domain [fused to MBP and referred to as R1(CTD)] dramatically decreased the ss DNA cleavage activity of the central domain. At the concentrations of R1(CD) used here, an approximately equimolar concentration of R1(CTD) resulted in nearly complete suppression of ss DNA cleavage activity (Fig. 4A). R1(CTD) was also capable of decreasing the DNA cleavage activity of R1(N+CD). However, at the lower concentrations of R1(N+CD) used in the ss DNA cleavage assays, it was necessary to use a >5-fold excess of R1-CTD to begin to observe the inhibition of cleavage activity (Fig. 4B).
FIG. 4.
The addition of the C-terminal domain diminishes ss DNA cleavage activity of the central domain in the presence and absence of RAG2. (A and B) DNA cleavage of a 5′-labeled ss WT 12-RSS (top) substrate by R1(CD) at 30 μg/ml (A) and R1(N+CD) at 2 μg/ml (B). To each protein, increasing concentrations of R1(CTD) from 1 to 30 μg/ml were added. The arrows indicate the lane in which R1(CTD) is equimolar with R1(CD) and 20-fold greater than R1(N+CD). (C) DNA cleavage activity on WT ss 12-RSS mediated by R1(CD) in the presence of R1(CTD) and/or core RAG2. Each lane contains R1(CD) at 30 μg/ml, lanes 2 and 4 contain equivalent concentrations of R1(CTD), and lanes 3 and 4 contain core RAG2 at 10 μg/ml. Products a, b, and c are labeled.
Significantly, R1(CTD) inhibits ss DNA cleavage of R1(CD), even with the addition of core RAG2 (Fig. 4C, lanes 3 to 4). Thus, the C-terminal domain effectively blocks the enhancement of the central domain's catalytic activity by core RAG2. The molecular basis for how the C-terminal domain inhibits ss DNA cleavage activity of the central domain is not yet known. The C-terminal domain could function either as a direct inhibitor by binding to the central domain and preventing access of DNA to the active site, or it could act indirectly by preferentially binding to the DNA substrate and displacing the central domain from the DNA.
Activity of the core RAG1 central domain on ds DNA substrates.
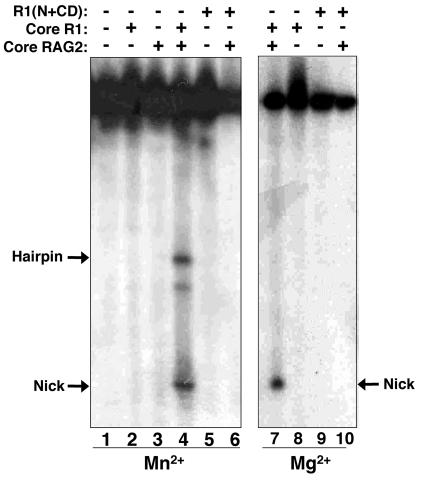
We next examined whether the central domain, alone or in the presence of RAG2, could cleave ds DNA substrates. Previous studies have indicated that small deletions and single point mutations located throughout RAG1 can decrease ds DNA cleavage activity (20, 41). Thus, not surprisingly, neither R1(N+CD) (Fig. 5, lanes 5, 6, 9, and 10) nor R1(CD) (data not shown) cleaved the ds 12-RSS substrate even in the presence of core RAG2. Furthermore, attempts to reconstitute DNA nicking and hairpin formation on the ds 12-RSS substrate by the addition of R1(CTD) to either R1(CD) or R1(N+CD) were unsuccessful, both in the absence and in the presence of core RAG2 (data not shown). This was not unexpected, given that R1(CTD) does not include residues 980 to 1008 from the C-terminal end of core RAG1. Previous attempts to mutate residues within this 29-residue region of core RAG1 yielded inactive protein (20). The inactivity on the ds substrates by R1(CD) and R1(N+CD) may be due to the fact that the ds DNA substrate does not become sufficiently distorted in the protein-DNA complex to facilitate catalytic activity of the RAG1 fragment. In contrast to the results with the separate domains, intact core RAG1 and core RAG2 cleaved ds 12-RSS, yielding a nicked product in the presence of Mg2+ (Fig. 5, lane 7) and both nicked and hairpin products in the presence of Mn2+ (Fig. 5, lane 4), as expected under the conditions used here.
FIG. 5.
Comparison of the ds DNA cleavage activities of RAG1 fragments. Cleavage of a ds WT 12-RSS substrate is shown. The proteins (each at 10 μg/ml) are indicated above each lane. The reactions in lanes 1 to 6 were in 5 mM Mn2+ and those in lanes 7 to 10 were in 5 mM Mg2+. The hairpin and nicked products are labeled.
Alleviation of the autoinhibitory function of the C-terminal domain in the presence of ds DNA.
To circumvent the requirement for a ds-to-ss DNA transition and yet still address our hypothesis that the inhibitory function of the C-terminal domain is suppressed with ds DNA, we utilized 3′ overhang substrates. Previously, it has been found that the RAG proteins cleave at or within 1 to 2 bases of the ds-ss junction in 3′ overhang substrates with no apparent sequence specificity (43). Here, we used a substrate in which the RSS was ss, with the sequence 5′ to the heptamer ds and the ds-ss junction at the normal V(D)J cleavage site (Fig. 6A). The substrate was radiolabeled at the 3′ end of the RSS-containing strand. We found that core RAG1 and core RAG2 cleaved at the ds-ss junction in Mn2+ (labeled product a in Fig. 6A), similar to previous results (43), suggesting that some ds region within a DNA substrate is sufficient to allow cleavage activity. In addition, we saw the formation of small amounts of product c from cleavage at the 5′ end of the nonamer (Fig. 6A), which was the preferred site of cleavage by R1(CD) (Fig. 2). This site is located 19 to 20 bases from the ds-ss junction within the ss DNA region of the 3′ overhang substrate. The core RAG proteins were not capable of cleaving this site in the entirely ss oligonucleotide (Fig. 1C, lanes 1 and 2). Thus, for cleavage to occur, only some regions of ds DNA in the substrate, which need not be directly adjacent to the cleavage site, are required.
FIG. 6.
DNA cleavage activity of the combined RAG1 domains versus that of the intact core protein on 3′ overhang substrates. (A) Activities of cores RAG1 and RAG2 alone and together. A schematic of the 3′ overhang substrate, with the top strand labeled with 32P at the 3′ end, is shown. The product resulting from cleavage at the ds-ss junction is labeled product a. A product corresponding to cleavage at the 5′ end of the nonamer is labeled product c. Each protein is at 10 μg/ml. (B) The identical substrate as in panel A was used. Each lane contains the indicated protein at 10 μg/ml. The products are labeled a, b, and c, with product a resulting from cleavage near the ds-ss junction.
To further test our model, we investigated whether the central domain could cleave within the ss regions of the 3′ overhang substrate. R1(CD) alone yielded cleavage products from the 3′ overhang substrate (Fig. 6B, lane 2), similar to what was observed with the entirely ss substrate that was radiolabeled at the 3′ end (Fig. 2B). In addition, R1(CD) produced a product within the ds region of the 3′ overhang substrate (marked with an asterisk in Fig. 6B). This cleavage site was not observed with the entirely ss substrate. However, as this activity was not affected by the addition of either R1(CTD) or core RAG2 and was not observed with the intact core RAG proteins, we did not examine it further in this study.
Next, we attempted to reconstitute the activity of intact core RAG1 on 3′ overhang substrates by using the central and C-terminal domains. For the reconstitution experiment of core RAG1 activity, it was possible to use R1(CD) rather than R1(N+CD), as NBR is not required for 3′ overhang removal (43). The addition of R1(CTD) to R1(CD) had variable effects on the levels of each of the cleavage products (Fig. 6B, lane 3). Use of R1(CTD) resulted in inhibition of the formation of products b and c, although they were not entirely eliminated. However, of the greatest significance, the level of product a resulting from cleavage at the ds-ss border was not reduced (it was even slightly increased) by the addition of R1(CTD). Overall, these results indicate that the inhibitory effects of the C-terminal domain can be blocked in the presence of ds DNA.
The addition of core RAG2 to the RAG1 domains yielded results similar to those with the RAG1 domains alone on the 3′ overhang substrate (Fig. 6B, lanes 4 to 5). These results demonstrate that core RAG2 is not required for cleavage with the separate RAG1 domains, even at the ds-ss junction. This is in contrast to the results with intact core RAG1, which requires core RAG2 for any cleavage activity to be observed. It is possible that the isolated central domain exists in a partially activated conformation, unlike the corresponding region in intact core RAG1, and thus core RAG2 is not absolutely required for the DNA cleavage activity of the isolated central domain, as it is with intact core RAG1. We propose that in the intact RAG1 protein two events must occur for DNA cleavage activity: (i) the inhibition of the C-terminal domain within the protein must be alleviated by the presence of ds DNA, and (ii) RAG2 must be present to activate cleavage by the central domain.
DISCUSSION
In this study, we demonstrate that the central domain of RAG1 is a catalytic domain with inherent ss DNA cleavage activity, which does not require, but is enhanced by, the addition of core RAG2. Given that it is likely that RAG1 and RAG2 binding induces base unpairing in the region of the RSS heptamer-coding flank border (6, 37), we propose that the resulting ss DNA efficiently serves as a binding target for the central domain of RAG1, which would then be capable of catalyzing the DNA nicking reaction.
This scenario does not include a catalytic role for E962, which is in contrast to previous results showing that mutation of E962 yields mostly inactive protein (23, 24). Possibly, E962 coordinates a second metal ion in the active site of the intact core RAG1 protein, which may be important in hairpin formation, a reaction we have been unable to successfully reconstitute using the separate domains (data not shown). Alternatively, the active-site metals may be coordinated by another glutamate residue in the central domain, such as E719, which may interact directly with divalent metal cations (26). Finally, as has been previously suggested (23), the active site in RAG1 may resemble restriction enzymes, which do not contain a third glutamate active-site residue. In this case, the E962 may function in another essential capacity, such as maintaining protein stability.
Our results show that the ss DNA cleavage activity of the isolated central domain is normally inhibited in intact core RAG1 by the C-terminal domain. Moreover, both intact core RAG proteins are required for cleavage of ds or mixed ss-ds DNA substrates. This is in contrast to recent findings that in a particular ss-ds substrate, core RAG1 alone could perform DNA cleavage (22). The restriction of RAG activity to, or adjacent to, ds DNA may be necessary since cleavage of ss DNA by the central domain can occur with low sequence specificity. RAG activity would then be restricted to the heptamer-coding flank border in V(D)J recombination and would also be prevented on regions of the genome with ss regions, such as at telomere ends, and during processes that transiently form ss DNA, including DNA repair, transcription, and replication. In this latter case, while RAG2 is degraded during the G1/S transition of the cell cycle (27), the autoregulation of RAG1 would still ensure prevention of very low levels of activity that may not be observed in our in vitro assays. Furthermore, as shown in Fig. 6A, preferred ss sites distal from ds-ss junctions can be cleaved in vitro by the core RAG proteins, which, if this occurs in vivo with the full-length proteins, may promote genomic instability.
We suggest that the ss DNA cleavage activity of the core RAG1 central domain may contribute to aberrant cleavage events that can result in chromosomal translocations. For example, a region in the Bcl-2 gene on chromosome 18 was recently shown to adopt ss DNA conformations that could readily be cleaved by the RAG proteins in vitro (36). This has important implications for the formation of the commonly observed translocation event between chromosomes 14 (at the immunoglobulin heavy-chain locus) and 18, which may be directly mediated by the RAG proteins (36). Additionally, the RAG proteins may mediate chromosomal translocations via a transposition mechanism. Two types of transposition reactions have been observed in vitro using the core RAG proteins. The first type, referred to as signal end transposition, inserts DNA fragments that contain 12- and 23-RSS at each end into relatively nonspecific DNA via a strand transfer reaction (1, 19). The insertion has been reported to occur more efficiently at target DNA that contains hairpins or other distorted DNA structures (25, 51). With DNA hairpins, it is likely that the hairpin tip contains some ss regions. While the distorted DNA structures may prove to be efficient substrates for the central domain of RAG1, it has yet to be shown how the isolated domain functions in the strand transfer reaction. Consequently, the second type of transposition reaction, referred to as inverse transposition (44), is of greater relevance here. In this reaction, the core RAG proteins first cleave at non-RSS DNA sequences, followed by insertion of the cleaved product into or near an RSS (44). This reaction effectively reverses the roles of donor and target DNA that occur in signal end transposition. Significantly, the sequence specificity for DNA nicking of the non-RSS DNA showed a slight preference for A-C-A sequences (44), similar to that observed here for the isolated central domain of core RAG1. Our results suggest that the region in RAG1 corresponding to the central domain could accomplish this cleavage activity. Both inverse and signal end transposition reactions have been speculated to contribute to the development of certain lymphomas (1, 19, 44). While it has been shown in vitro that the non-core region of RAG2 can inhibit the transposition reaction (10, 49, 50), another study has indicated that the full-length RAG proteins are capable of this aberrant activity (21). Furthermore, evidence for an in vivo transposition event has been reported recently (30). In conclusion, elucidating domain interactions during the DNA cleavage reactions mediated by RAG1 in normal V(D)J recombination, in comparison to the examples of aberrant cleavage reactions given above, will be an important step towards understanding the link between RAG protein regulation and potentially damaging chromosomal translocation events that lead to genomic instability.
Acknowledgments
We thank Ruby Rahman, Allison Cowan, and Rosemarie Simpson for technical assistance and LeAnn Godderz and Janeen Arbuckle for helpful comments.
This work was supported by a postdoctoral fellowship award to P.D. from the American Heart Association and by grants AI054467 from the National Institutes of Health and RPG-00-032-01-CIM from the American Cancer Society to K.K.R.
REFERENCES
- 1.Agrawal, A., Q. M. Eastman, and D. G. Schatz. 1998. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 394:744-751. [DOI] [PubMed] [Google Scholar]
- 2.Aidinis, V., D. C. Dias, C. A. Gomez, D. Bhattacharyya, E. Spanopoulou, and S. Santagata. 2000. Definition of minimal domains of interaction within the recombination-activating genes 1 and 2 recombinase complex. J. Immunol. 164:5826-5832. [DOI] [PubMed] [Google Scholar]
- 3.Akamatsu, Y., and M. A. Oettinger. 1998. Distinct roles of RAG1 and RAG2 in binding the V(D)J recombination signal sequences. Mol. Cell. Biol. 18:4670-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arbuckle, J. L., L. J. Fauss, R. Simpson, L. M. Ptaszek, and K. K. Rodgers. 2001. Identification of two topologically independent domains in RAG1 and their role in macromolecular interactions relevant to V(D)J recombination. J. Biol. Chem. 276:37093-37101. [DOI] [PubMed] [Google Scholar]
- 5.Brandt, V. L., and D. B. Roth. 2002. A recombinase diversified: new functions of the RAG proteins. Curr. Opin. Immunol. 14:224-229. [DOI] [PubMed] [Google Scholar]
- 6.Cuomo, C. A., C. L. Mundy, and M. A. Oettinger. 1996. DNA sequence and structure requirements for cleavage of V(D)J recombination signal sequences. Mol. Cell. Biol. 16:5683-5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Difilippantonio, M. J., C. J. McMahan, Q. M. Eastman, E. Spanopoulou, and D. G. Schatz. 1996. RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell 87:253-262. [DOI] [PubMed] [Google Scholar]
- 8.Eastman, Q. M., and D. G. Schatz. 1997. Nicking is asynchronous and stimulated by synapsis in 12/23 rule-regulated V(D)J cleavage. Nucleic Acids Res. 25:4370-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eastman, Q. M., I. J. Villey, and D. G. Schatz. 1999. Detection of RAG protein-V(D)J recombination signal interactions near the site of DNA cleavage by UV cross-linking. Mol. Cell. Biol. 19:3788-3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elkin, S. K., A. G. Matthews, and M. A. Oettinger. 2003. The C-terminal portion of RAG2 protects against transposition in vitro. EMBO J. 22:1931-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fugmann, S. D., A. I. Lee, P. E. Shockett, I. J. Villey, and D. G. Schatz. 2000. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu. Rev. Immunol. 18:495-527. [DOI] [PubMed] [Google Scholar]
- 12.Fugmann, S. D., and D. G. Schatz. 2001. Identification of basic residues in RAG2 critical for DNA binding by the RAG1-RAG2 complex. Mol. Cell 8:899-910. [DOI] [PubMed] [Google Scholar]
- 13.Fugmann, S. D., I. J. Villey, L. M. Ptaszek, and D. G. Schatz. 2000. Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol. Cell 5:97-107. [DOI] [PubMed] [Google Scholar]
- 14.Gellert, M. 2002. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu. Rev. Biochem. 71:101-132. [DOI] [PubMed] [Google Scholar]
- 15.Godderz, L. J., N. S. Rahman, G. M. Risinger, J. L. Arbuckle, and K. K. Rodgers. 2003. Self-association and conformational properties of RAG1: implications for formation of the V(D)J recombinase. Nucleic Acids Res. 31:2014-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomez, C. A., L. M. Ptaszek, A. Villa, F. Bozzi, C. Sobacchi, E. G. Brooks, L. D. Notarangelo, E. Spanopoulou, Z. Q. Pan, P. Vezzoni, P. Cortes, and S. Santagata. 2000. Mutations in conserved regions of the predicted RAG2 kelch repeats block initiation of V(D)J recombination and result in primary immunodeficiencies. Mol. Cell. Biol. 20:5653-5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haren, L., B. Ton-Hoang, and M. Chandler. 1999. Integrating DNA: transposases and retroviral integrases. Annu. Rev. Microbiol. 53:245-281. [DOI] [PubMed] [Google Scholar]
- 18.Hiom, K., and M. Gellert. 1997. A stable RAG1-RAG2-DNA complex that is active in V(D)J cleavage. Cell 88:65-72. [DOI] [PubMed] [Google Scholar]
- 19.Hiom, K., M. Melek, and M. Gellert. 1998. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell 94:463-470. [DOI] [PubMed] [Google Scholar]
- 20.Huye, L. E., M. M. Purugganan, M. M. Jiang, and D. B. Roth. 2002. Mutational analysis of all conserved basic amino acids in RAG-1 reveals catalytic, step arrest, and joining deficient mutants in the V(D)J recombinase. Mol. Cell. Biol. 22:3460-3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang, H., A. E. Ross, and S. Desiderio. 2004. Cell cycle-dependent accumulation in vivo of transposition-competent complexes between recombination signal ends and full-length RAG proteins. J. Biol. Chem. 279:8478-8486. [DOI] [PubMed] [Google Scholar]
- 22.Kim, D. R. 2003. Recombination activating gene 1 product alone possesses endonucleolytic activity. J. Biochem. Mol. Biol. 36:201-206. [DOI] [PubMed] [Google Scholar]
- 23.Kim, D. R., Y. Dai, C. L. Mundy, W. Yang, and M. A. Oettinger. 1999. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 13:3070-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landree, M. A., J. A. Wibbenmeyer, and D. B. Roth. 1999. Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 13:3059-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, G. S., M. B. Neiditch, R. R. Sinden, and D. B. Roth. 2002. Targeted transposition by the V(D)J recombinase. Mol. Cell. Biol. 22:2068-2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, W., F. C. Chang, and S. Desiderio. 2001. Rag-1 mutations associated with B-cell-negative SCID dissociate the nicking and transesterification steps of V(D)J recombination. Mol. Cell. Biol. 21:3935-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin, W. C., and S. Desiderio. 1994. Cell cycle regulation of V(D)J recombination-activating protein RAG-2. Proc. Natl. Acad. Sci. USA 91:2733-2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma, Y., U. Pannicke, K. Schwarz, and M. R. Lieber. 2002. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 108:781-794. [DOI] [PubMed] [Google Scholar]
- 29.McBlane, J. F., D. C. van Gent, D. A. Ramsden, C. Romeo, C. A. Cuomo, M. Gellert, and M. A. Oettinger. 1995. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell 83:387-395. [DOI] [PubMed] [Google Scholar]
- 30.Messier, T. L., J. P. O'Neill, S. M. Hou, J. A. Nicklas, and B. A. Finette. 2003. In vivo transposition mediated by V(D)J recombinase in human T lymphocytes. EMBO J. 22:1381-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mo, X., T. Bailin, and M. J. Sadofsky. 2001. A C-terminal region of RAG1 contacts the coding DNA during V(D)J recombination. Mol. Cell. Biol. 21:2038-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mo, X., T. Bailin, and M. J. Sadofsky. 1999. RAG1 and RAG2 cooperate in specific binding to the recombination signal sequence in vitro. J. Biol. Chem. 274:7025-7031. [DOI] [PubMed] [Google Scholar]
- 33.Mundy, C. L., N. Patenge, A. G. W. Matthews, and M. A. Oettinger. 2002. Assembly of the RAG1/RAG2 synaptic complex. Mol. Cell. Biol. 22:69-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagawa, F., K.-I. Ishiguro, A. Tsuboi, T. Yoshida, A. Ishikawa, T. Takemori, A. J. Otsuka, and H. Sakano. 1998. Footprint analysis of the RAG protein recombination signal sequence complex for V(D)J type recombination. Mol. Cell. Biol. 18:655-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peak, M. M., J. L. Arbuckle, and K. K. Rodgers. 2003. The central domain of core RAG1 preferentially recognizes single-stranded recombination signal sequence heptamer. J. Biol. Chem. 278:18235-18240. [DOI] [PubMed] [Google Scholar]
- 36.Raghavan, S. C., P. C. Swanson, X. Wu, C. L. Hsieh, and M. R. Lieber. 2004. A non-B-DNA structure at the Bcl-2 major breakpoint region is cleaved by the RAG complex. Nature 428:88-93. [DOI] [PubMed] [Google Scholar]
- 37.Ramsden, D. A., J. F. McBlane, D. C. van Gent, and M. Gellert. 1996. Distinct DNA sequence and structure requirements for the two steps of V(D)J recombination signal cleavage. EMBO J. 15:3197-3206. [PMC free article] [PubMed] [Google Scholar]
- 38.Rice, P. A., and T. A. Baker. 2001. Comparative architecture of transposase and integrase complexes. Nat. Struct. Biol. 8:302-307. [DOI] [PubMed] [Google Scholar]
- 39.Rodgers, K. K., Z. Bu, K. G. Fleming, D. G. Schatz, D. M. Engelman, and J. E. Coleman. 1996. A zinc-binding domain involved in the dimerization of RAG1. J. Mol. Biol. 260:70-84. [DOI] [PubMed] [Google Scholar]
- 40.Rodgers, K. K., I. J. Villey, L. Ptaszek, E. Corbett, D. G. Schatz, and J. E. Coleman. 1999. A dimer of the lymphoid protein RAG1 recognizes the recombination signal sequence and the complex stably incorporates the high mobility group protein HMG2. Nucleic Acids Res. 27:2938-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sadofsky, M. J., J. E. Hesse, J. F. McBlane, and M. Gellert. 1993. Expression and V(D)J recombination activity of mutated RAG-1 proteins. Nucleic Acids Res. 21:5644-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santagata, S., V. Aidinis, and E. Spanopoulou. 1998. The effect of Me2+ cofactors at the initial stages of V(D)J recombination. J. Biol. Chem. 273:16325-16331. [DOI] [PubMed] [Google Scholar]
- 43.Santagata, S., E. Besmer, A. Villa, F. Bozzi, J. S. Allingham, C. Sobacchi, D. B. Haniford, P. Vezzoni, M. C. Nussenzweig, Z.-Q. Pan, and P. Cortes. 1999. The RAG1/RAG2 complex constitutes a 3′ flap endonuclease: implications for junctional diversity in V(D)J and transpositional recombination. Mol. Cell 4:935-947. [DOI] [PubMed] [Google Scholar]
- 44.Shih, I. H., M. Melek, N. D. Jayaratne, and M. Gellert. 2002. Inverse transposition by the RAG1 and RAG2 proteins: role reversal of donor and target DNA. EMBO J. 21:6625-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spanopoulou, E., F. Zaitseva, F.-H. Wang, S. Santagata, D. Baltimore, and G. Panayotou. 1996. The homeodomain region of Rag-1 reveals the parallel mechanisms of bacterial and V(D)J recombination. Cell 87:263-276. [DOI] [PubMed] [Google Scholar]
- 46.Swanson, P. C. 2001. The DDE motif in RAG-1 is contributed in trans to a single active site that catalyzes the nicking and transesterification steps of V(D)J recombination. Mol. Cell. Biol. 21:449-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swanson, P. C. 2002. A RAG-1/RAG-2 tetramer supports 12/23-regulated synapsis, cleavage, and transposition of V(D)J recombination signals. Mol. Cell. Biol. 22:7790-7801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swanson, P. C., and S. Desiderio. 1999. RAG-2 promotes heptamer occupancy by RAG-1 in the assembly of a V(D)J initiation complex. Mol. Cell. Biol. 19:3674-3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swanson, P. C., D. Volkmer, and L. Wang. 2004. Full-length RAG-2, and not full-length RAG-1, specifically suppresses RAG-mediated transposition but not hybrid joint formation or disintegration. J. Biol. Chem. 279:4034-4044. [DOI] [PubMed] [Google Scholar]
- 50.Tsai, C. I., and D. G. Schatz. 2003. Regulation of RAG1/RAG2-mediated transposition by GTP and the C-terminal region of RAG2. EMBO J. 22:1922-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsai, C. L., M. Chatterji, and D. G. Schatz. 2003. DNA mismatches and GC-rich motifs target transposition by the RAG1/RAG2 transposase. Nucleic Acids Res. 31:6180-6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu, K., and M. R. Lieber. 2000. The nicking step in V(D)J recombination is independent of synapsis: implications for the immune repertoire. Mol. Cell. Biol. 20:7914-7921. [DOI] [PMC free article] [PubMed] [Google Scholar]