Abstract
Meiosis is the specialized cell division that generates gametes. In contrast to mitosis, molecular mechanisms and regulation of meiosis are much less understood. Meiosis shares mechanisms and regulation with mitosis in many aspects, but also has critical differences from mitosis. This review highlights these differences between meiosis and mitosis. Recent studies using various model systems revealed differences in a surprisingly wide range of aspects, including cell-cycle regulation, recombination, postrecombination events, spindle assembly, chromosome–spindle interaction, and chromosome segregation. Although a great degree of diversity can be found among organisms, meiosis-specific processes, and regulation are generally conserved.
Cells undergoing meiosis are unique in many respects (e.g., suppression of the S phase during the second round of cell division). The molecular mechanisms that underlie these critical events are beginning to be understood.
Meiosis is a special mode of cell division, which makes haploid cells from a diploid cell. It is essential for sexual reproduction in eukaryotes and diploid organisms and produces gametes, such as eggs and sperm. Sexual reproduction is thought to be essential for long-term survival of species, as it generates diversity and mixes the genetic materials within the species. This consists of two opposite processes: meiosis, which reduces chromosome numbers from diploid to haploid, and conjugation (fertilization), which restores the diploid state by fusion of two haploid cells. Meiosis generates diversity through two events: recombination and chromosome segregation. Missegregation during meiosis results in aneuploidy in progeny or fertilized eggs. In the case of humans, it is reported that 20% of all eggs are aneuploids, most of which are results of chromosome missegregation in oocytes (Hassold and Hunt 2001). This is a major cause of infertility, miscarriages, and birth defects, such as Down’s syndrome, in humans. Despite the medical importance, little is known about the molecular mechanisms of meiotic chromosome segregation in humans. Understanding meiosis is not only important for its own ends, but also provides unique insights into the fundamental regulation of mitosis. As many excellent reviews already cover specific aspects of meiosis, this review gives an overview by highlighting key meiotic events and molecular regulation distinct from mitosis.
CELL-CYCLE CONTROL
In eukaryotic mitotic cycles, chromosome replication and segregation alternate. This is essential for maintaining the genome stability. This is achieved by two-step regulation of replication by Cdk (Tanaka and Araki 2010). The first step, called licensing, allows Mcm2–7 to be recruited to form the prereplicative complex at replication origins only in G1 when Cdk activity is low. An increase in Cdk kinase activity, together with Cdc7 kinase activity in late G1, triggers initiation of DNA replication. As a high Cdk activity inhibits the formation of the prereplicative complex, the origin will not be licensed until the mitotic exit. This dual function of Cdk ensures only one firing from each replication fork in one mitotic cycle.
In contrast, meiosis consists of two divisions without an intervening S phase, which is essential for reducing the ploidy. Suppression of the intervening S phase is achieved by maintaining the Cdk activity sufficiently high between two meiotic divisions. In Xenopus oocytes, incomplete degradation of cyclin B and a low amount of the Wee1 kinase keeps the Cdk activity high. Artificial inactivation of Cdk1 after meiosis I results in DNA replication between the two divisions (Furuno et al. 1994; Iwabuchi et al. 2000; Nakajo et al. 2000). High Cdk1 activity inhibits the formation of the prereplicative complex by preventing binding of Mcm2–7 to replication origins. In Saccharomyces cerevisiae, a meiosis-specific protein kinase, Ime2, also contributes to phosphorylation of some of Cdk1 substrates to suppress replication between the two meiotic divisions (Holt et al. 2007).
PAIRING AND RECOMBINATION
Meiotic recombination exchanges the genetic materials between two homologous chromosomes. It is essential not only for exchanging genetic materials to generate diversity in offspring, but also for holding homologous chromosomes together through chiasma, to segregate chromosomes properly.
Homologous chromosomes pair along the whole length and this homologous paring is further stabilized by the formation of an elaborate structure, the synaptonemal complex. In yeast and mouse, meiotic recombination is required for proper synaptonemal complex formation (Loidl et al. 1994; Baudat et al. 2000; Romanienko and Camerini-Otero 2000), whereas in Drosophila and Caenorhabditis elegans, the synaptonemal complex can form independently of meiotic recombination (Dernburg et al. 1998; McKim et al. 1998).
Recombination mechanisms themselves are largely shared in both meiotic recombination and the homologous recombination repair process in the mitotic cell cycle, but there are crucial differences. In the case of meiosis, DNA double-strand breaks (DSBs) are obligatory rather than the result of accidental damage, as in the mitotic cell cycle. DSBs, which initiate meiotic recombination, are created by the conserved Spo11 endonuclease (Keeney et al. 1997). The sites of DSBs are not random, often clustering at meiotic recombination hot spots (Lichten and Goldman 1995). There is a line of evidence that the chromatin modifications are involved in the site selection of meiotic DSBs. In S. cerevisiae, methylation of histone 3 at lysine 4 (H3K4) coincides with sites of DSBs, and the H3K4 methyltransferase Set1 is required for DSB formation (Sollier et al. 2004; Borde et al. 2009). In mammalians, Prdm9, a H3K4 methyltransferase with a zinc finger domain, mediates the hot spot selection. The difference in the choice of hot spots among mouse strains was attributed to a difference in the amino acid sequence within the zinc finger domain (Baudat et al. 2010; Parvanov et al. 2010). In humans, the major Prdm9 isoform within the population was predicted and shown to specifically bind the known consensus sequence enriched near recombination hot spots (Baudat et al. 2010; Meyer et al. 2010). Furthermore, the allelic differences in the zinc finger domain are correlated to the usages of recombination hot spots in humans. Prdm9 is a fast-evolving protein in many animals, including humans (Oliver et al. 2010), and this rapid change is thought to counteract a loss of individual hot spots because of biased gene conversion during the recombination process (Nicolas et al. 1989).
The second difference is that the recombination partners are mainly homologous chromosomes in meiosis, whereas they are mainly sister chromatids in DNA repair during mitotic cycles (Kadyk and Hartwell 1992; Bzymek et al. 2010). This homolog bias in meiosis is crucial as recombination among sister chromatids would not be productive in terms of generating diversity or forming the chiasma that hold homologous chromosomes together during metaphase I. From studies in S. cerevisiae, the partner choice is thought to be mediated by strand exchange proteins (RecA homologs), Rad51 and Dmc1, which promote the invasion of single-stranded DNA into a double-stranded recombination partner. Rad51 is expressed both in mitotic cycles and meiosis and, on its own, promotes intersister chromatid recombination, whereas the meiosis-specific protein Dmc1, together with Rad51, promotes interhomolog recombination in meiosis (Cloud et al. 2012). In addition, interhomolog recombination is promoted in meiosis through suppression of Rad51 by two meiosis-specific factors: the kinase complex Red1/Hop1/Mek1 and the Rad51-interacting protein Hed1 (Busygina et al. 2008; Niu et al. 2009).
During recombination, a specific arrangement of chromosomes, called a bouquet, has been observed in a wide variety of organisms (Harper et al. 2004). In the bouquet arrangement, telomeres are attached to a specific area of the nuclear envelope. In the most well-studied organism, the fission yeast Schizosaccharomyces pombe, bouquet arrangement was shown to be associated with dynamic movement of the nucleus, which facilitates pairing and recombination (Fig. 1) (Chikashige et al. 1994). During fission yeast interphase, the spindle pole body (SPB) is associated with centromeres (Funabiki et al. 1993). At the onset of meiosis, the SPB switches its association from centromeres to telomeres (Chikashige et al. 1994). SUN and KASH domain proteins, together with Bqt1 and Bqt2, connect telomeres and cytoplasmic aster microtubules, which are organized by the meiosis-specific SPB protein Hrs1/Mcp6 (Saito et al. 2005; Tanaka et al. 2005; Chikashige et al. 2006). The dynein motor drives the oscillatory movement of the nucleus to facilitate homologous chromosome pairing (Yamamoto et al. 1999).
Figure 1.
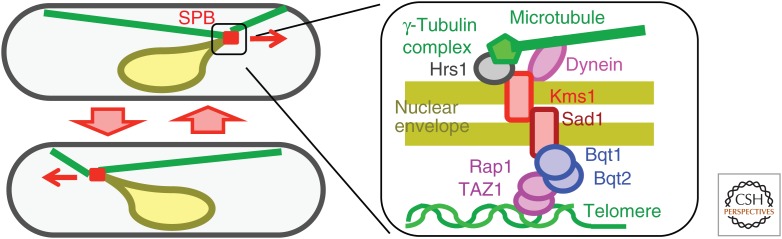
Bouquet formation and oscillatory nuclear movement in fission yeast meiosis. Clustering of telomeres and their linkage to the cytoskeleton enable oscillatory movement of the prophase nucleus and facilitate paring of homologous chromosomes. SPB, spindle pole body.
In C. elegans, the paring center near a telomere on each chromosome acts as the initiator of meiotic chromosome paring, and these pairing centers also interact with cytoplasmic dynein through links of SUN–KASH domain proteins, which span the nuclear envelope (Sato et al. 2009; Baudrimont et al. 2010; Wynne et al. 2012). Movement along the nuclear envelope, mediated by dynein, induces dynamic movement of pairing centers. In mouse spermatocytes, bouquet organization and microtubule-dependent nuclear movement were reported during early meiotic prophase (Scherthan et al. 1996; Morimoto et al. 2012). In addition, involvement of SUN–KASH domain proteins has been shown (Morimoto et al. 2012).
An interesting example is found in S. cerevisiae. Vigorous chromosome movement is observed in meiotic prophase I (Conrad et al. 2008; Koszul et al. 2008). Like other organisms, this chromosome movement is led by telomere cluster near spindle pole bodies, and a SUN domain protein is involved in this movement (Rao et al. 2011). Surprisingly, actin filaments, not microtubules, are responsible for this movement (Koszul et al. 2008).
POSTRECOMBINATION EVENTS
Compared with recombination and chromosome segregation, much less attention has been paid to the period between the two events in meiosis. However, this period is usually longest in meiosis. All mammalian oocytes arrest meiosis at birth until ovulation. This means that in human oocytes, arrest lasts up to 40 years. This prolonged arrest is linked to so-called maternal age effect in humans (Hassold and Hunt 2009). Maternal age effect is the phenomenon that the incidence of aneuploidy increases as the age of the mothers increases. The cause is still under intense discussion, but cohesin fatigue is one of the attractive hypotheses. In mitotic cycles, cohesin establishes at S phase and the same cohesin complex stays on chromosomes until mitosis (Uhlmann and Nasmyth 1998). If there is no new cohesin loading during the meiotic arrest, the same cohesin molecules have to keep chromatids together for decades. It is hypothesized that gradual loss of cohesin during the prolonged arrest probably increases the frequency of missegregation. Evidences suggest that cohesin does not turn over in mouse oocytes once it is established (Revenkova et al. 2010; Tachibana-Konwalski et al. 2010), and oocytes from old mothers have reduced cohesin on chromosomes in mouse (Lister et al. 2010).
During the postrecombination period, in some species, a compact cluster of chromosomes forms in the enlarged nucleus. In Drosophila oocytes, the structure was called the karyosome and forms soon after the completion of recombination (King 1970). Similar clustering of chromatin within the nucleus can be found within mammalian oocytes. In mouse oocytes, two types of chromatin organization were found in immature oocytes, which are often referred to as SN (surrounded nucleolus) and NSN (nonsurrounded nucleolus). In a nucleus with SN, meiotic chromosomes are clustered around the nucleolus with centromeres in proximity to the nucleolus. This clustered chromatin is also referred to as the karyosphere, and is also found in human oocytes (Parfenov et al. 1989). In mouse, oocytes with an SN configuration are more competent for further development after fertilization than ones with an NSN configuration (Zuccotti et al. 1998, 2002). From studies in Drosophila, it is proposed that clustering of meiotic chromosomes facilitates formation of one unified spindle (Lancaster et al. 2007). As oocytes have a large nucleus and cytoplasm and spindles assembled around chromosomes without centrosomes, chromosomes distant from each other may form separate spindles. Although chromosome clustering is a widespread phenomenon in oocytes, very few molecular studies have, so far, been performed on the molecular basis of this process. In Drosophila oocytes, karyosome formation requires the conserved kinase NHK-1 (Cullen et al. 2005; Ivanovska et al. 2005). A study identified barrier-to-autointegration factor (BAF), a linker protein between chromosomes and the nuclear envelope, as one of the critical substrates of NHK-1 in meiosis (Fig. 2) (Lancaster et al. 2007). It is proposed that the phosphorylation of BAF by NHK-1 is required for release of chromatin from the nuclear envelope to allow the karyosome formation. A further study showed that NHK-1 activity is suppressed by the meiotic recombination checkpoint to block nuclear reorganization, including karyosome formation, in response to unrepaired DSBs (Lancaster et al. 2010).
Figure 2.
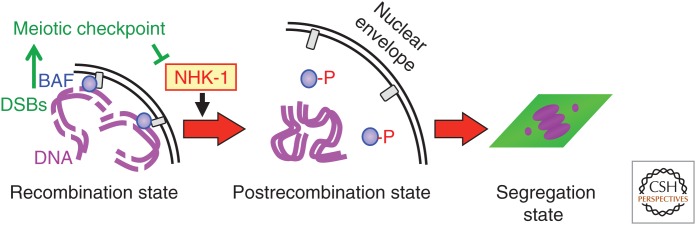
Formation of the karyosome in Drosophila oocytes. The conserved protein kinase NHK-1 phosphorylates barrier-to-autointegration factor (BAF), a linker between the nuclear envelope and chromatin, to release meiotic chromosomes from the nuclear envelope. The meiotic recombination checkpoint suppresses NHK-1 activity to keep the nucleus in the recombination state when DNA double-strand breaks (DSBs) are still present.
REDUCTIONAL AND EQUATIONAL CHROMOSOME SEGREGATION
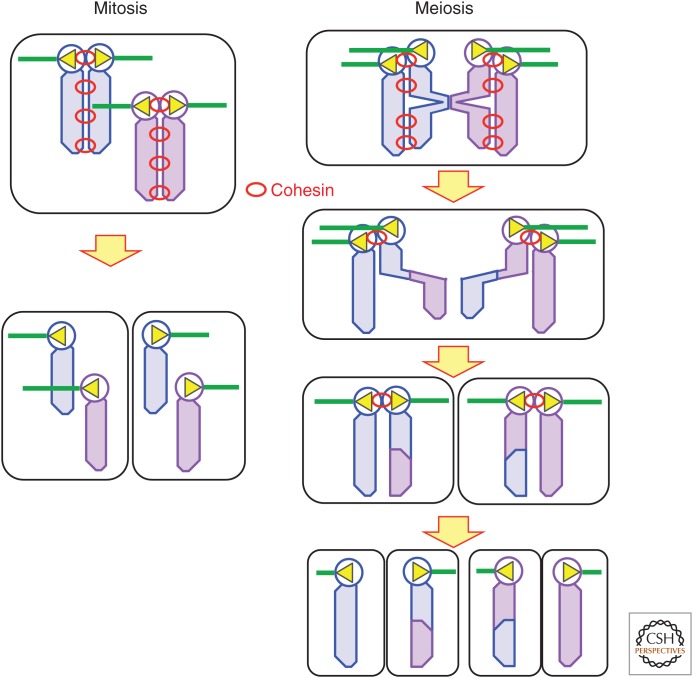
Homologous chromosomes are segregated in the first meiotic division, and sister chromatids are segregated in the second division. To achieve this, two major processes are specifically modified in meiosis in comparison with mitosis (Fig. 3).
Figure 3.
Reductional and equational chromosome segregation. Cohesin connects sister chromatids. In mitosis, sister kinetochores are attached to microtubules from the opposite poles. Cohesin connects sister chromatids and the removal of cohesin along chromosomes triggers sister chromatid separation. Homologous chromosomes behave independently. In meiosis I, sister kinetochores are attached to microtubules from the same pole. Homologous chromosomes are attached to the opposite poles and connected by chiasma. Destruction of cohesin from chromosome arms triggers homologous chromosome separation. Cohesin at centromeres is protected to provide a linkage among sister chromatids. In meiosis II, sister kinetochores are attached to microtubules from the opposite poles. Destruction of the centromeric cohesin triggers sister chromatid separation.
Monopolar Attachment of Sister Chromatids in Meiosis I
The first difference of meiosis from mitosis is the behavior of kinetochores to achieve bipolar attachment. In mitosis, sister kinetochores must attach to the opposite poles. In contrast, in meiosis I, sister kinetochores must attach to the same pole and homologous kinetochores must attach to the opposite poles. This is the key division that reduces the ploidy of cells in meiosis. In meiosis II, like mitosis, sister kinetochores must attach to the opposite poles.
In S. cerevisiae, the monopolin complex is responsible for monopolar orientation of sister kinetochores in meiosis I (Tóth et al. 2000; Rabitsch et al. 2003). The monopolin complex consists of casein kinase I and other regulatory subunits, and localizes to kinetochores in meiosis (Petronczki et al. 2006). Monopolin localization is dependent on Spo13, Polo kinase, and Cdc7 kinase (Clyne et al. 2003; Katis et al. 2004; Lee et al. 2004; Lo et al. 2008; Matos et al. 2008).
However, the involvement of the monopolin complex in monoorientation of sister kinetochores may be restricted to S. cerevisiae, which has a single microtubule attached to each kinetochore. In fission yeast, in which multiple microtubules attach to each kinetochore, the homologous complex of monopolin is required for preventing one kinetochore from attaching microtubules from opposite poles (so-called merotelic attachment) in mitosis (Gregan et al. 2007). Therefore, it is hypothesized that the molecular function of monopolin is to clamp microtubule attachment sites together between two sister kinetochores in the case of S. cerevisiae, and within a single kinetochore in mitosis in other organisms, which have multiple microtubules attached to one kinetochore (Gregan et al. 2007; Corbett et al. 2010). Instead, in fission yeast, mono-orientation of sister kinetochores is dependent on the meiosis-specific cohesin subunit Rec8, as well as the meiosis-specific protein Moa1, which localizes to kinetochores until metaphase I (Watanabe et al. 2001; Yokobayashi and Watanabe 2005). Rec8 and Moa1 interact with each other, but the molecular function of Moa1 remains to be understood. During meiosis I, the ability of the meiotic cohesin complex–containing Rec8 to localize at the core centromeres is necessary to promote mono-orientation of sister kinetochores, whereas the mitotic cohesin localizes at pericentrometic regions, not core centromeres, to promote biorientation of sister kinetochores (Sakuno et al. 2009). Therefore, it is hypothesized that linking two sister centromeres by cohesin brings meiosis I–specific kinetochore configuration.
Stepwise Removal of Cohesin
The second difference of meiosis from mitosis is stepwise removal of cohesin from chromosomes. Cohesin connects sister chromatids consisting of replicated DNA (Nasmyth and Haering 2009). In mitotic metaphase, cohesin resists the pulling forces acting on kinetochores toward the opposite poles. The cohesin complex is removed either by phosphorylation or cleavage of one of the subunits, Scc1. This removal triggers the separation of sister chromatids.
In contrast, in meiosis I, homologous chromosomes are connected by chiasma and pulled toward the opposite poles. This connection depends on cohesin localized among the sister chromatids distal to the chiasma. Removal of cohesin from chromosome arms abolishes the connection and triggers anaphase. The crucial difference from mitosis is that cohesin at centromeres must be protected in the metaphase/anaphase transition in meiosis I. This centromeric cohesin maintains a link among sister chromatids until anaphase II, when the remaining cohesin is removed. In most organisms, the meiosis-specific cohesin subunit Rec8 replaces Scc1 (Watanabe and Nurse 1999). A conserved protein, called Shugoshin (Sgo), is responsible for this centromere protection (Kitajima et al. 2004). Mei-S332 in Drosophila was the first identified member of Sgo. Mutants in mei-S332 showed precocious separation of sister chromatids in anaphase I, leading to missegregation of sister chromatid in the second meiotic division (Kerrebrock et al. 1992). Later, this was shown to be widely conserved in eukaryotes when a homolog of mei-S332, Sgo1, was identified in fission yeast as a protein that protects the meiotic cohesin subunit Rec8 from proteolysis in the centromeric regions in anaphase I (Kitajima et al. 2004). Both Rec8 and Sgo1 are expressed only in meiosis, and forced expression of both proteins in mitotic cells blocks nuclear division. Sgo recruits protein phosphatase 2A (PP2A) to centromeric regions and constantly dephosphorylates cohesin (Kitajima et al. 2006; Riedel et al. 2006). As phosphorylation of cohesin is required for cleavage, Sgo protects meiotic cohesin from cleavage in anaphase I. Sgo itself is recruited to centromeres by phosphorylation of histone 2A by Bub1 kinase (Kawashima et al. 2010).
In addition to the roles in meiosis, Sgo also has roles in ensuring the accuracy of chromosome segregation in mitosis (Yao and Dai 2012). Although the molecular mechanism is still under investigation, evidence showed that it recruits and regulates various proteins at centromeres, including PP2A, the chromosomal passenger complex (CPC), and the microtubule-depolymerizing kinesin MCAK (Tanno et al. 2010; Rivera et al. 2012). The identification and subsequent studies of Sgo are a good example of how the studies of meiosis have made crucial contributions to the understanding of mitosis.
ACENTROSOMAL SPINDLE FORMATION
A spindle in oocytes differs from a mitotic spindle in some key aspects. Remarkably, a spindle forms without centrosomes in the oocytes of many animals, including humans, mouse, Xenopus, Drosophila, and C. elegans (McKim and Hawley 1995). This is specific to the oocyte not meiosis in general, as spermatocytes still contain centrosomes that drive spindle formation. Centrosomes must be eliminated or inactivated during oogenesis, but the mechanism of this is not understood. Lack of centrosomes in oocytes raises a question as to how spindle microtubules are assembled. An in vitro spindle-assembly system in Xenopus extract played critical roles in solving the problem. It was shown that beads coated with random DNA can assemble a bipolar spindle in Xenopus extract (Heald et al. 1996). This indicates any DNA can recruit proteins that induce microtubule assembly. It revealed the central role of the Ran system in chromatin-mediated assembly of spindle microtubules (Gruss et al. 2001; Wiese et al. 2001). The Ran system was originally identified for nuclear transport, but, subsequently, identified for spindle assembly and nuclear envelope reassembly (Hetzer et al. 2002). Ran is a small G protein that can be switched between GTP- and GDP-binding forms (Fig. 4). A chromosome-associated protein, Rcc1, acts as a guanine nucleotide-exchanging factor (GEF), which converts Ran-GDP to Ran-GTP to generate the Ran-GTP gradient. Ran-GTP binds to importin by removing it from other binding proteins, including some “spindle-assembly factors,” which promote spindle assembly. Away from the chromosomes in which the Ran-GDP form is dominating, these spindle-assembly factors are kept inactive by binding to importin. Near the chromosomes in which Ran-GTP concentration is high, the spindle-assembly factors are released from importin to become active. These spindle-assembly factors include TPX2, NuMA, NuSAP, and HURP, and these collectively promote microtubule stabilization and bipolar spindle formation (Gruss et al. 2001; Wiese et al. 2001; Koffa et al. 2006; Ribbeck et al. 2006; Sillje et al. 2006).
Figure 4.
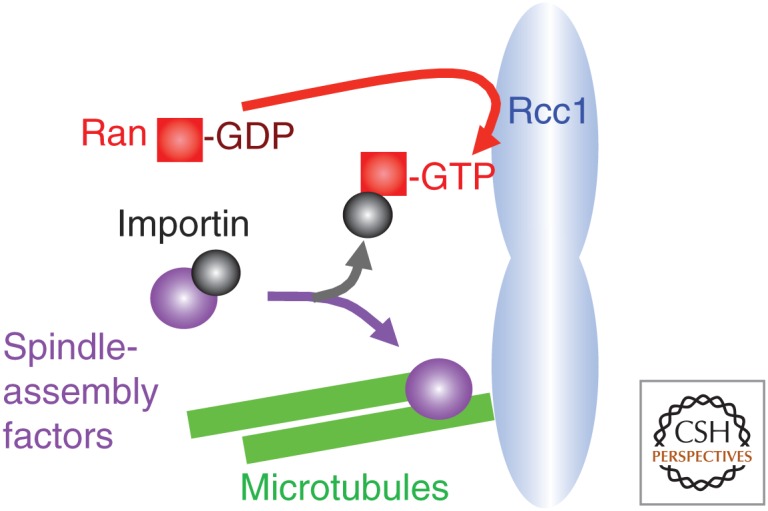
Chromosome-mediated spindle assembly through Ran. Ran-GDP is converted into Ran-GTP by chromosome-associated RCC1 to generate a Ran-GTP gradient. Near chromosomes, Ran-GTP activates spindle-assembly factors by removing importin from them. Microtubules and the spindle can be assembled only in proximity to the chromosomes.
The requirement of the Ran-GTP gradient-based mechanisms in chromosome-mediated acentrosomal spindle assembly is very clear in Xenopus extract, but it is less clear in living oocytes. Disrupting the Ran gradient by either expression of dominant negative or hyperactive forms of Ran did not prevent a spindle from forming around the chromosomes in mouse oocytes (Dumont et al. 2007). Similar observations have been made in Drosophila oocytes, although the gradient was not directly monitored (Cesario and McKim 2011). This indicates that chromosomes have alternative pathways or signals that induce microtubule assembly independently from Ran. The CPC-containing Aurora B kinase may act as an alternative pathway. First, in Xenopus egg extract, the CPC is essential for centrosome-independent spindle microtubule assembly (Sampath et al. 2004). Also, in Drosophila oocytes, it was shown that CPC is essential for spindle microtubule assembly (Colombie et al. 2008; Radford et al. 2012). Chromosomes activate Aurora B kinase independently of Ran and the activated kinase is then targeted to microtubules to promote spindle assembly (Tseng et al. 2010). The targets of the kinase activity include two microtubule-depolymerizing proteins, MCAK and Op18/stathmin (Andrews et al. 2004; Ohi et al. 2004; Gadea and Ruderman 2006).
Although it was known that the mitotic spindle forms without centrosomes in plants, it was, relatively, recently realized that the spindle can form in mitotic animal cells without centrosomes when they are artificially removed. In human cultured cells, when centrosomes were ablated using a laser, the spindle morphology and function were unaffected (Khodjakov et al. 2000). In Drosophila, inactivating essential centrosome components eliminated centrosomes from cells, but spindle formation and function is not disrupted (Basto et al. 2006). Furthermore, the flies lacking centrosomes develop with only a slight increase in the frequency of aneuploids.
As a mitotic spindle can form without centrosomes, a critical question is whether a meiotic spindle is simply the same as a mitotic spindle without centrosomes, or a spindle that is modified to cope with a lack of centrosomes? This is an unexplored question, but some evidence suggests the existence of oocyte-specific mechanisms to compensate for a lack of centrosomes. For example, in Drosophila mitosis, the γ-tubulin recruiting complex augmin is responsible for assembling most centrosome-independent spindle microtubules (Goshima et al. 2008). Therefore, a loss of the augmin complex, in conjunction with inactivation of centrosomes, results in a dramatic loss of spindle microtubules (Goshima et al. 2008; Wainman et al. 2009). In contrast, oocytes lacking the augmin complex (and centrosomes) still form robust spindles (Meireles et al. 2009). This suggests a meiosis-specific microtubule assembly pathway independent of centrosomes and augmin. Moreover, augmin shows meiosis-specific stable localization to acentrosomal spindle poles, suggesting that the meiosis-specific regulation of augmin may, in part, compensate for a lack of centrosomes in oocytes (Colombie et al. 2013).
ASYMMETRIC DIVISION OF OOCYTES
Meiosis produces four daughter haploid cells from one diploid oocyte. In the case of oogenesis, only one daughter becomes an egg and the others (polar bodies) will not participate in reproduction. Oocytes divide asymmetrically in each division to minimize a loss of the cytoplasm. For successful asymmetric division, the spindle must be positioned near the cell cortex and oriented perpendicularly to the cell cortex. Considerable studies on asymmetric divisions have been performed in mitosis, highlighting the critical roles of centrosomes and interaction between aster microtubules and the cell cortex (Knoblich 2010). Without centrosomes in oocytes, how does the meiotic spindle become oriented and positioned? Studies in mouse oocytes showed that instead of microtubules, the dynamic actin network plays a crucial role in the positioning of the meiotic spindle. The actin network in oocytes is formed by cooperative actions of the actin nucleators, Formin-2 (Fmn2), Spire 1, and Spire 2 (Azoury et al. 2008; Schuh and Ellenberg 2008; Pfender et al. 2011). Rab18a-positive vesicles serve as nodes of the network to regulate the density and myosin IVb-dependent dynamics (Holubcová et al. 2013). Transient destabilization of actin filaments caused by temporal degradation of Fmn2 is required for initial migration (Azoury et al. 2011).
Similarly, the meiosis II spindle needs to be positioned near the cortex. In mouse meiosis II, this is maintained by a flow of actin away from the spindle along the cortex and toward the spindle from the other side of the oocyte (Yi et al. 2011). This flow is driven by Arp2/3, N-WASP, and myosin II. A similar cytoplasmic flow was also observed in the late stage of the spindle migration in meiosis I.
INTERACTION BETWEEN THE SPINDLE AND CHROMOSOMES
Chromosome–microtubule interactions in oocytes may be “different” from those in mitosis. In mitosis, the main interaction is provided by kinetochores, which interact with dynamic microtubule ends. In the simplest model of mitosis, microtubules nucleated from centrosomes capture kinetochores and generate pulling forces (the “search and capture” model) (Kirschner and Mitchison 1986). When sister kinetochores are attached to microtubules from the opposite poles, chromosomes becomes congressed to the metaphase plate. The pulling forces acting between kinetochores and the opposite poles are resisted by cohesion among sister chromatids, and destruction of cohesin at the onset of anaphase triggers the movement of sister chromatids toward the poles. Although kinetochores are also important in meiosis, nonkinetochore interactions seem more prominent in oocytes than in mitotic cells.
In mouse, it has been shown that kinetochore-microtubule end-on attachment is not properly established until well after chromosome congression at the spindle equator (Brunet et al. 1999). Chromosomes move toward the spindle equator by sliding along the surface of the spindle without end-on attachment, leading to ring arrangement of chromosomes at the spindle equator (Kitajima et al. 2011). This congression is followed by trial-and-error establishment of bipolar end-on attachment of homologous kinetochores at the spindle equator. Full stable end-on attachment will not be achieved until several hours after nuclear envelope breakdown, and the delay of end-on attachment in oocytes appears to be caused by slow gradual increase of Cdk1 activity (Davydenko et al. 2013). An artificial premature increase of Cdk1 activity resulted in the premature establishment of attachment. As this also increased the lagging chromosomes in anaphase I, slow increase of Cdk1 activity is proposed to delay stable attachment until spindle bipolarity is established. It remains to be established how the chromosomes congress to the spindle equator without end-on microtubule attachment to kinetochores or how a gradual increase of Cdk delays the microtubule attachment to kinetochores.
Observations in C. elegans oocytes also indicated different contributions of kinetochores in meiosis to those in mitosis. First, microtubules appear to interact with chromosomes laterally during chromosome congression. This congression is at least partly mediated by the chromokinesin KLP-19, which localizes to the junction among the homologs (Wignall and Villeneuve 2009). Furthermore, inactivation of kinetochores by RNA interference (RNAi) resulted in less tight congression and misorientation of chromosomes relative to the spindle axis. Surprisingly, chromosomes without active kinetochores can separate during anaphase at a speed comparable with the wild type (Dumont et al. 2010). Anaphase chromosome movement seems to be driven by the elongation of spindle microtubules among separating homologous chromosomes. However, it should be noted that C. elegans centromeres are not restricted to small regions, as kinetochores are formed along proximal parts of chromosomes in meiosis (Dumont et al. 2010).
How do the chromosomes move without end-on attachment in oocytes? Even in mitosis, there is evidence of such forces acting on chromosomes. Polar ejection forces act on chromosome arms and are involved in chromosome congression at the metaphase plate (Rieder and Salmon 1994). When chromosomes were artificially cut, a chromosome fragment that lacked kinetochores moved toward the spindle equator (Rieder et al. 1986). Chromokinesins play a part in polar ejection forces, but interaction of chromosome arms with growing microtubule plus ends can also generate such forces. In the case of Drosophila oocytes, the chromokinesin Nod is thought to generate polar ejection forces (Theurkauf and Hawley 1992; Matthies et al. 1999). Nod is an immotile kinesin but can promote microtubule polymerization (Cui et al. 2005). In mouse oocytes, the chromokinesin Kid is dispensable for chromosome congression (Kitajima et al. 2011).
SPINDLE-ASSEMBLY CHECKPOINT
The spindle-assembly checkpoint is a mechanism to ensure the correct segregation of chromosomes and is crucial for genome stability. It monitors a lack of microtubule attachment to kinetochores and a lack of tension to block or delay anaphase onset through inhibition of anaphase-promoting complex/cyclosome (APC/C) (Lara-Gonzalez et al. 2012).
There are lines of evidence that suggest that the spindle-assembly checkpoint in meiosis is not robust as in mitosis. This is evident especially in oocytes, which display a high incidence of chromosome missegregation. In mouse oocytes, several studies show that anaphase I can initiate without all chromosomes achieving proper bipolar attachment, metaphase alignment, or interkinetochore tension (LeMaire-Adkins et al. 1997; Nagaoka et al. 2011; Kolano et al. 2012). In Xenopus oocytes, inhibition of spindle microtubules or bipolar spindle formation did not delay the onset of anaphase I (Shao et al. 2013). This lack of a robust spindle checkpoint in oocytes may be one of the reasons why meiosis in human oocytes shows a high level of chromosome missegregation.
Although tension among homologous chromosomes, not sister chromatids, has to be detected in meiosis I, the molecular mechanism may be shared with mitosis. During meiotic prometaphase I in yeast and mouse oocytes, the CPC is essential for releasing incorrect attachments (Kitajima et al. 2011; Meyer et al. 2013) to achieve the bipolar attachment of homologs. In Drosophila oocytes, it has been shown that the CPC is required for bipolar attachment (Resnick et al. 2009). Therefore, the requirement of the CPC in correcting erroneous attachment is universal in mitosis and meiosis, and conserved among eukaryotes.
CONCLUDING REMARKS
The study of meiosis has a long history, but far fewer studies have been performed on meiosis in comparison with mitosis, partly because of technical challenges. Although studies of meiosis often generated results that are largely extensions of what is already known in mitosis, some studies have revealed unexpected functions and regulations of meiosis. In some cases, they had impacts well beyond meiosis, especially on the understanding of mitosis. Recent studies have resulted in many important findings, and many more exciting discoveries are still waiting to come.
ACKNOWLEDGMENTS
I thank Robin Beaven for valuable comments. H.O. holds a Wellcome Trust Senior Research Fellowship in Basic Biomedical Science.
Footnotes
Editors: Mitsuhiro Yanagida, Anthony A. Hyman, and Jonathon Pines
Additional Perspectives on Mitosis available at www.cshperspectives.org
REFERENCES
- Andrews PD, Ovechkina Y, Morrice N, Wagenbach M, Duncan K, Wordeman L, Swedlow JR. 2004. Aurora B regulates MCAK at the mitotic centromere. Dev Cell 6: 253–268. [DOI] [PubMed] [Google Scholar]
- Azoury J, Lee KW, Georget V, Rassinier P, Leader B, Verlhac MH. 2008. Spindle positioning in mouse oocytes relies on a dynamic meshwork of actin filaments. Curr Biol 18: 1514–1519. [DOI] [PubMed] [Google Scholar]
- Azoury J, Lee KW, Georget V, Hikal P, Verlhac MH. 2011. Symmetry breaking in mouse oocytes requires transient F-actin meshwork destabilization. Development 138: 2903–2908. [DOI] [PubMed] [Google Scholar]
- Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW. 2006. Flies without centrioles. Cell 125: 1375–1386. [DOI] [PubMed] [Google Scholar]
- Baudat F, Manova K, Yuen JP, Jasin M, Keeney S. 2000. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol Cell 6: 989–998. [DOI] [PubMed] [Google Scholar]
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B. 2010. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327: 836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudrimont A, Penkner A, Woglar A, Machacek T, Wegrostek C, Gloggnitzer J, Fridkin A, Klein F, Gruenbaum Y, Pasierbek P, et al. 2010. Leptotene/zygotene chromosome movement via the SUN/KASH protein bridge in Caenorhabditis elegans. PLoS Genet 6: e1001219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borde V, Robine N, Lin W, Bonfils S, Géli V, Nicolas A. 2009. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J 28: 99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet S, Maria AS, Guillaud P, Dujardin D, Kubiak JZ, Maro B. 1999. Kinetochore fibers are not involved in the formation of the first meiotic spindle in mouse oocytes, but control the exit from the first meiotic M phase. J Cell Biol 146: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busygina V, Sehorn MG, Shi IY, Tsubouchi H, Roeder GS, Sung P. 2008. Hed1 regulates Rad51-mediated recombination via a novel mechanism. Genes Dev 22: 786–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzymek M, Thayer NH, Oh SD, Kleckner N, Hunter N, 2010. Double Holliday junctions are intermediates of DNA break repair. Nature 464: 937–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesario J, McKim KS. 2011. RanGTP is required for meiotic spindle organization and the initiation of embryonic development in Drosophila. J Cell Sci 124: 3797–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikashige Y, Ding DQ, Funabiki H, Haraguchi T, Mashiko S, Yanagida M, Hiraoka Y. 1994. Telomere-led premeiotic chromosome movement in fission yeast. Science 264: 270–273. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Tsutsumi C, Yamane M, Okamasa K, Haraguchi T, Hiraoka Y. 2006. Meiotic proteins bqt1 and bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell 125: 59–69. [DOI] [PubMed] [Google Scholar]
- Cloud V, Chan YL, Grubb J, Budke B, Bishop DK. 2012. Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science 337: 1222–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne RK, Katis VL, Jessop L, Benjamin KR, Herskowitz I, Lichten M, Nasmyth K. 2003. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat Cell Biol 5: 480–485. [DOI] [PubMed] [Google Scholar]
- Colombié N, Cullen CF, Brittle AL, Jang JK, Earnshaw WC, Carmena M, McKim K, Ohkura H. 2008. Dual roles of Incenp crucial to the assembly of the acentrosomal metaphase spindle in female meiosis. Development 135: 3239–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombié N, Głuszek AA, Meireles AM, Ohkura H. 2013. Meiosis-specific stable binding of augmin to acentrosomal spindle poles promotes biased microtubule assembly in oocytes. PLoS Genet 9: e1003562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad MN, Lee CY, Chao G, Shinohara M, Kosaka H, Shinohara A, Conchello JA, Dresser ME. 2008. Rapid telomere movement in meiotic prophase is promoted by NDJ1, MPS3, and CSM4 and is modulated by recombination. Cell 133: 1175–1187. [DOI] [PubMed] [Google Scholar]
- Corbett KD, Yip CK, Ee LS, Walz T, Amon A, Harrison SC. 2010. The monopolin complex crosslinks kinetochore components to regulate chromosome-microtubule attachments. Cell 142: 556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W, Sproul LR, Gustafson SM, Matthies HJ, Gilbert SP, Hawley RS. 2005. Drosophila Nod protein binds preferentially to the plus ends of microtubules and promotes microtubule polymerization in vitro. Mol Biol Cell 16: 5400–5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen CF, Brittle AL, Ito T, Ohkura H. 2005. The conserved kinase NHK-1 is essential for mitotic progression and unifying acentrosomal meiotic spindles in Drosophila melanogaster. J Cell Biol 171: 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydenko O, Schultz RM, Lampson MA. 2013. Increased CDK1 activity determines the timing of kinetochore-microtubule attachments in meiosis I. J Cell Biol 202: 221–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve AM. 1998. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94: 387–398. [DOI] [PubMed] [Google Scholar]
- Dumont J, Petri S, Pellegrin F, Terret ME, Bohnsack MT, Rassinier P, Georget V, Kalab P, Gruss OJ, Verlhac MH. 2007. A centriole- and RanGTP-independent spindle assembly pathway in meiosis I of vertebrate oocytes. J Cell Biol 176: 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont J, Oegema K, Desai A. 2010. A kinetochore-independent mechanism drives anaphase chromosome separation during acentrosomal meiosis. Nat Cell Biol 12: 894–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Hagan I, Uzawa S, Yanagida M. 1993. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol 121: 961–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuno N, Nishizawa M, Okazaki K, Tanaka H, Iwashita J, Nakajo N, Ogawa Y, Sagata N. 1994. Suppression of DNA replication via Mos function during meiotic divisions in Xenopus oocytes. EMBO J 13: 2399–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadea BB, Ruderman JV. 2006. Aurora B is required for mitotic chromatin-induced phosphorylation of Op18/Stathmin. Proc Natl Acad Sci 103: 4493–4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Mayer M, Zhang N, Stuurman N, Vale RD. 2008. Augmin: A protein complex required for centrosome-independent microtubule generation within the spindle. J Cell Biol 181: 421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregan J, Riedel CG, Pidoux AL, Katou Y, Rumpf C, Schleiffer A, Kearsey SE, Shirahige K, Allshire RC, Nasmyth K. 2007. The kinetochore proteins Pcs1 and Mde4 and heterochromatin are required to prevent merotelic orientation. Curr Biol 17: 1190–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss OJ, Carazo-Salas RE, Schatz CA, Guarguaglini G, Kast J, Wilm M, Le Bot N, Vernos I, Karsenti E, Mattaj IW. 2001. Ran induces spindle assembly by reversing the inhibitory effect of importin α on TPX2 activity. Cell 104: 83–93. [DOI] [PubMed] [Google Scholar]
- Harper L, Golubovskaya I, Cande WZ. 2004. A bouquet of chromosomes. J Cell Sci 117: 4025–4032. [DOI] [PubMed] [Google Scholar]
- Hassold T, Hunt P. 2001. To err (meiotically) is human: The genesis of human aneuploidy. Nat Rev Genet 2: 280–291. [DOI] [PubMed] [Google Scholar]
- Hassold T, Hunt P. 2009. Maternal age and chromosomally abnormal pregnancies: What we know and what we wish we knew. Curr Opin Pediatr 21: 703–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. 1996. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature 382: 420–425. [DOI] [PubMed] [Google Scholar]
- Hetzer M, Gruss OJ, Mattaj IW. 2002. The Ran GTPase as a marker of chromosome position in spindle formation and nuclear envelope assembly. Nat Cell Biol 4: E177–E184. [DOI] [PubMed] [Google Scholar]
- Holt LJ, Hutti JE, Cantley LC, Morgan DO. 2007. Evolution of Ime2 phosphorylation sites on Cdk1 substrates provides a mechanism to limit the effects of the phosphatase Cdc14 in meiosis. Mol Cell 25: 689–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holubcová Z, Howard G, Schuh M. 2013. Vesicles modulate an actin network for asymmetric spindle positioning. Nat Cell Biol 15: 937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovska I, Khandan T, Ito T, Orr-Weaver TL. 2005. A histone code in meiosis: The histone kinase, NHK-1, is required for proper chromosomal architecture in Drosophila oocytes. Genes Dev 19: 2571–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabuchi M, Ohsumi K, Yamamoto TM, Sawada W, Kishimoto T. 2000. Residual Cdc2 activity remaining at meiosis I exit is essential for meiotic M-M transition in Xenopus oocyte extracts. EMBO J 19: 4513–4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk LC, Hartwell LH. 1992. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132: 387–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katis VL, Matos J, Mori S, Shirahige K, Zachariae W, Nasmyth K. 2004. Spo13 facilitates monopolin recruitment to kinetochores and regulates maintenance of centromeric cohesion during yeast meiosis. Curr Biol 14: 2183–2196. [DOI] [PubMed] [Google Scholar]
- Kawashima SA, Yamagishi Y, Honda T, Ishiguro K, Watanabe Y. 2010. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science 327: 172–177. [DOI] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N. 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384. [DOI] [PubMed] [Google Scholar]
- Kerrebrock AW, Miyazaki WY, Birnby D, Orr-Weaver TL. 1992. The Drosophila mei-S332 gene promotes sister-chromatid cohesion in meiosis following kinetochore differentiation. Genetics 130: 827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, Cole RW, Oakley BR, Rieder CL. 2000. Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol 10: 59–67. [DOI] [PubMed] [Google Scholar]
- King RC. 1970. The meiotic behavior of the Drosophila oocyte. Int Rev Cytol 28: 125–168. [DOI] [PubMed] [Google Scholar]
- Kirschner M, Mitchison T. 1986. Beyond self-assembly: From microtubules to morphogenesis. Cell 45: 329–342. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Kawashima SA, Watanabe Y. 2004. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature 427: 510–517. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Sakuno T, Ishiguro K, Iemura S, Natsume T, Kawashima SA, Watanabe Y. 2006. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature 441: 46–52. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Ohsugi M, Ellenberg J. 2011. Complete kinetochore tracking reveals error-prone homologous chromosome biorientation in mammalian oocytes. Cell 146: 568–581. [DOI] [PubMed] [Google Scholar]
- Knoblich JA. 2010. Asymmetric cell division: Recent developments and their implications for tumour biology. Nat Rev Mol Cell Biol 11: 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koffa MD, Casanova CM, Santarella R, Köcher T, Wilm M, Mattaj IW. 2006. HURP is part of a Ran-dependent complex involved in spindle formation. Curr Biol 16: 743–754. [DOI] [PubMed] [Google Scholar]
- Kolano A, Brunet S, Silk AD, Cleveland DW, Verlhac MH. 2012. Error-prone mammalian female meiosis from silencing the spindle assembly checkpoint without normal interkinetochore tension. Proc Natl Acad Sci 109: E1858–E1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul R, Kim KP, Prentiss M, Kleckner N, Kameoka S. 2008. Meiotic chromosomes move by linkage to dynamic actin cables with transduction of force through the nuclear envelope. Cell 133: 1188–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster OM, Cullen CF, Ohkura H. 2007. NHK-1 phosphorylates BAF to allow karyosome formation in the Drosophila oocyte nucleus. J Cell Biol 179: 817–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster OM, Breuer M, Cullen CF, Ito T, Ohkura H. 2010. The meiotic recombination checkpoint suppresses NHK-1 kinase to prevent reorganisation of the oocyte nucleus in Drosophila. PLoS Genet 6: e1001179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Gonzalez P, Westhorpe FG, Taylor SS. 2012. The spindle assembly checkpoint. Curr Biol 22: R966–R980. [DOI] [PubMed] [Google Scholar]
- Lee BH, Kiburz BM, Amon A. 2004. Spo13 maintains centromeric cohesion and kinetochore coorientation during meiosis I. Curr Biol 14: 2168–2182. [DOI] [PubMed] [Google Scholar]
- LeMaire-Adkins R, Radke K, Hunt PA. 1997. Lack of checkpoint control at the metaphase/anaphase transition: A mechanism of meiotic nondisjunction in mammalian females. J Cell Biol 139: 1611–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten M, Goldman AS. 1995. Meiotic recombination hotspots. Annu Rev Genet 29: 423–444. [DOI] [PubMed] [Google Scholar]
- Lister LM, Kouznetsova A, Hyslop LA, Kalleas D, Pace SL, Barel JC, Nathan A, Floros V, Adelfalk C, Watanabe Y, et al. 2010. Age-related meiotic segregation errors in mammalian oocytes are preceded by depletion of cohesin and Sgo2. Curr Biol 20: 1511–1521. [DOI] [PubMed] [Google Scholar]
- Lo HC, Wan L, Rosebrock A, Futcher B, Hollingsworth NM. 2008. Cdc7-Dbf4 regulates NDT80 transcription as well as reductional segregation during budding yeast meiosis. Mol Biol Cell 19: 4956–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J, Klein F, Scherthan H. 1994. Homologous pairing is reduced but not abolished in asynaptic mutants of yeast. J Cell Biol 125: 1191–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J, Lipp JJ, Bogdanova A, Guillot S, Okaz E, Junqueira M, Shevchenko A, Zachariae W. 2008. Dbf4-dependent CDC7 kinase links DNA replication to the segregation of homologous chromosomes in meiosis I. Cell 135: 662–678. [DOI] [PubMed] [Google Scholar]
- Matthies HJ, Messina LG, Namba R, Greer KJ, Walker MY, Hawley RS. 1999. Mutations in the α-tubulin 67C gene specifically impair achiasmate segregation in Drosophila melanogaster. J Cell Biol 147: 1137–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim KS, Hawley RS. 1995. Chromosomal control of meiotic cell division. Science 270: 1595–1601. [DOI] [PubMed] [Google Scholar]
- McKim KS, Green-Marroquin BL, Sekelsky JJ, Chin G, Steinberg C, Khodosh R, Hawley RS. 1998. Meiotic synapsis in the absence of recombination. Science 279: 876–878. [DOI] [PubMed] [Google Scholar]
- Meireles AM, Fisher KH, Colombié N, Wakefield JG, Ohkura H. 2009. Wac: A new Augmin subunit required for chromosome alignment but not for acentrosomal microtubule assembly in female meiosis. J Cell Biol 184: 777–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RE, Kim S, Obeso D, Straight PD, Winey M, Dawson DS. 2013. Mps1 and Ipl1/Aurora B act sequentially to correctly orient chromosomes on the meiotic spindle of budding yeast. Science 339: 1071–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto A, Shibuya H, Zhu X, Kim J, Ishiguro K, Han M, Watanabe Y. 2012. A conserved KASH domain protein associates with telomeres, SUN1, and dynactin during mammalian meiosis. J Cell Biol 198: 165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, MacFie TS, McVean G, Donnelly P. 2010. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327: 876–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka SI, Hodges CA, Albertini DF, Hunt PA. 2011. Oocyte-specific differences in cell-cycle control create an innate susceptibility to meiotic errors. Curr Biol 21: 651–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajo N, Yoshitome S, Iwashita J, Iida M, Uto K, Ueno S, Okamoto K, Sagata N. 2000. Absence of Wee1 ensures the meiotic cell cycle in Xenopus oocytes. Genes Dev 14: 328–338. [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH. 2009. Cohesin: Its roles and mechanisms. Annu Rev Genet 43: 525–558. [DOI] [PubMed] [Google Scholar]
- Nicolas A, Treco D, Schultes NP, Szostak JW. 1989. An initiation site for meiotic gene conversion in the yeast Saccharomyces cerevisiae. Nature 338: 35–39. [DOI] [PubMed] [Google Scholar]
- Niu H, Wan L, Busygina V, Kwon Y, Allen JA, Li X, Kunz RC, Kubota K, Wang B, Sung P, et al. 2009. Regulation of meiotic recombination via Mek1-mediated Rad54 phosphorylation. Mol Cell 36: 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi R, Sapra T, Howard J, Mitchison TJ. 2004. Differentiation of cytoplasmic and meiotic spindle assembly MCAK functions by Aurora B–dependent phosphorylation. Mol Biol Cell 15: 2895–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver PL, Goodstadt L, Bayes JJ, Birtle Z, Roach KC, Phadnis N, Beatson SA, Lunter G, Malik HS, Pointing CP, 2010. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet 5: e1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfenov V, Potchukalina G, Dudina L, Kostyuchek D, Gruzova M. 1989. Human antral follicles: Oocyte nucleus and the karyosphere formation (electron microscopic and autoradiographic data). Gamete Res 22: 219–231. [DOI] [PubMed] [Google Scholar]
- Parvanov ED, Petkov PM, Paigen K. 2010. Prdm9 controls activation of mammalian recombination hotspots. Science 327: 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M, Matos J, Mori S, Gregan J, Bogdanova A, Schwickart M, Mechtler K, Shirahige K, Zachariae W, Nasmyth K. 2006. Monopolar attachment of sister kinetochores at meiosis I requires casein kinase 1. Cell 126: 1049–1064. [DOI] [PubMed] [Google Scholar]
- Pfender S, Kuznetsov V, Pleiser S, Kerkhoff E, Schuh M. 2011. Spire-type actin nucleators cooperate with Formin-2 to drive asymmetric oocyte division. Curr Biol 21: 955–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabitsch KP, Petronczki M, Javerzat JP, Genier S, Chwalla B, Schleiffer A, Tanaka TU, Nasmyth K. 2003. Kinetochore recruitment of two nucleolar proteins is required for homolog segregation in meiosis I. Dev Cell 4: 535–548. [DOI] [PubMed] [Google Scholar]
- Radford SJ, Jang JK, McKim KS. 2012. The chromosomal passenger complex is required for meiotic acentrosomal spindle assembly and chromosome biorientation. Genetics 192: 417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao HB, Shinohara M, Shinohara A. 2011. Mps3 SUN domain is important for chromosome motion and juxtaposition of homologous chromosomes during meiosis. Genes Cells 16: 1081–1096. [DOI] [PubMed] [Google Scholar]
- Resnick TD, Dej KJ, Xiang Y, Hawley RS, Ahn C, Orr-Weaver TL. 2009. Mutations in the chromosomal passenger complex and the condensin complex differentially affect synaptonemal complex disassembly and metaphase I configuration in Drosophila female meiosis. Genetics 181: 875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenkova E, Herrmann K, Adelfalk C, Jessberger R. 2010. Oocyte cohesin expression restricted to predictyate stages provides full fertility and prevents aneuploidy. Curr Biol 20: 1529–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K, Groen AC, Santarella R, Bohnsack MT, Raemaekers T, Köcher T, Gentzel M, Görlich D, Wilm M, Carmeliet G, et al. 2006. NuSAP, a mitotic RanGTP target that stabilizes and cross-links microtubules. Mol Biol Cell 17: 2646–2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Salmon ED. 1994. Motile kinetochores and polar ejection forces dictate chromosome position on the vertebrate mitotic spindle. J Cell Biol 124: 223–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Davison EA, Jensen LC, Cassimeris L, Salmon ED. 1986. Oscillatory movements of monooriented chromosomes and their position relative to the spindle pole result from the ejection properties of the aster and half-spindle. J Cell Biol 103: 581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel CG, Katis VL, Katou Y, Mori S, Itoh T, Helmhart W, Gálová M, Petronczki M, Gregan J, Cetin B, et al. 2006. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature 441: 53–61. [DOI] [PubMed] [Google Scholar]
- Rivera T, Ghenoiu C, Rodríguez-Corsino M, Mochida S, Funabiki H, Losada A. 2012. Xenopus Shugoshin 2 regulates the spindle assembly pathway mediated by the chromosomal passenger complex. EMBO J 31: 1467–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanienko PJ, Camerini-Otero RD. 2000. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell 6: 975–987. [DOI] [PubMed] [Google Scholar]
- Saito TT, Tougan T, Okuzaki D, Kasama T, Nojima H. 2005. Mcp6, a meiosis-specific coiled-coil protein of Schizosaccharomyces pombe, localizes to the spindle pole body and is required for horsetail movement and recombination. J Cell Sci 118: 447–459. [DOI] [PubMed] [Google Scholar]
- Sakuno T, Tada K, Watanabe Y. 2009. Kinetochore geometry defined by cohesion within the centromere. Nature 458: 852–858. [DOI] [PubMed] [Google Scholar]
- Sampath SC, Ohi R, Leismann O, Salic A, Pozniakovski A, Funabiki H. 2004. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell 118: 187–202. [DOI] [PubMed] [Google Scholar]
- Sato A, Isaac B, Phillips CM, Rillo R, Carlton PM, Wynne DJ, Kasad RA, Dernburg AF. 2009. Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell 139: 907–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H, Weich S, Schwegler H, Heyting C, Härle M, Cremer T. 1996. Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J Cell Biol 134: 1109–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M, Ellenberg J. 2008. A new model for asymmetric spindle positioning in mouse oocytes. Curr Biol 18: 1986–1992. [DOI] [PubMed] [Google Scholar]
- Shao H, Li R, Ma C, Chen E, Liu XJ. 2013. Xenopus oocyte meiosis lacks spindle assembly checkpoint control. J Cell Biol 201: 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silljé HH, Nagel S, Körner R, Nigg EA. 2006. HURP is a Ran-importin β-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr Biol 16: 731–742. [DOI] [PubMed] [Google Scholar]
- Sollier J, Lin W, Soustelle C, Suhre K, Nicolas A, Géli V, de La Roche Saint-André C. 2004. Set1 is required for meiotic S-phase onset, double-strand break formation and middle gene expression. EMBO J 23: 1957–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana-Konwalski K, Godwin J, van der Weyden L, Champion L, Kudo NR, Adams DJ, Nasmyth K. 2010. Rec8-containing cohesin maintains bivalents without turnover during the growing phase of mouse oocytes. Genes Dev 24: 2505–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Araki H. 2010. Regulation of the initiation step of DNA replication by cyclin-dependent kinases. Chromosoma 119: 565–574. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Kohda T, Yamashita A, Nonaka N, Yamamoto M. 2005. Hrs1p/Mcp6p on the meiotic SPB organizes astral microtubule arrays for oscillatory nuclear movement. Curr Biol 15: 1479–1486. [DOI] [PubMed] [Google Scholar]
- Tanno Y, Kitajima TS, Honda T, Ando Y, Ishiguro K, Watanabe Y. 2010. Phosphorylation of mammalian Sgo2 by Aurora B recruits PP2A and MCAK to centromeres. Genes Dev 24: 2169–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurkauf WE, Hawley RS. 1992. Meiotic spindle assembly in Drosophila females: Behavior of nonexchange chromosomes and the effects of mutations in the nod kinesin-like protein. J Cell Biol 116: 1167–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth A, Rabitsch KP, Gálová M, Schleiffer A, Buonomo SB, Nasmyth K. 2000. Functional genomics identifies monopolin: A kinetochore protein required for segregation of homologs during meiosis I. Cell 103: 1155–1168. [DOI] [PubMed] [Google Scholar]
- Tseng BS, Tan L, Kapoor TM, Funabiki H. 2010. Dual detection of chromosomes and microtubules by the chromosomal passenger complex drives spindle assembly. Dev Cell 18: 903–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Nasmyth K. 1998. Cohesion between sister chromatids must be established during DNA replication. Curr Biol 8: 1095–1101. [DOI] [PubMed] [Google Scholar]
- Wainman A, Buster DW, Duncan T, Metz J, Ma A, Sharp D, Wakefield JG. 2009. A new Augmin subunit, Msd1, demonstrates the importance of mitotic spindle-templated microtubule nucleation in the absence of functioning centrosomes. Genes Dev 23: 1876–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Nurse P. 1999. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature 400: 461–464. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Yokobayashi S, Yamamoto M, Nurse P. 2001. Pre-meiotic S phase is linked to reductional chromosome segregation and recombination. Nature 409: 359–363. [DOI] [PubMed] [Google Scholar]
- Wiese C, Wilde A, Moore MS, Adam SA, Merdes A, Zheng Y. 2001. Role of importin-β in coupling Ran to downstream targets in microtubule assembly. Science 291: 653–656. [DOI] [PubMed] [Google Scholar]
- Wignall SM, Villeneuve AM. 2009. Lateral microtubule bundles promote chromosome alignment during acentrosomal oocyte meiosis. Nat Cell Biol 11: 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynne DJ, Rog O, Carlton PM, Dernburg AF. 2012. Dynein-dependent processive chromosome motions promote homologous pairing in C. elegans meiosis. J Cell Biol 196: 47–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, West RR, McIntosh JR, Hiraoka Y. 1999. A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J Cell Biol 145: 1233–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Dai W. 2012. Shugoshins function as a guardian for chromosomal stability in nuclear division. Cell Cycle 11: 2631–2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi K, Unruh JR, Deng M, Slaughter BD, Rubinstein B, Li R. 2011. Dynamic maintenance of asymmetric meiotic spindle position through Arp2/3-complex-driven cytoplasmic streaming in mouse oocytes. Nat Cell Biol 13: 1252–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobayashi S, Watanabe Y. 2005. The kinetochore protein Moa1 enables cohesion-mediated monopolar attachment at meiosis I. Cell 123: 803–817. [DOI] [PubMed] [Google Scholar]
- Zuccotti M, Giorgi Rossi P, Martinez A, Garagna S, Forabosco A, Redi CA. 1998. Meiotic and developmental competence of mouse antral oocytes. Biol Reprod 58: 700–704. [DOI] [PubMed] [Google Scholar]
- Zuccotti M, Ponce RH, Boiani M, Guizzardi S, Govoni P, Scandroglio R, Garagna S, Redi CA. 2002. The analysis of chromatin organisation allows selection of mouse antral oocytes competent for development to blastocyst. Zygote 10: 73–78. [DOI] [PubMed] [Google Scholar]