Abstract
Mutations in the Caenorhabditis elegans gene smu-2 suppress mec-8 and unc-52 mutations. It has been proposed that MEC-8 regulates the alternative splicing of unc-52 transcripts, which encode the core protein of perlecan, a basement membrane proteoglycan. We show that mutation in smu-2 leads to enhanced accumulation of transcripts that skip exon 17, but not exon 18, of unc-52, which explains our finding that smu-2 mutations suppress the uncoordination conferred by nonsense mutations in exon 17, but not in exon 18, of unc-52. We conclude that smu-2 encodes a ubiquitously expressed nuclear protein that is 40% identical to the human RED protein, a component of purified spliceosomes. The effects of smu-2 mutation on both unc-52 pre-mRNA splicing and the suppression of mec-8 and unc-52 mutant phenotypes are indistinguishable from the effects of mutation in smu-1, a gene that encodes a protein that is 62% identical to human spliceosome-associated protein fSAP57. We provide evidence that SMU-2 protects SMU-1 from degradation in vivo. In vitro and in vivo coimmunoprecipitation experiments indicate that SMU-2 and SMU-1 bind to each other. We propose that SMU-2 and SMU-1 function together to regulate splice site choice in the pre-mRNAs of unc-52 and other genes.
The excision of introns from pre-mRNA is carried out by the spliceosome, a complex machine composed of five snRNAs and many—approximately 145 in humans (52)—different proteins. Understanding how the spliceosome regulates splicing and splice site choice is complicated by the phenomenon of alternative RNA splicing, in which a given transcript can be spliced in alternative ways to generate different mRNA isoforms, which may be translated into different protein products with different functions (2). In a few cases, alternative splicing factors that regulate the alternative splicing of specific pre-mRNA targets in a development- or tissue-specific manner are known. Well-studied examples include the Drosophila melanogaster Sxl, Tra, and Tra2 proteins, which regulate alternative splicing of genes involved in fly sex determination.
The Caenorhabditis elegans protein MEC-8 also behaves as a specific regulator of alternative splicing. It contains two RNA recognition motifs and promotes the accumulation of two alternatively spliced isoforms of unc-52 pre-mRNAs (29). mec-8 null mutants are viable and fertile but exhibit defects in mechanosensation, chemosensation, and embryogenesis (7, 28), which have been attributed to defects in the processing of pre-mRNA targets of MEC-8 action other than unc-52 (28, 43). The specific phenotypes conferred by mec-8 mutation along with the tissue-specific pattern of mec-8 expression (42) suggest that MEC-8 is a specific rather than general regulator of splicing, and no apparent orthologs of MEC-8 have been found in purified human spliceosomes.
MEC-8's action on unc-52 transcripts accounts for a synthetic lethal interaction exhibited by mec-8 mutations when they are combined with those for any of several otherwise-viable alleles of unc-52. The unc-52 gene encodes the C. elegans homolog of the protein core of mammalian perlecan, a heparan sulfate proteoglycan found in basement membranes (38). Null mutations in unc-52 prevent assembly of the myofilament lattice of body wall muscle during embryogenesis and result in paralysis and arrest at the twofold stage of embryonic elongation (38), a phenotype characteristic of nonfunctional muscle (49). Exons 16, 17, and 18 of unc-52, each of which encodes a single immunoglobulin repeat, are alternatively spliced to generate slightly different UNC-52 isoforms (34, 37, 38). Viable mutations in this alternatively spliced region, including nonsense mutations in exons 17 and 18, do not seem to affect embryogenesis or early larval development but cause later progressive loss of muscle attachments to the adjoining basement membrane, hypodermis, and cuticle, resulting in a late-onset paralysis (37). The phenotypic effect of the unc-52 viable mutations is greatly enhanced by loss-of-function mec-8 mutations, such that mec-8; unc-52(viable) embryos resemble unc-52 (null) embryos (28, 29). MEC-8 has been shown to be required to generate unc-52 mRNA isoforms that splice exon 15 to exon 19 and that splice exon 16 to exon 19 (29). Thus MEC-8 is needed to generate the isoforms that skip exons containing unc-52 viable mutations and that provide enough UNC-52 function for embryogenesis and early larval development of these unc-52 mutants.
We have taken a genetic approach to identifying new genes affecting alternative splicing. A temperature-sensitive mec-8 mutation was combined with a temperature-sensitive unc-52 viable mutation to yield a temperature-sensitive synthetic lethality. Recessive, loss-of-function suppressors of this synthetic lethality were identified as two genes, smu-1 and smu-2 (smu for suppressor of mec-8 and unc-52) (28). In addition to suppressing the mec-8; unc-52(viable) synthetic lethal interaction, smu-1 and smu-2 mutations suppress the late-onset paralysis conferred by certain unc-52 viable mutations and provide a weak bypass suppression of the pleiotropic effects of mec-8 mutations, suggesting that smu-1 and smu-2 affect the alternative splicing of a number of pre-mRNAs. Apart from mildly deleterious effects on mobility, growth, and brood size, smu-1 and smu-2 mutations confer no obvious phenotype on their own. The smu-1 gene was characterized molecularly and shown to encode a WD repeat protein with more than 60% identity to a human protein of unknown function (43). More recently, the human protein was shown to be a component of the spliceosome (52).
In this paper we describe the genetic and molecular characterization of smu-2. We have found that smu-2 mutations confer the same phenotype as mutations in smu-1 (43). smu-2 mutations interact with specific alleles of unc-52, and these interactions can be explained by changes in the relative abundance of unc-52 alternatively spliced transcripts. We have cloned the smu-2 gene and found that it encodes a highly conserved protein, the homolog of human RED, which has been isolated in purified human spliceosomes (35, 52). SMU-2 is a nuclear, ubiquitously expressed protein. We also show that SMU-2 interacts with SMU-1 in vitro and in vivo and that this interaction seems to be required to stabilize SMU-1. We propose that SMU-1 and SMU-2 bind to each other as components of the spliceosome and modulate splice site selection of many pre-mRNAs.
MATERIALS AND METHODS
Genetic methods, genes, and alleles.
Nematode culturing and genetics were performed as described previously (4, 45). Crosses involving unc-52(e669su250ts), which we refer to as unc-52(ts), were done at 25°C; all other crosses were done at 20°C. Except for RC301 (15), whose use is described in the next section, all strains were derived from the wild-type Bristol stock N2. Previously identified genes and mutations used in this work were as follows: LGI (linkage group I), mec-8(u218), smu-1(mn415), unc-101(m1); LGII, smu-2(mn416, mn610, mn611), unc-52(e1421, e669su250, e669, e1012, e444, e998), lin-31(n301); LGIII, unc-36(e251); LGV, him-5(e1490).
Genetic and physical mapping of smu-2.
To obtain a genetic map distance between smu-2 and lin-31, non-Unc-52 (recessively suppressed) progeny of smu-2(mn416) lin-31 unc-52(ts)/+ + unc-52(ts) hermaphrodites were picked and scored for the recessive Muv phenotype conferred by lin-31. Since lin-31 is incompletely penetrant, genotypic assignments were confirmed by scoring the phenotypes of self progeny of picked animals. Four Smu non-Lin-31 recombinants were obtained from a total of 475 non-Unc-52 animals, giving a smu-2-to-lin-31 map distance of about 1 centimorgan (cM) after correcting for growth at 25°C (39).
We used DNA dimorphisms between RC301 and N2 to situate smu-2 on the physical map (17). Four dimorphisms were identified by amplifying DNA by PCR and finding differences in the sizes of PCR products made from N2 and RC301 DNA. A fifth dimorphism, mnP5, was identified as a single nucleotide difference between sequenced PCR products. The primers used to identify the five dimorphisms, their locations on the physical map, and their corresponding differences in PCR size or nucleotide sequence are as follows. For mnP1 primer A was GGATTCTGGTTGTGATGACACG, primer B was GTATAGCCATTCCTCGATGTGG, the cosmid was ZC239, the N2 product was 1,009 bp, and the RC301 product was 900 bp; for mnP2 primer A was AGGAGCTGAGCAAGATTGCC, primer B was TATTGCACACCGAGGAGACC, the cosmid was F53C3, the N2 product was 867 bp, and there was no product for RC301; for mnP3 primer A was GAACGACCTAATAGTTTGTGCTCG, primer B was AAAGAGTCAACCTGAACTCGC, the cosmid was W09G10, the N2 product was 1,226 bp, and the RC301 product was ≈ 1,100 bp; for mnP4 primer A was TCACTATTCTCAGCGTTCTCTCC, primer B was CCAGTTGTCTTCACCTACGTCC; the yeast artificial chromosome (YAC) was Y49F6B, the N2 product was 1,442 bp, and the RC301 product was ≈ 1,375 bp; for mnP5 primer A was ACAGCCTTTATGTCGTTGGC, primer B was GTTCTGGAACATGATGCTGC, the cosmid was C16C8, and at position 40014 the N2 product was T and the RC301 product was C. Recombinants were mapped for the presence of these RC301 dimorphisms using single worm PCR assays (48). For mnP2, an unrelated PCR was done in the same tube as a control to ensure that DNA was present in the sample.
To map smu-2 with respect to the DNA dimorphisms, we first made a strain containing DNA sequence in the smu-2 region from RC301 coupled to unc-52(ts) by crossing RC301 males with lin-31 unc-52(ts) hermaphrodites and picking Unc-52 F2 progeny that did not segregate lin-31. We confirmed by PCR that this stock contained the five RC301 dimorphisms used in our mapping. To obtain recombinants for mapping, we crossed smu-2(mn416) lin-31 unc-52(ts)/+ males to the RC301-marked unc-52(ts) hermaphrodites, and from F1 Unc-52 cross progeny that segregated Muv non-Unc-52 self progeny, we picked non-Muv non-Unc-52 recombinants, genotype smu-2 unc-52(ts)/smu-2 lin-31 unc-52(ts). Homozygous smu-2 unc-52(ts) recombinants were recovered and scored with respect to the dimorphisms. The five dimorphisms are all situated left of lin-31 on the physical map in the left-to-right order as mnP1, mnP5, mnP4, mnP3, and mnP2. Out of 26 total recombinants, no crossovers occurred between mnP1 and mnP5, three occurred between mnP5 and mnP4, two occurred between mnP4 and mnP3, 16 occurred between mnP3 and mnP2, and 5 occurred between mnP2 and lin-31. Subsequent cloning of smu-2 confirmed that it lies between mnP5 and mnP4. We also mapped the left end point of the deficiency ccDf1, which deletes lin-31 and which complements smu-2. Arrested embryos of ccDf1/+ parents were scored for the presence of N2-type PCR products. We found that ccDf1 ends between mnP4 and mnP3.
Genetic interactions.
smu-2 was tested for interactions with a variety of unc-52 viable mutations, either alone or in combination with smu-1. To construct smu-2 unc-52 strains, lin-31 unc-52 mutants were generated for each unc-52 allele and crossed to homozygous smu-2 males. Cross progeny were allowed to produce self progeny, and Unc-52 non-Muv recombinants of genotype smu-2 unc-52/+ lin-31 unc-52 were picked, from which homozygous smu-2 unc-52 segregants were identified. For unc-52(e444) and unc-52(e998), where no suppression or enhancement of the Unc-52 phenotype was observed, the presence of the smu-2 mutation was confirmed by sequencing. To construct smu-1; smu-2 unc-52 triple mutants, unc-101; smu-2(mn416) unc-52 hermaphrodites were crossed to smu-1; unc-52 males. Non-Unc-101 Unc-52 progeny, genotype unc-101 +/+ smu-1; smu-2 unc-52/ + unc-52, were picked. In the next generation, non-Unc-52 progeny that segregated one-fourth Unc-101 progeny (unc-101/smu-1; smu-2 unc-52) were allowed to produce self progeny, and homozygous smu-1; smu-2 unc-52 segregants were identified. The presence of both smu-1 and smu-2(mn416) was confirmed by sequencing. To evaluate the extent of Unc-52 suppression, synchronous populations of worms were observed every 12 h throughout development until all worms were gravid adult hermaphrodites. The stage at which approximately 90% of the worms were paralyzed was recorded. For smu-2(mn416) unc-52(e1421) worms, we also counted the number of unhatched eggs and hatched larvae 24 h after removal of egg-laying hermaphrodites from a growth plate. smu-2 unc-52(e1421) animals were also scored for morphology (by differential interference contrast [DIC]microscopy), growth, and movement.
Cloning smu-2.
Transformation rescue of smu-2(mn416) was performed by injecting YAC DNA (100 ng/μl) or long-range PCR fragments (20 ng/μl) and the cosmid R1p16 (50 ng/μl), which rescues unc-36, into the gonads of smu-2(mn416) unc-52(ts); unc-36 young adult hermaphrodites, as described previously (32). Non-Unc-36 descendants of injected animals grown at 25°C were scored as adults for reappearance of the Unc-52 phenotype. YAC DNA was prepared as described previously (10), and long-range PCR products were made with the Expand long-template PCR system (Boehringer Mannheim). We also tested several DNA samples for smu-2 rescue in a mec-8; smu-2(mn416); unc-36 background by scoring for reversal of the suppression by smu-2 of the dye-filling defect conferred by mec-8 (28). Sequencing of smu-2 alleles was done with the Thermosequenase cycle sequencing kit (U.S. Biochemicals) or by the Advanced Genetics Analysis Center (University of Minnesota). A rescuing clone, pAS10, containing all of the smu-2 coding genomic sequence and 2 kb upstream of the predicted translational start site, was made by cloning the LR16F (CTGTGATGATGGTTGAAGGTGAC)/LR15R (GACAATGGCTGAAATGATCGATAGTTGG ) PCR product into the EcoRV site of pBluescript SK(−) (Stratagene). The PCR product LR16F/LR18R (CACGTTCTTTTTCACGTTCACGGC), which truncates the smu-2 gene where the mn610 mutation resides, also rescued smu-2. pAS14 was made by removing an NcoI/XhoI fragment from pAS10. pAS19 was created by inserting a NotI smu-2 genomic fragment from pAS10 into yk563h8, a full-length smu-2 cDNA. pAS21 was made by cutting and filling in the HindIII site of pAS19, thereby creating a frameshift mutation at amino acid 45. LR16F/LR15R(mn610) and LR16F/LR15R(mn416) were made by PCR using 16F and 15R primers with mn610 and mn416 mutant genomic DNA as the template. All cloning was done by standard molecular biology techniques (40).
dsRNA interference of smu-2.
Double-stranded RNA (dsRNA) interference was performed by injection of 1 mg of smu-2 dsRNA/ml, corresponding to nucleotides (nt) 1041 to 1721 of the smu-2 transcript. Sense and antisense RNAs were made with a T7/T3 Megascript in vitro transcription kit (Ambion) and purified by phenol-chloroform extraction in accordance with the manufacturer's protocol. Equivalent amounts of RNA were annealed in 3× IM buffer (20 mM KPO4 [pH 7.5], 3 mM K citrate, 2% polyethylene glycol 6000) at 68°C for 10 min and 37°C for 30 min. Progeny that were laid 24 h after injection were evaluated for embryonic lethality and the Unc-52 phenotype (at 25°C). In control experiments, 3× IM buffer was injected.
RNA isolation.
Semisynchronous larval populations were obtained by collecting embryos from bleached hermaphrodites (24) and allowing them to hatch on plates with food for 25 to 26 (L2 population) or 40 h (L4 population) at 20°C. For RNase protection experiments, unc-52(ts) and smu-2 unc-52(ts) larvae were grown at 25°C for 30 h to obtain L4 populations. Larvae were washed with M9 buffer (45) and frozen at −80°C. Fifty to 100 μl of concentrated worms was vortexed in 1.6 ml of Trizol (Life Technologies) with glass beads; the supernatant was mixed with 0.32 ml of chloroform, and the RNA was precipitated in 1 volume of isopropanol. RNA pellets were resuspended in 50 to 200 μl of diethyl pyrocarbonate-treated distilled H2O (dH2O). RNA samples for reverse transcription-PCR (RT-PCR) were treated with 1 μl of DNase RQ1 per 10 μg of RNA (Promega), extracted with phenol-chloroform, and resuspended in 10 to 15 μl of dH2O. RNA for smu-1::HA RT-PCR was obtained from mixed-stage populations and prepared as described above.
RT-PCR.
Reverse transcription of 1 to 2 μg of total RNA was performed with Superscript II reverse transcriptase (Life Technologies) in a 20-μl total volume by following the manufacturer's protocol; 60 μl of water was added to the reverse transcription reaction mixture, and 1 μl of this (or a 0.5 or 0.25 dilution) was used for PCR amplification, as described previously (43). PCR was performed with the unc-52 primers 16F (29) and 18/19R and the ama-1 primers A3 and B2 (43). Southern blots of PCR products were probed with 32P-end-labeled unc-52 16FB and ama-1 A2 oligonucleotides (43) and quantified with a phosphorimager (Molecular Dynamics) and ImageQuant software (Amersham Pharmacia Biotech). For each cDNA, PCR was performed with increasing cycle numbers (from 16 to 32 cycles) and increasing dilutions (1, 0.5, and 0.25 at 24 or 26 cycles) to evaluate the linearity of the experiments. PCR experiments using unc-52 primers were repeated with two independent sets of cDNAs from both smu-2 and wild-type L2 larvae and two independent sets of cDNAs from both smu-2 and wild-type L4 larvae. ama-1 primers were included in PCRs with one set of smu-2 and wild-type L2 cDNAs and one set of smu-2 and wild-type L4 cDNAs. Three samples which were within the linear range of the PCR were averaged to obtain a ratio of 16-18-19/16-17-18-19 transcripts and a ratio of 16-18-19/ama-1 transcripts, where appropriate, for each independent experiment. The mean 16-18-19/16-17-18-19 ratios for smu-2 and for the wild type were calculated from the four independent experiments, and the means were compared statistically with a paired t test. RT-PCR of smu-1::HA was performed with the smu-1 primers smu-1-for (GAAATATCAGGCCCAAGA) and HA-rev (CTCGAGGCACTGAGCAGC) and the ama-1 primers A3 and B2. smu-1::HA RT-PCRs were performed within the linear range of amplification.
RNase protection.
RNase protection was performed as previously described (43) with 5, 10, and 15 μg of total RNA, 2.5 μl of [32P]CTP (800 Ci/mmol, 20 mCi/ml), and an RPAIII kit (Ambion). The unc-52 16-18 RNA probe was generated from pCS168 linearized with HindIII (43) and transcribed with a T7 Maxiscript in vitro transcription kit (Ambion). In addition, a 267-bp ama-1 probe that protects nt 104 to 371 of the ama-1 cDNA was included in each experiment. To confirm the sizes of 16-18, 16-17, and 16-19 protected RNAs, sense RNAs corresponding to these RNA species were generated with a T7 Megascript in vitro transcription kit (Ambion) and the appropriate PCR templates. These sense RNAs produced protected bands of the expected sizes when hybridized to the unc-52 probe. Quantitative data were collected with a phosphorimager as for RT-PCR. The linearity of each RNase protection experiment was evaluated by using multiple concentrations of RNA (5, 10, and 15 μg) in each experiment. A ratio of 16-18/(16-17 plus 16-19) transcripts and of 16-18/ama-1 transcripts was obtained for each experiment by taking the mean from these three samples. The combined ratios from three experiments performed on two separate sets of RNA isolations that compared smu-2 unc-52 RNA to unc-52 RNA gave essentially the same results. Similar data were also obtained when comparing smu-2 to the wild type.
SMU-2 reporter strains.
Plasmid pAS17, which is a smu-2::gfp reporter that complements smu-2(mn416), was made by cloning an Eco01019I fragment from the vector pPD118.85 (A. Fire; www.ciwemb.edu) into the Eco01019I site of pAS10 so that all coding sequences remained in frame. The DNA was injected at a concentration of 20 ng/μl with 50 ng of R1p16 DNA/μl into smu-2(mn416); unc-36 hermaphrodites and chromosomally integrated, as mnIs33, by gamma irradiation (32). The mnIs33-bearing stock was outcrossed several times. To evaluate SMU-2::green fluorescent protein (GFP) expression, smu-2(mn416); unc-36; mnIs33[smu-2::gfp unc-36(+)] embryos and larvae were fixed in methanol-acetone as described previously (10) and washed once in phosphate-buffered saline (PBS) containing 0.02 μg of DAPI (4′,6′-diamidino-2-phenylindole)/ml and twice in PBS. With rare exceptions, DAPI fluorescence and GFP autofluorescence were observed in all cells. A smu-2::HA rescuing construct was made by replacing the natural stop codon of smu-2 with an XhoI site and inserting a sequence encoding three copies of an hemagglutinin (HA) epitope tag (46) flanked by XhoI sites. Immunofluorescence images of smu-2(mn416) unc-52(e669su250); unc-36; mnIs66[smu-2::HA; unc-36(+)] animals were collected as described below for smu-1::HA.
Western blots.
Western blotting was performed by standard procedures (40). Briefly, adult stage populations of worms were washed with 1 ml of water, and an equal amount of 2× sodium dodecyl sulfate (SDS) loading buffer was added to the worm pellet. The worms were flash-frozen in liquid nitrogen and immediately boiled for 5 min; 40 μl of this lysate was loaded in each lane. Western blots were blocked in blocking buffer (5% condensed nonfat milk, 1× PBS, 0.5%Tween), incubated overnight with the primary antibody, washed three times (5 min each) in 1× PBS-0.5% Tween, incubated for 2 h with the secondary antibody, washed three times (15 min each), and visualized with the ECL detection system (Amersham Pharmacia). The following antibodies were diluted in blocking buffer: rat anti-HA monoclonal antibody (MAb) 3F10 (1:500; Roche), mouse antiactin MAb C4 (1:20,000; ICN Biochemicals), horseradish peroxidase (HRP)-conjugated goat anti-rat (1:5,000; Pierce), and HRP-conjugated goat anti-mouse (1:5,000; Pierce).
Immunofluorescence.
Immunofluorescence studies of smu-1::HA (43) and smu-2::HA strains were performed on mixed-stage worms as described previously (11) using rat anti-HA MAb 3F10 diluted 1:200 (Roche) and Cy-3-conjugated goat anti-rat diluted 1:1,000 (Jackson Immunoresearch). L1 larvae were also fixed and stained with a methanol-acetone fixation-permeabilization protocol (28).
In vitro coimmunoprecipitation.
One microgram of circular plasmid DNA from pAS53 (smu-2::myc) and pAS59 (smu-1::no tag) was transcribed and translated in vitro in a 20-μl total volume with the TnT T7 quick-coupled transcription-translation system (Promega) as recommended by the manufacturer, with 1 μl of [35S]methionine (1,000 Ci/mmol at 10 mCi/ml; Amersham Pharmacia) and 1 μl of antimyc MAb 9E10 (Santa Cruz Biotechnology). To the in vitro-translated proteins, 30 μl of a 50% slurry of protein G-agarose beads (Sigma) and immunoprecipitation buffer E (20% glycerol, 20 mM HEPES [pH 7.9], 0.125 mM EDTA, 100 mM KCl, 0.1% NP-40, 0.5 mM dithiothreitol, and mini-EDTA-free protease inhibitor cocktail [Roche]) was added to make a final volume of 100 μl, and the mixture was incubated at 4°C for 1 h. The beads were then precipitated, washed three times in 200 μl of buffer E, resuspended in 1× SDS loading buffer, and boiled, and proteins were electrophoresed through an SDS-8% polyacrylamide gel. In all constructs, the vectors T7plinkTag and T7plink (9) were used for production of in vitro-translated proteins with and without the myc tag, respectively. pAS53 (smu-2::myc), pAS63 (smu-2::no tag), pAS59 (smu-1::no tag), pAS57 (smu-1::myc), and pAS55 (mec-8::myc) all contain the full coding regions for their respective genes. Control plasmids pDZ45 and pDZ46 (partial tra-1 clones in T7plink and T7plinkTag, which encode the N terminus through the DNA binding domain of TRA-1), were obtained from D. Zarkower, and plasmid pHG35 (full-length tim-1 clone in T7plink) was obtained from H. Gardner.
In vivo coimmunoprecipitation.
To obtain a strain containing both smu-2::HA and smu-1::gfp, animals of genotype smu-2(mn416) unc-52(ts); unc-36; mnIs66[smu-2::HA unc-36(+)] (strain SP2693) were crossed to smu-1; unc-36; mnIs31[smu-1::gfp; unc-36(+)] animals and descendants homozygous for both mnIs66 and mnIs31 were identified by antibody staining and fluorescence microscopy. To obtain a strain containing both smu-2::HA and sur-5::gfp, strain SP2693 was injected with 50 ng of sur-5::gfp/μl (50) and 100 ng of pRF4/μl (33). A resulting strain, genotype smu-2(mn416) unc-52(ts); unc-36; mnIs66[smu-2::HA unc-36(+)]; mnEx161[sur-5::gfp; rol-6(d)], which transmitted mnEx161 to approximately two-thirds of its progeny, was used for coimmunoprecipitation. To prepare embryo lysates, worms were grown for two generations. The embryos from bleached hermaphrodites (24) were washed three times in M9 buffer and once in HB buffer (50 mM HEPES, 1 mM EDTA, 1 mM EGTA, 100 mM NaCl, 100 mM KCl, 1 mM dithiothreitol, 4 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 5% glycerol, 0.3 μg of aprotinin/ml, 0.5 μg of pepstatin/ml, and mini-EDTA free protease inhibitor cocktail [Roche]) and were resuspended in 400 μl of HB buffer. Embryos were sonicated, centrifuged at 16,000 × g for 10 min (4°C) to remove cellular debris, and then cleared by centrifugation for 2 min at 16,000 × g. Approximately 900 μg of total worm protein was preincubated in 25 μl of 50% protein G-agarose beads (Sigma) plus 2 μg of mouse immunoglobulin G (Sigma) for 4 h at 4°C. The supernatants were then incubated with 5 μl of mouse 3E6 anti-GFP (QBiogene) antibody overnight at 4°C. Twenty-five microliters of protein G-agarose beads was added, incubated for 1 h, precipitated, washed three times in 500 μl of HB buffer, and resuspended in SDS loading buffer. Proteins were electrophoresed through an SDS-8% polyacrylamide gel, and Western blots were prepared as described above with rat anti-HA MAb 3F10 (1:1,000) or a chicken anti-GFP antibody (1:10,000) (Chemicon) with HRP-conjugated donkey anti-chicken (1:5,000; Jackson Immunoresearch). Western blots for coimmunoprecipitation experiments were visualized with SuperSignal ELISA Femto maximum-sensitivity substrate (Pierce).
RESULTS
Mutations in smu-2 and smu-1 exhibit identical patterns of interaction with different unc-52 mutations.
It was shown previously (28) that smu-2 mutations fully suppress the adult onset paralysis conferred by the temperature-sensitive unc-52 allele e669su250. This allele contains the e669 mutation, a premature stop in exon 17, and the point mutation su250 in intron 16 (Fig. 1A), which acts as a partial suppressor of e669 (30, 37). Whereas e669su250 animals become paralyzed as adults when grown at 25°C, smu-2 unc-52(e669su250) animals never become paralyzed. Mutations in smu-2 also weakly suppress the e669 mutation by itself: smu-2 unc-52(e669) animals became paralyzed about 24 h later than do unc-52(e669) animals (28). We tested the ability of smu-2 mutations to suppress additional unc-52 viable alleles (Table 1). We found that smu-2 mutations delayed the onset of paralysis conferred by the unc-52(e1012) mutation, another exon 17 nonsense mutation (37), by about 24 h. But smu-2 mutations did not suppress either the unc-52(e444) or unc-52(e998) mutation, which are nonsense mutations in exon 18 (37). We also found that mutation in smu-2 did not delay the adult onset paralysis conferred by the unc-52(e1421) mutation, which is a single base pair change in the 5′ splice site of intron 16 (37). On the contrary, we found that two-thirds of the smu-2 unc-52(e1421) embryos exhibited elongation defects similar to those observed in smu-1; unc-52(e1421) embryos (43); 37% of smu-2 unc-52(e1421) progeny arrested during embryogenesis, and another 29% hatched as abnormal larvae (Table 1). DIC microscopy showed that arrested embryos and hatched abnormal larvae had bulges and constrictions along the length of their bodies. The abnormal larvae grew more slowly and were less mobile than their siblings of the same age. The remaining 34% of smu-2 unc-52(e1421) progeny were normal in appearance and moved well until the adult stage, when Unc-52 paralysis occurs. The lethality of smu-2 unc-52(e1421) embryos resembles, but is less severe than, the lethality observed in unc-52 null mutants, which occurs at the twofold stage of embryogenesis (49). Thus the effect of smu-2 mutation on unc-52(e1421) can be thought of as an enhancement of the unc-52 reduced-function phenotype.
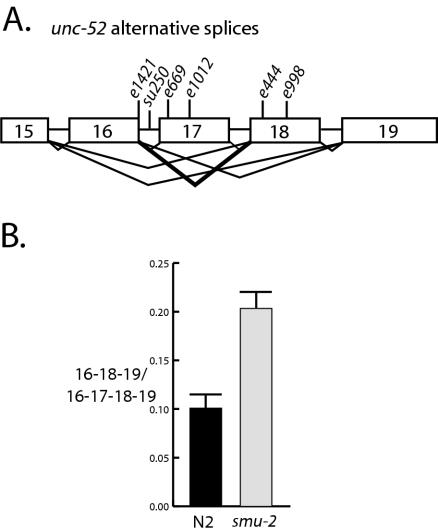
FIG. 1.
RT-PCR analysis of unc-52 transcripts in smu-2 mutants. (A) Depiction of the alternatively spliced region of unc-52. Boxes, exons 15 to 19; horizontal lines, introns. The locations of unc-52 viable mutations used in this study are indicated. The heavy lines from exon 16 to exon 18 indicate the alternatively spliced transcript that shows increased abundance in smu-2 mutants. (B) Results from four independent RT-PCR experiments performed on smu-2(mn416) and wild-type (N2) larvae. The mean ratios of 16-18-19/16-17-18-19 unc-52 transcripts were 0.100 ± 0.015 in wild-type (N2) larvae and 0.204 ± 0.018 in smu-2 larvae; these values were found to be significantly different in a paired t test (P = 0.025). These data indicate that there is approximately a twofold increase in the 16-18-19 transcript in smu-2 mutants.
TABLE 1.
Genetic interactions among smu-2, smu-1, and unc-52 mutations
| Genotypea | Developmental stage at onset of paralysis |
|---|---|
| unc-52(e669su250) | Adult |
| unc-52(e669) | Late L4 |
| unc-52(e1012) | Late L4 |
| unc-52(e444) | Early L4 |
| unc-52(e998) | Early L4 |
| unc-52(e1421) | Adult |
| smu-2(mn416) unc-52(e669su250) | None |
| smu-2(mn416) unc-52(e669) | Adult |
| smu-2(mn416) unc-52(e1012) | Adult |
| smu-2(mn416) unc-52(e444) | Early L4 |
| smu-2(mn416) unc-52(e998) | Early L4 |
| smu-2(mn416) unc-52(e1421) | Embryo; adultb |
| smu-1; unc-52(e669) | Adult |
| smu-1; unc-52(e1012) | Adult |
| smu-1; smu-2(mn416) unc-52(e669) | Adult |
| smu-1; smu-2(mn416) unc-52(e1012) | Adult |
Strains containing e669su250 were grown at 25°C. All others were grown at 20°C.
Thirty-seven percent of smu-2 unc-52(e1421) double mutants die as embryos, while 29% of the remaining larvae that hatch show abnormalities consistent with muscle defects. Those embryos that survive to adulthood with no or few abnormalities become uncoordinated at the adult stage in the same manner as unc-52(e1421) mutants.
smu-1 has been characterized both genetically and molecularly (43). Mutations in smu-1 interact with the unc-52 viable alleles identically to mutations in smu-2: they suppress the unc-52 alleles e669, e1012, and e669su250; they fail to suppress e444 and e998; and they enhance e1421. Mutations in smu-1 and smu-2 suppress the mechanosensory and chemosensory defects conferred by mec-8 mutations and the cold-sensitive partially penetrant embryonic lethality conferred by null mec-8 alleles (28). The similar phenotypes of smu-1 and smu-2 mutants suggest that these genes act in the same pathway. To address this question genetically, we constructed smu-1(null); smu-2(mn416) unc-52(e669 or e1012) triple mutants and evaluated their paralysis phenotypes. Suppression of unc-52 by smu-1; smu-2 mutations together appeared to be identical to suppression by either smu-1 or smu-2 mutations alone (Table 1). Furthermore, the smu-1; smu-2 double-mutant strains were no less healthy than either smu-1 or smu-2 single-mutant strains. These results are consistent with smu-1 and smu-2 acting in the same pathway.
Mutations in smu-2 affect the accumulation of alternatively spliced unc-52 transcripts.
Exons 16, 17, and 18 of unc-52 each encode a single immunoglobulin or NCAM repeat (38). Alternative splicing of the unc-52 transcript in this region leads to the synthesis of different protein isoforms with various numbers of these repeats, reflecting various patterns of exon skipping (Fig. 1A). The genetic interactions between the smu-2 and the unc-52 mutations that we have observed suggest that the smu-2 mutations suppress unc-52 nonsense mutations in exon 17 but not exon 18 by increasing the production of unc-52 transcripts that skip exon 17 but not exon 18. To test this hypothesis, we evaluated the levels of unc-52 transcripts in smu-2 mutants by semiquantitative RT-PCR and RNase protection. For RT-PCR analysis we measured the level of the unc-52 16-18-19 transcript with respect to the level of the unc-52 16-17-18-19 transcript, which is the major larval unc-52 transcript, and to the level of the ama-1 transcript. The abundance of the ama-1 transcript, which encodes the large subunit of RNA polymerase, is unchanged throughout development (18, 19) and serves as an internal control. To ensure that we were quantifying PCR products that were within the linear range of amplification, we analyzed the PCRs at increasing cycle numbers (16 cycles through 32 cycles) and at increasing dilutions of cDNA template (1, 0.5, and 0.25). Southern blots for each PCR experiment were probed with specific unc-52 or ama-1 probes and quantified by densitometry and ImageQuant software. Four independent RT-PCR experiments using RNA preparations from L2 and L4 larvae compared the abundance of unc-52 16-18-19 transcripts to that of 16-17-18-19 transcripts in smu-2 and wild-type animals, with essentially identical results. Combined data for all four experiments showed a statistically significant difference in the ratio of 16-18-19 transcripts to 16-17-18-19 transcripts in smu-2 mutants compared to the wild type, indicating about a twofold increase in the abundance of the unc-52 16-18-19 isoform in smu-2 mutants (Fig. 1B). In two experiments we measured the ratio of 16-18-19 transcripts to ama-1 transcripts, which also increased by about 1.7-fold in smu-2 mutants compared to the wild type (data not shown). By contrast, the ratio of 16-17-18-19 transcripts to ama-1 transcripts in smu-2 mutants did not change (data not shown). We also did not detect changes in any of the other unc-52 transcripts, such as 16-17-19 and 15-18-19, in smu-2 mutants (data not shown).
We performed RNase protection experiments to confirm the RT-PCR results. The unc-52 RNA probe contained 108 nt from pBluescript SK(−), the last 219 nt of exon 16, and the first 11 nt of exon 18 (Fig. 2A). 16-18-19 transcripts protected 230 nt of the probe, whereas 16-17-18-19 and 16-19 transcripts should protect 219 and 220 nt of the probe, respectively. Reconstruction experiments showed that the 219- and 220-nt bands did not reliably distinguish 16-17-18-19 transcripts from 16-19 transcripts, and so they were grouped together for analysis. A probe protected by ama-1 transcripts was also used as an internal control. Combined data from three independent RNA preparations showed a statistically significant increase in the level of 16-18-protected fragments in smu-2 unc-52(ts) mutants compared to unc-52(ts) animals when animals were grown at 25°C (Fig. 2B). An approximately twofold increase in the ratio of the 230-nt fragment (16-18) to the 219- to 220-nt bands (16-17 plus 16-19) or to the ama-1 band was observed in smu-2 unc-52(ts) animals compared to unc-52(ts) animals. As expected, no change was observed for the 219- to 220-nt band compared to the ama-1 band (data not shown). We repeated the experiment with N2 and smu-2 L4 RNA to eliminate any differential effects of the unc-52 mutation and found a similar increase in the 16-18 transcripts in smu-2 mutants (data not shown). We conclude from these and our RT-PCR analyses that the levels of 16-18-19 transcripts increase about twofold in smu-2 mutants, as has been observed in smu-1 mutants (43).
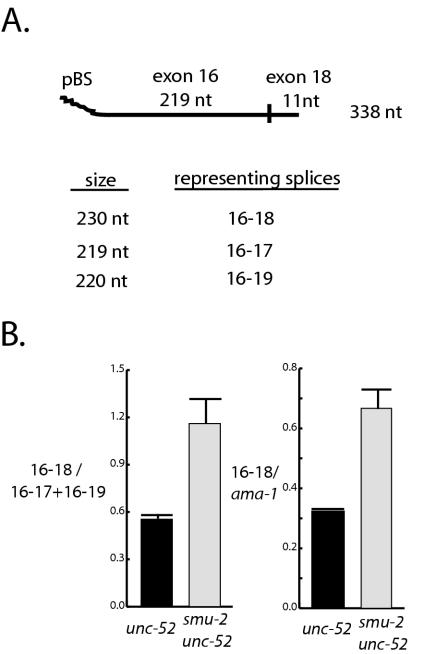
FIG. 2.
RNase protection analysis of unc-52 transcripts in smu-2 mutants. (A) Depiction of the unc-52 probe used for RNase protection experiments. The complete probe transcribed from the T7 promoter and including vector sequences is 338 nt long and contains 219 nt of exon 16 and 11 nt of exon 18. Probe lengths protected by different splice forms are indicated. (B) Data from three independent RNase protection experiments from unc-52 and smu-2 unc-52 larval RNA. The data from the 219- and 220-nt protected fragments, representing 16-17 and 16-19 splice forms, were grouped together because the fragments do not reliably distinguish these transcripts. The mean ratios for 16-18/(16-17 plus 16-19) transcripts was 0.545 ± 0.032 for unc-52 larvae and were 1.157 ± 0.156 for smu-2 unc-52 larvae; these values were found to be significantly different in a paired t test (P = 0.041). Mean ratios for 16-18/ama-1 transcripts were 0.323 ± 0.0002 for unc-52 animals and 0.667 ± 0.065 for smu-2 unc-52 animals; these values were also found to be significantly different in a paired t test (P = 0.034). These data indicate that there is about a twofold increase in the abundance of the 16-18 splice form in smu-2 mutants.
Molecular identification and characterization of smu-2.
We mapped smu-2 about 1 centimorgan to the left of the cloned gene lin-31. To refine the position of smu-2 on the physical map, we identified five sequence dimorphisms between strains N2 and RC301 in the smu-2 region. We then analyzed 26 crossovers between smu-2 and lin-31 in which the smu-2 lin-31 chromosome was derived from N2 and the recombining chromosome carried the RC301 sequence. We scored each crossover with respect to its inheritance of sequence dimorphisms (see Materials and Methods). The results placed smu-2 between dimorphisms mnP5 and mnP4 on the physical map (Fig. 3A). Using DNA from this region, we identified smu-2 by transformation rescue. Rescue was observed as the (unsuppressed) appearance of the Unc-52 phenotype at 25°C in smu-2(mn416) unc-52(ts) animals injected with candidate DNA. We were able to rescue smu-2 with the YAC Y37F3, the long-range PCR product LR13, and the long-range PCR product 16F/15R, which contains the single predicted gene Y49F6B.4 (Fig. 3B). This gene lies 18 kb to the right of mnP5 and 17.9 kb to the left of mnP4. To show that Y49F6B.4 is smu-2, we replaced genomic sequence after the first predicted intron with a full-length cDNA (yk563h8) corresponding to Y49F6B.4. This construct, which contains only 2 kb of genomic sequence upstream of the predicted translational start codon, rescued smu-2 (Fig. 3C). A frameshift mutation introduced in this construct at predicted codon 45 abolished rescue. We sequenced two alleles of smu-2 and found molecular lesions in the Y49F6B.4 gene (Fig. 4). mn416 has a 3′ splice site mutation in intron 1 (AG/AAA to AA/AAA). mn610 has a single base pair deletion in exon 6 that results in a frameshift after amino acid 407. A third allele, mn611, has a rearrangement, identified by Southern blotting and PCR (data not shown), in the last quarter of the smu-2 coding sequence. The analysis of several expressed sequence tags in sequence databases and our observation of a single band on Northern blots (data not shown) suggest that smu-2 encodes a single protein. Sequencing of yk563h8 confirmed the predicted intron-exon boundaries. Our experiments involving 5′ rapid amplification of cDNA ends showed that the transcription start site is located 77 bases upstream of the AUG start codon (position 160113 of Y49F6B) and that the transcripts are not trans-spliced to SL1 or SL2 (data not shown).
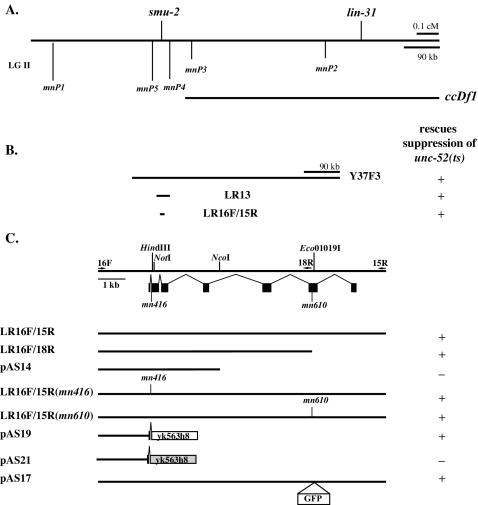
FIG. 3.
Mapping and cloning smu-2. The scales in panels A and B are identical. (A) Genetic map position of smu-2 with respect to the lin-31 gene and DNA sequence dimorphisms between strains N2 and RC301, which were used in mapping. The left end point of ccDf1, representing a deficiency that deletes lin-31 and complements smu-2, was mapped between dimorphisms mnP3 and mnP4. (B and C) Transformation rescue data for smu-2 with YAC DNA and long-range PCR products. + indicates that at least one line that rescued smu-2, as assayed by reversal of unc-52 suppression, was obtained; − indicates that several lines that did not rescue smu-2 were obtained. (C) The exon-intron structure of smu-2 is indicated below the restriction map. Arrows, locations of primers used to generate long-range PCR products. Eco01019I, NotI, and HindIII restriction sites used to construct pAS17, pAS19, and pAS21, respectively, are indicated. yk563h8 is a full-length smu-2 cDNA. pAS21 has a frameshift mutation introduced at the HindIII site (light gray box). The placement of the GFP gene for plasmid pAS17 is shown. smu-2(mn416) has a point mutation located at the 3′ splice site of intron 1. smu-2(mn610) has a single base pair deletion in the middle of exon 6. LR16F/15R(mn416) and LR16F/15R(mn610) are PCR products that were generated from genomic DNA made from the respective smu-2 mutants.
FIG. 4.
Alignment of SMU-2 and its homologs. The alignment was adapted from individual pairwise alignments made with the ClustalW program. Similar residues are boxed; identical residues in two or more homologs are shaded. A complete cDNA of the Drosophila homolog, CG18005, was sequenced by us. A cDNA corresponding to the Arabidopsis homolog, At2g26460, was sequenced by The Institute for Genomic Research. The RED domain of human RED is underlined. The positions of the mn416 and mn610 mutations and the insertion site for GFP are indicated.
Database searches revealed that SMU-2 is a highly conserved protein with homologs in vertebrates, invertebrates such as D. melanogaster, and plants such as Arabidopsis thaliana. The N terminus of a Schizosaccharomyces pombe protein also shows limited similarity to SMU-2. SMU-2 is 39.8% identical to a human protein that has been isolated by two groups from purified human spliceosomes and identified by mass spectrometry (35, 52). The human protein has been called IK factor (22, 35) and, more recently, RED (1, 52). RED has been shown to localize to the nucleus and to be present in all tissue types (1). RED was named after a domain rich in arginine (R), aspartic acid (E), and glutamic acid (D) residues. In the alignment of SMU-2 with its human, Drosophila, and Arabidopsis homologs, it is apparent that the RED domain itself is not highly conserved (Fig. 4). SMU-2 contains a series of RD repeats in this region, but it also contains several serine residues and an extensive region that is rich in aspartic acid and lysine residues (an EK domain). Surprisingly, the Drosophila and Arabidopsis homologs do not contain any semblance of a RED domain. Secondary structure predictions and examination of amino acid charge distribution also fail to find conserved features in this region for all four proteins. Although SMU-2 is predicted to contain a coiled-coil domain in this region, the others are not.
The smu-2 null phenotype.
Our molecular analysis of three smu-2 mutations revealed that none is a candidate null mutation. Indeed, we have shown that PCR products generated from either mn416 or mn610 DNA can rescue the smu-2 mutant phenotype (Fig. 3C), presumably owing to overexpression by the multicopy arrays that are generated from the injected DNA (32). For mn416, which has a 3′ splice site mutation, splicing to exon 2 may sometimes occur, and, for mn610, which has a frameshift mutation affecting the last one-third of the protein, overexpression of the truncated protein may rescue smu-2. Phenotypically, all three smu-2 mutants appear to be nearly wild type, although they, much like smu-1 mutants, have a tendency to coil their bodies and have slightly reduced brood sizes compared to N2 animals. We have determined that smu-2(mn416)/ccDf11 animals, in which ccDf11 deletes smu-2, mimic smu-2(mn416) homozygotes phenotypically, both with respect to their viability and ability to suppress unc-52(ts), suggesting that the smu-2 null phenotype is viable. To investigate this point further, we performed dsRNA interference on both unc-52 mutants and smu-2(mn416) mutants using smu-2 dsRNA. smu-2 RNA interference (RNAi) suppressed unc-52(ts) very efficiently: 99% of the progeny of injected unc-52(ts) animals were suppressed (Table 2). To ensure that we were decreasing the levels of smu-2 mRNA, we injected a strain that contained a rescuing smu-2::gfp reporter construct (pAS17) and observed that all GFP fluorescence was gone in embryos from injected animals (data not shown). In all of our injections of wild-type, unc-52, or smu-2(mn416) animals with smu-2 dsRNA, we never observed any embryonic lethality above that seen in control animals (Table 2) or any other obvious mutant phenotype.
TABLE 2.
smu-2 RNAi of unc-52 and smu-2 mutant strains
| Genotype | % of progeny of injected animals with phenotype:
|
|
|---|---|---|
| Embryonic lethalityb | Non-Unc-52 | |
| unc-52(e669su250)a | 4.2 (17/408) | 1.5 (6/391) |
| smu-2(RNAi) unc-52(e669su250) | 2.2 (10/453) | 99.1 (439/443) |
| smu-2(mn416)a | 2.9 (11/376) | Not applicable |
| smu-2(RNAi mn416) | 3.8 (20/530) | Not applicable |
Injected with 3× IM buffer.
Numbers in parentheses are ratios of the numbers of embryos with the phenotype to the total numbers of embryos scored from injected hermaphrodites.
Localization and expression pattern of SMU-2.
We made a rescuing construct, pAS17, which produces a fusion protein in which GFP is inserted in the last quarter of the SMU-2 protein (Fig. 3C and 4). We found that SMU-2::GFP is a nuclear protein (Fig. 5). SMU-2::GFP was observed at all stages of development including early embryogenesis and in oocytes (Fig. 5). To show that SMU-2 is ubiquitously expressed, we costained embryos and larvae carrying the integrated smu-2::gfp array with the DNA stain DAPI and observed SMU-2::GFP in essentially every cell that was stained by DAPI (Fig. 5D and E). Unlike most transgenes in C. elegans, the smu-2::gfp transgene was expressed in the germ line (Fig. 5F and G). To confirm this result with another SMU-2-expressing transgene, we constructed a rescuing transgene that placed a HA tag at the very end of the protein. This construct also led to nuclear expression in all cells, including the germ line (data not shown). We occasionally observed that SMU-2::GFP localized to a subnuclear compartment that we believe is the nucleolus because of its morphology and its increased size in ncl-1 mutants, which have enlarged nucleoli. However, nucleolar localization was dependent on the health of the animal. Animals suffering from prolonged exposure to sodium azide showed a dramatic increase in nucleolar localization. We rarely observed nucleolar localization of SMU-2::HA. We suppose that SMU-2 is not normally localized to the nucleolus. We did not observe any other consistent pattern of subnuclear localization.
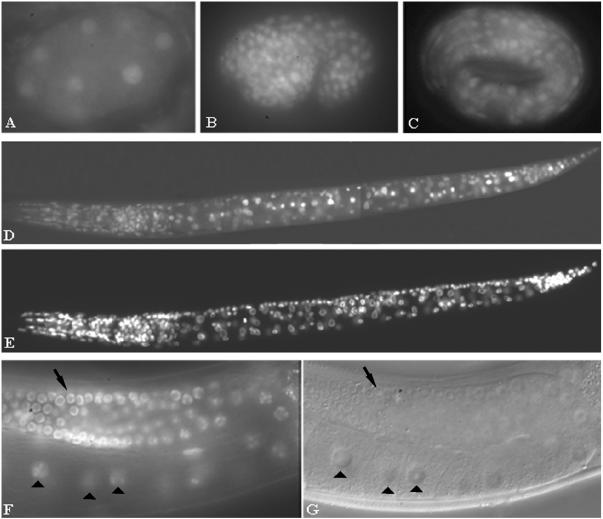
FIG. 5.
smu-2::gfp expression patterns. (A to D) SMU-2::GFP is nuclear and ubiquitously expressed throughout development. (A) A 10-cell embryo. (B) Comma-stage embryo. (C) Threefold-stage embryo. (D) L1 larva. (E) DAPI staining of the nuclei in larva of panel D. (F and G) SMU-2::GFP is expressed in the germ line and oocytes. (F) A portion of the gonad in a young adult worm with nuclear expression in the germ line (arrow) and oocytes (arrowheads). (G) DIC image of the worm in panel F.
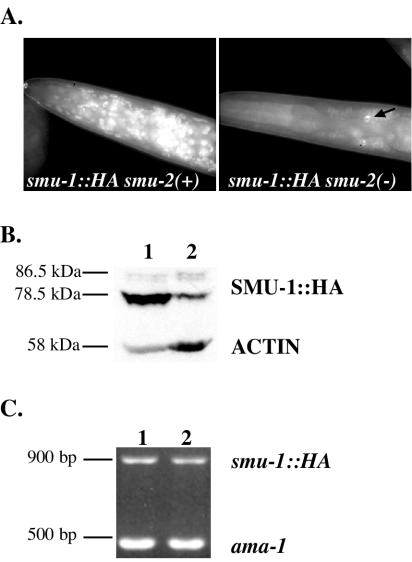
smu-2 mutation affects the accumulation of SMU-1.
When either integrated array smu-1::gfp or smu-1::HA (43) was present in a homozygous smu-2(mn416) background, the expression of the corresponding SMU-1 fusion protein was substantially reduced (Fig. 6). For each smu-1 transgene, limited nuclear expression in a few anterior neurons and hypodermal nuclei in the smu-2 animals was still observed (Fig. 6A). We analyzed the abundance of SMU-1::HA by Western blotting and saw that the major SMU-1::HA protein, migrating at 78.5 kDa, was significantly reduced in smu-2 animals (Fig. 6B). To rule out the possibility that smu-2 regulates smu-1 transcription or transcript stability, we performed RT-PCR on these strains to measure smu-1::HA transcript levels. We observed no changes in smu-2 mutants compared to smu-2(+) animals (Fig. 6C). We suggest that the wild-type SMU-2 protein is required to prevent SMU-1 degradation. SMU-1 seems not to be required for SMU-2 stability, however, because we did not observe any change in smu-2::gfp expression in smu-1 mutants (data not shown).
FIG. 6.
Effect of smu-2 mutation on expression of smu-1::HA. (A) Anti-HA immunofluorescence of SMU-1::HA in smu-2(+) and smu-2(mn416) genetic backgrounds. HA staining is dramatically reduced in smu-2 mutants, although staining is observed in some head neurons (arrow). (B) Western analysis of SMU-1::HA in smu-2(+) (lane 1) and smu-2(mn416) animals (lane 2). The major SMU-1::HA protein migrating at 78.5 kDa is markedly reduced in smu-2 mutants. Both the 78.5- and 86.5-kDa bands are specific to the smu-1::HA transgene since they do not appear in N2 animals (not shown). An antiactin antibody was used to monitor overall protein levels. (C) RT-PCR analysis of smu-1::HA transcripts in smu-2(+) (lane 1) and smu-2(mn416) (lane 2) animals. ama-1 was used as an internal control.
Evidence that SMU-2 and SMU-1 directly interact.
Genetic evidence suggests that smu-2 and smu-1 act together; both proteins are ubiquitously expressed in the nucleus, and SMU-1 accumulation depends on SMU-2. We therefore sought evidence of a SMU-1/SMU-2 physical interaction by performing in vitro and in vivo coimmunoprecipitation experiments.
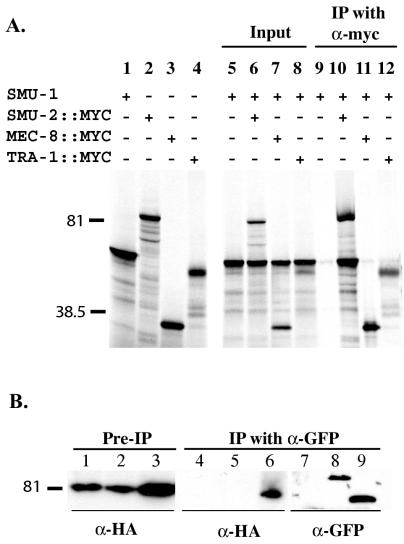
SMU-2::MYC tagged protein and untagged SMU-1 were labeled with [35S]methionine by coupled in vitro transcription and translation and immunoprecipitated with an antimyc antibody. As shown in Fig. 7, we consistently observed that SMU-2::MYC was able to coimmunoprecipitate SMU-1. SMU-1 was not coimmunoprecipitated by other nuclear proteins, such as MEC-8 and TRA-1, the latter a transcription factor that regulates C. elegans sex determination (14, 51) (Fig. 7A). In similar experiments we saw that SMU-2::MYC was unable to coimmunoprecipitate other nuclear proteins, such as untagged TRA-1 and untagged TIM-1, a protein that regulates chromosome cohesion (8) (data not shown).
FIG. 7.
In vitro and in vivo coimmunoprecipitation of SMU-1 and SMU-2. (A) SMU-1 was transcribed and translated in vitro with either SMU-2::MYC, TRA-1::MYC, or MEC-8::MYC by using [35S]methionine, and the reaction mixture was immunoprecipitated with an antimyc MAb. Proteins were separated by SDS-polyacrylamide gel electrophoresis. Lanes 1 to 4, translation of SMU-1, SMU-2::MYC, MEC-8::MYC, and TRA-1::MYC, respectively; lanes 5 to 8, 1/10 of the input from each total reaction volume; lanes 9 to 12, proteins immunoprecipitated by the antimyc antibody. Labeled proteins were visualized by autoradiography. (B) Protein lysates were made from embryos containing smu-2::HA (lanes 1, 4, and 7), both smu-2::HA and sur-5::gfp (lanes 2, 5, and 8), and both smu-2::HA and smu-1::gfp (lanes 3, 6, and 9). Western blots of proteins separated by SDS-polyacrylamide gel electrophoresis were prepared. Lanes 1 to 3, 1/40 of the total protein lysate before coimmunoprecipitation; lanes 4 to 9, 1/2 of the total proteins that were immunoprecipitated with anti-GFP MAb 3E6. Lanes 1 to 6 were probed with the anti-HA MAb, and lanes 7 to 9 were probed with chicken anti-GFP.
To show that SMU-1 and SMU-2 interact in vivo, we performed coimmunoprecipitation experiments on worms expressing tagged SMU-1 and SMU-2 proteins. Embryo lysates were prepared from strains containing smu-2::HA alone, both smu-2::HA and smu-1::gfp, and both smu-2::HA and sur-5::gfp. Lysates were immunoprecipitated with an anti-GFP antibody, and immunoprecipitates were analyzed for SMU-2::HA. SUR-5::GFP is a nuclear protein of unknown function (50) and served as a control to show that SMU-2 does not interact nonspecifically with nuclear proteins or with GFP. We found that SMU-1::GFP and SMU-2::HA consistently coimmunoprecipitated but that SUR-5::GFP and SMU-2::HA did not (Fig. 7B).
DISCUSSION
smu-2 regulates the alternative splicing of unc-52 transcripts.
We have shown that recessive mutations in smu-2 lead to enhanced accumulation of unc-52 transcripts in which exon 17, but not exon 18, has been skipped. Loss-of-function mutations in smu-1 have the same consequence (43). It was concluded for smu-1 that this accumulation is due to enhanced skipping of exon 17 during splicing rather than to differential effects on mRNA stability (43). The earlier arguments, which need not be repeated here, also apply to smu-2 and are consistent with both our molecular identification of SMU-2 as a homolog of a human spliceosome-associated protein and the recent identification (52) of a human spliceosome-associated protein with 62% amino acid identity to SMU-1.
The enhanced formation of unc-52 transcripts that skip exon 17 but not exon 18 supports the idea that smu-2 mutations suppress the late-onset paralysis conferred by nonsense mutations in exon 17 but not exon 18 by increasing the abundance of functional UNC-52 protein in the extracellular matrix between body wall muscle and hypodermis, where it is needed to help anchor the muscle to the hypodermis and cuticle (16, 49). As animals increase in size during larval and early-adult stages, muscle anchorages must be added to prevent the tearing away of muscle that leads to paralysis in unc-52(viable) mutants (34, 37). It is not surprising that the suppression of unc-52(e669) and unc-52(e1012) is only partial, because the effect of smu-2 mutations on splicing is fairly small, a twofold increase in 16-18 transcripts. The complete suppression of unc-52(e669su250) at the restrictive temperature of 25°C is also not surprising because the su250 mutation, a single base pair change in the middle of intron 16 (37), promotes an increase in the accumulation of 16-18 transcripts even at the restrictive temperature (43).
A somewhat different mechanism is required to explain the interaction that we have seen between a smu-2 loss-of-function mutation and the unc-52(e1421) mutation, a 5′ splice site mutation in intron 16. The double mutant exhibits a partially penetrant embryonic lethality that is not seen with either single mutant but which is similar (albeit milder) to the phenotype produced by null unc-52 alleles. The animals that survive embryogenesis unscathed suffer from late-onset paralysis at the same time that single-mutant unc-52(e1421) animals do. We presume that the e1421 mutation blocks a significant increase in 16-18 splice isoforms that smu-2 mutation would otherwise promote during larval development. On the other hand, smu-2 mutation apparently leads to a significant decrease in an embryonically important splice form. One possibility, suggested previously for the effect of a smu-1 mutation (43), is that smu-2 mutation leads to a decrease in the 15-19 splice isoform, which is prominent in wild-type embryos (43) (data not shown). Since formation of splice isoforms that contain exon 16, including the embryonically abundant 16-19 isoform, is compromised in e1421, the level of functional unc-52 transcripts in smu-2 unc-52(e1421) mutants could be inadequate to ensure normal embryonic development. Lethal interactions between the smu-2 mutation and the remaining viable unc-52 mutations may not occur because a decrease in the 15-19 splice isoform could be compensated for by an increase in the 16-19 splice isoform; the formation of both 15-19 and 16-19 isoforms is promoted by MEC-8 during embryogenesis (29, 34, 42).
smu-2 probably affects the splicing of additional transcripts.
It was shown previously that smu-2 mutations suppress, generally weakly, three distinct phenotypes or phenes conferred by mutations in mec-8: defects in mechanosensation, chemosensation, and embryogenesis at low temperature (28). It was concluded that these phenes are independent of unc-52 action and are probably due to defects in the processing of additional pre-mRNA targets of MEC-8 action (29, 34). It was also concluded that the smu-2 loss-of-function mutations exert their suppressing effects on the mec-8 mutant phenes by providing a weak bypass function and not by partial restoration of MEC-8 activity (28), since all mec-8 mutations that were tested, including those associated with presumed null alleles u74, u456, and u391, are partially suppressed by a smu-2 mutation (28). The u74 mutation results in an amino acid substitution at a highly conserved glycine residue in the first RRM domain of MEC-8, the u456 mutation is a deletion that removes most of this domain, and the u391 mutation is a complex rearrangement that produces no detectable MEC-8 (10, 42). The finding that loss of SMU-2 activity results in a partial bypass function led to the hypothesis that suppression by smu-2 is due to loosened restrictions on wild-type patterns of splicing such that the loss of SMU-2 function leads to a low level of altered splice site choice that can partially compensate for the loss of MEC-8. Our results here on the effects of smu-2 mutation on the splicing of unc-52 pre-mRNA support this picture and strengthen our supposition that SMU-2 affects the splicing of additional MEC-8 targets, and perhaps non-MEC-8 targets as well. SMU-2's ubiquitous expression would be consistent with this picture.
On the other hand, if general splicing were grossly disturbed, we would expect smu-2 mutants to be inviable, whereas they show only mild impairment in fertility and mild locomotory defects. None of our three mutant alleles has a null mutation, but putting a deletion mutation opposite that of one of our mutant alleles did not lead to a more severe phenotype, and smu-2(RNAi), which we showed was effective in reducing smu-2 expression and in suppressing an unc-52 mutation, had no effect on viability. We think it likely that the smu-2 null phenotype is viable, as is the smu-1 null phenotype (43), and that SMU-2 has mild effects on splicing, such as we saw for unc-52 pre-mRNA splicing.
SMU-2 and SMU-1 bind to each other.
Mutations in smu-1 and smu-2 behave identically in their patterns of suppression of unc-52 and mec-8 mutations and have identical effects on the pattern of unc-52 splicing. Furthermore, smu-1; smu-2 double mutants are phenotypically indistinguishable from each of the single mutants. These genetic arguments suggest that SMU-1 and SMU-2 exert their effects together, each being required for their combined wild-type function in controlling splicing. We also showed that mutation in smu-2 decreases markedly the abundance of SMU-1 protein without altering the level of smu-1 transcript, suggesting that SMU-2 is required to stabilize SMU-1. Consistent with this view is our finding, by both in vitro and in vivo assays, that SMU-1 and SMU-2 bind to each other. SMU-1 is a WD repeat protein (43). The canonical WD repeat protein, Gβ, is stabilized through a coiled-coil interaction at its N terminus with its partner Gγ (5, 41).
SMU-2 and SMU-1 are highly conserved spliceosome-associated proteins.
We have found that SMU-2 is very similar to a mammalian protein named RED. SMU-2 and RED share amino acid sequence throughout their entire lengths, with 40% overall identity. RED appears to be a ubiquitously expressed nuclear protein (1), as is SMU-2. RED has been identified by two groups as a component of the human spliceosome (35, 52). SMU-2 is clearly its closest worm homolog. RED homologs are present in several other species, such as D. melanogaster and A. thaliana, but not in the budding yeast Saccharomyces cerevisiae, which does not carry out alternative splicing.
The RED protein is named for its outstanding feature, the RED domain, a region of residues with alternating charge—arginines alternating with glutamate or aspartate. The corresponding region in SMU-2 lacks glutamate but contains arginine alternating with aspartate or, occasionally, serine, making what we might refer to as an RD-RS hybrid domain. Other components of the spliceosome contain RD domains (44), including the U1 snRNP component U1 70K. The spliceosome also contains several proteins with RS domains, including the SR and SR-related protein families (13). The RS domains in SR proteins have a high degree of serine phosphorylation, which results in alternating charge, as in the RED and RD domains. RS domains are to some extent functionally interchangeable modules (47), and, in one case, an RS domain was replaced with an RD domain that provided the same function (6). The RS domain of SR proteins mediates protein-protein interaction (13). The function of the RED domain has not been studied, but the RED domain's similarity to an RS domain suggests that it too mediates protein-protein interaction. The Arabidopsis and Drosophila homologs seem to lack a RED domain altogether. Although we have performed secondary structure analyses on the sequences in the region of the RED domain in these homologs, we have not been able to find any conserved features among them.
The molecular identification of smu-1 indicated that it encoded a protein that was 62% identical to a predicted human protein of unknown function (43). SMU-1 is, like SMU-2, a ubiquitously expressed nuclear protein. More recently, the predicted human SMU-1 homolog was found to be a component of the human spliceosome and was named fSAP57 for functional-spliceosome-associated protein 57 (52). SMU-1 is the only closely related worm homolog of fSAP57. In view of the high conservation of both SMU-1/fSAP57 and SMU-2/RED, we propose that SMU-1 and SMU-2 in worms and fSAP57 and RED in humans work together in spliceosomes to mediate splice site choice.
How do SMU-2 and SMU-1 work?
According to the picture that emerges, SMU-1 and SMU-2 work together to modulate, modestly, some but not all splice site choices. For unc-52 transcripts, SMU-1 and SMU-2 are needed to decrease about twofold the skipping of exon 17 but do not affect the splicing of neighboring exons such as skipping of exon 18 (data not shown). This suggests that SMU-1 and SMU-2 discriminate among different RNA sequence elements. A single nucleotide change, the su250 mutation, in the middle of intron 16 that alters the frequency of exon 17 skipping seems to lie in a cis-acting regulatory sequence (43). Regulatory sequences in both exons and introns that have either positive or negative effects on splicing have been identified in other systems (2). The discrimination of RNA sequence by SMU-1 and SMU-2 may well be indirect, involving other spliceosomal components, particularly since neither SMU-1 nor SMU-2 contains an apparent RNA binding motif. In addition, neither protein is expressed tissue specifically, and it seems unlikely that they regulate alternative splicing in a tissue-specific fashion. We suggest that they contribute to the wild-type fidelity of the spliceosome's action, such that their absence leads to alterations in splice site choice, which are only slightly harmful to the fitness of the organism.
Only a very small proportion of all spliceosomal proteins have had their functions tested in vivo by analysis of mutants. Fly SRp55 mutants (36) and mouse SRp20 mutants (20) are inviable, and RNAi has shown that SF1 (31), SF2/ASF (26, 53), and the U2 snRNP-specific proteins SAP49 (12), U2AF35, and U2AF65 (53) are essential in C. elegans. But RNAi treatment of SRm160 and five different SR protein genes in C. elegans (21, 26) resulted in no obvious phenotype, although subtle effects on splicing, as we have seen in smu-1 and smu-2 mutants, were not assayed. It has been suggested that many of the SR genes that gave no phenotype when treated singly by RNAi serve redundant functions because, when certain combinations of them were treated simultaneously by RNAi, lethal phenotypes were obtained (21, 26, 27). Targeting different combinations of SR genes for RNAi, however, yielded different developmental abnormalities, suggesting that different family members do not provide precisely overlapping functions. We imagine that inactivation of each spliceosomal protein may have an effect, if only a subtle one, and that the strong effects of multiple gene inactivation may be the consequence of multiple splicing errors. Of course there may also be synergistic effects in a multiply compromised spliceosome.
We do not know how SMU-1 and SMU-2 modulate splicing. Splice site choice seems often to be regulated by controlling the binding to the pre-mRNA of factors that influence assembly of early spliceosomal complexes. For example, the SR-related protein SRm160, which lacks RNA binding motifs (as do SMU-1 and SMU-2), is thought to act as a coactivator for certain splices by bridging interactions between other splicing factors, including U1 and U2 snRNP components (3). SMU-1 and SMU-2 might act similarly to favor inclusion of exon 17 of the unc-52 transcript. They could do this either by promoting certain early complexes or by blocking others. A negative mode of action might be carried out by a region of SMU-2 that is rich in lysine and aspartic acid residues, an EK domain. The EK domain of SR-related protein SRrp86 was recently shown to regulate negatively both constitutive and alternative splicing (25). Finally, it is possible that SMU-1 and SMU-2 modulate a later catalytic step in the splicing reaction, as recently found for the Drosophila Sxl protein (23).
Acknowledgments
We thank Heather Gardner and Dave Zarkower for tim-1 and tra-1 constructs, Y. Kohara for cDNA clones, and Todd Starich and John Yochem for much good advice. We give an especially big thanks to Caroline Spike for many useful discussions and technical guidance. Some nematode strains were provided by the Caenorhabditis Genetics Center.
This work was supported by U.S. National Institutes of Health (NIH) research grants GM22387 (R.K.H.) and GM56367 (J.E.S.). A.K.S. was a recipient of a doctoral dissertation fellowship from the University of Minnesota and a fellowship from NIH training grant HD07480.
REFERENCES
- 1.Assier, E., H. Bouzinba-Segard, M. C. Stolzenberg, R. Stephens, J. Bardos, P. Freemont, D. Charron, J. Trowsdale, and T. Rich. 1999. Isolation, sequencing and expression of RED, a novel human gene encoding an acidic-basic dipeptide repeat. Gene 230:145-154. [DOI] [PubMed] [Google Scholar]
- 2.Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72:291-336. [DOI] [PubMed] [Google Scholar]
- 3.Blencowe, B. J., R. Issner, J. A. Nickerson, and P. A. Sharp. 1998. A coactivator of pre-mRNA splicing. Genes Dev. 12:996-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenner, S. 1974. The genetics of Caenorhabditis elegans Genetics 77:71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bubis, J., and H. G. Khorana. 1990. Sites of interaction in the complex between beta- and gamma-subunits of transducin. J. Biol. Chem. 265:12995-12999. [PubMed] [Google Scholar]
- 6.Cartegni, L., and A. R. Krainer. 2003. Correction of disease-associated exon skipping by synthetic exon-specific activators. Nat. Struct. Biol. 10:120-125. [DOI] [PubMed] [Google Scholar]
- 7.Chalfie, M., and M. Au. 1989. Genetic control of differentiation of the Caenorhabditis elegans touch receptor neurons. Science 243:1027-1033. [DOI] [PubMed] [Google Scholar]
- 8.Chan, R. C., A. Chan, M. Jeon, T. F. Wu, D. Pasqualone, A. E. Rougvie, and B. J. Meyer. 2003. Chromosome cohesion is regulated by a clock gene paralogue TIM-1. Nature 423:1002-1009. [DOI] [PubMed] [Google Scholar]
- 9.Dalton, S., and R. Treisman. 1992. Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell 68:597-612. [DOI] [PubMed] [Google Scholar]
- 10.Davies, A. G., C. A. Spike, J. E. Shaw, and R. K. Herman. 1999. Functional overlap between the mec-8 gene and five sym genes in Caenorhabditis elegans. Genetics 153:117-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finney, M., and G. Ruvkun. 1990. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell 63:895-905. [DOI] [PubMed] [Google Scholar]
- 12.Fujita, M., T. Kawano, K. Terashima, Y. Tanaka, and H. Sakamoto. 1998. Expression of spliceosome-associated protein 49 is required for early embryogenesis in Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 253:80-84. [DOI] [PubMed] [Google Scholar]
- 13.Graveley, B. R. 2000. Sorting out the complexity of SR protein functions. RNA 6:1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hodgkin, J. 1987. A genetic analysis of the sex-determining gene, tra-1, in the nematode Caenorhabditis elegans. Genes Dev. 1:731-745. [DOI] [PubMed] [Google Scholar]
- 15.Hodgkin, J. 1993. Molecular cloning and duplication of the nematode sex-determining gene, tra-1. Genetics 133:543-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hresko, M. C., B. D. Williams, and R. H. Waterston. 1994. Assembly of body wall muscle and muscle cell attachment structures in Caenorhabditis elegans. J. Cell Biol. 124:491-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jakubowski, J., and K. Kornfeld. 1999. A local, high-density, single-nucleotide polymorphism map used to clone Caenorhabditis elegans cdf-1. Genetics 153:743-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeon, M., H. F. Gardner, E. A. Miller, J. Deshler, and A. E. Rougvie. 1999. Similarity of the C. elegans developmental timing protein LIN-42 to circadian rhythm proteins. Science 286:1141-1146. [DOI] [PubMed] [Google Scholar]
- 19.Johnstone, I. L., and J. D. Barry. 1996. Temporal reiteration of a precise gene expression pattern during nematode development. EMBO J. 15:3633-3639. [PMC free article] [PubMed] [Google Scholar]
- 20.Jumaa, H., G. Wei, and P. J. Nielsen. 1999. Blastocyst formation is blocked in mouse embryos lacking the splicing factor SRp20. Curr. Biol. 9:899-902. [DOI] [PubMed] [Google Scholar]
- 21.Kawano, T., M. Fujita, and H. Sakamoto. 2000. Unique and redundant functions of SR proteins, a conserved family of splicing factors, in Caenorhabditis elegans development. Mech. Dev. 95:67-76. [DOI] [PubMed] [Google Scholar]
- 22.Krief, P., Y. Augery-Bourget, S. Plaisance, M. F. Merck, E. Assier, V. Tanchou, M. Billard, C. Boucheix, C. Jasmin, and B. Azzarone. 1994. A new cytokine (IK) down-regulating HLA class II: monoclonal antibodies, cloning and chromosome localization. Oncogene 9:3449-3456. [PubMed] [Google Scholar]
- 23.Lallena, M. J., K. J. Chalmers, S. Llamazares, A. I. Lamond, and J. Valcarcel. 2002. Splicing regulation at the second catalytic step by Sex-lethal involves 3′ splice site recognition by SPF45. Cell 109:285-296. [DOI] [PubMed] [Google Scholar]
- 24.Lewis, J. A., and J. T. Fleming. 1995. Basic culture methods, p. 3-29. In H. F. Epstein and D. C. Shakes (ed.), Caenorhabditis elegans: modern biological analysis of an organism. Academic Press, San Diego, Calif.
- 25.Li, J., D. C. Barnard, and J. G. Patton. 2002. A unique glutamic acid-lysine (EK) domain acts as a splicing inhibitor. J. Biol. Chem. 277:39485-39492. [DOI] [PubMed] [Google Scholar]
- 26.Longman, D., I. L. Johnstone, and J. F. Caceres. 2000. Functional characterization of SR and SR-related genes in Caenorhabditis elegans. EMBO J. 19:1625-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Longman, D., T. McGarvey, S. McCracken, I. L. Johnstone, B. J. Blencowe, and J. F. Caceres. 2001. Multiple interactions between SRm160 and SR family proteins in enhancer-dependent splicing and development of C. elegans. Curr. Biol. 11:1923-1933. [DOI] [PubMed] [Google Scholar]
- 28.Lundquist, E. A., and R. K. Herman. 1994. The mec-8 gene of Caenorhabditis elegans affects muscle and sensory neuron function and interacts with three other genes: unc-52, smu-1 and smu-2. Genetics 138:83-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lundquist, E. A., R. K. Herman, T. M. Rogalski, G. P. Mullen, D. G. Moerman, and J. E. Shaw. 1996. The mec-8 gene of C. elegans encodes a protein with two RNA recognition motifs and regulates alternative splicing of unc-52 transcripts. Development 122:1601-1610. [DOI] [PubMed] [Google Scholar]
- 30.Mackenzie, J. M., Jr., R. L. Garcea, J. M. Zengel, and H. F. Epstein. 1978. Muscle development in Caenorhabditis elegans: mutants exhibiting retarded sarcomere construction. Cell 15:751-762. [DOI] [PubMed] [Google Scholar]
- 31.Mazroui, R., A. Puoti, and A. Kramer. 1999. Splicing factor SF1 from Drosophila and Caenorhabditis: presence of an N-terminal RS domain and requirement for viability. RNA 5:1615-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mello, C., and A. Fire. 1995. DNA transformation, p. 451-482. In H. F. Epstein and D. C. Shakes (ed.), Caenorhabditis elegans: modern biological analysis of an organism. Academic Press, San Diego, Calif.
- 33.Mello, C. C., J. M. Kramer, D. Stinchcomb, and V. Ambros. 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10:3959-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mullen, G. P., T. M. Rogalski, J. A. Bush, P. R. Gorji, and D. G. Moerman. 1999. Complex patterns of alternative splicing mediate the spatial and temporal distribution of perlecan/UNC-52 in Caenorhabditis elegans. Mol. Biol. Cell 10:3205-3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neubauer, G., A. King, J. Rappsilber, C. Calvio, M. Watson, P. Ajuh, J. Sleeman, A. Lamond, and M. Mann. 1998. Mass spectrometry and EST-database searching allows characterization of the multi-protein spliceosome complex. Nat. Genet. 20:46-50. [DOI] [PubMed] [Google Scholar]
- 36.Ring, H. Z., and J. T. Lis. 1994. The SR protein B52/SRp55 is essential for Drosophila development. Mol. Cell. Biol. 14:7499-7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rogalski, T. M., E. J. Gilchrist, G. P. Mullen, and D. G. Moerman. 1995. Mutations in the unc-52 gene responsible for body wall muscle defects in adult Caenorhabditis elegans are located in alternatively spliced exons. Genetics 139:159-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rogalski, T. M., B. D. Williams, G. P. Mullen, and D. G. Moerman. 1993. Products of the unc-52 gene in Caenorhabditis elegans are homologous to the core protein of the mammalian basement membrane heparan sulfate proteoglycan. Genes Dev. 7:1471-1484. [DOI] [PubMed] [Google Scholar]
- 39.Rose, A. M., and D. L. Baillie. 1979. A mutation in Caenorhabditis elegans that increases recombination frequency more than threefold. Nature 281:599-600. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 41.Schmidt, C. J., and E. J. Neer. 1991. In vitro synthesis of G protein beta gamma dimers. J. Biol. Chem. 266:4538-4544. [PubMed] [Google Scholar]
- 42.Spike, C. A., A. G. Davies, J. E. Shaw, and R. K. Herman. 2002. MEC-8 regulates alternative splicing of unc-52 transcripts in C. elegans hypodermal cells. Development 129:4999-5008. [DOI] [PubMed] [Google Scholar]
- 43.Spike, C. A., J. E. Shaw, and R. K. Herman. 2001. Analysis of smu-1, a gene that regulates the alternative splicing of unc-52 pre-mRNA in Caenorhabditis elegans. Mol. Cell. Biol. 21:4985-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Staknis, D., and R. Reed. 1995. Members of a family of proteins (the RD family) detected by a U1 70K monoclonal antibody are present in spliceosomal complexes. Nucleic Acids Res. 23:4081-4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sulston, J., and J. Hodgkin. 1988. Methods, p. 587-606. In W. B. Wood (ed.), The nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 46.Tyers, M., G. Tokiwa, R. Nash, and B. Futcher. 1992. The Cln3-Cdc28 kinase complex of S. cerevisiae is regulated by proteolysis and phosphorylation. EMBO J. 11:1773-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, J., S. H. Xiao, and J. L. Manley. 1998. Genetic analysis of the SR protein ASF/SF2: interchangeability of RS domains and negative control of splicing. Genes Dev. 12:2222-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Williams, B. D., B. Schrank, C. Huynh, R. Shownkeen, and R. H. Waterston. 1992. A genetic mapping system in Caenorhabditis elegans based on polymorphic sequence-tagged sites. Genetics 131:609-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams, B. D., and R. H. Waterston. 1994. Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J. Cell Biol. 124:475-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yochem, J., T. Gu, and M. Han. 1998. A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics 149:1323-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zarkower, D., and J. Hodgkin. 1993. Zinc fingers in sex determination: only one of the two C. elegans Tra-1 proteins binds DNA in vitro. Nucleic Acids Res. 21:3691-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou, Z., L. J. Licklider, S. P. Gygi, and R. Reed. 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419:182-185. [DOI] [PubMed] [Google Scholar]
- 53.Zorio, D. A., and T. Blumenthal. 1999. U2AF35 is encoded by an essential gene clustered in an operon with RRM/cyclophilin in Caenorhabditis elegans. RNA 5:487-494. [DOI] [PMC free article] [PubMed] [Google Scholar]