Abstract
Src family kinases regulate multiple cellular processes including proliferation and oncogenesis. C-terminal Src kinase (Csk) encodes a critical negative regulator of Src family kinases. We demonstrate that the Drosophila melanogaster Csk ortholog, dCsk, functions as a tumor suppressor: dCsk mutants display organ overgrowth and excess cellular proliferation. Genetic analysis indicates that the dCsk−/− overgrowth phenotype results from activation of Src, Jun kinase, and STAT signal transduction pathways. In particular, blockade of STAT function in dCsk mutants severely reduced Src-dependent overgrowth and activated apoptosis of mutant tissue. Our data provide in vivo evidence that Src activity requires JNK and STAT function.
Normal development requires strict spatial and temporal control of cellular processes such as proliferation and differentiation for properly sized and functioning organisms to form. This control is achieved through a network of signal transduction pathways that coordinate developmental events between cells, tissues, and organs. Inappropriate activation of these signaling networks can cause diseases such as oncogenesis in which individual cells respond to aberrant internal cues to overproliferate. Src family cytoplasmic tyrosine kinases (SFKs) play important roles within these networks to regulate both developmental events and disease states, but the exact roles of SFKs in these events remain ambiguous.
Humans and mice have at least eight SFKs, including Src, Fyn, and Yes. SFKs are composed of a tyrosine kinase domain, an SH2 domain, an SH3 domain, and a regulatory C-terminal region. They can be activated by receptor tyrosine kinases, cytokine receptors, G protein-coupled receptors, and integrins (reviewed in reference 70). SFK activation can cause cell cycle entry, cytoskeletal rearrangements, and alterations in cell adhesion (reviewed in reference 1). Mammalian tissue culture models have identified numerous downstream effectors of SFK functions; these include signaling molecules in the Ras/extracellular signal-regulated kinase (ERK), Jun kinase, Jak/STAT, and Rac/Rho pathways (70). SFK activities have not been well explored in vivo, however, in part due to functional redundancy among SFKs. For example, src−/− mice show only subtle osteoclast defects while src−/−; fyn−/−; yes−/− mouse embryos show early lethality and multiple developmental anomalies including neural tube defects and dramatically reduced size (37, 43, 63). Fibroblasts derived from src−/−; fyn−/−; yes−/− mice show reduced proliferation, suggesting that some of the phenotypes of compound knockouts are caused by proliferative defects during development (37). However, the precise roles of Src, Fyn, and Yes in the cell cycle during development remain unknown.
SFKs are maintained in an inactive state through tyrosine phosphorylation of their C-terminal region by the negative regulator C-terminal Src kinase (Csk), which itself is closely related to SFKs (reviewed in reference 6). Deletion or mutation of the Csk target site leads to up-regulation of SFK activity (38, 52). Mammals have two Csk family members, Csk and Chk. Mice deficient for Csk show hyperactivation of SFKs and a striking embryonic phenotype also characterized by early lethality, neural tube defects, and reduced size (30, 49). Surprisingly, csk−/− fibroblasts do not show increased proliferation, which conflicts with data indicating that increased SFK activity leads to cell cycle entry. This could reflect functional compensation by Chk, which also negatively regulates SFKs (26, 58). The redundancy between multiple SFKs and Csk as well as the early lethality of Csk and compound SFK knockouts has impeded detailed evaluation of SFK function in developing tissues.
Abnormal constitutive activation of SFKs has been implicated in oncogenesis (reviewed in reference 32). Numerous human tumors possess activated SFKs, but SFK mutations have been found in only a fraction of tumors. Some human colon cancers harbor mutations that abolish the ability of the C-terminal domain to inhibit Src kinase activity (31). The transforming v-Src oncogene shows deletion of the Csk target site (17). Since SFKs can be abnormally activated through disregulation of the C-terminal region, reduced Csk family kinase activity could promote oncogenesis. Yet, the role of Csk and/or Chk in tumors is controversial or unclear. Large deletions within the region of chromosome 15 that harbors Csk have been observed in colon cancers, the tumor types that commonly show elevated SFK activity (2, 7, 14, 19, 56), but no specific loss-of-function Csk mutations have been found in tumors to date. Reduced Csk expression and function is correlated with Src activation in primary hepatocellular tumors, primary colorectal tumors, and colon carcinoma cell lines (12, 45). However, others have reported elevated Csk in tumors with high SFK activity (73). In addition, csk−/− primary mouse fibroblasts do not show a transformed phenotype (30, 49). Perhaps mutations in other loci, such as chk, are required to reveal a tumor suppressor function for Csk (26, 58). A detailed exploration of Csk function in vivo is required to understand its role in disease, but again, such studies have been impeded by the early lethality of csk−/−mice.
The imaginal disks of Drosophila melanogaster provide a powerful model system for the study of signal transduction. Imaginal discs share several properties with mammalian epithelial tissues: both are composed of epithelial cells that must maintain proportional growth, differentiation, and renewal to form functional tissues and organs. Cells within imaginal discs undergo proliferation and differentiation in response to molecular pathways that have been highly conserved across species and that function in oncogenesis. For example, studies of the eye imaginal disc have provided important evidence that the Ras and Jak/STAT signal transduction pathways are crucial for normal growth, proliferation, and differentiation (5, 25, 35, 44, 62). Recent genetic analyses of Drosophila tumor suppressor mutations have led to new insights about known human tumor suppressors and identification of new putative human tumor suppressors such as lats and salvador (34, 53, 67, 74).
The Drosophila genome contains two SFKs, Src42A and Src64B, that are functionally similar to their mammalian counterparts (20, 66). Src42A and Src64B loss-of-function mutations disrupt cytoskeletal regulation within developing oocytes and embryos (20, 68). Yet the full repertoire of SFK functions remains to be elucidated in Drosophila. Src42A and Src64B are regulated by a Csk-like activity in flies, but until now, the gene responsible for that activity was unknown (42). In this report, we present the cloning and characterization of the Drosophila Csk ortholog, dCsk. Loss of dCsk function led primarily to overgrowth phenotypes in developing tissues such as the eye; genetic data indicated that excess proliferation was due to up-regulation of SFKs. We provide evidence that this overgrowth required the JNK and STAT signal transduction pathways. Reducing STAT function prevented growth and normal differentiation of dCsk mutant tissue, instead provoking dCsk−/− cells to undergo apoptosis. Our data provide in vivo evidence for a Src-dependent proapoptotic pathway triggered by reduced STAT function. They are consistent with results from Stewart et al. (64), published during submission of the manuscript of this report. Together, these results connect SFK signaling to the cell cycle and suggest an approach for restraining its proliferative potential.
MATERIALS AND METHODS
Fly stocks and genetics.
Flies were grown at 25°C. Fly stocks were obtained from the Bloomington Stock Center unless otherwise noted. S030003 and S017909 were from the Szeged Stock Center. Src64BP1 was a gift from M. Simon. Stat92Ej6C8 was a gift from S. Hou. Src42ASu1 and Src42A18-2 were gifts from X. Lu. To create EGUF clones, we established y w: ey-Gal4 UAS-FLP/+; FRT82B GMR-hid l(3)CL-R/FRT82B dCsk flies by standard crosses; w; FRT82B GMR-hid l(3)CL-R/FRT82B Ubi-GFPnlsS65T flies were utilized as controls for minor artifacts inherent in the EGUF system.
Genomic and EST analysis.
The sequence flanking the j1D8 and S030003 P-element insertions was generated and mapped by the Berkeley Drosophila Genome Project and Szeged Stock Center, respectively. The following CG17309 expressed sequence tags (ESTs) were obtained from the Berkeley Drosophila Genome Project and fully sequenced: LD36541, LP09923, GH10267, LD22810, and LD33364. Sequences were assembled, compared, and analyzed with BLAST, MultAlin, PROSCAN, and Genestream.
Rescue and reversion.
To create the heat shock-inducible dCsk transgene hs-dCsk, the LD22810 cDNA was cloned into pPCaSpeR-hs, and stable insertions were created. dCskj1D8, dCskS030003, and dCskS017909 were extensively outcrossed to remove observed background mutations. w; hs-dCsk/+; dCsk/dCsk and w;+/+; dCsk/dCsk embryos were collected for 3 to 4 days in vials. Larvae were heat shocked at 37°C for 30 min every 10 to 16 h to induce dCsk expression. For reversion, S017909 and S030003 were excised by standard crosses; over 10 independent excisions were scored for reversion of lethality. j1D8 failed to excise.
Larval and pupal body size measurements.
Embryos were collected for 4 h, and larvae were grown at similar densities. For mass measurements, larvae were cleaned and weighed in groups of 15 to 20 on a Mettler AE50 balance. A minimum of three groups was measured for each genotype at each time point. The average body mass was calculated by determining the average of the sum of the average body mass per group. Values for each time point were normalized to the average mass of wild-type control larvae. For pupal measurements, pupae were photographed and relative length measurements were taken from printed enlargements. Values were normalized to those of wild-type pupae.
Clonal analysis and flow cytometry.
Flow cytometry was performed generally as described previously (51). Dissociated imaginal disc cells were run on a Cytomation MoFlo cytometer. Data were analyzed in Summit, version 3.1 (Cytomation). For analysis of loss-of-function clones, the genotypes were y w hs-FLP/+: FRT82B Ubi-GFPnlsS65T/FRT82B dCskj1D8 and y w hs-FLP/+; FRT82B Ubi-GFPnlsS65T/FRT82B dCskS030003. Clones were induced by heat shock at 48 and 72 h after embryo deposition (AED) and dissected at 120 h. Green fluorescent protein (GFP)-positive and -negative tissues were used to control for GFP detection and to set gates. Experiments were repeated at least three times.
Histology, immunohistochemistry, and scanning electron microscopy (SEM).
In situ hybridization was performed as described previously (69) with a probe to the 5′ end of both dCsk transcripts bounded by NcoI and BsgI sites. Negative controls lacked probe. Digoxigenin was detected with an alkaline phosphatase-conjugated antibody (Boehringer Mannheim).
For adult sections, heads were fixed in 1% glutaraldehyde-2% osmium tetroxide-phosphate-buffered saline (PBS), dehydrated, washed, and incubated for 4 h in 1:1 propylene oxide Durcupan ACM resin, overnight in 100% resin, and finally at 65°C to harden. Serial sections were stained with 0.5% methylene blue-0.1% toluidine blue. Digital photographs were taken on a Zeiss Axioplan.
For immunohistochemistry, tissue was fixed for 20 min in 4% paraformaldehyde with 1× PBS or 1× PEM and stains were performed in 1× PBS, 10% fetal bovine serum, and 0.3% Triton X-100. For antibodies, affinity-purified anti-Stat92E was used at 1:500 (15), anti-phospho-histone H3 (Upstate Biotechnology) was used at 1:200, anti-armadillo was used at 1:5, and 22C10 and active caspase 7 (New England Biolabs) were used at 1:4 and 1:50, respectively. Secondary antibodies were conjugated to Alexa Red or Green (Molecular Probes). For dCsk mitotic clones, we used ey-FLP/+; FRT82B Ubi-GFPnlsS65T/FRT82B dCsk. Digital photographs were taken on a Zeiss Axioplan. Confocal micrographs were taken by using the Zeiss LSM 510 system. To address cell autonomous growth, we did not rely on direct scoring of clonal patches within the eye disc in part because we were not able to reliably distinguish the boundaries of clones with single-cell resolution with the reagents available.
To estimate mitotic activity in larval discs, we examined printed enlargements of phosphohistone stains of EGUF discs. We controlled for tissue mass by counting phosphohistone-positive nuclei within a quadrant of fixed size such that we recorded the number of positive nuclei within identically sized fields of tissue for each genotype. Nuclei were counted in 3 quadrants per disc, and the average number of mitotic nuclei per quadrant was determined. For pupal discs, the total number of phosphohistone-positive cells were counted per disc.
For SEM, adult flies were fixed in 95% ethanol, rehydrated, treated with 1% osmium tetroxide, dried, and sputter coated. Ommatidia were counted on printed enlargements of SEMs. For dCskj1D8 EGUF clones, estimates of ommatidia were made by using SEMs of the entire eye plus separate SEMs to visualize folds.
RESULTS
dCsk encodes a negative regulator of growth and proliferation.
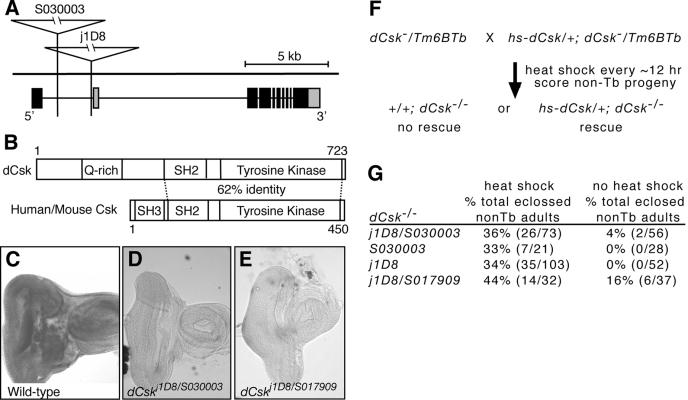
In a screen for mutations that genetically modify an overexpressed, oncogenic form of the Ret receptor tyrosine kinase in Drosophila (unpublished data), we identified three transposable P-elements that enhanced the activated Ret phenotype. Fly lines j1D8, S030003, and S017909 contain P-element insertions within the CG17309 locus (Fig. 1A). We fully sequenced 5 of 50 known CG17309 ESTs and determined that CG17309 encodes two nearly identical predicted proteins that differ only at the N terminus. The predicted proteins contain a tyrosine kinase domain and an SH2 domain that, together, show the highest homology with Csk family kinases (Fig. 1B). In fact, CG17309 proteins show a higher homology to Csk orthologs from other species such as mice, Xenopus sp., and hydra than to any other Drosophila tyrosine kinase. They also contain a glutamine-rich region in place of the SH3 domain found in mammalian Csk proteins (Fig. 1B). Consistent with other members of the Csk family, CG17309 proteins lack an N-terminal myristoylation signal and lack a C-terminal negative regulatory tyrosine present in SFKs. Also, CG17309 proteins lack plextrin homology and Tec homology domains, which distinguish them from the closely related Tec-Btk family tyrosine kinases. Previous analyses of the Drosophila genome have concluded that CG17309 encodes the sole Drosophila Csk ortholog (48). Based on these data and data presented below, we will refer to this locus as the Drosophila Csk ortholog, or dCsk, and the three insertion lines as dCskj1D8, dCskS030003, and dCskS017909 (see also reference 64).
FIG. 1.
Mutations in dCsk. (A) Diagram of the dCsk locus. Grey boxes illustrate alternative 5′ and 3′ exons. j1D8 and S030003 are inserted within the first intron of the predominant dCsk transcript. j1D8 is 75 bp upstream and S030003 is 2.3 kb upstream of the first exon of the second dCsk transcript. S017909 was not sequenced, but it maps to 86E, the cytological region that contains dCsk (18). (B) Predicted domain structure of dCsk protein and comparison between dCsk and mammalian Csk. The protein identity for the SH2 kinase domain is indicated. (C to E) In situ hybridization of third instar eye-antennal imaginal discs with a DNA probe to dCsk shows broad staining (C) that is lost in dCsk mutant tissue (D and E). dCsk mutant tissue was photographed at twice the exposure to visualize the tissue. (F and G) Diagram and results of rescue experiments with a heat shock-induced dCsk transgene (hs-dCsk). Only half of the dCsk−/− progeny per sample received the hs-dCsk transgene. We were unable to independently score for the presence or absence of the transgene, so all dCsk−/− larvae and pupae were monitored for rescue. Therefore, a maximum of 50% of the total dCsk−/− progeny have the potential to be rescued per sample. Rescued dCsk mutants were almost always viable normal adults. Heat shock of wild-type control animals only nominally affected adult eclosure (data not shown). Tb marks nonmutant (Balancer) chromosomes and indicates phenotypically normal heterozygous flies.
All three dCsk−/− mutant lines are lethal and displayed a stronger phenotype in trans to a deficiency. dCskj1D8 exhibited the earliest lethal phase, dying within 6 to 18 h after pupariation, a lethal phase similar to dCskj1D8 placed in trans to a deficiency, illustrating that dCskj1D8 is a strong hypomorphic mutation. Excision of the dCskS030003 and dCskS017909 insertions reverted their lethality and/or noncomplementation with dCskj1D8. In situ hybridization indicated that dCsk mRNA is ubiquitously expressed within developing larval tissues (Fig. 1C). dCskj1D8, dCskS030003, and dCskS017909 mutant tissues showed reduced dCsk expression by in situ hybridization (Fig. 1D and E). Finally, heat shock-induced expression of a dCsk cDNA rescued the lethality and mutant phenotypes in all three dCsk alleles (Fig. 1F and G). By itself, ectopic, ubiquitous dCsk overexpression had no detectable effect on the adult phenotype (data not shown). These data demonstrate that all three P-element insertions disrupt the dCsk locus.
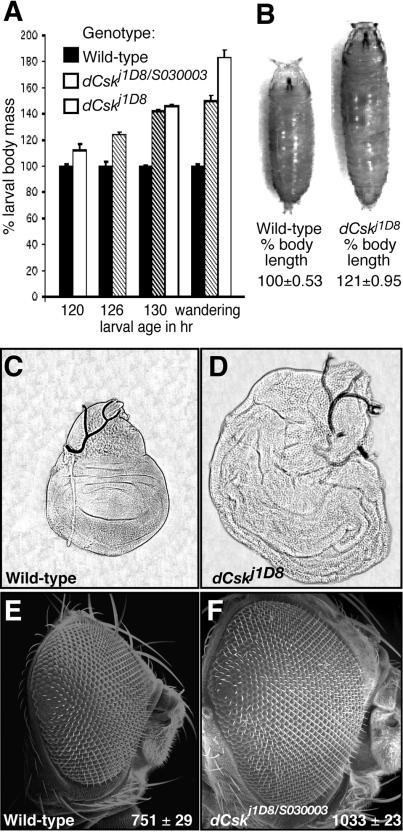
During fly development, embryos hatch to progress through three larval stages followed by pupation and metamorphosis. dCsk mutants occasionally survived through later pupal development, allowing for characterization of dCsk−/− larvae and pupae. The most striking phenotype of dCsk mutants was their increased body size relative to wild-type animals (Fig. 2A and B). Early third instar dCsk−/− larvae weighed 30% more than age-matched wild-type larvae and eventually grew to weigh 84% more than wild-type larvae due to a prolonged larval stage in which they continued to feed and grow long after wild-type controls had formed a prepupa (Fig. 2A). dCsk−/− pupae displayed a 21% increase in body length versus controls (Fig. 2B). Wandering dCsk mutant larvae showed enlargement of tissues such as the brain, ventral ganglion, and salivary glands and enlargement of the wing, leg, and eye imaginal discs (Fig. 2C and D) (data not shown). Delayed pupariation is frequently observed in Drosophila growth control mutants that show large larval body size and imaginal disc overgrowth; this delay may reflect defective coordination of larval growth and endocrine signals for metamorphosis (10, 11, 78).
FIG. 2.
Mutations in dCsk cause increased body and organ size. (A) Graph of average larval mass during third instar (final larval stage). Values are normalized to wild-type larval mass for each time point. Larval age is noted in hours AED. A minimum of 40 larvae were measured for each genotype at each time point. Unstaged wandering third-instar wild-type larvae were compared to >160-h dCskj1D8 wandering larvae. Bars indicate standard errors. (B) Wild-type and dCsk mutant pupae with relative length measurements. Values are normalized to wild-type body length. A minimum of 35 pupae was measured for each genotype. (C and D) Phase contrast images of wing imaginal discs from wandering third instar larvae. Panel D shows a dCskj1D8 mutant disc photographed at the same magnification as a wild-type disc (C). (E and F) SEMs of a wild-type adult eye and a dCskJ1D8/S030003 pharate adult eye taken at the same magnification. Inset values show the average number of ommatidia per genotype (n = 3 for each).
Pharate adults are animals that attain a nearly adult morphology but die within the pupal case. The eyes and heads of the occasional dCskj1D8/S030003 and dCskS030003 mutants that survived as pharate adults were frequently enlarged, and posterior ommatidia were sometimes misaligned (Fig. 2E and F). Histological sections indicated that individual mutant ommatidia were morphologically normal (data not shown), but dCsk mutant eyes contained more ommatidia than wild-type controls (Fig. 2E and F). Rarely, the eyes were replaced with duplicated antennae (data not shown). In addition, the wings and legs were severely malformed, the notum was sometimes split, and the head, legs, and notum often contained cuticle outgrowths (data not shown).
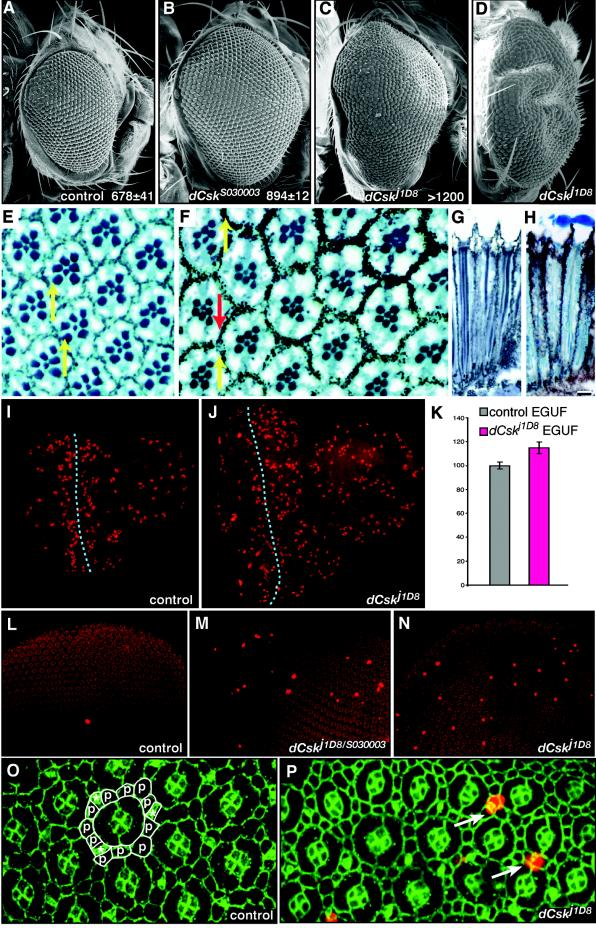
To resolve the origin of the retinal defects, we utilized the EGUF system to generate whole-eye clones in which all adult eye tissue is homozygous for dCsk mutations in an otherwise heterozygous animal (65). This approach permitted us to isolate dCsk activity within the retina from effects of the prolonged larval stage; flies with eyes homozygous for dCsk mutations developed along a normal time course. dCsk−/− EGUF clones were also enlarged in comparison to controls, with some dCskj1D8 clones so enlarged that the eyes became malformed to pack onto a normally sized head (Fig. 3A to D). Occasionally, dCsk−/− EGUF clones resulted in antennal duplication and cuticle overgrowth, phenotypes that recapitulated defects seen in dCsk−/− pharate adults (data not shown). These data indicate that dCsk regulates organ size within the developing eye.
FIG. 3.
dCsk mutants show an increased proliferation. (A to D) SEMs of EGUF eye clones FRT82B Ubi-GFP control (A), dCskS030003 (B), and dCskj1D8 (C) and rear view of severely overgrown dCskj1D8 EGUF clone (D). Inset values show the average number of ommatidia per eye per genotype (n = 3 or more). (E) Adult retinal sections of control EGUF eyes. The normal complement of photoreceptor neurons can be confirmed by the 7 rhabdomeres within each ommatidium. Note that each rhabdomere array forms a trapezoid that points upward (yellow arrows). (F) Sections of adult dCskj1D8 EGUF eye clones show normal ommatidia. However, some ommatidia have reversed planar polarity, as assessed by rhabdomere assays (red arrow). (G and H) Longitudinal sections of an adult EGUF control retina (G) and a dCskj1D8 retina (H) show that dCskj1D8 ommatidia are normal in depth as well as width. Lenses were lost from some ommatidia during sectioning. (I and J) Larval eye-antennal imaginal discs from EGUF clones stained with anti-phospho-histone to highlight mitotic nuclei. The anterior is shown toward the right. For each,the eye disc is to the left and the rounded portion of tissue to the right is the antennal disc. The dotted line marks the position of the morphogenetic furrow. The very large phospho-histone-positive nuclei posterior to the furrow are within the overlying peripodial membrane and not located in the eye disc proper. Most phospho-histone-positive nuclei posterior to the furrow are within the eye disc and are part of the second mitotic wave. Note the increased size of the dCsk eye disc and the increased number of phosphohistone-positive nuclei anterior to the furrow in panel J. (K) Analysis of mitosis anterior to the morphogenetic furrow in eye imaginal discs. The number of mitotic nuclei anterior to the furrow was quantified, and results were controlled for tissue mass (see Materials and Methods). Results are normalized to those of control EGUF tissue. Bars represent standard errors. (L to N) Pupal eye discs at 26 h after puparium formation stained with anti-phospho-histone (bright red) to highlight mitotic nuclei. All nuclei stain faintly (background red) with the antiphosphohistone antibody. (O and P) Pupal eye discs at 26 h after puparium formation stained with anti-phospho-histone antibody (red) to mark mitotic cells and anti-armadillo antibody (green) to mark cell boundaries. Each individual ommatidium is composed of 8 photoreceptors (located basally out of the plane of focus), 4 overlying cone cells, and 2 primary pigment cells. (O) Cells that make up the interommatidial lattice surrounding a single unit ommatidium are outlined in white; lattice cells include bristle groups (*) and secondary and tertiary pigment cells (p). (P) The excess mitotic cells in dCskj1D8 tissue are located within the interommatidial lattice (arrows) as determined by nuclear position and cell positions.
The enlarged dCsk−/− EGUF eyes contained an increased number of ommatidia (Fig. 3A to C). Histological sections showed that dCsk mutant EGUF eyes contained some ommatidia with planar polarity inversions (Fig. 3E and F). Yet, cells within dCsk−/− adult ommatidia were otherwise normal in morphology and size (Fig. 3E to H), indicating that eye overgrowth occurred by the addition of extra cells during eye imaginal disc development rather than by increased ommatidial or cell size. Retinal cell proliferation occurs almost exclusively within the embryonic and larval eyes, and the observed extra cells most likely derive from excess proliferation during these stages. Importantly, previous studies show that blocking apoptosis does not affect eye size (28, 40). Consistent with overproliferation, late larval-stage eye-antennal imaginal discs from dCsk−/− EGUF clones were enlarged compared to age-matched controls and showed an increase in proliferating cells (Fig. 3I to K).
Genotypically, dCsk−/− EGUF eyes could be become enlarged either because dCsk−/− cells grow faster than normal cells or because dCsk−/− cells fail to exit the cell cycle properly and continue to proliferate when wild-type cells would not. To explore this issue, we took advantage of the tight control of mitoses during larval and pupal eye development. In the larval eye, the morphogenetic furrow is a region of cell cycle coordination in which no mitotic cells are normally present. Yet, mitotic cells were sometimes observed within the morphogenetic furrow in dCsk−/− EGUF larval eye tissue (see also reference 64) (data not shown). The presence of mitotic cells in this region suggests that dCsk mutant cells fail to exit the cell cycle. The early pupal retina normally undergoes very little if any proliferation, restricted only to the precursor cells of the interommatidial bristles. Only an average of 2 mitotic cells were observed in wild-type tissue (2 ± 0.40, mean ± standard error [SE], n = 15). However, we observed an increase in mitotic cells in both dCskj1D8/S030003 whole mutant and dCskj1D8 EGUF pupal retinas (15 ± 4.2 mitotic cells, mean ± SE, n = 15) relative to wild-type controls, demonstrating that dCsk mutant cells do not normally exit the cell cycle (Fig. 3L to N). These proliferating cells were located within the interommatidial lattice, and some appeared to be bristle precursor cells (Fig. 3O and P). dCsk−/− mutant pupal retinas contained excess interommatidial lattice cells (Fig. 3O and P), some of which are likely derived from increased lattice proliferation. Staining for apical cell profiles revealed that dCsk−/− tissues had an average of 16 cells surrounding a single ommatidial cluster (16 ± 0.48, mean ± SE, n = 15) while control tissues showed an average of 12 cells surrounding a single cluster (12 ± 0.21, mean ± SE, n = 15).
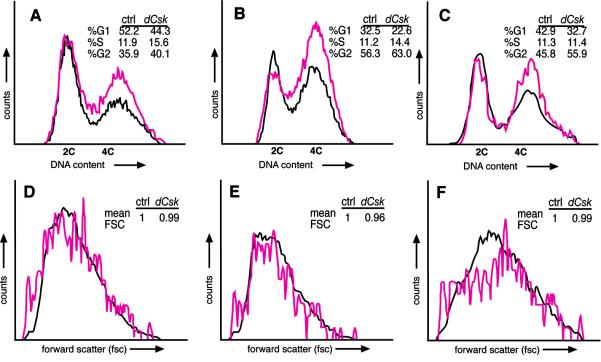
To further explore the role of dCsk in cell proliferation, we utilized flow cytometry analysis in whole eyes and Flp-FRT-generated clones. First, our analysis demonstrated that dissociated cells from whole dCsk mutant eye-antennal and wing disks consistently exhibited a decrease in the G0-G1 population and an increase in the G2-M population compared to cells from age-matched control tissues (Fig. 4A and B). We found these differences in cell cycle profiles in mutant larvae over a range of ages, from 120 to 130 h AED. Similar results were observed with dCsk−/− EGUF larval eyes (data not shown). To assess whether the defects observed with dCsk mutants are cell autonomous, we used the Flp-FRT system to generate mutant clones within the eye. To rigorously score the effects on individual cells (see also Materials and Methods), we dissociated the cells and used flow cytometry analysis to distinguish the dCsk homozygous clonal cells from their wild-type and heterozygous neighbors. dCsk mutant clones contained an increased G2-M population and a decreased G0-G1 population relative to surrounding control tissue (Fig. 4C), a cell cycle defect indicative of increased proliferation. These results are consistent with a similar analysis of the wing (64). Forward scatter measurements confirmed that dCsk homozygous clonal cells and their neighbors were the same average cell size even in different phases of the cell cycle (Fig. 4D to F). Together, these data argue that dCsk controls tissue growth cell autonomously by negatively regulating cellular proliferation without affecting cell size, although we cannot rule out subtle nonautonomous effects.
FIG. 4.
Flow cytometry analysis shows a cell-autonomous proliferative defect in dCsk mutant cells. (A and B) Flow cytometry analysis on wild-type (black) and dCskj1D8/S030003 mutants (pink). The DNA content of eye-antennal disc cells (A) and wing disk cells (B) from 126- to 128-h-AED larvae is shown. We observed similar differences between wild-type and dCsk mutant cell cycle profiles in more than seven repeat experiments. (C) DNA content of dCskj1D8 mutant cells from mitotic clones and neighboring wild-type and heterozygous cells from eye-antennal tissue (120 h AED). Forward scatter (FSC) profiles of eye-antennal cells from panel C (D) and separate analysis of cells with 2C (E) and 4C (F) DNA content. FSC is a relative measurement of cell size (51). The mean FSC of dCsk mutant cells in relation that of to control (ctrl) cells is indicated.
dCsk acts in opposition to the Src and Jun N-terminal kinase (JNK) pathways.
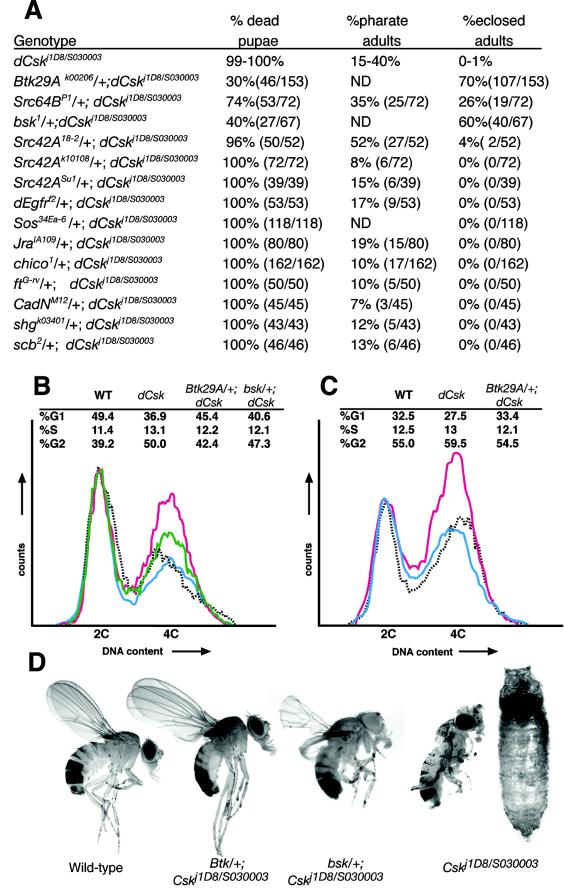
We utilized a dCskj1D8/S030003 trans-heterozygote combination, which shows an intermediate phenotype, to test candidate loci for an in vivo role in dCsk function (Fig. 5A). We crossed null or strongly hypomorphic alleles of candidate genes into dCskj1D8/S03003 mutants and monitored lethal phase and phenotypes. Several candidate genes, such as members of the Ras pathway, failed to genetically interact with dCsk (Fig. 5A). The dCsk phenotype was suppressed by mutations in the Drosophila Src ortholog Src64B (Fig. 5A). Normally, 10 to 40% of developing dCskj1D8/S030003 flies survived to the pharate stage, and only 0 to 1% eclosed (emerged) as live adults from their pupal cases. Removing one copy of Src64B led to fully 61% surviving to become either pharate adults or live adults, and 26% of all Src64BPI/+; dCskj1D8/S030003 flies eclosed as live adults from their pupal cases. The eclosed adults often displayed wing and leg defects and typically died within 24 to 48 h (data not shown). Mutations in the Src ortholog Src42A weakly suppressed dCsk phenotypes: 56% of dCsk mutants either eclosed as live adults or survived to the pharate stage when one copy of Src42A was removed by using the Src42A18-2 allele (Fig. 5A).
FIG. 5.
Reduced Src, Btk29A, and bsk function suppresses the dCsk phenotype. (A) Genetic interactions between dCskj1D8/S030003 and candidate effector genes. Control dCskj1D8/S030003 animals were grown alongside each experimental group. Observed pharate adults are included in the total count of dead pupae. Results for dCsk controls are presented as ranges observed in separate groups of animals (>35 animals per control group). The bsk2 allele was tested and displayed an effect similar to that of bsk1 (data not shown). dEgfr encodes the epidermal growth factor receptor ortholog; Sos encodes a Ras GTPase exchange factor; Jra encodes the c-jun ortholog; chico encodes the insulin receptor substrate ortholog; ft, CadN, and shg encode cadherin orthologs; and scb encodes an α-integrin subunit. The dEgfr, Sos, Jra, and ft alleles used are nulls, and the chico, Btk29A, Src64B, bsk, Src42A18-2, Src42ASu1, CadN, and scb alleles used are strong hypomorphs. The dEgfr, Sos, and Jra alleles are all known genetic modifiers of receptor tyrosine kinase and Ras pathways (21, 39). ND, not determined. (B and C) FACS analysis for DNA content of eye-antennal imaginal disc cells (B) and wing imaginal discs (C) from wild-type (WT) (dashed grey), dCskj1D8/S030003 (pink), Btk29Ak00206/+; dCskj1D8/S030003 (blue), and bsk1/+; dCskj1D8/S030003 (green) 128-h-AED larvae. (D) Photographs of adult flies demonstrating that Btk29Ak00206 and bsk1 mutations rescue dCskj1D8/S030003 mutants. The fly on the far right is a rare dCskj1D8/S030003 pharate adult; note the curled legs, the uninflated wings, and the slightly enlarged eye.
The Btk29A locus encodes the sole Tec-Btk family kinases in the Drosophila genome, which function downstream of fly Src kinases such as Src64B (3, 24, 57). Mutations in Btk29A strongly suppressed dCsk phenotypes: 70% of Btk29A−/+; dCskj1D8/S030003 flies fully eclosed as nearly normal adults and exhibited only mild wing defects (Fig. 5A and D). These Btk29A−/+; dCskj1D8/S030003flies were otherwise viable and fertile. In addition, reduced Btk29A function also noticeably suppressed the increased body size and prolonged larval phase observed in dCsk mutants (data not shown), although previous studies had not implicated Btk29A, Src64B, or Src42A in larval growth control or pupation. Btk29A also mediated the cell cycle defects observed in dCsk mutants: fluorescence-activated cell sorter (FACS) analysis of dissociated wing and eye antennal imaginal discs derived from Btk29A−/+; dCskj1D8/S030003 larvae indicated that removal of a copy of Btk29A suppressed the increase in G2-M cells observed in dCsk mutants (Fig. 5B and C).
The JNK signaling pathway has also been identified as a mediator of Src signaling in both mammals and Drosophila (68). Consistent with this data, removing one copy of the JNK ortholog basket (bsk) also suppressed the dCsk phenotype. 60% of bsk1/+; dCskj1D8/S030003 flies formed viable adults that fully or partially eclosed (Fig. 5A). Similar to Src64BPI/+; dCskj1D8/S030003 survivors, bsk1/+; dCskj1D8/S030003 adults exhibited leg and wing defects and died shortly after eclosion (Fig. 5D). bsk1/+; dCskj1D8/S030003 larvae and pupae also showed suppression of the increased body size and prolonged larval phase observed in dCskj1D8/S030003 mutants (data not shown). FACS analysis indicated that bsk1/+; dCskj1D8/S030003 larval eye-antennal discs have an increased G0-G1 and decreased G2-M population relative to +/+; dCskj1D8/S030003 discs (Fig. 5B), demonstrating that mutations in bsk suppress the cell cycle defects caused by loss of dCsk.
dCsk negatively regulates Jak/Stat signaling.
Another pathway linked to Src signaling in mammalian tissue culture models is the Jak/Stat signal transduction pathway. Src can directly phosphorylate and activate STAT3 in vitro, and STAT3 function and activation are required for Src transforming activity in multiple tissue culture cell lines (8, 13, 75). In the Drosophila eye, the Jak/Stat pathway controls proliferation and planar polarity. The Drosophila Jak/Stat pathway is composed of the ligand Unpaired (Upd), the receptor Domeless, the single Jak ortholog Hopscotch (Hop), and the single STAT ortholog Stat92E. Mutations that reduce the activity of hop or upd reduce the growth of the eye (5, 44), whereas gain-of-function mutations in Upd and Hop cause increased cellular proliferation within the eye and other tissues (4, 27). Overexpression of PIAS, a negative regulator of Stat92E, reduces growth of the eye (5). Loss-of-function mutations in hop and gain-of-function mutations in upd also disrupt planar polarity within the eye (76).
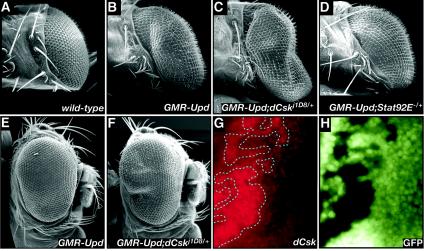
Recent work has demonstrated that Upd overexpression in the eye leads to a STAT-dependent overgrowth phenotype and ommatidial polarity defects very similar to those seen in our dCsk−/− EGUF clones (Fig. 6B) (4, 15). In particular, Upd overexpression within the eye caused increased eye size by increasing the number of ommatidia and elevated cellular proliferation within undifferentiated cells anterior to the morphogenetic furrow, all without affecting ommatidial size or cell size (4); this phenotype is reminiscent of defects observed in dCsk−/− EGUF clones. Removing one copy of Stat92E suppressed the Upd overexpression phenotype, demonstrating that the Upd phenotype was sensitive to alterations in Jak/Stat function (Fig. 6D). Removing one copy of dCsk enhanced eye overgrowth caused by Upd overexpression, demonstrating that dCsk negatively regulates the Jak/Stat pathway in this paradigm (Fig. 6B, C, E, and F).
FIG. 6.
dCsk negatively regulates Jak/Stat activity. (A to F) SEMs showing dorsal views and/or lateral views of eyes from the wild type (A) and overexpression of Upd in a wild-type background (B and E), a dCskj1D8/+ background (C and F), and a Stat92E−/+ background (D). The posterior region of the GMR-Upd; dCskj1D8/+ eye in panels C and F is enlarged relative to GMR-Upd;+/+ in panels B and E. (G and H) Stat92E expression (red) is increased in dCskS030003 clones (clones designated by a dotted line have a lack of GFP expression).
One indicator of Drosophila Jak/Stat activity is Stat92E transcript and protein levels: upd and hop mutant flies show decreased Stat92E transcript and protein expression and Upd overexpression in the eye and elsewhere leads to increased Stat92E transcript and protein (15, 33, 77) Previous studies have concluded that increased levels of Stat92E protein identifies cells receiving an Upd and/or Jak signal (33). Cells fully mutant for dCsk showed a clear elevation in Stat92E protein levels relative to wild-type or heterozygous eye tissue (Fig. 6G and H). This increase indicates that the Jak/Stat pathway is up-regulated in dCsk mutants and that this up-regulation may provoke some of the cellular defects observed in dCsk−/− eyes.
dCsk phenotype requires Stat92E function.
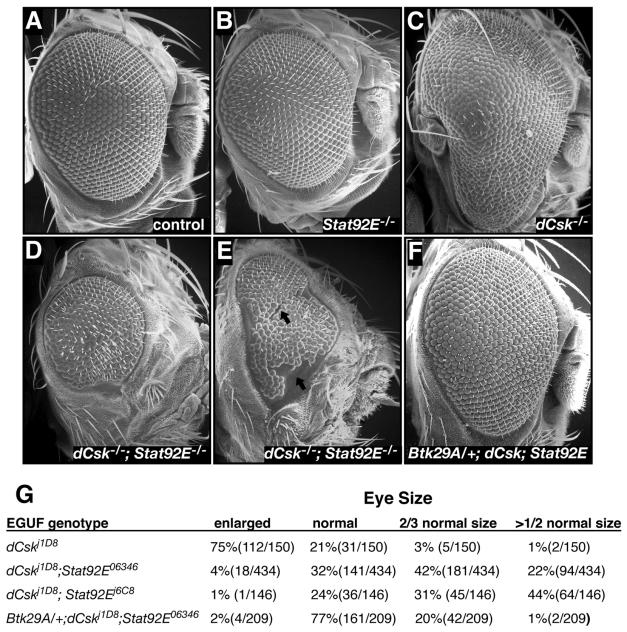
To further explore the role of Stat92E in dCsk function, we utilized the EGUF system to create eyes fully mutant for both dCsk and Stat92E; the presence of both loci on the same chromosomal arm simplified this approach. Eyes mutant for Stat92E alone were mostly normal, showing a slight reduction in size, some misaligned ommatidia and, infrequently, missing antennal structures (Fig. 7B). Genotypically, dCskj1D8; Stat92E06346 EGUF eyes were consistently and often significantly smaller than either dCskj1D8 or Stat92E06346 EGUF eyes alone, demonstrating a block in the overgrowth phenotype (Fig. 7A to E and G). In addition, dCskj1D8; Stat92E06346 adult eyes were frequently fragmented, with scars and/or patches of eye tissue separated by patches of cuticle (Fig. 7E), suggesting that mutant tissue underwent localized programmed cell death during development. dCskj1D8; Stat92E06346 EGUF eyes also showed loss of antennal structures and head cuticle malformations. The cuticle malformations were present on animals with small and scarred eyes, suggesting that these malformations are secondary to retinal defects. All of these observations were confirmed in dCskj1D8; Stat92Ej6C8 EGUF clones, which demonstrated an even higher penetrance of eye tissue loss (Fig. 7G).
FIG. 7.
dCsk phenotypes require Stat92E function. (A to F) SEMs of EGUF eyes comparing the control (A) and Stat92E06346 (B), dCskj1D8 (C), dCskj1D8; Stat92E06346 (D and E), and Btk29Ak00206/+; dCskj1D8; Stat92E06346 (F). Note the scarring and fragmentation seen in dCskJ1D8; Stat92E06346 adult eyes (arrows in panel E). (G) Penetrance of EGUF phenotypes per eye of the genotypes indicated. Some dCskJ1D8; Stat92E−/− EGUF eyes were actually the size of a pinpoint, and some showed cuticle scarring that split the eye field.
To determine whether the defects we observed in dCskj1D8; Stat92E−/− EGUF clones were Src dependent, we removed one copy of Btk29A in dCskj1D8; Stat92E06346 EGUF clones. If the reduced eye size of dCskj1D8; Stat92E EGUF clones is due to Src hyperactivation, then reduced Btk29A function should rescue the dCskj1D8; Stat92E EGUF phenotype; if, however, the dCskj1D8; Stat92E−/− phenotype is caused by nonspecific synthetic lethality, then it should not be sensitive to the loss of a single copy of Btk29A. Consistent with the former possibility, reduced Btk29A suppressed and rescued the dCskj1D8; Stat92E06346 EGUF phenotype to a more normal phenotype (Fig. 7F and G). In particular, 64% of all adult dCskj1D8; Stat92E06346 eyes were two-thirds or less of normal size while only 21% of all adult eyes from Btk29Ak00206/+; dCskj1D8; Stat92E06346 mutant eyes were similarly reduced. Additionally, 77% of the Btk29Ak00206/+; dCskj1D8; Stat92E06346 eyes were normal or nearly normal in size, whereas only 32% of dCskj1D8; Stat92E06346 EGUF clones were similarly normal. Indeed, most Btk29Ak00206/+; dCskj1D8; Stat92E06346 EGUF clones looked very similar to Stat92E−/− EGUF clones, as both genotypes showed some misaligned ommatidia and, occasionally, missing antennal structures.
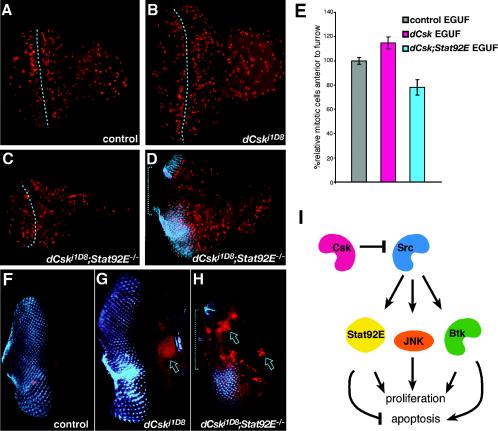
To determine the developmental origin of the dCskj1D8; Stat92E−/− EGUF phenotype, we examined eye-antennal imaginal discs. dCskj1D8; Stat92E06346 mutant larval eye-antennal discs frequently showed reduced sizes relative to controls, Stat92E−/−, or dCsk−/− EGUF clones (Fig. 8A to D). dCskj1D8; Stat92E06346 EGUF clones also frequently showed a reduction or absence of developing antennal tissues. dCskj1D8; Stat92E06346 EGUF clones also showed reduced mitoses anterior to the morphogenetic furrow compared to control or dCskj1D8 clones (Fig. 8E). In addition, dCskj1D8; Stat92E06346 mutant larval eye tissue often exhibited patchy expression of neural markers within the developing eye field, and these regions also showed decreased proliferation relative to control or dCskj1D8 tissue (Fig. 8D). Regions with reduced neural development harbored cells with abnormal and pyknotic nuclei as visualized by 4′,4′-diamidino-2-phenylindole (DAPI) staining (data not shown), suggesting that cells within the eye were undergoing apoptosis. Flow cytometry experiments indicated that dCskj1D8; Stat92E06346 larval eye tissue contained many apoptotic cells (data not shown). Consistent with these data, dCskj1D8; Stat92E06346 mutant larval eye tissue often exhibited increased programmed cell death and tissue loss within the developing eye field (Fig. 8F to H). This apoptosis primarily occurred in regions with reduced neural marker expression, indicating that defective neural differentiation may occur as a consequence of excessive apoptosis during development. Such extensive apoptosis is likely to disrupt the propagation of normal developmental processes in the eye field, accounting for much of the tissue loss and scarring observed in adult dCskj1D8; Stat92E−/− EGUF clones. In summary, reducedStat92E activity inhibited SFK-mediated overgrowth in dCsk mutant tissue by reducing cell proliferation and promoting apoptotic cell death.
FIG. 8.
dCsk; Stat92E mutant tissue shows reduced proliferation and increased apoptosis. (A to D) Phospho-histone staining (red spots) designates mitotic nuclei in developing larval eye-antennal discs. The anterior is toward the right. The dotted line marks the morphogenetic furrow. (A) FRT82B Ubi-GFP control; (B) dCskj1D8 EGUF eye discs are larger and contain somewhat higher numbers of mitoses anterior to the furrow; (C) dCskj1D8; Stat92E06346 EGUF eye discs are smaller with somewhat fewer mitotic cells anterior to the furrow; (D) a dCskj1D8; Stat92E06346 EGUF eye disc showing a region that fails to express the 22C10 neural marker (blue). Note the large field nearly devoid of mitotic cells (bracket) in dCskj1D8; Stat92E06346 tissue (D) within and anterior to a region that would normally express 22C10. (E) Analysis of mitosis anterior to the morphogenetic furrow in eye imaginal discs. The number of mitotic nuclei anterior to the furrow was quantified, and results were controlled for tissue mass (see Materials and Methods). Results are normalized to those of control EGUF tissue. Bars represent standard errors. (F to H) Photoreceptor cell differentiation and cell death were visualized with 22C10 (blue) and anti-active caspase 7 (red) antibodies. Little cell death is present in the control (F); the emerging eye field is apparent as organized photoreceptors in the posterior half of the eye disc. Note the increased size of the dCskJ1D8 disc (G); the flare in 22C10 staining is due to a fold in the optic stalk. (G) dCskJ1D8 EGUF clones often contain a region of cell death anterior to the eye field (arrow) but little within. (H) dCskJ1D8; Stat92E06346 EGUF eye discs contain, in addition, substantial cell death within the eye field (arrows). Bracket indicates a portion of the eye field devoid of developing photoreceptors. (I) Diagram depicting the function of dCsk within developing eye epithelia. dCsk inhibits growth and proliferation by inhibiting Src signaling. Src signaling acts through Btk29A, bsk (JNK), and Stat92E activity to induce tissue overgrowth and excess proliferation. Stat92E function is required for the survival of dCsk mutant tissue. Excess Src-Btk activity may activate proapoptotic pathways that are revealed when Stat92E function is reduced.
DISCUSSION
Csk family kinases encode critical negative regulators of SFKs. In this report, we demonstrate that Drosophila dCsk is a vital negative regulator of growth and proliferation. Loss of dCsk activity leads to overgrowth of multiple tissues, and this overgrowth requires the functions of Src-Btk, JNK, and STAT signal transduction pathways (Fig. 8I). In another recent report, dCsk is linked to signaling from the Lats tumor suppressor (64). Together, these results provide support for the long-suspected role of human Csk as tumor suppressors.
Partial reduction of Src64B, Src42A, or Btk29A activity suppressed the dCsk−/− phenotype, providing functional data to support the view that the imaginal disc overgrowth, defective larval and pupal development, and lethality of dCsk−/− mutants results from inappropriate activation of the Src-Btk signal transduction pathways. Mutations in Btk29A more strongly suppressed dCsk phenotypes than either Src42A or Src64B mutations, perhaps reflecting that (i) Src paralogs act redundantly to each other in Drosophila as in mammals (68) and (ii) Btk29A has previously been shown to act downstream of SFKs in flies and in mammals (24, 57, 59, 68). We provide in vivo evidence that loss of Csk function hyperactivates Btk to drive cell cycle entry in development, demonstrating that Tec-Btk family kinases are critical to SFK-mediated proliferation. Our data raise the possibility that partial reduction of Tec-Btk kinase activity could reduce proliferation in other cellular contexts in which overgrowth is driven by hyperactivated SFKs, such as in colon tumors.
Tissue culture models show that constitutively activated SFK signal transduction modulates the function of numerous downstream effector molecules and pathways. Using a loss-of-function approach to identify effectors that mediate the dCsk overgrowth phenotypes, we failed to implicate some of these pathways in dCsk function. For example, SFKs up-regulate the SOS-Ras-ERK pathway in multiple tissue culture studies and Drosophila overexpression models (for examples, see references 41, 60, and 66). However, although dRas1 signaling is active throughout retinal development, reduced dEGFR, Sos, and Jra (c-jun) gene dosage failed to affect the dCsk phenotype. dCsk mutations also failed to modify a hypermorphic allele of dEGFR (data not shown). Levels of doubly phosphorylated and activated ERK appeared unaltered in dCsk−/− tissue (data not shown). Moreover, the dCsk phenotype failed to phenocopy defects caused by Ras pathway hyperactivation. For example, constitutively active dRas1 causes increased cell size and patterning defects in the developing imaginal discs (25, 35, 36, 54), defects that were not observed in dCsk mutant eye tissues. These data argue that not every signal transduction pathway implicated in SFK tissue culture models necessarily functions as predicted within a developing epithelial tissue.
Our studies emphasized the importance of two signaling pathways in dCsk and SFK function. Since certain defects in dCsk−/− animals, such as a split notum, resembled those of hep (JNKK) mutants, we suspected that JNK pathway activity was involved in dCsk function (68). Phenotypic and FACS analysis established that reduced JNK (bsk) function suppressed the phenotypes and cell cycle defects caused by loss of dCsk. These results confirm studies indicating that JNK functions downstream of the Src-Btk pathway in Drosophila and mammalian tissue culture cells (50). Components of the JNK pathway are required for Src-dependent cellular transformation (22, 46, 72), but the exact role of JNK in these cells is unknown. Importantly, our data show that the JNK pathway mediates proliferative responses to Src signaling in vivo. Further work will be needed to precisely understand its role in proliferation.
Our genetic studies also highlighted the importance of the Jak/Stat signal transduction pathway. dCsk proved a negative regulator of Jak/Stat signaling; for example, dCsk mutant tissues show up-regulation of Stat92E protein, a hallmark of Jak/Stat activation in Drosophila. Stat92E, the sole Drosophila STAT ortholog, is most similar to mammalian STAT3. In mammalian cells, Src directly phosphorylates and activates STAT3 and STAT3 function and activation are required for Src transforming activity (8, 13, 75). Conversely, overexpression of Csk blocks STAT3 activation in v-Src transformed fibroblasts (55). Activating mutations in STAT3 can also promote oncogenesis in mice (9). However, the physiological significance of these interactions within developing epithelia remains unclear.
dCsk; Stat92E double mutant clones revealed that blockade of STAT function in dCsk mutants severely reduced Src-dependent overgrowth and promoted apoptosis of mutant tissue. dCsk−/−; Stat92E−/− EGUF adult eyes are nearly identical to phenotypes caused by overexpression of Dacapo, the fly ortholog of the cdk inhibitor p21, and PTEN, a negative regulator of cell proliferation and growth (29, 61, 71). Importantly, removing Stat92E function in dCsk mutant tissue led to a synthetic small eye phenotype and did not simply rescue the dCsk−/− proliferative phenotype. This outcome distinguishes Stat92E from mutations in Src64B, Btk29A, or bsk, which rescued dCsk-mediated defects toward a normal phenotype. The loss of tissue in dCsk−/−; Stat92E−/− clones indicates that Src-Btk signaling provokes apoptosis in the absence of Stat92E function (Fig. 8I). Consistent with this interpretation, reduced Btk29A function rescued the dCsk−/−; Stat92E−/− EGUF phenotype to a more normal phenotype, demonstrating that the reduced growth and increased apoptosis observed in the dCsk−/−; Stat92E−/− tissues is indeed Src-Btk pathway dependent.
Our data suggest the existence of a Src-dependent proapoptotic pathway that is normally suppressed by STAT (Fig. 8I). One possible component of this pathway is JNK, given that JNK signaling is an important activator of apoptosis in both flies and mammals (16). Perhaps Src-dependent hyperactivation of Bsk (JNK) in dCsk−/−; Stat92E−/− tissue contributes to cell death in the absence of proliferative and/or survival signals provided by Stat92E. However, a number of other candidate pathways may also mediate this response. The further characterization and identification of these pathways may have important implications for interceding in Src-mediated oncogenesis.
Together, these observations indicate that, in tissue that contains hyperactive Src or reduced Csk, blocking STAT function is sufficient to trigger apoptosis and decrease proliferation in the absence of any further mutations or interventions. Reduced STAT3 function can promote apoptosis within breast and prostate cancer cells that show elevated SFK activity, but the molecular pathways driving apoptosis in these cells are unknown (23, 47). These cells may require survival signals provided by STAT3 to counteract apoptosis due to chromosomal abnormalities or other defects. Alternatively, these cells may die because of proapoptotic signals provided by hyperactive SFKs in the absence of STAT3 function. Our data argue that the latter may be true, which suggests the intriguing possibility that therapeutic blockade of STAT function in tumors with activated Src may actively provoke Src-dependent apoptosis and growth arrest in tumor tissues.
Acknowledgments
We thank Bill Eades of the Siteman Cancer Center Flow Cytometry Core Lab for assistance with FACS analysis, Mike Veith for assistance with SEMs, and Scott Portman for assistance with injections.
This research was supported by a grant from the National Cancer Institute (5R01CA084309-03) to R.L.C. and an NIH training grant to R.D.R.
REFERENCES
- 1.Abram, C. L., and S. A. Courtneidge. 2000. Src family tyrosine kinases and growth factor signaling. Exp. Cell Res. 254:1-13. [DOI] [PubMed] [Google Scholar]
- 2.Aligayer, H., D. D. Boyd, M. M. Heiss, E. K. Abdalla, S. A. Curley, and G. E. Gallick. 2002. Activation of Src kinase in primary colorectal carcinoma: an indicator of poor clinical prognosis. Cancer 94:344-351. [DOI] [PubMed] [Google Scholar]
- 3.Baba, K., A. Takeshita, K. Majima, R. Ueda, S. Kondo, N. Juni, and D. Yamamoto. 1999. The Drosophila Bruton's tyrosine kinase (Btk) homolog is required for adult survival and male genital formation. Mol. Cell. Biol. 19:4405-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bach, E., S. Vincent, M. P. Zeidler, and N. Perrimon. 2003. A sensitized genetic screen to identify novel regulators and components of the Drosophila JAK-STAT pathway. Genetics 165:1149-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Betz, A., N. Lampen, S. Martinek, M. W. Young, and J. E. Darnell, Jr. 2001. A Drosophila PIAS homologue negatively regulates stat92E. Proc. Natl. Acad. Sci. USA 98:9563-9568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bjorge, J. D., A. Jakymiw, and D. J. Fujita. 2000. Selected glimpses into the activation and function of Src kinase. Oncogene 19:5620-5635. [DOI] [PubMed] [Google Scholar]
- 7.Bolen, J. B., A. Veillette, A. M. Schwartz, V. DeSeau, and N. Rosen. 1987. Activation of pp60c-src protein kinase activity in human colon carcinoma. Proc. Natl. Acad. Sci. USA 84:2251-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bromberg, J. F., C. M. Horvath, D. Besser, W. W. Lathem, and J. E. Darnell, Jr. 1998. Stat3 activation is required for cellular transformation by v-src. Mol. Cell. Biol 18:2553-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bromberg, J. F., M. H. Wrzeszczynska, G. Devgan, Y. Zhao, R. G. Pestell, C. Albanese, and J. E. Darnell, Jr. 1999. Stat3 as an oncogene. Cell 98:295-303. [DOI] [PubMed] [Google Scholar]
- 10.Bryant, P. J., and P. Levinson. 1985. Intrinsic growth control in the imaginal primordia of Drosophila, and the autonomous action of a lethal mutation causing overgrowth. Dev. Biol. 107:355-363. [DOI] [PubMed] [Google Scholar]
- 11.Buratovich, M. A., and P. J. Bryant. 1997. Enhancement of overgrowth by gene interactions in lethal(2)giant discs imaginal discs from Drosophila melanogaster. Genetics 147:657-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cam, W. R., T. Masaki, Y. Shiratori, N. Kato, T. Ikenoue, M. Okamoto, K. Igarashi, T. Sano, and M. Omata. 2001. Reduced C-terminal Src kinase activity is correlated inversely with pp60(c-src) activity in colorectal carcinoma. Cancer 92:61-70. [DOI] [PubMed] [Google Scholar]
- 13.Cao, X., A. Tay, G. R. Guy, and Y. H. Tan. 1996. Activation and association of Stat3 with Src in v-Src-transformed cell lines. Mol. Cell. Biol. 16:1595-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cartwright, C. A., M. P. Kamps, A. I. Meisler, J. M. Pipas, and W. Eckhart. 1989. pp60c-src activation in human colon carcinoma. J. Clin. Investig. 83:2025-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, H. W., X. Chen, S. W. Oh, M. J. Marinissen, J. S. Gutkind, and S. X. Hou. 2002. mom identifies a receptor for the Drosophila JAK/STAT signal transduction pathway and encodes a protein distantly related to the mammalian cytokine receptor family. Genes Dev. 16:388-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, Y. R., and T. H. Tan. 2000. The c-Jun N-terminal kinase pathway and apoptotic signaling. Int. J. Oncol. 16:651-662. [DOI] [PubMed] [Google Scholar]
- 17.Cooper, J. A., K. L. Gould, C. A. Cartwright, and T. Hunter. 1986. Tyr527 is phosphorylated in pp60c-src: implications for regulation. Science 231:1431-1434. [DOI] [PubMed] [Google Scholar]
- 18.Deak, P., M. M. Omar, R. D. Saunders, M. Pal, O. Komonyi, J. Szidonya, P. Maroy, Y. Zhang, M. Ashburner, P. Benos, C. Savakis, I. Siden-Kiamos, C. Louis, V. N. Bolshakov, F. C. Kafatos, E. Madueno, J. Modolell, and D. M. Glover. 1997. P-element insertion alleles of essential genes on the third chromosome of Drosophila melanogaster: correlation of physical and cytogenetic maps in chromosomal region 86E-87F. Genetics 147:1697-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Angelis, P. M., O. P. Clausen, A. Schjolberg, and T. Stokke. 1999. Chromosomal gains and losses in primary colorectal carcinomas detected by CGH and their associations with tumour DNA ploidy, genotypes and phenotypes. Br. J. Cancer 80:526-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dodson, G. S., D. J. Guarnieri, and M. A. Simon. 1998. Src64 is required for ovarian ring canal morphogenesis during Drosophila oogenesis. Development 125:2883-2892. [DOI] [PubMed] [Google Scholar]
- 21.Dong, X., L. Tsuda, K. H. Zavitz, M. Lin, S. Li, R. W. Carthew, and S. L. Zipursky. 1999. ebi regulates epidermal growth factor receptor signaling pathways in Drosophila. Genes Dev. 13:954-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan, G., S. E. Merritt, M. Kortenjann, P. E. Shaw, and L. B. Holzman. 1996. Dual leucine zipper-bearing kinase (DLK) activates p46SAPK and p38mapk but not ERK2. J. Biol. Chem. 271:24788-24793. [DOI] [PubMed] [Google Scholar]
- 23.Garcia, R., T. L. Bowman, G. Niu, H. Yu, S. Minton, C. A. Muro-Cacho, C. E. Cox, R. Falcone, R. Fairclough, S. Parsons, A. Laudano, A. Gazit, A. Levitzki, A. Kraker, and R. Jove. 2001. Constitutive activation of Stat3 by the Src and JAK tyrosine kinases participates in growth regulation of human breast carcinoma cells. Oncogene 20:2499-2513. [DOI] [PubMed] [Google Scholar]
- 24.Guarnieri, D. J., G. S. Dodson, and M. A. Simon. 1998. SRC64 regulates the localization of a Tec-family kinase required for Drosophila ring canal growth. Mol. Cell 1:831-840. [DOI] [PubMed] [Google Scholar]
- 25.Halfar, K., C. Rommel, H. Stocker, and E. Hafen. 2001. Ras controls growth, survival and differentiation in the Drosophila eye by different thresholds of MAP kinase activity. Development 128:1687-1696. [DOI] [PubMed] [Google Scholar]
- 26.Hamaguchi, I., N. Yamaguchi, J. Suda, A. Iwama, A. Hirao, M. Hashiyama, S. Aizawa, and T. Suda. 1996. Analysis of CSK homologous kinase (CHK/HYL) in hematopoiesis by utilizing gene knockout mice. Biochem. Biophys. Res. Commun. 224:172-179. [DOI] [PubMed] [Google Scholar]
- 27.Harrison, D. A., R. Binari, T. S. Nahreini, M. Gilman, and N. Perrimon. 1995. Activation of a Drosophila Janus kinase (JAK) causes hematopoietic neoplasia and developmental defects. EMBO J. 14:2857-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hay, B. A., T. Wolff, and G. M. Rubin. 1994. Expression of baculovirus P35 prevents cell death in Drosophila. Development 120:2121-2129. [DOI] [PubMed] [Google Scholar]
- 29.Huang, H., C. J. Potter, W. Tao, D. M. Li, W. Brogiolo, E. Hafen, H. Sun, and T. Xu. 1999. PTEN affects cell size, cell proliferation and apoptosis during Drosophila eye development. Development 126:5365-5372. [DOI] [PubMed] [Google Scholar]
- 30.Imamoto, A., and P. Soriano. 1993. Disruption of the csk gene, encoding a negative regulator of Src family tyrosine kinases, leads to neural tube defects and embryonic lethality in mice. Cell 73:1117-1124. [DOI] [PubMed] [Google Scholar]
- 31.Irby, R. B., W. Mao, D. Coppola, J. Kang, J. M. Loubeau, W. Trudeau, R. Karl, D. J. Fujita, R. Jove, and T. J. Yeatman. 1999. Activating SRC mutation in a subset of advanced human colon cancers. Nat. Genet. 21:187-190. [DOI] [PubMed] [Google Scholar]
- 32.Irby, R. B., and T. J. Yeatman. 2000. Role of Src expression and activation in human cancer. Oncogene 19:5636-5642. [DOI] [PubMed] [Google Scholar]
- 33.Johansen, K. A., D. D. Iwaki, and J. A. Lengyel. 2003. Localized JAK/STAT signaling is required for oriented cell rearrangement in a tubular epithelium. Development 130:135-145. [DOI] [PubMed] [Google Scholar]
- 34.Kango-Singh, M., R. Nolo, C. Tao, P. Verstreken, P. R. Hiesinger, H. J. Bellen, and G. Halder. 2002. Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development 129:5719-5730. [DOI] [PubMed] [Google Scholar]
- 35.Karim, F. D., H. C. Chang, M. Therrien, D. A. Wassarman, T. Laverty, and G. M. Rubin. 1996. A screen for genes that function downstream of Ras1 during Drosophila eye development. Genetics 143:315-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karim, F. D., and G. M. Rubin. 1998. Ectopic expression of activated Ras1 induces hyperplastic growth and increased cell death in Drosophila imaginal tissues. Development 125:1-9. [DOI] [PubMed] [Google Scholar]
- 37.Klinghoffer, R. A., C. Sachsenmaier, J. A. Cooper, and P. Soriano. 1999. Src family kinases are required for integrin but not PDGFR signal transduction. EMBO J. 18:2459-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kmiecik, T. E., and D. Shalloway. 1987. Activation and suppression of pp60c-src transforming ability by mutation of its primary sites of tyrosine phosphorylation. Cell 49:65-73. [DOI] [PubMed] [Google Scholar]
- 39.Kockel, L., J. Zietlinger, L. M. Staszewski, M. Mlodzik, and D. Bohmann. 1997. Jun in Drosophila development: redundant and nonredundant functions and regulation by two MAPK signal transduction pathways. Genes Dev. 11:1748-1758. [DOI] [PubMed] [Google Scholar]
- 40.Kurada, P., and K. White. 1998. Ras promotes cell survival in Drosophila by downregulating hid expression. Cell 95:319-329. [DOI] [PubMed] [Google Scholar]
- 41.Kussick, S. J., K. Basler, and J. A. Cooper. 1993. Ras1-dependent signaling by ectopically-expressed Drosophila src gene product in the embryo and developing eye. Oncogene 8:2791-2803. [PubMed] [Google Scholar]
- 42.Kussick, S. J., and J. A. Cooper. 1992. Overexpressed Drosophila src 64B is phosphorylated at its carboxy-terminal tyrosine, but is not catalytically repressed, in cultured Drosophila cells. Oncogene 7:2461-2470. [PubMed] [Google Scholar]
- 43.Lowe, C., T. Yoneda, B. F. Boyce, H. Chen, G. R. Mundy, and P. Soriano. 1993. Osteopetrosis in Src-deficient mice is due to an autonomous defect of osteoclasts. Proc. Natl. Acad. Sci. USA 90:4485-4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo, H., H. Asha, L. Kockel, T. Parke, M. Mlodzik, and C. R. Dearolf. 1999. The Drosophila Jak kinase hopscotch is required for multiple developmental processes in the eye. Dev. Biol. 213:432-441. [DOI] [PubMed] [Google Scholar]
- 45.Masaki, T., M. Okada, M. Tokuda, Y. Shiratori, O. Hatase, M. Shirai, M. Nishioka, and M. Omata. 1999. Reduced C-terminal Src kinase (Csk) activities in hepatocellular carcinoma. Hepatology 29:379-384. [DOI] [PubMed] [Google Scholar]
- 46.Minden, A., A. Lin, F. X. Claret, A. Abo, and M. Karin. 1995. Selective activation of the JNK signaling cascade and c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell 81:1147-1157. [DOI] [PubMed] [Google Scholar]
- 47.Mora, L. B., R. Buettner, J. Seigne, J. Diaz, N. Ahmad, R. Garcia, T. Bowman, R. Falcone, R. Fairclough, A. Cantor, C. Muro-Cacho, S. Livingston, J. Karras, J. Pow-Sang, and R. Jove. 2002. Constitutive activation of Stat3 in human prostate tumors and cell lines: direct inhibition of Stat3 signaling induces apoptosis of prostate cancer cells. Cancer Res. 62:6659-6666. [PubMed] [Google Scholar]
- 48.Morrison, D. K., M. S. Murakami, and V. Cleghon. 2000. Protein kinases and phosphatases in the Drosophila genome. J. Cell Biol. 150:F57-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nada, S., T. Yagi, H. Takeda, T. Tokunaga, H. Nakagawa, Y. Ikawa, M. Okada, and S. Aizawa. 1993. Constitutive activation of Src family kinases in mouse embryos that lack Csk. Cell 73:1125-1135. [DOI] [PubMed] [Google Scholar]
- 50.Nagao, M., Y. Kaziro, and H. Itoh. 1999. The Src family tyrosine kinase is involved in Rho-dependent activation of c-Jun N-terminal kinase by Galpha12. Oncogene 18:4425-4434. [DOI] [PubMed] [Google Scholar]
- 51.Neufeld, T. P., A. F. de la Cruz, L. A. Johnston, and B. A. Edgar. 1998. Coordination of growth and cell division in the Drosophila wing. Cell 93:1183-1193. [DOI] [PubMed] [Google Scholar]
- 52.Piwnica-Worms, H., K. B. Saunders, T. M. Roberts, A. E. Smith, and S. H. Cheng. 1987. Tyrosine phosphorylation regulates the biochemical and biological properties of pp60c-src. Cell 49:75-82. [DOI] [PubMed] [Google Scholar]
- 53.Potter, C. J., G. S. Turenchalk, and T. Xu. 2000. Drosophila in cancer research. An expanding role. Trends Genet. 16:33-39. [DOI] [PubMed] [Google Scholar]
- 54.Prober, D. A., and B. A. Edgar. 2000. Ras1 promotes cellular growth in the Drosophila wing. Cell 100:435-446. [DOI] [PubMed] [Google Scholar]
- 55.Ram, P. T., C. M. Horvath, and R. Iyengar. 2000. Stat3-mediated transformation of NIH-3T3 cells by the constitutively active Q205L Galphao protein. Science 287:142-144. [DOI] [PubMed] [Google Scholar]
- 56.Rooney, P. H., A. Boonsong, J. A. McKay, S. Marsh, D. A. Stevenson, G. I. Murray, S. Curran, N. E. Haites, J. Cassidy, and H. L. McLeod. 2001. Colorectal cancer genomics: evidence for multiple genotypes which influence survival. Br. J. Cancer 85:1492-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roulier, E. M., S. Panzer, and S. K. Beckendorf. 1998. The Tec29 tyrosine kinase is required during Drosophila embryogenesis and interacts with Src64 in ring canal development. Mol. Cell 1:819-829. [DOI] [PubMed] [Google Scholar]
- 58.Samokhvalov, I., J. Hendrikx, J. Visser, A. Belyavsky, D. Sotiropolous, and H. Gu. 1997. Mice lacking a functional chk gene have no apparent defects in the hematopoietic system. Biochem. Mol. Biol. Int. 43:115-122. [DOI] [PubMed] [Google Scholar]
- 59.Saouaf, S. J., S. Mahajan, R. B. Rowley, S. A. Kut, J. Fargnoli, A. L. Burkhardt, S. Tsukada, O. N. Witte, and J. B. Bolen. 1994. Temporal differences in the activation of three classes of non-transmembrane protein tyrosine kinases following B-cell antigen receptor surface engagement. Proc. Natl. Acad. Sci. USA 91:9524-9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schlaepfer, D. D., and T. Hunter. 1996. Evidence for in vivo phosphorylation of the Grb2 SH2-domain binding site on focal adhesion kinase by Src-family protein-tyrosine kinases. Mol. Cell. Biol. 16:5623-5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Secombe, J., J. Pispa, R. Saint, and H. Richardson. 1998. Analysis of a Drosophila cyclin E hypomorphic mutation suggests a novel role for cyclin E in cell proliferation control during eye imaginal disc development. Genetics 149:1867-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simon, M. A., D. D. Bowtell, G. S. Dodson, T. R. Laverty, and G. M. Rubin. 1991. Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell 67:701-716. [DOI] [PubMed] [Google Scholar]
- 63.Soriano, P., C. Montgomery, R. Geske, and A. Bradley. 1991. Targeted disruption of the c-src proto-oncogene leads to osteopetrosis in mice. Cell 64:693-702. [DOI] [PubMed] [Google Scholar]
- 64.Stewart, R. A., D.-M. Li, H. Huang, and T. Xu. 2003. A genetic screen for modifiers of the lats tumor suppressor gene identifies C-terminal Src kinase as a regulator of cell proliferation in Drosophila. Oncogene 22:6436-6444. [DOI] [PubMed] [Google Scholar]
- 65.Stowers, R. S., and T. L. Schwarz. 1999. A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics 152:1631-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takahashi, F., S. Endo, T. Kojima, and K. Saigo. 1996. Regulation of cell-cell contacts in developing Drosophila eyes by Dsrc41, a new, close relative of vertebrate c-src. Genes Dev. 10:1645-1656. [DOI] [PubMed] [Google Scholar]
- 67.Tapon, N., K. F. Harvey, D. W. Bell, D. C. Wahrer, T. A. Schiripo, D. A. Haber, and I. K. Hariharan. 2002. salvador promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 110:467-478. [DOI] [PubMed] [Google Scholar]
- 68.Tateno, M., Y. Nishida, and T. Adachi-Yamada. 2000. Regulation of JNK by Src during Drosophila development. Science 287:324-327. [DOI] [PubMed] [Google Scholar]
- 69.Tautz, D., and C. Pfeifle. 1989. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98:81-85. [DOI] [PubMed] [Google Scholar]
- 70.Thomas, B. J., K. H. Zavitz, X. Dong, M. E. Lane, K. Weigmann, R. L. Finley, Jr., R. Brent, C. F. Lehner, and S. L. Zipursky. 1997. roughex down-regulates G2 cyclins in G1. Genes Dev. 11:1289-1298. [DOI] [PubMed] [Google Scholar]
- 71.Tseng, A. S., and I. K. Hariharan. 2002. An overexpression screen in Drosophila for genes that restrict growth or cell-cycle progression in the developing eye. Genetics 162:229-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Turkson, J., T. Bowman, J. Adnane, Y. Zhang, J. Y. Djeu, M. Sekharam, D. A. Frank, L. B. Holzman, J. Wu, S. Sebti, and R. Jove. 1999. Requirement for Ras/Rac1-mediated p38 and c-Jun N-terminal kinase signaling in Stat3 transcriptional activity induced by the Src oncoprotein. Mol. Cell. Biol. 19:7519-7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Watanabe, N., S. Matsuda, S. Kuramochi, J. Tsuzuku, T. Yamamoto, and K. Endo. 1995. Expression of C-terminal src kinase in human colorectal cancer cell lines. Jpn. J. Clin. Oncol. 25:5-9. [PubMed] [Google Scholar]
- 74.Xu, T., W. Wang, S. Zhang, R. A. Stewart, and W. Yu. 1995. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development 121:1053-1063. [DOI] [PubMed] [Google Scholar]
- 75.Yu, C. L., D. J. Meyer, G. S. Campbell, A. C. Larner, C. Carter-Su, J. Schwartz, and R. Jove. 1995. Enhanced DNA-binding activity of a Stat3-related protein in cells transformed by the Src oncoprotein. Science 269:81-83. [DOI] [PubMed] [Google Scholar]
- 76.Zeidler, M. P., N. Perrimon, and D. I. Strutt. 1999. Polarity determination in the Drosophila eye: a novel role for unpaired and JAK/STAT signaling. Genes Dev. 13:1342-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zheng, Z., and S. X. Hou. 2002. Dynamic expression of STAT92E protein and function in JAK/STAT signal transduction pathway in Drosophila development. Annu. Drosoph. Res. Conf. 43:512B. [Google Scholar]
- 78.Zitnan, D., F. Sehnal, and P. J. Bryant. 1993. Neurons producing specific neuropeptides in the central nervous system of normal and pupariation-delayed Drosophila. Dev. Biol. 156:117-135. [DOI] [PubMed] [Google Scholar]