Abstract
p53 promotes apoptosis in response to death stimuli by transactivation of target genes and by transcription-independent mechanisms. We recently showed that wild-type p53 rapidly translocates to mitochondria in response to multiple death stimuli in cultured cells. Mitochondrial p53 physically interacts with antiapoptotic Bcl proteins, induces Bak oligomerization, permeabilizes mitochondrial membranes, and rapidly induces cytochrome c release. Here we characterize the mitochondrial p53 response in vivo. Mice were subjected to γ irradiation or intravenous etoposide administration, followed by cell fractionation and immunofluorescence studies of various organs. Mitochondrial p53 accumulation occurred in radiosensitive organs like thymus, spleen, testis, and brain but not in liver and kidney. Of note, mitochondrial p53 translocation was rapid (detectable at 30 min in thymus and spleen) and triggered an early wave of marked caspase 3 activation and apoptosis. This caspase 3-mediated apoptosis was entirely p53 dependent, as shown by p53 null mice, and preceded p53 target gene activation. The transcriptional p53 program had a longer lag phase than the rapid mitochondrial p53 program. In thymus, the earliest apoptotic target gene products PUMA, Noxa, and Bax appeared at 2, 4, and 8 h, respectively, while Bid, Killer/DR5, and p53DinP1 remained uninduced even after 20 h. Target gene induction then led to further increase in active caspase 3. Similar biphasic kinetics was seen in cultured human cells. Our results suggest that in sensitive organs mitochondrial p53 accumulation in vivo occurs soon after a death stimulus, triggering a rapid first wave of apoptosis that is transcription independent and may precede a second slower wave that is transcription dependent.
The basis for p53's striking apoptotic and tumor suppressor activity lies in its gene's pleiotropism, which involves transcription-dependent and transcription-independent functions. p53 responds to a broad range of death stimuli by rapid stabilization and activation. p53-mediated cell death primarily signals through the mitochondrial pathway, while the death receptor pathway plays a minor role (42). One important mechanism through which p53 mediates its biological response is transcriptional activation of proapoptotic target genes and transrepression of prosurvival proteins. Although p53 apoptotic effectors can localize to the cell membrane (Killer/DR5, CD95, and PERP) (1, 30, 35), or the cytoplasm (PIGs and PIDD) (19, 36), some notable p53 target gene products such as the BH3-only proteins Noxa (32) and PUMA (31, 47), Bax (29), and p53AIP1 (33) reside and/or act at the mitochondria.
It was previously shown that in response to a death stimulus such as DNA damage or hypoxia, a fraction of the stabilized p53 rapidly translocates to mitochondria in primary, immortal, and transformed cultured cells (23, 27, 39). To study the functional consequences of this phenomenon, exogenous p53 was forcibly targeted to mitochondria in p53 null cancer cells and mitochondrial p53 was shown to be sufficient to launch apoptosis and suppress colony formation in a transcription-independent fashion (23, 27). Moreover, in response to death stimuli, endogenous mitochondrial p53 forms inhibitory complexes with endogenous antiapoptotic Bclxl and Bcl2 proteins. Purified wild-type p53 protein induces oligomerization of Bak and permeabilization of the outer mitochondrial membrane and strongly promotes cytochrome c release in vitro. With the use of computational and genetic approaches, it was determined that the p53 DNA binding domain is involved in p53-Bclxl complex formation. Conversely, tumor-derived transactivation-deficient missense mutants concomitantly lose or compromise their ability to interact with Bclxl and to promote cytochrome c release. Thus, tumor-derived p53 mutations may represent “double hits,” eliminating the transcriptional as well as the direct mitochondrial functions of p53 (27). Based on these tissue culture studies, we proposed that mitochondrial translocation of p53 triggers a rapid proapoptotic response that jump-starts and amplifies the slower transcription-based response, which requires a certain ramping time typically in the range of 4 to 8 h after p53 induction.
The stress response to γ irradiation (γIR) in the mouse is well studied. It is known to be organ specific and largely or completely p53 dependent (11, 18, 20-22, 25, 26). For example, p53 null mice are completely protected from apoptosis in response to γIR and doxorubicin (Adriamycin) in radiosensitive organs like thymi and spleens (18, 20) (see Fig. 4B). Here we sought to determine the participation of the direct mitochondrial p53 pathway in the physiologic apoptotic response in vivo. We find that in sensitive target organs, mitochondrial p53 translocation can trigger a rapid first wave of caspase 3 activation and cell death, followed by a slower transcription-dependent p53 death wave. Our results are the first evidence that direct mitochondrial p53 activity indeed contributes to the physiological apoptotic response in an animal and suggest that the mitochondrial pathway may participate in tumor suppression in vivo.
FIG. 4.
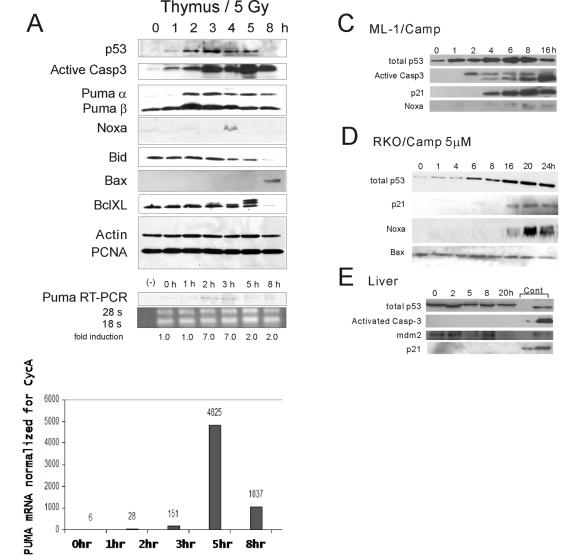
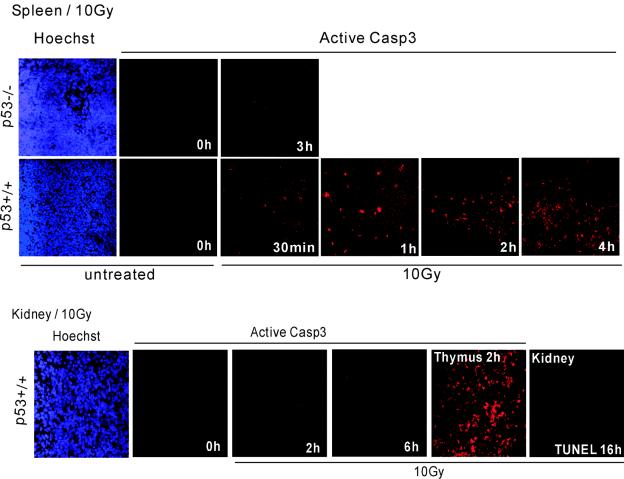
Stress-induced mitochondrial p53 accumulation coincided with a burst of early caspase 3 activation and cell death that preceded the production of apoptotic p53 target gene products in vivo. (A) Mice were subjected to 5-Gy γIR or left untreated. At 0, 1, 2, 3, 4, 5, 8, and 20 h, thymi were harvested. (A, top panels) Whole-cell homogenates were immediately prepared and immunoblotted with antibodies against p53 (UM1), activated caspase 3 (Active Casp3), PUMA α and β, Noxa, Bid, DR5/Killer, p53DINP1, Bax, and Bclxl. The latter are apoptotic target gene products that are induced by p53, except Bclxl, which is transrepressed by p53 (44). Actin and PCNA were used to adjust for loading of equal amounts of protein. Total cellular p53 stabilization started at 1 h. Cleaved caspase 3 was already generated by 1 h, and levels increased at 2 and 3 h and reached their peak at 5 h. In contrast, PUMA α and β protein induction began at 2 h while Noxa was only faintly and transiently detectable at 4 h. Bax was induced very late at 8 h. Bid, Killer/DR5, and p53DINP1 (data not shown) remained uninduced throughout. (A, bottom panels) Semiquantitative RT-PCR of PUMA transcripts from thymi treated or left untreated as described above. PUMA α and β induction also began at 2 h. Real-time RT-PCR confirmed this result. (B) Early caspase 3 activation and cell death after γIR in vivo. p53+/+ mice and p53−/− mice were subjected to 5-Gy (thymus) or 10-Gy (spleen and kidney) γIR or left untreated. At the indicated times, organs were harvested and snap-frozen sections were immunostained with an antibody that specifically recognizes the 17- to 19-kDa fragment of cleaved caspase 3 (the same antibody mentioned in the legend to Fig. 4A), followed by TRITC-donkey anti-rabbit antibody. Serial sections were also stained for TUNEL. Caspase 3 activation and cell death in thymus and spleen were strictly p53 dependent. Figure S1 (see the supplementary material available at http://www.path.sunysb.edu/faculty/umoll/default.htm) shows the same result in thymus after a dose of 10 Gy. On the other hand, kidney undergoes a strict p53-dependent arrest response (22). Thymus at 2 h serves here as the positive control for caspase 3 staining. Nuclei of the untreated (0 h) sections were counterstained with Hoechst. (C and D) Kinetic relationships between p53 mitochondrial translocation and target gene activation in human tumor cells. Wild-type p53-harboring ML-1 (C) and RKO (D) cells were treated with 5 μM camptothecin (Camp). At the indicated times, whole-cell lysates were prepared and immunoblotted. Active caspase 3 (Casp 3) appears as a 19- and 17-kDa doublet in ML-1 cells and as a 17-kDa protein in mouse tissues. Equal amounts of total protein per lane were loaded in the immunoblots shown in panels C and D. (E) Mice were subjected to 5-Gy γIR or left untreated. At 0, 2, 5, 8, and 20 h, livers were harvested and whole-cell homogenates were prepared and immunoblotted with antibodies against p53 (UM1), activated caspase 3, p21WAF1, and mdm2. With this treatment, the liver shows essentially no p53 induction and lacks any trace of an apoptotic or arrest response (22). Cont, doxorubicin-treated MEFs were used as positive controls.
MATERIALS AND METHODS
Animals and treatment.
Thirty-six healthy p53 wild-type mice (C57BL/6 and 129; 6 to 12 weeks old; Charles River Inc.) and p53 null mice (homozygous for p53N12-M; Taconic Farms) were obtained. Mice were subjected to whole-body γIR with either 5 or 10 Gy in a Cs137 γ-irradiator (1.07 Gy/min; GammaCell 40; Atomic Energy of Canada Ltd.). Alternatively, the topoisomerase II inhibitor etoposide (10 mg/kg of body weight; Sigma) was administered intravenously by tail vein injection. Both untreated and treated mice were sacrificed at the indicated time intervals (0, 0.5, 1, 2, 3, 4, 5, 6, 8, and 20 h postexposure) by CO2 asphyxiation. Target organs (thymi, spleens, livers, kidneys, testes, and brains) were immediately harvested and either used to make touch preparations, snap-frozen in liquid nitrogen, or washed in ice-cold phosphate-buffered saline (PBS) buffer containing protease inhibitor (Roche) and quickly processed for subcellular fractionation. For etoposide treatment, normal mice were treated by tail vein injection with 10 mg of etoposide/kg or left untreated. Mitochondria were prepared from thymi and spleens and analyzed as described above.
Tissue culture.
Primary human mammary epithelial cells (HMEC) were purchased from Clonetics and cultured short term for three additional passages in special media according to the manufacturer's instructions. Mouse embryo fibroblasts (MEFs) were prepared from 14-day-old 129 mouse embryos and passaged three times. Cells were treated with doxorubicin (0.34 μM) for 4 or 6 h or mock treated, and mitochondria were prepared as described below. Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assays (Roche) were used to assess apoptosis as previously described (27).
Mitochondrial fractionation.
Freshly harvested organs were finely minced with a scalpel and incubated in Ca-reticulocyte standard buffer cell lysis buffer (1.5 mM CaCl2, 10 mM NaCl, 10 mM Tris [pH 7.6]) to let the cells swell. After 10 min, the tissues were subjected to a glass dounce homogenizer (loose fit; Wheaton) to break open the cell membranes. An aliquot of the resulting crude (whole) cell homogenate was stored short term in sample buffer at −20°C until further use. The remainder of the crude cell homogenate was centrifuged at 855 × g for 5 to 10 min at 4°C to remove nuclei and any intact cells. The supernatant was centrifuged two more times for maximum clearance. From this postnuclear cytosol, mitochondria were isolated by discontinuous sucrose gradient centrifugation as previously described (23, 28). Briefly, the supernatant was laid over a sucrose step gradient (1.0 M over 1.5 M sucrose) and centrifuged at 26,000 rpm (85,000 × g; Beckman SW 41 rotor) for 35 min at 4°C. Mitochondria sedimented as a white band at the 1.0 M-1.5 M interphase and were carefully extracted from the side with a syringe. For cultured cells, ML-1 and RKO cells (both with wild-type p53) were grown in 10% fetal bovine serum-RPMI medium and 10% fetal bovine serum-Dulbecco modified Eagle medium, respectively. Cells were either left untreated or treated with topoisomerase I inhibitor camptothecin (5 μM; Sigma) for the indicated times and processed for mitochondrial isolation as described above. Protein concentrations of crude cell homogenate and isolated mitochondria were determined by Bio-Rad protein assay.
Western blotting.
Equal amounts of total protein from mitochondrial and crude cell homogenates (typically 5 to 15 μg per lane, depending on the organ) were resolved on sodium dodecyl sulfate-polyacrylamide gel electorphoresis gels, transferred onto nitrocellulose membranes (ECL; Amersham), and immunoblotted with a monoclonal antibody against mouse p53 (PAb 246; Biosource), polyclonal antibodies against mouse p53 (CM5 [Vector] and UM1 [raised against recombinant mouse wild-type p53]), human p53 (DO-1), human or mouse PCNA (PC-10), mouse IκB-α (C-21), and mouse p21 (sc6246) (all from Santa Cruz), and polyclonal antibodies against human p21 (Ab-1; Oncogene), human or mouse mitochondrial mthsp70 (clone JG1; Affinity Bioreagents), human or mouse cytochrome oxidase IV (clone 1A12; Molecular Probes), and human or mouse cleaved caspase 3 (Asp 175; catalog no. 9661; Cell Signaling Technologies). Crude cell homogenates were also immunoblotted with the following p53 target gene products: Bid (catalog no. 2003; recognizes full-length Bid; Cell Signaling), PUMA (Abcam), Noxa (Ab-1 [Oncogene] and IMG349 [Imgenex]), Bax (Ab-1 [Oncogene] and clone 2D2 [NeoMarker]), BclXL/xs (Cell Signaling), death receptor 5 (recognizes Killer/DR5), PIG3, and p53DINP1 (all Biovision Research products); mdm2 (clone IF2; Oncogene); kdel (Stressgen); calreticulin (Affinity Bioreagents); and vimentin and actin (Neomarker). Blots were developed with ECL chemiluminescence (Amersham).
RT-PCR.
Mice were subjected to 5 Gy of γIR or left untreated. At 0, 1, 2, 3, 4, 5, and 8 h, thymi were harvested and total RNA was immediately extracted by Trizol. Semiquantitative reverse transcription (RT)-PCR was performed using PUMA gene-specific primers s5′-ATGGCGGACGACCTCAAC-3′ and as5′-AGTCCCATGAAGAGATTGTACATGAC-3′ (14) and 100 ng of RNA in the Titan one-step kit (Roche). Products were analyzed by 10% polyacrylamide gel electrophoresis, visualized with SYBR Green, and quantitated by phosphorimaging. Total RNA (100 ng) was reverse transcribed using the OmniScript RT kit (QIAGEN) and random primers. Copy numbers for PUMA were determined by real-time RT-PCR using the SYBR Green dye method (QIAGEN) and primers as described above. 28S rRNA was used to normalize samples.
Immunofluorescence.
The harvested organs were bisected and used to make touch and smear preparations (for thymus and spleen only) or snap-frozen in liquid N2 for cryosectioning (5 μm). Cells and tissues were air dried and fixed with ice-cold methanol-acetone (1:1). Slides were blocked in 10% normal goat serum for 20 min, followed by incubation overnight at 4°C with the following primary antibodies: CM5 (Vector), UM1, and PAb 246 (Biosource International) for p53; cleaved caspase 3 (Cell Signaling Technologies); Grp75 (raised in rabbit against the C terminus of mitochondrial hsp70); and anti-mouse immunoglobulin G and anti-rabbit immunoglobulin G for negative controls. After washing, slides were incubated in fluorescein isothiocyanate (FITC)- and/or tetramethyl rhodamine isocyanate (TRITC)-conjugated secondary antibody (diluted in 2% normal goat serum; Jackson Immunoresearch Laboratory) for 1 h at room temperature. In some cases, smears consisting of live thymocytes (floating in PBS, with no air drying) were double stained. To this end, cells were overlaid with MitoTracker Red CM-H2XRos solution (diluted in PBS according to the instructions of the manufacturer [Molecular Probes]) for 15 to 30 min at 37°C. Excess solution was removed and cells were fixed in freshly made paraformaldehyde, gently washed in PBS, and processed for p53 immunofluorescence. In most cases, nuclei were counterstained with Hoechst 33342 or TO-PRO 3 (Molecular Probes) before being viewed with a confocal laser microscope (ZEISS LSM 510 META).
RESULTS
DNA damage induces rapid accumulation of mitochondrial p53 in radiosensitive tissues in vivo.
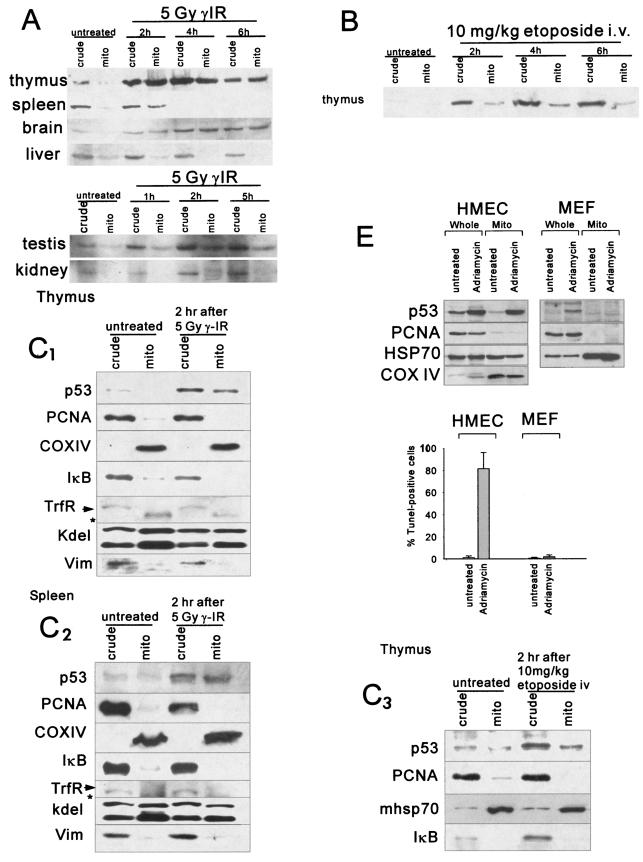
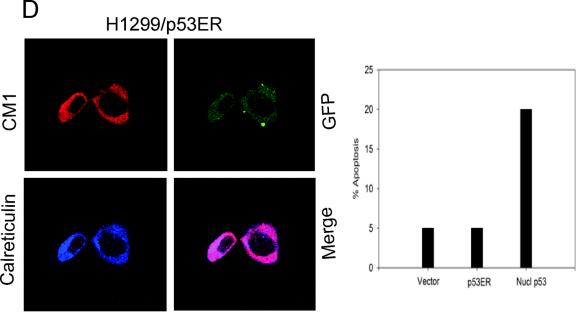
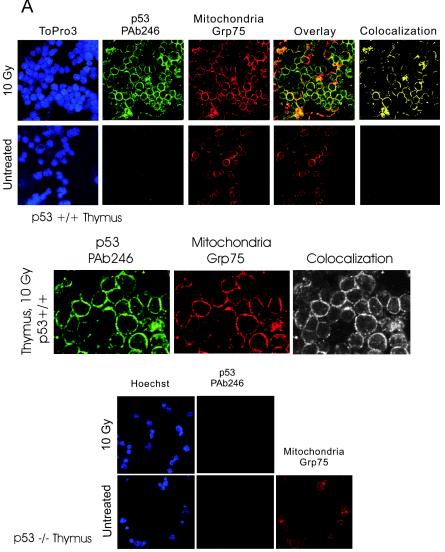
It was previously demonstrated that a fraction of stress-induced wild-type p53 protein rapidly translocates to mitochondria during p53-dependent death but not during p53-independent death or during p53-mediated cell cycle arrest. This was the case in all tested immortal and transformed cell lines of mouse and human origin, as well as in isolated primary mouse thymocytes (23, 27, 39). Mechanistically, p53 mitochondrial translocation results in Bak oligomerization, outer membrane permeabilization, cytochrome c release, caspase 3 activation, and subsequent apoptosis (23, 27). To determine whether the mitochondrial pathway also participates in the physiologic apoptotic p53 response in an animal, we studied the induction of mitochondrial p53 in various organs of normal mice after whole-body irradiation or intravenous etoposide administration. Mice subjected to γIR (5 and 10 Gy) or mock treated were sacrificed after various time intervals, and thymi, spleens, brains, livers, testes, and kidneys were removed. Mitochondria were rapidly purified, and p53 accumulation was characterized by immunoblotting. Figure 1A illustrates the various tissue responses to γIR within the first 5 to 6 h after damage. All organs exhibited a p53 response to γIR, as indicated by an induction of total cellular (crude) p53, albeit at different degrees (Fig. 1A and C; see also Fig. 3A and 4A and E). This organ profile of γIR-mediated p53 induction in vivo in the mouse is consistent with earlier results (11, 18, 21, 22). Of note, in addition to the overall cellular induction, p53 rapidly accumulated at the mitochondria in thymi, spleens, and testes and, somewhat surprisingly, in brains. This suggests that the mitochondrial p53 pathway participates in the p53 stress response of radiosensitive organs. In contrast, no mitochondrial p53 accumulation was detectable in kidneys and livers, indicating that this pathway does not participate in the p53 response of these radioresistant organs. In fact, in kidney the response to γIR is a p53-mediated cell cycle arrest but not apoptosis (22). In liver the induction of cellular p53 after γIR was minimal (Fig. 1A; see also Fig. 4E), in agreement with earlier reports (22). Mitochondrial p53 translocation also occurred in response to therapeutic doses of etoposide, a clinically used topoisomerase inhibitor. Induction of total cellular p53 and mitochondrial translocation were apparent in thymus and spleen at 2 h after etoposide treatment (Fig. 1B and data not shown). The purity of mitochondrial preparations from organs after γIR and etoposide treatments was assessed by immunoblotting with antibodies against the abundant nuclear and cytosolic marker proteins PCNA and IκB, respectively, as well as the plasma membrane marker transferrin receptor and the cytoskeletal marker vimentin (Fig. 1C). As shown in Fig. 1C, the mitochondrial fractions from organs were essentially free of nuclear, cytoplasmic, plasma membrane, and cytoskeletal contamination. Mitochondrial enrichment was seen with markers COX IV (an integral inner-membrane protein) or mthsp70 (a mitochondrial matrix protein). The only significant contamination present in our mitochondrial fractions was from the endoplasmic reticulum (ER), detected by the kdel antibody against Grp78 and Grp94 (Fig. 1C). Repeated attempts to purify mitochondrial fractions by using different fractionation protocols to remove ER membranes were not successful (data not shown). However, we do not think that the ER is a source of nonspecific p53 tracking with mitochondria for the following reason: p53 has not been reported to be localized to the ER, although ER stress induces cytoplasmic—but not ER—localization and degradation of p53 (37). To exclude any possibility that p53, although not reported to localize to the ER, could launch apoptosis from the ER, we deliberately targeted p53 to the ER by fusing the p53 C terminus to the ER leader sequence of human cytochrome b5. As expected, this so-called p53ER fusion protein, when transfected into p53 null H1299 cells, localized to the ER and nuclear membranes, as verified by colocalization with ER protein calreticulin (Fig. 1D). Importantly, however, p53ER was devoid of any apoptotic ability in p53 null H1299 and SaOS-2 cells (Fig. 1D). Thus, we conclude that the ER contamination does not falsely contribute to mitochondrial p53 detection and has no negative impact on the functional interpretation of our mitochondrial p53 data.
FIG. 1.
Rapid mitochondrial p53 accumulation in response to γIR in thymus, spleen, testis, and brain but not in liver and kidney. (A) Normal mice were subjected to 5-Gy whole-body γIR or left untreated. After the indicated time intervals, mitochondria (mito) were purified from various organs. Crude (whole-cell) lysates and mitochondrial fractions were characterized by immunoblotting with the anti-mouse p53 antibody CM5. Equal amounts (5 to 15 μg per lane) of total protein from crude lysates and mitochondrial fractions were loaded for each organ. (B) Mitochondrial p53 accumulation in response to etoposide. Normal mice were treated with 10 mg of etoposide/kg by tail vein injection or left untreated. Mitochondria and whole-cell lysates were prepared from thymi at the indicated times and analyzed as described above. Equal amounts (15 μg) of total protein per lane were loaded. (C) Mitochondrial fractions were free of nuclear and cytoplasmic contamination. Shown are results for thymi and spleens from untreated and treated (2 h after γIR or intravenous [i.v.] treatment with etoposide at 10 mg/kg) normal mice. The purity of mitochondrial fractions from organs was assessed by immunoblotting with antibodies against the nuclear and cytoplasmic marker proteins PCNA and IκB, the plasma membrane marker transferrin receptor (TrfR), the cytoskeletal marker vimentin (Vim), and the ER marker kdel. Mitochondrial enrichment was seen with mitochondrial markers COX IV (inner membrane bound) or mthsp70 (mitochondrial matrix). Equal amounts (5 μg) of total protein per lane were loaded, except in the case of vimentin (50 μg). Mitochondrial fractions from organs contained ER. (D, left panels) To exclude the possibility that p53, although not reported to localize to the ER, could launch apoptosis from the ER, p53 was deliberately targeted to the ER by fusion of its C terminus with the ER leader sequence of human cytochrome b5. This p53ER fusion protein, when transfected into p53 null H1299 cells, localized to the ER, as shown by colocalization with calreticulin. CM1, p53 antibody; GFP, green fluorescent protein. (D, right panel) However, p53ER was devoid of apoptotic ability in p53 null H1299 cells as well as in SaOS-2 cells (data not shown). Nucl p53, nuclear p53. (E) Mitochondrial p53 accumulation in HMEC. HMEC were left untreated or treated with doxorubicin (0.34 μM), which led to massive apoptosis within 24 h as determined by TUNEL. Mitochondria and whole-cell lysates were prepared from an aliquot after 6 h of exposure to doxorubicin. Equal amounts (5 μg) of total protein per lane were loaded. p53 did not translocate to mitochondria during p53-mediated cell cycle arrest in primary MEFs. MEFs were left untreated or treated with doxorubicin (0.34 μM), leading to cellular p53 accumulation and cell cycle arrest within 24 h (data not shown). Mitochondria and whole-cell lysates were prepared from an aliquot after 4 h. Immunoblots with equal amounts (5 μg) of total protein loaded per lane are shown as in panel C.
FIG. 3.
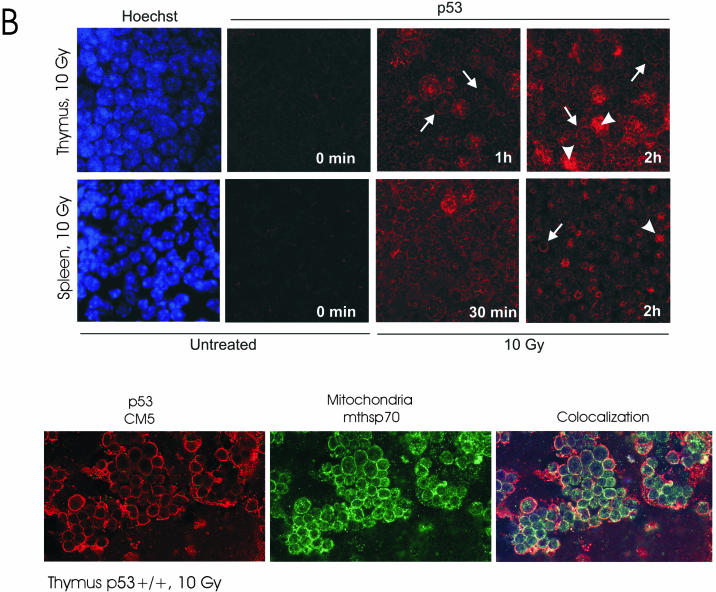
Mitochondrial translocation of p53 was detectable as early as 30 min after γIR in thymus and spleen. (A) Mice were subjected to 5-Gy γIR or left untreated. After the indicated times, spleens and thymi were removed for kinetic studies. Mitochondria (mito) were isolated by fractionation and immunoblotted for p53 (with CM5) and purity markers as described in the legend to Fig. 1C. p53 started to accumulate at mitochondria as early as 30 min after stress in both organs. (B, top panels) Likewise, mitochondrial p53 accumulation was detected by immunofluorescence at 30 and 60 min postirradiation in spleen and thymus, respectively, as indicated by predominantly perinuclear punctate p53 staining (example cells are indicated by arrows). In contrast, marked nuclear p53 accumulation became increasingly prominent starting at 2 h postirradiation (examples are indicated by arrowheads). (B, bottom panels) Tissue smears were prepared from thymus 1 h after 10-Gy irradiation in vivo and double-stained with polyclonal anti-p53 antibody CM5 (in red) and monoclonal anti-mthsp70 (in green). (C) Human RKO cells (colorectal carcinoma cells expressing wild-type p53) were treated with a low dose (5 μM) of camptothecin for 0 to 4 h prior to mitochondrial isolation. Immunoblots of crude and mitochondrial lysates (5 μg of total protein per lane) for p53 (DO1) and the indicated markers are shown. As in thymus and spleen in mouse, mitochondrial p53 accumulation started to be detectable at 30 min poststress.
We next extended these observations to primary cells in culture that represented additional cell types. Consonant with the mitochondrial p53 behavior in some organs in vivo, HMEC also exhibited p53 translocation to mitochondria when checked 6 h after treatment with doxorubicin, followed by subsequent massive apoptosis (Fig. 1E, top left and bottom panels). In contrast, MEFs failed to undergo mitochondrial p53 translocation after doxorubicin treatment despite total cellular p53 induction and subsequently underwent cell cycle arrest but not apoptosis (Fig. 1E, right top and bottom panels). This restriction of mitochondrial p53 translocation to an apoptotic response in primary cells recapitulates the behavior of leukemic ML-1 cells, which undergo mitochondrial p53 translocation and subsequent apoptosis only after apoptotic stimuli but not after cell cycle arrest stimuli (23).
Detection of stress-induced mitochondrial p53 in thymus and spleen by confocal immunofluorescence.
To directly visualize mitochondrial p53 translocation during the tissue response in vivo, we next performed confocal immunofluorescence microscopy on untreated and irradiated thymus and spleen. Frozen sections, smears, or touch preparations were used, and the focus was on events shortly after treatment. As seen in Fig. 2A, top panels, untreated thymocytes exhibited no detectable staining with the mouse p53-specific monoclonal antibody PAb 246. In contrast, shortly after DNA damage, irradiated thymocytes exhibited prominent, finely punctated perinuclear ring staining with PAb 246 (Fig. 2A, top panels). An enlarged view is shown in Fig. 2A, middle panels. This punctated perinuclear p53 pattern far exceeded in intensity a concomitant faint nuclear staining. Importantly, perinuclear p53 largely colocalized to mitochondria, as indicated by the superimposable pattern obtained with an antibody against the mitochondrial heat shock protein Grp75 (Fig. 2A, top and middle panels). The same colocalization was also seen with MitoTracker-stained thymocytes (data not shown). To confirm the specificity of PAb 246 staining and to rule out any potential cytoplasmic cross-reactivity in irradiated tissues, we prepared identical samples from p53 null mice. As seen in Fig. 2A, bottom panels, neither untreated nor treated p53 null thymocytes generated any detectable signal with the p53 antibody while mitochondria were readily seen with Grp75 immunostaining.
FIG. 2.
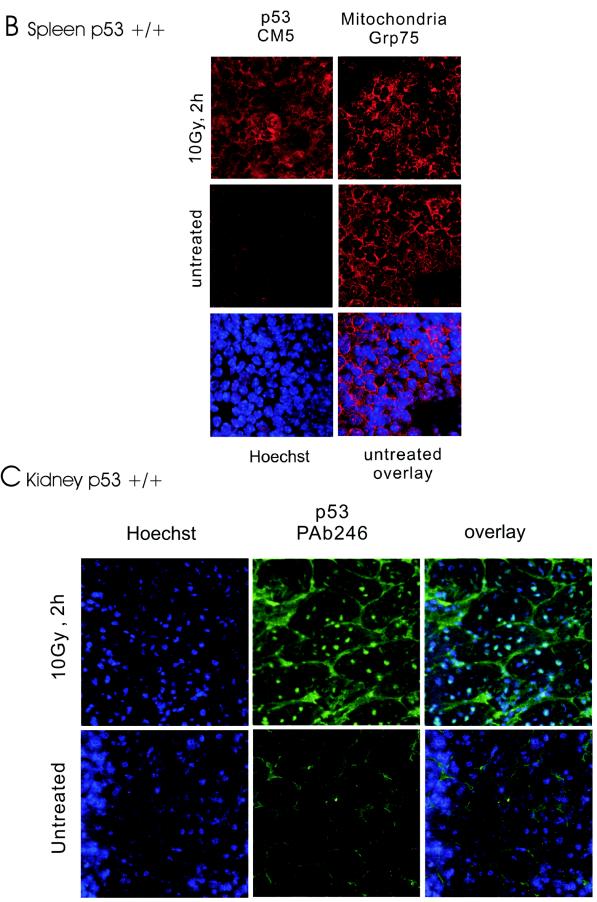
Detection of stress-induced mitochondrial p53 accumulation in vivo by confocal immunofluorescence. (A, top panels) Colocalization of p53 and mitochondrial marker protein Grp75 (mthsp70) in irradiated thymus. Normal mice were subjected to 10-Gy γIR (top row) or left untreated (bottom row). After 2 h, tissue smears were prepared from thymi and double-stained with monoclonal anti-p53 antibody PAb 246 followed by FITC-goat anti-mouse antibody and a rabbit anti-Grp75 antibody followed by TRITC-donkey anti-rabbit antibody. Untreated cells exhibited no detectable staining with PAb 246. In contrast, all irradiated thymocytes exhibited prominent, finely punctated perinuclear ring staining, which exceeded the concomitant faint nuclear staining at this time point. Cytoplasmic p53 largely colocalized with mitochondria (yellow only). Nuclei were counterstained with Hoechst or ToPro3. (A, middle panels) Magnified region from Fig. 2A, top panels, for visualization of punctate staining. (A, bottom panels) Specificity of the p53 antibody. p53−/− mice were treated or left untreated, and thymocytes were stained with PAb 246 followed by FITC-goat anti-mouse antibody. No signal was detectable with the p53 antibody. Images in all figures were captured by a Zeiss confocal microscope model 510 SM at identical acquisition and image processing parameters. (B) Mitochondrial p53 accumulation in irradiated spleen. Normal mice were subjected to 10-Gy γIR or left untreated. After 2 h, spleens were removed and touch preparation slides were made and stained with polyclonal anti-p53 CM5 or anti-Grp75 antibodies, followed by TRITC-donkey anti-rabbit antibody. In untreated spleen, CM5 gave minimal background staining. In contrast, treated spleen showed finely punctate, perinuclear p53 accumulation similar to the Grp75 pattern and consistent with mitochondrial localization. Weak nuclear p53 staining was discerned as well. Nuclei of untreated cells were counterstained with Hoechst and overlaid with Grp75 to emphasize the mitochondrial pattern. (C) Nuclear p53 in irradiated kidney. Mice were subjected to 10-Gy γIR or left untreated. After 2 h, kidneys were removed and frozen sections were stained with anti-p53 antibody PAb 246, followed by FITC-goat anti-mouse antibody. Nuclei were counterstained with Hoechst. The p53 antibody shows nonspecific tubular basement membrane staining in untreated and treated cells (large green rings). In treated cells, only nuclear p53 accumulation was detectable.
A result similar to that seen in thymus was seen in spleen (Fig. 2B). To confirm the validity of previous findings, we turned to another p53 antibody. With polyclonal mouse anti-p53 CM5, prominent perinuclear staining was observed in spleen 2 h postirradiation (Fig. 2B, top left panel). Concomitant faint nuclear staining was focally present as well. In contrast, untreated spleen showed only weak background staining (middle left panel), while the mitochondrial staining pattern obtained with Grp75 was independent of the absence or presence of irradiation (Fig. 2B, right column). Thus, these data suggest mitochondrial p53 accumulation after irradiation in spleen. Immunofluorescence of testis was inconclusive with both p53 antibodies due to high nonspecific background staining of cells lining the seminiferous tubules. Taking these findings together, we see in vivo evidence for γIR-induced mitochondrial translocation of p53 in radiosensitive organs by two independent methods, i.e., single-cell-based immunofluorescence and biochemical fractionation.
In contrast, the radioresistant kidney exhibited only nuclear but no mitochondrial p53 accumulation by subcellular fractionation. Consistent with this result, immunofluorescence of irradiated kidney showed exclusive nuclear p53 staining (Fig. 2C, top panels), while untreated kidney failed to exhibit specific p53 staining (Fig. 2C, bottom panels). In both cases, we see nonspecific staining of tubular basement membranes with PAb 246.
Mitochondrial translocation of p53 is detectable as early as 30 min after γIR in thymus and spleen.
It was previously demonstrated for cultured ML-1 leukemia cells that mitochondrial p53 translocation occurs within 1 h after a death stimulus, preceding mitochondrial events such as membrane depolarization, caspase activation, and cytochrome c release (23). Also, cultured null cells expressing mitochondrially targeted, transcriptionally inactive p53 as the only source of p53 readily undergo apoptosis (23, 27). It was further shown that addition of exogenous p53 protein to purified mitochondria is sufficient to induce Bak oligomerization and cytochrome c release in vitro (27). In sum, these data allowed for the hypothesis that p53's direct mitochondrial action may lead to an early first wave of apoptosis, preceding a second wave that is caused by apoptotic p53 response genes such as the PUMA (31, 47), Noxa (32), and Bax (29) genes and others. Of note, all known apoptotic p53 response genes are expressed in a tissue-specific and probably stress-specific manner (11). For example, according to an in situ hybridization study, thymus induces mainly Noxa, and to a lesser extent PUMA, transcripts in response to γIR while spleen induces mostly PUMA but only very few Noxa mRNAs and liver induces no apoptotic targets at all (11). To assess the relative timing of p53's mitochondrial activity within the physiological p53 stress response, we determined the kinetics of p53 translocation in vivo. As seen in Fig. 3A, mitochondrial p53 accumulation was detected by cell fractionation as early as 30 min after γIR in thymus and spleen. This rapid time course resembles the kinetics for cultured RKO cells, a human colon adenocarcinoma line, which started to accumulate p53 at mitochondria at 30 min, leading to significant levels at 4 h (Fig. 3C). Consistent with this finding, mitochondrial p53 accumulation was also detected by immunofluorescence at 30 and 60 min postirradiation in spleen and thymus, respectively, as indicated by predominantly perinuclear punctate p53 staining at these early time points (Fig. 3B). In contrast, marked nuclear p53 accumulation became prominent in thymus and spleen starting at 2 h postirradiation (Fig. 3B, top panels). Taken together, these data support the notion of a biphasic wave of apoptosis, with the mitochondrial p53 action preceding the nuclear p53 action.
Stress-induced mitochondrial p53 accumulation coincides with a burst of early caspase 3 activation and cell death that precedes the induction of apoptotic p53 target gene products in vivo.
As discussed above, mitochondrial p53 started to accumulate as early as 30 min postirradiation in spleen and thymus in vivo. Given the rapid and direct apoptogenic activity of p53 in cultured cells, we asked whether the production of its penultimate biochemical effector, i.e., cleaved caspase 3, is temporally correlated in vivo as well. Using immunoblotting and immunofluorescence with an antibody specific for the activated form of caspase 3, we found that this is in fact the case. This antibody recognized the large 17- to 19-kDa fragment but did not recognize full-length procaspase 3 or other cleaved caspases. A careful kinetic analysis of p53 induction and caspase 3 activation was performed by immunoblotting on total lysates from thymic tissues after 5-Gy irradiation. In irradiated thymus, total cellular p53 was in the earliest stages of induction by 1 h (Fig. 4A, top panel). Importantly, however, by 1 h caspase 3 activation had already begun (Fig. 4A, second panel). Moreover, immunofluorescence analysis confirmed that by 1 h poststress, caspase 3 activation was already ongoing at low levels in the majority of thymocytes (Fig. 4B, thymus). Between 1 and 2 h poststress, caspase 3 activation then continued so that at 2 h a fraction of thymocytes expressed high levels of activated caspase 3 (Fig. 4B, thymus). Thus, at 2 h poststress caspase 3 activation in thymus was already fully ongoing, which coincided with the maximum mitochondrial p53 accumulation in this organ (Fig. 1A). Of note, early caspase 3 activation in thymus led to early apoptosis, as indicated by significant TUNEL positivity at 2 h (Fig. 4B, thymus). A similar wave of early caspase 3 activation and apoptosis in thymus was seen after 10-Gy irradiation (see Fig. S1, middle panel, in the supplementary material available at http://www.path.sunysb.edu/faculty/umoll/default.htm). Starting at 1 h after 10-Gy irradiation, low-level caspase 3 staining was widely present, and at 2 h significant TUNEL positivity was seen. The latter was also reflected after hematoxylin and eosin staining by the appearance of small purple apoptotic bodies that were absent in untreated thymus, albeit the sensitivity of the hematoxylin and eosin staining assay was the lowest among those of the three assays. Overall, irradiation-induced thymocyte death was very fast, since at 8 h poststress, the vast majority of thymocytes had undergone apoptosis, as indicated by homogeneous TUNEL staining (Fig. 4B, thymus). Of note, total cellular p53 levels in thymus reached their peak only at 3 h (Fig. 4A, top panel), while caspase 3 activation reached its peak 2 h later, at 5 h.
Similar to our results with thymus, caspase 3 activation in spleen started at 30 min poststress as indicated by clearly detectable individual positive cells (Fig. 4B, spleen). The number of apoptotic cells then steadily increased thereafter until 4 h poststress, when the majority of cells expressed cleaved caspase 3. Importantly, γIR-induced caspase 3 activation in thymus and spleen was completely p53 dependent at these early time points, since p53 null mice failed to show detectable cleaved caspase in these organs (Fig. 4B). In contrast, in p53+/+ kidney, which underwent strictly nuclear p53 accumulation (Fig. 2C) and was reported to undergo cell cycle arrest (22), no active caspase 3 staining was detectable even 6 h after 10-Gy irradiation (Fig. 4B, kidney) and TUNEL remained negative even at 16 h poststress. This result also serves as another specificity control for the above thymus and spleen data.
To further elucidate the contribution of the mitochondrial p53 pathway to the total apoptotic p53 response in vivo in mice, it was critical to characterize the temporal relationship between mitochondrial p53 accumulation and ensuing caspase 3 activation and cell death and the appearance of p53 transcriptional activity. We therefore did an extensive kinetic analysis of the induction of relevant apoptotic p53 target genes in thymus after DNA damage. To ensure direct comparability of transcriptional kinetics with the immunofluorescence data, we used 5-Gy γIR. Of note, the p53 transcriptional response had a longer lag phase than the rapid mitochondrial response (Fig. 4A). This is consistent with the increased time required for transcription and translation, as well as the possibly coordinated induction of proapoptotic targets and synchronous repression of prosurvival targets (reviewed in reference 25). Among the six relevant targets that we profiled in thymus, PUMA was the earliest and physiologically most relevant apoptotic product in irradiation-induced thymus apoptosis, and PUMA first started to be induced at the transcript and protein levels at 2 h (Fig. 4A). PUMA induction was later followed by minimal and transient induction of Noxa at 4 h and by very late Bax induction at 8 h. Interestingly, Killer/DR5, Bid, and p53DinP1 (34, 40) remained uninduced even after 20 h (Fig. 4A and data not shown). Conversely, levels of the prosurvival protein Bclxl, which is transcriptionally repressed by p53 (44), had declined by 8 h. Thus, PUMA, Noxa, and Bax proteins began to accumulate after the initial wave of p53-dependent caspase activation which occurred at 1 h and led to an early wave of cell death. Of note, PUMA induction coincided with an additional increase in caspase 3 cleavage, generating higher levels of this key apoptotic mediator once transcription was initiated (Fig. 4A).
A similar biphasic kinetics of the early p53-dependent caspase 3 activation that precedes the subsequent p53-dependent target gene activation was seen in cultured tumor cells. It was previously shown that wild-type p53-harboring ML-1 cells undergo p53 translocation within 1 h of treatment with 5 μM camptothecin (23). Consonant with this data, ML-1 cells that received a death stimulus via treatment with 5 μM camptothecin exhibited an early first phase of caspase 3 activation at 2 h (Fig. 4C), while the earliest evidence of transcriptional activation was seen only at 4 h with p21 protein induction (10), followed at 6 h by minimal and transient induction of Noxa protein, that peaked at 8 h but then declined thereafter (Fig. 4C). Once transcription had been initiated, a late second phase of additional caspase activation started at 8 h and peaked at 16 h, likely triggered by the compound mitochondrial action of several p53 target genes such as the Noxa, PUMA, Bax, and p53AIP1 genes (29, 31-33, 47) and others. Moreover, RKO cells that started to accumulate mitochondrial p53 as early as 30 min after exposure to 5 μM camptothecin (Fig. 3C) showed an even more delayed p53 transcriptional response (Fig. 4D). The induction of p21 and Noxa proteins coincided with the peak of total cellular p53 levels and became detectable only at 16 h, while Bax and PIG3 (36) remained completely uninduced (Fig. 4D and data not shown). Thus, although there were some cell- and/or stress-type specific variations in the individual kinetics, a common picture emerges from mouse thymus in vivo and human tumor cells in culture, converging on a biphasic p53 response pattern in radiosensitive tissues and cells.
On the opposite end of the spectrum from sensitive tissues that readily undergo p53-dependent apoptosis, such as thymus and spleen, stands the radioresistant liver. Liver did not mount an effective p53 response and activated neither its nuclear nor its mitochondrial pathway. After γIR, this organ completely lacked any mitochondrial p53 pathway (Fig. 1A), as well as a transcriptional p53 pathway, as indicated by the lack of induction of target genes such as those for p21 and mdm2 in vivo (Fig. 4E). These data are entirely consistent with the reported lack of p53 activation in the liver of transgenic LacZ-p53 reporter mice after γIR (18, 22). Kidney, on the other hand, fell in between these extremes by mounting a p53 response that was restricted to transcriptional activation of p53 targets and cell cycle arrest, at least at this dosage (10 Gy) (Fig. 2C and 4B, kidney), consistent with previous reports (22).
In summary, our results show that in radiosensitive organs such as thymus and spleen mitochondrial p53 accumulation occurs within 30 min after a death stimulus in vivo and can lead to a rapid first wave of a caspase-mediated proapoptotic p53 response that is transcription independent, followed by a second wave that is transcription dependent.
DISCUSSION
The apoptotic response to DNA damage in the mouse has been well characterized. It has been clearly established that p53 activity is the prime determinant of radiation and drug sensitivity in vivo. In radiosensitive organs such as thymus, spleen, and small intestine, activation of caspase 3 and subsequent apoptosis after γIR and treatment with DNA-damaging drugs are largely or completely p53 dependent (8, 11, 18, 20, 21, 25, 26, 32; this study). In contrast, radioresistant organs such as liver, kidney, lung, and skeletal muscle do not activate caspase 3 and apoptosis after 5- to 10-Gy γIR (8, 11, 18, 20; this study). However, the precise mechanism by which p53 kills cells has been unclear, largely because the basis for p53's potent apoptotic activity lies in the p53 gene's pleiotropism, which involves transcription-dependent and -independent functions. Moreover, the pattern of p53-induced apoptotic target gene expression is highly selective and complex and clearly shows cell and tissue specificity. For example, the key effector of apoptosis in colon and the red pulp of spleen is Bid while in thymus and the white pulp of spleen it is PUMA (11, 40) (see below). Bid and PUMA have been shown to trigger the mitochondrial apoptotic pathway. They cause permeabilization of the outer membrane and release of activators of caspases and nucleases such as cytochrome c, SMAC/Diablo, Omi/HtrA2, AIF, and endonuclease G.
An important question in γIR-induced cell death is the relative contribution made by individual apoptogenic p53 target gene products. Although about a dozen different p53 target genes that can mediate apoptosis when forcibly overexpressed have been characterized, it remains unclear whether any single transcriptional target is critical. Genes whose products act directly at the mitochondria like the BH3-only proteins PUMA, Noxa, and Bid or the BH123 protein Bax stand out. However, with knockout mice lacking the PUMA, Noxa, and Bax genes now available, it has become clear that none of these knockouts has the power to produce a phenocopy of p53−/− mice (16, 43, 45). Instead, each gene captures only an aspect of the pleiotropic p53 gene action at best. Moreover, each of them exhibits considerable tissue specificity in its action. Surprisingly, thymocytes from Bax−/− knockout mice and Noxa−/− knockout mice as well as those from Bax−/−-Noxa−/− double knockout mice are capable of undergoing apoptosis after DNA damage as efficiently as wild-type thymocytes (43, 45), clearly indicating that in thymocytes Bax and Noxa are fully dispensable for DNA damage-induced death. Bax-deficient mice develop only benign B- and T-cell hyperplasias and no tumors (16). On the other hand, Noxa does make an important contribution in gut stem cells. Noxa−/− mice are tumor free when unchallenged but show resistance to X ray-induced gastrointestinal death due to impaired apoptosis of the epithelial cells in the crypts of the small intestines (43). Moreover, Noxa−/− MEFs exhibit partial resistance to E1A-induced apoptosis in response to DNA damage by doxorubicin (43) or etoposide (45), and this resistance is further enhanced by Bax deficiency (43). Thus, Noxa contributes to p53-mediated death in fibroblasts and crypt intestinal cells but plays no role in thymocytes.
PUMA−/− thymocytes fail to be a phenocopy of p53−/− thymocytes, and PUMA knockout mice remain completely free of tumors, including the predominant T-cell lymphomas that are so typical of p53 knockout mice (14, 45). Yet PUMA is an important mediator of p53-induced normal thymocyte death after challenge (Fig. 4A) (11, 14, 45). Recent studies report that ex vivo-cultured PUMA−/− thymocytes exhibit partial protection from γIR after 1.25 to 2.5 Gy, showing impaired apoptosis (14, 45). Of note, however, and important for this discussion, protection became apparent only after prolonged observation at 16 to 72 h, while little difference between wild-type and PUMA−/− thymocytes existed within the first 8 h after damage. However, as widely reported in the literature and reproduced in our own hands, γIR-induced thymocyte death at doses of up to 10 Gy in vivo and in vitro is characterized by its surprising speed. In fact, the rate of apoptosis in thymocytes in general is one of the fastest among those in mammalian cells, resulting in killing of >70% of thymocytes at 8 to 10 h (8, 20; this study). For example, we detected massive thymocyte death in animals within the first 5 h after 5-Gy γIR (Fig. 4B), in agreement with in vitro findings by Clarke et al. (8), who report death of 45% of cultured thymocytes at 8 h after 2.5-Gy γIR, and Lowe et al. (20), who report death of 60% at 10 h after 5-Gy γIR and death of 90% at 20 h after a dose of only 1 Gy. Intriguingly, PUMA deficiency also provides a much stronger protective effect against non-p53-mediated thymocyte death, such as that mediated by the protein kinase C inhibitor staurosporine and, most strikingly, the phorbol ester phorbol myristate acetate, consistent with previous data (12, 45). Taken together, these data support the notion that PUMA plays a significant role in later phases of thymocyte death. Furthermore, PUMA may be more critical for p53-independent than for p53-dependent thymocyte death. Nevertheless, with regard to transcriptional mechanisms of p53-mediated death in the thymus, PUMA clearly is the single most important target among all known p53 targets. Indeed, we found that in thymus in vivo, PUMA protein was the earliest product, first induced at 2 h, while Noxa was only weakly and transiently induced at 4 h. This is in reasonable agreement with data for cultured thymocytes, where PUMA and Noxa mRNA transcripts are detected at 5 and 6 h after γIR, respectively (45). On the other hand, Bax protein appeared only at 8 h, while Bid, Killer/DR5, and p53DinP1 failed to be induced altogether (Fig. 4A).
Over the years, a transcription-independent proapoptotic activity has been ascribed to p53, yet insight into its mechanism of action was lacking (2-4, 6, 9, 13, 17, 24, 38, 41, 46). We recently identified a novel p53 death pathway in which p53 has a rapid and direct transcription-independent action at the mitochondria. This action is specific for p53-mediated apoptosis and does not occur during p53-mediated cell cycle arrest. To examine whether this pathway is physiologically significant in vivo, we asked two questions. First, is there evidence for p53 mitochondrial translocation in the live animal after DNA damage? Second, if so, what is the kinetic relationship between p53 mitochondrial translocation, caspase 3 activation, and death on the one hand and the p53 transcriptional program on the other hand? A mitochondrial p53 action that precedes the transcriptional p53 action would be strong physiologic evidence for p53's independent but synergistic death function. Using cell fractionation and immunofluorescence on organs, we show here that in radiosensitive organs such as thymus and spleen, mitochondrial p53 translocation started within 30 min after an apoptotic insult and was rapidly followed by an early burst of caspase 3 activation and programmed cell death. Similarly, mitochondrial p53 accumulation was also induced by 2 h after γIR in testis and brain. In contrast, induction or suppression of critical p53 target gene products in thymus and spleen was not detected until later, i.e., at 2 h for PUMA, at 4 h for Noxa, and at 8 h for Bax and Bclxl. This induction then led to further increase of cleaved caspase 3, beyond the already elevated levels triggered by the early mitochondrial phase. Very similar biphasic kinetics was seen in cultured human tumor cells such as ML-1 and RKO cells, which also exhibited the mitochondrial p53 program (Fig. 3C and 4C and D). In contrast, the same stress failed to elicit mitochondrial p53 translocation in radioresistant organs such as kidney and liver, where stress-induced p53 was confined to the nucleus and caspase 3 activation and apoptosis remained undetectable (Fig. 1A, 2C, and 4B and E). Together, these findings strongly suggest that p53's direct proapoptotic action at the mitochondria is physiologically significant and contributes to the rapid stress response of radiosensitive tissues in vivo. It is worth noting that crypt cells within the small intestine, which are a small epithelial subcompartment within the mucosa but represent the tissue stem cells, are extremely radiosensitive and undergo p53-dependent apoptosis within 4.5 h after 1-Gy γIR (25, 26). It is conceivable that mitochondrial p53 translocation contributes to their rapid apoptotic response as well. However, neither fractionation (a bulk assay) nor immunostaining of frozen sections (due to lack of available high quality mouse-specific p53 antibodies which work on paraffin-embedded tissues) currently affords us the resolution and sensitivity needed to test this hypothesis in the gut. Interestingly, irradiated brain, which is considered relatively radioresistant (18), shows stress-induced mitochondrial p53 translocation (Fig. 1A) and also exhibits p53 transcriptional activity, as indicated by LacZ-p53 reporter mice (18).
Interestingly, transactivation-deficient mutant p53 is competent in promoting apoptosis in some instances (4, 6, 13, 38, 46) but not in others (5, 15, 17). For example, the human 22/23QS allele, which is broadly transactivation and transrepression defective, is apoptotically very active when overexpressed in human p53 null tumor cells, consistent with a compensating mitochondrial p53 action (2, 6, 7, 17). On the other hand, mice homozygous for the equivalent 25/26QS allele are reported to be phenocopies of p53 null mice in γIR-induced death of embryonic stem cells and thymocytes and in transformability of MEFs by E1A plus ras (5, 15). While the latter result may suggest that the mitochondrial pathway by itself is insufficient for p53-dependent apoptosis and tumor suppressor activity in vivo in the mouse, it is interesting that the 25/26QS mutant's ability to translocate to mitochondria has been found to be compromised (7). Moreover, the possibility has not been ruled out that primary cells cannot tolerate high steady-state levels of the 25/26QS mutant protein unless they acquire additional mutations in the p53 gene itself or in the downstream pathway that render them resistant to cell death.
In summary, we showed that mitochondrial p53 translocation triggers a rapid wave of caspase 3 activation and cell death during the physiologic death response of radiosensitive organs in vivo. This direct mitochondrial p53 death jump-starts and amplifies the slower transcription-based p53 response and may be one of the distinguishing features between radiosensitive and radioresistant organs.
Acknowledgments
This work has been supported by grants from the National Institutes of Health (CA060664) and the American Cancer Society (CCG-105595) to U.M.M.
We thank Petr Pancoska for technical help.
REFERENCES
- 1.Attardi, L. D., E. E. Reczek, C. Cosmas, E. G. Demicco, M. E. McCurrach, S. W. Lowe, and T. Jacks. 2000. PERP, an apoptosis-associated target of p53, is a novel member of the PMP-22/gas3 family. Genes Dev. 14:704-718. [PMC free article] [PubMed] [Google Scholar]
- 2.Baptiste, N., P. Friedlander, X. Chen, and C. Prives. 2002. The proline-rich domain of p53 is required for cooperation with anti-neoplastic agents to promote apoptosis of tumor cells. Oncogene 21:9-21. [DOI] [PubMed] [Google Scholar]
- 3.Bennett, M., K. Macdonald, S. W. Chan, J. P. Luzio, R. Simari, and P. Weissberg. 1998. Cell surface trafficking of Fas: a rapid mechanism of p53-mediated apoptosis. Science 282:290-293. [DOI] [PubMed] [Google Scholar]
- 4.Caelles, C., A. Helmberg, and M. Karin. 1994. p53-dependent apoptosis in the absence of transcriptional activation of p53-target genes. Nature 370:220-223. [DOI] [PubMed] [Google Scholar]
- 5.Chao, C., S. Saito, J. Kang, C. W. Anderson, E. Appella, and Y. Xu. 2000. p53 transcriptional activity is essential for p53-dependent apoptosis following DNA damage. EMBO J. 19:4967-4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, X., L. J. Ko, L. Jayaraman, and C. Prives. 1996. p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev. 10:2438-2451. [DOI] [PubMed] [Google Scholar]
- 7.Chipuk, J. E., T. Kuwana, L. Bouchier-Hayes, N. M. Droin, D. D. Newmeyer, M. Schuler, and D. R. Green. 2004. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science 303:1010-1014. [DOI] [PubMed] [Google Scholar]
- 8.Clarke, A. R., C. A. Purdie, D. J. Harrison, R. G. Morris, C. C. Bird, M. L. Hooper, and A. H. Wyllie. 1993. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature 362:849-852. [DOI] [PubMed] [Google Scholar]
- 9.Ding, H. F., Y. L. Lin, G. McGill, P. Juo, H. Zhu, J. Blenis, J. Yuan, and D. E. Fisher. 2000. Essential role for caspase-8 in transcription-independent apoptosis triggered by p53. J. Biol. Chem. 275:38905-38911. [DOI] [PubMed] [Google Scholar]
- 10.el-Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817-825. [DOI] [PubMed] [Google Scholar]
- 11.Fei, P., E. J. Bernhard, and W. S. el-Deiry. 2002. Tissue-specific induction of p53 targets in vivo. Cancer Res. 62:7316-7327. [PubMed] [Google Scholar]
- 12.Han, J., C. Flemington, A. B. Houghton, Z. Gu, G. P. Zambetti, R. J. Lutz, L. Zhu, and T. Chittenden. 2001. Expression of bbc3, a pro-apoptotic BH3-only gene, is regulated by diverse cell death and survival signals. Proc. Natl. Acad. Sci. USA 98:11318-11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haupt, Y., S. Rowan, E. Shaulian, K. H. Vousden, and M. Oren. 1995. Induction of apoptosis in HeLa cells by trans-activation-deficient p53. Genes Dev. 9:2170-2183. [DOI] [PubMed] [Google Scholar]
- 14.Jeffers, J. R., E. Parganas, Y. Lee, C. Yang, J. Wang, J. Brennan, K. H. MacLean, J. Han, T. Chittenden, J. N. Ihle, P. J. McKinnon, J. L. Cleveland, and G. P. Zambetti. 2003. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell 4:321-328. [DOI] [PubMed] [Google Scholar]
- 15.Jimenez, G. S., M. Nister, J. M. Stommel, M. Beeche, E. A. Barcarse, X. Q. Zhang, S. O'Gorman, and G. M. Wahl. 2000. A transactivation-deficient mouse model provides insights into Trp53 regulation and function. Nat. Genet. 26:37-43. [DOI] [PubMed] [Google Scholar]
- 16.Knudson, C. M., K. S. Tung, W. G. Tourtellotte, G. A. Brown, and S. J. Korsmeyer. 1995. Bax-deficient mice with lymphoid hyperplasia and male germ cell death. Science 270:96-99. [DOI] [PubMed] [Google Scholar]
- 17.Kokontis, J. M., A. J. Wagner, M. O'Leary, S. Liao, and N. Hay. 2001. A transcriptional activation function of p53 is dispensable for and inhibitory of its apoptotic function. Oncogene 20:659-668. [DOI] [PubMed] [Google Scholar]
- 18.Komarova, E. A., M. V. Chernov, R. Franks, K. Wang, G. Armin, C. R. Zelnick, D. M. Chin, S. S. Bacus, G. R. Stark, and A. V. Gudkov. 1997. Transgenic mice with p53-responsive lacZ: p53 activity varies dramatically during normal development and determines radiation and drug sensitivity in vivo. EMBO J. 16:1391-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin, Y., W. Ma, and S. Benchimol. 2000. Pidd, a new death-domain-containing protein, is induced by p53 and promotes apoptosis. Nat. Genet. 26:122-127. [DOI] [PubMed] [Google Scholar]
- 20.Lowe, S. W., E. M. Schmitt, S. W. Smith, B. A. Osborne, and T. Jacks. 1993. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature 362:847-849. [DOI] [PubMed] [Google Scholar]
- 21.MacCallum, D. E., P. A. Hall, and E. G. Wright. 2001. The Trp53 pathway is induced in vivo by low doses of gamma radiation. Radiat. Res. 156:324-327. [DOI] [PubMed] [Google Scholar]
- 22.MacCallum, D. E., T. R. Hupp, C. A. Midgley, D. Stuart, S. J. Campbell, A. Harper, F. S. Walsh, E. G. Wright, A. Balmain, D. P. Lane, and P. A. Hall. 1996. The p53 response to ionising radiation in adult and developing murine tissues. Oncogene 13:2575-2587. [PubMed] [Google Scholar]
- 23.Marchenko, N. D., A. Zaika, and U. M. Moll. 2000. Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J. Biol. Chem. 275:16202-16212. [DOI] [PubMed] [Google Scholar]
- 24.Matas, D., A. Sigal, P. Stambolsky, M. Milyavsky, L. Weisz, D. Schwartz, N. Goldfinger, and V. Rotter. 2001. Integrity of the N-terminal transcription domain of p53 is required for mutant p53 interference with drug-induced apoptosis. EMBO J. 20:4163-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merritt, A. J., T. D. Allen, C. S. Potten, and J. A. Hickman. 1997. Apoptosis in small intestinal epithelia from p53-null mice: evidence for a delayed, p53-independent G2/M-associated cell death after gamma-irradiation. Oncogene 14:2759-2766. [DOI] [PubMed] [Google Scholar]
- 26.Merritt, A. J., C. S. Potten, C. J. Kemp, J. A. Hickman, A. Balmain, D. P. Lane, and P. A. Hall. 1994. The role of p53 in spontaneous and radiation-induced apoptosis in the gastrointestinal tract of normal and p53-deficient mice. Cancer Res. 54:614-617. [PubMed] [Google Scholar]
- 27.Mihara, M., S. Erster, A. Zaika, O. Petrenko, T. Chittenden, P. Pancoska, and U. M. Moll. 2003. p53 has a direct apoptogenic role at the mitochondria. Mol. Cell 11:577-590. [DOI] [PubMed] [Google Scholar]
- 28.Mihara, M., and U. M. Moll. 2003. Detection of mitochondrial localization of p53. Methods Mol. Biol. 234:203-209. [DOI] [PubMed] [Google Scholar]
- 29.Miyashita, T., S. Kitada, S. Krajewski, W. A. Horne, D. Delia, and J. C. Reed. 1995. Overexpression of the Bcl-2 protein increases the half-life of p21Bax. J. Biol. Chem. 270:26049-26052. [DOI] [PubMed] [Google Scholar]
- 30.Muller, M., S. Wilder, D. Bannasch, D. Israeli, K. Lehlbach, M. Li-Weber, S. L. Friedman, P. R. Galle, W. Stremmel, M. Oren, and P. H. Krammer. 1998. p53 activates the CD95 (APO-1/Fas) gene in response to DNA damage by anticancer drugs. J. Exp. Med. 188:2033-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakano, K., and K. H. Vousden. 2001. PUMA, a novel proapoptotic gene, is induced by p53. Mol. Cell 7:683-694. [DOI] [PubMed] [Google Scholar]
- 32.Oda, E., R. Ohki, H. Murasawa, J. Nemoto, T. Shibue, T. Yamashita, T. Tokino, T. Taniguchi, and N. Tanaka. 2000. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 288:1053-1058. [DOI] [PubMed] [Google Scholar]
- 33.Oda, K., H. Arakawa, T. Tanaka, K. Matsuda, C. Tanikawa, T. Mori, H. Nishimori, K. Tamai, T. Tokino, Y. Nakamura, and Y. Taya. 2000. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 102:849-862. [DOI] [PubMed] [Google Scholar]
- 34.Okamura, S., H. Arakawa, T. Tanaka, H. Nakanishi, C. C. Ng, Y. Taya, M. Monden, and Y. Nakamura. 2001. p53DINP1, a p53-inducible gene, regulates p53-dependent apoptosis. Mol. Cell 8:85-94. [DOI] [PubMed] [Google Scholar]
- 35.Owen-Schaub, L. B., L. S. Angelo, R. Radinsky, C. F. Ware, T. G. Gesner, and D. P. Bartos. 1995. Soluble Fas/APO-1 in tumor cells: a potential regulator of apoptosis? Cancer Lett. 94:1-8. [DOI] [PubMed] [Google Scholar]
- 36.Polyak, K., Y. Xia, J. L. Zweier, K. W. Kinzler, and B. Vogelstein. 1997. A model for p53-induced apoptosis. Nature 389:300-305. [DOI] [PubMed] [Google Scholar]
- 37.Qu, L., S. Huang, D. Baltzis, A. M. Rivas-Estilla, O. Pluquet, M. Hatzoglou, C. Koumenis, Y. Taya, A. Yoshimura, and A. E. Koromilas. 2004. Endoplasmic reticulum stress induces p53 cytoplasmic localization and prevents p53-dependent apoptosis by a pathway involving glycogen synthase kinase-3beta. Genes Dev. 18:261-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Regula, K. M., and L. A. Kirshenbaum. 2001. p53 activates the mitochondrial death pathway and apoptosis of ventricular myocytes independent of de novo gene transcription. J. Mol. Cell. Cardiol. 33:1435-1445. [DOI] [PubMed] [Google Scholar]
- 39.Sansome, C., A. Zaika, N. D. Marchenko, and U. M. Moll. 2001. Hypoxia death stimulus induces translocation of p53 protein to mitochondria. Detection by immunofluorescence on whole cells. FEBS Lett. 488:110-115. [DOI] [PubMed] [Google Scholar]
- 40.Sax, J. K., P. Fei, M. E. Murphy, E. Bernhard, S. J. Korsmeyer, and W. S. el-Deiry. 2002. BID regulation by p53 contributes to chemosensitivity. Nat. Cell Biol. 4:842-849. [DOI] [PubMed] [Google Scholar]
- 41.Schuler, M., E. Bossy-Wetzel, J. C. Goldstein, P. Fitzgerald, and D. R. Green. 2000. p53 induces apoptosis by caspase activation through mitochondrial cytochrome c release. J. Biol. Chem. 275:7337-7342. [DOI] [PubMed] [Google Scholar]
- 42.Schuler, M., and D. R. Green. 2001. Mechanisms of p53-dependent apoptosis. Biochem. Soc. Trans. 29:684-688. [DOI] [PubMed] [Google Scholar]
- 43.Shibue, T., K. Takeda, E. Oda, H. Tanaka, H. Murasawa, A. Takaoka, Y. Morishita, S. Akira, T. Taniguchi, and N. Tanaka. 2003. Integral role of Noxa in p53-mediated apoptotic response. Genes Dev. 17:2233-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sugars, K. L., V. Budhram-Mahadeo, G. Packham, and D. S. Latchman. 2001. A minimal Bcl-x promoter is activated by Brn-3a and repressed by p53. Nucleic Acids Res. 29:4530-4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Villunger, A., E. M. Michalak, L. Coultas, F. Mullauer, G. Bock, M. J. Ausserlechner, J. M. Adams, and A. Strasser. 2003. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science 302:1036-1038. [Online.] [DOI] [PubMed] [Google Scholar]
- 46.Wagner, A. J., J. M. Kokontis, and N. Hay. 1994. Myc-mediated apoptosis requires wild-type p53 in a manner independent of cell cycle arrest and the ability of p53 to induce p21waf1/cip1. Genes Dev. 8:2817-2830. [DOI] [PubMed] [Google Scholar]
- 47.Yu, J., L. Zhang, P. M. Hwang, K. W. Kinzler, and B. Vogelstein. 2001. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol. Cell 7:673-682. [DOI] [PubMed] [Google Scholar]