Abstract
Glucocorticoids (GCs) are commonly prescribed drugs, but their anti-inflammatory benefits are mitigated by metabolic side effects. Their transcriptional effects, including tissue-specific gene activation and repression, are mediated by the glucocorticoid receptor (GR), which is known to bind as a homodimer to a palindromic DNA sequence. Using ChIP-exo in mouse liver under endogenous corticosterone exposure, we report here that monomeric GR interaction with a half-site motif is more prevalent than homodimer binding. Monomers colocalize with lineage-determining transcription factors in both liver and primary macrophages, and the GR half-site motif drives transcription, suggesting that monomeric binding is fundamental to GR's tissue-specific functions. In response to exogenous GC in vivo, GR dimers assemble on chromatin near ligand-activated genes, concomitant with monomer evacuation of sites near repressed genes. Thus, pharmacological GCs mediate gene expression by favoring GR homodimer occupancy at classic palindromic sites at the expense of monomeric binding. The findings have important implications for improving therapies that target GR.
Glucocorticoids (GCs) are cholesterol-derived steroid hormones that control metabolic and homeostatic processes essential for mammals. Natural GCs and their synthetic analogs are among the most widely prescribed drugs in the world because of their anti-inflammatory and immunosuppressive properties that are important for the treatment of rheumatoid arthritis, cerebral edema, allergic reactions, and asthma, and they are utilized to prevent organ transplant rejection and graft-versus-host disease (Ito et al. 2006a,b; Kirwan and Power 2007). Unfortunately, the development of major metabolic side effects, including diabetes, obesity, hypertension, osteoporosis, and muscle atrophy, can cause treatment to end (Rosen and Miner 2005). Both natural and synthetic GCs bind and activate GR (NR3C1), a nuclear receptor transcription factor (TF) ubiquitously expressed in the body (Heitzer et al. 2007; Evans and Mangelsdorf 2014). Upon ligand binding, GR translocates from the cytoplasm to the nucleus where it can either activate or repress the expression of target genes in a cell-type-specific manner. GR can regulate transcription by binding as a homodimer to its palindromic recognition sequence, termed a GC response element (GRE), yet the molecular mechanisms leading to transcriptional activation versus repression are unclear. Better understanding of this is the focus of intense effort, as the clinical benefits of GC treatment are thought to be due to the transcriptional repression of cytokines and chemokines in immune cells, whereas the unwanted side effects may be linked to the activation of metabolic genes in other tissues (Beck et al. 2009).
A popular model proposes that DNA binding of the GR homodimer transactivates expression, whereas protein–protein interactions tethering monomeric GR to other DNA-bound TFs, such as NFKB1 or JUN-FOS heterodimers (AP-1), mediate transrepression (Glass and Saijo 2010). Early support came from studies of GRdim mice initially developed to separate transactivation and transrepression functions. These mice carry an amino acid substitution in the D-loop of the DNA-binding domain of GR (A465T), which was designed to reduce GR homodimer formation and DNA binding (Reichardt et al. 1998). In agreement with the transactivation mechanism, GRdim mice show a reduced ability to activate transcription in the liver in response to exogenous ligands (Frijters et al. 2010). However, later studies were inconsistent with expectations for transrepression by revealing that GRdim mice exhibit a diminished response to GC treatment in inflammatory paradigms such as TNF-induced inflammation (Vandevyver et al. 2012), LPS- and CLP-induced sepsis (Kleiman et al. 2012; Silverman et al. 2013), antigen-induced rheumatoid arthritis (Baschant et al. 2011, 2012), allergic contact dermatitis (Kleiman and Tuckermann 2007), and experimental autoimmune encephalomyelitis, a mouse model for multiple sclerosis (Schweingruber et al. 2014). This was in part due to the inability of GRdim to induce GR-dependent anti-inflammatory genes (Vandevyver et al. 2012). Although its DNA-binding properties are compromised relative to GR (Gebhardt et al. 2013; Watson et al. 2013), the inability of GRdim to dimerize and bind DNA has been challenged by in vitro studies (Jewell et al. 2012; Presman et al. 2014; Sedwick 2014). Complicating matters further, recent cistromic analyses in primary macrophages (Uhlenhaut et al. 2013) and the liver (Grøntved et al. 2013) have revealed similar enrichment of GR at recognition sequences near both ligand-activated and ligand-repressed genes, suggesting factors other than GR occupancy as important regulatory determinants. As a whole, these studies highlight the need for improved understanding of GR binding to native chromatin and how it pertains to transactivation and transrepression.
To this end, and to gain insight into the impaired hepatic gene expression and anti-inflammatory responses of GRdim mice, we compared chromatin immunoprecipitation (ChIP) with deep sequencing (ChIP-seq), ChIP with lambda exonuclease digestion and sequencing (ChIP-exo), and transcriptomic analyses in liver tissue isolated from wild-type (WT) and GRdim mice. Our study reveals extensive genomic occupancy of monomeric and dimeric GR that is specified by DNA motifs and colocalized TFs. Exogenous GC treatment of mice uncovers a mechanism whereby GR dimers transactivate at the expense of monomers, resulting in both increased and decreased GC-mediated gene expression.
Results
GR, but not GRdim, occupies the palindromic motif as a dimer at a minority of genomic sites in mouse liver
To compare genomic occupancy by GR and GRdim under physiological conditions, we performed ChIP-seq in liver, where GR regulates diverse processes ranging from hepatic neonatal growth to glucose and lipid homeostasis (Rose and Herzig 2013). Liver tissue was isolated from WT and GRdim mice during the morning and evening to control for potential circadian effects of serum corticosterone levels superimposed over an ultradian rhythm, stress effects, and local GC production by hydroxysteroid 11-beta dehydrogenase 1 (Conway-Campbell et al. 2012). Biological replicates revealed highly correlated occupancy for GR at 6 a.m. and 6 p.m. in either WT or GRdim mice, with stronger occupancy at 6 p.m. (Supplemental Fig. 1). De novo motif analyses revealed enrichment for different GR motifs (Fig. 1A), and comparison of GR binding sites in WT versus GRdim mice revealed a bifurcated distribution (Fig. 1B), suggesting that GR and GRdim occupy two classes of sites categorized by relative binding strength. We identified 2110 sites that preferentially bind GR versus GRdim, termed WT-selective, and a major class of 11,108 common sites that are occupied similarly by GR and GRdim (Supplemental Fig. 2). Nearly indistinguishable behavior was observed for the WT-selective and common sites at 6 a.m. and 6 p.m. (Supplemental Fig. 3). These data demonstrate that GRdim binds the genome in vivo, and the absence of substantial GRdim-specific peaks indicates that it occupies a subset of GR binding sites.
Figure 1.

GR, but not GRdim, occupies the canonical palindromic motif as a dimer in liver. (A) Top-ranked de novo motifs from HOMER for the GR cistromes from WT and GRdim mice. See Supplemental Material for a comprehensive list of motifs. (B) Scatter plots comparing sequence tags from 14,940 GR ChIP-seq peaks with at least two reads per million (RPM) in any condition in livers isolated from WT and GRdim mice killed at either 6 a.m. (top) or 6 p.m. (bottom). Blue and red highlight WT-selective and common sites, respectively. (C) GR binding upstream of the tyrosine aminotransferase (Tat) gene. The 5′ ends of forward- and reverse-stranded sequence tags are indicated in red and blue, respectively, for the ChIP-exo tracks. Tracks are RPM normalized. (D) Distance distribution for opposite-stranded peaks with at least 0.2 RPM from GR ChIP-exo in liver isolated at 6 a.m. from WT and GRdim mice is shown for WT-selective sites. The number of peak pairs and prominent peak distances are indicated. Schematic of opposite-stranded peaks is shown at top. (E) GR ChIP-exo for WT-selective sites in liver isolated at 6 a.m. MEME top-ranked de novo sequence with a hit count of at least 5% is shown at the top. See Supplemental Material for a full list of motifs. Average profiles (middle) and density heatmaps (bottom) of the raw sequence tags are shown for both mouse models. Red and blue indicate the 5′ ends of the forward- and reverse-stranded tags, respectively.
To define the occupancy properties of GR and GRdim in more detail, we performed ChIP-exo using biological replicates from WT and GRdim mice to map sites at a higher resolution than afforded by ChIP-seq. The technique uses lambda exonuclease to trim DNA to the point where a crosslinked TF blocks further enzymatic activity (Mymryk and Archer 1994). This creates a 5′ border at a fixed distance from the TF, and sequencing of the digestion products can yield an unambiguous determination of the bound sequence positioned between opposite-stranded peak pairs (Rhee and Pugh 2011). Figure 1C shows the GR ChIP-exo profile at a WT-selective site in liver compared to conventional ChIP-seq data, with discrete ChIP-exo peaks positioned within the larger ChIP-seq peak. For an unbiased interrogation of the data, we determined the distance between opposite-stranded peaks across the genome and plotted the frequency distribution to examine whether a particular spacing emerged from the background that is reflective of GR occupancy. Although the peak-to-peak distance for GR could not be predicted in advance without previously reported ChIP-exo results, enrichment of its motif within peak pairs spaced by the predominate distance would be indicative of binding. Interestingly, ChIP-exo peak pairs separated by 11 and 31 base pairs (bp) were frequently observed at WT-selective sites in WT mice at both 6 a.m. (Fig. 1D) and 6 p.m. (Supplemental Fig. 4A). Two peak pairs can result from a population of bound dimers if only the left or right monomer is crosslinked to the DNA because the exonuclease block is thought to occur at the site of crosslinking. This requires that only a small fraction of molecules becomes crosslinked, which is consistent with the low efficiencies observed for formaldehyde treatment. Moreover, it could be accentuated when dimerization is promoted by DNA interaction, which may be the case for GR (Robblee et al. 2012; Hudson et al. 2013). In support of dimer binding, de novo examination of the 6 a.m. regions protected from exonuclease digestion revealed that the palindromic GR motif at 81% of the sites was flanked by two peak pairs to yield four exonuclease borders in WT but not GRdim mice (Fig. 1E). No other motif was present at ≥5% of the peak pairs, and similar results were observed with 6 p.m. sites (Supplemental Fig. 4B). In agreement with peak pairs that fall outside of the GR motif, the exonuclease-cut sites spaced by 31 bp reside at regions with no substantial nucleotide bias (Supplemental Fig. 4C), suggesting that DNA sequence does not influence the ChIP-exo patterning. Together, these data indicate that GR binds its palindromic motif as a dimer in vivo and that GRdim lacks this ability.
GR and GRdim monomers occupy the liver genome at half-site motifs and tethered sites
Remarkably, the sites commonly bound by GR and GRdim comprise a major subset of the GR liver cistrome yet lack association with the palindromic motif. ChIP-exo at these sites identified peak pairs separated by 11 and 25 bp that emerged from the background in both WT and GRdim mice at 6 a.m. (Fig. 2A) and 6 p.m. (Supplemental Fig. 5A). De novo examination of peak pairs with short spacing (5–15 bp) identified the GR half-site and FOXA motifs as the only sequences present in at least 5% of the sites (Fig. 2B; Supplemental Fig. 5B). Consistent with the low frequency of short-spaced peak pairs, a minority of the common sites has peak pairs flanking the GR half-site motif at a fixed distance (Fig. 2C; Supplemental Fig. 5C,D). This could suggest that GR and GRdim occupy most of the common sites independently of the half-site motif. However, it is also possible that nuclear receptor monomers may not perform well in the ChIP-exo assay. In agreement with this, reexamination of a ChIP-exo study with the estrogen receptor (Serandour et al. 2013) revealed tighter peak pairs flanking the full- versus the half-site motifs (Supplemental Fig. 6). Moreover, in support of sequence-specific binding, the GR half-site motif is centrally enriched at ChIP-seq peaks from common sites (Fig. 2D), and its frequency within a peak scales with GR-occupancy strength (Fig. 2E). As a whole, the data indicate that GR and GRdim occupy the genome through sequence-specific interaction with the half-site motif, but experimental limitations preclude a direct assessment of its prevalence.
Figure 2.

GR and GRdim occupy the liver genome at the half-site motif. (A) The distance distribution for opposite-stranded peaks with at least 0.2 RPM from GR ChIP-exo in liver isolated at 6 a.m. is shown for sites commonly bound in WT and GRdim mice, with the number of peak pairs and prominent peak distances indicated. (B) MEME de novo sequences from 6 a.m. common site peak pairs separated by 5–15 bp and with a hit count of at least 5%. See Supplemental Material for a comprehensive list of motifs. (C) GR ChIP-exo at 6 a.m. common sites, with average profiles and density heatmaps for the half-site motif shown for both mouse models. Red and blue indicate the 5′ ends of the forward- and reverse-stranded tags, respectively. (D) Distribution of the GR-half motif relative to the center of GR ChIP-seq peaks for the common sites subdivided into quartiles. (E) Plot of GR-binding strength relative to the number of half-site motifs present in ChIP-seq peaks from common sites.
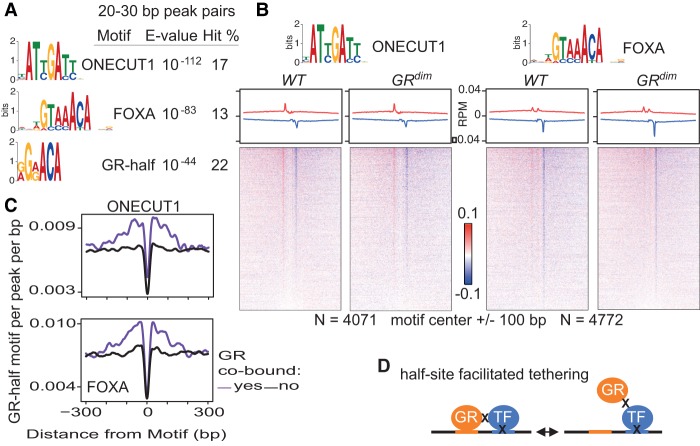
Examination of peak pairs with 20- to 30-bp spacing revealed centrally enriched ONECUT1, FOXA, and GR-half motifs at both 6 a.m. (Fig. 3A) and 6 p.m. (Supplemental Fig. 5B) for the common sites. Presence of the ONECUT1 and FOXA motifs raised the possibility of formaldehyde crosslinking between GR or GRdim and DNA-crosslinked ONECUT1 or a FOXA factor(s). To address this, we used published ChIP-seq data sets for ONECUT1 (Laudadio et al. 2012) and FOXA2 (Li et al. 2012) in liver to examine the GR ChIP-exo reads at common sites that are also bound by ONECUT1 or FOXA2. Opposite-stranded peaks flank the ONECUT1 motif with the same spacing found by ONECUT1 ChIP-exo (Wang et al. 2014), while an asymmetric profile of positioned peaks observed previously for FOXA2 ChIP-exo (Serandour et al. 2013) is present at the FOXA motif (Fig. 3B; Supplemental Fig. 5D). Their signal strength is low, most likely reflecting transient protein–protein interactions, yet their tight positioning indicates occupancy. Intriguingly, GR half-site motifs are located nearby the ONECUT1 and FOXA motifs (Fig. 3C), suggesting that GR recruitment is mediated and/or stabilized through sequence-specific interaction with the DNA. DNA crosslinking of both GR and a neighboring TF is unlikely to explain these results because this would preclude formation of a peak pair of uniform length with enrichment for a centered motif. It is interesting that the motifs for HNF4A and CEBP proteins are highly enriched by ChIP-seq but not by ChIP-exo. GR half-site motifs are located nearby the HNF4A and CEBP motifs at cobound common sites (Supplemental Fig. 7A), but positioned peak pairs are not prominent (Supplemental Fig. 7B). These data could reveal selectivity for GR–TF interactions in liver, such that GR preferentially associates with FOXA and ONECUT1 versus HNF4A and CEBP TFs. However, it is formally possible that GR interacts with each of these factors but that HNF4A and CEBP TFs do not produce robust ChIP-exo profiles. The general applicability of ChIP-exo remains to be determined given that only a handful of studies on a small number of TFs have been published to date. As a whole, the data indicate that GR and GRdim monomers occupy the liver genome via sequence-specific binding to the GR half-site motif and through interaction with DNA-bound TFs. The latter, termed half-site-facilitated tethering, is mediated and/or stabilized by binding to the half site (Fig. 3D). Half-site-facilitated tethering may occur generally, as examination of previously described genomic sites in pituitary-derived AtT-20 cells with proposed tethering of GR to STAT3 (Langlais et al. 2012) shows enrichment for the GR half-site motif (Supplemental Fig. 8).
Figure 3.
GR and GRdim occupy the liver genome at tethered sites. (A) MEME de novo sequences from 6 a.m. common site peak pairs separated by 20–30 bp and with a hit count of at least 5%. See Supplemental Material for a comprehensive list of motifs. (B) GR ChIP-exo at 6 a.m. common sites, with average profiles and density heatmaps for the ONECUT1 (left) and FOXA (right) motifs shown for both mouse models. GR common sites cobound by ONECUT1 or FOXA2 were interrogated. Red and blue indicate the 5′ ends of the forward- and reverse-stranded tags, respectively. (C) Distribution of the GR half-site motif relative to neighboring motifs at common sites cobound by ONECUT1 (top) or FOXA2 (bottom). Results for ONECUT1 and FOXA2 liver sites without GR are shown for comparison. (D) Half-site-facilitated tethering. The GR half-site motif is represented in orange. X indicates formaldehyde crosslinking between proteins or protein–DNA. Two formaldehyde crosslinking events between GR and a DNA-crosslinked TF are necessary to detect sites where GR appears bound to noncanonical motifs.
GR monomers bind near lineage-determining TFs in liver
GR ChIP-seq and ChIP-exo from WT and GRdim mice implicate functions for hepatic lineage TFs, prompting us to investigate whether GR dimers and monomers differentially colocalize with them. De novo motif analyses revealed that the dimeric (WT-selective) ChIP-seq peaks are most enriched for the palindromic GR motif, while the monomeric (common) sites contain the recognition sequences for liver TFs along with the GR half site (Fig. 4A). When directly compared to each other as foreground and background data sets, the dimeric sites showed strong enrichment for only the palindromic GR motif, while the monomeric sites were modestly enriched for the HNF4A motif as the top-ranked sequence (see Supplemental Material). This indicates that the GR-palindromic motif distinguishes dimeric from monomeric sites and implies that liver TFs colocalize with monomeric sites more than dimeric. Consistent with the latter, interrogation of published ChIP-seq data sets revealed stronger co-occupancy of the hepatic TFs HNF4A, FOXA2, CEBPB, and ONECUT1 at monomeric versus dimeric sites (Fig. 4B) and that the fraction of monomeric, but not dimeric, sites increases along with the number of colocalizing TFs (Fig. 4C). Collectively, the data suggest that monomeric versus dimeric occupancy has a greater dependency on liver-determining TFs.
Figure 4.

Monomeric GR colocalizes with lineage-determining TFs in liver. (A) HOMER de novo motif analyses for the dimeric and monomeric GR binding sites from liver ChIP-seq. The four top-ranked lineage TF motifs are shown relative to the top-ranked GR sequence. See Supplemental Material for a comprehensive list of motifs. (B) Box plots interrogating the co-occupancy of liver TFs at GR binding sites. (C) Distribution of dimer and monomer GR binding sites relative to colocalized HNF4A, CEBPB, ONECUT1, and/or FOXA2. All TF combinations were examined.
GRdim partitions the GR cistrome similarly in primary macrophages and liver
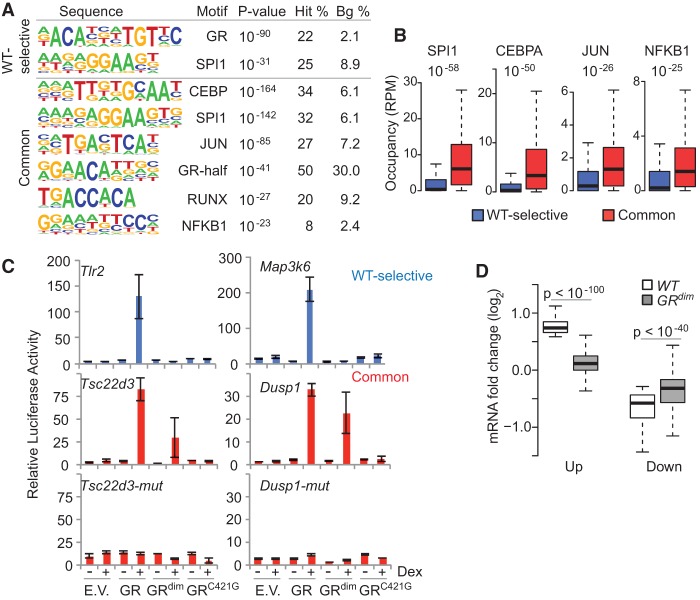
To investigate the general nature of our findings, we performed GR ChIP-seq in primary bone marrow–derived macrophages isolated from WT and GRdim mice and activated with lipopolysaccharide (LPS). Sites commonly bound by both GR and GRdim make up the majority of the GR cistrome and are enriched for myeloid-specific and inflammatory-acting TF motifs in addition to the GR half site, whereas WT-selective sites are most enriched for the palindromic GR motif (Fig. 5A; Supplemental Fig. 9). Direct comparison to each other as foreground and background data sets showed modest enrichment for the CEBP motif at common sites and strong enrichment for only the palindromic GR motif at WT-selective sites (see Supplemental Material). In addition, examination of published ChIP-seq data sets showed that the co-occupancy of macrophage TFs with GR is stronger at common versus WT-selective sites (Fig. 5B). Together, these findings indicate similar properties for GR binding sites in macrophages and liver. To explore a transcriptional function for monomeric sites, we placed several WT-selective and common GR binding sites into luciferase reporters and assayed their activity in response to cotransfection of GR or GRdim (Fig. 5C). While GR drives more transcription than GRdim at the WT-selective sites, GRdim compares favorably at the common regions, agreeing with earlier studies demonstrating in vitro activity for GRdim (Adams et al. 2003; Meijsing et al. 2009; Jewell et al. 2012). Mutation of the GR half-site sequence within the common regions destroys GR and GRdim activity, demonstrating that transcriptional activation by monomeric GR requires a half-site motif. To test this in a more physiological setting, we compared the expression of genes regulated by a GR ligand in LPS-stimulated WT macrophages to that in GRdim macrophages. As shown in Figure 5D, ligand has a reduced ability to modulate induced and repressed genes in GRdim macrophages, consistent with earlier findings in liver (Frijters et al. 2010). Collectively, the data suggest comparable functions for GR monomers and dimers in primary macrophages and liver.
Figure 5.
GRdim partitions the GR cistrome in primary macrophages. (A) Top-ranked de novo motifs from HOMER for the WT-selective and common GR binding sites from ChIP-seq in primary macrophages. Comprehensive motif results are reported in the Supplemental Material. (B) Box plots interrogating the co-occupancy of macrophage TFs at GR binding sites. (C) Regions named after the nearest gene that include WT-selective and common sites were assayed by a luciferase reporter in the absence (−) or presence (+) of dexamethasone (Dex) and cotransfection of empty vector (E.V.), GR, GRdim, or the DNA-binding mutant GRC421G. (D) Response of WT-regulated genes (up or down) from LPS- and dex-treated macrophages in GRdim-derived macrophages under the same treatment.
Exogenous GC redistributes GR from monomeric to dimeric sites at ligand-regulated genes
To explore potential differences between GR monomers and dimers at pharmacological exposure to GC, we examined GR occupancy in liver isolated at 6 a.m. from WT and GRdim mice treated for 24 h with prednisolone, a synthetic GC used to treat a wide range of inflammatory and auto-immune conditions (Czock et al. 2005). Most GR binding sites were unaffected by prednisolone (Supplemental Fig. 10A), yet occupancy was increased at 889 sites and decreased at another 626 sites in WT mice, with minimal changes occurring at these sites in GRdim mice (Fig. 6A). Encouragingly, the top-ranked gene ontology (GO) categories associated with the gained sites involve glucose metabolism (Fig. 6A; Supplemental Fig. 10B), consistent with GR function in liver (Rose and Herzig 2013). Gained sites are strongly enriched for the GR palindromic motif and a robust dimer signature in WT but not GRdim mice (Fig. 6B; Supplemental Fig. 10C), indicating that the exogenous ligand primarily stimulates dimeric occupancy. In addition to GR, CEBPB is also recruited to the gained sites (Fig. 6C), agreeing with earlier work demonstrating collaborative binding interactions between GR and CEBPB (Grøntved et al. 2013) and indicating that the genomic occupancy of multiple TFs can be indirectly affected by ligand. Lost sites, in contrast, are highly enriched for the GR half motif. Indeed, modest association with the STAT motif agrees with our finding of GR half-site motifs near sites of interactions between GR and STAT TFs in published data sets from liver (Tronche et al. 2004) and AtT-20 cells (Langlais et al. 2012), supporting the model of half-site-facilitated tethering.
Figure 6.

Glucocorticoids (GCs) redistribute GR from monomeric to dimeric sites at regulated genes. (A) Density heatmap of ChIP-seq reads from GR peaks at 6 a.m. with at least 2 RPM in any condition. Prednisolone (pred) treatment of WT and GRdim mice spanned 24 h, and regulated sites in WT mice have at least a twofold difference compared with untreated controls. Top-ranked GO terms for the gained and lost sites are indicated on the left. MEME top-ranked de novo sequences are indicated on the right. See Supplemental Material for a comprehensive list of motifs. (B) GR ChIP-exo at gained sites in WT mice with or without Pred treatment. Average profiles (top) and density heatmaps (bottom) show the 5′ ends of the forward-stranded (red) and reverse-stranded (blue) sequence tags. (C) Box plot comparing CEBPB occupancy at sites with increased GR binding (gained) after Pred treatment. (D) Scatter plot comparing sequence tags from GR ChIP-seq in WT liver with and without Pred treatment, with sites containing up- or down-regulated RNAPII occupancy in response to Pred highlighted in red or blue, respectively. (E) Fraction of Pred-regulated genes with a transcription start site within 100 kb of a gained or lost GR binding site (top) or a non-Pred-regulated dimeric or monomeric GR binding site (bottom).
We performed ChIP-seq for RNA polymerase II (RNAPII), which is recruited to active enhancers on a global scale (Szutorisz et al. 2005; Koch et al. 2008; Kim et al. 2010), to explore a potential transcriptional function for the prednisolone-regulated sites. A direct correlation was found between occupancy changes to RNAPII and GR in response to prednisolone. Increased and decreased RNAPII associates with increased and decreased GR, respectively (Fig. 6D; Supplemental Fig. 10D), suggesting that GR occupancy modulates the activity of these regions. To investigate their association with gene expression, we mapped gained or lost sites to prednisolone-regulated genes (Frijters et al. 2010). Figure 6E shows that prednisolone-induced genes were enriched more than sixfold for gained versus lost sites, suggesting that induced occupancy by GR dimers drives activated transcription. Moreover, dimer sites without prednisolone-regulated occupancy were also colocalized with increased RNAPII (Fig. 6D) and enriched near induced genes (Fig. 6E), suggesting multiple mechanisms mediate prednisolone's effects. Repressed genes had a greater tendency to harbor lost sites, suggesting that prednisolone stimulates GR dimers to activate particular genes at the expense of others that lose monomers (Supplemental Fig. 10E). GRdim redistribution is diminished relative to GR, likely due to its inability to form dimers on chromatin, and provides a rationale for the reduced ability of exogenous GC to regulate liver genes in GRdim versus WT mice (Frijters et al. 2010).
Discussion
GR has been a focus of extensive study for decades, yet how it activates some genes while repressing others remains an open question. The two main mechanisms of transactivation and transrepression propose that the monomeric/dimeric state of the receptor determines its transcriptional function such that GR dimers activate through binding of inverted repeats, whereas monomers repress by tethering to DNA-bound TFs. Our data reveal a strong association between dimeric GR occupancy and activated gene expression on a genome-wide scale in both liver tissue and primary macrophages. However, the discovery of monomeric occupancy near transcriptionally active genes, and the ability of monomeric-binding sites to drive transcription in vitro, indicate an expanded transactivation mechanism incorporating GR monomers. In support of this, similar monomeric function has recently been described in vitro for human U2OS osteosarcoma cell lines carrying stably integrated GR alleles (Schiller et al. 2014). Revision of the transrepression mechanism may also be in order to include binding of GR monomers to half sites. Direct binding to so-called negative GREs by GR monomers has been proposed to mediate repression (Surjit et al. 2011; Hudson et al. 2013), but we find little interaction between GR and this sequence motif, consistent with earlier studies (Grøntved et al. 2013; Uhlenhaut et al. 2013) and indicating that this scenario does not play a prominent role in macrophages or liver. Rather, we observe ligand-dependent redistribution of GR and RNAPII from monomeric to dimeric GR binding sites in liver. The model supposes that GR and RNAPII are available in limiting amounts so that gain of occupancy at one set of sites leads to loss at another. Note that prednisolone, whose potency is four to five times that of corticosterone, was delivered at a pharmacological dosage that exceeds even the highest physiological GC level by at least two orders of magnitude (Herrmann et al. 2009). Thus, we believe that exogenous GC, at pharmacological levels, is required for redistribution of GR from monomeric sites that are occupied under physiological levels of hormone.
Sequence-specific binding by GR monomers is compatible with tethering as a mode of action. Tethering was originally described using reporter assays with constructs lacking GR motifs and overexpressed TFs. It is thought to occur when GR targets a DNA-bound TF to indirectly bind DNA, and is generally invoked to explain ChIP-seq peaks lacking the expected motif for the immunoprecipitated TF. However, tethered sites often reside at regions bound by multiple TFs such that an unambiguous determination of the tethering factor is not possible. Using the superior resolution of ChIP-exo, we identify a FOXA protein(s) and ONECUT1 as GR tethering partners in liver. That a GR half-site motif is positioned nearby these sites suggests a role for sequence-specific binding. We favor a model termed half-site-facilitated tethering, where sequence-specific interaction of GR monomers to half-site motifs promotes and/or stabilizes transient contacts between monomers and nearby TFs. These are captured by formaldehyde treatment, but whether the protein–protein crosslinks are formed with monomers on or off the DNA cannot be determined by our data. However, little or no evidence for tethering was found near dimer-binding sites, which are bound more tightly due to cooperative effects between monomers (Gebhardt et al. 2013; Hudson et al. 2013; Watson et al. 2013), suggesting that longer residence times on the DNA prevent transient interactions by reducing the time spent sampling sites and supporting the idea that some of the tethered sites are formed when monomers are not bound to half sites. Consistent with this, single molecule tracking approaches have revealed extensive nonspecific chromatin binding of TFs through either protein–DNA or protein–protein processes when not bound to target recognition sequences (Chen et al. 2014).
Monomer-binding sites display greater cell-type specificity and enrichment for lineage-determining TFs relative to dimer sites. There may be multiple underlying causes for this behavior that involve GC content of the DNA and open chromatin effects. However, we suggest that the simplest explanation is that GR monomers are more dependent on the cobinding of neighboring TFs than dimers, indicating an important role in tissue-specific gene regulation. The estrogen receptor alpha (ESR1) has been shown to occupy two classes of binding sites in vivo, with the first characterized by high-affinity ESR1 motifs bound in multiple cell types and the second by the absence of canonical ESR1 motifs and cell-specific occupancy with tissue-selective TFs (Joseph et al. 2010; Gertz et al. 2013). The second class shares properties with GR monomer sites, suggesting that ESR1 and possibly other steroid receptors occupy the genome as monomers in vivo. As monomeric binding lacks the stabilizing forces employed by the dimer on DNA, its strong colocalization with other TFs may reflect a need for assisted loading onto genomic sites (Voss et al. 2011). Indeed, GR tethering partners differ across cell types and are enriched for lineage-determining TFs (Kassel and Herrlich 2007), which may be explained by half-site-facilitated tethering.
The ability of GRdim to rescue the essential function(s) of GR suggests that GR monomers are biologically important. Early interpretations proposed no DNA-binding functions for GRdim (Reichardt et al. 1998), but our data show that GRdim occupies a majority of sites comprising the GR cistromes in a primary cell type and native tissue and displays little or no selective binding. Recently, the inability of GRdim to form homodimers has been challenged through forced expression of GFP- or YFP-tagged GRdim and monitoring dimerization by immunoprecipitation (Jewell et al. 2012) or a number and brightness assay (Presman et al. 2014). We detect little, if any, dimeric occupancy for GRdim, indicating that it interacts with the genome primarily as a monomer under physiological conditions. In fact, the absence of chromatin-bound dimers in GRdim mice likely contributes to the phenotype of these animals. Thus, both monomers and dimers play important transcriptional roles, which may provide flexibility for GR, enabling a single molecule to differentially modulate multiple gene expression programs in the same cell and in multiple cell types.
Methods
Animal care
GRdim mice were back-crossed for at least four generations to the BALB/c background. Twelve-week-old male GRdim and GR+/+ littermate controls were maintained in a 12-h dark–light cycle. For ChIP-seq in liver, 6 a.m. and 6 p.m. tissues were collected at the beginning of the light and dark phases, respectively. A third group of mice was treated for 24 h with subcutaneous applied prednisolone pellets (12 mg/kg; Innovative Research of America). All mice were kept under standardized conditions with water and food ad libitum in a specific pathogen-free animal facility at the Leibniz Institute for Age Research (Fritz Lipmann Institute, Jena) and at the University of Ulm. Animal experiments were performed using procedures in accordance with the Thüringer Landesamt für Lebensmittelsicherheit und Verbraucherschutz and the Regierungspräsidium in Tübingen, Germany.
Cell and tissue preparation
Bone-marrow–derived macrophages were isolated and differentiated in culture as previously described (Barish et al. 2005). Cells were treated overnight with 1 µM dexamethasone (Dex; Sigma) and/or LPS (100 ng/mL, Sigma) for 3 h. Liver tissue isolated from deceased mice was immediately frozen in liquid nitrogen. To process for ChIP, the tissue was thawed, minced, and crosslinked in 1% formaldehyde/PBS. Cells were disrupted and nuclei purified by Dounce homogenization in a hypotonic buffer (20 mM HEPES/NaOH at pH 7.5, 0.25 M sucrose, 3 mM MgCl2, 0.2% NP-40, 3 mM 2-mercaptoethanol, complete protease inhibitor cocktail). To prepare ChIP extracts, nuclei were suspended in SDS lysis buffer (50 mM HEPES/NaOH at pH 7.5, 1% SDS, 10 mM EDTA, complete protease inhibitor), incubated for 10 min at 4°C, and subjected to microtip probe sonication under conditions optimized for IP efficiency.
ChIP-seq and ChIP-exo
ChIP-seq in primary macrophages was performed as described earlier (Uhlenhaut et al. 2013). ChIP in liver was performed with sonicated, nuclear extract prepared from formaldehyde-crosslinked tissue. Each experimental condition, e.g., genotype (WT or GRdim), time of harvest (6 a.m. or 6 p.m.), treatment (with or without prednisolone), was assayed in duplicate with separate mice for ChIP-seq, and the same biological replicates were examined by ChIP-exo. The following antibodies were used: GR (PA1-511A, Pierce; sc-1004, Santa Cruz), RNAPII (sc-899, sc-9001, Santa Cruz), and CEBPB (sc-150, Santa Cruz). ChIP-seq libraries were produced and sequenced according to Illumina protocols as previously described (Steger et al. 2010), while ChIP-exo libraries were prepared according to Serandour et al. (2013). Sequencing was performed with multiplexed libraries. Details for ChIP-seq and ChIP-exo data processing are reported in the Supplemental Material.
RNA-seq
RNA isolation from primary macrophages was carried out after overnight incubation with dexamethasone and 6-h LPS treatment using Qiagen RNeasy kits. Libraries were made per manufacturer's instructions using Illumina TruSeq sample prep kits and sequenced using an Illumina HiSeq.
Luciferase reporter assays
Luciferase assays were carried out as described elsewhere (Uhlenhaut et al. 2013). In short, candidate enhancers were amplified from mouse genomic DNA using oligos that added 5′ XhoI and 3′ HindIII or BglII restriction sites. Oligo sequences are reported in the Supplemental Information. Cis-regulatory elements were cloned into pGL4.23 (Promega). After transfection, CV-1 cells were treated overnight with 1 µM Dex or ethanol, and reporter activity was determined with Promega's ONE-Glo system.
Data access
Genome-wide data sets from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE59752 (liver), GSE59764 (macrophage ChIP-seq), and GSE68160 (macrophage RNA-seq).
Supplementary Material
Acknowledgments
We thank Sabine Schmidt, Sebastian Schäfer, Anja Bauerfeind, Susanne Blachut, the MDC Genomics Core, Franziska Greulich, and Sonja Steinbach for their contributions to the macrophage experiments. We thank the animal facility staff of the Fritz-Lipmann Institute and the University of Ulm and in particular Sara Holly, Tobias Rappold, Dominique Galendo, and Petra Kirsch. We also thank the Functional Genomics Core of the Penn Diabetes Center (DK19525) for deep sequencing. This work was supported by the Deutsche Forschungsgemeinschaft through grants UH 275/1-1 (N.H.U.) and Tu220/3, Tu220/6/Sp1468 Immunobone (J.T.); European Molecular Biology Organization ALTF 686-2010 (N.H.U.); European Union FP7 BRAINAGE (J.T.); and National Institute of Diabetes and Digestive and Kidney Diseases grants R21 DK098769 (K.J.W.), R01 DK45586 (M.A.L.), and R01 DK098542 (D.J.S.).
Author contributions: H.W.L. performed the computational analyses. N.H.U. (macrophage) and D.J.S. (liver) performed genomics experiments. A.R., J.W., and S.H. performed animal work. N.H. and K.J.W. provided support. M.A.L. facilitated the collaboration. H.W.L., N.H.U., M.A.L., J.T., and D.J.S. wrote the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.188581.114.
References
- Adams M, Meijer OC, Wang J, Bhargava A, Pearce D. 2003. Homodimerization of the glucocorticoid receptor is not essential for response element binding: activation of the phenylethanolamine N-methyltransferase gene by dimerization-defective mutants. Mol Endocrinol 17: 2583–2592. [DOI] [PubMed] [Google Scholar]
- Barish GD, Downes M, Alaynick WA, Yu RT, Ocampo CB, Bookout AL, Mangelsdorf DJ, Evans RM. 2005. A Nuclear Receptor Atlas: macrophage activation. Mol Endocrinol 19: 2466–2477. [DOI] [PubMed] [Google Scholar]
- Baschant U, Frappart L, Rauchhaus U, Bruns L, Reichardt HM, Kamradt T, Bräuer R, Tuckermann JP. 2011. Glucocorticoid therapy of antigen-induced arthritis depends on the dimerized glucocorticoid receptor in T cells. Proc Natl Acad Sci 108: 19317–19322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baschant U, Lane NE, Tuckermann J. 2012. The multiple facets of glucocorticoid action in rheumatoid arthritis. Nat Rev Rheumatol 8: 645–655. [DOI] [PubMed] [Google Scholar]
- Beck IME, Vanden Berghe W, Vermeulen L, Yamamoto KR, Haegeman G, De Bosscher K. 2009. Crosstalk in inflammation: the interplay of glucocorticoid receptor-based mechanisms and kinases and phosphatases. Endocr Rev 30: 830–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhang Z, Li L, Chen B-C, Revyakin A, Hajj B, Legant W, Dahan M, Lionnet T, Betzig E, et al. 2014. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell 156: 1274–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway-Campbell BL, Pooley JR, Hager GL, Lightman SL. 2012. Molecular dynamics of ultradian glucocorticoid receptor action. Mol Cell Endocrinol 348: 383–393. [DOI] [PubMed] [Google Scholar]
- Czock D, Keller F, Rasche FM, Häussler U. 2005. Pharmacokinetics and pharmacodynamics of systemically administered glucocorticoids. Clin Pharmacokinet 44: 61–98. [DOI] [PubMed] [Google Scholar]
- Evans RM, Mangelsdorf DJ. 2014. Nuclear receptors, RXR, and the big bang. Cell 157: 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frijters R, Fleuren W, Toonen EJM, Tuckermann JP, Reichardt HM, van der Maaden H, van Elsas A, van Lierop M-J, Dokter W, de Vlieg J, et al. 2010. Prednisolone-induced differential gene expression in mouse liver carrying wild type or a dimerization-defective glucocorticoid receptor. BMC Genomics 11: 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt JCM, Suter DM, Roy R, Zhao ZW, Chapman AR, Basu S, Maniatis T, Xie XS. 2013. Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nat Methods 10: 421–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz J, Savic D, Varley KE, Partridge EC, Safi A, Jain P, Cooper GM, Reddy TE, Crawford GE, Myers RM. 2013. Distinct properties of cell-type-specific and shared transcription factor binding sites. Mol Cell 52: 25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, Saijo K. 2010. Nuclear receptor transrepression pathways that regulate inflammation in macrophages and T cells. Nat Rev Immunol 10: 365–376. [DOI] [PubMed] [Google Scholar]
- Grøntved L, John S, Baek S, Liu Y, Buckley JR, Vinson C, Aguilera G, Hager GL. 2013. C/EBP maintains chromatin accessibility in liver and facilitates glucocorticoid receptor recruitment to steroid response elements. EMBO J 32: 1568–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzer MD, Wolf IM, Sanchez ER, Witchel SF, DeFranco DB. 2007. Glucocorticoid receptor physiology. Rev Endocr Metab Disord 8: 321–330. [DOI] [PubMed] [Google Scholar]
- Herrmann M, Henneicke H, Street J, Modzelewski J, Kalak R, Buttgereit F, Dunstan CR, Zhou H, Seibel MJ. 2009. The challenge of continuous exogenous glucocorticoid administration in mice. Steroids 74: 245–249. [DOI] [PubMed] [Google Scholar]
- Hudson WH, Youn C, Ortlund EA. 2013. The structural basis of direct glucocorticoid-mediated transrepression. Nat Struct Mol Biol 20: 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Chung KF, Adcock IM. 2006a. Update on glucocorticoid action and resistance. J Allergy Clin Immunol 117: 522–543. [DOI] [PubMed] [Google Scholar]
- Ito K, Getting SJ, Charron CE. 2006b. Mode of glucocorticoid actions in airway disease. ScientificWorldJournal 6: 1750–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell CM, Scoltock AB, Hamel BL, Yudt MR, Cidlowski JA. 2012. Complex human glucocorticoid receptor dim mutations define glucocorticoid induced apoptotic resistance in bone cells. Mol Endocrinol 26: 244–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph R, Orlov YL, Huss M, Sun W, Kong SL, Ukil L, Pan YF, Li G, Lim M, Thomsen JS, et al. 2010. Integrative model of genomic factors for determining binding site selection by estrogen receptor-α. Mol Syst Biol 6: 456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassel O, Herrlich P. 2007. Crosstalk between the glucocorticoid receptor and other transcription factors: molecular aspects. Mol Cell Endocrinol 275: 13–29. [DOI] [PubMed] [Google Scholar]
- Kim T-K, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirwan J, Power L. 2007. Glucocorticoids: action and new therapeutic insights in rheumatoid arthritis. Curr Opin Rheumatol 19: 233–237. [DOI] [PubMed] [Google Scholar]
- Kleiman A, Tuckermann JP. 2007. Glucocorticoid receptor action in beneficial and side effects of steroid therapy: lessons from conditional knockout mice. Mol Cell Endocrinol 275: 98–108. [DOI] [PubMed] [Google Scholar]
- Kleiman A, Hübner S, Rodriguez Parkitna JM, Neumann A, Hofer S, Weigand MA, Bauer M, Schmid W, Schütz G, Libert C, et al. 2012. Glucocorticoid receptor dimerization is required for survival in septic shock via suppression of interleukin-1 in macrophages. FASEB J 26: 722–729. [DOI] [PubMed] [Google Scholar]
- Koch F, Jourquin F, Ferrier P, Andrau J-C. 2008. Genome-wide RNA polymerase II: not genes only! Trends Biochem Sci 33: 265–273. [DOI] [PubMed] [Google Scholar]
- Langlais D, Couture C, Balsalobre A, Drouin J. 2012. The Stat3/GR interaction code: predictive value of direct/indirect DNA recruitment for transcription outcome. Mol Cell 47: 38–49. [DOI] [PubMed] [Google Scholar]
- Laudadio I, Manfroid I, Achouri Y, Schmidt D, Wilson MD, Cordi S, Thorrez L, Knoops L, Jacquemin P, Schuit F, et al. 2012. A feedback loop between the liver-enriched transcription factor network and miR-122 controls hepatocyte differentiation. Gastroenterology 142: 119–129. [DOI] [PubMed] [Google Scholar]
- Li Z, Tuteja G, Schug J, Kaestner KH. 2012. Foxa1 and Foxa2 are essential for sexual dimorphism in liver cancer. Cell 148: 72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. 2009. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science 324: 407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mymryk JS, Archer TK. 1994. Detection of transcription factor binding in vivo using lambda exonuclease. Nucleic Acids Res 22: 4344–4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presman DM, Ogara MF, Stortz M, Alvarez LD, Pooley JR, Schiltz RL, Grøntved L, Johnson TA, Mittelstadt PR, Ashwell JD, et al. 2014. Live cell imaging unveils multiple domain requirements for in vivo dimerization of the glucocorticoid receptor. PLoS Biol 12: e1001813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt HM, Kaestner KH, Tuckermann J, Kretz O, Wessely O, Bock R, Gass P, Schmid W, Herrlich P, Angel P, et al. 1998. DNA binding of the glucocorticoid receptor is not essential for survival. Cell 93: 531–541. [DOI] [PubMed] [Google Scholar]
- Rhee HS, Pugh BF. 2011. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell 147: 1408–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robblee JP, Miura MT, Bain DL. 2012. Glucocorticoid receptor-promoter interactions: energetic dissection suggests a framework for the specificity of steroid receptor-mediated gene regulation. Biochemistry 51: 4463–4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose AJ, Herzig S. 2013. Metabolic control through glucocorticoid hormones: an update. Mol Cell Endocrinol 380: 65–78. [DOI] [PubMed] [Google Scholar]
- Rosen J, Miner JN. 2005. The search for safer glucocorticoid receptor ligands. Endocr Rev 26: 452–464. [DOI] [PubMed] [Google Scholar]
- Schiller BJ, Chodankar R, Watson LC, Stallcup MR, Yamamoto KR. 2014. Glucocorticoid receptor binds half sites as a monomer and regulates specific target genes. Genome Biol 15: 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweingruber N, Fischer HJ, Fischer L, van den Brandt J, Karabinskaya A, Labi V, Villunger A, Kretzschmar B, Huppke P, Simons M, et al. 2014. Chemokine-mediated redirection of T cells constitutes a critical mechanism of glucocorticoid therapy in autoimmune CNS responses. Acta Neuropathol 127: 713–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedwick C. 2014. Wanted: a new model for glucocorticoid receptor transactivation and transrepression. PLoS Biol 12: e1001814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serandour AA, Brown GD, Cohen JD, Carroll JS. 2013. Development of an Illumina-based ChIP-exonuclease method provides insight into FoxA1-DNA binding properties. Genome Biol 14: R147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman MN, Mukhopadhyay P, Belyavskaya E, Tonelli LH, Revenis BD, Doran JH, Ballard BE, Tam J, Pacher P, Sternberg EM. 2013. Glucocorticoid receptor dimerization is required for proper recovery of LPS-induced inflammation, sickness behavior and metabolism in mice. Mol Psychiatry 18: 1006–1017. [DOI] [PubMed] [Google Scholar]
- Steger DJ, Grant GR, Schupp M, Tomaru T, Lefterova MI, Schug J, Manduchi E, Stoeckert CJ Jr, Lazar MA. 2010. Propagation of adipogenic signals through an epigenomic transition state. Genes Dev 24: 1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M, Ganti KP, Mukherji A, Ye T, Hua G, Metzger D, Li M, Chambon P. 2011. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell 145: 224–241. [DOI] [PubMed] [Google Scholar]
- Szutorisz H, Dillon N, Tora L. 2005. The role of enhancers as centres for general transcription factor recruitment. Trends Biochem Sci 30: 593–599. [DOI] [PubMed] [Google Scholar]
- Tronche F, Opherk C, Moriggl R, Kellendonk C, Reimann A, Schwake L, Reichardt HM, Stangl K, Gau D, Hoeflich A, et al. 2004. Glucocorticoid receptor function in hepatocytes is essential to promote postnatal body growth. Genes Dev 18: 492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenhaut NH, Barish GD, Yu RT, Downes M, Karunasiri M, Liddle C, Schwalie P, Hübner N, Evans RM. 2013. Insights into negative regulation by the glucocorticoid receptor from genome-wide profiling of inflammatory cistromes. Mol Cell 49: 158–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandevyver S, Dejager L, Van Bogaert T, Kleyman A, Liu Y, Tuckermann J, Libert C. 2012. Glucocorticoid receptor dimerization induces MKP1 to protect against TNF-induced inflammation. J Clin Invest 122: 2130–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss TC, Schiltz RL, Sung M-H, Yen PM, Stamatoyannopoulos JA, Biddie SC, Johnson TA, Miranda TB, John S, Hager GL. 2011. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell 146: 544–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Chen J, Wang C, Uusküla-Reimand L, Chen K, Medina-Rivera A, Young EJ, Zimmermann MT, Yan H, Sun Z, et al. 2014. MACE: model based analysis of ChIP-exo. Nucleic Acids Res 42: e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson LC, Kuchenbecker KM, Schiller BJ, Gross JD, Pufall MA, Yamamoto KR. 2013. The glucocorticoid receptor dimer interface allosterically transmits sequence-specific DNA signals. Nat Struct Mol Biol 20: 876–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.