Abstract
Transcription factors are abundant Sumo targets, yet the global distribution of Sumo along the chromatin and its physiological relevance in transcription are poorly understood. Using Saccharomyces cerevisiae, we determined the genome-wide localization of Sumo along the chromatin. We discovered that Sumo-enriched genes are almost exclusively involved in translation, such as tRNA genes and ribosomal protein genes (RPGs). Genome-wide expression analysis showed that Sumo positively regulates their transcription. We also discovered that the Sumo consensus motif at RPG promoters is identical to the DNA binding motif of the transcription factor Rap1. We demonstrate that Rap1 is a molecular target of Sumo and that sumoylation of Rap1 is important for cell viability. Furthermore, Rap1 sumoylation promotes recruitment of the basal transcription machinery, and sumoylation of Rap1 cooperates with the target of rapamycin kinase complex 1 (TORC1) pathway to promote RPG transcription. Strikingly, our data reveal that sumoylation of Rap1 functions in a homeostatic feedback loop that sustains RPG transcription during translational stress. Taken together, Sumo regulates the cellular translational capacity by promoting transcription of tRNA genes and RPGs.
Nutrient availability is a major challenge to cellular homeostasis. Changes in nutrient availability activate signal transduction pathways that rewire cell metabolism and mobilize new energy sources (De Virgilio and Loewith 2006). The rapamycin-sensitive target of rapamycin complex 1 (TORC1) is a kinase complex that coordinates the cellular nutrient response (Loewith et al. 2002). Under nutrient-rich conditions, TORC1 inhibits catabolism while promoting anabolic processes like protein synthesis (De Virgilio and Loewith 2006).
The rate of protein synthesis is dependent on the rate of ribosome synthesis, which requires transcription of ribosomal protein genes (RPGs) (De Virgilio and Loewith 2006). RPGs are among the most intensively transcribed genes; 50% of all RNA polymerase II (RNAPII) initiation events occur at the 138 RPG promoters in the yeast genome (Warner 1999). The promoter-bound transcription factor Rap1 plays a key role in transcription of nearly all RPGs. Rap1 is required for recruitment of Ifh1, Fhl1, Hmo1, Sfp1, and TFIID, which promote recruitment of RNAPII (Schawalder et al. 2004; Wade et al. 2004; Garbett et al. 2007; Knight et al. 2014). Additionally, Rap1 mediates transcription of about 180 non-RPGs, including glycolytic genes (Lieb et al. 2001), and it binds telomeric regions to regulate telomere length and to maintain a repressed chromatin state (Shore 1997).
RPG transcription is strongly dependent on nutrient availability, and TORC1 activates RPG transcription by promoting phosphorylation of Sfp1 and Ifh1, leading to their recruitment to RPG promoters (Schawalder et al. 2004; Wade et al. 2004; Cai et al. 2013). Inhibiting TORC1 with rapamycin results in rapid dephosphorylation and release from the promoter of Sfp1 and Ifh1, thereby inhibiting transcription of RPGs. In contrast, localization of Rap1 to RPG promoters is not affected by TORC1 (Wade et al. 2004). Despite the fact that localization of Rap1 to chromatin is dynamic (Lickwar et al. 2012), it is currently not known whether the cell regulates the transcriptional activity of Rap1 (Schawalder et al. 2004).
Nutrient starvation elicits a stress response in yeast (Simpson and Ashe 2012), and stress induces sumoylation of various proteins (Zhou et al. 2004; Tempe et al. 2008; Jentsch and Psakhye 2013). Proteomic studies have found that transcription factors (TFs) are major Sumo substrates, although the functional consequences of most sumoylation events remain unknown (Wohlschlegel et al. 2004; Hannich et al. 2005). Current consensus is that sumoylation of TFs generally represses transcription (Gill 2005). For instance, sumoylation of the repressor complex Tup1-Ssn6 inhibits transcription (Texari et al. 2013). Furthermore, sumoylation of histones inhibits transcription, although the physiological relevance remains unknown (Nathan et al. 2006). Recent studies indicate that Sumo can also have a positive role (Rosonina et al. 2010). Surprisingly, a comprehensive analysis of the genome-wide association of Sumo with chromatin has not yet been performed in S. cerevisiae, and the physiological relevance of sumoylation of the vast majority of TFs remains unclear.
In this study, we analyze the genome-wide localization of Sumo on chromatin and describe a molecular mechanism by which Sumo promotes transcription of RPGs.
Results
Genome-wide analysis of Sumo reveals that Sumo localizes to genes involved in translation
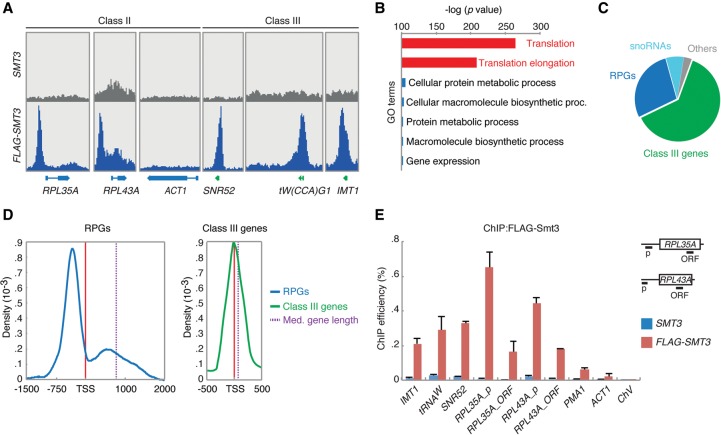
To better understand the function of Sumo in transcription, we performed Sumo ChIP-seq experiments with a strain expressing FLAG-tagged SMT3 (the yeast gene encoding Sumo), using untagged cells as a negative control (Supplemental Fig. S1A). We identified 670 significant Sumo peaks corresponding to 631 unique ORFs (Fig. 1A; Supplemental Table S1, tab1), including several genes previously found to be enriched for Sumo, such as CDC19, ADH1, PMA1, and the GAL1/10 promoter, but not ACT1 (Fig. 1A; Supplemental Fig. S1B; Rosonina et al. 2010; Texari et al. 2013). GO analysis showed strong overrepresentation of genes involved in mRNA translation (Fig. 1B; Supplemental Table S2). However, visual inspection of this data set revealed that the peak calling software had inaccurately assigned several Sumo peaks to dubious ORFs that were in close proximity to genuine ORFs (Supplemental Fig. S1C). Using more stringent filters with a higher peak-calling cutoff score, combined with visual inspection of all the Sumo peaks (see Methods), we exchanged these dubious ORFs with the genuine Sumo-containing ORFs. Although this resulted in removal of several genes with low levels of Sumo, including CDC19, PMA1, and ADH1 (Supplemental Table S1, tab2), we decided to focus on this stringent data set for the rest of our study, accepting that higher confidence comes at the cost of decreased sensitivity. This high-confidence Sumo data set contained 423 Sumo peaks at 395 unique genes, and consisted of 246 RNAPIII-transcribed tRNA genes, 110 RPGs, 12 protein-coding genes, and 27 noncoding RNAs, like snoRNAs (Fig. 1C; Supplemental Table S1). Together, these data show that in log-phase cells, RNAPIII-transcribed genes and RPGs are major targets of Sumo.
Figure 1.
Sumo is enriched at class III genes and RPGs. (A) Snapshots of the Sumo ChIP-seq experiment with FLAG-tagged Sumo (FLAG-SMT3) and untagged control cells (SMT3). (B,C) GO analysis of the ChIP-seq data set reveals highly significant overrepresentation of genes involved in translation, such as RPGs and class III genes. (D) Metagene analysis of Sumo distribution patterns at RPGs and class III genes. (E) Validation of ChIP-seq data by conventional ChIP-qPCR. Error bars indicate SEM of three independent experiments.
Metagene analysis of Sumo localization patterns revealed that RPGs had a strong Sumo peak several hundred base pairs upstream of the transcription start site (TSS), followed by a significant amount of Sumo toward the 5′ end of the ORF, whereas the RNAPIII-transcribed tRNA genes had a single, sharp Sumo peak right at the TSS (Fig. 1D).
The genome-wide results were confirmed by targeted ChIP-qPCR on selected genes (Fig. 1E). We also performed these experiments in the temperature-sensitive ubc9-1 mutant, which expresses a destabilized form of the E2 conjugase Ubc9, resulting in a strong reduction in cellular protein sumoylation even at a permissive temperature of 30°C (Supplemental Fig. S1D; Betting and Seufert 1996; Rosonina et al. 2010). We carried out these experiments at 30°C, because the restrictive temperature of 37°C causes a stress response accompanied with increased global sumoylation (Tempe et al. 2008), which could confound our results. Importantly, Sumo levels at the promoter of RPL35A and at tDNAW (which encodes tRNA-tryptophan, tRNAW) were dramatically reduced in this mutant (Supplemental Fig. S1E), and this reduction was fully rescued by a plasmid containing WT UBC9 (Supplemental Fig. S1F), demonstrating that our data are specific and not artifacts caused by “hyper-ChIPable” chromatin (Teytelman et al. 2013).
In conclusion, these results show that Sumo primarily localizes to class III genes and RPGs.
TORC1 and Sumo pathways cooperate to promote transcription of RPG and tRNA genes
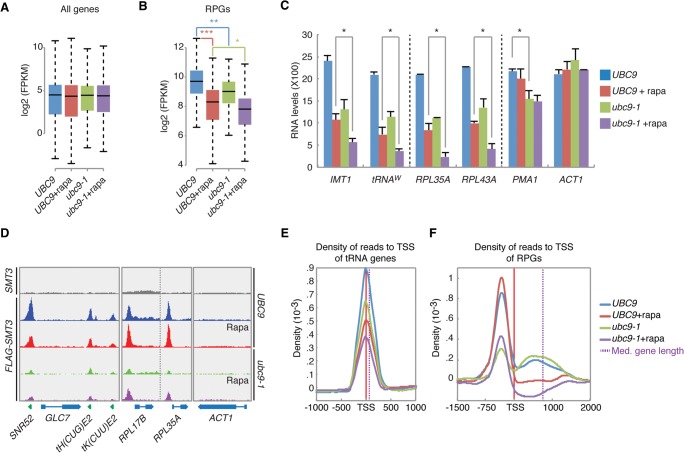
To test whether Sumo is important for transcription, we performed RNA-seq experiments using wild-type (WT) and ubc9-1 cells (Supplemental Table S3). Globally, the relative expression levels of the transcriptome were unaffected in the ubc9-1 mutant (Fig. 2A). In sharp contrast, expression of almost all RPGs was reduced in ubc9-1 mutants (Fig. 2B; Supplemental Fig. S2A). These data show that Ubc9 positively regulates the expression of RPGs.
Figure 2.
Ubc9 and TORC1 cooperate in regulation of transcription. (A) The global transcriptome is not affected by either Ubc9 or TORC1. WT cells and ubc9-1 mutants were treated for 30 min with 100 nM rapamycin (rapa) or with DMSO before RNA-seq. Potential differences between groups were tested with a two-sided Mann-Whitney U test. (B) Transcription of RPGs is regulated by both TORC1 and Ubc9. Cells were treated and RNA analyzed as in A. Differences between groups were tested as in A. (*) P = 0.008192; (**) P = 7.182 × 10−6; (***) P = 1.691 × 10−15. (C) Targeted qPCR experiment. Error bars, SEM of three independent experiments. (*) P < 0.01. (D) Snapshots of Sumo ChIP-seq in WT cells and ubc9-1 mutants treated with DMSO or rapamycin. (E,F) Metagene analysis of Sumo patterns at tDNA (E) and RPGs (F).
tRNA genes and RPGs are regulated by TORC1 (Lempiainen and Shore 2009; Wei and Zheng 2010); therefore, we hypothesized that Ubc9 cooperates with TORC1 to regulate their transcription. We performed whole-transcriptome analysis of WT and ubc9-1 cells after treatment with the TORC1-specific inhibitor rapamycin. No effect was observed on global transcription (Fig. 2A; Supplemental Fig. S2B). As expected, transcription of RPGs was specifically inhibited by rapamycin; however, RPG transcription was significantly further reduced in rapamycin-treated ubc9-1 mutants (Fig. 2B). The RNA-seq data were validated by targeted qPCR experiments, which also included the tRNAs IMT1, and tRNAW (Fig. 2C). These experiments confirm our ChIP-seq data and show that TORC1 and Ubc9 have additive effects on transcription of tDNA and RPGs. In contrast, rapamycin did not affect PMA1 expression, and the ubc9-1 mutation had only a relatively minor effect on expression of PMA1, which is consistent with our finding that Sumo levels are relatively low at this gene (Supplemental Fig. S1B). Furthermore, expression of ACT1, which is neither controlled by TORC1 nor enriched for Sumo (Fig. 1A), was completely unaffected by rapamycin and the ubc9-1 mutation (Fig. 2C). These data demonstrate that TORC1 and Ubc9 cooperate specifically in transcription of tRNA genes and RPGs.
To determine whether TORC1 affects sumoylation at chromatin, we performed a Sumo ChIP-seq experiment in WT and ubc9-1 cells treated with rapamycin. Because TORC1 is known to promote transcription of RPGs and tRNA genes, and because we found that Sumo also positively affects transcription of these genes, we expected that rapamycin treatment would either have no effect or that it would lead to decreased Sumo levels. Indeed, rapamycin caused decreased sumoylation at tRNA genes in WT cells and a further decrease in ubc9-1 mutants (Fig. 2D,E). However, contrary to our expectations, rapamycin treatment resulted in increased Sumo levels at the promoter of RPGs but decreased sumoylation in the body of the genes (Fig. 2D,F). This rapamycin-dependent pattern was similar in ubc9-1 mutants, although the overall level of Sumo was reduced. These ChIP-seq data were validated by targeted ChIP-qPCR, confirming that TORC1 differentially affects the sumoylation patterns of RPGs and tRNA genes (Supplemental Fig. S2C).
Finally, we studied the dynamics of Sumo levels at tRNA genes and RPGs during rapamycin treatment and found that Sumo levels at tDNAW rapidly decreased upon rapamycin treatment, which resulted in a reduction of tRNAW levels (Supplemental Fig. S2D). In contrast, Sumo levels at the promoter region of RPL35A progressively increased, whereas RPL35A expression levels were negatively affected by rapamycin (Supplemental Fig. S2E).
Taken together, these data reveal a dynamic function for TORC1 and Ubc9-Sumo in transcription of RPGs and tRNA genes. The data also reveal that although both TORC1 and Sumo are important for expression of RPGs (Fig. 2B), inhibition of TORC1 results in a paradoxical increase in Sumo levels at RPG promoters (Fig. 2F; Discussion).
Sumoylation at RPGs occurs at Rap1 binding sites
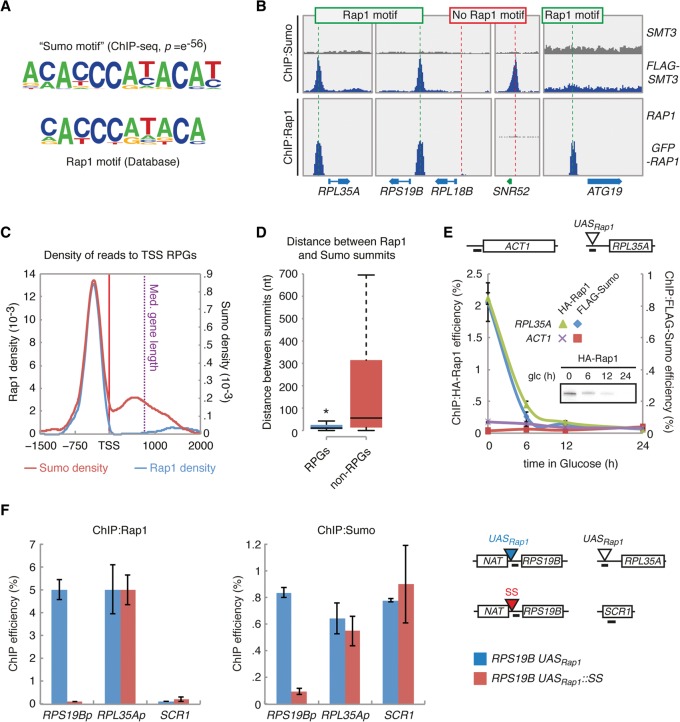
To gain insight into the mechanism by which Sumo regulates transcription, we determined whether any particular DNA sequence motif occurred within peaks of Sumo enrichment using our Sumo-ChIP-seq data set (Fig. 3A, top). When we compared this sequence to known TF consensus motifs, we found that it corresponded to the consensus sequence of the transcription factor Rap1 (Fig. 3A, bottom). Interestingly, several reports have indicated that Rap1 may be a Sumo target, although its relevance for transcription was never explored (Wohlschlegel et al. 2004; Denison et al. 2005; Hang et al. 2011; Lickwar et al. 2012; Albuquerque et al. 2013; Lescasse et al. 2013). Encouraged by these findings, we decided to focus on sumoylation of Rap1 at RPG promoters.
Figure 3.
Sumoylation at RPG promoters requires the presence of Rap1. (A) The Sumo consensus motif is highly similar to the Rap1 DNA binding motif. (B) Snapshots of the Rap1 and Sumo ChIP-seq experiments. (C) Perfect overlap between Sumo peaks and Rap1 peaks at RPG promoters. (D) Box plot showing the nucleotide (nt) distance between Sumo and Rap1 peaks at RPGs and non-RPGs. Median values are 11 and 55.5 nt for RPGs and non-RPGs, respectively (Mann-Whitney U test; W = 3823, P = 3.837 × 10−5). (E) Recruitment of Sumo depends on Rap1. Cells expressing Rap1 from the GAL1 promoter were grown to log phase in the presence of galactose, washed, and incubated in glucose. Levels of endogenous HA-Rap1 and FLAG-Sumo at the RPL35A promoter were analyzed by ChIP. Primer pair locations are indicated above the graph. (Inset) Level of Rap1 by Western blotting. Error bars, SEM. (F) The Rap1 binding site at the RPS19B promoter was either left intact (RPS19B_UASRAP1) or replaced with scrambled sequence (RPS19B_UASRAP1SS; see Supplemental Methods for construction details). Insertion of scrambled sequence resulted in loss of Rap1 at RPS19B (left panel) and prevented recruitment of Sumo (right panel). Error bars, SEM.
First, we performed a Rap1 ChIP-seq experiment (Supplemental Table S4) and found excellent correlation with a previous study that mapped Rap1 (overlap of 424 out of 429 genes) (Lickwar et al. 2012). The Rap1 binding sites were then aligned with the genomic sites enriched for Sumo. Strikingly, 95% of RPGs showed perfect colocalization of promoter-bound Rap1 and Sumo (Fig. 3B,C). A small number of RPGs does not contain a Rap1 consensus motif (Lieb et al. 2001). Our ChIP-seq data showed that Sumo was undetectable at RPGs that did not recruit Rap1, including RPL18B (Fig. 3B; Supplemental Fig. S3A). The presence of Sumo at the Rap1 binding site was specific for RPGs, because non-RPGs that also recruit Rap1, like ATG19 (Lieb et al. 2001), either did not contain any Sumo (Fig. 3B) or they had a Sumo peak that did not align with the Rap1 peak (Fig. 3D; Supplemental Fig. S3B), such as SNR47 (Supplemental Fig. S3C). This indicates that other TFs must be sumoylated at these genes, which is the scope of future studies. Taken together, these data strongly suggest that Rap1 is a Sumo target at RPG promoters.
To demonstrate that Rap1 is essential for sumoylation at RPG promoters, we performed Rap1 depletion experiments. The endogenous RAP1 promoter was replaced with the GAL1 promoter, which is active in the presence of galactose but rapidly repressed when cells are transferred to glucose. Indeed, incubation in glucose resulted in depletion of Rap1 and its disappearance from the promoter of RPL35A (Fig. 3E), strongly decreasing RPL35A mRNA levels (Supplemental Fig. S3D). Strikingly, Sumo also disappeared from this region with kinetics indistinguishable from those of Rap1 (Fig. 3E), showing that Rap1 is required for sumoylation at RPG promoters. Indeed, mutating the Rap1 binding site at the promoter of RPS19B prevented the recruitment of Sumo (Fig. 3F), whereas recruitment of Sumo at RPL35A (with intact Rap1 binding sites) as well as SCR1 (an RNAPIII-transcribed gene which recruits Sumo independently of Rap1) was unaffected. Together, these data demonstrate that the presence of Sumo at RPG promoters specifically depends on the presence of Rap1.
Sumoylation of Rap1 is required for transcription of RPGs
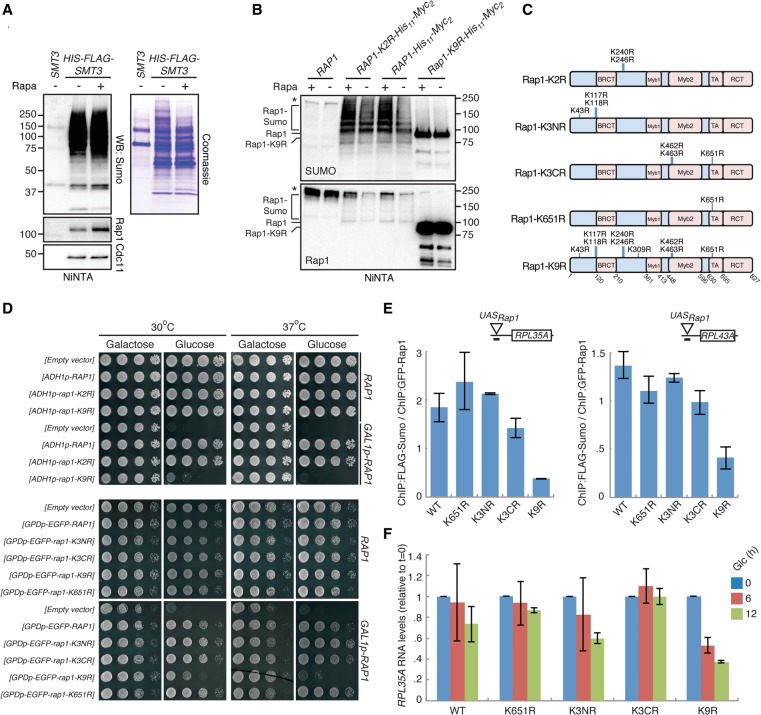
Next, we wanted to identify targets of Sumo with an unbiased experimental approach. Using HIS6-FLAG-tagged Sumo, we first purified Sumo from formaldehyde-fixed cells with FLAG antibodies (semi-native conditions) (Supplemental Fig. S3E) or from cells lysed with TCA using the HIS6 tag (denaturing conditions; see Methods). Analysis of the eluates by mass spectrometry (MS) showed that Rap1 was present in the Sumo-containing protein fractions (Supplemental Table S5). We also performed this analysis with cells treated with DMSO or rapamycin and found that the presence of Rap1 in the Sumo-containing protein fractions was more pronounced upon treatment with rapamycin (Supplemental Fig. S3F). To directly show that Rap1 is sumoylated, HIS6-tagged Sumo was purified under denaturing conditions followed by Western blotting with Rap1-specific antibodies. This confirmed that Rap1 is a Sumo target and that Rap1 sumoylation increases after rapamycin treatment (Fig. 4A), whereas total Rap1 levels remained unchanged (Supplemental Fig. S3G). As a control, rapamycin did not affect sumoylation of the septin Cdc11, a well-known Sumo target (Johnson and Blobel 1999). Similar data were obtained in a reciprocal experiment in which purified HIS11-Myc2-tagged Rap1 was analyzed by Western blotting with Sumo antibodies (Fig. 4B). These data demonstrate that Rap1 is a direct target of Sumo and that its sumoylation status increases upon inhibition of TORC1.
Figure 4.
Sumoylation of Rap1 is required for efficient RPG expression. (A) Sumoylation of Rap1 increases after rapamycin treatment. Cells were treated for 30 min with 100 nM rapamycin, and HIS6-FLAG-tagged Sumo was purified under denaturing conditions. Levels of Rap1 and Cdc11 in eluates were analyzed by Western blotting (left). Total protein levels in the eluates were visualized by Coomassie staining (right). (B) WT cells transformed with plasmids harboring HIS11-Myc2-tagged versions of RAP1, rap1-K2R, or rap1-K9R were treated with 100 nM rapamycin for 30 min. Rap1, Rap1-K2R, and Rap1-K9R were purified under denaturing conditions and visualized by Western blotting with Rap1 antibodies. (C) rap1 mutants used in this study. (D) Expression of rap1-K9R as the sole source of Rap1 results in severe growth defects. Cells expressing RAP1 from its endogenous promoter (“RAP1”) or from the GAL1/10 promoter (“GAL1_p-RAP1”) were transformed with an empty vector or with plasmids containing either RAP1, rap1-K2R, rap1-K9R, rap1-K3NR, rap1-K3CR, or rap1-K651R under control of the ADH1 or the GPD promoter, as indicated. Tenfold dilutions were spotted on plates containing either galactose or glucose and incubated for 2 d. (E,F) Rap1 sumoylation is important for transcription. FLAG-Sumo cells expressing endogenous RAP1 from the GAL1/10 promoter were transformed with plasmids harboring wild-type RAP1 (WT) or the rap1-K3NR, K3CR, K651R, or K9R alleles. Cells were grown to log phase in galactose, washed, and incubated in glucose for the indicated times, after which Sumo levels at the RPL35A and RPL43A promoters were analyzed by ChIP (E), and immature (i.e., unspliced) RNA levels of RPL35A were determined by qPCR (F). Error bars, SEM.
Recently, sumoylation of lysines K240 and K246 in Rap1 (“Rap1-K2R”) (see Fig. 4C for an overview of rap1 mutants used in this study) was shown to be important for telomere homeostasis (Lescasse et al. 2013). However, we found that mutation of these sites did not affect Rap1 sumoylation (Fig. 4B), indicating that other lysines must be sumoylated. Sumoylation-site prediction software (Zhao et al. 2014) predicted a total of nine sumoylation sites (K43, K117, K118, K240, K246, K309, K462, K463, and K651). We mutated these nine lysines to arginine (“rap1-K9R”) and found that many sumoylation events were almost completely prevented (note that this mutant migrates slightly faster than WT Rap1) (Fig. 4B). We also studied the growth rate of this mutant using the aforementioned Rap1 depletion strain, in which HA-tagged RAP1 is expressed from the GAL1 promoter. This depletion strain was transformed with a construct harboring various GFP-tagged rap1 mutants under control of the glucose-inducible GPD promoter. As shown in Figure 4D, only expression of the rap1-K9R mutant caused a growth defect at 30°C and inviability at 37°C. In an attempt to narrow down the sumoylation sites, we made three additional rap1 mutants (rap1-K651R, rap1-K(3N)R, and rap1-K(3C)R), but none of these mutants displayed a detectable growth defect (Fig. 4D). These data indicate that Rap1 is sumoylated on multiple lysines with partially redundant functions.
We studied the effect of these rap1 mutants on RPG expression using the Rap1 depletion system. Cells were grown to log phase in galactose and transferred to glucose-containing medium (Supplemental Fig. S3H), after which the levels of Rap1 and Sumo at the RPL35A and RPL43A promoters were analyzed (Fig. 4E; Supplemental Fig. S3I,J). Interestingly, although Rap1-K9R efficiently localized to RPG promoters (Supplemental Fig. S3J), Sumo levels were diminished and RPG expression was strongly reduced (Fig. 4E,F), whereas little or no effect on RPG expression was observed in rap1-K651R, rap1-K(3N)R, and rap1-K(3C)R mutants (Fig. 4F).
Taken together, these data show that Rap1 sumoylation is important for RPG expression and cell viability.
Sumoylation of Rap1 promotes recruitment of the basal transcription machinery
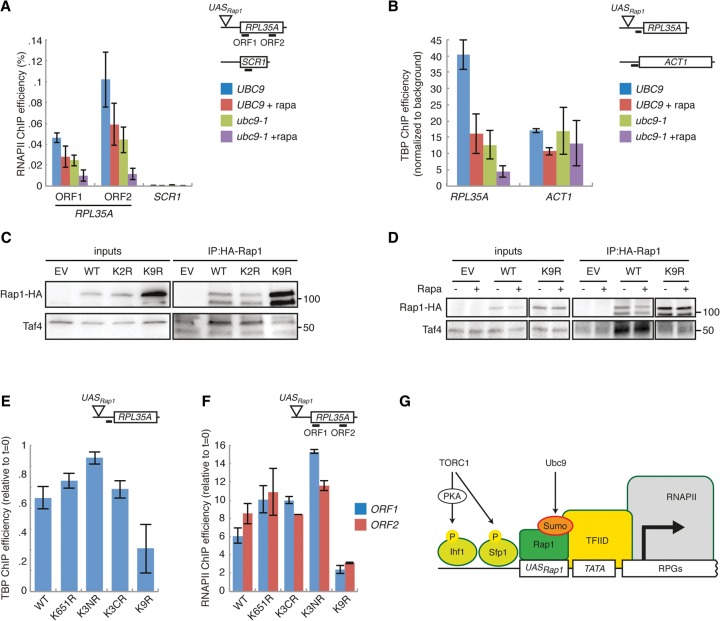
Because recruitment of RNAPII to RPGs depends on the interaction between Rap1 and TFIID (Garbett et al. 2007), we hypothesized that sumoylation of Rap1 is important for recruitment of TFIID and RNAPII. We performed an RNAPII ChIP-seq experiment and found that the levels of RNAPII at RPGs were indeed strongly reduced in ubc9-1 mutants and nearly undetectable after rapamycin treatment, whereas RNAPII levels at non-RPGs remained unchanged (Supplemental Fig. S4A). These data were confirmed by targeted ChIP-qPCR (Fig. 5A). Interestingly, recruitment of the TFIID component TBP to RPG promoters was also strongly reduced under these conditions, suggesting that Rap1 sumoylation mediates recruitment of TFIID (Fig. 5B). We tested this hypothesis directly by coimmunoprecipitation experiments. Although the TFIID subunit Taf4 efficiently coimmunoprecipitated with HA-tagged Rap1 in WT cells, almost no Taf4 coimmunoprecipitated with HA-Rap1-K9R (Fig. 5C). The interaction between sumoylated Rap1 and TFIID did not depend on TORC1 (Fig. 5D), which is consistent with our previous data indicating that Ubc9 and TORC1 act in parallel pathways. Finally, we analyzed recruitment of TFIID and RNAPII to RPGs in cells expressing nonsumoylatable Rap1-K9R as the sole source of Rap1. In accordance with the data obtained with the ubc9-1 mutant, recruitment of TFIID and RNAPII was strongly reduced in these cells (Fig. 5E,F). We conclude that sumoylation of Rap1 mediates efficient recruitment of the basal transcription machinery.
Figure 5.
Sumoylation of Rap1 mediates recruitment of the basal transcription machinery. (A) The level of RNAPII at RPL35A depends on the combined activity of TORC1 and Ubc9. Log-phase cells were treated with 100 nM rapamycin for 30 min, after which the level of RNAPII at RPL35A and SCR1 was analyzed by ChIP. The location of primer pairs is indicated. Error bars, SEM. (B) TFIID recruitment depends on TORC1 and Ubc9. Log-phase cells were treated as in A, and the level of TBP at RPL35A and ACT1 was determined by ChIP. Error bars, SEM. (C) Rap1 sumoylation promotes the interaction with TFIID. WT cells transformed with plasmids expressing HA-tagged wild-type RAP1, rap1-K2R, or rap1-K9R were subjected to immunoprecipitation with anti-HA beads followed by Western blotting with anti-Taf4 antibodies. (D) TORC1 activity does not modulate the interaction between Rap1 and TFIID. Cells treated with 100 nM rapamycin (30 min) were processed as in C. (E,F) Sumoylation of Rap1 promotes recruitment of TFIID and RNAPII to RPL35A. ChIPs were performed as in Figure 4E using antibodies against TBP (E) and RNAPII (F). Error bars, SEM. (G) Model of our findings. Sumoylation of Rap1 by Ubc9 promotes RPG transcription in parallel to TORC1.
Discussion
TFs are among the most abundant targets of Sumo, yet the global distribution of Sumo and physiological relevance of TF sumoylation are poorly understood. Here, we show that Sumo mainly localizes to pro-growth genes like tRNA genes and RPGs. The distribution of Sumo strongly resembles the genomic distribution of Sumo in mammalian cells (Neyret-Kahn et al. 2013). There have been conflicting reports as to how Sumo affects transcription at these genes in mammals; one study reported that Sumo promotes their transcription (Liu et al. 2012), whereas another study found that Sumo is inhibitory (Neyret-Kahn et al. 2013). The reason for these differences is unclear, but could be technical (Neyret-Kahn et al. 2013). These two studies did not identify the Sumo target at these genes, making it difficult to interpret their different findings. In contrast, we found that in S. cerevisiae, Sumo has a clear stimulatory effect on transcription of RPGs, and we unraveled the molecular mechanism.
We discovered that sumoylation of Rap1 is required for efficient transcription of RPGs. Sumoylation of K240/246 of Rap1 was recently shown to be important for telomere homeostasis (Lescasse et al. 2013). However, we found that preventing sumoylation of these two lysine residues, as well as various combinations of other N-terminal and C-terminal residues, had no effect on Rap1 sumoylation, RPG transcription, and cell viability. Only in the rap1-K9R mutant was sumoylation almost completely abolished, indicating that Rap1 is sumoylated on multiple lysines with (partially) redundant functions, which is commonly observed for Sumo and ubiquitin substrates (Fischer et al. 2011; Psakhye and Jentsch 2012). Furthermore, Rap1-K9R protein levels appeared to be increased, which could be the result of decreased protein turnover; Sumo and ubiquitin can target the same lysine residues in their substrates (Ulrich 2005), and therefore it is possible that one or several of these lysines may be involved in ubiquitin-mediated proteasomal degradation of Rap1. This is the topic of future studies.
Although Rap1 is a major Sumo target at RPG promoters, we cannot exclude the possibility that other TFs are also sumoylated. For instance, high-throughput MS studies have found that Hmo1 and Ifh1 may be Sumo targets (Wohlschlegel et al. 2004; Zhou et al. 2004). Furthermore, in addition to promoter regions, we also detected Sumo in the body of RPGs, where it likely contributes to transcription; several RPGs that contained neither Rap1 nor Sumo in their promoters did contain Sumo toward the 5′ end of the ORF, and their expression was reduced in the ubc9-1 mutant. Here, the relevant Sumo target still needs to be identified. A good candidate is RNAPII, which was previously shown to be sumoylated; although the exact physiological significance remains unclear (Wohlschlegel et al. 2004; Zhou et al. 2004; Chen et al. 2009). Histones may also be Sumo substrates (Nathan et al. 2006). However, histone sumoylation has been associated with inhibition of transcription (Nathan et al. 2006), whereas we found that Sumo has a positive function at the majority of its target genes (Fig. 2B; Supplemental Table S3; Supplemental Fig. S2A), making it unlikely that histones are Sumo targets at these genes.
How Sumo regulates tRNA genes remains unclear. We and others have found that various components of RNAPIII, as well as mammalian Maf1 (an RNAPIII inhibitor), are Sumo substrates (Supplemental Table S5; Wohlschlegel et al. 2004; Rohira et al. 2013).
Whereas Sumo clearly activates RPGs and tDNA genes, we also found that Sumo inhibits transcription of a number of genes (Supplemental Table S3). Although this class of genes was not the focus of our study, it is worth mentioning that at least some of these genes contain binding sites for the Tup1-Ssn6 repressor complex, such as the GAL1 promoter; and sumoylation of Ssn6 was recently shown to be important for transcriptional repression of GAL1 (Texari et al. 2013). The negative effect of Sumo on transcription is the topic of future studies. Furthermore, Sumo also plays a role in chromosome duplication and DNA repair (Tempe et al. 2008), but our ChIP-seq experiments likely did not identify these events because we used unsynchronized, log-phase cells, causing the Sumo signal to disappear in the background.
Sumoylation of Rap1 stimulates its ability to recruit the basal transcription machinery. Exactly how Sumo promotes the interaction of Rap1 with TFIID is the subject of ongoing studies. Most Sumo-interacting proteins bind Sumo through low-affinity Sumo interaction motifs (SIMs) (Psakhye and Jentsch 2012). Potential SIMs are abundant in TFIID components, making the identification of functional SIMs a challenge.
In addition to recruiting TFIID, sumoylation of Rap1 may affect RPG expression through other mechanisms. For instance, Rap1 was recently shown to impact cytoplasmic decay of RPL30 mRNA (Bregman et al. 2011). Sumo levels are high at the Rap1 binding site of RPL30 (not shown). Thus, while sumoylation of Rap1 promotes RPG transcription through recruitment of TFIID, it is possible that it simultaneously restrains the expression levels of RPGs by inducing the cytoplasmic degradation of RPG mRNA.
Finally, we found that TORC1 and Ubc9 are both required for full transcription of RPGs (see Fig. 5G for a model). Paradoxically, inhibiting TORC1 resulted in increased Rap1 sumoylation at RPGs (Fig. 2F; Supplemental Fig. S2C,E), yet inhibiting Rap1 sumoylation strongly decreased RPG transcription (Figs. 2B, 4F). Why would the cell activate the Ubc9-Sumo-Rap1 pathway under conditions that inhibit TORC1 activity? We hypothesize that the Ubc9-Sumo pathway provides positive feedback on RPG transcription to maintain cellular homeostasis. A homeostatic feedback loop has previously been reported for TORC1 (Urban et al. 2007); TORC1 activity increases when protein synthesis is inhibited by cycloheximide (Loewith and Hall 2011). Inhibiting protein synthesis with cycloheximide triggers a potent stress response (Searle et al. 1975; Tempe et al. 2008); indeed, reducing protein translation by cycloheximide induced a global increase in protein sumoylation as well as increased sumoylation of Rap1 (Supplemental Fig. S4B,C; see Supplemental Fig. S4D for a model).
In conclusion, we found that Sumo preferentially localizes to genes involved in cell growth and proliferation, and that it is required for transcription of these genes. Furthermore, Rap1 is a major target of Sumo at RPGs, and the Ubc9-Sumo-Rap1 pathway cooperates with TORC1 to promote RPG transcription. Finally, sumoylation of Rap1 stimulates RPG transcription by recruiting the basal transcription machinery.
Methods
Yeast strains and plasmids
S. cerevisiae strains were grown in appropriate media, depending on the experiment/genotype. Strains were derived directly from either the S288c strains RDKY3615 (Chen and Kolodner 1999) or BY4741 using standard gene-replacement methods or intercrossing (see Supplemental Table S6 for strains and plasmids).
ChIP, ChIP-seq, and RNA-seq
ChIP and ChIP-seq (cells fixed with 1% [wt/vol] formaldehyde for 30 min) and RNA-seq were performed as described previously (Zimmermann et al. 2011; Chymkowitch et al. 2012) with minor modifications (see Supplemental Material).
Sequencing data analysis
For all ChIP-seq and RNA-seq analyses, we used the S. cerevisiae genome sequence and associated annotation (R64-1-1.75) downloaded from Ensembl (Flicek et al. 2014). ChIP-seq reads were mapped using Bowtie 2 version 2.2.3 using default parameters (Langmead and Salzberg 2012). Generated and mapped reads are shown in Supplemental Table S1. SAM files were sorted and indexed using SAMtools (Li et al. 2009). Visualization of read density was done in the integrative genome viewer (IGV) (Robinson et al. 2011).
MACS2 version 2.0.10 was used for peak calling (Zhang et al. 2008). For Rap1 samples, automatic detection of fragment length was used. This could not be obtained for Sumo; instead, a fragment length of 200 was used. We filtered peaks called by MACS2 using control samples with no tag to identify a cutoff at which few peaks were called in the control samples. For Rap1, a log10 P-value of 50 was used. Still, a few peaks remained in the control sample, which we filtered out unless they had a twice as high score in the ChIP-sample. BEDTools (Quinlan and Hall 2010) was used to identify overlapping peaks, and for all intersections, a 1-bp overlap requirement was used. For analysis of the Sumo ChIP-seq data, a log10 P-value of 6 was used, resulting in a data set of 631 genes (Supplemental Table S1). Peaks were then annotated according to genomic location and the closest overlapping gene using ChIPpeakAnno (Zhu et al. 2010) and biomaRt (Durinck et al. 2009). We used the middle of the peak to measure the distance to TSSs. Manual inspection of the resulting Sumo ChIP-seq data set revealed a significant number of ORFs with relatively low Sumo signals. These were absent when the data set was filtered with a more stringent log10 P-value of 20. However, this stringent data set still contained several ORFs (particularly dubious/hypothetical ORFs) that were in close proximity to either tRNA genes or promoter regions of ORFs with high Sumo levels, especially RPGs. In most of these cases, visual inspection revealed that the software had improperly assigned Sumo peaks to the wrong ORF, because the algorithm assigns the peak to the TSS of the ORF that is most closely positioned to the summit of the peak, which is incorrect in a minority of cases (e.g., YGR164W, YDR448W, YDR022C, YBR085W, YPR131C, and YMR196C-A) (see Supplemental Fig. S1C). These overlapping dubious/hypothetical ORFs were removed from the data set, whereas the genes that were originally not called by the software due to such wrongful assignment were added back to the data set [for the examples shown in Supplemental Fig. S1C, these were tR(UCU)G2, RPS17B, tV(UAC)D, RPL19A, RPS23B, and RPL36A]. This resulted in a high-confidence data set consisting of a total number of 395 genes that are highly enriched for Sumo (Supplemental Table S1).
Functional annotation of associated genes was performed in the functional annotation tool found at http://www.yeastgenome.org/. See Supplemental information for further details.
RNA preparation and reverse transcription
Total RNA purification and reverse transcription were performed as previously described (Chymkowitch et al. 2012).
Sumo pull-down under denaturing conditions
Pull-downs in denaturing conditions were performed as previously described (Sacher et al. 2005) with minor modifications (see Supplemental Experimental Procedures).
Cell lysis, immunoprecipitation, and Western blotting
IP and Western blotting was performed as previously described (Sacher et al. 2005; Kats et al. 2009; Chymkowitch et al. 2012). All buffers contained N-Ethylmaleimide to prevent desumoylation.
Data access
All high-throughput sequencing data have been submitted to the European Nucleotide Archive (ENA; http://www.ebi.ac.uk/ena). See Supplemental Table S7 for sample accession numbers. The ChIP-seq study accession number is PRJEB7579, and the study unique name is ena-STUDY-Oslo Genomics Core Facility-21-10-2014-13:44:44:043-43. The RNA-seq study accession number is PRJEB7579, and the study unique name is ena-STUDY-Oslo Genomics Core Facility-21-10-2014-13:44:44:043-43.
Supplementary Material
Acknowledgments
We gratefully acknowledge Drs. W. Seufert, H. Zhou, and S. Alberti for reagents; M. Bjørås and P. Collas for infrastructure and discussions; and Dr. J. Krijgsveld for mass spectrometry services. This project is supported by grants to J.M.E. from the Norwegian Research Council (221694) and the Norwegian Cancer Society (project number 3311782). P.C. is supported by a grant from the Norwegian Research Council (221920).
Author contributions: J.M.E. conceived the project. P.C., A.N.P., and J.M.E. designed and performed experiments. Mass spectrometry was performed by C.J.K. H.A. and S.L. performed computational analysis of genome-wide data sets. P.C. and J.M.E. wrote the paper with input from all other authors.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.185793.114.
Freely available online through the Genome Research Open Access option
References
- Albuquerque CP, Wang G, Lee NS, Kolodner RD, Putnam CD, Zhou H. 2013. Distinct SUMO ligases cooperate with Esc2 and Slx5 to suppress duplication-mediated genome rearrangements. PLoS Genet 9: e1003670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betting J, Seufert W. 1996. A yeast Ubc9 mutant protein with temperature-sensitive in vivo function is subject to conditional proteolysis by a ubiquitin- and proteasome-dependent pathway. J Biol Chem 271: 25790–25796. [DOI] [PubMed] [Google Scholar]
- Bregman A, Avraham-Kelbert M, Barkai O, Duek L, Guterman A, Choder M. 2011. Promoter elements regulate cytoplasmic mRNA decay. Cell 147: 1473–1483. [DOI] [PubMed] [Google Scholar]
- Cai L, McCormick MA, Kennedy BK, Tu BP. 2013. Integration of multiple nutrient cues and regulation of lifespan by ribosomal transcription factor Ifh1. Cell Rep 4: 1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Kolodner RD. 1999. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat Genet 23: 81–85. [DOI] [PubMed] [Google Scholar]
- Chen X, Ding B, LeJeune D, Ruggiero C, Li S. 2009. Rpb1 sumoylation in response to UV radiation or transcriptional impairment in yeast. PLoS One 4: e5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chymkowitch P, Eldholm V, Lorenz S, Zimmermann C, Lindvall JM, Bjørås M, Meza-Zepeda LA, Enserink JM. 2012. Cdc28 kinase activity regulates the basal transcription machinery at a subset of genes. Proc Natl Acad Sci 109: 10450–10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Virgilio C, Loewith R. 2006. Cell growth control: little eukaryotes make big contributions. Oncogene 25: 6392–6415. [DOI] [PubMed] [Google Scholar]
- Denison C, Rudner AD, Gerber SA, Bakalarski CE, Moazed D, Gygi SP. 2005. A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol Cell Proteomics 4: 246–254. [DOI] [PubMed] [Google Scholar]
- Durinck S, Bullard J, Spellman PT, Dudoit S. 2009. GenomeGraphs: integrated genomic data visualization with R. BMC Bioinformatics 10: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer ES, Scrima A, Böhm K, Matsumoto S, Lingaraju GM, Faty M, Yasuda T, Cavadini S, Wakasugi M, Hanaoka F, et al. 2011. The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell 147: 1024–1039. [DOI] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, et al. 2014. Ensembl 2014. Nucleic Acids Res 42: D749–D755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbett KA, Tripathi MK, Cencki B, Layer JH, Weil PA. 2007. Yeast TFIID serves as a coactivator for Rap1p by direct protein-protein interaction. Mol Cell Biol 27: 297–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G. 2005. Something about SUMO inhibits transcription. Curr Opin Genet Dev 15: 536–541. [DOI] [PubMed] [Google Scholar]
- Hang LE, Liu X, Cheung I, Yang Y, Zhao X. 2011. SUMOylation regulates telomere length homeostasis by targeting Cdc13. Nat Struct Mol Biol 18: 920–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannich JT, Lewis A, Kroetz MB, Li SJ, Heide H, Emili A, Hochstrasser M. 2005. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J Biol Chem 280: 4102–4110. [DOI] [PubMed] [Google Scholar]
- Jentsch S, Psakhye I. 2013. Control of nuclear activities by substrate-selective and protein-group SUMOylation. Annu Rev Genet 47: 167–186. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Blobel G. 1999. Cell cycle–regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J Cell Biol 147: 981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kats ES, Enserink JM, Martinez S, Kolodner RD. 2009. The Saccharomyces cerevisiae Rad6 post replication repair and Siz1/Srs2 homologous recombination-inhibiting pathways process DNA damage that arises in asf1 mutants. Mol Cell Biol 29: 5226–5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight B, Kubik S, Ghosh B, Bruzzone MJ, Geertz M, Martin V, Dénervaud N, Jacquet P, Ozkan B, Rougemont J, et al. 2014. Two distinct promoter architectures centered on dynamic nucleosomes control ribosomal protein gene transcription. Genes Dev 28: 1695–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempiainen H, Shore D. 2009. Growth control and ribosome biogenesis. Curr Opin Cell Biol 21: 855–863. [DOI] [PubMed] [Google Scholar]
- Lescasse R, Pobiega S, Callebaut I, Marcand S. 2013. End-joining inhibition at telomeres requires the translocase and polySUMO-dependent ubiquitin ligase Uls1. EMBO J 32: 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickwar CR, Mueller F, Hanlon SE, McNally JG, Lieb JD. 2012. Genome-wide protein–DNA binding dynamics suggest a molecular clutch for transcription factor function. Nature 484: 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb JD, Liu X, Botstein D, Brown PO. 2001. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet 28: 327–334. [DOI] [PubMed] [Google Scholar]
- Liu HW, Zhang J, Heine GF, Arora M, Gulcin Ozer H, Onti-Srinivasan R, Huang K, Parvin JD. 2012. Chromatin modification by SUMO-1 stimulates the promoters of translation machinery genes. Nucleic Acids Res 40: 10172–10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R, Hall MN. 2011. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189: 1177–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P, Hall MN. 2002. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell 10: 457–468. [DOI] [PubMed] [Google Scholar]
- Nathan D, Ingvarsdottir K, Sterner DE, Bylebyl GR, Dokmanovic M, Dorsey JA, Whelan KA, Krsmanovic M, Lane WS, Meluh PB, et al. 2006. Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications. Genes Dev 20: 966–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyret-Kahn H, Benhamed M, Ye T, Le Gras S, Cossec JC, Lapaquette P, Bischof O, Ouspenskaia M, Dasso M, Seeler J, et al. 2013. Sumoylation at chromatin governs coordinated repression of a transcriptional program essential for cell growth and proliferation. Genome Res 23: 1563–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psakhye I, Jentsch S. 2012. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 151: 807–820. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohira AD, Chen CY, Allen JR, Johnson DL. 2013. Covalent small ubiquitin-like modifier (SUMO) modification of Maf1 protein controls RNA polymerase III-dependent transcription repression. J Biol Chem 288: 19288–19295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosonina E, Duncan SM, Manley JL. 2010. SUMO functions in constitutive transcription and during activation of inducible genes in yeast. Genes Dev 24: 1242–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher M, Pfander B, Jentsch S. 2005. Identification of SUMO-protein conjugates. Methods Enzymol 399: 392–404. [DOI] [PubMed] [Google Scholar]
- Schawalder SB, Kabani M, Howald I, Choudhury U, Werner M, Shore D. 2004. Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature 432: 1058–1061. [DOI] [PubMed] [Google Scholar]
- Searle J, Lawson TA, Abbott PJ, Harmon B, Kerr JF. 1975. An electron-microscope study of the mode of cell death induced by cancer-chemotherapeutic agents in populations of proliferating normal and neoplastic cells. J Pathol 116: 129–138. [DOI] [PubMed] [Google Scholar]
- Shore D. 1997. Telomere length regulation: getting the measure of chromosome ends. Biol Chem 378: 591–597. [PubMed] [Google Scholar]
- Simpson CE, Ashe MP. 2012. Adaptation to stress in yeast: to translate or not? Biochem Soc Trans 40: 794–799. [DOI] [PubMed] [Google Scholar]
- Tempe D, Piechaczyk M, Bossis G. 2008. SUMO under stress. Biochem Soc Trans 36: 874–878. [DOI] [PubMed] [Google Scholar]
- Texari L, Dieppois G, Vinciguerra P, Contreras MP, Groner A, Letourneau A, Stutz F. 2013. The nuclear pore regulates GAL1 gene transcription by controlling the localization of the SUMO protease Ulp1. Mol Cell 51: 807–818. [DOI] [PubMed] [Google Scholar]
- Teytelman L, Thurtle DM, Rine J, van Oudenaarden A. 2013. Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc Natl Acad Sci 110: 18602–18607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich HD. 2005. Mutual interactions between the SUMO and ubiquitin systems: a plea of no contest. Trends Cell Biol 15: 525–532. [DOI] [PubMed] [Google Scholar]
- Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O, Wanke V, Anrather D, Ammerer G, Riezman H, et al. 2007. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell 26: 663–674. [DOI] [PubMed] [Google Scholar]
- Wade JT, Hall DB, Struhl K. 2004. The transcription factor Ifh1 is a key regulator of yeast ribosomal protein genes. Nature 432: 1054–1058. [DOI] [PubMed] [Google Scholar]
- Warner JR. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24: 437–440. [DOI] [PubMed] [Google Scholar]
- Wei Y, Zheng XS. 2010. Maf1 regulation: a model of signal transduction inside the nucleus. Nucleus 1: 162–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel JA, Johnson ES, Reed SI, Yates JR III. 2004. Global analysis of protein sumoylation in Saccharomyces cerevisiae. J Biol Chem 279: 45662–45668. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Xie Y, Zheng Y, Jiang S, Liu W, Mu W, Liu Z, Zhao Y, Xue Y, Ren J. 2014. GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res 42: W325–W330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Ryan JJ, Zhou H. 2004. Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J Biol Chem 279: 32262–32268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu LJ, Gazin C, Lawson ND, Pagès H, Lin SM, Lapointe DS, Green MR. 2010. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 11: 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann C, Chymkowitch P, Eldholm V, Putnam CD, Lindvall JM, Omerzu M, Bjørås M, Kolodner RD, Enserink JM. 2011. A chemical-genetic screen to unravel the genetic network of CDC28/CDK1 links ubiquitin and Rad6-Bre1 to cell cycle progression. Proc Natl Acad Sci 108: 18748–18753. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.