Abstract
Melanogenesis is the process that regulates skin and eye pigmentation. Albinism, a genetic disease causing pigmentation defects and visual disorders, is caused by mutations in genes controlling either melanin synthesis or melanosome biogenesis. Here we show that a common transcriptional control regulates both of these processes. We performed an analysis of the regulatory region of Oa1, the murine homolog of the gene that is mutated in the X-linked form of ocular albinism, as Oa1's function affects melanosome biogenesis. We demonstrated that Oa1 is a target of Mitf and that this regulatory mechanism is conserved in the human gene. Tissue-specific control of Oa1 transcription lies within a region of 617 bp that contains the E-box bound by Mitf. Finally, we took advantage of a virus-based system to assess tissue specificity in vivo. To this end, a small fragment of the Oa1 promoter was cloned in front of a reporter gene in an adeno-associated virus. After we injected this virus into the subretinal space, we observed reporter gene expression specifically in the retinal pigment epithelium, confirming the cell-specific expression of the Oa1 promoter in the eye. The results obtained with this viral system are a preamble to the development of new gene delivery approaches for the treatment of retinal pigment epithelium defects.
Albinism is caused by a defect in the pigmentation of the skin and the eye. Some forms of oculocutaneous albinism (OCA) can result from impaired melanin synthesis (5). Other forms of albinism, such as ocular albinism type 1 (OA1), are caused by the defective biogenesis of melanosomes, the organelles in which melanin is synthesized. In the skin of OA1 patients, melanin is in fact synthesized and accumulates in melanosomes, but the organelles are much larger than normal and are therefore called macromelanosomes (27, 37). The gene that is mutated in patients affected by OA1 was identified previously (7), and the protein was characterized as an integral melanosomal membrane protein (29). The function of the OA1 protein is still not well understood; OA1 was reported to bind G proteins on the cytoplasmic side of the melanosome (30), but its function as a G-protein-coupled receptor is still unclear. Histological analyses of skin melanocytes and retinal pigment epithelium (RPE) from patients suggest that OA1 has a function in melanosome formation, mainly because the melanosomes have an abnormal size (27, 37). In a mouse model of the disease, mice also develop giant melanosomes, and the study of the murine phenotype permitted a definition of macromelanosomes as being derived from single melanosomes that do not stop growing (17). Thus, when the Oa1 gene is mutated in both humans and mice, melanosome biogenesis is abnormal. The murine homologue of OA1 was cloned (6, 26) and the pattern of expression was studied at several developmental stages and in the adult (32). Oa1 starts to be expressed in the outer layer of the eye cup at embryonic day 10.5 (E10.5), and transcription in the RPE can be detected at all stages until adulthood. Oa1 mRNA was also found in skin melanocytes. The same study revealed that the expression is tissue specific and restricted to pigment cells. Similar to that of genes involved in different forms of OCA, Oa1 expression starts before the deposition of melanin. The transcriptional regulation of the genes encoding the enzymes synthesizing melanin, such as tyrosinase (TYR) and tyrosinase-related proteins 1 and 2 (TRP1 and TRP2), was studied, and some common regulators were identified. We therefore asked whether the same transcriptional regulators also control the expression of genes involved in melanosome biogenesis, such as OA1.
The basic helix-loop-helix-leucine zipper (bHLH-LZ) factor microphthalmia transcription factor (Mitf) plays a crucial role in the development and differentiation of pigment cells in both the skin and the eye (36). More than 20 different Mitf mutations have been described for mice. They all result in a deficiency in skin and ear melanocytes and in defects in eye size and pigmentation (24). Mutations of the MITF gene in humans are associated with Waardenburg syndrome type 2 (33, 34) and albinism-deafness (Tietz) syndrome (2). MITF has been shown to activate the expression of melanocyte-specific genes such as TYR and TRP1 through an evolutionarily conserved 11-bp sequence termed the M-box (22). The M-box contains a core CATGTG E-box motif recognized by the members of the bHLH-LZ family of transcription factors. A differential control of TRP2 regulation by MITF was defined for skin melanocytes and RPE (31). Otherwise, an indirect transcriptional control by MITF was shown for MATP (encoding membrane-associated transporter protein; also called AIM1) expression (12). We were interested in the identification of new MITF target genes for multiple reasons: (i) to better understand how the mechanism of pigmentation is regulated in melanocytes and RPE and (ii) to identify new melanocytic markers that may play a role in melanoma survival and prognosis (36).
Here we present an analysis of the murine Oa1 transcription regulatory region, its evolutionary conservation in the human gene, and a demonstration that Oa1 is an MITF target gene. Analyses of the transactivating activity in several constructs containing different segments of the Oa1 5′ region identified positive elements that regulate the tissue-specific expression of this gene. We show that an E-box bound by Mitf is a key element for the tissue-specific transcription of Oa1. This regulatory mechanism is also conserved during evolution. Finally, we took advantage of an adeno-associated virus (AAV) vector containing the bp −562 to +55 Oa1 genomic region upstream of a reporter gene to evaluate the ability of this sequence to drive tissue-specific expression in the eye. Injection of the AAV into the subretinal space demonstrated that the −562- to +55-bp element is sufficient to confer tissue specificity in vivo.
MATERIALS AND METHODS
Reagents.
Hydrocortisone, human transferrin, putrescine, l-glutamine, bovine insulin, triiodothyronine, cholera toxin, phenylmethylsulfonyl fluoride, aprotinin, pepstatin, and leupeptin were purchased from Sigma-Aldrich. RPMI medium, Dulbecco's modified Eagle's medium (DMEM), Aim-V medium, fetal bovine serum (FBS), trypsin-EDTA, nonessential amino acids, and minimum essential medium (MEM) vitamins were purchased from Invitrogen Life Technologies. The FuGENE6 reagent was purchased from Roche. Synthetic oligonucleotides were purchased from MWG AG BIOTECH.
Cell cultures.
B16-F10 murine melanoma cells (10) and NIH 3T3 fibroblast cells were grown at 37°C in 5% CO2 in DMEM supplemented with 10% FBS, penicillin (100 U/ml), and streptomycin (50 μg/ml). The B16-F10 medium was supplemented with 1 mM sodium pyruvate, nonessential amino acids, and MEM vitamins.
ARPE-19 human RPE cells were purchased from the American Type Culture Collection (Manassas, Va.) and grown in DMEM-F-12 supplemented with 10% FBS, 2.8 × 10−8 M hydrocortisone, 3 × 10−7 M bovine insulin, 6.3 × 10−8 M human transferrin, 2.4 × 10−6 M putrescine, 2 × 10−3 M l-glutamine, and 200 pM cholera toxin at 37°C in 10% CO2.
MNT-1 pigmented human melanoma cells were grown in DMEM and 10% Aim-V medium supplemented with 20% FBS, nonessential amino acids, and 1 mM sodium pyruvate.
RNA isolation and reverse transcription-PCR (RT-PCR).
RPEs were dissected from wild-type and Mitfmi-enu198 (14) adult mice, and the total RNAs were purified by the use of TRIzol reagent (Gibco-BRL) according to the manufacturer's instructions. Five micrograms of total RNA was reverse transcribed by using random hexamer primers and Superscript II reverse transcriptase (Gibco-BRL). cDNAs from wild-type and Mitfmi-enu198 mice were PCR amplified by using either Oa1-, Tyr-, Trp1-, or Trp2-specific primers, as follows: for Oa1, OA1forward, 5′-GTGTGAGAGGGGCCTGGACCA-3′, and OA1reverse, 5′-ATAAACCATGTGGTCCTAGCT-3′; for Tyr, TYRforward, 5′-ATTGATTTTGCCCATGAAGC-3′, and TYRreverse, 5′-CCCAGATCCTTGGATGTTATG-3′; for Trp1, TRP1forward, 5′-ACTGACCCTTGTGGCTCATC-3′, and TRP1reverse, 5′-GAGAAATCCACATCCCCAAA-3′; and for Trp2, TRP2forward, 5′-CATCTGTGGATTTCTAGAGG-3′, and TRP2reverse, 5′-AGGATGGCCGGCTTCTTC-3′.
In order to normalize the cDNA preparations, we amplified the same samples with the following specific primers for glyceraldehyde-3-phosphate dehydrogenase (Gapdh): GAPDHforward, 5′-ACAGTCCATGCCATCACTGCC-3′, and GAPDHreverse, 5′-GCCTGCTTCACCACCTTCTTG-3′. Amplification was performed by using Taq Gold polymerase (Perkin Elmer, Norwalk, Conn.) for 35 cycles of 94°C for 30 s, 55 or 58°C for 60 s, and 72°C for 60 s.
Plasmids and DNA constructs.
A fragment of the mouse Oa1 promoter (bp −3,373 to +55 relative to the transcription start site) was obtained from the 9N2 3.8 clone (6) by NdeI and AgeI endonuclease digestion and was inserted into the PGL-3 basic vector (Promega) upstream of the luciferase reporter gene to generate the plasmid pOa−3373/+55.
A series of unidirectional truncations from the 5′ end of pOa−3373/+55 (pOa−1783/+55, pOa−1282/+55, pOa−854/+55, pOa−562/+55, and pOa−454/+55) were generated by endonuclease digestion with KpnI and PstI, EcoRV, HindIII, BglII, and SwaI, respectively. In addition, the constructs pOa−378/+55, pOa−140/+55, and pOa−11/+55 were generated by PCRs, using pOa−454/+55 as a template, primers 5′-AGGAGCCAGGCTTAGTGGAT-3′, 5′-CTCTGCATATCCAGCATTAG-3′, and 5′-CAGCCCAGCACCTGATCAGG-3′, respectively, as forward primers, and primer 5′-CCCTCACCAGCCCAGCAC-3′ as a reverse primer. The PCR fragments were cloned into the PGL-3 basic vector.
The entire coding sequence of mouse Mitf-M was obtained by RT-PCR with the B16-F10 total RNA, PFU polymerase (Promega), and the following primers: 5′-ATGCTGGAAATGCTAGAATACA-3′ and 5′-CTAACACGCATGCTCCGTTT-3′. The conditions were 32 cycles of 95°C for 30 s, 56°C for 30 s, and 68°C for 2.30 min. The Mitf-M cDNA was cloned into the pRC/CMV expression vector (Invitrogen).
Since PCR amplification can introduce base mutations, all of the constructs reported above were checked by sequence analysis.
Nuclear extracts and electrophoretic mobility shift assays (EMSAs).
Nuclear extracts were prepared from B16-F10 and MNT-1 cells. Each cell monolayer was scraped into 2 ml of cold phosphate-buffered saline and recovered by centrifugation at 400 × g for 5 min at 4°C. The pellets were resuspended in 4 volumes of hypotonic buffer A (10 mM HEPES-KOH [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol [DTT]), mixed, and incubated for 15 min on ice. After the addition of Nonidet NP-40 to a final concentration of 0.62%, the samples were centrifuged for 10 min at 20,800 × g, and the pellets were resuspended in 2.5 volumes (relative to the original cell pellets) of ice-cold buffer B (20 mM HEPES-KOH [pH 7.9], 10% [vol/vol] glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.5 mM DTT, 0.2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 mM NaVO4, 5 mM NaF, 2.5 μg of aprotinin/ml, 1 mM leupeptin, and 1 mM pepstatin) and rocked vigorously at 4°C. The extracts were centrifuged for 5 min at 15,300 × g at 4°C. The supernatants (nuclear extracts) were stored in aliquots at −70°C. The final protein concentrations were determined by the Bradford method (Bio-Rad).
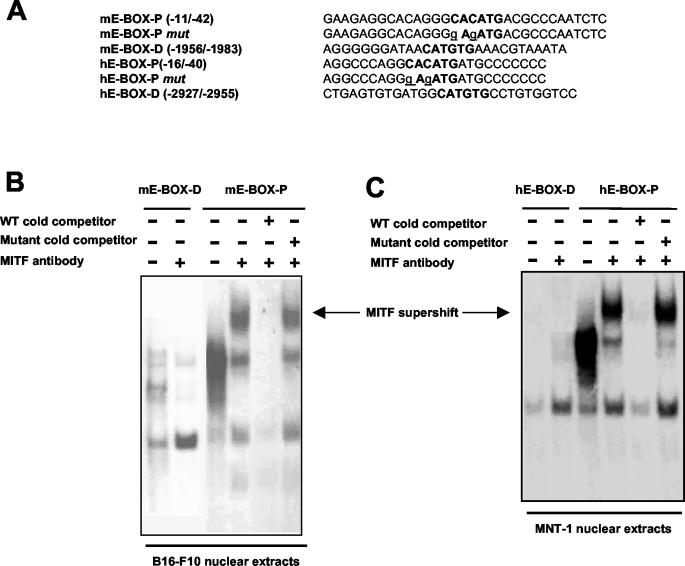
EMSAs were performed with the following double-stranded synthetic oligonucleotides (E-box elements are underlined): mE-BOX-P for positions −11 to −42, 5′-GAAGAGGCACAGGGCACATGACGCCCAATCTC-3′; and mE-BOX-D for positions −1956 to −1983, 5′-AGGGGGGATAACATGTGAAACGTAAATA-3′. Human OA1-specific oligonucleotides containing the homologous genomic regions were prepared by using the following sequences: hE-BOX-P for positions −15 to −40, 5′-AGGCCCAGGCACATGATGCCCCCCC-3′; and hE-BOX-D for positions −2927 to −2955, 5′-CTGAGTGTGATGGCATGTGCCTGTGGTCC-3′.
Oligonucleotides were end labeled with T4 polynucleotide kinase and [γ-32P]ATP. As controls for binding specificity, E-boxes were mutagenized as shown in Fig. 4A. Five micrograms of each nuclear extract was preincubated for 30 min on ice in binding buffer containing 10 mM Tris (pH 7.5), 100 mM KCl, 1 mM DTT, 1 mM EDTA, 5% glycerol, 80 μg of salmon sperm DNA/ml, 3 μg of poly(dI-dC), 2 μg of bovine serum albumin, 2 mM MgCl2, and 2 mM spermidine. A 32P-labeled probe (20,000 to 40,000 cpm) was added to the reaction and further incubated for 15 min at room temperature. When indicated, a 50-fold excess of unlabeled competitor oligonucleotides was added during preincubation. Supershift experiments were performed by pretreatment of the nuclear extract with a C5 anti-Mitf monoclonal antibody (15) in binding reaction buffer for 15 min at room temperature before the addition of the labeled probe. DNA-protein complexes were resolved by electrophoresis in a 4% polyacrylamide (37.5:1 acrylamide-bisacrylamide) gel in TBE buffer (22.5 mM Tris-borate, 0.5 mM EDTA, pH 8) at 4°C for 5 h at 110 V. The gel was dried and exposed on a Kodak film.
FIG. 4.
Mitf binds the E-box close to the transcription start site of the Oa1 gene. (A) Sequences of oligonucleotides used for EMSAs. E-boxes are shown in bold, and the introduced mutations are shown in lowercase and underlined. (B) Gel shift analysis was performed with a B16-F10 murine melanoma nuclear extract. Competition experiments were performed by adding a 50-fold excess of unlabeled homologous (WT) or mutated oligonucleotides. The arrow indicates the supershift detected in the presence of an anti-Mitf monoclonal antibody. (C) Gel shift analysis of human OA1 promoter performed with nuclear extracts of MNT-1 human melanoma cells. Competition experiments were performed by adding a 50-fold excess of unlabeled homologous (WT) or mutated oligonucleotides. The arrow indicates the supershift detected in the presence of an anti-Mitf monoclonal antibody.
Transfection and luciferase assays.
Different cell lines were seeded in 12-well plates (7.0 × 104 per well), cultured overnight, and transfected with 0.5 μg of DNA by use of the FuGENE6 transfection reagent (Roche). The ratio of DNA to FuGENE6 was 1:3, which resulted in similar transfection efficiencies in all cell types used. The pRL-CMV vector (10 ng per well; Promega) driving Renilla reniformis luciferase was included in each transfection as a control to normalize the transcriptional activity of the Oa1 promoter reporter fragments. The expression construct pRC/CMV-Mitf-M (0.1, 0.2, or 0.3 μg) and/or empty pRC/CMV alone, added to the transfection in order to maintain equal amounts of DNA, was included in cotransfection experiments. The preparation of cell lysates and the measurement of luciferase activity were performed by using the Dual Reporter assay system (Promega) according to the manufacturer's instructions and were quantified with a Lumino (STRATEC Electronic) luminometer. Each transfection point was repeated multiple times with different preparations of DNA. The data, expressed as means and standard deviations, were derived from at least four or five independent experiments, as reported in the figure legends.
Site-directed mutagenesis.
Nucleotide substitutions were introduced into each E-box element of the Oa1 promoter region by DNA amplification with a QuikChange site-directed mutagenesis kit (Stratagene) used according to the manufacturer's instructions. The pOa−3373/+55mut1 construct was obtained by using the primer 5′-GGAAGAGGCACAGGGgAgATGACGCCCAATCTC-3′ (lowercase characters indicate base substitutions in the E-box element [underlined]). The pOa−3373/+55mut2 construct was obtained by using the prim-er 5′-GGAGAAGGGGGGATAACATcTcAAACGTAAATAAA-3′. ThepOa−3373/+55mut1-mut2 construct, containing mutations in both E-box sites, was obtained by subcloning the SmaI-SmaI fragment from pOa−3373/+55mut2 into pOa−3373/+55mut1.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation was performed as previously described (23), using material prepared from 501mel melanoma cells. The OA1 promoter region was amplified by using the primers 5′-CTCCTCCGCCCGCCCAAGCATCACCTC-3′ and 5′-GTTCCAACCCGCGGGCCTCTCGTCCTCA-3′. The downstream control region was amplified by using the primers 5′-CGCGCCACCATGCCCGACTAAT-3′ and 5′-GCATGGTGGCGTGTGGCTGTA-3′.
AAV production.
The Oa1 genomic sequence (bp −562 to +55) was amplified from genomic DNA or from the pOa−3373/+55mut1 construct by using 5′-GCTAGCGATCTCACTGTGTAGTGTTAGCTGGCATGG-3′ as a forward primer and 5′-CTGCAGCCGGTCTCTGGTTTGCGGAACC-3′ as a reverse primer (NheI and PstI cloning sites are underlined). Amplification was performed with Taq Advantage DNA PCR mix (BD Bioscience) for 30 cycles at 95°C for 30 s, 68°C for 60 s, and 68°C for 60 s. The PCR products were cloned into the pAAV2.1eGFP vector (3) at the NheI and PstI restriction sites. These constructs were called pAAV2.1-Oa1-EGFP and pAAV2.1-Oa1mut-EGFP, and the fidelity of Taq amplification was controlled by sequence analysis. AAV2/5 viruses were produced by triple transfection of the pAAV2.1-Oa1-EGFP vector (or pAAV2.1-Oa1mut-EGFP or pAAV2.1-CMV-EGFP), the pack 2/5 vector (16), and an adenovirus helper plasmid (3) into 293 cells (American Type Culture Collection). The AAV2/5 viral particles were purified by CsCl gradients, and their titer was assessed by real-time PCR (16).
Subretinal injection.
The use of animals for this work was done in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. Subretinal injections in adult C57/BL6 mice and indirect ophthalmoscopy were performed as described previously (9, 21). One month after injection, the mice were perfused with 4% paraformaldehyde and their eyes were harvested. After equilibration in 30% sucrose, the eyes were embedded in 7.5% gelatin and 12-μm-thick cryosections were collected on SuperfrostPlus slides. The slides were covered with Vectashield coverslips (Vector Laboratories) and photographed through a rhodamine filter (Axioplan; Zeiss). Images were acquired with an AxioCam MRc camera (Carl Zeiss), using Axiovision 3.0 software (Carl Zeiss), with a resolution of 1,030 by 1,300 pixels.
RESULTS
Identification of putative binding sites for known transcription factors in Oa1 gene.
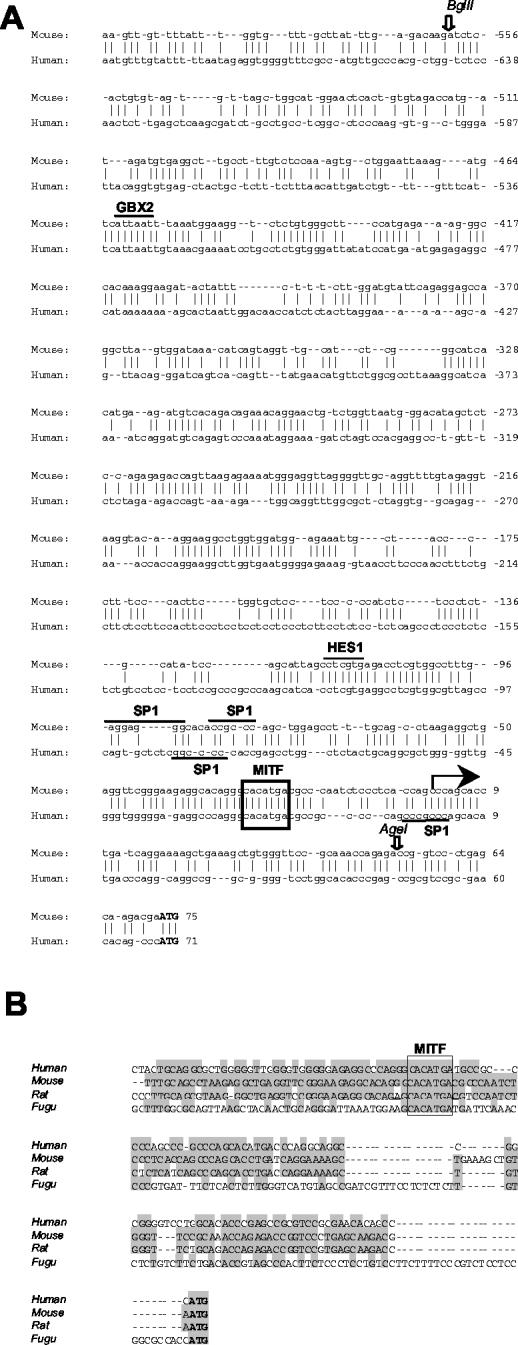
The 3.4-kb region upstream of the murine Oa1 start codon was cloned and sequenced as previously reported (clone 9N2 3.8) (6). The transcription start site of the Oa1 gene was assumed to be 72 bp upstream of the ATG codon, as previously reported (6) and based on a BLAST analysis of murine expressed sequence tags from the database. We compared the murine genomic sequence with the 5′ region of the human OA1 gene obtained from public databases, using BLAST and Vista as informatic tools. An in silico analysis revealed 70% identity in the first 180 bases upstream of the murine transcription start site and 59% identity when we extended the analysis to 700 bp (Fig. 1A). This high homology between the mouse and human genes indicates that the genomic region immediately upstream of the transcription start site is well conserved during evolution. We used the TRANSFAC Professional 6.4 program to identify putative binding sites for known transcription factors in the 3.4-kb region. This allowed the identification of transcription factor binding sites that are conserved in the murine and human sequences. Sp1 binding sites were identified in the mouse and human sequences (underlined in Fig. 1A), while no TATA boxes were found. An E-box at position −28 in the mouse sequence (boxed in Fig. 1A) which was conserved in the human sequence was also found in the rat and fugu genomes (Fig. 1B). A second E-box was identified at position −1972 in the mouse gene and at position −2942 in the human gene (data not shown). The TRANSFAC Professional 6.4 program revealed the presence of a binding site for Hes1, a bHLH transcriptional repressor factor that was previously shown to be involved in neuronal differentiation, in the murine sequence (19). Interestingly, this site is completely conserved in the human sequence (Fig. 1A). Finally, a putative binding site for Gbx2, a homeobox protein, is completely conserved in the mouse and human genes (Fig. 1A). These preliminary data suggest that this genomic region, which is well conserved during evolution, may contain binding elements for transcription factors that control Oa1-specific expression in pigment cells. E-boxes are particularly interesting because they bind bHLH-LZ transcription factors. Interestingly, the highly conserved E-box positioned at −28 presented the sequence CACATGA, in which the A at the 3′ position was reported to favor the specific binding of Mitf (1).
FIG. 1.
Conservation of Oa1 regulatory region during evolution. (A) Sequence comparison between the murine and human Oa1 genomic regions upstream of the ATG codon (in bold). Bioinformatic analysis of the murine Oa1 promoter identified putative binding sites for known transcription factors. Binding sites that are conserved in the murine and human sequences are indicated. The large arrow indicates the transcription start site in the mouse gene. Open arrows indicate restriction sites used for the generation of construct pOa−562/+55. (B) The first 68 bp upstream of the ATG codon of the murine Oa1 gene were compared by Clustal methods to the same genomic region in the human, rat, and fugu genomes. Bases that are conserved in the mouse and other species are shaded. The E-box is boxed.
In vitro analysis of Oa1 promoter defines cell-specific transcriptional control.
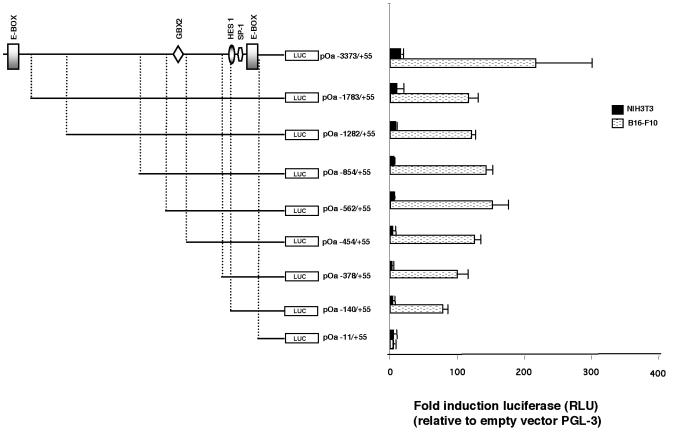
To define the minimal genomic sequence that confers basic expression and tissue specificity on the Oa1 gene, we cloned nine fragments of different sizes of the murine Oa1 promoter upstream of the luciferase reporter gene. To test the tissue-specific activity of the promoter, we transiently transfected these plasmids into either pigmented murine melanoma cells (B16-F10) or nonpigmented fibroblasts (NIH 3T3). The activities of these promoter regions were measured by a luciferase assay and were compared to that of the empty vector, which was considered to be a fold induction of 1, for each experiment. Similar studies were also performed with another pigmented cell type that was not derived from a tumor, called Melan-a cells. We did not find differences in the transactivating profiles of the Oa1 promoter in B16-F10 melanoma cells and Melan-a melanocytes (data not shown). However, higher transactivating activities were measured in the B16-F10 melanoma cell line, probably due to the facts that Melan-a melanocytes are derived from primary cultures and that the expression levels of some melanosome-specific transcription factors are less abundant in these cells. In fact, RT-PCR and Western blotting analysis of Mitf expression detected lower levels of this factor in the Melan-a culture than in the B16-F10 culture (data not shown).
The highest transactivating activity was measured in cells transfected with the pOa−3373/+55 murine genomic fragment, and a deletion of 1.6 kb decreased the activity to 75% (construct pOa−1783/+55) (Fig. 2). Bioinformatic studies identified an E-box in the 1.6-kb region (bp −3373 to −1783). When we then analyzed several deletion derivatives of the pOa−1783/+55 construct, we found that the transcriptional activities were maintained at similar levels up to construct pOa−562/+55. After that, further deletion caused a progressive decrease in the promoter activity.
FIG. 2.
Promoter analysis of Oa1 gene. (Left) Murine Oa1 promoter fragments were cloned upstream of the firefly luciferase reporter gene in the pGL-3-Basic vector. A schematic representation of putative transcription factor binding sites is shown for the longest element analyzed. (Right) Firefly luciferase activities were normalized with the R. reniformis luciferase that was cotransfected in each transfection experiment. The means and standard deviations from at least seven independent experiments are shown. RLU, relative light units.
An analysis of the data obtained by transfection studies of the deletion constructs in pigmented and nonpigmented cells demonstrated that the tissue-specific regulation of Oa1 in pigmented cells lies within the first 140 bp upstream of the mRNA transcription start site (Fig. 2). In fact, this element still shows differential expression in pigmented cells compared to the activity observed in fibroblasts. Interestingly, the E-box (at position −28), which can be bound by the bHLH transcription factor Mitf, is contained within this region.
Oa1 expression depends upon Mitf in vitro and in vivo.
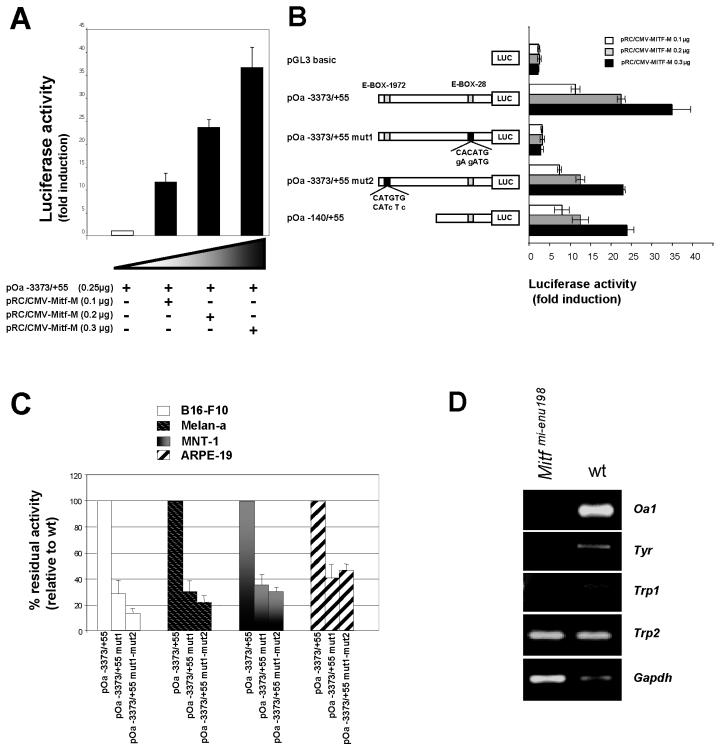
Previous studies demonstrated that Mitf plays a key role in the transcriptional regulation of pigmentation genes (36). We therefore focused our attention on the Mitf binding site contained in the 140-bp region. For this purpose, we cloned a Mitf-M cDNA into a eukaryotic expression vector by RT-PCR of RNA purified from B16-F10 cells. To determine the expression of Mitf-transfected cells, we performed Western blotting analysis, using a monoclonal antibody that was specific for Mitf. Immunohistochemistry in Cos-7 cells confirmed that the transcription factor was localized inside the nucleus (data not shown). We cotransfected NIH 3T3 fibroblasts, which do not normally express Mitf, with the pOa−3373/+55 promoter construct and increasing amounts of the Mitf expression plasmid. In these experiments, the Oa1 promoter showed a dose-responsive activity to Mitf (Fig. 3A). Similarly, we measured the transactivation of the pOa−140/+55 construct by Mitf (Fig. 3B). Transactivation of the Oa1 promoter was completely lost when we introduced two single base mutations into the −28 E-box (CACATG to GAGATG), as shown in Fig. 3B (pOa−3373/+55mut1). On the other hand, a dose response to Mitf was preserved in cotransfection experiments with a pOa−3373/+55 construct in which two single base mutations were introduced into the E-box localized at position −1972 (pOa−3373/+55mut2 in Fig. 3B). These in vitro data suggest that the E-box positioned 28 bp from the RNA transcription start site plays a key role in the response to Mitf, in contrast to the distal E-box.
FIG. 3.
Mitf transactivation of Oa1 promoter. (A) Cotransfection experiments in NIH 3T3 cells with pOa−3373/+55 construct and increasing doses of Mitf expression vector showing dose response to Mitf. (B) Mutagenesis of two bases in the E-box at position −28 (pOa−3373/+55mut1) completely abolishes the response to Mitf in vitro. Mutagenesis of two bases in the E-box at position −1972 (pOa−3373/+55mut2) does not impair the response to Mitf. (C) Wild-type and mutagenized promoter constructs were analyzed in different pigmented cell lines. The luciferase activity measured for the pOa−3373/+55 wild-type construct was considered to be 100%. (D) RT-PCR analysis of Oa1 expression in Mitf mutant mice (Mitfmi-enu198) showing that Oa1 transcription depends on Mitf in the eye, as was also found for Tyr and Trp1, but not for Trp2.
To better understand the tissue specificity of this Mitf binding site, we analyzed the contribution of this E-box to the transcriptional activity of Oa1 in different pigmented cell types. For this purpose, we transfected murine melanoma cells (B16-F10), murine melanocytes (Melan-a), human melanoma cells (MNT-1), and human RPE cells (ARPE-19) with either the wild-type pOa−3373/+55 construct or the promoter with a mutation in one or both E-boxes. Mutation of the −28 E-box (pOa−3373/+55mut1) had a strong effect on reporter expression in pigmented cells. In fact, expression was reduced to 30% in mouse and human melanoma cells and to 40% in RPE cells (Fig. 3C). The mutation of both E-boxes (pOa−3373/+55mut1-mut2) did not show a further reduction (no statistically significant difference could be measured) compared to the single mutation of the −28 E-box (Fig. 3C). Therefore, the −28 E-box plays a key role in the response of the Oa1 promoter to Mitf in pigmented cells.
To demonstrate that Oa1 expression is indeed dependent on Mitf in vivo, we performed RT-PCR experiments on RNAs from Mitf mutant RPE. We harvested RPE from the Mitfmi-enu198 mutant mouse, in which there is an Asp207Gly substitution in the DNA binding domain of the Mitf protein (14). We could not detect Oa1 mRNA in the RPE of these mice (Fig. 3D). The Tyr and Trp1 genes were also not expressed in the Mitfmi-enu198 mutant RPE, while Trp2, which is known to not be dependent on Mitf in the eye (31), was normally expressed (Fig. 3D).
Mift directly binds the Oa1 promoter.
The in vitro and in vivo data suggested that Mitf controls Oa1 expression but did not demonstrate that this effect is due to direct binding to the Oa1 promoter. In fact, another albinism gene, Matp, which probably controls melanosome biogenesis (11, 20) as does Oa1, shows a reduction in expression in Mitf mutant mice (8). However, Mitf does not bind to the Matp promoter, and therefore the effect seen in Mitf mutant mice may be indirect or mediated by a distinct enhancer element (12). To address this issue with the Oa1 promoter, we performed EMSAs to define whether Mitf directly binds the E-box positioned 28 bp from the transcription start site. For these experiments, we used B16-F10 nuclear extracts that expressed high levels of Mitf. We tested the binding of Mitf to an oligonucleotide that contained the −28 E-box (Fig. 4A, mE-BOX-P). When we added an anti-Mitf monoclonal antibody to the binding reaction, we detected a supershift of the Mitf-DNA complex (Fig. 4B, arrow), indicating a specific binding of Mitf to the oligonucleotide. The binding could be competed by the wild-type cold oligonucleotide but not by the mutated oligonucleotide (Fig. 4, mE-BOX-P mut). We also tested the ability of Mitf to bind the −1972 E-box (Fig. 4A, mE-BOX-D), but we never detected a specific binding by supershift experiments (Fig. 4B). The specific binding of MITF to the E-box proximal to the transcription start site of the human sequence (Fig. 4A, hE-BOX-P) was detected by using human melanoma cell (MNT-1) nuclear extracts (Fig. 4C). As was seen for the murine sequence, a supershift of the MITF-DNA complex was detected in the presence of the anti-Mitf antibody and the binding could be specifically competed by cold wild-type oligonucleotides. The distal E-box at position −2942 in the human genomic sequence (Fig. 4A, hE-BOX-D) did not bind to MITF in vitro (Fig. 4C). The EMSAs, together with transactivation studies, suggested that Mitf specifically binds the Oa1 genomic region at position −28, but not at the distal E-box 1,972 bp from the transcription start site.
Mitf transcriptional control of Oa1 is conserved in humans.
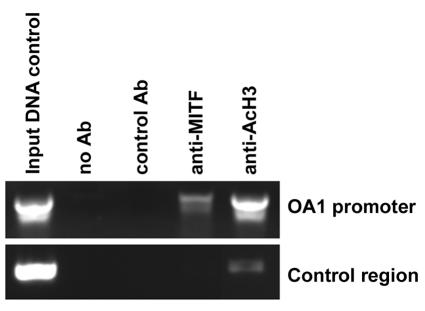
Our in vitro and in vivo studies in the mouse showed that Mitf positively controls Oa1 transcription. EMSAs suggested that this regulation is also conserved in the human OA1 gene. In order to confirm that MITF directly binds to the human OA1 promoter, we performed chromatin immunoprecipitation studies. Chromatin from 501mel human melanoma cells was immunoprecipitated with an anti-MITF antibody. We tested by PCR the binding of MITF to the E-box proximal to the human transcription start site (7). We specifically amplified the OA1 gene from cross-linked chromatin isolated from 501mel cells and immunoprecipitated it with an anti-MITF antibody (Fig. 5). Immunoprecipitation of the same chromatin preparation with an anti-AcH3 antibody further demonstrated that this is a transcriptionally active region (Fig. 5). These data confirmed that the E-box proximal to the transcription start site was bound by MITF in vivo and that the MITF transcriptional control of OA1 expression has been conserved throughout evolution.
FIG. 5.
Chromatin immunoprecipitation. Chromatin immunoprecipitation was performed with material from 501mel cells with the primers specified in Materials and Methods. A region residing in the first intron was amplified as a control.
The small 617-bp element is able to drive tissue-specific expression of Oa1 in vivo.
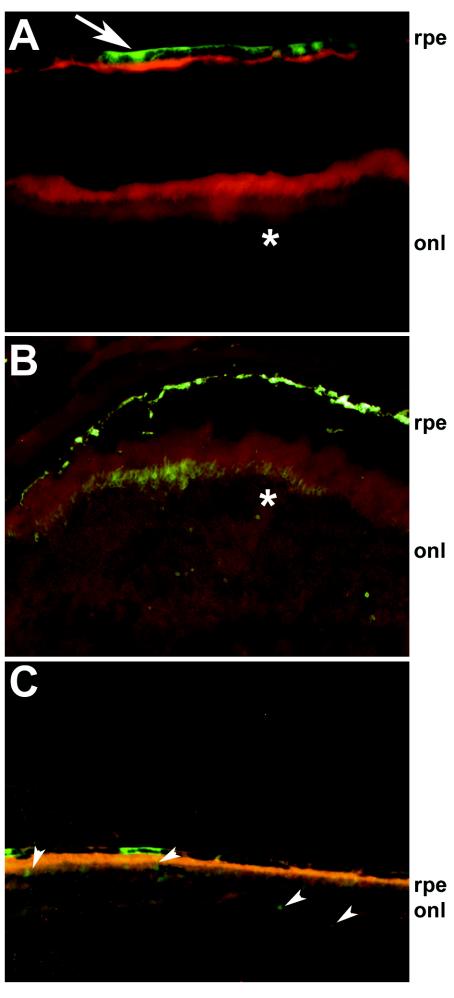
The promoter studies using in vitro cell lines indicated that the pOa−562/+55 element contains all of the information needed for the physiological and tissue-specific expression of Oa1. In fact, studies with several deletions of the Oa1 promoter showed a decrease in transactivating potential starting with the pOa−562/+55 construct. This element also had a strong activity in pigmented cells compared to that in fibroblasts. We therefore defined this construct as the smallest element that is fully active in the control of tissue specificity. In order to evaluate whether the small genomic region that we identified could be used in the future to drive the efficient tissue-specific expression of Oa1 in misexpression experiments, we cloned the −562 to +55 sequence upstream of a reporter gene (enhanced green fluorescent protein [EGFP]) in an AAV vector (AAV2/5-Oa1-EGFP). We chose the AAV2/5 serotype because members of our laboratory previously showed that this virus is able to transduce both RPE cells, which express Oa1 normally, and neural retina cells, which do not express Oa1 (4). If the −562 to +55 Oa1 promoter sequence is able to confer tissue specificity, then the expression of EGFP will be restricted just to RPE cells and no expression should be detected in neural retinal cells. Mice (n = 10 6-week-old male C57/BL6 mice) were injected subretinally with 1 to 3 μl of AAV2/5-Oa1-EGFP (corresponding to 1.6 × 109 to 4.8 × 109 genome copies) or AAV2/5-CMV-EGFP (corresponding to 1 × 109 to 3 × 109 genome copies) in the right and left eye, respectively. Two and four weeks after vector administration, the animals were analyzed by indirect ophthalmoscopy to detect EGFP expression (9). In 6 of 10 left eyes (injected with AAV2/5-CMV-EGFP) and in none of the right eyes (injected with AAV2/5-Oa1-EGFP), green fluorescence was ophthalmoscopically evident in the fundi. Four weeks after injection, the animals were sacrificed and their eyes were enucleated, fixed, and cryosectioned. A histological analysis of the sections showed robust EGFP expression in the RPE and photoreceptors of the retinas injected with the cytomegalovirus (CMV)-containing vector (Fig. 6B). Each eye that was positive by ophthalmoscopic analysis showed EGFP expression at the histological level. EGFP expression was specifically present in the RPE but not in the photoreceptor cells of the AAV2/5-Oa1-EGFP-treated retinas (5 of 10 injected eyes) (Fig. 6A, arrow), although it was weaker than in the AAV2/5-CMV-EGFP-treated retinas. The weak RPE-specific expression driven by the Oa1 promoter explained our inability to detect the fluorescence by indirect ophthalmoscopy in live animals. Therefore, while the AAV2/5 virus infected both RPE and photoreceptor cells, the −562 to +55 Oa1 promoter sequence conferred tissue specificity, driving expression of the reporter gene in RPE cells only.
FIG. 6.
Tissue-specific expression driven by Oa1 promoter in vivo. C57/BL6 wild-type mice were injected subretinally with either AAV2/5-Oa1-EGFP (A), AAV2/5-CMV-EGFP (B), or AAV2/5-Oa1mut-EGFP (C). While the CMV promoter drives expression in RPE and photoreceptor cells (B, asterisk), the Oa1 promoter drives specific expression in RPE cells only (A, arrow), not in photoreceptor cells (A, asterisk). Mutagenesis of the Mitf binding site reduces the expression driven by the Oa1 promoter in the RPE and drives a low expression level in the photoreceptor layer (C, arrowheads). onl, outer nuclear layer (photoreceptor cells).
Finally, using the same injection protocol, we injected AAV2/5-Oa1mut-EGFP, in which the −562 to +55 sequence was mutagenized at the E-box (position −28), into mouse retinas. Expression driven by the mutagenized promoter was lower in RPE cells than that driven by the wild-type promoter (Fig. 6C). Furthermore, we also noticed a low expression level in the photoreceptor cell layer (Fig. 6C, arrowheads).
DISCUSSION
Loss of function of the OA1 gene causes X-linked ocular albinism, a genetic disease with a prevalence of 1 in 50,000 births. The peculiar feature in OA1 patients is the fact that skin and RPE melanosomes do not appear to be depigmented, as is often seen in other forms of albinism, but they have an abnormally large size (27, 37). The OA1 protein is localized mostly in premelanosomes, the precursors of these organelles, but it is also found on the membranes of mature melanosomes (28, 29). Based on this evidence, OA1 is believed to be important for the biogenesis of the organelles and not for melanin deposition. Melanin production and deposition are regulated by enzymes that catalyze melanin synthesis (TYR, TRP1, and TRP2). This highlights the importance of undertaking studies aimed at elucidating the molecular basis of albinism by the identification of common pathways that regulate the expression of genes involved in melanin synthesis and melanosome organellogenesis.
For this purpose, we analyzed the promoter region of the Oa1 gene. Here we have presented in vitro and in vivo data that strongly associate Mitf with Oa1 transcription. Mitf was previously shown to play a key role in the tissue-specific transcription of several pigmentation genes. Similarly, we found that Mitf is necessary for Oa1 expression in the RPE, as seen by RT-PCR studies of Mitfmi-enu198 mutant mice. We also showed that the Mitf binding element is conserved between the mouse and human genes, suggesting an important and evolutionarily conserved regulation of Oa1 expression. Interestingly, bioinformatic analysis revealed that the Mitf binding site is also conserved in more evolutionarily distant species, such as fugu.
Mitf is a highly tissue-specific transcription factor, with its expression being restricted to a few cell types (25). The tissue-specific regulation of Oa1 transcription probably resides in the Mitf E-box located 28 bp from the transcription start site. The mutation of two bases in this binding site caused a significant reduction in expression in pigmented cells, while it had no significant effect in NIH 3T3 cells. The direct binding of Mitf to the −28 E-box in the Oa1 promoter was demonstrated by in vitro and in vivo experiments confirming the direct regulation of Oa1 transcription by the bHLH-LZ factor. A second, more distal E-box was identified at position −1972. Our experiments ruled out the possibility that this is a second site for Mitf. Our data are in full agreement with previous observations that the A at the 3′ end of the 6-bp palindrome (E-box) favors the binding of Mitf (1). In fact, the presence of this A in the sequence was found to be completely conserved in the proximal (bp −28) murine and human E-boxes, while it is not present in either the human or murine distal E-box.
Deletion studies with the Oa1 promoter revealed that the most important elements for the regulation of Oa1 transcription and for tissue specificity are located within the first 562 bp upstream of the mRNA start site. In fact, we did not measure significant differences in transcriptional activity between the constructs pOa−1783/+55 and pOa−562/+55. The Mitf E-box resides within this sequence, but other transcription factors that bind this same genomic region must have an important role in transcriptional regulation. In fact, mutating the −28 E-box did not completely shut off transcription in pigmented cells. Bioinformatic analysis highlighted Sp1 binding sites located close to the Mitf element. This transcription factor may control the basal expression of the Oa1 gene. The first 562 bp also contain a binding site for Hes1 that is conserved in the human sequence. Hes1 is a bHLH gene that is regulated by the Notch signaling pathway (18). Hes1 is expressed in neural retinal progenitors and acts as a repressor to negatively regulate neurogenesis (35). The presence of a binding site for Hes1 in the Oa1 promoter suggests that Hes1 may also be involved in repressing the expression of RPE-specific genes in the neural retina. Further studies will be required to understand the functional role of these elements in the regulation of Oa1 transcription.
Our in vitro and in vivo studies identified a 617-bp genomic fragment (pOa−562/+55) of the Oa1 gene which contains elements for basal and tissue-specific transcription. In vitro studies showed that this sequence is able to confer specific expression in pigmented cells. This same small genomic fragment was tested in vivo by intraocular AAV delivery and was found to specifically control expression in the RPE. When we compared the EGFP signals in transduced eyes, the expression detected from the Oa1 promoter was lower than that from the CMV promoter (Fig. 6). This was an expected result since in situ hybridization studies previously found that Oa1 is expressed at low levels in the RPE compared to other pigmentation genes (32). Interestingly, we found that the tissue specificity was lost when we mutated the Mitf binding site. In fact, we detected a low promoter activity in the RPE but also detected a low expression level in the photoreceptor cell layer. These results suggest that Mitf is necessary for expression in the RPE, as demonstrated by the lack of Oa1 transcription in Mitf mutant mice; however, it is not sufficient because a low expression level was still activated when we prevented Mitf from binding to the promoter. The low expression level in photoreceptor cells suggests that the E-box, bound by Mitf in pigmented cells, may also be bound by a bHLH-LZ factor that acts as a repressor in neural retinal cells. Therefore, binding of this factor to the E-box will be prevented by mutagenesis and expression will be ectopically activated in these cells. A similar competitive activity was previously suggested for the bHLH protein ITF2 (13). The lower expression level in the RPE from the mutagenized promoter than from the wild-type promoter was expected based on in vitro data showing a reduction to 30 to 40% of the wild-type activity but not a complete loss of expression. Furthermore, the expression of EGFP cannot be quantified in vivo due to the high stability of this protein. Therefore, we believe that the promoter element identified here may ensure the specific and physiological expression of Oa1 in pigmented cells in vivo. These are fundamental requirements for gene delivery experiments. This genomic region has additional qualities for gene transfer approaches; for example, the 617-bp sequence is small, which is an important characteristic for a promoter that is to be used in viral systems with size constraints. All of these characteristics make the Oa1 promoter a new tool for somatic gene transfer in animal models of RPE-specific diseases. In fact, it prevents undesired expression in the photoreceptor cells and drives specific expression in the RPE cell layer only. To prove the effectiveness of the promoter, we are undertaking phenotype rescue experiments with Oa1 null mice (17), using this newly identified element to control the physiological expression of recombinant Oa1 in vivo.
In conclusion, our study of the dependence of Oa1 expression on Mitf establishes a link between genes involved in melanin synthesis and those involved in melanosome biogenesis. In fact, Mitf regulates the expression of genes controlling both of these processes. In addition, these data are of potential medical relevance for the study of the pathogenesis of albinism, for the study of melanoma, and for future RPE gene therapy efforts.
Acknowledgments
This work was supported in part by research grant 1-FY01-117 from the March of Dimes Birth Defects Foundation, by research grants from Fondazione Telethon and the Vision of Children Foundation to V.M., by research grant 1R01EY015136-01 from NEI to A.B., and by grant AR43369 from NIAMS to D.E.F.
We thank J. Favor for Mitf mutant mice, R. Giavazzi for the B16-F10 melanoma cell line, C. Tacchetti and C. Valetti for the MNT-1 melanoma cell line, and G. Diez-Roux, M. Zollo, and A. D'Angelo for discussions of the data.
REFERENCES
- 1.Aksan, I., and C. R. Goding. 1998. Targeting the microphthalmia basic helix-loop-helix-leucine zipper transcription factor to a subset of E-box elements in vitro and in vivo. Mol. Cell. Biol. 18:6930-6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amiel, J., P. M. Watkin, M. Tassabehji, A. P. Read, and R. M. Winter. 1998. Mutation of the MITF gene in albinism-deafness syndrome (Tietz syndrome). Clin. Dysmorphol. 7:17-20. [PubMed] [Google Scholar]
- 3.Auricchio, A., M. Hildinger, E. O'Connor, G. P. Gao, and J. M. Wilson. 2001. Isolation of highly infectious and pure adeno-associated virus type 2 vectors with a single-step gravity-flow column. Hum. Gene Ther. 12:71-76. [DOI] [PubMed] [Google Scholar]
- 4.Auricchio, A., G. Kobinger, V. Anand, M. Hildinger, E. O'Connor, A. M. Maguire, J. M. Wilson, and J. Bennett. 2001. Exchange of surface proteins impacts on viral vector cellular specificity and transduction characteristics: the retina as a model. Hum. Mol. Genet. 10:3075-3081. [DOI] [PubMed] [Google Scholar]
- 5.Barsh, G. S. 1996. The genetics of pigmentation: from fancy genes to complex traits. Trends Genet. 12:299-305. [DOI] [PubMed] [Google Scholar]
- 6.Bassi, M. T., B. Incerti, D. J. Easty, E. V. Sviderskaya, and A. Ballabio. 1996. Cloning of the murine homologue of the ocular albinism type 1 (OA1) gene: sequence, genomic structure and expression analysis in pigment cells. Genome Res. 6:880-885. [DOI] [PubMed] [Google Scholar]
- 7.Bassi, M. T., M. V. Schiaffino, A. Renieri, F. De Nigris, L. Galli, M. Bruttini, M. Gebbia, A. A. B. Bergen, R. A. Lewis, and A. Ballabio. 1995. Cloning of the gene for ocular albinism type 1 from the distal short arm of the X chromosome. Nat. Genet. 10:13-19. [DOI] [PubMed] [Google Scholar]
- 8.Baxter, L. L., and W. J. Pavan. 2002. The oculocutaneous albinism type IV gene Matp is a new marker of pigment cell precursors during mouse embryonic development. Mech. Dev. 116:209-212. [DOI] [PubMed] [Google Scholar]
- 9.Bennett, J., D. Duan, J. F. Engelhardt, and A. M. Maguire. 1997. Real-time, noninvasive in vivo assessment of adeno-associated virus-mediated retinal transduction. Investig. Ophthalmol. Vis. Sci. 38:2857-2863. [PubMed] [Google Scholar]
- 10.Chirivi, R. G., A. Garofalo, M. J. Crimmin, L. J. Bawden, A. Stoppacciaro, P. D. Brown, and R. Giavazzi. 1994. Inhibition of the metastatic spread and growth of B16-BL6 murine melanoma by a synthetic matrix metalloproteinase inhibitor. Int. J. Cancer 58:460-464. [DOI] [PubMed] [Google Scholar]
- 11.Costin, G. E., J. C. Valencia, W. D. Vieira, M. L. Lamoreux, and V. J. Hearing. 2003. Tyrosinase processing and intracellular trafficking is disrupted in mouse primary melanocytes carrying the underwhite (uw) mutation. A model for oculocutaneous albinism (OCA) type 4. J. Cell Sci. 116:3203-3212. [DOI] [PubMed] [Google Scholar]
- 12.Du, J., and D. E. Fisher. 2002. Identification of Aim-1 as the underwhite mouse mutant and its transcriptional regulation by MITF. J. Biol. Chem. 277:402-406. [DOI] [PubMed] [Google Scholar]
- 13.Furumura, M., S. B. Potterf, K. Toyofuku, J. Matsunaga, J. Muller, and V. J. Hearing. 2001. Involvement of ITF2 in the transcriptional regulation of melanogenic genes. J. Biol. Chem. 276:28147-28154. [DOI] [PubMed] [Google Scholar]
- 14.Hallsson, J. H., J. Favor, C. Hodgkinson, T. Glaser, M. L. Lamoreux, R. Magnusdottir, G. J. Gunnarsson, H. O. Sweet, N. G. Copeland, N. A. Jenkins, and E. Steingrimsson. 2000. Genomic, transcriptional and mutational analysis of the mouse microphthalmia locus. Genetics 155:291-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hemesath, T. J., E. R. Price, C. Takemoto, T. Badalian, and D. E. Fisher. 1998. MAP kinase links the transcription factor microphthalmia to c-Kit signalling in melanocytes. Nature 391:298-301. [DOI] [PubMed] [Google Scholar]
- 16.Hildinger, M., A. Auricchio, G. Gao, L. Wang, N. Chirmule, and J. M. Wilson. 2001. Hybrid vectors based on adeno-associated virus serotypes 2 and 5 for muscle-directed gene transfer. J. Virol. 75:6199-6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Incerti, B., K. Cortese, A. Pizzigoni, E. M. Surace, S. Varani, M. Coppola, G. Jeffery, M. Seeliger, G. Jaissle, D. C. Bennett, V. Marigo, M. V. Schiaffino, C. Tacchetti, and A. Ballabio. 2000. Oa1 knock-out: new insights on the pathogenesis of ocular albinism type 1. Hum. Mol. Genet. 9:2781-2788. [DOI] [PubMed] [Google Scholar]
- 18.Jarriault, S., C. Brou, F. Logeat, E. H. Schroeter, R. Kopan, and A. Israel. 1995. Signalling downstream of activated mammalian Notch. Nature 377:355-358. [DOI] [PubMed] [Google Scholar]
- 19.Kageyama, R., T. Ohtsuka, and K. Tomita. 2000. The bHLH gene Hes1 regulates differentiation of multiple cell types. Mol. Cell 10:1-7. [DOI] [PubMed] [Google Scholar]
- 20.Lehman, A. L., W. K. Silvers, N. Puri, K. Wakamatsu, S. Ito, and M. H. Brilliant. 2000. The underwhite (uw) locus acts autonomously and reduces the production of melanin. J. Investig. Dermatol. 115:601-606. [DOI] [PubMed] [Google Scholar]
- 21.Liang, F. Q., V. Anand, A. Maguire, and J. Bennett. 2000. Intraocular delivery of recombinant virus. Methods Mol. Med. 47:125-139. [DOI] [PubMed] [Google Scholar]
- 22.Lowings, P., U. Yavuzer, and C. R. Goding. 1992. Positive and negative elements regulate a melanocyte-specific promoter. Mol. Cell. Biol. 12:3653-3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGill, G. G., M. Horstmann, H. R. Widlund, J. Du, G. Motyckova, E. K. Nishimura, Y. L. Lin, S. Ramaswamy, W. Avery, H. F. Ding, S. A. Jordan, I. J. Jackson, S. J. Korsmeyer, T. R. Golub, and D. E. Fisher. 2002. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell 109:707-718. [DOI] [PubMed] [Google Scholar]
- 24.Moore, K. J. 1995. Insight into the microphthalmia gene. Trends Genet. 11:442-448. [DOI] [PubMed] [Google Scholar]
- 25.Nakayama, A., M. T. Nguyen, C. C. Chen, K. Opdecamp, C. A. Hodgkinson, and H. Arnheiter. 1998. Mutations in microphthalmia, the mouse homolog of the human deafness gene MITF, affect neuroepithelial and neural crest-derived melanocytes differently. Mech. Dev. 70:155-166. [DOI] [PubMed] [Google Scholar]
- 26.Newton, J. M., S. J. Orlow, and G. S. Barsh. 1996. Isolation and characterization of a mouse homolog of the X-linked ocular albinism (OA1) gene. Genomics 37:219-225. [DOI] [PubMed] [Google Scholar]
- 27.O'Donnell, F. E. J., G. W. J. Hambrick, W. R. Green, W. J. Iliff, and D. L. Stone. 1976. X-linked ocular albinism: an oculocutaneous macromelanosomal disorder. Arch. Ophthalmol. (Chicago) 94:1883-1892. [DOI] [PubMed] [Google Scholar]
- 28.Samaraweera, P., B. Shen, J. M. Newton, G. S. Barsh, and S. J. Orlow. 2001. The mouse ocular albinism 1 gene product is an endolysosomal protein. Exp. Eye Res. 72:319-329. [DOI] [PubMed] [Google Scholar]
- 29.Schiaffino, M. V., C. Baschirotto, G. Pellegrini, S. Montalti, C. Tacchetti, M. De Luca, and A. Ballabio. 1996. The ocular albinism type 1 (OA1) gene product is a membrane glycoprotein localized to melanosomes. Proc. Natl. Acad. Sci. USA 93:9055-9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schiaffino, M. V., M. d'Addio, A. Alloni, C. Baschirotto, C. Valetti, K. Cortese, C. Puri, M. T. Bassi, C. Colla, M. De Luca, C. Tacchetti, and A. Ballabio. 1999. Ocular albinism: evidence for a defect in an intracellular signal transduction system. Nat. Genet. 23:108-112. [DOI] [PubMed] [Google Scholar]
- 31.Smith, S. B., B. K. Zhou, and S. J. Orlow. 1998. Expression of tyrosinase and the tyrosinase related proteins in the Mitfvit (vitiligo) mouse eye: implications for the function of the microphthalmia transcription factor. Exp. Eye Res. 66:403-410. [DOI] [PubMed] [Google Scholar]
- 32.Surace, E. M., B. Angeletti, A. Ballabio, and V. Marigo. 2000. Expression pattern of the ocular albinism type 1 (OA1) gene in the murine retinal pigment epithelium. Investig. Ophthalmol. Vis. Sci. 41:4333-4337. [PubMed] [Google Scholar]
- 33.Tassabehji, M., V. E. Newton, X. Z. Liu, A. Brady, D. Donnai, M. Krajewska-Walasek, V. Murday, A. Norman, E. Obersztyn, W. Reardon, et al. 1995. The mutational spectrum in Waardenburg syndrome. Hum. Mol. Genet. 4:2131-2137. [DOI] [PubMed] [Google Scholar]
- 34.Tassabehji, M., V. E. Newton, and A. P. Read. 1994. Waardenburg syndrome type 2 caused by mutations in the human microphthalmia (MITF) gene. Nat. Genet. 8:251-255. [DOI] [PubMed] [Google Scholar]
- 35.Tomita, K., M. Ishibashi, K. Nakahara, S. L. Ang, S. Nakanishi, F. Guillemot, and R. Kageyama. 1996. Mammalian hairy and enhancer of split homolog 1 regulates differentiation of retinal neurons and is essential for eye morphogenesis. Neuron 16:723-734. [DOI] [PubMed] [Google Scholar]
- 36.Widlund, H. R., and D. E. Fisher. 2003. Microphthalmia-associated transcription factor: a critical regulator of pigment cell development and survival. Oncogene 22:3035-3041. [DOI] [PubMed] [Google Scholar]
- 37.Wong, L., F. E. O'Donnell, Jr., and W. R. Green. 1983. Giant pigment granules in the retinal pigment epithelium of a fetus with X-linked ocular albinism. Ophthalm. Paediatr. Genet. 2:47-65. [Google Scholar]