Abstract
It was recently demonstrated that during apoptosis, active caspase 9 and caspase 3 rapidly accumulate in the mitochondrion-enriched membrane fraction (D. Chandra and D. G. Tang, J. Biol. Chem.278:17408-17420, 2003). We now show that active caspase 8 also becomes associated with the membranes in apoptosis caused by multiple stimuli. In MDA-MB231 breast cancer cells treated with etoposide (VP16), active caspase 8 is detected only in the membrane fraction, which contains both mitochondria and endoplasmic reticulum (ER), as revealed by fractionation studies. Immunofluorescence microscopy, however, shows that procaspase 8 and active caspase 8 predominantly colocalize with the mitochondria. Biochemical analysis demonstrates that both procaspase 8 and active caspase 8 are localized mainly on the outer mitochondrial membrane (OMM) as integral proteins. Functional analyses with dominant-negative mutants, small interfering RNAs, peptide inhibitors, and Fas-associated death domain (FADD)- and caspase 8-deficient Jurkat T cells establish that the mitochondrion-localized active caspase 8 results mainly from the FADD-dependent and tumor necrosis factor receptor-associated death domain-dependent mechanisms and that caspase 8 activation plays a causal role in VP16-induced caspase 3 activation and cell death. Finally, we present evidence that the OMM-localized active caspase 8 can activate cytosolic caspase 3 and ER-localized BAP31. Cleavage of BAP31 leads to the generation of ER- localized, proapoptotic BAP20, which may mediate mitochondrion-ER cross talk through a Ca2+-dependent mechanism.
Caspases play a crucial role in the execution of the apoptotic process (46). Caspases are synthesized as inactive zymogens, which become proteolytically cleaved during apoptosis to generate active enzymes. Caspase 8 acts as the most upstream caspase in apoptotic signaling initiated by Fas (CD95), tumor necrosis factor (TNF) receptor (TNFR), and TNFR-related death receptors including TRAIL (TNF-related apoptosis-inducing ligand) receptors (TRAIL-R1 and TRAIL-R2) and DRs (death receptors; e.g., DR3) (1, 5, 36, 38, 41). Engagement of Fas by Fas ligand directly recruits the adaptor protein Fas-associated death domain (FADD), procaspase 8, procaspase 10, and c-FLIP to the death-inducing signaling complex (DISC) (1, 41). By contrast, engagement of TNFR1 by TNF-α does not directly recruit FADD or caspase 8 to the activated receptor (19). Instead, TNFR1-mediated apoptosis appears to proceed via two sequential signaling complexes (37). The plasma membrane-bound complex I is rapidly formed upon receptor activation and contains TNFR1, adaptor protein TRADD (TNFR-associated death domain), death domain-containing kinase RIP1, and TRAF-2, leading to NF-κB activation (37). Then, complex I leaves the receptor and forms a different, long-lived complex, complex II, which localizes mainly in the cytosol and contains apoptotic proteins FADD, caspase 8, and caspase 10 in addition to TRADD, RIP1, and TRAF-2 (37). The activation of complex II results in cell death (37). Thus, TNF-α induces the complex II-mediated apoptosis only when the complex I-initiated prosurvival signal (i.e., NF-κB) fails to be activated (37). How procaspase 8 is activated in the DISC or complex II is not entirely clear but may involve proximity-induced dimerization and autocatalytic cleavage (1, 3, 36).
Stimulation with chemotherapeutic drugs, trophic factor deprivation, oxidative stress, cytotoxic chemicals, and ionizing or UV radiation also leads to caspase 8 activation, which may represent either an initiating or a secondary amplifying event in apoptosis (4, 12, 15, 18, 21, 22, 26-28, 51, 59, 60, 62, 63). Activated caspase 8 in these apoptotic systems either directly activates the executioner caspase, caspase 3 (47, 55), or cleaves the BH3-only protein Bid, generating the truncated form of Bid, t-Bid (29, 31). t-Bid then translocates to the outer mitochondrial membrane (OMM) and promotes oligomerization of Bax or Bak to facilitate release of cytochrome c and other apoptogenic proteins from the intermembrane space (IMS) of the mitochondria into the cytosol. The released cytochrome c, in the presence of (d)ATP, binds to and activates adaptor protein Apaf-1, which in turn recruits caspase 9, leading to the formation of apoptosome, activation of caspase 9, and subsequent activation of executioner caspases, caspase 3, 6, or 7 (61). In addition to caspase 3 and Bid, caspase 8 has also been shown to cleave RIP (30), plectin (54), caspase 7 (39), caspase 9 (35), and BAP31, an endoplasmic reticulum (ER)-specific protein (40).
Apart from their cytosolic residence, various procaspases and active caspases, including caspases 9 and 3, are also localized in other subcellular compartments, such as mitochondria (reference 10 and references therein). Procaspase 8, which is transcribed as two major isoforms (i.e., p55/53) (48), predominantly colocalizes with the mitochondria in MCF7 breast cancer cells (53, 54). Similarly, in human fibroblasts, mouse striatal cells, and COS-1 cells, procaspase 8 has been shown to localize in the IMS, inner membrane, and matrix of the mitochondria (43). It is thought that, upon apoptosis induction, procaspase 8 stored in the mitochondria, like other procaspases, is released and becomes activated in the cytosol. Recently, a novel procaspase 8 isoform, procaspase 8L, has been shown to be peripherally associated with the cytosolic face of the ER and its processing appears to be regulated through an association with BAP31 (6).
Recently, our laboratory found that apoptosis induced by many stimuli involves an early mitochondrial activation characterized by rapid up-regulation of the mitochondrial respiratory chain (MRC) proteins and other mitochondrially localized non-MRC proteins (9). A cardinal feature of this mitochondrial activation-dependent apoptotic pathway (or MADAP) is the early up-regulation and accumulation of cytochrome c in the mitochondria, which precedes its release (9). Furthermore, in the MADAP, activated caspases 9 and 3 rapidly translocate to the mitochondria, perhaps helping the systematic dismantling of the organelle (10). Here, we continue to utilize these apoptotic models (9, 10) and show that active caspase 8 also becomes associated with the mitochondria in response to multiple apoptotic stimuli. By focusing on MDA-MB231 breast cancer cells treated with genotoxic drug etoposide (i.e., VP16), we further characterize the origin, localization, and potential biological functions of the membrane-associated active caspase 8.
MATERIALS AND METHODS
Cells and reagents.
GM701 human fibroblasts, MDA-MB231 (breast), PC3, Du145, and LNCaP (prostate) cancer cells were maintained as described previously (9, 10). Jurkat T leukemic cells, FADD-deficient and caspase 8-deficient Jurkat T cells, B lymphoid cells, BJAB-Mock and BJAB FADD-DN (BJAB cells stably transfected with dominant-negative [DN] FADD) were cultured as described previously (60) in RPMI 1640 medium supplemented with 10% fetal bovine serum. Du145 cells stably transfected with ER-targeted enhanced yellow fluorescent protein (EYFP) (Du145-ER-EYFP) were created in P. Daniel's lab and cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. The primary antibodies used were as follows: rabbit polyclonal anti-caspase 3 (Biomol, Plymouth Meeting, Pa.); monoclonal anti-caspase 6 and 7, -cytochrome c, -FADD, -TRADD, and -Na+/K+-ATPase and rabbit polyclonal anti-caspase 8 and -Apaf-1 (BD PharMingen, San Diego, Calif.); rabbit polyclonal anti-Bak, -caspase 10, and -Fas and goat polyclonal anti-lamin A/B (Santa Cruz Biotechnology, Santa Cruz, Calif.); rabbit polyclonal anti-caspase 9 and goat polyclonal anti-lactate dehydrogenase (anti-LDH) (Chemicon, Temecula, Calif.); monoclonal anti-caspase 8 (Oncogene Research Products, San Diego, Calif.); monoclonal anti-COX II (cytochrome c oxidase subunit II) and -COX IV (Molecular Probes, Eugene, Oreg.); rabbit polyclonal anti-poly(ADP-ribose) polymerase (Roche, Indianapolis, Ind.); rabbit polyclonal anti-voltage-dependent anion channel (anti-VDAC) (Calbiochem, San Diego, Calif.); monoclonal antiactin (ICN, Aurora, Ohio). Rabbit polyclonal anti-BAP31 and anti-caspase 8L and Bid antibodies were courtesies of G. Shore and X. Wang, respectively. All secondary antibodies, i.e., goat, donkey, or sheep anti-mouse, -rabbit, or -goat immunoglobulin G (IgG) conjugated to horseradish peroxidase, fluorescein isothiocyanate, or rhodamine, biotinylated goat anti-mouse antibodies, and enhanced chemiluminescence reagents were acquired from Amersham Biosciences (Piscataway, N.J.). Streptavidin conjugated to Alexa Fluor 594 or 488 and mitochondrial (i.e., MitoTracker) and ER (brefeldin A, BODIPY 558/568 conjugates) dyes were purchased from Molecular Probes. Fluorogenic caspase substrates DEVD-7-amino-4-trifluoromethyl-coumarin (AFC), IETD-AFC, and LEHD-AFC; general caspase inhibitor z-VAD-fmk; caspase 3/7 inhibitor z-DEVD-fmk; and recombinant active caspases 8 (p18/p11), 9 (p35/p12), and 3 (p17/p12) were bought from Biomol. The caspase 8 inhibitor z-IETD-fmk was purchased from BD PharMingen. Ru360 and thapsigargin (TG) were obtained from Calbiochem. All other chemicals were purchased from Sigma (St. Louis, Mo.) unless specified otherwise.
Subcellular fractionation and Western blotting.
The basic procedure was described previously (2, 9, 10, 23). Briefly, cells were treated with various chemicals, inhibitors, or vehicle (ethanol or dimethyl sulfoxide) as a control. At the end of treatment, cells were harvested, washed twice in ice-cold phosphate-buffered saline, and resuspended in 600 μl of homogenizing buffer (20 mM HEPES-KOH [pH 7.5], 10 mM KCl, 1.5 mM MgCl2, 1 mM sodium EDTA, 1 mM sodium EGTA, and 1 mM dithiothreitol [DTT]) containing 250 mM sucrose and protease inhibitors. After a 30-min incubation on ice, cells were homogenized in the same buffer with a glass Pyrex homogenizer (type A pestle, 140 strokes). Unbroken cells, large plasma membrane pieces, and nuclei were removed by centrifugation at 1,000 × g for 5 min at 4°C. The resulting supernatant was centrifuged at 10,000 × g at 4°C for 20 min to obtain the pellet, which we designated the membrane fraction or membranes (2, 16). The remaining supernatant was subjected to centrifugation at 100,000 × g for 1 h at 4°C to obtain the supernatant (i.e., S100) and the pellet, which we designated the cytosol and microsomes, respectively (2, 16). In some experiments, we also utilized the 10,000 × g supernatant, i.e., the 10k supernatant. The membrane fractions or microsomes were washed (three times) in homogenizing buffer and then solubilized in TNC buffer (10 mM Tris-acetate [pH 8.0], 0.5% NP-40, 5 mM CaCl2) containing protease inhibitors. The protein concentration was determined by using the Micro-BCA kit (Pierce, Rockford, Ill.).
For Western blotting, various amounts of membrane or cytosolic proteins were loaded on a sodium dodecyl sulfate-15% polyacrylamide gel. After gel electrophoresis and protein transfer, the membrane was sequentially probed, stripped, and then reprobed for various molecules, which are indicated in Results.
Preparation of mitochondria by sucrose gradient purification.
The crude mitochondrial (i.e., 10,000 × g) pellet obtained from differential centrifugation was washed (three times) with homogenizing buffer and then suspended in the same buffer. This mitochondrial suspension was layered on top of a discontinuous sucrose gradient consisting of 20 ml of 1.2 M sucrose, 10 mM HEPES-NaOH, and 1 mM EDTA (pH 7.5) and 17 ml of 1.6 M sucrose, 10 mM HEPES-NaOH, and 1 mM EDTA (pH 7.5). The sample was centrifuged (Beckman XL-90 ultracentrifuge) at 27,000 rpm for 2 h at 4°C. The mitochondrion-enriched organelles were recovered at the interface of 1.2 and 1.6 M sucrose and washed in homogenizing buffer (43). The final pellet was solubilized in TNC buffer.
Quantification of apoptosis, IETDase, LEHDase, and DEVDase activity measurement.
Apoptotic nuclei were scored under a fluorescence microscope upon staining with 4′,6′-diamidino-2-phenylindole (DAPI) (10, 57). For caspase activity measurement, cells were washed twice in phosphate-buffered saline and the whole-cell lysate was made in the lysis buffer {50 mM HEPES, 1% Triton X-100, 0.1% 3-[(cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 1 mM DTT, and 0.1 mM EDTA}. Then, 100 μg of whole-cell lysate, membrane, or cytosol was added to a reaction mixture containing 50 μM fluorogenic peptide substrates (i.e., acetyl-IETD-AFC [Ac-IETD-AFC], Ac-DEVD-AFC, or Ac-LEHD-AFC), 50 mM HEPES (pH 7.4), 10% glycerol, 0.1% CHAPS, 100 mM NaC1, 1 mM EDTA, and 10 mM DTT, in a total volume of 100 μl and incubated at 37°C for 1 h. Production of AFC was monitored in a spectrofluorimeter with an excitation wavelength of 400 nm and an emission wavelength of 505 nm. The results were presented as activation versus that of the control (9, 10).
Fluorescence microscopy.
Immunofluorescent staining was performed as previously described (9, 10). Various cells, either treated or untreated, were fixed (4% paraformaldehyde), permeabilized (1% Triton X-100), and then stained with a monoclonal antibody to caspase 8 or control antibody MOPC 21. Coverslips were then incubated with biotinylated goat anti-mouse IgG (1:500) and then with streptavidin-conjugated Alexa Fluor 488 (1:500). In some experiments, cells were double stained for caspase 8 and mitochondria (with MitoTracker Orange CMTMRos [25 nM]) or ER (with either brefeldin A-BODIPY 558/568 or anti-BAP31 antibody). When anti-BAP31 antibody was used for double staining, cells were first stained for caspase 8 as described above. Cells were then postblocked in 10% goat whole serum for 30 min at 37°C, incubated with anti-BAP31 antibody for 2 h at 37°C, and finally incubated with goat anti-rabbit IgG conjugated to Alexa Fluor 594 (1:500) for 1 h at 37°C. For BAP31 and ER colocalization, either Du145-ER-EYFP cells were used directly in BAP31 staining or Du145 or MDA-MB231 cells were first transfected with the ER-EYFP plasmid and then stained for BAP31. In some cases, mitochondria and ER were also stained live with organelle-specific dyes for 15 min. In all cases, cell nuclei were labeled with DAPI (9, 10). Stained slides were observed on a BX40 Olympus epifluorescence microscope. Images were captured with MagnaFire software and processed with Adobe Photoshop.
Transient transfection of DN caspase 8, caspase 9, FADD, or TRADD plasmid or Nex-enhanced green fluorescent protein (Nex-EGFP).
MDA-MB231 cells, plated 1 day earlier on 10-cm-diameter culture dishes to achieve 50 to 60% confluence, were transfected with 5 μg of various vectors by using FuGENE 6 (10). Twenty-four hours after transfection, cells were treated with VP16 for up to 48 h. At the end of treatment, cells were scored for apoptosis (600 to 700 cells were counted for each condition) or harvested for Western blotting of caspase 3 or for activity measurements.
siRNA downregulation of caspase 8.
Caspase 8 small interfering RNAs (siRNAs) (target 1, 5′-AAAGGGAACTTCAGACACCAG-3′; target 2, 5′-AAAAGCAAACCTCGGGGATAC-3′) and scrambled siRNAs were synthesized by using the Silencer siRNA construction kit (Ambion, Austin, Tex.) according to the manufacturer's instructions. LNCaP cells, plated 1 day earlier on six-well culture dishes to achieve 50 to 60% confluence, were transfected with siRNAs (50 nM) by using siPORT lipid (Ambion). Twenty-four hours after transfection, cells were treated with VP16 for up to 48 h. Samples were analyzed for apoptosis or caspase 3 cleavage.
Proteinase K digestion of membrane-bound organelles.
The basic experimental procedure was described previously (10). Briefly, freshly isolated membranes (100 μg) were incubated in homogenizing buffer (without protease inhibitors) alone or in the presence of proteinase K (0.1 μg/ml) only or proteinase K plus Triton X-100 (1% final concentration). After a 10-min incubation on ice, 2 μl of phenylmethylsulfonyl fluoride (100 mM) was added to terminate proteolysis followed by the addition of 6× loading buffer. Samples were boiled for 5 min and analyzed by Western blotting.
High-salt or alkali extraction of membranes.
The membrane pellet (100 μg) was suspended in either 0.5 M NaCl or 0.1 M Na2CO3 (pH 11.5) and incubated on ice for 20 min (10). At the end of the incubation, the sample was centrifuged at 100,000 × g for 1 h. The resulting pellet was then lysed in TNC buffer. Both the supernatant and the pellet were then subjected to Western blotting.
Cell-free reconstitution experiments.
The basic experimental procedure was described previously (10). All cell-free reactions were performed in homogenizing buffer in a total volume of 100 μl. Freshly isolated cytosol or membranes were incubated at 37°C for 4 h with recombinant active caspase 8, 9, or 3 (at a final concentration of 1 μM) in the presence or absence of individual caspase inhibitors. In some cases, either NP-40 or TNC buffer was used to lyse the mitochondria. At the end, samples containing coincubated mitochondria were centrifuged at 10,000 × g for 20 min to obtain the membrane pellet. Following two washes in homogenizing buffer, the mitochondrial pellet, together with the supernatant, was subjected to Western blotting. To determine whether the mitochondrially localized active caspase 8 can cleave or activate caspases or BAP31, purified cytosol, the 10,000 × g (i.e., 10k) supernatant, or the mitochondria (lysed or unlysed) from VP16 (10 μM)-treated or untreated cells were coincubated in various combinations for 4 h at 37°C. After coincubation, the pelleted mitochondria were washed twice with homogenizing buffer and then equal amounts of proteins were used in Western blotting.
RESULTS
Active caspase 8 is detected exclusively in the mitochondrion-ER-enriched membrane fraction in MDA-MB231 cells treated with etoposide (VP16).
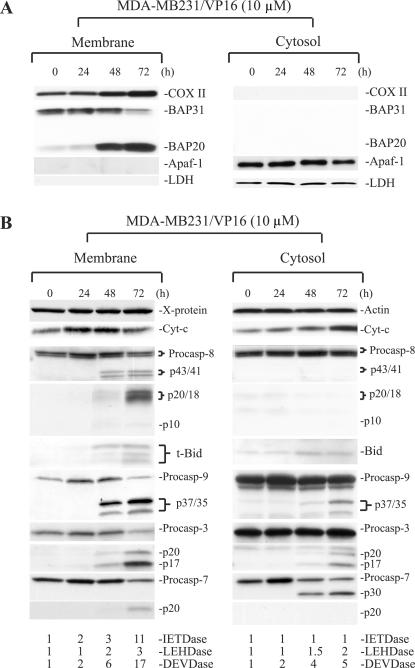
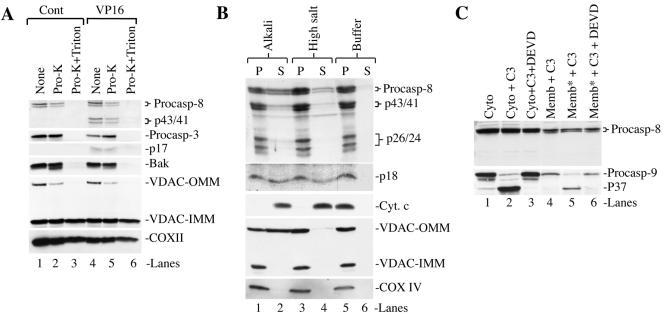
To determine the subcellular localizations of active caspase 8, we utilized, as a model, MDA-MB231 breast cancer cells treated with the DNA-damaging agent VP16. We first characterized the qualities of the cytosolic and membrane fractions. As shown in Fig. 1A, COX II, a mitochondrial inner membrane protein, was detected only in the membrane fraction, whereas LDH, a cytosolic glycolytic enzyme, was detected only in the cytosol. Another cytosolic protein, Apaf-1, was also detected only in the cytosol, as previously observed (10). To further characterize the membrane fraction, we reprobed the blot for nuclear lamins A and B, plasma membrane-specific Na+/K+-ATPase, or ER protein BAP31, a 28-kDa outer membrane integral protein (7, 40). The results revealed the presence of BAP31 and its cleavage product, BAP20, only in the membrane fraction (Fig. 1A) (note that the anti-BAP31 antibody frequently detected a lower band representing the p27 BAP29 protein) (7). Nuclear lamins were not detected in either fraction (data not shown), consistent with previous observations (2). Na+/K+-ATPase was also not detected in appreciable amounts in either fraction (data not shown). These results suggest that (i) the membrane and cytosolic fractions do not have cross-contaminations, (ii) the membrane fraction contains both mitochondria and ER, and (iii) the membrane fraction contains no significant nuclear or plasma membranes.
FIG. 1.
Association of active caspase 8 exclusively with the membrane fraction. (A) MDA-MB231 cells treated with VP16 for the time intervals indicated were harvested to prepare the membrane (i.e., the 10,000 × g pellet) and cytosolic (i.e., S100) fractions, which were used in Western blotting for the molecules indicated. In these experiments, 35 μg of cytosol or 60 μg of membrane was loaded into each lane. Procaspases and their corresponding cleavage products were shown. Data are representative of the results from three to four repeat experiments. In panel B, cell fractionation and Western blotting were performed as described for panel A. Cytosol and membrane fractions were also used to measure caspase 9 (LEHDase), caspase 3/7 (DEVDase), or caspase 8 (IETDase) activity. Data are representative of the results from two to three repeat experiments. The X protein, which was detected by the anti-cytochrome c antibody (9, 10), was used as a loading control for the membrane fraction. Cyt-c, cytochrome c. Note that in this and all of the following figures, procaspase 8 (Procasp-8) was detected by using the polyclonal anti-caspase 8 antibody, whereas the p20 and p18 bands were detected by using the monoclonal anti-caspase 8 antibody.
MDA-MB231 cells stimulated with VP16 undergo mitochondrial activation-dependent apoptosis characterized by an early up-regulation of MRC as well as mitochondrially localized non-MRC proteins (9). Indeed, COX II was up-regulated in VP16-treated MDA-MB231 cells (Fig. 1A). In addition to COX II (Fig. 1A), treated MDA-MB231 cells also showed increased levels of cytochrome c, which was up-regulated at 24 h, at which time increased protein levels were seen in both the mitochondrial and cytosolic fractions (Fig. 1B), as observed with other MADAP models (9). Together, these results suggest that, as in other apoptotic systems involving early mitochondrial activation (9), VP16-induced MDA-MB231 cell death is accompanied by an early up-regulation of MRC proteins such as cytochrome c and COX II.
Later during apoptosis, active caspase 9 (p37/35; the p35 band is derived from the apoptosome activation, whereas the p37 band is derived from caspase 3 cleavage) and caspase 3 (p20 and p17) fragments and activities were detected in both the cytosolic and membrane fractions at 48 h posttreatment (Fig. 1B) when obvious poly(ADP-ribose) polymerase cleavage was observed and ∼60% of the cells became apoptotic (9). Procaspase 7 was also localized in both the membrane and cytosolic fractions, which became activated in response to VP16. Interestingly, the intermediate cleavage product, p30, was detected only in the cytosol, whereas the active cleavage product, p20, was detected only in the membranes (Fig. 1B). Consistent with caspase activation, VP16-induced apoptosis of MDA-MB231 cells was inhibited by the general caspase inhibitor z-VAD (data not shown). In contrast to procaspases 9, 3, and 7, procaspase 6 was not cleaved (data not shown).
Strikingly different from active caspases 9, 3, and 7, active caspase 8 fragments and caspase 8 activity were detected only in the membrane fraction (Fig. 1B). The procaspase 8 gene is transcribed as two main isoforms, i.e., p55/53 procaspase 8a and procaspase 8b (48). Upon activation, the initial cleavage at Asp374 generates two cleavage intermediates, p43/p41 and p12, with the latter being rapidly converted to p10 (36, 48). Afterwards, an additional cleavage at Asp261 produces the active enzyme subunit p20/p18 and the inactive p26/p24 prodomain (36, 48). In VP16-treated MDA-MB231 cells, although the p55/p53 procaspase 8 bands were detected in both the cytosol and the membranes, p43/41 and p20/p18 were observed only in the membrane fraction (Fig. 1B). Supporting the Western blotting data, substrate cleavage assays confirmed the presence of caspase 8 (the IETDase) activity only in the membrane fraction, in contrast to caspase 9 (the LEHDase) or caspase 3/7 (the DEVDase) activities, which were detected in both compartments (Fig. 1B). Interestingly, at 72 h, all 4 procaspases in the membrane fraction were decreased with a corresponding increase in their cleavage products and enzymatic activities (Fig. 1B), raising the possibility that the membrane-associated procaspases might be activated in situ.
One of the main substrates of caspase 8 is ∼22-kDa Bid, which is cleaved to ∼15-kDa t-Bid, which translocates to the mitochondria to promote cytochrome c release and amplify caspase 8-mediated apoptosis. In addition to caspase 8, other caspases (such as caspase 3) and noncaspase proteases (e.g., calpain and granzyme B) can also cleave Bid to generate a series of ≤15-kDa truncated products (8, 13, 15, 33, 51, 65). In VP16-treated MDA-MB231 cells, low levels of 15-kDa t-Bid and smaller degradation products were seen in the membrane fraction around 48 h posttreatment, which became more obvious by 72 h (Fig. 1B).
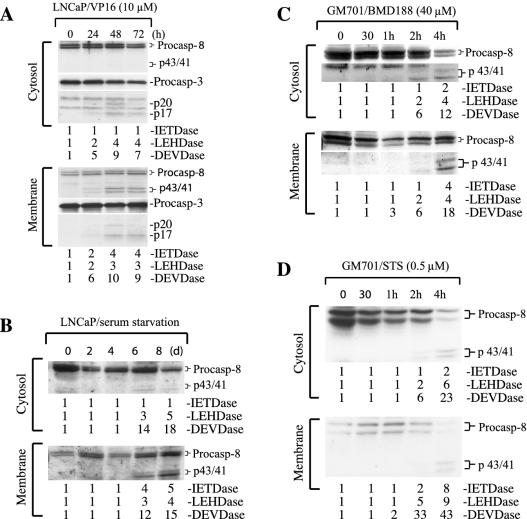
To determine whether association of active caspase 8 with the mitochondrion-ER-enriched membrane fraction is unique to MDA-MB231 cells, we treated LNCaP prostate cancer cells with VP16. The results similarly revealed a predominant association of active caspase 8 with the membrane fraction, although caspase 9 and caspase 3 activities were detected in both the cytosol and membrane fractions (Fig. 2A). To determine whether caspase 8 association with the membranes is a more general phenomenon, we studied its activation and distribution in LNCaP cells subjected to serum starvation or GM701 fibroblasts treated with BMD188, a hydroxamic compound that requires the MRC function for its apoptotic activity (23), or with staurosporine (STS), a promiscuous protein kinase inhibitor. As shown in Fig. 2B to D, caspase 8 cleavage and activity were detected in the membrane fractions in all three apoptosis systems. In starved LNCaP cells, caspase 8 activity was observed mainly in the membrane fraction, whereas caspase 9 and caspase 3 activities were seen in both fractions (Fig. 2B). By contrast, in GM701 cells treated with BMD188 or STS, caspase 8 activity was detected in both the cytosol and the membranes but only after activation of caspases 9 and 3 (Fig. 2C and D).
FIG. 2.
Association of active caspase 8 with the membrane fraction in multiple apoptotic systems. (A) LNCaP cells were treated with VP16 for the time intervals indicated. At the end of treatment, cells were harvested, fractionated, and used in Western blotting and activity measurement as described in the legend to Fig. 1. (B to D) Cytosol and membrane fractions prepared from LNCaP cells subjected to serum starvation (B) or GM701 fibroblasts treated with BMD188 (C) or STS (D) were used to measure caspase activities or in Western blotting of caspase 8. Data are representative of the results from two to three repeat experiments. Procasp, procaspase.
Caspase 8 localizes primarily in the mitochondria.
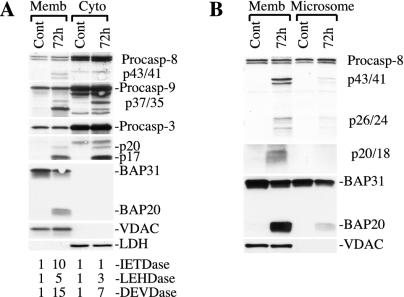
The above results suggest that association of active caspase 8 with the mitochondrion-ER-enriched membranes is a common phenomenon in the apoptotic process. Next, we sought to clarify where active caspase 8 might be localized, the mitochondria, ER, or both, and where in these organelles. As demonstrated in Fig. 1A, the membranes prepared with the differential centrifugation method, an approach widely used by investigators (for examples, see references 8, 26, 29, 31, 45, 49, 51, 53, 54, 64, and 65) to obtain the mitochondrial or mitochondrion-enriched fraction, contained both mitochondria and ER. Therefore, we first attempted to separate the two organelles by density gradient centrifugation, a strategy utilized previously (10, 16, 42, 56) to prepare highly enriched mitochondria. As shown in Fig. 3A, active caspase 8 was again only detected in the membrane fraction, whereas active caspases 9 and 3 were detected in both fractions. As expected, the mitochondrion-specific protein VDAC was detected only in the membrane fraction, whereas LDH was detected only in the cytosol (Fig. 3A). Nevertheless, BAP31 was still detected in the membrane (Fig. 3A). These results suggest that the gradient centrifugation approach failed to separate the mitochondria from the ER.
FIG. 3.
Association of active caspase 8 with the mitochondrion-ER-enriched membranes. (A) Cytosol (Cyto) or sucrose gradient-purified membranes (Memb) prepared from control (Cont) or VP16-treated (10 μM for 72 h) MDA-MB231 cells were used in Western blotting (60 μg/lane) for the molecules indicated or in caspase activity measurements. (B) The membrane or microsome fraction was used in Western blotting (60 μg/lane) for the molecules indicated. Data are representative of the results from at least two repeat experiments. Procasp, procaspase.
In an alternative approach, we prepared the microsomal fraction (i.e., the 100,000 × g pellet), which is highly enriched with ER but largely devoid of mitochondria (2, 16), and examined caspase 8 association in this preparation. As shown in Fig. 3B, the ER-enriched microsomes had little mitochondria, as evidenced by a minimal amount of VDAC. Compared to the mitochondrion-ER-enriched membrane, which contained abundant active caspase 8 (evidenced by the presence of p20/p18), the microsomal fraction contained barely detectable caspase 8 cleavage products, although it possessed significant amounts of procaspase 8 (Fig. 3B). These results suggest that most active caspase 8 in VP16-stimulated MDA-MB231 cells might localize in the mitochondria. Interestingly, BAP31, an ER-specific protein, was detected in both mitochondrion-ER-enriched membranes and ER-enriched microsomes (Fig. 3B), suggesting that a significant percentage of ER intimately interacts and thus cofractionates with the mitochondria. However, the proapoptotic cleavage product of BAP31, BAP20 (7), was detected mostly in the mitochondrion-ER membrane fraction (Fig. 3B).
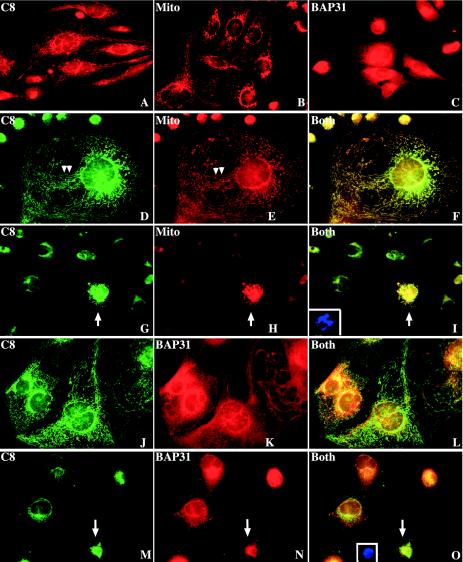
To further elucidate the subcellular localization of caspase 8, we carried out immunofluorescence labeling. To avoid any potential cross-interference from different fluorophores, we first performed a single-labeling experiment. In untreated MDA-MB231 cells, procaspase 8 was present in the cytoplasm and, more prominently, in distinct perinuclear networks composed of rod-shaped, worm-like organelles (Fig. 4A), which resembled mitochondria, as identified by MitoTracker labeling (Fig. 4B). This staining pattern was distinct from that of ER, which was identified by staining with either brefeldin A-BODIPY 558/568 conjugates (data not shown) or anti-BAP31 antibody (Fig. 4C). BAP31 staining showed typical perinuclear ER networks (Fig. 4C) and good colocalization with the ER-targeted EYFP (data not shown). Indeed, double-labeling experiments revealed very good colocalization of caspase 8 and the mitochondria in untreated MDA-MB231 cells (Fig. 4D to F). In VP16-treated apoptotic cells, caspase 8 labeling became more intense and still colocalized with the fragmented mitochondria (Fig. 4G to I). In contrast, caspase 8 did not show obvious colocalization with the BAP31-labeled ER in either control (Fig. 4J to L) or VP16-treated (Fig. 4M to O) MDA-MB231 cells. In apoptotic cells, caspase 8 labeling was still different from the ER staining and the two did not significantly overlap (Fig. 4M to O).
FIG. 4.
Caspase 8 localizes in the mitochondria in MDA-MB231 cells. (A to C) Individual staining of caspase 8 (C8) with the monoclonal anti-caspase 8 antibody (A), mitochondria (Mito) with Mitotracker Red CMXRos (B), or ER (BAP31) with the rabbit anti-BAP31 antibody (C). (D to O) Untreated MDA-MB231 cells (D to F and J to L) or cells treated with VP16 (10 μM for 48 h) (G to I and M to O) were double-labeled for caspase 8 (D, G, J, and M) and mitochondria (E and H) or ER (K and N). The micrographs in panels F, I, L, and O are respective overlays (Both). The insets in panels I and O depict apoptotic nuclei for the cells marked by the arrows. The arrowheads in panels D and E illustrate obvious distribution patterns of caspase 8, mitochondria, and ER along cytoskeletons. Original magnification, ×400. Note that, in panels D to F and J to L, several well-spread cells are shown to clearly reveal the colocalizations. The micrographs shown are representative of >500 images analyzed.
Collectively, the fluorescence microscopy and the biochemical data suggest that, in MDA-MB231 cells, procaspase 8 is distributed in both the cytosol and the mitochondria and active caspase 8 is localized mainly in the mitochondria. Similar mitochondrial localization of caspase 8 was also observed by fluorescence microscopy in several other systems, including starved LNCaP cells and BMD188-treated PC3 cells (data not shown).
Caspase 8 localizes mainly on the OMM as integral proteins.
Subsequently, we explored the submitochondrial localization of caspase 8 in MDA-MB231 cells by using two approaches. In the first, we treated the mitochondrion-enriched membranes from control or VP16-stimulated MDA-MB231 cells with proteinase K. As shown in Fig. 5A, both procaspase 3 and the p17 active band were not affected (lanes 1 and 2 versus lanes 4 and 5). However, when the membrane fraction was treated with proteinase K in the presence of 1% Triton X-100 (to solubilize the membranes), both bands were completely degraded (Fig. 5A, lanes 3 and 6). These results were consistent with earlier observations (10) that the membrane-associated procaspase 3 and activated caspase 3 are localized in the IMS of the mitochondria. The OMM protein Bak was partially degraded by proteinase K alone and completely degraded by proteinase K plus Triton X-100 (Fig. 5A). VDAC, which normally concentrates in the contact sites and spans both the inner mitochondrial membrane (IMM) and the OMM (12), was detected as two distinct bands in MDA-MB231 cells (Fig. 5A). The upper band, VDAC-OMM, represented the VDAC oligomers on the OMM, behaved like the OMM-localized Bak and was partially degraded by proteinase K alone and completely degraded by proteinase K plus Triton X-100 (Fig. 5A). The lower band, VDAC-IMM, represented the VDAC monomer on the IMM, behaved like the IMM-localized COX II and was completely resistant to proteinase K digestion, even in the presence of Triton X-100 (Fig. 5A). The degradation pattern of caspase 8 (both procaspase and cleaved forms) was similar to those of Bak and VDAC-OMM, i.e., partial degradation by proteinase K alone and complete degradation by proteinase K plus Triton X-100. Triton X-100 (1%) alone did not degrade any of these proteins (data not shown).
FIG. 5.
Caspase 8 is localized mainly on the OMM and is not activated by caspase 3. (A) Both procaspase 8 (Procasp-8) and active caspase 8 are localized on the OMM. The membrane fractions prepared from control (Cont) or VP16 (10 μM)-stimulated MDA-MB231 cells were treated with proteinase K (pro-K) alone or proteinase K plus Triton X-100 and then used in Western blotting for the molecules indicated. (B) Freshly purified mitochondrial membranes from VP16-treated MDA-MB231 cells (10 μM for 72 h) were subjected to either alkali treatment or high-salt extraction. At the end, samples were centrifuged to collect pellets (P) or supernatants (S) and used in Western blotting for the molecules indicated. Cyt. c, cytochrome c. (C) Active caspase 3 does not activate caspase 8 in MDA-MB231 cells. One hundred micrograms of cytosol (Cyto) or membranes (Memb), which were either unlysed or lysed with NP-40 (1% final concentration; indicated by an asterisk), prepared from untreated MDA-MB231 cells was incubated with or without recombinant active caspase 3 (C3) (1 μM) in a total volume of 100 μl in the absence or presence of DEVD-CHO (10 μM; 37°C for 4 h). At the end of the incubation, samples (40 μl each) were used in Western blotting for caspase 8 or caspase 9. Data are representative of the results from two to three repeat experiments.
In the second approach, we subjected the mitochondrial membranes to alkali and high-salt extraction, which would strip off some of the proteins on the OMM and also cause the release of the IMS proteins (10). As shown in Fig. 5B, the OMM-integral protein VDAC-OMM was partially extracted, in particular, by alkali extraction, whereas the VDAC-IMM, like the COX IV protein localized on the IMM, was not affected by either treatment (lanes 2 and 4). By contrast, the IMS protein cytochrome c was completely released by either extraction method (Fig. 5B). Under the same conditions, some procaspase 8 and active caspase 8 (i.e., p18) behaved like the VDAC-OMM and were partially extracted (Fig. 5B, lanes 2 and 4 versus lane 6), whereas intermediate fragments, i.e., p43/41 and p26/24, were mostly associated with the membranes (lanes 1 to 6).
Together, the results from these two sets of experiments suggest that, in MDA-MB231 cells, procaspase 8 and its cleaved products, in particular, p18, appear to mostly reside on the OMM as integral proteins as evidenced by their protease sensitivity and extraction patterns similar to those of OMM-integral proteins such as Bak and VDAC-OMM. On the other hand, it is possible that a small fraction of procaspase 8 and active caspase 8 may also be localized in the IMS based on their partial release by alkali extraction.
Caspase 8 was not activated by caspase 3 in MDA-MB231 cells.
Next, we investigated the potential origin of mitochondrially localized active caspase 8. We adopted a previously described approach (10) and utilized recombinant active caspases 3 and 9 to address whether they can cleave the membrane-associated or, as a control, cytosolic procaspase 8. As shown in Fig. 5C (lanes 1 to 3), exogenous caspase 3 readily cleaved the cytosolic procaspase 9 to generate the p37 fragment, which was inhibited by DEVD. Active caspase 3 also cleaved the membrane-associated caspase 9 but only when the organelles were lysed (Fig. 5C, lanes 4 to 6), consistent with previous observations that the membrane-associated caspase 9 (as well as caspase 3) is localized in the IMS of the mitochondria (10). The caspase 3-mediated cleavage of IMS-localized procaspase 9 was similarly inhibited by DEVD (Fig. 5C, lane 6). Recombinant active caspase 3 also led to processing and activation of endogenous caspase 3 (data not shown) (10). Surprisingly, recombinant active caspase 3 failed to process procaspase 8 in either cytosol or mitochondria (Fig. 5C). Similarly, recombinant active caspase 9 failed to process procaspase 8, although it readily activated caspase 3 (data not shown). These results altogether suggest that, in MDA-MB231 cells, both cytosolic and membrane-bound procaspase 8 are not appreciably cleaved by caspase 3.
Procaspase 8 is activated through FADD/TRADD-dependent mechanisms, and active caspase 8 plays a causal role in VP16-induced cell death.
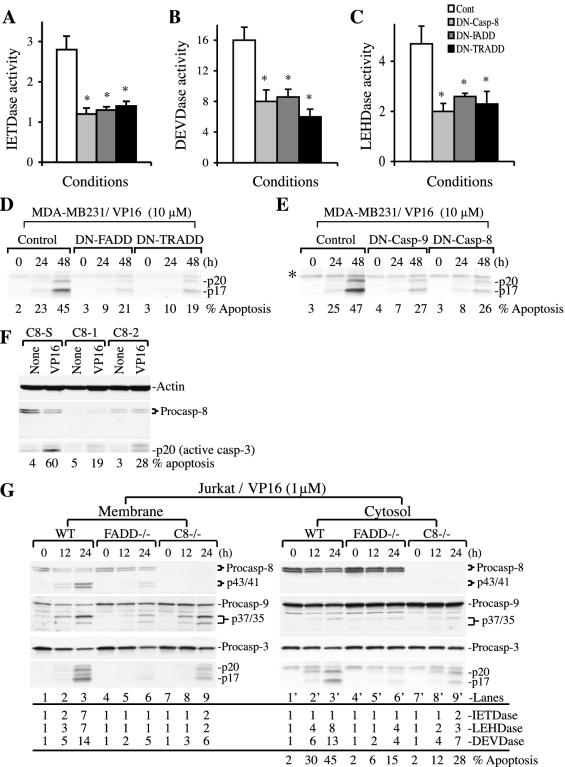
To determine whether caspase 8 activation in VP16-stimulated MDA-MB231 cells involves adaptor proteins, we transfected cells with DN constructs to TRADD or FADD (20). As shown in Fig. 6A, DN-FADD and DN-TRADD significantly inhibited caspase 8 activity, suggesting that VP16 treatment led to caspase 8 activation through FADD/TRADD-dependent mechanisms. Note that in these experiments, caspase activities, which were measured at 48 h post-VP16 treatment, were similar to those in preceding experiments (e.g., the results shown in Fig. 1B). DN-FADD and DN-TRADD also inhibited caspase 9 and caspase 3 activation as well as apoptosis (Fig. 6B to D), suggesting that FADD/TRADD-activated caspase 8 may play a causal role in activating caspase 9 and caspase 3 and in VP16-induced MDA-MB231 cell apoptosis. To further explore this point, we transfected cells with DN caspase 8 (6) or, as a control, DN caspase 9 (52) prior to VP16 treatment. The results revealed that DN caspase 8 inhibited not only caspase 8 activity (Fig. 6A) but also caspase 9 (Fig. 6C) and caspase 3 (Fig. 6B and E) activation and cell death (Fig. 6E), thus substantiating the idea that caspase 8 activation contributes to VP16-induced caspase 3 activation and MDA-MB231 cell death. As expected, DN caspase 9 also inhibited caspase 3 activation and cell death (Fig. 6E).
FIG. 6.
FADD/TRADD-dependent activation of caspase 8 and a critical role for caspase 8 in VP16-induced MDA-MB231 cell death. (A to C) DN-FADD and DN-TRADD inhibit caspase activation. Log-phase MDA-MB231 cells transfected with either DN-FADD or DN-TRADD were treated with VP16 (10 μM for 48 h). Whole-cell lysates were used to measure caspase activities as detailed in Materials and Methods. The transfection efficiency was 20 to 25%, as revealed by transfection of green fluorescent protein vectors. Data represent means ± standard deviations. *, P < 0.05 (Student's t test). (D) DN-FADD and DN-TRADD inhibit VP16-induced caspase 3 activation and apoptosis. Log-phase MDA-MB231 cells were transfected with DN-FADD or DN-TRADD. Twenty-four hours later, cells were treated with VP16 for 48 h. At the end, whole-cell lysates (60 μg/lane) were used in Western blotting of caspase 3. Apoptosis was quantified by DAPI staining. (E) DN caspase 9 or DN caspase 8 inhibits VP16-induced caspase 3 activation and apoptosis. Whole-cell lysates (60 μg/lane) prepared from MDA-MB231 cells treated for various conditions (as for panel D) were used in Western blotting of caspase 3. Apoptosis was quantified by DAPI staining. The asterisk indicates a nonspecific band. (F) LNCaP cells transfected with two siRNAs for caspase 8 (C8-1 and C8-2) or scrambled siRNA (C8-S). Twenty four hours after transfection, cells were treated with VP16 (10 μM for 48 h). At the end of treatment, apoptosis was quantified and whole-cell lysates (60 μg/lane) were used for Western blotting. (G) Wild-type, FADD−/−, or caspase 8−/− Jurkat cells treated with VP16 (1 μM) for 12 or 24 h were used to prepare membrane and cytosol fractions. Equal amounts of proteins (60 μg/lane) were used in Western blotting for the molecules indicated. Caspase activities were also determined by utilizing 50 μg of cytosol and membrane fractions. The percentages of apoptosis were determined as described in Materials and Methods. Cont, control; Procasp, procaspase; WT, wild type.
To provide further support for caspase 8 involvement, we pretreated MDA-MB231 cells with caspase 8 inhibitor, IETD-fmk, prior to VP16 stimulation. Concentrations of IETD as low as 4 μM reduced both membrane-associated caspase 8 activation and Bid cleavage and inhibited the IETDase activity, and IETD at 30 μM demonstrated stronger inhibitory effects (data not shown). IETD also significantly inhibited the activation of both caspases 9 and 3 (data not shown), consistent with the results obtained with the DN constructs. Finally, we designed two caspase 8 siRNAs and a scrambled control siRNA. When these siRNAs were transfected into LNCaP cells, caspase 8 siRNAs but not the scrambled siRNA downregulated caspase 8 (Fig. 6F). More importantly, downregulation of caspase 8 inhibited caspase 3 activation (i.e., p20) as well as apoptosis (Fig. 6F).
To provide direct evidence for a causal involvement of FADD and caspase 8 in VP16-induced cell death, we used Jurkat cells deficient for FADD and caspase 8. Like MDA-MB231 cells, treatment of Jurkat cells with VP16 (1 μM) resulted in association of active caspase 8 exclusively with the membrane fraction (Fig. 6G, lanes 1 to 3 versus lanes 1′ to 3′). VP16-induced caspase 8 cleavage, caspase activities, and cell death were significantly inhibited in FADD-deficient Jurkat cells compared to wild-type cells (Fig. 6G, lanes 5 and 6 versus lanes 2 and 3), thus confirming an essential role for FADD in VP16-induced caspase 8 activation. Caspase 3 activation in both the membrane fraction and the cytosol was also inhibited in FADD−/− cells (Fig. 6G), thus verifying a critical role for FADD in mediating caspase 3 activation upon VP16 treatment. Indeed, we also observed reduced caspase 3 activation in VP16-treated BJAB lymphoma cells stably expressing DN-FADD (data not shown). FADD deficiency also resulted in decreased caspase 9 cleavage products, mainly p37 (Fig. 6G, lane 6 versus lane 3), which results from caspase 3 activity, consistent with reduced caspase 3 activation in VP16-treated FADD−/− Jurkat cells.
Procaspase 8 and its cleaved products, as expected, were not detected in caspase 8-deficient Jurkat cells (Fig. 6G, lanes 7 to 9 and 7′ to 9′). Caspase 8 deficiency led to reduced caspase 3 activation in both mitochondria (Fig. 6G, lanes 7 to 9) and the cytosol (Fig. 6G, lanes 7′ to 9′), thus confirming a critical role for caspase 8 in VP16-induced caspase 3 activation. Interestingly, caspase 8 deficiency caused less inhibition of caspase 3 activation than FADD deficiency (Fig. 6G), suggesting that other FADD-activated caspases, e.g., caspase 10, may also be involved in activating caspase 3 in response to VP16 stimulation. Also, caspase 8 deficiency did not result in a significant reduction in the p37 active caspase 9 fragment (Fig. 6G, lanes 7 to 9 versus lanes 1 to 3), suggesting that the residual caspase 3 activity may be sufficient to cleave caspase 9.
Altogether, the results presented in Fig. 6 establish critical roles for caspase 8 and FADD in VP16-induced caspase 3 activation and cell death.
Biological functions of OMM-associated active caspase 8.
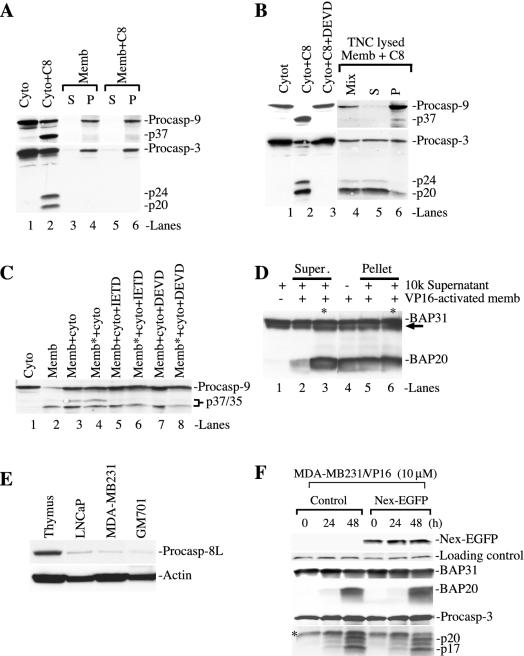
Next, we explored the potential biological functions of the membrane-associated caspase 8. The preceding experiments indicate that caspase 8 may participate in VP16-induced apoptosis by cleaving Bid (data not shown) and caspase 3 (Fig. 6). Using cell-free assays, we addressed, in a more direct manner, whether the OMM-localized active caspase 8 could degrade caspase 3 and perhaps some other substrates, e.g., BAP31. To set up the experimental system, we first determined the effect of recombinant active caspase 8. As shown in Fig. 7A and B, the recombinant active caspase 8 effectively degraded the procaspase 9 in the cytosol to generate the p37 band (Fig. 7A and B, lanes 2), which was inhibited by DEVD (Fig. 7B, lane 3), suggesting that the p37 caspase 9 band resulted from the caspase 8-activated caspase 3. Indeed, recombinant active caspase 8 robustly activated caspase 3 in the cytosol (Fig. 7A and B, lanes 2). Interestingly, DEVD also inhibited caspase 3 cleavage in the cytosol (Fig. 7B, lane 3), probably because DEVD-fmk at the concentration used (i.e., 10 μM) also inhibited caspase 8 activity (58). When similar experiments were carried out with unlysed membranes prepared from the untreated MDA-MB231 cells, recombinant active caspase 8 did not cleave either procaspase 3 or procaspase 9 (Fig. 7A, lanes 3 to 6), consistent with these latter enzymes being localized in the IMS of the mitochondria (10). Indeed, when the recombinant caspase 8 was incubated with the membranes lysed by TNC buffer containing NP-40, robust cleavage of caspase 3 was observed (Fig. 7B, lanes 4 to 6). Caspase 3 cleavage products fractionated preferentially in the TNC-solubilized supernatants (Fig. 7B, lane 5). There was also a low level of procaspase 9 processing to generate the p37 band, which fractionated preferentially in the pellet (Fig. 7B, lane 6). Collectively, these results indicate that (i) recombinant active caspase 8 can readily activate the cytosolic procaspase 3, (ii) activated caspase 3 in turn cleaves procaspase 9 to generate the p37 active caspase 9 that may in turn activate more procaspase 3, thus forming a positive feedback amplification loop, and (iii) the caspase 8 → caspase 3 → caspase 9 → caspase 3 activation cascade may also occur to the mitochondrially localized procaspase 9 and procaspase 3.
FIG. 7.
Active caspase 8 can degrade caspase 3 and BAP31. (A and B) Cell-free experiments with recombinant active caspase 8. In panel A, 100 μg of cytosol (Cyto) (lane 1 to 2) or membranes (Memb) (lanes 3 to 6) was incubated with recombinant active caspase 8 (C8) (2 μM) or buffer alone in a total volume of 100 μl for 4 h at 37°C. For lanes 3 to 6, samples were centrifuged at the end of incubation to obtain the supernatant (S) and pellet (P). The pellet was then resuspended in 100 μl of TNC buffer. All samples (40 μl/lane) were used in Western blotting for caspase 9 or 3. (B) The experiments were done as described for panel A except that, in lanes 4 to 6, the membranes were solubilized with TNC buffer containing NP-40. At the end of incubation, half of the sample was centrifuged to obtain the supernatant and pellet and the other half (Mix) was directly used in Western blotting. In lane 3, DEVD-fmk was used at 10 μM. (C) Cell-free experiments with the VP16-activated membrane. One hundred micrograms of membrane fraction from VP16-treated (10 μM for 48) MDA-MB231 cells, either unlysed or lysed (indicated by an asterisk), was incubated with the 100 μg of untreated cytosol in the absence or presence of 10 μM IETD-fmk or DEVD-fmk for 4 h at 37°C. At the end of incubation, equal amounts of proteins (40 μg/lane) were directly used in the Western blotting for caspase 9. (D) The membrane fraction isolated from the VP16-stimulated (10 μM for 72 h) MDA-MB231 cells cleaves BAP31. One hundred micrograms each of 10k supernatant and VP16-activated membrane fraction were either incubated alone or coincubated (4 h at 37°C). At the end of incubation, samples were centrifuged to obtain the supernatants (Super.) or pellet. Equal amounts of proteins (40 μg/lane) were then used in the Western blotting for BAP31. For lanes 3 and 6, NP-40 was added at a final concentration of 1% (indicated by an asterisk). The arrow indicates the p27 BAP protein. +, present; −, absent. (E) Low levels of caspase 8L expression in cultured human cells. Equal amounts of whole-cell lysates (100 μg/lane) prepared from mouse thymuses or from the cells indicated were used in Western blotting for caspase 8 with the polyclonal caspase 8L-specific antibody. The blot was reprobed for actin. (F) Caspase 8L is not involved in VP16-induced BAP31 cleavage or apoptosis. MDA-MB231 cells were either untransfected (control) or transfected with a DN caspase 8L construct (Nex-EGFP). Whole-cell lysates (60 μg/ml) were used in Western blotting for the molecules indicated. Nex-EGFP was detected by using a polyclonal anti-green fluorescent protein vector. The loading control is a nonspecific band detected by the anti-green fluorescent protein antibody. The asterisk indicates a nonspecific band detected by the anti-caspase 3 antibody. Procasp, procaspase.
Subsequently, we tested whether the active caspase 8 on the OMM could also degrade cytosolic procaspase 3. To this end, we prepared the mitochondria from the VP16-treated MDA-MB231 cells and coincubated them with the cytosol from untreated MDA-MB231 cells. As shown in Fig. 7C, the untreated cytosol had only procaspase 9 (lane 1) and the VP16-stimulated membrane fraction had an obvious p35 band and a faint caspase 3-derived p37 caspase 9 band (lane 2). Coincubation of the activated membrane with the cytosol resulted in a significantly increased p37 band (Fig. 7C, lane 3), which was inhibited by both IETD (Fig. 7C, lane 5) and DEVD (Fig. 7C, lane 7), suggesting that the enhanced p37 band resulted from caspase 8-activated caspase 3. Lysis of the membranes did not significantly increase the intensity of the p37 band (Fig. 7C, lane 4), consistent with active caspase 8 being localized mostly on the OMM. As expected, both IETD and DEVD inhibited the p37 caspase 9 in the lysed membranes coincubated with the cytosol (Fig. 7C, lanes 6 and 8).
Next, we coincubated the VP16-activated mitochondria, which contained active caspase 8 on the OMM, with the 10k supernatant that contained some ER, which normally would fractionate in the microsomes upon centrifugation at 100,000 × g (e.g., the results shown in Fig. 3B), and sought to determine whether BAP31, an ER-specific substrate of caspase 8 (6, 7, 40), could be cleaved. As shown in Fig. 7D, the 10k supernatant alone had only BAP31 (lane 1), whereas the VP16-activated membranes alone had both BAP31 and cleaved BAP20 (lane 4). Coincubation of the VP16-activated membranes with the 10k supernatant resulted in the generation of BAP20 in the supernatant (Fig. 7D, lane 2), which was significantly increased by the presence of detergent (Fig. 7D, lane 3), because both caspase 8 and BAP31 were membrane-integral proteins and the solubilization of the membranes increased the accessibility of caspase 8 to BAP31. As controls, the amounts of BAP20 in the pellets following coincubation did not change significantly whether the detergent was present or not (Fig. 7D, compare lanes 5 and 6 with lane 4). Since BAP31 can only be cleaved by caspase 8 (6, 7, 40), these results suggest that the active caspase 8 on the OMM is capable of cleaving the BAP31 on the outer ER membrane.
Recently, a novel procaspase 8 isoform, procaspase 8L, which has an N-terminal 59-amino-acid extension (i.e., the Nex domain) compared to the p55 procaspase 8, has been shown to be peripherally associated with the cytosolic face of the ER in some cells, and its processing appears to be regulated through an association with BAP31 (6). To determine whether procaspase 8L may be involved in VP16-induced BAP31 cleavage and apoptosis, we first determined the levels of procaspase 8L in various cells by using a procaspase 8L-specific antibody (6). As shown in Fig. 7E, cultured human cells generally express very low levels of procaspase 8L compared to mouse thymuses, which expressed much higher levels. To determine whether this low level of procaspase 8L may be involved in VP16-induced BAP31 cleavage and apoptosis, we transfected the MDA-MB231 cells with a vector encoding EGFP-tagged Nex (Nex-EGFP), which functions as a DN to procaspase 8L (6), prior to VP16 stimulation. The results demonstrated that Nex-EGFP did not affect the VP16-induced BAP31 cleavage or caspase 3 activation (Fig. 7F).
Potential mitochondrion-ER cross talk in VP16-induced cell death.
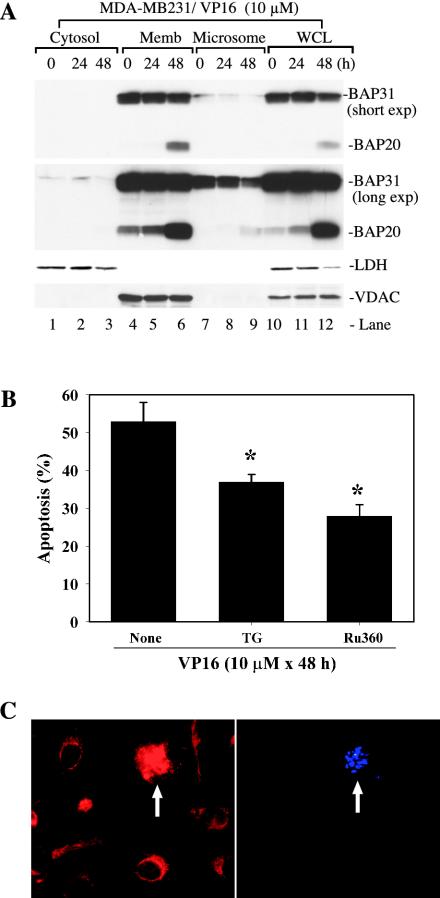
Our fractionation experiments suggest that there exist two populations of ER: one that cofractionates with the mitochondria and another that fractionates in the microsomes (Fig. 3B). Interestingly, the BAP31 in the ER that cofractionated with the mitochondria seemed to be preferentially cleaved (Fig. 3B). To corroborate this finding, we carried out detailed time course studies. The results indicated that the majority of BAP31 was detected in the mitochondrion-enriched membrane fraction and that the BAP31 in this population of ER was preferentially degraded to BAP20 (Fig. 8A, compare lanes 4 to 6 to lanes 7 to 9). As expected, LDH was detected only in the cytosol, whereas VDAC was detected only in the mitochondrion-enriched membrane fraction, and both proteins were detected in the whole-cell lysate (Fig. 8A). Since caspase 8 was mainly on the OMM and BAP31 was mainly on the ER outer membrane (6, 7, 40), these results (Fig. 3B and 8A) suggest that there may exist a mitochondrion-ER interaction leading to functional cross talks between the two organelles.
FIG. 8.
Potential mitochondrion-ER cross talk in VP16-induced apoptosis. (A) BAP31 was preferentially cleaved in the population of ER that cofractionated with the mitochondria. Vehicle-treated or VP16-treated MDA-MB231 cells were used to obtain various fractions indicated. Equal amounts of proteins (40 μg/lane) were used in Western blotting for the molecules indicated. Two different exposures (i.e., short and long exp.) for BAP31 were shown. Note that only VDAC-OMM was shown for the sake of simplicity. Memb, membrane; WCL, whole-cell lysate. (B) Depletion of ER Ca2+ store or inhibition of mitochondrion uptake of Ca2+ inhibited VP16-induced apoptosis. MDA-MB231 cells were pretreated with either vehicle (ethanol) or 50 nM TG or 50 μM Ru360 for 1 h. After removing these chemicals, cells were treated with VP16 (10 μM) for 48 h. At the end of treatment, cells were scored for apoptosis by DAPI staining. On average, 250 to 400 cells were counted for each condition and data represent means ± standard deviations. *, P < 0.01 (Student's t test). (C) Mitochondrial fragmentation during VP16-induced apoptosis. MDA-MB231 cells treated with VP16 (10 μM for 48 h) were labeled live with Mitotracker and DAPI. The arrows indicate DAPI-positive (i.e., apoptotic) cells that show fragmentation of the mitochondria. The micrographs shown are representative of ∼400 hundred images analyzed.
One way that mediates the ER-mitochondrion cross talk is through BAP20, the BAP31 cleavage product that localizes specifically in the ER (7). BAP20 has been shown to induce the ER to release Ca2+, which is taken up by the mitochondrion, leading to mitochondrial fragmentation and subsequent cell death (7). To examine whether such mechanisms may exist in VP16-stimulated MDA-MB231 cells, we first utilized a low concentration of TG to deplete the ER Ca2+ store (7, 49). TG is an irreversible inhibitor of the ER outer membrane pump SERCA, whose function is to maintain the concentration of ER Ca2+ several orders of magnitude above that of the cytosol. Although high concentrations of TG are known to kill cells, low concentrations of TG appear to slowly empty the ER Ca2+ store without inducing obvious cell death (7, 49). As shown in Fig. 8B, pretreatment of MDA-MB231 cells with 50 nM TG (7) significantly inhibited VP16-induced cell death.
The Ca2+ released from the ER induced by BAP20 is rapidly taken up by the mitochondria, leading to physical and functional compromises in the organelles (7). To determine whether the mitochondrial uptake of Ca2+ contributed to the VP16-induced cell death, we made use of Ru360, an oxygen-bridged dinuclear ruthenium amine complex that specifically inhibits the mitochondrial Ca2+ uptake (7, 34). As shown in Fig. 8B, pretreatment of MDA-MB231 cells with Ru360 resulted in an even stronger inhibition of VP16-induced apoptosis than that of TG. Mitochondrial Ca2+ uptake leads to mitochondrial fission or fragmentation (7). Indeed, we observed that the majority (i.e., 95% of ∼800 cells analyzed) of the VP16-treated, apoptotic MDA-MB231 cells showed prominent mitochondrial fragmentation, as shown in Fig. 8C (also Fig. 4H).
DISCUSSION
Active caspase 8 localizes mainly on the OMM.
One of the most interesting findings we have made in the present study is the association of active caspase 8 exclusively with the membranes during apoptosis. In multiple cell systems, including prostate (LNCaP and PC3) and breast (MDA-MB231) cancer cells, fibroblasts (GM701), and Jurkat T cells, procaspase 8, similar to procaspase 9 and procaspase 3 (10), localizes in both the mitochondria and the cytosol. However, upon apoptotic stimulation, active caspase 8 (both cleavage products and activity) is detected nearly exclusively in the mitochondrion-enriched membrane fraction in some cell systems, e.g., VP16-treated MDA-MB231, LNCaP, or Jurkat cells or serum-starved LNCaP cells. Although the mitochondrion-enriched membranes also contain a significant amount of ER, fluorescence microscopy reveals clear colocalization of caspase 8 with the mitochondria. In some cells, procaspase 8L has been shown to associate with the ER through interactions with BAP31 and Bcl-2/Bcl-XL (6, 40). In our cell systems, procaspase 8L is expressed at very low levels, and upon fluorescence microscopy, no significant colocalization of caspase 8 or caspase 8L with the ER is observed. Altogether, these observations suggest that, at least in the cells we have studied, the majority of the membrane-associated (pro)caspase 8 localizes on the mitochondria but not the ER.
Subsequent biochemical characterizations indicate that both mitochondrial procaspase and active caspase 8 in MDA-MB231 cells show protease sensitivity and alkali or high-salt extraction patterns very similar to the typical OMM proteins, such as Bak and VDAC-OMM, but different from the IMS (i.e., cytochrome c) or IMM (e.g., COX II, COX IV, and VDAC-IMM) proteins. These observations led us to conclude that the mitochondrial caspase 8 localizes mainly on the OMM as an integral protein. However, it remains possible that some caspase 8 may also be present in the IMS, as suggested by its partial release by alkali extraction. These conclusions are overall consistent with those of earlier studies showing various caspase 8 submitochondrial localizations in different cell types (43, 53, 54). Interestingly, the mitochondrion-associated procaspases and active caspases 9 and 3 seem to be localized only in the IMS (10; this study). At present, it is unclear how caspase 8 and other caspases are targeted to the different compartments of the mitochondria, as no obvious mitochondrial targeting sequences have been identified in these proteins. Recently, it has been found that, in Bcl-XL-protected MCF7 breast cancer cells overexpressing Fas, large quantities of active caspase 8 produced by DISC activation are sequestered (and thus inactivated) on the OMM, presumably by association with the protein bidirectional apoptosis regulator, or BAR (53). This interaction is thought to be mediated by the BAR protein functioning as a bridging molecule (i.e., forming the Bcl-XL-BAR-caspase 8 complex). Although MDA-MB231 cells express significant amounts of Bcl-XL on the mitochondria and VP16 treatment up-regulates Bcl-2 (but not Bcl-XL) (unpublished data), the Bcl-XL-BAR-caspase 8 mechanism (53) may not be involved, as the OMM-localized caspase 8 in VP16-treated MDA-MB231 cells is enzymatically active and can robustly degrade substrates.
Caspase 8 is activated through FADD/TRADD-dependent mechanisms and plays a critical role in VP16-induced caspase 3 activation and cell death.
Caspase 8 is activated in perhaps most apoptotic systems, including cell death induced by various chemotherapeutic drugs. Some drugs may rapidly up-regulate death receptors (e.g., DR5 and Fas) or their ligands (e.g., Fas ligand, TRAIL, and TNF-α), thereby directly activating the death receptor-mediated activation of procaspase 8 (1, 4, 12, 22, 27, 28, 63). By contrast, other drugs may indirectly activate procaspase 8 through apoptosome-activated caspases, in particular, caspase 3 (12, 15, 59). Therefore, in these latter apoptotic systems, caspase 8 activation functions primarily as a positive feedback amplification mechanism. In several apoptotic models that have been studied (9, 10; this study), caspase 8 activation occurs either around the same time or after other caspases are activated, consistent with the general concept that caspase 8 activation may represent either an initiating or a secondary amplifying event in death signaling.
Several pieces of evidence suggest that caspase 8 activation may represent an early and critical event in etoposide-induced apoptosis. First, caspase 8 activation occurs around the same time that caspases 9, 3, and 7 are activated. Second, caspase 8 activation is not downstream of caspase 3. It has been shown that recombinant murine active caspase 3 can robustly cleave murine procaspase 8 (58) and that anticancer drug-induced caspase 8 activation may be mediated by caspase 3 (12, 15, 59). Nevertheless, we find that even recombinant active caspase 3 does not activate procaspase 8 in MDA-MB231 cells, although it readily cleaves procaspases 9 and 3. One potential explanation is that caspase 3 does not activate caspase 6, as caspase 3-mediated activation of caspase 8 appears to require caspase 6 (11, 50). In support of this idea, caspase 6 is not activated in VP16-treated MDA-MB231 or LNCaP cells. Third, DN caspase 8, IETD, and siRNAs to caspase 8 all inhibit activation of caspase 3 and cell death. Finally, caspase 8-deficient Jurkat cells show reduced caspase 3 activation and death sensitivity to VP16. Incomplete inhibition of apoptosis in caspase 8-deficient Jurkat cells can be explained by the involvement of related caspases, such as caspase 10 (25).
VP16-induced caspase 8 activation appears to be mediated via FADD-dependent mechanisms, as it can be inhibited by DN-FADD in MDA-MB231 and BJAB cells. More direct evidence comes from FADD−/− Jurkat cells, which demonstrate greatly reduced caspase 8 activation in response to VP16. The residual amount of caspase 8 activation in the VP16-stimulated FADD−/− Jurkat cells (Fig. 6G) may have resulted from other adaptor proteins, such as TRADD. Indeed, DN-TRADD also partially inhibits VP16-induced caspase 8 activation and apoptosis in MDA-MB231 cells. These results strongly suggest that FADD/TRADD-mediated signals and caspase 8 activation sit upstream of caspase 3 activation in VP16-treated cells (Fig. 9). In support of this, caspase 3 activation is significantly reduced in FADD-deficient and, less dramatically, in caspase 8-deficient Jurkat cells. By contrast, FADD or caspase 8 deficiency does not lead to a significant inhibition of apoptosome-derived caspase 9 activation (i.e., p35). These observations collectively support our model (Fig. 9) in which VP16 stimulation activates, in parallel, both apoptosome-mediated activation of caspase 9 and FADD/TRADD-mediated activation of caspase 8 (and perhaps caspase 10). However, the FADD-caspase 8 pathway seems to play a much more prominent role in caspase 3 activation. Our results are consistent with some studies showing that anticancer drugs (including etoposide) can up-regulate or activate the death receptor pathway (12, 22, 27, 28, 63). On the other hand, our results and conclusions seemingly do not accord with those of some other studies (15, 59, 64), which suggest that caspase 8 does not play an initiating role or does not contribute to VP16 (or some other anticancer drug)-induced apoptosis. Although the precise reasons for these discrepancies are presently unclear, possible explanations may include different concentrations of VP16 (VP16 concentrations as high as 50 to 100 μM were used in some studies) or different cell types used.
FIG. 9.
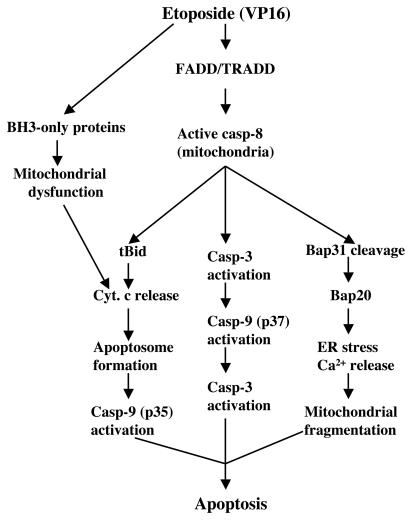
Schematic illustrating apoptotic signaling pathways in MDA-MB231 cells upon stimulation by etoposide. See text for details. casp, caspase; Cyt. c, cytochrome c.
An interesting and also important question is whether procaspase 8 is first activated in the cytosol through DISC and then the activated caspase 8 translocates to the OMM or whether the OMM-localized procaspase 8 becomes activated in the OMM through a DISC-like complex. Although there is some circumstantial evidence that caspase 8 can be activated independent of the CD95 receptor-ligand interaction (64), the answer to this question awaits further experimentation.
Potential roles for membrane-associated caspase 8 in cleaving substrates and mediating mitochondrion-ER cross talk.
What could be the advantages for deploying active caspase 8 on the OMM? Conceptually, this distribution pattern could increase the accessibility and efficiency of caspase 8 to its substrates localized in different compartments, including cytosols and organelles. The first potential substrate of the active caspase 8 is Bid (Fig. 9), which is cleaved into multiple ≤15-kDa fragments in etoposide-stimulated MDA-MB231 cells. The deployment of active caspase 8 on the OMM could presumably couple cleavage of cytosolic Bid and insertion of active Bid fragments into the mitochondria to induce cytochrome c release, thus facilitating the apoptosome pathway. Part of the caspase 8 effect on Bid cleavage may be mediated through the caspase 8-activated caspase 3, as the latter has been shown to efficiently degrade Bid (15, 51). Some of the <15-kDa Bid fragments may have been derived from other proteases, such as calpain, granzyme B, or serine proteases (8, 21, 33). Recently, JNK has been shown to induce caspase 8-independent cleavage of Bid at a distinct site to generate 21-kDa jBid, which translocates to the mitochondria, preferentially releases Smac/DIABLO to disrupt the TRAF2-cIAP1 complex, and facilitates caspase 8 activation (14). Throughout our experiments, we failed to detect jBid, thus ruling out the involvement of this pathway.
The second potential substrate for the OMM-localized caspase 8 may be cytosolic procaspase 3, which upon activation, can activate caspase 9, which in turn activates more caspase 3, thus forming a positive amplification loop (Fig. 9). The third potential substrate for the OMM-localized caspase 8 may be ER-resident BAP31, which is prominently cleaved into proapoptotic BAP20 in VP16-treated cells (Fig. 9). As BAP20 can cause severe ER stress by inducing Ca2+ release and increased mitochondrial fission (7), these observations suggest the following signaling pathway upon etoposide stimulation: active caspase 8 on the OMM → cleavage of BAP31 to BAP20 on the ER → ER Ca2+ release → mitochondrial uptake of excessive Ca2+ → mitochondrial fission → compromise of mitochondrial integrity and function → contribution to apoptosis. In support, it seems that only BAP31 localized on the population of the ER that cofractionates with the mitochondria becomes preferentially cleaved to generate BAP20. Moreover, the blocking of ER Ca2+ release or mitochondrial Ca2+ uptake inhibits VP16-induced cell death. Furthermore, most VP16-stimulated apoptotic MDA-MB231 cells show prominent mitochondrial fragmentation. Finally, this scheme is consistent with recent demonstrations of close mitochondrion-ER interactions and a critical role for ER in apoptosis signaling (7, 17, 24, 32, 44, 49, 66).
In summary, the present study suggests that (i) some apoptotic stimuli induce an early activation of procaspase 8 in a FADD/TRADD-dependent manner, (ii) active caspase 8 becomes associated mainly with the OMM, (iii) the OMM-localized active caspase 8 may cleave substrates in both the cytosol and the ER outer membrane, and (iv) active caspase 8 on the OMM may mediate mitochondrion-ER cross talk through the cleavage of BAP31. Our ongoing work will focus on the molecular mechanisms of the FADD/TRADD-mediated activation of procaspase 8 in relation to the mitochondrial localization, which will undoubtedly improve our understanding of drug-induced cancer cell death and benefit cancer chemotherapy.
Acknowledgments
We thank M. King for providing GM701 cells; Biomide Corporation for BMD188; X. Wang for antibody against Bid; G. Shore for anti-Bap31 and -procaspase 8L antibodies, DN caspase 8, and Nex-EGFP; Y. Lazebnik for DN caspase 9; C. Vincenz for DN-FADD and DN-TRADD; and members of the Tang lab for generous support and helpful discussion. We also thank S. Bratton for insightful discussions and for critically reading the manuscript.
This work was supported in part by NIH National Cancer Institute grant CA 90297, American Cancer Society grant RSG MGO-105961, Department of Defense grant DAMD17-03-1-0137, and NIEHS Center grant ES07784. D.C. was supported by Department of Defense Postdoctoral Traineeship Award DAMD17-02-0083. B.B. is a graduate student in the GSBS program.
REFERENCES
- 1.Ashkenazi, A., and V. M. Dixit. 1998. Death receptors: signaling and modulation. Science 281:1305-1308. [DOI] [PubMed] [Google Scholar]
- 2.Bhatia, B., C. J. Maldonado, S. Tang, D. Chandra, R. D. Klein, D. Chopra, S. B. Shappell, P. Yang, R. A. Newman, and D. G. Tang. 2003. Subcellular localization and tumor-suppressive functions of 15-lipoxygenase 2 (15-LOX2) and its splice variants. J. Biol. Chem. 278:25091-25100. [DOI] [PubMed] [Google Scholar]
- 3.Boatright, K. M., M. Renatus, F. L. Scott, S. Sperandio, H. Shin, I. M. Pedersen, J. E. Ricci, W. A. Edris, D. P. Sutherlin, D. R. Green, and G. S. Salvesen. 2003. A unified model for apical caspase activation. Mol. Cell 11:529-541. [DOI] [PubMed] [Google Scholar]
- 4.Boesen-de Cock, J. G., A. D. Tepper, E. de Vries, W. J. van Blitterswijk, and J. Borst. 1999. Common regulation of apoptosis signaling induced by CD95 and the DNA-damaging stimuli etoposide and gamma-radiation downstream from caspase-8 activation. J. Biol. Chem. 274:14255-14261. [DOI] [PubMed] [Google Scholar]
- 5.Boldin, M. P., T. M. Goncharov, Y. V. Goltsev, and D. Wallach. 1996. Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor-induced cell death. Cell 85:803-815. [DOI] [PubMed] [Google Scholar]
- 6.Breckenridge, D. G., M. Nguyen, S. Kuppig, M. Reth, and G. C. Shore. 2002. The procaspase-8 isoform, procaspase-8L, recruited to the BAP31 complex at the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 99:4331-4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breckenridge, D. G., M. Stojanovic, R. C. Marcellus, and G. C. Shore. 2003. Caspase cleavage product of BAP31 induces mitochondrial fission through endoplasmic reticulum calcium signals, enhancing cytochrome c release to the cytosol. J. Cell Biol. 160:1115-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cartron, P. F., P. Juin, L. Oliver, S. Martin, K. Meflah, and F. M. Vallette. 2003. Nonredundant role of Bax and Bak in Bid-mediated apoptosis. Mol. Cell Biol. 23:4701-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandra, D., J. W. Liu, and D. G. Tang. 2002. Early mitochondrial activation and cytochrome c up-regulation during apoptosis. J. Biol. Chem. 277:50842-50854. [DOI] [PubMed] [Google Scholar]
- 10.Chandra, D., and D. G. Tang. 2003. Mitochondrially localized active caspase-9 and caspase-3 result mostly from translocation from the cytosol and partly from caspase-mediated activation in the organelle. Lack of evidence for Apaf-1-mediated procaspase-9 activation in the mitochondria. J. Biol. Chem. 278:17408-17420. [DOI] [PubMed] [Google Scholar]
- 11.Cowling, V., and J. Downward. 2002. Caspase-6 is the direct activator of caspase-8 in the cytochrome c-induced apoptosis pathway: absolute requirement for removal of caspase-6 prodomain. Cell Death Differ. 9:1046-1056. [DOI] [PubMed] [Google Scholar]
- 12.Debatin, K. M., D. Poncet, and G. Kroemer. 2002. Chemotherapy: targeting the mitochondrial cell death pathway. Oncogene 21:8786-8803. [DOI] [PubMed] [Google Scholar]
- 13.Degli Esposti, M., G. Ferry, P. Masdehors, J. A. Boutin, J. A. Hickman, and C. Dive. 2003. Post-translational modification of Bid has differential effects on its susceptibility to cleavage by caspase 8 or caspase 3. J. Biol. Chem. 278:15749-15757. [DOI] [PubMed] [Google Scholar]
- 14.Deng, Y., X. Ren, L. Yang, Y. Lin, and X. Wu. 2003. A JNK-dependent pathway is required for TNFalpha-induced apoptosis. Cell 115:61-70. [DOI] [PubMed] [Google Scholar]
- 15.Engels, I. H., A. Stepczynska, C. Stroh, K. Lauber, C. Berg, R. Schwenzer, H. Wajant, R. U. Janicke, A. G. Porter, C. Belka, M. Gregor, K. Schulze-Osthoff, and S. Wesselborg. 2000. Caspase-8/FLICE functions as an executioner caspase in anticancer drug-induced apoptosis. Oncogene 19:4563-4573. [DOI] [PubMed] [Google Scholar]
- 16.Evans, W. H. 1992. Isolation and characterization of membranes and cell organelles, p. 233-270. In D. Rickwood (ed.), Preparative centrifugations: a practical approach. The IRL Press, Oxford, United Kingdom.
- 17.Filippin, L., P. J. Magalhaes, G. Di Benedetto, M. Colella, and T. Pozzan. 2003. Stable interactions between mitochondria and endoplasmic reticulum allow rapid accumulation of calcium in a subpopulation of mitochondria. J. Biol. Chem. 278:39224-39234. [DOI] [PubMed] [Google Scholar]
- 18.Fulda, S., E. Meyer, C. Friesen, S. A. Susin, G. Kroemer, and K. M. Debatin. 2001. Cell type specific involvement of death receptor and mitochondrial pathways in drug-induced apoptosis. Oncogene 20:1063-1075. [DOI] [PubMed] [Google Scholar]
- 19.Harper, N., M. Hughes, M. MacFarlane, and G. M. Cohen. 2003. Fas-associated death domain protein and caspase-8 are not recruited to the tumor necrosis factor receptor 1 signaling complex during tumor necrosis factor-induced apoptosis. J. Biol. Chem. 278:25534-25541. [DOI] [PubMed] [Google Scholar]
- 20.Hsu, H., H. B. Shu, M. G. Pan, and D. V. Goeddel. 1996. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell 84:299-308. [DOI] [PubMed] [Google Scholar]
- 21.Huang, H. L., L. W. Fang, S. P. Lu, C. K. Chou, T. Y. Luh, and M. Z. Lai. 2003. DNA-damaging reagents induce apoptosis through reactive oxygen species-dependent Fas aggregation. Oncogene 22:8168-8177. [DOI] [PubMed] [Google Scholar]
- 22.Johnston, J. B., A. F. Kabore, J. Strutinsky, X. Hu, J. T. Paul, D. M. Kropp, B. Kuschak, A. Begleiter, and S. B. Gibson. 2003. Role of the TRAIL/APO2-L death receptors in chlorambucil- and fludarabine-induced apoptosis in chronic lymphocytic leukemia. Oncogene 22:8356-8369. [DOI] [PubMed] [Google Scholar]
- 23.Joshi, B., L. Li, B. G. Taffe, Z. Zhu, S. Wahl, H. Tian, E. Ben-Josef, J. D. Taylor, A. T. Porter, and D. G. Tang. 1999. Apoptosis induction by a novel anti-prostate cancer compound, BMD188 (a fatty acid-containing hydroxamic acid), requires the mitochondrial respiratory chain. Cancer Res. 59:4343-4355. [PubMed] [Google Scholar]
- 24.Karbowski, M., and R. J. Youle. 2003. Dynamics of mitochondrial morphology in healthy cells and during apoptosis. Cell Death Differ. 10:870-880. [DOI] [PubMed] [Google Scholar]
- 25.Kischkel, F. C., D. A. Lawrence, A. Tinel, H. LeBlanc, A. Virmani, P. Schow, A. Gazdar, J. Blenis, D. Arnott, and A. Ashkenazi. 2001. Death receptor recruitment of endogenous caspase-10 and apoptosis initiation in the absence of caspase-8. J. Biol. Chem. 276:46639-46646. [DOI] [PubMed] [Google Scholar]
- 26.Kuwana, T., J. J. Smith, M. Muzio, V. Dixit, D. D. Newmeyer, and S. Kornbluth. 1998. Apoptosis induction by caspase-8 is amplified through the mitochondrial release of cytochrome c. J. Biol. Chem. 273:16589-16594. [DOI] [PubMed] [Google Scholar]
- 27.Lacour, S., O. Micheau, A. Hammann, V. Drouineaud, J. Tschopp, E. Solary, and M. T. Dimanche-Boitrel. 2003. Chemotherapy enhances TNF-related apoptosis-inducing ligand DISC assembly in HT29 human colon cancer cells. Oncogene 22:1807-1816. [DOI] [PubMed] [Google Scholar]
- 28.LeBlanc, H., D. Lawrence, E. Varfolomeev, K. Totpal, J. Morlan, P. Schow, S. Fong, R. Schwall, D. Sinicropi, and A. Ashkenazi. 2002. Tumor-cell resistance to death receptor-induced apoptosis through mutational inactivation of the proapoptotic Bcl-2 homolog Bax. Nat. Med. 8:274-281. [DOI] [PubMed] [Google Scholar]
- 29.Li, H., H. Zhu, C. J. Xu, and J. Yuan. 1998. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 94:491-501. [DOI] [PubMed] [Google Scholar]
- 30.Lin, Y., A. Devin, Y. Rodriguez, and Z. G. Liu. 1999. Cleavage of the death domain kinase RIP by caspase-8 prompts TNF-induced apoptosis. Genes Dev. 13:2514-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo, X., I. Budihardjo, H. Zou, C. Slaughter, and X. Wang. 1998. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell 94:481-490. [DOI] [PubMed] [Google Scholar]
- 32.Mandic, A., J. Hansson, S. Linder, and M. C. Shoshan. 2003. Cisplatin induces endoplasmic reticulum stress and nucleus-independent apoptotic signaling. J. Biol. Chem. 278:9100-9106. [DOI] [PubMed] [Google Scholar]
- 33.Mandic, A., K. Viktorsson, L. Strandberg, T. Heiden, J. Hansson, S. Linder, and M. C. Shoshan. 2002. Calpain-mediated Bid cleavage and calpain-independent Bak modulation: two separate pathways in cisplatin-induced apoptosis. Mol. Cell. Biol. 22:3003-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matlib, M. A., Z. Zhou, S. Knight, S. Ahmed, K. M. Choi, J. Krause-Bauer, R. Phillips, R. Altschuld, Y. Katsube, N. Sperelakis, and D. M. Bers. 1998. Oxygen-bridged dinuclear ruthenium amine complex specifically inhibits Ca2+ uptake into mitochondria in vitro and in situ in single cardiac myocytes. J. Biol. Chem. 273:10223-10231. [DOI] [PubMed] [Google Scholar]
- 35.McDonnell, M. A., D. Wang, S. M. Khan, M. G. Vander Heiden, and A. Kelekar. 2003. Caspase-9 is activated in a cytochrome c-independent manner early during TNFalpha-induced apoptosis in murine cells. Cell Death Differ. 10:1005-1015. [DOI] [PubMed] [Google Scholar]
- 36.Medema, J. P., C. Scaffidi, F. C. Kischkel, A. Shevchenko, M. Mann, P. H. Krammer, and M. E. Peter. 1997. FLICE is activated by association with the CD95 death-inducing signaling complex (DISC). EMBO J. 16:2794-2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Micheau, O., and J. Tschopp. 2003. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 114:181-190. [DOI] [PubMed] [Google Scholar]
- 38.Muzio, M., A. M. Chinnaiyan, F. C. Kischkel, K. O'Rourke, A. Shevchenko, J. Ni, C. Scaffidi, J. D. Bretz, M. Zhang, R. Gentz, M. Mann, P. H. Krammer, M. E. Peter, and V. M. Dixit. 1996. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death-inducing signaling complex. Cell 85:817-827. [DOI] [PubMed] [Google Scholar]
- 39.Muzio, M., G. S. Salvesen, and V. M. Dixit. 1997. FLICE induced apoptosis in a cell-free system. Cleavage of caspase zymogens. J. Biol. Chem. 272:2952-2956. [DOI] [PubMed] [Google Scholar]
- 40.Ng, F. W., M. Nguyen, T. Kwan, P. E. Branton, D. W. Nicholson, J. A. Cromlish, and G. C. Shore. 1997. p28 Bap31, a Bcl-2/Bcl-XL- and procaspase-8-associated protein in the endoplasmic reticulum. J. Cell Biol. 139:327-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peter, M. E., and P. H. Krammer. 2003. The CD95(APO-1/Fas) DISC and beyond. Cell Death Differ. 10:26-35. [DOI] [PubMed] [Google Scholar]
- 42.Petit, P. X., J. E. O'Connor, D. Grunwald, and S. C. Brown. 1990. Analysis of the membrane potential of rat- and mouse-liver mitochondria by flow cytometry and possible applications. Eur. J. Biochem. 194:389-397. [DOI] [PubMed] [Google Scholar]
- 43.Qin, Z. H., Y. Wang, K. K. Kikly, E. Sapp, K. B. Kegel, N. Aronin, and M. DiFiglia. 2001. Pro-caspase-8 is predominantly localized in mitochondria and released into cytoplasm upon apoptotic stimulation. J. Biol. Chem. 276:8079-8086. [DOI] [PubMed] [Google Scholar]
- 44.Reddy, R. K., C. Mao, P. Baumeister, R. C. Austin, R. J. Kaufman, and A. S. Lee. 2003. Endoplasmic reticulum chaperone protein GRP78 protects cells from apoptosis induced by topoisomerase inhibitors: role of ATP binding site in suppression of caspase-7 activation. J. Biol. Chem. 278:20915-20924. [DOI] [PubMed] [Google Scholar]
- 45.Ricci, J. E., R. A. Gottlieb, and D. R. Green. 2003. Caspase-mediated loss of mitochondrial function and generation of reactive oxygen species during apoptosis. J. Cell Biol. 160:65-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salvesen, G. S., and V. M. Dixit. 1997. Caspases: intracellular signaling by proteolysis. Cell 91:443-446. [DOI] [PubMed] [Google Scholar]
- 47.Scaffidi, C., S. Fulda, A. Srinivasan, C. Friesen, F. Li, K. J. Tomaselli, K. M. Debatin, P. H. Krammer, and M. E. Peter. 1998. Two CD95 (APO-1/Fas) signaling pathways. EMBO J. 17:1675-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scaffidi, C., J. P. Medema, P. H. Krammer, and M. E. Peter. 1997. FLICE is predominantly expressed as two functionally active isoforms, caspase-8/a and caspase-8/b. J. Biol. Chem. 272:26953-26958. [DOI] [PubMed] [Google Scholar]
- 49.Scorrano, L., S. A. Oakes, J. T. Opferman, E. H. Cheng, M. D. Sorcinelli, T. Pozzan, and S. J. Korsmeyer. 2003. BAX and BAK regulation of endoplasmic reticulum Ca2+: a control point for apoptosis. Science 300:135-139. [DOI] [PubMed] [Google Scholar]
- 50.Slee, E. A., M. T. Harte, R. M. Kluck, B. B. Wolf, C. A. Casiano, D. D. Newmeyer, H. G. Wang, J. C. Reed, D. W. Nicholson, E. S. Alnemri, D. R. Green, and S. J. Martin. 1999. Ordering the cytochrome c-initiated caspase cascade: hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner. J. Cell Biol. 144:281-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Slee, E. A., S. A. Keogh, and S. J. Martin. 2000. Cleavage of BID during cytotoxic drug and UV radiation-induced apoptosis occurs downstream of the point of Bcl-2 action and is catalysed by caspase-3: a potential feedback loop for amplification of apoptosis-associated mitochondrial cytochrome c release. Cell Death Differ. 7:556-565. [DOI] [PubMed] [Google Scholar]
- 52.Srinivasula, S. M., M. Ahmad, Y. Guo, Y. Zhan, Y. Lazebnik, T. Fernandes-Alnemri, and E. S. Alnemri. 1999. Identification of an endogenous dominant-negative short isoform of caspase-9 that can regulate apoptosis. Cancer Res. 59:999-1002. [PubMed] [Google Scholar]
- 53.Stegh, A. H., B. C. Barnhart, J. Volkland, A. Algeciras-Schimnich, N. Ke, J. C. Reed, and M. E. Peter. 2002. Inactivation of caspase-8 on mitochondria of Bcl-xL-expressing MCF7-Fas cells: role for the bifunctional apoptosis regulator protein. J. Biol. Chem. 277:4351-4360. [DOI] [PubMed] [Google Scholar]
- 54.Stegh, A. H., H. Herrmann, S. Lampel, D. Weisenberger, K. Andra, M. Seper, G. Wiche, P. H. Krammer, and M. E. Peter. 2000. Identification of the cytolinker plectin as a major early in vivo substrate for caspase 8 during CD95- and tumor necrosis factor receptor-mediated apoptosis. Mol. Cell. Biol. 20:5665-5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stennicke, H. R., J. M. Jurgensmeier, H. Shin, Q. Deveraux, B. B. Wolf, X. Yang, Q. Zhou, H. M. Ellerby, L. M. Ellerby, D. Bredesen, D. R. Green, J. C. Reed, C. J. Froelich, and G. S. Salvesen. 1998. Pro-caspase-3 is a major physiologic target of caspase-8. J. Biol. Chem. 273:27084-27090. [DOI] [PubMed] [Google Scholar]
- 56.Susin, S. A., H. K. Lorenzo, N. Zamzami, I. Marzo, C. Brenner, N. Larochette, M. C. Prevost, P. M. Alzari, and G. Kroemer. 1999. Mitochondrial release of caspase-2 and -9 during the apoptotic process. J. Exp. Med. 189:381-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang, D. G., L. Li, D. P. Chopra, and A. T. Porter. 1998. Extended survivability of prostate cancer cells in the absence of trophic factors: increased proliferation, evasion of apoptosis, and the role of apoptosis proteins. Cancer Res. 58:3466-3479. [PubMed] [Google Scholar]
- 58.Van de Craen, M., W. Declercq, I. Van den brande, W. Fiers, and P. Vandenabeele. 1999. The proteolytic procaspase activation network: an in vitro analysis. Cell Death Differ. 6:1117-1124. [DOI] [PubMed] [Google Scholar]
- 59.Varfolomeev, E. E., M. Schuchmann, V. Luria, N. Chiannilkulchai, J. S. Beckmann, I. L. Mett, D. Rebrikov, V. M. Brodianski, O. C. Kemper, O. Kollet, T. Lapidot, D. Soffer, T. Sobe, K. B. Avraham, T. Goncharov, H. Holtmann, P. Lonai, and D. Wallach. 1998. Targeted disruption of the mouse caspase 8 gene ablates cell death induction by the TNF receptors, Fas/Apo1, and DR3 and is lethal prenatally. Immunity 9:267-276. [DOI] [PubMed] [Google Scholar]
- 60.von Haefen, C., T. Wieder, F. Essmann, K. Schulze-Osthoff, B. Dorken, and P. T. Daniel. 2003. Paclitaxel-induced apoptosis in BJAB cells proceeds via a death receptor-independent, caspases-3/-8-driven mitochondrial amplification loop. Oncogene 22:2236-2247. [DOI] [PubMed] [Google Scholar]
- 61.Wang, X. 2001. The expanding role of mitochondria in apoptosis. Genes Dev. 15:2922-2933. [PubMed] [Google Scholar]
- 62.Wang, X., S. W. Ryter, C. Dai, Z. L. Tang, S. C. Watkins, X. M. Yin, R. Song, and A. M. Choi. 2003. Necrotic cell death in response to oxidant stress involves the activation of the apoptogenic caspase-8/bid pathway. J. Biol. Chem. 278:29184-29191. [DOI] [PubMed] [Google Scholar]
- 63.Wen, J., N. Ramadevi, D. Nguyen, C. Perkins, E. Worthington, and K. Bhalla. 2000. Antileukemic drugs increase death receptor 5 levels and enhance Apo-2L-induced apoptosis of human acute leukemia cells. Blood 96:3900-3906. [PubMed] [Google Scholar]
- 64.Wesselborg, S., I. H. Engels, E. Rossmann, M. Los, and K. Schulze-Osthoff. 1999. Anticancer drugs induce caspase-8/FLICE activation and apoptosis in the absence of CD95 receptor/ligand interaction. Blood 93:3053-3063. [PubMed] [Google Scholar]
- 65.Yi, X., X. M. Yin, and Z. Dong. 2003. Inhibition of Bid-induced apoptosis by Bcl-2. tBid insertion, Bax translocation, and Bax/Bak oligomerization suppressed. J. Biol. Chem. 278:16992-16999. [DOI] [PubMed] [Google Scholar]
- 66.Zong, W. X., C. Li, G. Hatzivassiliou, T. Lindsten, Q. C. Yu, J. Yuan, and C. B. Thompson. 2003. Bax and Bak can localize to the endoplasmic reticulum to initiate apoptosis. J. Cell Biol. 162:59-69. [DOI] [PMC free article] [PubMed] [Google Scholar]