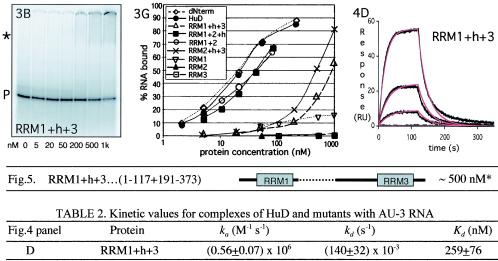
Volume 20, no. 13, p. 4765-4722, 2000. Recently, upon reinitiating studies focused on the role of HuD RRM2 in RNA binding, we obtained results that differed from the ones published. Extensive analysis of plasmids, and a variety of old and new protein preparations, revealed that although the construct was correct, previously the wrong plasmid had been used for a series of protein preparations. Tryptic digestion indicates that what was thought to be an RRM1+h+3 mutant HuD protein was in fact an RRM3 deletion mutant instead, yielding a protein of almost identical size but different domain composition. We have repeated all the analyses reported in the original paper with the correct protein (see corrected figure panels below). The key conclusion of the paper remains unaltered. (All three RRM domains play distinct roles in RNA binding. While RRM1 is the most important domain, RRM3 is required to form a stable HuD/RNA complex. The role of RRM3 in binding is masked in steady-state analyses of a domain 3 deletion mutant because its removal increases both the on and off rates of the complex.) However, our conclusion regarding the role of RRM2 needs to be revised. While we originally thought its role was distinct but similar to that of RRM3, it is clear from the data below that RRM2 plays a unique role in RNA binding that is more similar to that of RRM1. Removal of this domain strongly affects RNA binding ability, as seen by gel shift analysis (Fig. 3B and G and 5). The more accurate kinetic analysis (Fig. 4D; Table 2) shows that it weakens association by ∼7-fold and increases the dissociation rate by almost 50-fold, resulting in a loss of affinity of over 2 orders of magnitude. (By comparison, as described in the original paper, RRM1 deletion decreases association ∼20-fold and increases dissociation 100-fold, resulting in a 2,000-fold loss in affinity.) We sincerely apologize for any inconvenience our error may have caused.
. 2004 Aug;24(15):6888. doi: 10.1128/MCB.24.15.6888.2004
HuD RNA Recognition Motifs Play Distinct Roles in the Formation of a Stable Complex with AU-Rich RNA
Sungmin Park
1, David G Myszka
1, Michael Yu
1, Sarah J Littler
1, Ite A Laird-Offringa
1
Sungmin Park
1Norris Cancer Center, University of Southern California Keck School of Medicine, Los Angeles, California 90089-9176, and Huntsman Cancer Institute, University of Utah School of Medicine, Salt Lake City, Utah 84132
Find articles by Sungmin Park
David G Myszka
1Norris Cancer Center, University of Southern California Keck School of Medicine, Los Angeles, California 90089-9176, and Huntsman Cancer Institute, University of Utah School of Medicine, Salt Lake City, Utah 84132
Find articles by David G Myszka
Michael Yu
1Norris Cancer Center, University of Southern California Keck School of Medicine, Los Angeles, California 90089-9176, and Huntsman Cancer Institute, University of Utah School of Medicine, Salt Lake City, Utah 84132
Find articles by Michael Yu
Sarah J Littler
1Norris Cancer Center, University of Southern California Keck School of Medicine, Los Angeles, California 90089-9176, and Huntsman Cancer Institute, University of Utah School of Medicine, Salt Lake City, Utah 84132
Find articles by Sarah J Littler
Ite A Laird-Offringa
1Norris Cancer Center, University of Southern California Keck School of Medicine, Los Angeles, California 90089-9176, and Huntsman Cancer Institute, University of Utah School of Medicine, Salt Lake City, Utah 84132
Find articles by Ite A Laird-Offringa
1Norris Cancer Center, University of Southern California Keck School of Medicine, Los Angeles, California 90089-9176, and Huntsman Cancer Institute, University of Utah School of Medicine, Salt Lake City, Utah 84132
PMCID: PMC444871
This corrects the article "HuD RNA Recognition Motifs Play Distinct Roles in the Formation of a Stable Complex with AU-Rich RNA" in volume 20 on page 4765.