Abstract
An IgG2 monoclonal antibody (mAb) solution was subjected to stirring, generating high concentrations of nanometer and subvisible particles, which were then successfully size enriched into different size bins by low speed centrifugation or a combination of gravitational sedimentation and Fluorescence-Activated Cell Sorting (FACS). The size-fractionated mAb particles were assessed for their ability to elicit the release of cytokines from a population of donor-derived human peripheral blood mononuclear cells (PBMC) at two phases of the immune response. Fractions enriched in nanometer-sized particles showed a lower response than those enriched in micron-sized particles in this assay. Particles of 5–10 μm in size displayed elevated cytokine release profiles compared to other size ranges. Stir-stressed mAb particles had amorphous morphology, contained protein with partially altered secondary structure, elevated surface hydrophobicity (compared to controls), and trace levels of elemental fluorine. FACS size-enriched the mAb particle samples, yet did not notably alter the overall morphology or composition of particles as measured by Microflow imaging, Transmission Electron Microscopy, and Scanning Electron Microscopy-Energy Dispersive X-ray Spectroscopy. The utility and limitations of FACS for size separation of mAb particles and potential of in-vitro PBMC studies to rank order the immunogenic potential of various types of mAb particles is discussed.
Keywords: proteins, protein aggregation, particles, monoclonal antibody, IgG, immune response, immunogenicity, PBMC, in-vitro
INTRODUCTION
The presence of aggregates in recombinant protein therapeutics is a serious concern due to their potential immunogenicity risk in patients.1–10 Some notable clinical examples of immunogenicity include Factor VIII7, human growth hormone (hGH),8 and most likely erythropoietin (EPO).2,3 Unfortunately, exactly how aggregates produce an immune response, and the extent of their impact in in-vivo responses, is not fully understood. Aggregates can arise at all steps of protein production: fermentation, expression, purification, formulation, filling, transport, storage, and administration.11,12 Since each protein has distinct physicochemical properties, it may aggregate differently depending on the environmental stresses and solution conditions, which in turn, may expose various epitopes that may be more immune reactive than others. For example, with monoclonal antibodies (mAbs), different solution conditions can lead to formation of different types and sizes of aggregates and particles.13–15 Various environmental stresses including interfaces, temperatures, freeze-thaw, containers, pH, ionic strength, excipients, and concentration can all lead to protein aggregation.16–22
Our knowledge of the immunogenic potential of protein therapeutics is confounded by the fact that in addition to the presence of aggregates, immunogenicity risk can be contributed by factors that may arise from many patient related factors such as genetics, age, disease-related factors, and other concomitant medications the patient may be taking. The route of administration, dose, and the frequency of administration are also important factors. Product related factors such as non-human T-cell epitope content in sequence, origin of the active substance, process related contaminants, and formulation can impact immunogenicity as well. 23–26 The presence of so many variables complicates our understanding of the immunogenic potential of various therapeutic proteins. Therefore, determining the potential impact of aggregates by themselves on immunogenicity is very challenging. Despite these obstacles, a combination of in silico predictions 27–30, in vitro cell-based assays27,31–33, and in vivo transgenic animal models34–38 are currently being extensively evaluated to assess the relative potential of different protein aggregates and particles to elicit immune responses in humans.
One particularly promising in vitro approach is the use of human Peripheral Blood Mononuclear Cells (PBMC) derived from a healthy donor population with heterozygous MHC genotypes to screen antigens or mAb candidates for their propensity to stimulate certain features of the immune system.1 In-vitro models have been previously used to examine the immunogenicity of recombinant human erythropoietin-alpha39, recombinant Factor VIII39 and IgG mAbs40 with a certain degree of success. The correlation of clinical immunogenicity with the in vitro PBMC output was also investigated for an immunogenic human fusion protein. Subjects that induced IFN-gamma secreting T cells in PBMC were associated with a higher magnitude of immune response (as detected by binding and neutralizing anti-drug antibody) in clinical trials.31 These in vitro assays are relatively cheaper and faster to perform than in vivo animal model-based studies, incorporate the diversity of the genetic polymorphism of alleles in the representative human population, can be used to screen a large number of candidate biotherapeutic mAbs during early development, and can be employed to compare the relative immunogenicity of different pharmaceuticals.27,31–33
Most of the studies to date have focused on evaluation of immune activation by challenging with heterogeneous sized aggregates. However, there is still a gap where the actual aggregate size associated with the immune size has not been thoroughly addressed. Previously, three different IgG mAbs, including the one used in this study, were stressed by a variety of different methods and the resulting particles were characterized in terms of size, particle counts, conformation, morphology, and reversibility.13,41 Different types of aggregates formed depending on the type of stress applied.13 These different stressed mAbs were tested in an in vitro system to compare their relative response in human PBMC.40 It was found that aggregates, prepared by stirring all three mAbs, displayed the highest response compared to aggregates generated by all the other stresses tested.40 These results indicated that the presence of a high number of 2-10 μm particles (which were partially reversible, and retained some folded structure) in the stir-induced sample may be responsible for the increased PBMC response.40
The purpose of this study was to further examine the impact of protein particle size on signature cytokine secretions of human PBMC at different stages of the immune response.40 An IgG2 monoclonal antibody (mAb2) solution was stirred to generate protein particles of varying sizes. The mAb2 particles were then separated into various size enriched populations using (1) low speed centrifugation to enrich for nanometer vs. micron sized particles, and (2) Fluorescence Activated Cell Sorting (FACS) to separate enriched fractions of micron-sized protein particles. FACS is a promising approach for protein particle characterization, as was recently described by Rombach-Riegraf et al 42 for size separation, and for related applications such as to detect and differentiate subvisible protein particles from silicone oil protein particles. 43–45,46 The stirring induced mAb particles were also physically characterized, before and after FACS separation, in terms of their particle number, size distribution, mass distribution, morphology and composition using a combination of techniques including Microflow imaging (MFI) with radar chart analysis, FTIR microscopy, and multiple electron microscopy analyses (TEM and SEM-EDS).
MATERIALS AND METHODS
Materials
The IgG2 mAb, subsequently referred to as mAb2, was supplied by Amgen Inc. at 10.1 mg/mL. This mAb has been used previously to characterize the immune potential of different mAbs and their aggregates.40,47 The antibody was highly purified48 and endotoxin levels were confirmed to be in an acceptable range (< 1.00 EU/mL) for use in these cell based assays. The stock mAb2 solution was stored at 4 °C prior to use.
Generation of Aggregates
The stock mAb2 was diluted to 1 mg/mL with 10 mM sodium acetate pH 5.0 (A5 buffer) and stressed by stirring (referred to as stir-20h) as described previously by Joubert et al.13 Stirring stress was chosen since previous work showed that stirring of this IgG mAb produced aggregates that induced the highest response from PBMC.40 Additionally, for biophysical comparison purposes, a 1 mg/mL solution of mAb2 was heated extensively at 90°C for 30 min (labeled heat control).
Size separation of mAb particles
Two techniques were used to enrich different size populations of stirring induced particles: (1) slow speed centrifugation, and (2) gravitational settling of the samples combined with Fluorescence Activated Cell Sorting (FACS), subsequently referred to as “FACS”. For method (1), 1 mL of stir-20h mAb2 sample was centrifuged at 2500 × g using a Baxter Hereaus Biofuge 15 Model 3604 (Deerfield, IL) for 5 min. The supernatant was removed and the pellet was resuspended in one mL of A5 buffer. For method (2), 5 mL of stir-20h mAb2 sample was gently pipetted into 12×75mm polypropylene tubes and allowed to settle in a disturbance-free environment for 3 h at room temperature. After 3 h, 200 uL aliquots were removed carefully from the top to the bottom using a pipetman with gel loading tips and collected into eppendorf tubes. The last aliquot in the tube (referred to as “bottom”) contained a greater number of larger micron-sized particles and this sample was then further size fractionated by FACS as described below.
Fluorescence Activated Cell Sorting (FACS)
A Beckman Coulter MoFlo XDP (Brea, CA) was set up as described in the instrumentation manual with calibrations done according to the manufacturer’s specifications. A flow cytometry size calibration kit from Invitrogen (Molecular Probes, Carlsbad, CA) was used for general size correlation between reference beads and protein particles. The bottom sample was filtered through a 70 μm nylon mesh filter into a 12 × 75 mm polypropylene tube, and then diluted 15–20x with A5 buffer (called “pre-sort”). Four polypropylene tubes, each containing 200 uL of A5 were placed at 4°C in a sterile FACS sorting chamber. Samples were FACS sorted under the purify mode using a ceramic 70 μm nozzle tip at a pressure of 60 psi and at a speed of 5000–10,000 events/sec. The gain and sensitivity values of the detectors were optimized to maximize the protein particle detection and threshold. The charge plates were set to 4000V. Intellisort I was used to determine drop delay using flow check fluorospheres. All experiments used phosphate buffered saline (PBS) at pH 7.2, as the sheath fluid, daily prepared using deionized water (Labconco, Kansas City, MO) under aseptic conditions. The sheath fluid passed through an inline 0.22 μm filter just prior to mixture with pre-sort. Endotoxin levels were tested in each part of the instrument and were found to be at an acceptable range after the rigorous cleaning procedure as described below. Summit software version 5.2 (Beckman Coulter) was used for data collection and data analysis was performed using Kaluza Flow Analysis (Beckman Coulter).
Endotoxin cleaning and testing
Removing Endotoxins
Prior to using the stir-stressed mAb samples in the in-vitro cell-based assay, the samples were tested for endotoxin. High levels of endotoxin (>10 EU/mL) were present in the FACS-sorted samples despite sterilization of all instrument components and use of aseptic procedures. Therefore a protocol was developed to remove endotoxins by washing the instrument and all of its components with various solutions in the following sequence: 1) 10% bleach for 2 hours 2) deionized (Labconco) water for 2 hours 3) 70% ethanol for 2 hours 4) deionized water for 2 hours, 5) 1% Triton-X 114 (Sigma, St. Louis, MO) for 2 hours, 6) deionized water for 3 hours 7) approximately 1 mg/mL polymyxin B sulfate solution (Sigma, St. Louis, MO) for 3 hours, and 8) final flushing with deionized water for 3 hours.
Endotoxin Testing
All protein samples and buffers were assessed for endotoxin levels by a LAL (Limulus amebocyte lysate) test with the Charles River Endosafe®-PTS™(Charles River, Wilmington, MA) system prior to being used in the biological assay. The analysis was performed according to the manufacturer’s instructions.
Particle counting and sizing of mAb particles
HIAC-Royco Liquid Particle Counter
HIAC/Royco liquid particle counter model 9703 with a HRLD-150 sensor and PharmSpec software PharmSpec (HACH Ultra Analytics, Grants Pass, OR) was used for obtaining particle counts and size distribution in some experiments. The HIAC method is described in detail by Joubert et al.13,40
MicroFlow Imaging (MFI)
Particle images, size distribution, and counts were obtained using MFI System DPA4200 (Protein Simple, Santa Clara, CA). For analysis, stir-stressed mAb2 samples, generated at 1mg/mL, were diluted 100x in A5 buffer. Unstressed, buffer, and FACS sorted samples were diluted 25x prior to MFI analysis. After obtaining a clean base line, 300 μL of each sample was loaded using the peristaltic pump. No notable differences were observed in degassed versus non-treated samples (data not shown), so samples were not degassed for these studies.
MFI data analysis
Microflow imaging particle concentration data are displayed as either bar charts or radar charts, all corrected for dilution factors. Additionally, a plot of % Particles vs Sample (size bin) is displayed. The percentage of particles as detected by MFI was calculated by dividing the concentration of particles of a certain size range by the total number of particles in all size ranges. This percentage is referred to as “enrichment” throughout this manuscript. MFI morphology radar charts were generated in the size ranges 2–5, 5–10, and >10 μm to evaluate changes in particle intensity and aspect ratios as described previously.15,49 Finally, MFI particle data were used to calculate the estimated mass of protein within the particles, in the following four size bins: 1–2, 2–5, and 5–10, >10μm, using the ellipsoid volume method recently described.50
Resonant Mass Measurement (RMM)
A Particle Metrology System (Affinity Biosensors, Santa Barbara, California) was used with a Hi-Q micro sensor to quantify submicron and small micron particles from 0.2 to 1.85 μm. Sensor calibration and reference solution preparations were done according to the manufacturer’s instructions. After a clean frequency trace was obtained, protein sample was loaded for 30 s, with a stop trigger of 200 particles. This limit of detection was empirically determined and used throughout the study. A density value for mAb of 1.41 g/mL was used.
Biological testing of mAb particles using PBMC (in vitro comparative immunogenicity assessment assay, IVCIA)
Isolating Peripheral Blood Mononuclear Cells (PBMC)
PBMC from healthy human donors were obtained from Amgen’s environmental health and safety department. PBMC were isolated according to the procedure described previously.40
Challenging PBMC with protein samples and controls
Peripheral blood mononuclear cells from up to eight human donors were plated and acclimatized in 96-well culture plates as described previously.40 Acclimatized cells were then challenged with control (unstressed), and stir-20h mAb2 samples at equal volume, equal protein concentration at 40 μg/ml (total protein in solution determined by OD280 measurements), or equal particle number (based on light obscuration or MFI measurements). Negative controls, consisting of buffers isolated by FACS or medium-treated cells, and positive controls, such as lipopolysaccharide (LPS) or phytohemagglutinin (PHA) were also tested. LPS and PHA were controls at the early or late-stage immune response, respectively. Samples were incubated for either 20 h or 7 days (in separate culture plates), to assess an early stage or late stage immune response, respectively, as described by Joubert et al.40
Meso-Scale Discovery Assay
A 96-well human cytokine electro-chemiluminescence assay kit K151AYB-1 (Meso-Scale Discovery, Gaithersburg, MD) was used, according to the manufacturer’s instructions, to quantify the concentration of monocyte chemotactic protein-1 (MCP-1) that was secreted by PBMC in response to the samples. The 96 well plates were analyzed using an MSD Sector Imager 6000 instrument (Meso Scale Discovery, Gaithersburg, MD) and MSD Discovery Workbench Software 4.0 (Meso Scale Discovery). SoftMax Pro was used for data analysis to convert the output luminescent units into protein concentration (pg/mL) using a standard curve. Responses were presented in terms of concentration of MCP-1 secreted and percentage of responding donors. To determine the percentage of responding donors, the stimulation index (SI) was also calculated. The SI was calculated by dividing the amount of cytokine detected (pg/mL) in the sample of interest by the cytokine detected in the unstressed sample. For these studies, a response was considered to be positive if the SI ≥ 2.0 (i.e., response of the PBMC to the stressed sample was at least two fold higher than the control). The percentage of donors (% donors) that responded was calculated by taking the total number of donors that had an SI ≥ 2.0 as a percentage of the total number of donors tested. The cutoff of SI ≥ 2.0 was determined by statistical analysis as described elsewhere.40
Multiplex Cytokine Analysis
Quantification of cytokines released from PBMC was performed using a multiplex plate format using Milliplex (EMD Millipore, Billerica, MA) human panel kits and a Luminex Multiplexing instrument (specifically a Luminex FLEXMAP 3D instrument from EMD Millipore) as described by Joubert et al.40 For analysis of the PBMC supernatants at the early phase (after 20h incubation), the following cytokines were monitored: IL-10, IL-1ra, IL-1α, IL-1β, IL-6, IL-8, MCP-1, MIP-1α, MIP-1β, TNF-α, and TNF-β. For analysis of the PBMC supernatants at the late phase (after 7 days of incubation), the following cytokines were monitored: IFN-γ, IL-10, IL-12p40, IL-12p70, IL-13, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, and TNF-α. A robust PBMC response was difficult to obtain due to the dilute nature of the FACS purified samples. Therefore, the concentration of cytokines released by the PBMC was directly reported and the percentage of responding donors was calculated differently than for the MSD results. The cytokine response was analyzed as follows: for each cytokine monitored, the number of donors, who responded the highest (by releasing the largest amount of that same cytokine) to a specific size-enriched particle-containing sample was determined. The number of donors that responded highest to this size bin was then taken as a percentage of the total number of donors that were tested. In all cases, negative controls such as buffer isolated by FACS and medium-treated cells showed minimal response and positive controls such as LPS and/or PHA showed a very high response (SI≫2.0; data not shown). In all cases, PBMC responses induced by the mAb samples were much lower than those induced by the LPS or PHA positive controls (data not shown).
Biophysical characterization of mAb particles
Extrinsic Fluorescence Spectroscopy
8-Anilino-1-naphthalene sulfonate (8,1-ANS) was used to study changes in the accessibility of apolar regions in the protein samples. An Agilent 8453 (Santa Clara, CA) was used with baseline and light scattering corrections.51 Fluorescence data were obtained as described previously.15 A spectrum of buffer containing ANS was subtracted from each spectrum prior to data analysis using Microsoft Excel software. ANS peak positions and intensities were monitored and compared to that of the control samples to compare relative similarities or differences in surface hydrophobicity among samples.
Fourier Transform Infrared Spectroscopy/Microscopy
The control, heat, and stir-20h mAb2 samples in solution were analyzed with a Bruker Tensor 27 FTIR Spectrometer and Bio-ATR cell. Two hundred fifty-six scans were recorded from 600 to 4000 cm−1 with a resolution of 4 cm−1 as described in detail in Telikepalli et al.15 For analyzing mAb particles in the bottom sample, 3 μm gold filters (Pall Corporation, Port Washington, NY) were used to capture and wash protein particles and then analyzed by FTIR microscopy as described in detail elsewhere.15
SDS-PAGE
Each sample other than the FACS sorted samples was dissolved in NuPAGE LDS sample buffer (Life Technologies, Carlsbad, California) with and without 50 mM dithiothreitol (BioRad Laboratories, Hercules, CA) and incubated at 90°C for 5 min. Approximately 3 μg of each sample was separated on a 3–8% Tris-acetate gel using Tris-acetate running buffer (Life Technologies) for 65 min at 150V. A Hi-Mark unstained molecular weight ladder was used as a reference (Life Technologies). Protein bands were visualized by staining with Colloidal Coomassie (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions.
Transmission Electron Microscopy (TEM)
Six microliters of each sample was placed onto Lacey Carbon 300 Mesh Copper grids (Ted Pella, Redding, California) and allowed to sit for 2 min, with the excess wiped off by a Kimwipe. Stir-20h and bottom samples were diluted 100x with A5 buffer prior to being loaded onto the TEM grids. FACS sorted samples, heat-stressed, unstressed control, and buffers were loaded directly onto the TEM grids with no dilution. The grids were then placed into filtered 2% uranyl acetate for 2 min and extra stain was removed with a Kimwipe. The wet grids were air-dried for several min prior to being examined by TEM. Samples were imaged on an FEI Technai F20 XT Field Emission Transmission Electron Microscope (Hillsboro, OR) using 200 kV electron acceleration voltage. Images were captured at a standardized, normative electron dose and at a constant defocus value from the carbon-coated surfaces.52
Scanning Electron Microscopy (SEM)/Energy Dispersive X-ray Spectroscopy (EDS)
A FEI Versa 3D Dual Beam Scanning Electron Microscope/Focused Ion Beam (Hillsboro, OR) with an XMAX silicon drift detector (Oxford Instruments, Abingdon, Oxfordshire, UK) was used to obtain information regarding the surface morphology, elemental composition, and distribution of elements of the protein samples. SEM data was obtained at an acceleration voltage of 7 kV and a spot size of 4.0 using an Everthart Thornely (ET) detector for image collection. Elemental mapping and energy spectra were acquired and processed with AZtecEnergy software (Oxford Instruments, UK). For sample preparation, a small square piece of ruby red mica sheet (Electron Microscopy Sciences, Hatfield, PA) was mounted onto a standard SEM pin stub specimen mount (Ted Pella, Redding, CA). The grids prepared for TEM analysis were placed on top of the mica and a thin coating (3 nm) of electrically conductive material (gold) was deposited on the sample by a low vacuum sputter coater (Quorum Technology, Laughton, UK).
RESULTS
Initial comparisons of nanometer vs. micron sized mAb particles in a PBMC assay
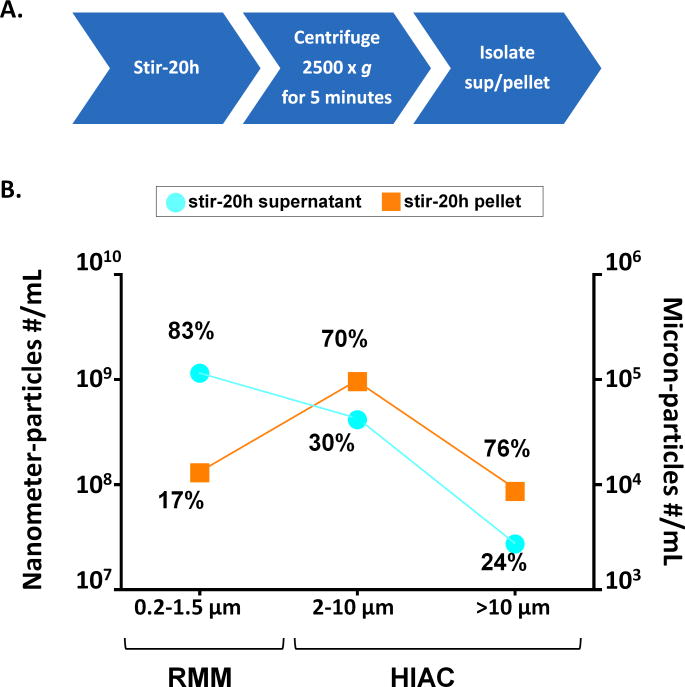
As an initial experiment, the IgG2 mAb (mAb2) in 10 mM acetate buffer, pH 5 (A5 buffer) was stirred for 20h and the particles generated were fractionated by slow speed centrifugation to separate the supernatant and pellet, containing enriched nanometer and micron-sized particles, respectively. Figure 1 shows the size distribution of nanometer and micron-sized particles present in the supernatant and pellet components as measured by HIAC and resonant mass measurement (RMM). There was almost a 10-fold higher number of smaller nanometer-sized particles (0.2-1.5 μm) in the supernatant than in the pellet, which contained more of the micron-sized particles (2-150 μm). The percentage values shown in Figure 1 were calculated by dividing the number of particles in either the supernatant or pellet by the total number of particles present in both supernatant and pellet in a particular size range. Hence the percentages signify the enrichment of a given size range in the samples. For example, 83% of all the particles detected in the 0.2–1.5 μm range are in the supernatant, while only 17% of particles in this size range are in the pellet. In addition, 70% of the particles detected in the 2–10 μm range are in the pellet, but only 30% of the particles in this size range are in the supernatant. To get a better estimate for the precision of each technique, relative standard deviations (RSD) were calculated by testing the stir-stressed mAb2 multiple times (N=10) on RMM; the RSD over the size range 0.2 – 1.5 μm was about 30%. Likewise, the RSD values from particle concentration measurements from HIAC were determined from multiple testing results from another mAb solution containing protein particles, which showed RSD values of 1% and 8% for the 2–10 μm particles, and for particles greater than 10 μm, respectively.53.
Figure 1.
Separation of nanometer-sized vs. micron-sized stirring-induced particles of mAb2 by low speed centrifugation. A) Schematic of experimental setup, and B) particle number and size distributions of the fractionated samples are shown in three different size ranges. The concentration of smaller particles (0.2–1.5 μm) for both supernatant and pellet was obtained by Resonant Mass Measurement (RMM; estimated RSD ~30 %, see text). The concentration of particles 2 μm to greater than 10 μm was obtained from light obscuration measurements (HIAC; estimated RSD 1%-8%, see text). The percentages were calculated by dividing the number of particles in either the supernatant or pellet by the total number of particles present in both supernatant and pellet at a particular size range.
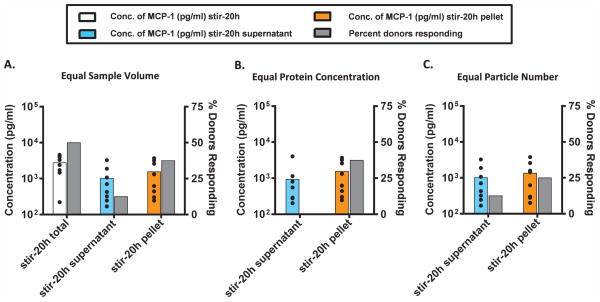
The fractionated samples were then analyzed for their cytokine response using an in vitro comparative immunogenicity assessment (IVCIA) assay,40 as described in the Materials and Methods section. To accomplish this, we tested the propensity of the supernatant (enriched in nanometer sized particles) and the resuspended pellet (enriched in micron-sized particles) to elicit a response in PBMC by monitoring the level of MCP-1 secreted. MCP-1 was chosen as a representative cytokine since we previously identified this cytokine as part of the cytokine signature that is released by PBMC in response to aggregates at the early phase.40 PBMC from eight naïve healthy human donors were challenged with the mAb2 stir-20h total, supernatant, and pellet samples; the secretion of MCP-1 was assessed at the early phase (20 h) by an electrochemiluminescence assay. These initial results are shown in Figure 2. The Y-axis on the left and corresponding colored bars represent the average concentration of MCP-1 detected in response to the different aggregated samples, while the Y-axis on the right and the corresponding gray bars show the percentage of donors that responded at least two fold higher to the aggregated samples above the unstressed mAb sample (SI ≥ 2.0). The dots represent the amount of cytokine secreted by each individual donor, and point out the distribution of responses across the population tested. Figure 2 highlights the variability of MCP-1 expression levels for individual donors as well as the variability in responsiveness to the different mAb samples. Individual donors have an inherent level of responsiveness that is dependent on their immune status and baseline inflammatory state. It should be noted that the overall measure of immune responsiveness to a challenge was a cumulative output of both the concentration of the cytokine secreted (colored bars) and the percentage of donors that responded (gray bars).
Figure 2.
Peripheral blood mononuclear cells (PBMC) from 8 human donors were incubated with solutions of mAb2 particles (see Figure 1) and examined for the release of MCP-1. MCP-1 was measured by electro-chemiluminescence at the early phase (20 h incubation) at (A) equal sample volume, (B) equal protein concentration, and (C) equal particle numbers. The average concentration (n=8, colored bars) of MCP-1 and percentage of donors that responded (gray bars) to the aggregated mAb (two fold above the unstressed mAb2) is shown. The black dots represent the concentration of MCP-1 secreted by each individual donor. Different aggregate samples are shown horizontally as follows; stir-20h total (white), stir-20h supernatant (enriched in nanometer size particles; light blue), and stir-20h pellet (enriched in micron sized particles; orange); see Figure 1. The media and buffer-stressed controls responded far below the threshold, and the LPS positive control responded much more intensely (SI≫2.0) than the aggregated protein.
The stir-20h total, supernatant, and pellet samples had different properties that might be important for causing a response in the assay, and so they were tested at equal volume, equal protein concentration and equal particle number in the assay, one at a time (Figure 2). When the three samples were challenged at equal sample volume, the stir-20h total sample (no fractionation) showed the highest response with respect to both the magnitude of secreted MCP-1 and the number of responding donors (Figure 2A). This sample also had the largest number of particles (it contained all the particles present in both the supernatant and pellet fractions before fractionation) so it was not surprising that it induced the largest response. The stir-20h pellet showed a higher response than the corresponding supernatant in terms of magnitude of cytokine secretion and number of responding donors. Similarly, at equal protein concentration (Figure 2B) and equal particle number (Figure 2C), the stir-20h pellet sample induced a higher response than the supernatant for both magnitude of cytokine secreted and the number of responding donors. At equal protein concentration, even though MCP-1 was detected when PBMC were challenged with the supernatant sample, none of the 8 donors responded two fold higher than to the unstressed sample (Figure 2B). The percentage of responding donors is a particularly useful indicator of immune response since it is measured above the donor’s response to the unstressed mAb so it takes into account the inherent responsiveness of the individual. In all three cases (Figure 2A–C), the pellet sample induced a higher percentage of responding donors than the supernatant, regardless of whether the samples were tested at equal volume (38% vs 13%), equal protein concentration (38% vs 0%), or equal particle number (25% vs 13%), suggesting that micron sized particles enhance the response of PBMC in vitro to a greater extent than nanometer sized particles at the early phase. The level of cytokine secretion induced by the mAb2 samples was, however, much lower than that induced by the positive control (lipopolysaccharide, LPS; data not shown).
To further test the wider applicability of these trends, the supernatant and pellet fractions of a second IgG2 monoclonal antibody were also tested in the IVCIA assay (data not shown). For both mAbs, the pellet fraction induced a higher response than the supernatant fraction, indicating that micron size particles induce a more robust response as compared to nanometer sized particles at the early phase.
Size enrichment of various micron sized particles using FACS
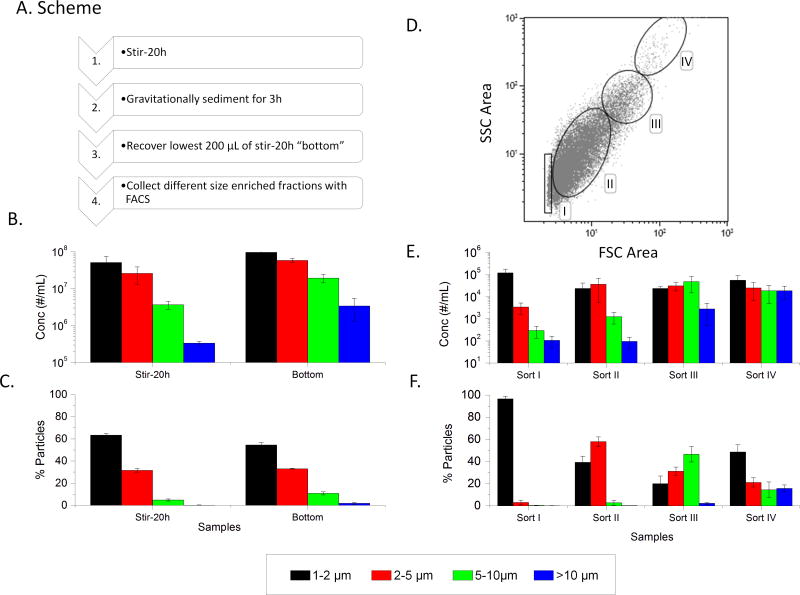
On the basis of the IVCIA data from evaluating stirring induced nanometer vs. micron sized mAb particles, it was observed that the larger micron sized particles showed higher relative responses in the donor population used (see above). We therefore set out to develop a method to size fractionate the micron sized mAb particles for further analysis. A wide variety (>20 conditions) of settling, centrifugation and filtration experiments were evaluated to size fractionate micron-sized, stirring-induced mAb2 particles with limited success (data not shown). After much trial and error experimentation, a methodology was identified that utilized a combination of gravitational settling under specific conditions combined with FACS, to enrich distinct micron sized particle populations (Figure 3A).
Figure 3.
Enrichment of different micron sized mAb2 particle populations using FACS separation. (A) Flowchart of experimental steps. (B) Subvisible particle counts in stirred sample (stir-20h) and bottom fraction after gravitational settling (bottom sample) as measured by MFI. (C) The percentage of particles in each size bin for these same two samples. (D) Upon FACS sorting the bottom sample, a two dimensional dot plot of response from forward vs side scattering signals (FSC Area vs. SSC Area) is generated with gatings labeled Sort I–IV, (E) Sorts I–IV were analyzed for subvisible particle distribution with MFI. (F) The percentage of particles in each size bin were determined. The graphs represent the average of three separate experiments (N=3) with the error bars representing one standard deviation.
The stir-20h mAb2 sample was gravitationally sedimented for 3h under specific conditions (see Methods section). The bottom fraction (referred to as the “bottom” sample) was then collected. The particle concentration and enrichment factor of the bottom sample as measured by MFI, in comparison to the original stir stressed mAb2 sample, is shown in Figures 3B and 3C, respectively. The bottom sample was then passed through the FACS and the forward scattering area (FSC) vs. side scattering (SSC) area dot plots were obtained as shown in Figure 3D. Each dot represents a counted particle and the FSC signal is generally considered to be a good indicator of particle size.54 Based on some initial correlations between FACS gating schemes and MFI sizing data (data not shown), gating schemes were optimized for best size enrichment and are shown in Figure 3D as I, II, III, and IV. Based on these four gatings, the stir-20h bottom samples were sorted and collected. These four sorted samples (I, II, III, and IV) were collected and analyzed by MFI as shown in Figures 3E and 3F. Sort 1 contains an enriched number of 1-2 μm particles, sort II contains an enriched number of 2-5 μm particles, Sort III an enriched number of 5–10 μm particles, and Sort IV an enriched number of greater than 10 μm particles (>10 μm); all sizes were calculated based on equivalent circular diameter (ECD). Comparing MFI enrichment results in Figure 3C with Figure 3F show a distinct enrichment in all four size ranges with good reproducibility as indicated by error bars. Comparing MFI counts in Figure 3B with Figure 3E, however, show a large decrease in particle concentration after FACS separation, due to an extensive dilution of the samples of over ~1000 fold. In this case, the FACS analysis had to be run numerous times (~20 runs over several months) to collect a sufficient volume of sample, for further analysis. Since so many runs were required for use in the PBMC assay, the FACS samples had to be collected and frozen after each run.
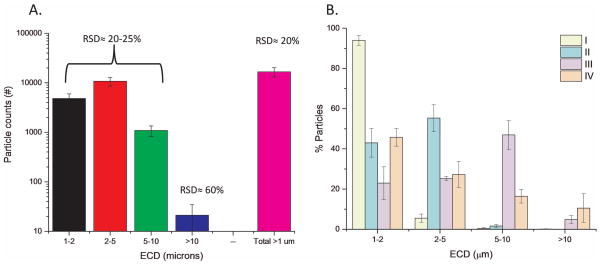
For logistical reasons, FACS samples had to undergo a total of two freeze thaw (F/T) cycles prior to being evaluated in the IVCIA assay. The frozen FACS sorted samples from each individual FACS run were thawed, pooled with other similar sorts, aliquoted and frozen again. It was these pooled frozen samples that were then subsequently thawed for biophysical and FACS analysis. These FACS sorted samples contained predominantly subvisible particles in PBS buffer, since the mAb particles had been largely separated from free protein by the FACS. For example, total protein levels were below detection in these samples as measured by OD 280 nm measurements, SDS-PAGE, or with a colorimetric bicinchoninic acid (BCA) assay (data not shown). Therefore, protein levels within the subvisible mAb particles could only be estimated by calculation from MFI data.50 To initially characterize the size measurements with these samples, one of the FACS sorted samples that underwent two F/T cycles, was examined by MFI (N=9) to examine the precision of the MFI results with these FACS samples. RSD values between 20–25% were obtained for particle counts in the first three size bins (1–2, 2–5, and 5–10 μm), 60% for the largest size bin analyzed, and 20% for the total MFI particle counts (Figure 4A). These RSD values give the relative error associated with each of these particle counts in these different size ranges. Additionally, the FACS sort pools (Sort I, II, III, IV) that underwent 2 F/T cycles were directly analyzed by MFI (N=3). The particle enrichment factor for each of these four FACS samples is maintained after two F/T treatments (See results in Figure 3F vs. Figure 4B).
Figure 4.
Determination of relative standard deviation percentage (RSD %) of MFI particle count and determination of enrichment factors for FACS separated samples of mAb2 particles. (A) Representative FACS sorted sample of stir-induced aggregated mAb2, obtained as outlined in Figure 3 and having undergone two freeze-thaw cycles, was used for repeat MFI testing to determine RSD % as a function of particle size bins (N=9 with the error bars representing one standard deviation), and (B) enrichment analysis of FACS sorted samples (Sort I, II, III, IV) after undergoing two freeze-thaw cycles. Data are an average of three separate experiments (N=3) with the error bars representing one standard deviation.
Table 1 summarizes subvisible particle number, size and mass characteristics, based on MFI analysis, for each of the pools of the four FACS sort samples (Sort I, II, III, and IV) that underwent two F/T cycles. The total particle concentration is shown including estimated ranges (based on twice the calculated RSD values from Figure 4). In addition, the particle concentration in each of the four size ranges (including enrichment values and estimated ranges) are shown. It is apparent here that the experimentally determined total particle concentration results (column 3) falls within the range expected based on RSD calculations (column 4) for nearly all of the FACS Sorts. In addition, the calculated mass of the protein particles in each size range, based on MFI morphology measurements and assumptions of protein particle density as described by Kalonia et al, 2014,50 is shown in Table 1.
Table 1.
Summary of Various Characteristics of the FACS-Sorted Samples I–IV used in the IVCIA Assay are shown. The average particle concentration, experimental concentration range obtained from N = 2, and estimated concentration range based on 2RSD from N = 9 from Figure 4 measurements using the same protein are shown. In addition, the average concentration and range of particles in each of the four size bins (1–2, 2–5, 5–10, >10 μm), their corresponding enrichment values, mass of protein in each bin (as calculated from Kalonia et al.50), and the range of mass of protein within those size bins is shown.
| Sorts | Total Particle Concentration (#) | Size Bin | Particle Concentration and Enrichment by Size Bin | Calculated Particle Mass by Size Bin | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Average Concentration (#/mL) | Experimental Concentration Range (#/mL) N = 2 | Estimated Concentration Range Based on 2RSD (#/mL) N = 9 | Equivalent Circular Diameter (μ m) | Average Concentration (#/mL) | Average Enrichment (%) | Concentration Range (#/mL) | Enrichment Range (%) | Average Mass Protein (ng) | Mass Range Protein (ng) | |
| I | 71,300 | 61,100–81,400 | 49,000–110,000 | 1–2 | 67,400 | 94 | 56,000–79,000 | 92–97 | 2.9 | 2.4–3.3 |
| 2–5 | 3500 | 5 | 2700–4400 | 3–7 | 1.5 | 1.0–1.9 | ||||
| 5–10 | 280 | 0 | 60–500 | 0–1 | 2.1 | 0.5–3.6 | ||||
| >10 | 100 | 0 | 50–150 | 0 | 5.1 | 2.4–7.8 | ||||
| II | 31,000 | 25,500–36,500 | 22,000–51,000 | 1–2 | 14,400 | 47 | 12,700–16,000 | 44–50 | 1 | 0.8–1.2 |
| 2–5 | 16,300 | 52 | 12,400–20,100 | 49–55 | 9.6 | 6.8–12 | ||||
| 5–10 | 400 | 1 | 360–380 | 1 | 1.3 | 1.2–1.4 | ||||
| >10 | 20 | 0 | 13–30 | 0 | 7.5 | 1.0–14 | ||||
| III | 110,400 | 85,900–135,000 | 81,000–190,000 | 1–2 | 30,000 | 28 | 25,200–34,700 | 26–29 | 1.7 | 1.4–2.0 |
| 2–5 | 28,200 | 26 | 23,000–33,800 | 25–26 | 28 | 21–36 | ||||
| 5–10 | 48,000 | 43 | 35,300–60,700 | 41–45 | 345 | 241–449 | ||||
| >10 | 4200 | 4 | 2900–5600 | 3–4 | 119 | 90–147 | ||||
| IV | 46,900 | 32,000–62,000 | 37,000–87,000 | 1–2 | 21,200 | 46 | 16,100–26,200 | 42–51 | 1.2 | 0.9–1.5 |
| 2–5 | 14,300 | 31 | 10,300–18,300 | 30–32 | 11 | 7.3–14 | ||||
| 5–10 | 8000 | 16 | 4100–12,000 | 13–19 | 71 | 32–110 | ||||
| >10 | 3400 | 7 | 1400–5500 | 4–9 | 103 | 45–162 | ||||
IVCIA testing of FACS size-enriched populations of micron sized mAb particles
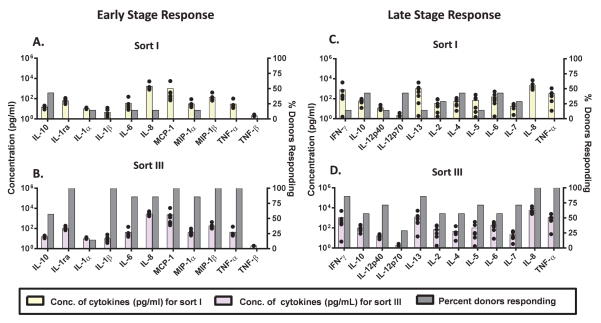
The four FACS samples described in Table 1 (Sort I, II, III, and IV) were next evaluated in the IVCIA assay. PBMC from 7 human donors were challenged with equal volumes and similar particle counts of each of these four FACS samples. The response to these FACS enriched micron-sized mAb particles was monitored at two different times, in the early stage (20 h incubation) and late stage (7 days incubation). Multiplex cytokine analysis was used to assess two different panels of cytokines that were specific for each stage of the incubation. The resulting cytokine profiles are shown in Figures 5 and 6. A head-to-head comparison of the PBMC response to Sort I (containing enriched 1–2 μm particles) and Sort III (containing enriched 5–10 μm particles) is shown in Figure 5 at both the early phase (20 h, Figures 5A, 5B) and late phase (7 days, Figures 5C, 5D). Since the overall particle counts in each Sort sample were low, the PBMC responses observed were also low (compared to results shown in Figure 2).
Figure 5.
Peripheral blood mononuclear cells (PBMC) from 7 human donors were incubated with mAb2 particle samples collected from the FACS separation and analyzed for cytokine release. Sort I (containing enriched 1–2 μm particles) (A, C) and Sort III (containing enriched 5–10 μm particles) (B, D) from FACS separation as well as the relevant controls were tested for the release of cytokines by multiplex cytokine analysis at the early phase (20 h) (A, B) and late phase (7 day) (C, D). FACS samples were added to the PBMC culture at equal volume and similar particle concentration (high concentration; > 20,000+ particles/mL in culture). The average concentration across donors (N=7, colored bars) of the 11 cytokines tested at the early phase (A, B) and 12 cytokines tested at the late phase (C, D) is shown. The percentage of responding donors (gray bars) represents the number of the 7 donors tested that responded higher (by magnitude of secreted cytokine) to each sort. The black dots represent the concentration of cytokines secreted by each individual donor. The LPS-and PHA-positive controls responded much more intensely (SI≫2.0) than the aggregated protein samples (data not shown).
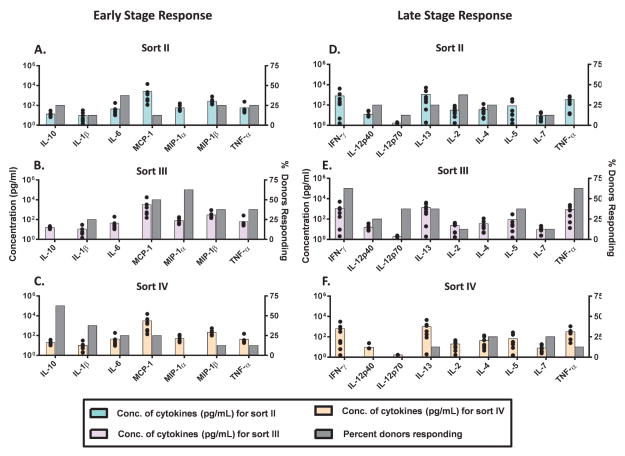
Figure 6.
Peripheral blood mononuclear cells (PBMC) from 8 human donors were incubated with FACS isolated mAb2 particle samples (containing very little soluble aggregates): sort II (enriched in 2–5 μm particles) (A, D), sort III (enriched for 5–10 μm particles) (B, E), and sort IV (enriched for particles > 10 μm) (C, F) and analyzed for their cytokine release. The FACS samples were added to the PBMC culture at similar particle numbers (low concentration; >9,000+ particles/mL in culture). These FACS samples as well as the relevant controls were tested for the release of signature cytokines by multiplex cytokine analysis at the early phase (20h) and late phase (7 days). The average concentration across donors (N=8, colored bars) of representative cytokines tested at the early and at the late phases is shown. The percentage of responding donors (gray bars) represents the number of the 8 donors tested that responded higher (by magnitude of secreted cytokines) to each sort. The black dots represent the concentration of cytokines secreted by each individual donor.
Comparison of results in Figures 5A with 5B show that the concentrations of cytokines released (colored bars corresponding to the left-hand Y-axis) by PBMC in response to Sort I vs. Sort III are similar. However, the percentage of donors that responded (shown on the right hand Y-axis) to Sort III is higher as compared to Sort I. Although this result shows that most of the donors responded more to the particles in Sort III than Sort I, it is important to keep in mind that this was not a dramatic response and the donors responded only marginally higher overall. Similar observations can be made by comparing results in Figures 5C with 5D at the late phase of the PBMC response, which shows additional T-cell effector cytokines that are more specific to the late phase of the immune response. This subtle difference between Sort I and Sort III is thought to be partly due to the low particle counts, which prevented robust cytokine release. However, it is apparent that more donors consistently responded to the sort III sample than to the sort I sample.
Since Sort I showed a lower response than Sort III (Figure 5), it was not assessed further. Sorts II, III, and IV were further assessed (Figure 6). PBMC from 8 human donors were challenged with similar particle counts of Sort II, III, and IV and the resulting cytokine profiles are shown in Figure 6 at both the early phase (Figure 6A, 6B, 6C) and the late phase (Figure 6D, 6E, 6F). Lower numbers of particles were added to the PBMC culture for this comparison (as compared to Figure 5) as some of the sorts were more dilute and so less particles were added to keep the comparison equal among the Sort II, III and IV samples. Therefore, the overall response, in terms of cytokine concentration, was lower in Figure 6 as compared to Figure 5 probably due to the more dilute nature of the samples tested in Figure 6. Despite these limitations, a trend of a greater number of responding donors across the different cytokines was observed for the sort III sample (enriched in 5–10 μm particles) for the PBMC at both the early and late phases of this in vitro assay (Fig. 6).
Biophysical characterization of FACS size-enriched populations of micron-sized mAb particles
The sort I–IV size enriched samples from the FACS contained subvisible particles, but an undetectable amount of free protein in solution (as detected by OD280, SDS-PAGE or BCA assay), so this provided an opportunity to characterize protein particles without the presence of IgG2 mAb in solution. Because these FACS samples had a low mAb particle concentration (and thus low protein concentration), only a limited number of physical assays could be used (see below). Therefore, the IgG2 mAb sample generated just prior to FACS separation was also characterized (stirring followed by gravitational settling and selection of the “bottom” sample) to better understand the nature of the micron-sized particles going into the FACS separation. These samples contained both protein particles and soluble IgG2 mAb.
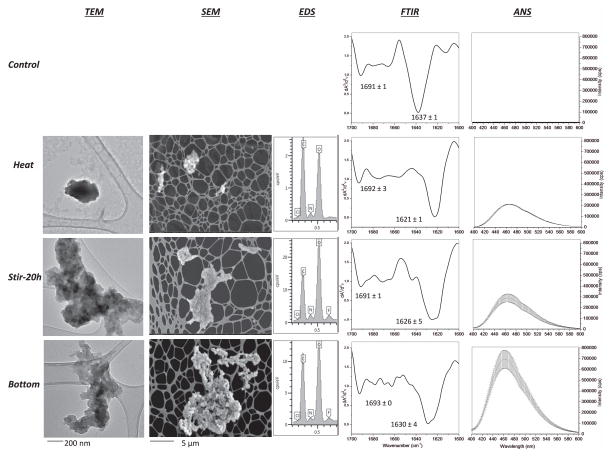
The biophysical analysis of the starting material for FACS analysis focused on four samples: two controls (unstressed and extensively heated mAb2 solutions as negative and positive controls, respectively), the stirred antibody solution (stir-20h), and the “bottom” sample from the stirred sample after gravitational settling (i.e., the starting material for the FACS separation). Only the heat-stressed control contained high molecular weight aggregates containing reducible covalent, disulfide linkages, which were not present in the other three samples as measured by SDS-PAGE (Supplementary Fig S1). Figure 7 shows TEM, SEM, EDS, FTIR, and ANS fluorescence spectroscopy data for the four samples. The unstressed control (Figure 7; first row) contained few particles and thus could not be imaged by TEM and SEM-EDS (see Supplemental Figures S2 and S3). The unstressed mAb2 solution contains antibody with largely native secondary structure (intramolecular beta sheets) with characteristic positions of the two peaks at 1691 cm−1 and 1637 cm−1, as determined by second-derivative FTIR spectroscopy. It also shows virtually no extrinsic fluorescence intensity indicating limited exposure of apolar regions for ANS binding.
Figure 7.
Biophysical analysis of mAb2 solutions containing stirring-induced particles that were used as starting material for FACS separation. Control samples (unstressed and heat denatured) and two stirred samples (Stir-20h, Bottom) of mAb2 were characterized for morphology by SEM and TEM, composition by EDS, overall secondary structure by solution FTIR and FTIR microscopy, and surface hydrophobicity by extrinsic fluorescence using 1,8-ANS as indicated in this Figure. Representative TEM and SEM images are shown at two resolutions (the unstressed control had virtually no particles so no TEM and SEM images are shown). For FTIR analysis, shifts in two peaks for intramolecular beta sheet content (occurring between 1620 to 1640 cm−1 and 1690 to 1700 cm−1) were monitored in the second derivative FTIR spectra. The peak minima are shown for each sample (N=3 with the error bars representing one standard deviation).
Similar biophysical analysis was then performed on the subvisible particle containing samples of mAb2. Results from heat stressed, stir-20h, and the bottom samples are shown in Rows 2, 3 and 4 of Figure 7, respectively. These particles were observed to be amorphous in nature by TEM and SEM and contained predominantly carbon, nitrogen, and oxygen by EDS analysis (as expected for protein). All protein samples contained trace levels of elemental chlorine, presumably bound to the mAb protein during exposure to NaCl solutions during purification. Interestingly, the stir-20h samples also contain fluorine, probably originating from the Teflon coated stir-bar, while the heated particles did not contain fluorine (unstressed and heat stressed samples also lacked fluorine; see Supplemental Figures S2 and S3). The heat control samples contained protein with structurally altered conformation, containing mAb with predominantly intermolecular beta sheets, as indicated by the 1621 cm−1 peak by FTIR, as well as increase in surface hydrophobicity relative to the unstressed control. The major peak in FTIR spectra of the particles isolated from the two stirred samples (stir-20h and bottom samples) was between the two control samples, even though there was some increased variability in peak position. The bottom sample showed the largest increase in ANS intensity indicating a relatively increased surface hydrophobicity (or aggregation) of protein within this sample (last row in Figure 7).
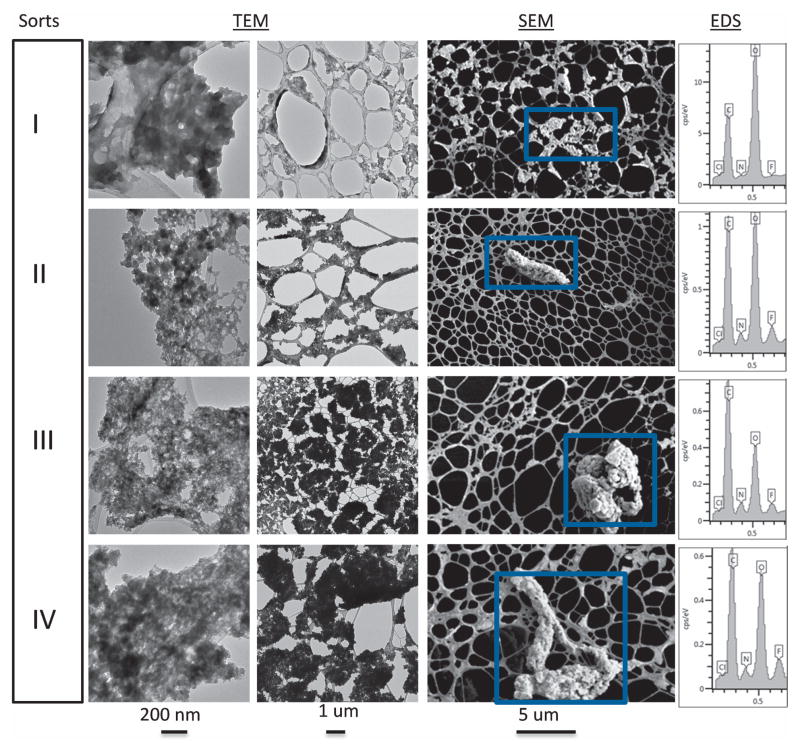
Thus, there is some level of structural alteration of the protein contained in the stir-induced protein particles as well as increase in surface hydrophobicity relative to the unstressed and heated controls. The bottom sample was the starting material for the FACS separation, leading to collection of Sorts I, II, III, and IV containing size enriched micron sized particles. The protein particles after FACS separation (Sorts I, II, III, and IV) were quite dilute compared to the samples described above, but were in sufficient number to be visualized by TEM and SEM (Figure 8). Representative TEM images at two resolutions (200 nm and 1 μm) with protein particles represented by the darker gray regions are shown, while the lighter, smoother, gray regions are due to the TEM grids used for sample preparation. Representative SEM images (5 μm resolution) are shown, and the protein particles appear lighter with unique shapes and are highlighted by the blue boxes. These SEM images provide a unique visual representation of the morphology of the protein particles compared to TEM. Aside from their size, the protein particles from the four FACS samples (Sorts I, II, III, IV) are similar morphologically. This is especially apparent in the 1 and 5 μm resolution TEM and SEM images, respectively. The proteins within the blue boxes in the SEM images were analyzed for chemical composition by EDS (last column). Similar to the starting material used in the FACS separation, the FACS separated particles contain C, N, O, and trace amounts of chlorine and fluorine. Elemental mapping of these particles confirmed that the fluorine was present only in the particles and not in the background or control samples (Supplemental Figure S4).
Figure 8.
Representative TEM (200 nm and 1 μm resolution) and SEM (5 μm resolution) images of mAb2 particles of four different FACS sorted samples (Sort I, II, III, IV). The SEM particle images (representative graphs shown here), selected by the blue boxes, were also analyzed for their elemental composition by EDS as shown in far right column of the figure.
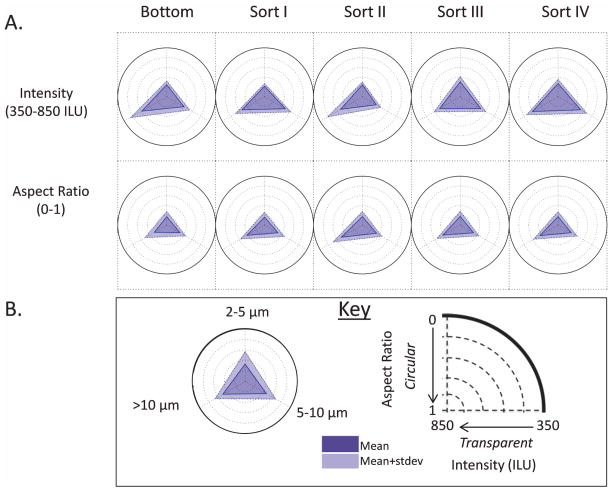
In addition, MFI data were used to compare the morphology of these purified particles by constructing radar charts.15,49,55 These radar charts are a data visualization technique, which facilitates comparisons of a specific morphological characteristic in a large number of samples such as their aspect ratio and transparency (intensity). The radar charts shown in Figure 9 focus on the morphological nature of the particles and do not display results on the concentration of particles in the different size bins (which are summarized in Table 1). For example, MFI morphology radar chart analysis of the Sort I, II, III, and IV FACS samples, and the bottom mAb2 sample (starting material for FACS separation) is shown in Figure 9. As shown in the key in Figure 9B, size bins start at 2–5 μm at the top of the circle and increase clockwise up to > 10 μm. The outermost circle has four more concentric circles within it and consists of a triangle, located in the center, pointing to three size bins. The triangle can get larger or smaller within the circle towards one or more size bin. Depending on whether the intensity parameter or aspect ratio parameter are being analyzed, the expanding triangle shows that the particles in that size bin are becoming elongated (aspect ratio; going from 1 to 0) or more opaque (intensity; going from 850 to 350 ILU).
Figure 9.
Radar plots of MFI morphology data (aspect ratio and intensity; refer to the key in B) for subvisible particles of mAb2 present in the starting material for FACS (bottom sample) and FACS sorted samples (sorts I-IV, see text) are shown in A. Radar plots show MFI morphology data distributions of the micron sized particles that fall within each of the three size bins shown in the key of B. The data shown are the average of three separate experiments (N = 3) and the error represents one standard deviation.
To visualize whether a change occurred in a specific morphological parameter, the reader can compare the position of the triangle’s vertex across five concentric circles, which represent a measurement value of aspect ratio or intensity. Since each vertex of the triangle represents a different size bin, the expanding or contracting of each vertex towards the smallest to largest concentric circle, respectively, indicates that a change in a morphological parameter of particles within that size bin has occurred. From Figure 9A, no differences were observed in intensity or aspect ratio of FACS sorted mAb particles in the 2–5 μm size range although these small sizes approach the limit of MFI resolution for morphology parameters (around 4 μm). In the upper row of triangles in Figure 9, the vertex of the triangles pointing to 2–5 μm for all 5 triangles (corresponding to intensity parameter) are in the same position. This means that the transparency of the 2–5 μm particles present in each of these five samples is similar to one another. A similar observation can be made for the second row of radar charts (corresponding to the aspect ratio). There is no change in the aspect ratio of the 2–5 μm particles present in each of these samples. As a visual reference, the reader can use movement between the concentric circles to gauge changes in morphological parameters. For the 5–10 μm size bin, particles present in Sort I, III, and IV samples appear slightly more opaque than similarly sized particles in the bottom and Sort II samples. This can be seen in the upper panel of Figure 9A, where the triangle’s vertex corresponding to this size bin of the Sort I, III and IV radar charts are slightly more expanded towards the outermost concentric circle as compared to the similar vertex in the bottom and Sort II samples. Particles larger than 10 μm in the bottom and Sort II samples appear more opaque than similarly sized particles in Sort I, III, and IV. In terms of aspect ratio, the FACS sorted samples showed more elongated particles in the largest size bin (>10μm) compared to the bottom sample. Overall, Figure 9 helps determine that the morphological differences (aspect ratio and transparency, especially) arising from the FACS separation are relatively minor, and overall, the morphological parameters of the protein particles in the four FACS samples were similar.
DISCUSSION
This work evaluated the immune activation potential of protein aggregates of varying size by examining size-enriched populations of stirring-induced mAb2 particles in an in-vitro cell based assay, known as IVCIA, with PBMC from human donors. As an initial set of experiments, we showed that micron sized protein particles enhanced cytokine secretion in the IVCIA assay to a greater extent than nanometer-sized particles. To determine which size subset of micron sized aggregates induced the greatest response, we utilized a combination of gravitational settling and FACS separation to obtain different micron size-enriched protein particles from a stirred solution of mAb2. FACS sorting resulted in isolated protein particles, with no detectable soluble protein in samples as measured by OD 280nm, SDS-PAGE or BCA analysis, offering the opportunity for additional biophysical characterization of isolated protein particles (in addition to testing these particles in the IVCIA assay).
The mAb2 particles induced by stirring were shown, compared to untreated and heat stressed controls, to contain an amorphous morphology with protein within the particles having some structural alterations in their overall secondary structure and increased surface hydrophobicity, but retaining some of the native higher-order structures as well (Figure 7). The FACS separation results for these mAb2 particles are consistent with those of Nishi et al. in that this technique involves a relatively mild condition that does not disrupt particles to a detectable extent.45 In this work, even after two freeze-thaw cycles, significant size-enrichment of mAb particles is maintained within the FACS sorted samples. This result indicates either these aggregates are irreversible, or that the conditions used to enrich and isolate them do not disrupt the reversible interactions. Previous characterization of aggregates of mAb2 generated by stirring demonstrated some reversibility upon dilution, supporting the second conclusion.40
Fluorescence-activated cell sorting separates particles by imparting a charge on the droplet containing them. From our analysis of morphology of the FACS sorted samples, although minor differences from MFI radar chart analysis were noted, it appears FACS does not dramatically alter the overall nature of particles before and after FACS separation. The small differences observed in the radar plots may actually reflect the different amounts of particles in different size bins (Table 1). Thus, when evaluating the average aspect ratio or intensity in a size bin, one also needs to consider the number of particles present in that bin. A combination of TEM, SEM-EDS and MFI analysis of the morphology and composition of mAb2 particles before and after FACS separation showed no notable differences (Figure 7, 8, 9). Interestingly, particles generated by stirring contained fluorine as measured by SEM-EDS, and this element was not present in the buffer and control mAb2 (unstressed and heat stressed) samples indicating that it was originating from the stir-stress (Teflon-coated stir bars were used in this study) and carried with the particles through the FACS separation. The presence of this element in protein particles and its impact on PBMC responses is a subject for investigation in future work. This result also highlights that generating particles under accelerated conditions (i.e., extensive stirring with a Teflon coated stir bar) does not necessarily mimic mAb particle formation under real-time storage conditions. In fact, fluorine was not observed in control or heat-stressed mAb2 samples.
There are some practical limitations of the FACS technique for preparative isolation of protein particles that need to be improved in the future. First, FACS isolation results in significant dilution of the particle sample (~1000X in these studies) resulting in low particle yields (compare Figure 3B to Figure 3E). In fact, to prepare sufficient amounts of size-enriched mAb particle samples for biophysical and biological testing in this work, many months of almost continuous FACS runs were required. This in turn required freezing of individual sorts from individual runs, followed by subsequent thawing, pooling and refreezing of particle preparations to obtain the Sorts I, II, III, and IV evaluated in this work. Even then, low particle counts obtained (< 10,000 particles/mL) approached the lower limit of particles required for the IVCIA assay and consequently led to low PBMC responses. In addition, it is also difficult to further concentrate the FACS separated particles without loss of material or inadvertently altering their size distribution due to additional processing. Although Rombach-Riegraf et al. showed that resorting IgG particle samples by FACS increases particle purity within distinct size bins42, this was not a viable option in our work since it would have dramatically decreased particle concentration even more, making the samples unsuitable for the in-vitro assay or biophysical characterization. Based on these considerations, further work is needed to develop a particle purification technique that can more effectively obtain sufficient quantities of highly concentrated size-enriched protein particle samples for use in these in-vitro assays.
The dilution from the FACS could result in the loss of reversible aggregates, so that the samples assessed after FACS analysis might be primarily irreversible aggregates. This should present a “worse case” for immunological potential assessment, but might not reflect the original aggregate population. It was found that even after two freeze-thaws, the FACS sorted samples retained a similar distribution of subvisible particle counts, indicating that they are in a fairly stable state where they do not appear to either dissociate or aggregate, indicating potential irreversibility. SDS-PAGE gels showed that the “bottom” sample, used as the starting material for FACS separation, did not contain covalently linked high molecular weight aggregates; unfortunately, due to the dilute nature of the FACS samples, they could not be assessed by this technique. It is possible that the bottom sample contained either covalently or non-covalently linked aggregates that dissociated upon sample preparation. In that case, the aggregates should be fairly reversible. However, more work needs to be done to further characterize the reversibility nature of these FACS sorted samples.
These IVCIA results are consistent with other studies that have examined the role of the size of subvisible particles in eliciting an immune response.35,40,56–63 For example, stressed conditions that generated high levels of subvisible particles elicited ADA titers in various mouse models with heat stressed allergens57, human growth hormone 58, and recombinant human interferon alpha 2b.35 Further evidence suggests that larger sized subvisible particles are more likely to elicit various immune responses. For example, when high pressure was applied to aggregates of human or murine growth hormone, subvisible particle counts declined along with a corresponding decrease in immunogenicity in mice. 58,64 These results agree with prior work using protein coated spheres as a model system for protein aggregates of different sizes where antibody-coated 5 μm microspheres induced a greater response in (1) the IVCIA assay compared to heterogeneous stress-induced aggregate populations40, and (2) in a Xeno-het mouse model system compared to antibody-coated nanospheres (20–50 nm).47 In this work, 5–10 μm sized protein particles displayed elevated levels of cytokine responses compared to the other sized mAb2 particles tested in the same IVCIA assay. In addition, the fraction enriched in high numbers of nanometer sized particles induced a lower response than the fraction enriched in micron sized particles in the IVCIA assay, indicating that nanometer sized particles may not be the preferred size range for inducing an immune response.
The role of structural integrity of protein molecules within particles on immune responses has also been widely studied. 35,40,56,57,59,61–63,65 Results from one of our laboratories have previously shown that aggregated solutions with high numbers of 2–10 μm particles and the retention of at least some folded protein structure caused the highest PBMC immune response.40 In another study, Filipe et al. subjected a fully human IgG1 to various stresses and found that the stress that formed the most micron sized particles, which were mostly native in structure, actually did not show a high immune response.59 Instead copper-catalyzed aggregates, containing little to no visible and micron sized particles, low surface hydrophobicity, and large changes in secondary and tertiary structures were the ones that showed the highest ADA response in transgenic mice.59 This aggregate type was shown to contain a high level of copper and histidine oxidation,13,14 a modification not found in other types of stress treated aggregates. These results highlight the importance of aggregate type in inducing an immune response, which may be more important than size, amount, or morphology of the aggregates.59 In our work, the stirring induced mAb2 particles contained protein structural alterations in secondary and tertiary structure as shown by FTIR microscopy and extrinsic ANS fluorescence spectroscopy, respectively. Compared to the heat control, the stirred samples displayed some retention of native higher-order structures, which may be necessary for the observed PBMC response.
The inter-relationship of protein particle size, number and mass presents a challenge in determining the most important factors in generating an immune response. Rombach-Riegraf et al showed that the mass of protein present in subvisible particles may be an important property.61 They saw a positive correlation between protein mass in subvisible particles, generated by stressing an IgG mAb, and the amount of antigen processed and presented by the dendritic cells (DC). The more subvisible particles there are, the greater the mass of protein, and consequently, the greater the presentation of peptide by DC, which can influence DC maturation and activation of CD4+ T cells61. While we did not design the IVCIA assay to examine the effect of protein mass of the FACS sorted samples, we subsequently estimated the weight of protein particles in Sorts I–IV using MFI morphology data using a new method from one of our laboratories.50 As shown in Table 1, Sort III, which showed the greatest response in the IVCIA assay, contained both the highest number of particles and the highest mass of protein present in the 5–10 μm size range (Table 1). Even then, roughly 25% of the total protein mass in this sample was estimated to be from the particles larger than 10 μm. Additionally, even though Sort IV contained a significant mass of protein calculated from the 5–10 μm particles, this sample showed a lower relative response in this PBMC in-vitro assay. Sort IV contained an enriched number of particles greater than 10 μm, and consequently most of the mass of the protein was present in these particles. This highlights that both protein mass and number of particles are important factors for consideration in monitoring cytokine responses in this assay.
In summary, we observed that size-enriched fractions of stirring-induced mAb2 particles, especially in the 5–10 μm range, elicited a relatively greater cytokine response in a number of PBMC donors in the in-vitro assay. However, many questions remain. In terms of physical characteristics of the mAb aggregates, it would be beneficial to use the methods established in this work to perform additional case studies with FACS size-enriched mAb particle populations (e.g., with different mAbs and different stresses and thus different physicochemical characteristics) to determine if the trends in size and immune potential observed in this work are generally observed across different types of mAb aggregates. In terms of biological readouts, even though this PBMC in-vitro model avoids the challenges associated with animal models, accounts for human genetic diversity, and mimics administration of the drug, it still requires a large number of particles, perhaps much more than clinically relevant, to obtain a strong signal.40 Additionally, it is important to note that the cytokines monitored in this study serve as biomarkers of early and late phases of the immune response in this in-vitro system. They are useful tools to assess the relative response of a diverse human population of PBMC and to rank responses relative to different types of aggregates. However, how the PBMC cytokine responses correlate with various in-vivo immune processes or immunogenicity is currently unknown due to the complexities arising from the pleiotropic nature of cytokines and the complexity of the immune system where the T cell driven B cell response and consequent signaling and antibody formation cannot be reproduced in-vitro. A combination of this in-vitro assay with other in-silico and in-vivo models, however, should result in enhanced predictive value in determining the relative immunogenic potential of different therapeutics.
Supplementary Material
Supplementary Figure S1. (A) Nonreduced and (B) reduced SDS-PAGE gels of control mAb2 (unstressed), heat denatured mAb2, and two stirred mAb2 samples (stir-20h, bottom) in A5 buffer. The bottom sample is the starting material for the FACS separation (see text). MW standards are shown with indicated values in kDa. Please note the additional samples in the gel correspond to the supernatant and pellet samples of stirred mAb solutions from the slow speed centrifugation approach described in the Methods.
Supplementary Figure S2. Representative TEM images at three size scales of buffer (sodium acetate buffer, pH 5), control mAb2 (unstressed), heat denatured mAb2, and two mAb2 stirred samples (stir-20h, bottom) in A5 buffer. The bottom sample is the starting material for the FACS separation (see text).
Supplementary Figure S3. Representative EDS spectra and SEM images of buffer (sodium acetate buffer, pH 5), unstressed, heat denatured, and two stirred samples (stir-20h, bottom) of mAb2 in A5 buffer. The bottom sample is the starting material for the FACS separation (see text). Regions in the SEM images (indicated by the white boxes or shapes) were selected for elemental composition determination. Elements in greatest abundance are shown on the right hand side of the figure in ratio form. The numerator depicts the number of times an element is detected in a region while the denominator depicts the total number of regions analyzed. All samples show background levels of carbon and oxygen, uranium, copper, aluminum, silicon, and gold coming from the sample grids, sample studs, and coating required for acquiring proper images.
Supplementary Figure S4. Representative EDS spectra and SEM images of bottom and FACS sorted samples (I, II, III, IV) are shown. Regions in the SEM images (indicated by the white shapes) were selected for elemental composition determination. Elements in greatest abundance are shown on the right hand side of the figure in ratio form. The numerator depicts the number of times an element is detected in a region while the denominator depicts the total number of regions analyzed. All samples show background levels of carbon and oxygen, uranium, copper, aluminum, silicon, and gold coming from the sample grids, sample studs, and coating required for acquiring proper images.
Acknowledgments
The authors wish to thank Cavan Kalonia for assistance with calculating the mass of protein in subvisible particle, and John Ferbas and Keith Kelley for useful discussions regarding FACS. The authors would also like to thank Dr. Sunil David for helpful discussions on endotoxin removal. The University of Kansas authors would like to acknowledge Amgen for financial support for these studies. Additional financial support was provided by the Kansas Biosciences Authority, and ST financial support was provided by NIH biotechnology training grant 5-T32-GM008359.
Footnotes
SUPPORTING INFORMATION AVAILABLE
This article contains supplementary material available from the authors upon request or via the Internet at http://onlinelibrary.wiley.com
References
- 1.Baker MP, Reynolds HM, Lumicisi B, Bryson CJ. Immunogenicity of protein therapeutics: The key causes, consequences and challenges. Self Nonself. 2010;1(4):314–322. doi: 10.4161/self.1.4.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boven K, Knight J, Bader F, Rossert J, Eckardt KU, Casadevall N. Epoetin-associated pure red cell aplasia in patients with chronic kidney disease: solving the mystery. Nephrology, dialysis, transplantation: official publication of the European Dialysis and Transplant Association - European Renal Association. 2005;20(Suppl 3):iii33–40. doi: 10.1093/ndt/gfh1072. [DOI] [PubMed] [Google Scholar]
- 3.Casadevall N. What is antibody-mediated pure red cell aplasia (PRCA)? Nephrology, dialysis, transplantation: official publication of the European Dialysis and Transplant Association - European Renal Association. 2005;20(Suppl 4):iv3–8. doi: 10.1093/ndt/gfh1088. [DOI] [PubMed] [Google Scholar]
- 4.Chirino AJ, Ary ML, Marshall SA. Minimizing the immunogenicity of protein therapeutics. Drug Discov Today. 2004;9(2):82–90. doi: 10.1016/S1359-6446(03)02953-2. [DOI] [PubMed] [Google Scholar]
- 5.Claman HN. Tolerance to a Protein Antigen in Adult Mice and the Effect of Nonspecific Factors. Journal of Immunology. 1963;91:833–839. [PubMed] [Google Scholar]
- 6.Gamble CN. The role of soluble aggregates in the primary immune response of mice to human gamma globulin. International Archives of Allergy and applied. Immunology. 1966;30(5):446–455. doi: 10.1159/000229829. [DOI] [PubMed] [Google Scholar]
- 7.Goudemand J, Rothschild C, Demiguel V, Vinciguerrat C, Lambert T, Chambost H, Borel-Derlon A, Claeyssens S, Laurian Y, Calvez T FVIII-LFB Recombinant FVIII study groups . Influence of the type of factor VIII concentrate on the incidence of factor VIII inhibitors in previously untreated patients with severe hemophilia A. Blood. 2006;107(1):46–51. doi: 10.1182/blood-2005-04-1371. [DOI] [PubMed] [Google Scholar]
- 8.Moore WV, Leppert P. Role of aggregated human growth hormone (hGH) in development of antibodies to hGH. The Journal of Clinical Endocrinology and Metabolism. 1980;51(4):691–697. doi: 10.1210/jcem-51-4-691. [DOI] [PubMed] [Google Scholar]
- 9.Wang W, Roberts CJ, editors. Aggregation of Therapeutic Proteins. Hoboken, NJ: John Wiley & Sons; 2010. p. 486. [Google Scholar]
- 10.Wang W, Singh SK, Li N, Toler MR, King KR, Nema S. Immunogenicity of protein aggregates--concerns and realities. International Journal of Pharmaceutics. 2012;431(1–2):1–11. doi: 10.1016/j.ijpharm.2012.04.040. [DOI] [PubMed] [Google Scholar]
- 11.Mahler HC, Friess W, Grauschopf U, Kiese S. Protein aggregation: pathways, induction factors and analysis. Journal of Pharmaceutical Sciences. 2009;98(9):2909–2934. doi: 10.1002/jps.21566. [DOI] [PubMed] [Google Scholar]
- 12.Cromwell ME, Hilario E, Jacobson F. Protein aggregation and bioprocessing. The AAPS Journal. 2006;8(3):E572–579. doi: 10.1208/aapsj080366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joubert MK, Luo Q, Nashed-Samuel Y, Wypych J, Narhi LO. Classification and characterization of therapeutic antibody aggregates. The Journal of Biological Chemistry. 2011;286(28):25118–25133. doi: 10.1074/jbc.M110.160457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo Q, Joubert MK, Stevenson R, Ketchem RR, Narhi LO, Wypych J. Chemical modifications in therapeutic protein aggregates generated under different stress conditions. The Journal of Biological Chemistry. 2011;286(28):25134–25144. doi: 10.1074/jbc.M110.160440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Telikepalli SN, Kumru OS, Kalonia C, Esfandiary R, Joshi SB, Middaugh CR, Volkin DB. Structural characterization of IgG1 mAb aggregates and particles generated under various stress conditions. Journal of Pharmaceutical Sciences. 2014;103(3):796–809. doi: 10.1002/jps.23839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwura T, Fukuda J, Yamazaki K, Arisaka F. Conformational stability, reversibility and heat-induced aggregation of alpha-1-acid glycoprotein. Journal of Biochemistry. 2014 doi: 10.1093/jb/mvu050. [DOI] [PubMed] [Google Scholar]
- 17.Liu L, Braun LJ, Wang W, Randolph TW, Carpenter JF. Freezing-induced perturbation of tertiary structure of a monoclonal antibody. Journal of Pharmaceutical Sciences. 2014;103(7):1979–1986. doi: 10.1002/jps.24013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sahin E, Grillo AO, Perkins MD, Roberts CJ. Comparative effects of pH and ionic strength on protein-protein interactions, unfolding, and aggregation for IgG1 antibodies. Journal of Pharmaceutical Sciences. 2010;99(12):4830–4848. doi: 10.1002/jps.22198. [DOI] [PubMed] [Google Scholar]
- 19.Sharma B. Immunogenicity of therapeutic proteins. Part 2: impact of container closures. Biotechnology Advances. 2007;25(3):318–324. doi: 10.1016/j.biotechadv.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Shire SJ, Shahrokh Z, Liu J. Challenges in the development of high protein concentration formulations. Journal of Pharmaceutical Sciences. 2004;93(6):1390–1402. doi: 10.1002/jps.20079. [DOI] [PubMed] [Google Scholar]
- 21.Wang W. Protein aggregation and its inhibition in biopharmaceutics. International Journal of Pharmaceutics. 2005;289(1–2):1–30. doi: 10.1016/j.ijpharm.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 22.Wiesbauer J, Prassl R, Nidetzky B. Renewal of the air-water interface as a critical system parameter of protein stability: aggregation of the human growth hormone and its prevention by surface-active compounds. Langmuir: the ACS Journal of Surfaces and Colloids. 2013;29(49):15240–15250. doi: 10.1021/la4028223. [DOI] [PubMed] [Google Scholar]
- 23.Schellekens H. Factors influencing the immunogenicity of therapeutic proteins. Nephrology, Dialysis, Transplantation: Official Publication of the European Dialysis and Transplant Association - European Renal Association. 2005;20(Suppl 6):vi3–9. doi: 10.1093/ndt/gfh1092. [DOI] [PubMed] [Google Scholar]
- 24.Ponce R, Abad L, Amaravadi L, Gelzleichter T, Gore E, Green J, Gupta S, Herzyk D, Hurst C, Ivens IA, Kawabata T, Maier C, Mounho B, Rup B, Shankar G, Smith H, Thomas P, Wierda D. Immunogenicity of biologically-derived therapeutics: assessment and interpretation of nonclinical safety studies. Regulatory Toxicology and Pharmacology: RTP. 2009;54(2):164–182. doi: 10.1016/j.yrtph.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 25.Schellekens H. Immunogenicity of therapeutic proteins: clinical implications and future prospects. Clinical Therapeutics. 2002;24(11):1720–1740. doi: 10.1016/s0149-2918(02)80075-3. discussion 1719. [DOI] [PubMed] [Google Scholar]
- 26.Jahn EM, Schneider CK. How to systematically evaluate immunogenicity of therapeutic proteins-regulatory considerations. N Biotechnol. 2009;25(5):280–286. doi: 10.1016/j.nbt.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 27.Johnson R, Jiskoot W. Models for evaluation of relative immunogenic potential of protein particles in biopharmaceutical protein formulations. Journal of Pharmaceutical Sciences. 2012;101(10):3586–3592. doi: 10.1002/jps.23248. [DOI] [PubMed] [Google Scholar]
- 28.Perry LC, Jones TD, Baker MP. New approaches to prediction of immune responses to therapeutic proteins during preclinical development. Drugs R&D. 2008;9(6):385–396. doi: 10.2165/0126839-200809060-00004. [DOI] [PubMed] [Google Scholar]
- 29.Koren E, De Groot AS, Jawa V, Beck KD, Boone T, Rivera D, Li L, Mytych D, Koscec M, Weeraratne D, Swanson S, Martin W. Clinical validation of the “in silico” prediction of immunogenicity of a human recombinant therapeutic protein. Clinical Immunology. 2007;124(1):26–32. doi: 10.1016/j.clim.2007.03.544. [DOI] [PubMed] [Google Scholar]
- 30.Tatarewicz SM, Wei X, Gupta S, Masterman D, Swanson SJ, Moxness MS. Development of a maturing T-cell-mediated immune response in patients with idiopathic Parkinson’s disease receiving r-metHuGDNF via continuous intraputaminal infusion. Journal of Clinical Immunology. 2007;27(6):620–627. doi: 10.1007/s10875-007-9117-8. [DOI] [PubMed] [Google Scholar]
- 31.Wullner D, Zhou L, Bramhall E, Kuck A, Goletz TJ, Swanson S, Chirmule N, Jawa V. Considerations for optimization and validation of an in vitro PBMC derived T cell assay for immunogenicity prediction of biotherapeutics. Clinical Immunology. 2010;137(1):5–14. doi: 10.1016/j.clim.2010.06.018. [DOI] [PubMed] [Google Scholar]
- 32.Jaber A, Baker M. Assessment of the immunogenicity of different interferon beta-1a formulations using ex vivo T-cell assays. Journal of Pharmaceutical and Biomedical Analysis. 2007;43(4):1256–1261. doi: 10.1016/j.jpba.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 33.Stickler M, Rochanayon N, Razo OJ, Mucha J, Gebel W, Faravashi N, Chin R, Holmes S, Harding FA. An in vitro human cell-based assay to rank the relative immunogenicity of proteins. Toxicological Sciences: an Official Journal of the Society of Toxicology. 2004;77(2):280–289. doi: 10.1093/toxsci/kfh021. [DOI] [PubMed] [Google Scholar]
- 34.Brinks V, Jiskoot W, Schellekens H. Immunogenicity of therapeutic proteins: the use of animal models. Pharmaceutical Research. 2011;28(10):2379–2385. doi: 10.1007/s11095-011-0523-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hermeling S, Aranha L, Damen JM, Slijper M, Schellekens H, Crommelin DJ, Jiskoot W. Structural characterization and immunogenicity in wild-type and immune tolerant mice of degraded recombinant human interferon alpha2b. Pharmaceutical Research. 2005;22(12):1997–2006. doi: 10.1007/s11095-005-8177-9. [DOI] [PubMed] [Google Scholar]
- 36.Braun A, Kwee L, Labow MA, Alsenz J. Protein aggregates seem to play a key role among the parameters influencing the antigenicity of interferon alpha (IFN-alpha) in normal and transgenic mice. Pharmaceutical Research. 1997;14(10):1472–1478. doi: 10.1023/a:1012193326789. [DOI] [PubMed] [Google Scholar]
- 37.Stewart TA, Hollingshead PG, Pitts SL, Chang R, Martin LE, Oakley H. Transgenic mice as a model to test the immunogenicity of proteins altered by site-specific mutagenesis. Molecular Biology & Medicine. 1989;6(4):275–281. [PubMed] [Google Scholar]
- 38.Whiteley PJ, Lake JP, Selden RF, Kapp JA. Tolerance induced by physiological levels of secreted proteins in transgenic mice expressing human insulin. The Journal of Clinical Investigation. 1989;84(5):1550–1554. doi: 10.1172/JCI114331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaitonde P, Balu-Iyer SV. In vitro immunogenicity risk assessment of therapeutic proteins in preclinical setting. Drug Design & Discovery Methods in Molecular Biology. 2011;716:267–280. doi: 10.1007/978-1-61779-012-6_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joubert MK, Hokom M, Eakin C, Zhou L, Deshpande M, Baker MP, Goletz TJ, Kerwin BA, Chirmule N, Narhi LO, Jawa V. Highly aggregated antibody therapeutics can enhance the in vitro innate and late-stage T-cell immune responses. The Journal of Biological Chemistry. 2012;287(30):25266–25279. doi: 10.1074/jbc.M111.330902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Narhi LO, Schmit J, Bechtold-Peters K, Sharma D. Classification of protein aggregates. Journal of Pharmaceutical Sciences. 2012;101(2):493–498. doi: 10.1002/jps.22790. [DOI] [PubMed] [Google Scholar]
- 42.Rombach-Riegraf V, Allard C, Angevaare E, Matter A, Ossuli B, Strehl R, Raulf F, Bluemel M, Egodage K, Jeschke M, Koulov AV. Size fractionation of microscopic protein aggregates using a preparative fluorescence-activated cell sorter. Journal of Pharmaceutical Sciences. 2013;102(7):2128–2135. doi: 10.1002/jps.23532. [DOI] [PubMed] [Google Scholar]
- 43.Espargaro A, Sabate R, Ventura S. Thioflavin-S staining coupled to flow cytometry. A screening tool to detect in vivo protein aggregation. Molecular Bio Systems. 2012;8(11):2839–2844. doi: 10.1039/c2mb25214g. [DOI] [PubMed] [Google Scholar]
- 44.Mach H, Bhambhani A, Meyer BK, Burek S, Davis H, Blue JT, Evans RK. The use of flow cytometry for the detection of subvisible particles in therapeutic protein formulations. Journal of Pharmaceutical Sciences. 2011;100(5):1671–1678. doi: 10.1002/jps.22414. [DOI] [PubMed] [Google Scholar]
- 45.Nishi H, Mathas R, Furst R, Winter G. Label-free flow cytometry analysis of subvisible aggregates in liquid IgG1 antibody formulations. Journal of Pharmaceutical Sciences. 2014;103(1):90–99. doi: 10.1002/jps.23782. [DOI] [PubMed] [Google Scholar]
- 46.Ludwig DB, Trotter JT, Gabrielson JP, Carpenter JF, Randolph TW. Flow cytometry: a promising technique for the study of silicone oil-induced particulate formation in protein formulations. Analytical Biochemistry. 2011;410(2):191–199. doi: 10.1016/j.ab.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bi V, Jawa V, Joubert MK, Kaliyaperumal A, Eakin C, Richmond K, Pan O, Sun J, Hokom M, Goletz TJ, Wypych J, Zhou L, Kerwin BA, Narhi LO, Arora T. Development of a human antibody tolerant mouse model to assess the immunogenicity risk due to aggregated biotherapeutics. Journal of Pharmaceutical Sciences. 2013;102(10):3545–3555. doi: 10.1002/jps.23663. [DOI] [PubMed] [Google Scholar]
- 48.Shukla AA, Hubbard B, Tressel T, Guhan S, Low D. Downstream processing of monoclonal antibodies--application of platform approaches. Journal of chromatography B, Analytical Technologies in the Biomedical and Life Sciences. 2007;848(1):28–39. doi: 10.1016/j.jchromb.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 49.Kalonia C, Kumru OS, Kim JH, Middaugh CR, Volkin DB. Radar chart array analysis to visualize effects of formulation variables on IgG1 particle formation as measured by multiple analytical techniques. Journal of Pharmaceutical Sciences. 2013;102(12):4256–4267. doi: 10.1002/jps.23738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kalonia C, Kumru OS, Prajapati I, Mathaes R, Engert J, Zhou S, Middaugh CR, Volkin DB. Calculating the mass of subvisible protein particles with improved accuracy using micro-flow imaging data. Journal of Pharmaceutical Sciences. 2015;104(2):536–547. doi: 10.1002/jps.24156. [DOI] [PubMed] [Google Scholar]
- 51.Mach H, Middaugh CR. Ultraviolet spectroscopy as a tool in therapeutic protein development. Journal of Pharmaceutical Sciences. 2010;100(4):1214–1227. doi: 10.1002/jps.22385. [DOI] [PubMed] [Google Scholar]
- 52.Kumru OS, Liu J, Ji JA, Cheng W, Wang YJ, Wang T, Joshi SB, Middaugh CR, Volkin DB. Compatibility, physical stability, and characterization of an IgG4 monoclonal antibody after dilution into different intravenous administration bags. Journal of Pharmaceutical Sciences. 2012;101(10):3636–3650. doi: 10.1002/jps.23224. [DOI] [PubMed] [Google Scholar]
- 53.Jiao N. Personal Communication. 2014.
- 54.Shapiro HM. Practical Flow Cytometry. New Jersey: Wiley; 2003. p. 681. [Google Scholar]
- 55.Kim JH, Iyer V, Joshi SB, Volkin DB, Middaugh CR. Improved data visualization techniques for analyzing macromolecule structural changes. Protein Science. 2012;21(10):1540–1553. doi: 10.1002/pro.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hermeling S, Schellekens H, Maas C, Gebbink MF, Crommelin DJ, Jiskoot W. Antibody response to aggregated human interferon alpha2b in wild-type and transgenic immune tolerant mice depends on type and level of aggregation. Journal of Pharmaceutical Sciences. 2006;95(5):1084–1096. doi: 10.1002/jps.20599. [DOI] [PubMed] [Google Scholar]
- 57.Johansen P, Senti G, Martinez Gomez JM, Wuthrich B, Bot A, Kundig TM. Heat denaturation, a simple method to improve the immunotherapeutic potential of allergens. European Journal of Immunology. 2005;35(12):3591–3598. doi: 10.1002/eji.200535076. [DOI] [PubMed] [Google Scholar]
- 58.Fradkin AH, Carpenter JF, Randolph TW. Immunogenicity of aggregates of recombinant human growth hormone in mouse models. Journal of Pharmaceutical Sciences. 2009;98(9):3247–3264. doi: 10.1002/jps.21834. [DOI] [PubMed] [Google Scholar]
- 59.Filipe V, Jiskoot W, Basmeleh AH, Halim A, Schellekens H, Brinks V. Immunogenicity of different stressed IgG monoclonal antibody formulations in immune tolerant transgenic mice. mAbs. 2012;4(6):740–752. doi: 10.4161/mabs.22066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fradkin AH, Mozziconacci O, Schoneich C, Carpenter JF, Randolph TW. UV photodegradation of murine growth hormone: chemical analysis and immunogenicity consequences. European Journal of Pharmaceutics and Biopharmaceutics. 2014;87(2):395–402. doi: 10.1016/j.ejpb.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rombach-Riegraf V, Karle AC, Wolf B, Sorde L, Koepke S, Gottlieb S, Krieg J, Djidja MC, Baban A, Spindeldreher S, Koulov AV, Kiessling A. Aggregation of human recombinant monoclonal antibodies influences the capacity of dendritic cells to stimulate adaptive T-cell responses in vitro. Public Library of Science One. 2014;9(1):e86322. doi: 10.1371/journal.pone.0086322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Beers MM, Gilli F, Schellekens H, Randolph TW, Jiskoot W. Immunogenicity of recombinant human interferon beta interacting with particles of glass, metal, and polystyrene. Journal of Pharmaceutical Sciences. 2012;101(1):187–199. doi: 10.1002/jps.22744. [DOI] [PubMed] [Google Scholar]
- 63.van Beers MM, Sauerborn M, Gilli F, Brinks V, Schellekens H, Jiskoot W. Oxidized and aggregated recombinant human interferon beta is immunogenic in human interferon beta transgenic mice. Pharmaceutical Research. 2011;28(10):2393–2402. doi: 10.1007/s11095-011-0451-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fradkin AH, Carpenter JF, Randolph TW. Glass particles as an adjuvant: a model for adverse immunogenicity of therapeutic proteins. Journal of Pharmaceutical Sciences. 2011;100(11):4953–4964. doi: 10.1002/jps.22683. [DOI] [PubMed] [Google Scholar]
- 65.Seong SY, Matzinger P. Hydrophobicity: an ancient damage-associated molecular pattern that initiates innate immune responses. Nature Reviews Immunology. 2004;4(6):469–478. doi: 10.1038/nri1372. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. (A) Nonreduced and (B) reduced SDS-PAGE gels of control mAb2 (unstressed), heat denatured mAb2, and two stirred mAb2 samples (stir-20h, bottom) in A5 buffer. The bottom sample is the starting material for the FACS separation (see text). MW standards are shown with indicated values in kDa. Please note the additional samples in the gel correspond to the supernatant and pellet samples of stirred mAb solutions from the slow speed centrifugation approach described in the Methods.
Supplementary Figure S2. Representative TEM images at three size scales of buffer (sodium acetate buffer, pH 5), control mAb2 (unstressed), heat denatured mAb2, and two mAb2 stirred samples (stir-20h, bottom) in A5 buffer. The bottom sample is the starting material for the FACS separation (see text).
Supplementary Figure S3. Representative EDS spectra and SEM images of buffer (sodium acetate buffer, pH 5), unstressed, heat denatured, and two stirred samples (stir-20h, bottom) of mAb2 in A5 buffer. The bottom sample is the starting material for the FACS separation (see text). Regions in the SEM images (indicated by the white boxes or shapes) were selected for elemental composition determination. Elements in greatest abundance are shown on the right hand side of the figure in ratio form. The numerator depicts the number of times an element is detected in a region while the denominator depicts the total number of regions analyzed. All samples show background levels of carbon and oxygen, uranium, copper, aluminum, silicon, and gold coming from the sample grids, sample studs, and coating required for acquiring proper images.
Supplementary Figure S4. Representative EDS spectra and SEM images of bottom and FACS sorted samples (I, II, III, IV) are shown. Regions in the SEM images (indicated by the white shapes) were selected for elemental composition determination. Elements in greatest abundance are shown on the right hand side of the figure in ratio form. The numerator depicts the number of times an element is detected in a region while the denominator depicts the total number of regions analyzed. All samples show background levels of carbon and oxygen, uranium, copper, aluminum, silicon, and gold coming from the sample grids, sample studs, and coating required for acquiring proper images.