Abstract
Bioassay-directed fractionation of an antiproliferative ethanol extract of the roots of Ocotea macrocarpa (Lauraceae) afforded the new butanolide macrocarpolide A (1), and the two new secobutanolides macrocarpolides B (2) and C (3), together with the known butanolides linderanolide B (4) and isolinderanolide (5). The structure elucidation of all compounds was carried out based on NMR and mass spectroscopic data analyses. The absolute configurations of all compounds isolated were determined by comparison of their optical rotation values with those found in literature. Compounds 1–5 showed good antiproliferative activities against the A2780 ovarian cell line, with IC50 values of 2.57 ± 0.12 (1), 1.98 ± 0.23 (2), 1.67 ± 0.05 (3), 2.43 ± 0.41 (4), and 1.65 ± 0.44 µM (5), respectively.
Keywords: Antiproliferative activity, Ocotea macrocarpa, Butanolide, Lauraceae
As a part of the Madagascar International Cooperative Biodiversity Group (ICBG) program,2ab an ethanol extract of the roots of Ocotea macrocarpa was found to have moderate activity against the A2780 ovarian cancer cell line (IC50 3.9 µg/ml). This extract was thus selected for further evaluation for the presence of novel anticancer agents. The plant genus Ocotea, the largest member of the Lauraceae family, comprises approximately 350 species that are distributed throughout tropical and subtropical climates. Most species are found in the Americas from Mexico to Argentina, seven species are found in Africa, one specie is found in the Canary Islands, and about 34 recognized species are found in Madagascar.3ab Some species are used in traditional medicine, including for treatment of fever and malaria.4 Chemical investigations on various Ocotea species have led to the isolation of a wide range of secondary metabolites including alkaloids, flavonoids, lignans, and terpenoids, many of which exhibited interesting antiproliferative, antifungal, antiherpetic, antiinflammatory, and antimicrobial activities.5abcdefg
Bioassay-guided isolation of an extract of the roots of Ocotea macrocarpa produced five bioactive compounds: one new butanolide (1), two new secobutanolides (2 and 3), and two known butanolides, linderanolide B (4)6 and isolinderanolide (5).7 The structures of the known compounds were determined by a comparison of their 1H NMR and mass spectra data with literature data, together with a comparison of their optical rotation values with the literature values.
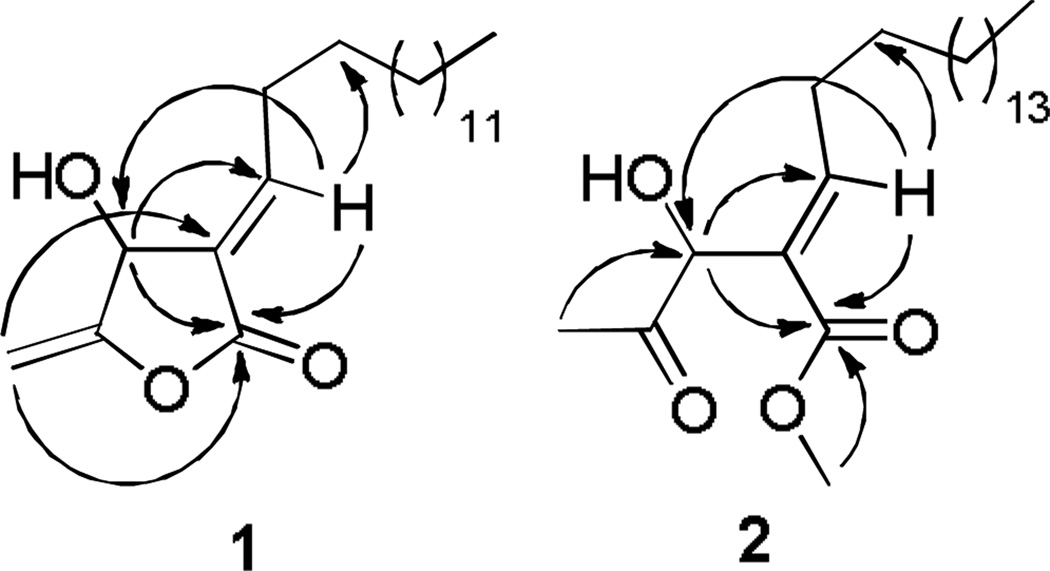
Compound 1 was isolated as a colorless oil. The molecular formula was determined to be C20H34O3 by HRESIMS ([M + H]+, m/z 323.2586, cal. for C20H35O3+ 323.2581). The IR exhibited the characteristic absorption bands at 3450 cm−1 for a hydroxyl group, and 1760 and 1700 cm−1 for an α,β-unsaturated-γ-lactone.8 The UV spectrum of 1 had an absorption maximum at 226 nm. The IR, UV and 1H NMR spectroscopic data of 1 were comparable to those of 4 and 5, suggesting that 1 had the same β-hydroxy-γ-methylene-α,β-unsaturated γ-lactone skeleton. The proton signal at δH 7.10 (dt, J = 7.8, 2.0 Hz, 1H, H-1′) in 1 differed significantly from the corresponding signals in 4 and 5 at δH 6.68 (td, J= 7.8, 2.0 Hz, 1H, H-1′), suggesting the E configuration for Δ3(1′) in 1.89 The 1H NMR spectrum of 1 also displayed resonances assignable to two exomethylene protons appearing at δH 4.96 and δH 4.72 (dd, J = 2.8, 1.4 Hz, each 1H, H2-6), one oxymethine at δH 5.26 (brs, 1H, H-4), and two deshielded methylene protons at δH 2.50 and δH 2.43 (dt, J = 14.8, 7.2 Hz, each 1H, H2-2′). The positions of these protons were assigned from HMBC experimentation (Fig. 2). The exocyclic olefinic signals at δH 4.96 and δH 4.72 (H2-6) were correlated with both a quaternary carbon at δC 157.8 (C-5) and a methine carbon at δC 66.7 (C-4). Carbon 5 also correlated with the oxymethine signal at δH 5.26 (H-4). Furthermore, clear long range correlations between both the oxymethine proton at δH 7.10 (H-1′) to the carbonyl carbon at δC 166.1 (C-2) were observed in the HMBC spectrum.
Figure 2.
Key HMBC correlations of 1 and 2.
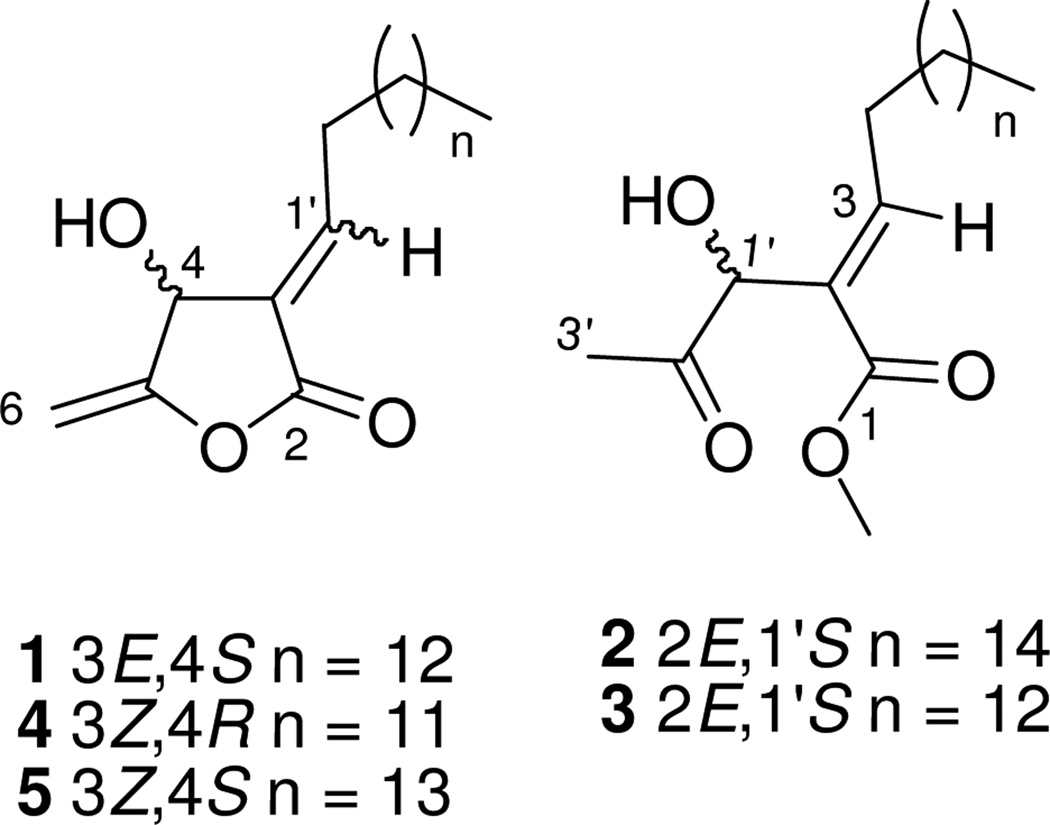
In addition, a broad peak at δH 1.25–1.31 (28H, H-3′–14′) and a triplet at δH 0.88 (J = 7.0 Hz, H-15′) were attributed to the methylene protons in a long alkyl chain and the terminal methyl group in 1, respectively. Compound 1 showed an [α]21D value of −11.11 (c 0.27, MeOH), indicating the S configuration at C-4 as described for previously reported butanolides.9,10ab The complete assignments of all protons and carbons of 1 (Table 1) were accomplished by further interpretation of its HMBC and HSQC spectra. Thus, the structure of 1 was elucidated as (3E,4S)-4-hydroxy-5-methylene-3-pentadecylidene-dihydro-furan-2-one, and named macrocarpolide A.
Table 1.
1H and 13C NMR data for compound 1.a
| Posn | δHb | δcc |
|---|---|---|
| 2 | 166.1 (C) | |
| 3 | 127.4 (C) | |
| 4 | 5.26 brs | 66.7 (CH) |
| 5 | 157.8 (C) | |
| 6 | 4.96 dd (2.8, 1.4) | 91.5 (CH2) |
| 4.72 dd (2.8, 1.4) | ||
| 1′ | 7.10 dt (7.8, 2.0) | 150.3 (CH) |
| 2′ | 2.50 dt (14.6, 7.7) | 29.8 (CH2) |
| 2.43 dt (14.6, 7.7) | ||
| 3′ | 1.25–1.31 | 28.3 (CH2) |
| 4′ | 1.25–1.31 | 29.8–29.5 (CH2) |
| 5′ | 1.25–1.31 | 29.8–29.5 (CH2) |
| 6′ | 1.25–1.31 | 29.8–29.5 (CH2) |
| 7′ | 1.25–1.31 | 29.8–29.5 (CH2) |
| 8′ | 1.25–1.31 | 29.8–29.5 (CH2) |
| 9′ | 1.25–1.31 | 29.8–29.5 (CH2) |
| 10′ | 1.25–1.31 | 29.8–29.5 (CH2) |
| 11’ | 1.25–1.31 | 29.8–29.5 (CH2) |
| 12′ | 1.25–1.31 | 29.8–29.5 (CH2) |
| 13′ | 1.25–1.31 | 32.1 (CH2) |
| 14′ | 1.25–1.31 | 22.8 (CH2) |
| 15′ | 0.88 t (7.0) | 14.3 (CH3) |
Assignments based on analysis of 2D NMR spectra.
Data (δ) measured at 500 MHz; brs = broad singlet, dd= doublet of doublets, dt = doublet of triplets. J values are in Hz and are omitted if the signals overlapped as multiplets. The overlapped signals were assigned from HSQC and HMBC spectra without designating multiplicity.
Data (δ) measured at 125 MHz; CH3, CH2, CH, and C multiplicities were determined by HSQC experiment.
Compound 2, a colorless oil, had a molecular formula of C23H42O4, as deduced from its HRESIMS spectrum (m/z 383.3157 [M+H]+, calcd. for C23H43O4+, 383.3156). The IR spectrum of 2 showed absorption bands characteristic of hydroxyl (3458 cm−1), ester (1734 cm−1), and ketone (1715 cm−1) groups. The UV absorption at 222 nm together with its IR and 1H NMR spectroscopic data indicated a secobutanolide skeleton.9,10b Comparison of the 1H NMR spectroscopic data of 2 with those of 1 revealed that the 1H NMR of 2 exhibited additional signals at δH 3.73 (s, 3H, 1-OMe) and δH 2.15 (s, 3H, H-3′), but lacked the signals at δH 4.96 and δH 4.72 in 1. This fact confirmed the presence of a methoxy and an acetyl group, and the absence of the α,β-unsaturated-γ-lactone ring in 2. In the HMBC spectrum, protons of the acetyl group at δH 2.15 (H-3′) showed correlations to an oxymethine group at δC 73.5 (C-1′). The methoxy protons at δH 3.73 (1-OMe) correlated with a carbonyl carbon at δC 166.7 (C-1), and the olefinic proton at δH 7.08 (t, J= 7.7 Hz, H-3) exhibited cross peaks with both the oxymethine carbon (δC 73.5, C-1′) and the carbonyl carbon (δC 166.7, C-1). Those correlations confirmed the assignment of a secobutanolide skeleton to 2. By the same analysis used to characterize compound 1, the deshielded methylene group of 2 was assigned at C-4 by the HMBC correlation between δH 2.35 (q, J = 7.6 Hz, 2H, H-4) and the quaternary olefinic carbon at δC 129.9 (C-2). Furthermore, the presence of an E trisubstituted double bond was evident from the characteristic chemical shift of the olefinic proton at δH 7.08 (H-3), compared to that of known compounds with a Z conformation (δH 6.69).9,10b
The positive optical activity (+2.23, c 2.24, MeOH) of 2 indicated that C-1′ possessed the S configuration.11abc Similarly to 1, the complete assignments of all protons and carbons of 2 (Table 1) were accomplished by further interpretation of its HMBC and HSQC spectra. From the above data, compound 2 was assigned as (2E)-2-[(1S)-1-hydroxy-2-oxo-propyl]-nonadec-2-enoic acid methyl ester, and named macrocarpolide B.
The molecular formula of compound 3 (C21H38O4, HRESIMS m/z: 355.2856 [M+H]+, calcd. for C21H39O4+, 355.2843) differed from that of 2 by C2H4, suggesting a two-carbon deletion in the side chain. Analysis of the UV, IR and 1H NMR spectra revealed 3 to be a similar secobutanolide to 2, with the same E geometry of the trisubstituted double bond [δH 7.08 (t, J = 7.0 Hz, 1H, H-3)], but with two carbons less in the alkyl chain. Similarly to 2, the S configuration at C-1′ was deduced by the positive optical rotation value of +2.27 (c 0.88, MeOH).11abc The complete assignments of all protons and carbons of 3 (Table 2) were accomplished by interpretation of its HMBC and HSQC spectra. Therefore, compound 3 was assigned as (2E)-2-[(1S)-1-hydroxy-2-oxo-propyl]-heptadec-2-enoic acid methyl ester, and named macrocarpolide C.
Table 2.
1H and 13C NMR data for compounds 2 and 3.a
| 2 | 3 | |||
|---|---|---|---|---|
| Posn | δHb | δcc | δHb | δcc |
| 1 | 166.7 (C) | 166.7 (C) | ||
| 2 | 129.9 (C) | 129.9 (C) | ||
| 3 | 7.08 t (7.7) | 149.3 (CH) | 7.08 t (7.0) | 149.3 (CH) |
| 4 | 2.35 q (7.6) | 28.9 (CH2) | 2.35 q (7.6) | 28.9 (CH2) |
| 5 | 1.25–1.31 | 28.4 (CH2) | 1.26 br s | 28.4 (CH2) |
| 6 | 1.25–1.31 | 29.8–29.5 (CH2) | 1.26 br s | 29.8–29.5 (CH2) |
| 7 | 1.25–1.31 | 29.8–29.5 (CH2) | 1.26 br s | 29.8–29.5 (CH2) |
| 8 | 1.25–1.31 | 29.8–29.5 (CH2) | 1.26 br s | 29.8–29.5 (CH2) |
| 9 | 1.25–1.31 | 29.8–29.5 (CH2) | 1.26 br s | 29.8–29.5 (CH2) |
| 10 | 1.25–1.31 | 29.8–29.5 (CH2) | 1.26 br s | 29.8–29.5 (CH2) |
| 11 | 1.25–1.31 | 29.8–29.5 (CH2) | 1.26 br s | 29.8–29.5 (CH2) |
| 12 | 1.25–1.31 | 29.8–29.5 (CH2) | 1.26 br s | 29.8–29.5 (CH2) |
| 13 | 1.25–1.31 | 29.8–29.5 (CH2) | 1.26 br s | 29.8–29.5 (CH2) |
| 14 | 1.25–1.31 | 29.8–29.5 (CH2) | 1.26 br s | 29.8–29.5 (CH2) |
| 15 | 1.25–1.31 | 29.8–29.5 (CH2) | 1.26 br s | 32.1 (CH2) |
| 16 | 1.25–1.31 | 29.8–29.5 (CH2) | 1.26 br s | 29.8–29.5 (CH2) |
| 17 | 1.25–1.31 | 32.1 (CH2) | 0.88 t (7.0) | 14.3 (CH3) |
| 18 | 1.25–1.31 | 22.8 (CH2) | ||
| 19 | 0.88 t (7.0) | 14.3 (CH3) | ||
| 1′ | 4.90 brd (4.9) | 73.5 (CH) | 4.90 brs | 73.5 (CH) |
| 2′ | 206.2 (C) | 206.2 (C) | ||
| 3′ | 2.15 s | 25.0 (CH3) | 2.15 s | 25.0 (CH3) |
| 1-OMe | 3.73 s | 52.2 (CH3) | 3.73 s | 52.2 (CH3) |
Assignments based on analysis of 2D NMR spectra.
Data (δ) measured at 500 MHz; brs = broad singlet, dd= doublet of doublets, dt = doublet of triplets. J values are in Hz and are omitted if the signals overlapped as multiplets. The overlapped signals were assigned from HSQC and HMBC spectra without designating multiplicity.
Data (δ) measured at 125 MHz; CH3, CH2, CH, and C multiplicities were determined by HSQC experiment.
Compounds 1–5 showed good antiproliferative activities against the drug-sensitive A2780 ovarian cell line12 as previously described13 using paclitaxel (IC50 0.073 ± 0.015 µM) as the positive control. Their IC50 values were 2.57 ± 0.12 (1), 1.98 ± 0.23 (2), 1.67 ± 0.05 (3), 2.43 ± 0.41 (4), and 1.65 ± 0.44 µM (5). The similar IC50 values for the five compounds suggests that they have a similar mechanism of action, possibly as Michael acceptors.
Supplementary Material
Figure 1.
Structures of compounds 1–5.
Acknowledgments
This project was supported by the Fogarty International Center, the National Cancer Institute, the National Institute of Allergy and Infectious Diseases, the National Institute of Mental Health, the National Institute on Drug Abuse, the National Heart Lung and Blood Institute, the National Center for Complementary and Alternative Medicine, the Office of Dietary Supplements, the National Institute of General Medical Sciences, the Biological Sciences Directorate of the National Science Foundation, and the Office of Biological and Environmental Research of the U.S. Department of Energy under Cooperative Agreement U01 TW00313 with the International Cooperative Biodiversity Groups. This project was also supported by the National Research Initiative of the Cooperative State Research, Education and Extension Service, USDA, Grant #2008-35621-04732. This support is gratefully acknowledged. Work at Virginia Tech was supported by the National Science Foundation under Grant CHE-0722638 for the purchase of the Agilent 6220 mass spectrometer. We thank Mr. B. Bebout for obtaining the mass spectra. Fieldwork essential for this project was conducted under a collaborative agreement between the Missouri Botanical Garden and the Parc Botanique et Zoologique de Tsimbazaza and a multilateral agreement between the ICBG partners, including the Centre National d’Application des Recherches Pharmaceutiques. We thank Stéphan Rakotonandrasana, Richard Randrianaivo, Armand Andriatsarafara, and Nambinintsoa Rakotonjanahary for assistance with plant collection, and we gratefully acknowledge courtesies extended by the Government of Madagascar (Ministère des Eaux et Forêts).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary data
Supplementary data associated with this article, consisting of experimental procedures and 1H NMR spectra for compounds 1–5, can be found, in the online version, at http://dx.doi.org/10.1016/j.tetlet.XXXX.YY.ZZZ
References and notes
- 1.Biodiversity Conservation and Drug Discovery in Madagascar, Part 63. For Part 62, see Liu Y, Young K, Rakotondraibe LH, Brodie PJ, Wiley JD, Cassera MB, Callmander MW, Rakotondrajaona R, Rakotobe E, Rasamison VE, TenDyke K, Shen Y, Kingston DGI. Antiproliferative Compounds from Cleistanthus boivinianus from the Madagascar Dry Forest. J. Nat. Prod. doi: 10.1021/np501020m. submitted for publication December 17, 2014.
- 2.(a) Harinantenaina L, Brodie PJ, Maharavo J, Bakary G, TenDyke K, Shen Y, Kingston DGI. Bioorg. Med. Chem. 2013;21:2912. doi: 10.1016/j.bmc.2013.03.078. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kingston DGI. J. Org. Chem. 2008;73:3975. doi: 10.1021/jo800239a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Rohwer JG. Syst. Bot. 2000;25:60. [Google Scholar]; (b) van der Werff H. Adansonia, sér. 3. 2013;35:235. [Google Scholar]
- 4.Coelho de Souza G, Haas APS, von Poser GL, Schapoval EES, Elisabetsky E. J. Ethnopharmacol. 2004;90:135. doi: 10.1016/j.jep.2003.09.039. [DOI] [PubMed] [Google Scholar]
- 5.(a) Destryana RA, Young DG, Woolley CL, Huang T-C, Wu H-Y, Shih W-L. J. Am. Oil Chem. Soc. 2014;91:1531. [Google Scholar]; (b) de Camargo MJ, Miranda MLD, Kagamida CM, Rodrigues ED, Garcez FR, Garcez WS. Quim. Nova. 2013;36:1008. [Google Scholar]; (c) Garett R, Romanos MTV, Borges RM, Santos MG, Rocha L, da Silva AJR. Rev. Bras. Farmacogn. 2012;22:306. [Google Scholar]; (d) Yamaguchi MU, Garcia FP, Cortez DAG, Ueda-Nakamura T, Filho BPD, Nakamura CV. Antonie Van Leeuwenhoek. 2011;99:507. doi: 10.1007/s10482-010-9516-3. [DOI] [PubMed] [Google Scholar]; (e) Garcez FR, da Silva AFG, Garcez WS, Linck G, Matos MdFC, Santos ECS, Queiroz LMM. Planta Med. 2011;77:383. doi: 10.1055/s-0030-1250401. [DOI] [PubMed] [Google Scholar]; (f) Castro RD, Lima EO. Rev. Bras. Plant. Med. 2011;13:203. [Google Scholar]; (g) Cuca LE, Leon P, Coy ED. Chem. Nat. Compd. 2009;45:179. [Google Scholar]
- 6.Seki K, Sasaki T, Wano S, Haga K, Kaneko R. Phytochemistry. 1995;40:1175. [Google Scholar]
- 7.Anderson JE, Ma W, Smith DL, Chang CJ, McLaughlin JL. J. Nat. Prod. 1992;55:71. doi: 10.1021/np50079a011. [DOI] [PubMed] [Google Scholar]
- 8.Cheng H-I, Lin W-Y, Duh C-Y, Lee K-H, Tsai I-L, Chen I-S. J. Nat. Prod. 2001;64:1502. doi: 10.1021/np0102298. [DOI] [PubMed] [Google Scholar]
- 9.Cheng M-J, Tsai I-L, Lee S-J, Jayaprakasam B, Chen I-S. Phytochemistry (Elsevier) 2005;66:1180. doi: 10.1016/j.phytochem.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 10.(a) Martinez VJC, Yoshida M, Gottlieb OR. Phytochemistry. 1981;20:459. [Google Scholar]; (b) Tsai I-L, Hung C-H, Duh C-Y, Chen I-S. Planta Med. 2002;68:142. doi: 10.1055/s-2002-20260. [DOI] [PubMed] [Google Scholar]
- 11.(a) Chen C-Y, Chen C-H, Wong C-H, Liu Y-W, Lin Y-S, Wang Y-D, Hsui Y-R. J. Nat. Prod. 2007;70:103. doi: 10.1021/np060425k. [DOI] [PubMed] [Google Scholar]; (b) Kuo S-Y, Hsieh T-J, Wang Y-D, Lo W-L, Hsui Y-R, Chen C-Y. Chem. Pharm. Bull. 2008;56:97. doi: 10.1248/cpb.56.97. [DOI] [PubMed] [Google Scholar]; (c) Tanaka H, Nakamura T, Ichino K, Ito K. Phytochemistry. 1989;28:1905. [Google Scholar]
- 12.Louie KG, Behrens BC, Kinsella TJ, Hamilton TC, Grotzinger KR, McKoy WM, Winker MA, Ozols RF. Cancer Res. 1985;45:2110. [PubMed] [Google Scholar]
- 13.Cao S, Brodie PJ, Miller JS, Randrianaivo R, Ratovoson F, Birkinshaw C, Andriantsiferana R, Rasamison VE, Kingston DGI. J. Nat. Prod. 2007;70:679. doi: 10.1021/np060627g. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.