Abstract
Electrotransfection is a technique utilized for gene delivery in both preclinical and clinical studies. However, its mechanisms are not fully understood. The goal of this study was to investigate specific pathways of endocytosis involved in electrotransfection. In the study, three different human cell lines (HEK293, HCT116, and HT29) were either treated with ice cold medium postelectrotransfection or endocytic inhibitors prior to electrotransfection. The inhibitors were pharmacological agents (chlorpromazine, genistein, and amiloride) or different small interfering RNA (siRNA) molecules that could knockdown expression of clathrin heavy chain (CLTC), caveolin-1, and Rab34, respectively. The reduction in gene expressions was confirmed with western blot analysis at 48-72h post-siRNA treatment. It was observed that treatments with either ice cold medium, chlorpromazine, or genistein resulted in significant reductions in electrotransfection efficiency (eTE) in all three cell lines, compared to the matched controls, but amiloride treatment had insignificant effects on eTE. For cells treated with siRNA, only CLTC knockdown resulted in eTE reduction for all three cell lines. Together, these data demonstrated that the clathrin-mediated endocytosis played an important role in electrotransfection.
Introduction
Electrotransfection is a gene delivery technique that relies on application of pulsed electric fields to facilitate gene transport into cells. It is also referred to as electroporation and electric field-mediated gene delivery in the literature.1–3 Effective electrotransfection is hinged upon overcoming a series of major physiological barriers—from the site of plasmid DNA (pDNA) administration to its ultimate destination in the nucleus of target cells.4 One of the major barriers encountered in gene delivery is the plasma membrane. Mechanisms by which an electric field facilitates pDNA transport across this barrier are still speculative and poorly characterized. Previous studies have suggested diffusion, electro-osmosis, and electrophoresis as potential mechanisms.5,6 Of these three possibilities, electrophoresis has been subjected to the most investigation. Klenchin et al.7 utilized a monolayer of cells cultured on a porous film to show a 10-fold increase in electrotransfection efficiency (eTE) when pulses with a polarity that promote pDNA electrophoresis toward the monolayer were applied versus pulses with the opposite polarity. Bureau et al.8 corroborated these findings by showing a significantly greater eTE in vivo when a series of pulses consisting of one short, high-voltage “electroporating” pulse followed by four long, low-voltage electrophoresis-inducing pulses were used, compared to using a single high-voltage pulse or four low-voltage pulses alone. Results from these studies support the notion that electrophoresis has a substantial effect on pDNA delivery across the cell membrane and consequently, on the ultimate transfection efficiency. On the other hand, contradictory findings were demonstrated by Liu et al.,9 who applied electric pulse sequences with either alternating polarity or consistent polarity to tissues, and showed that the pulse polarity had no effects on transgene expression in vivo. If electrophoresis was the dominant mechanism for transmembrane transport of pDNA, the pulse sequence with consistent polarity should yield a higher level of transgene expression. The same authors also observed insignificant changes in transfection efficiency when the electrophoretic effect was diminished by supplementing the buffer with 10% Ficoll to increase the viscosity of pDNA solution or with Mg2+ to reduce the negative charge of pDNA.9
More recently, investigators have observed that the applied electric fields enable the formation of stable complexes between pDNA and the plasma membrane in the region facing the cathode when the field magnitude exceeds a threshold;10–13 and that eTE can be significantly reduced if cells are pretreated with inhibitors of endocytosis.14 The observations indicate that pDNA is not internalized immediately after electrically pulsing the cells, and suggest that electrotransfection is a two-step process: (i) formation of stable complexes between pDNA and the plasma membrane, induced by an applied electric field, and (ii) adsorptive endocytosis of membrane-bound pDNA, which is likely to occur after the application of pulsed electric fields.
The goal of this study was to determine specific pathways of endocytosis that are involved in electrotransfection of cells. A common approach to investigation of endocytosis is to use pharmacological inhibitors that can transiently block specific pathways. For example, chlorpromazine (CPZ) inhibits clathrin-mediated endocytosis through anchoring clathrin and adaptor protein 2 (AP2) complex to endosomes, thereby preventing the assembly of coated pits at the inner surface of the plasma membrane.15 Genistein is a tyrosine kinase inhibitor that inhibits caveolae-dependent endocytosis by preventing (i) actin depolymerization in the local cortical cytoskeleton, which must precede internalization of caveolar vesicles, and (ii) recruitment of dynamin 2, which is required for the scission of vesicles from cell membrane.16 Amiloride has been used to inhibit macropinocytosis although the mechanisms of inhibition are still uncertain. It has been shown that amiloride causes acidification in the submembranous region at the site of macropinocytosis through inhibition of Na+/H+ exchangers,17 and that GTPases involved in actin remodeling are highly sensitive to submembranous pH. Hence, amiloride may inhibit macropinocytosis by lowering pH at these sites, leading to impaired actin polymerization, which is crucial for macropinocytosis.17 However, one potential problem with using the pharmacological inhibitors described above is that they may reduce eTE via endocytosis-independent mechanisms.18 Therefore, it is important to independently confirm the results with other approaches to inhibition of endocytosis, such as siRNA-mediated knockdown of proteins involved in endocytosis.
Results
Effects of low temperature on electrotransfection efficiency
Exposure of cells to ice-cold medium (0–4 °C) is a commonly used method for nonspecific inhibition of endocytosis. Therefore, we treated cells with ice-cold medium, and investigated effects of treatment on endocytosis of pDNA. In experimental design, we expected the endocytosis to occur only within a short period (< 40 minutes) post the application of electric pulses.14 Considering that heat transfer in medium was not instantaneous, we chose to directly add ice-cold or room-temperature medium to cells immediately after electrotransfection. With the experimental protocol described in the Materials and Methods section, we observed that a significant amount of pDNA uptake was found in all three cell types treated with room-temperature medium, whereas none or a small amount of pDNA molecules were detected in the cells exposed to ice-cold medium (see Figure 1). No pDNA was detected in the nonpulsed control groups. For HCT116, HT29, and HEK293 cells exposed to the room-temperature medium (i.e., the control cells), the eTE levels were 67.4, 18.9, and 50.6% (see Figure 2a), respectively. Exposure of cells to the ice-cold medium significantly reduced the eTE (see Figure 2b), and slightly reduced the cell viability (Figure 2c). These data demonstrated that the pDNA uptake was temperature-sensitive during the first 30-minute recovery period of cells, suggesting that the pDNA uptake was mediated by endocytosis. To identify specific pathways of endocytosis that might be involved in electrotransfection, we investigated effects of cell treatment with endocytic inhibitors on eTE.
Figure 1.
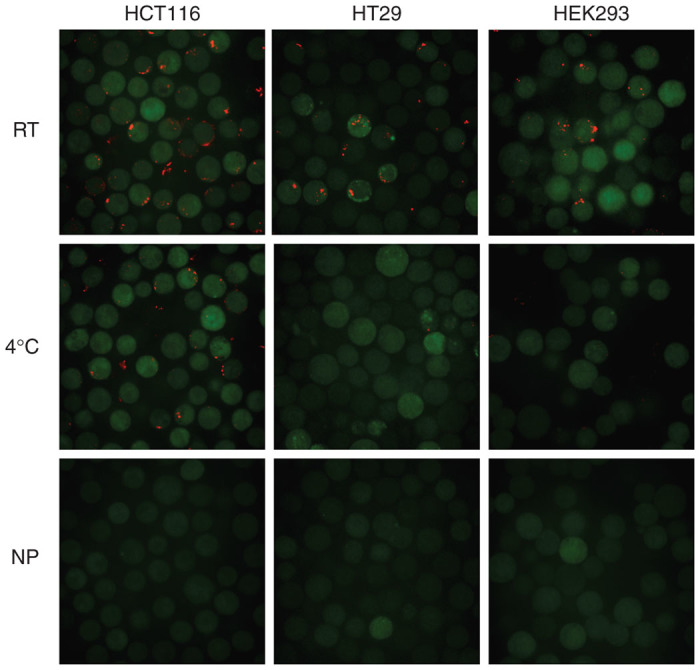
Effect of temperature on plasmid DNA (pDNA) uptake. Immediately after electrotransfection, HEK293, HT29, and HCT116 cells were treated with room-temperature (RT) or ice-cold (0–4 °C) medium for 30 minutes, and further incubated at 37 °C for 1 hour for cells to attach to the surface. The pDNA was labeled with rhodamine (red), and cells were stained with CellTracker Green CMFDA. A nonpulsed (NP) group was included as a negative control.
Figure 2.
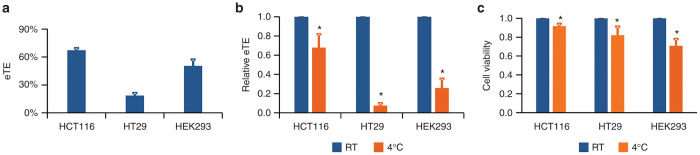
Effect of temperature on electrotransfection efficiency and cell viability. The experimental conditions were the same as those described in the legend of Figure 1. At 24 hours postelectrotransfection, the electrotransfection efficiency (eTE) was quantified with flow-cytometry, and the cell viability was quantified with the MTT assay. Panel a shows the eTE of cells in the room-temperature (RT) groups (i.e., the controls). The difference in eTE between any two cell lines were statistically significant (n = 5, P < 0.05, Mann–Whitney U-test). Panel b shows the relative eTE normalized by the data in the corresponding RT groups (n = 4). Panel c shows the relative cell viability normalized by the data in the corresponding RT groups (n = 8). *P < 0.05 (Wilcoxon Signed-Rank test).
Effects of pharmacological inhibitors on electrotransfection efficiency
Cells were treated with CPZ, genistein, and amiloride at different concentrations for 1 hour prior to electrotransfection with a green fluorescence protein (GFP)-encoding plasmid. The CPZ treatment significantly decreased eTE in all three cell lines (P < 0.05), compared to matched controls with an equivalent volume of DMSO vehicle (Figure 3a). Pretreatment of the cells with genistein led to a significant reduction in eTE in HEK293 cells (P < 0.05) but not in other cell lines (Figure 3b). Amiloride treatment had insignificant effects on eTE in all tested cell samples (P > 0.05) (Figure 3c), except for HEK293 cells treated with amiloride at a higher concentration (2.5 mmol/l).
Figure 3.
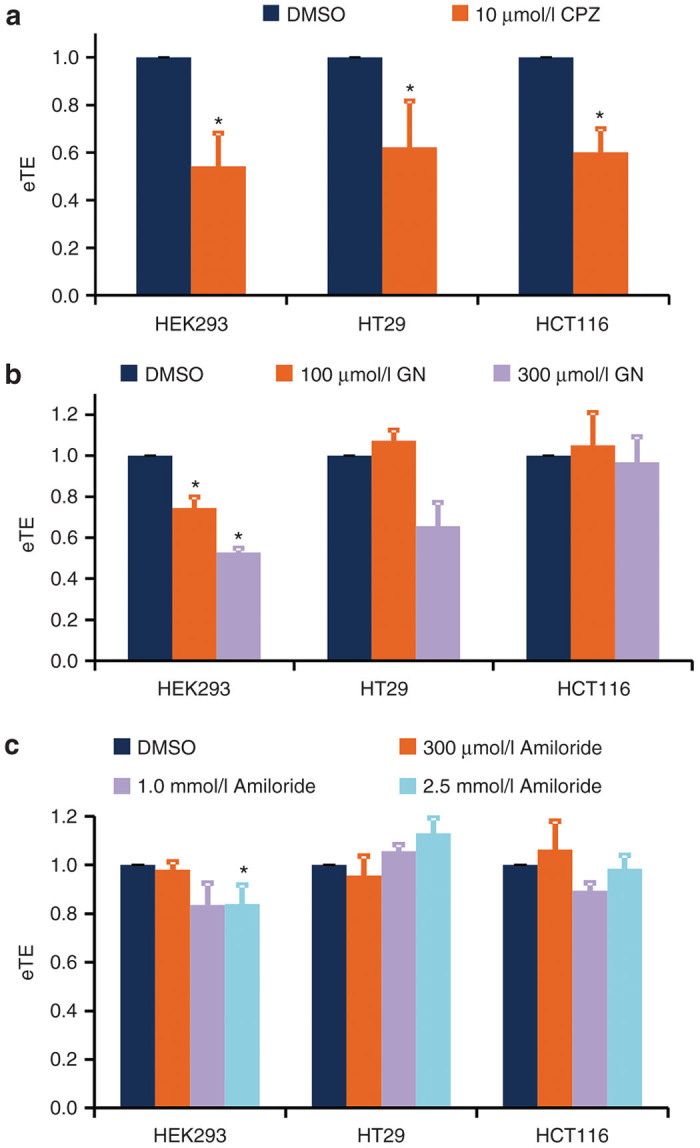
Effects of pharmacological inhibitor treatments on electrotransfection efficiency. HEK293, HT29, and HCT116 cells were pretreated with (a) chlorpromazine (CPZ), (b) genistein (GN), and (c) amiloride at different concentrations for 1 hour. The control groups were treated with an equivalent volume of DMSO. Electrotransfection efficiency was quantified at 24 hours postelectrotransfection. All data were normalized by the matched controls. *P < 0.05 (n = 4–6, Mann–Whitney U-test).
Effect of gene knockdown on electrotransfection efficiency
To confirm the results shown in Figure 3, we also investigated the dependence of eTE on expressions of three proteins: clathrin heavy chain (CLTC), caveolin-1 (CAV-1), and Rab34, which could affect clathrin-mediated endocytosis, caveolae-dependent endocytosis, and macropinocytosis, respectively. In experiments, cells were transfected either with two specific small interfering RNA (siRNA) (siRNA-1 and siRNA-2) molecules directed against two different nucleotide sequences within the encoding gene (see Table 1), or with nonspecific siRNA duplexes with comparable GC content (i.e., the negative control siRNA). Western blot analysis showed that expressions of all three proteins were reduced at 48–72 hours post-siRNA transfection (see Figure 4a). The extent of reduction was >90% for CLTC and Rab34 in all cell lines, except for Rab34 in HT29 cells that was undetectable even in control cells. Reverse transcription-polymerase chain reaction (RT-PCR) analysis confirmed that Rab34 was not expressed in this cell line (Figure 4b). The extent of CAV-1 reduction varied with the cell line. It was >90% in HT29 cells, 78 or 83% in HCT116 cells, and 33 or 49% in HEK-293 cells, depending on the sequence of siRNA used in the study.
Table 1. Specific siRNA sequences for different target genes.
| Target gene | siRNA sequence | GC content | |
|---|---|---|---|
| CLTC | #1 | 5′-CCGGAAATTTGATGTCAATACTTCA-3′ | Low |
| #2 | 5′-GAGTGCTTTGGAGCTTGTCTGTTTA-3′ | Low | |
| CAV-1 | #1 | 5′-CCCACTCTTTGAAGCTGTTGGGAAA-3′ | Med |
| #2 | 5′-TCCGCATCAACTTGCAGAAAGAAAU-3′ | Low | |
| Rab34 | #1 | 5′-AATTCTCGGACATTCTCACCAGTGA-3′ | Low |
| #2 | 5′-AATCGTTCCATCTCGAAGTCCACTC-3′ | Med | |
siRNA, small interfering RNA.
Figure 4.
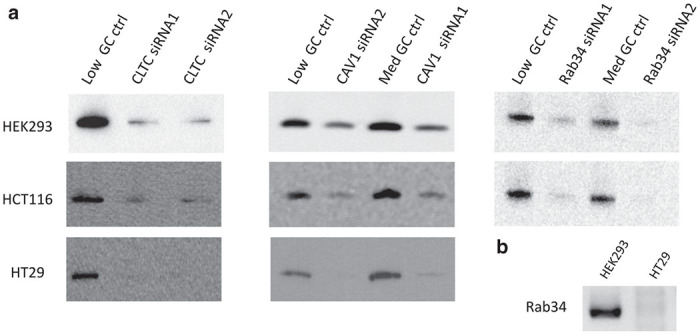
Analysis of gene expression levels in cells after silencing with specific small interfering RNA (siRNA) shown in Table 1. (a) HEK293, HT29, and HCT116 cells were treated with either two different siRNA oligos directed against CLTC, CAV-1, or Rab34, respectively, or with control siRNA with similar GC contents. Western blot analysis was performed at 48–72 hours after siRNA treatment. Please note that Rab34 protein was undetectable in untreated HT29 cells (data not shown). (b) Levels of Rab34 mRNA were determined with RT-PCR in untreated HEK293 and HT29 cells. The result showed that mRNA for Rab34 was detectable in HEK293 cells but undetectable in HT29 cells.
Having confirmed the knockdown of protein levels via western blot analysis, HEK293, HCT116, and HT29 cells were then subjected to the same siRNA transfection protocols. At 48–72 hours post-siRNA treatments, the cells were electrotransfected with a GFP-encoding plasmid; the eTE was quantified after 24 hours. The experimental results are shown in Figure 5. The CLTC knockdown resulted in a significant reduction in eTE in all three cell lines (P < 0.05) (Figure 5a). However, neither CAV-1 nor Rab34 knockdown could significantly decrease eTE (Figure 5b,c).
Figure 5.
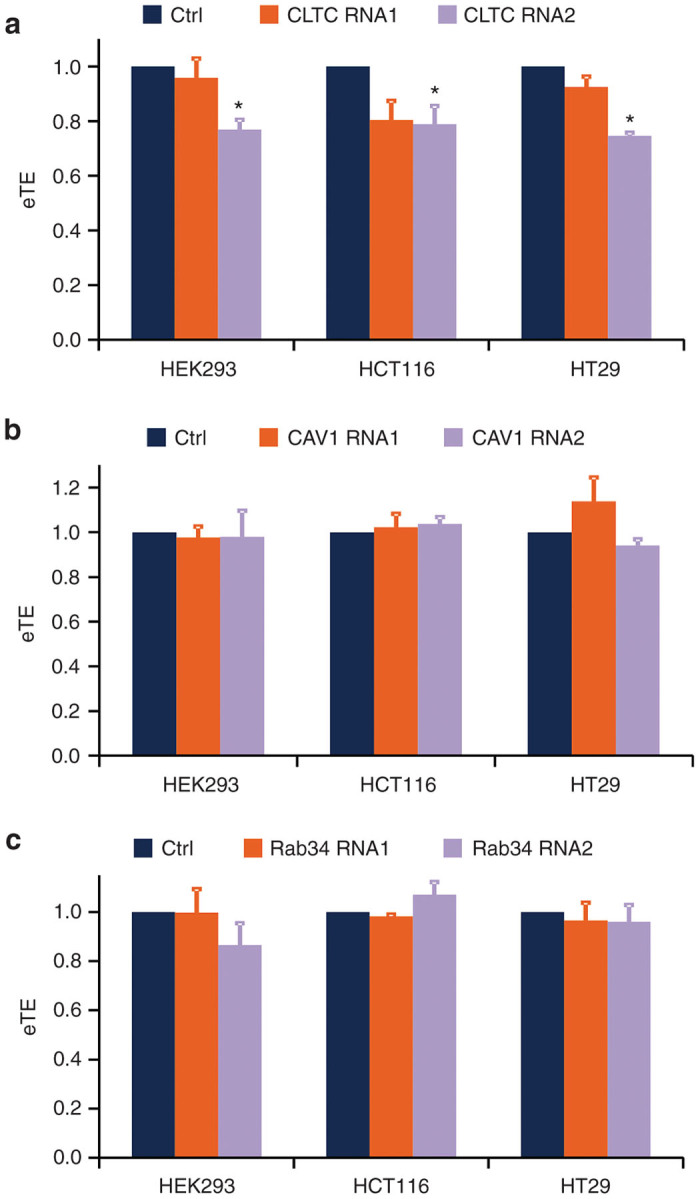
Effect of small interfering RNA (siRNA) treatment on electrotransfection efficiency. HEK293, HT29, and HCT116 cells were pretreated with either two different siRNA oligos against (a) CLTC, (b) CAV-1, or (c) Rab34, or with control siRNA with similar GC contents. At 48–72 hours post-treatment, the cells were electrotransfected, and the electrotransfection efficiency was quantified at 24 hours after incubation. All data were normalized by the matched controls. *P < 0.05 (n = 4–6, Mann–Whitney U-test).
Discussion
The aim of this study was to determine specific endocytic pathways that were involved in electrotransfection of cells. Our data showed that both pDNA uptake and eTE were sensitive to the medium temperature after electric pulsing of cells. The data also demonstrated that pretreatment of cells with endocytic inhibitors or siRNA could significantly reduce eTE. The reduction was caused specifically by inhibitors for clathrin-medicated endocytosis, suggesting that this endocytic pathway was more important than others, at least for the three cell lines tested in our study.
Previous studies have suggested that application of pulsed electric fields facilitates the interactions of pDNA with the cell membrane, and that the membrane bound pDNA is internalized by cells through endocytosis.12–14 To further demonstrate the cellular uptake of pDNA via endocytosis, we first treated cells with the ice-cold medium, a commonly accepted method for inactivating endocytosis.19,20 Our data showed that the treatment significantly reduced the electric pulse-mediated pDNA uptake and the expression of the reporter gene (i.e., GFP). The reductions were not due to cell killing effect because the ice-cold medium treatment had only minor influences on cell viability (see Figure 2c), which has also been observed in a previous study,21 suggesting that the reductions were caused by the inactivation of endocytosis.
Another interesting observation in our study was that the decreases in pDNA uptake and eTE, caused by the treatment of cells with the ice-cold medium, were smaller in HCT116 cells than in the other two cell lines, implying that endocytosis was faster in this cell line. Similarly, we observed in a separate experiment that postapplication of electric pulses, incubation of cells on ice, rather than directly mixing the cells with the ice-cold medium, could significantly reduce eTE in HT29 and HEK293 cells, but not in HCT116 cells (data not shown). To understand this observation, we need to consider rate of heat transfer to cells. When the cells in the cuvette were incubated on ice, the mode of heat transfer was conduction, which was significantly slower, compared to the rate of cell cooling via mixing pulsed cells with the ice-cold medium. If the endocytosis of pDNA was fast in HCT116 cells, it might have already happened before the temperature of the cells incubated on ice was reduced to ~4 °C.
The experiment discussed above had two limitations. One was that the cold temperature might nonspecifically block intracellular processes that were not involved in gene delivery but could influence transgene expression; another was that it did not provide information on endocytic pathways involved in electrotransfection. To confirm the effects of electro-endocytosis and identify specific pathways that mediated electrotransfection, we used pharmacological inhibitors and siRNA to block the pathways of endocytosis (i.e., macropinocytosis and those mediated by clathrin or caveolin) prior to electrotransfection, and investigated how the treatments affected eTE.
The use of pharmacological inhibitors is a well-established approach for studying endocytic pathways involved in the internalization of ligands, biomarkers, and nonviral gene carriers, such as cationic liposomes and polymers.18,22–24 A variety of chemical inhibitors of endocytosis have been developed and screened to selectively target different endocytic pathways, especially the three best-characterized ones: clathrin-mediated endocytosis, caveolae-dependent endocytosis, and macropinocytosis. Elucidating the means of internalization and routes of intracellular trafficking of gene carriers has provided insightful information regarding the efficiency of different endocytic pathways in leading to ultimate gene expression. This information can be utilized to design and/or modify properties of genes/gene carriers to either target certain pathways or circumvent certain endocytic obstacles/barriers. We have thus adopted this strategy to study the internalization of naked pDNA when electrotransfection is used as the gene delivery method. CPZ, genistein, and amiloride are well-established, specific inhibitors of clathrin-mediated endocytosis, caveolae-dependent endocytosis, and macropinocytosis, respectively. Our results exhibited a significant decrease in eTE in the three cell lines, HEK293, HCT116, and HT29, when cells were treated with CPZ at 10 μmol/l prior to electrotransfection, while minor or no change in eTE was observed after genistein or amiloride treatment (see Figure 3). These data implied that clathrin-mediated endocytosis was the dominant pathway of pDNA internalization in all three cell lines.
Although endocytic drug pretreatment did substantially diminish eTE, it could not completely inhibit electrotransfection for any of the cell lines tested in the study. There are several possible explanations for the incomplete inhibition. One is that the drug treatments, although effective in targeting specific endocytic pathways, do not completely inhibit or stop their activities. As a result, some residual degree of pDNA uptake via clathrin-mediated endocytosis could still exist in CPZ-treated cells, accounting for the remaining electrotransfection efficiency. The second possibility is related to a limitation in the experimental protocol. Monolayers of cells were treated with the pharmacological inhibitors of endocytosis, whereas electrotransfection was performed with cells suspended in pDNA solution. The preparation of cell suspension from the monolayers and the required incubation of cells on ice took ~20–25 minutes. This time lag could be adequately long to allow for significant drug clearance from the cells, and thus partial recovery of the targeted endocytic pathways. Drug clearance could be a reason for the impairment but not complete inhibition of pDNA endocytosis by the time when electrotransfection was performed. Furthermore, any recovery of endocytosis that occurred within a few hours post the application of electric pulses could also contribute to the cellular uptake of pDNA.14 The third possibility is that there are alternative pathways for endocytosis, which were not targeted by the specific endocytic inhibitors used in this study. In fact, there exist poorly characterized clathrin- and caveolae-independent, nonmacropinocytic pathways.25 Although poorly understood, these endocytic pathways still play crucial roles in many cellular functions and may contribute to uptake, intracellular trafficking, and ultimate gene expression in electrotransfected cells. Finally, it was possible that an unknown fraction of pDNA molecules observed in cells were internalized through electric pulse-induced defects in the membrane, which was independent of endocytic pathways. For HT29 cells, this fraction was likely to be small since its eTE was less than 20% after the cells were treated with the ice-cold medium for 30 minutes.
Pharmacological inhibition provides a fast, easy-to-use strategy for transient impairment of targeted endocytic pathways. While it offers many advantages, it also suffers from certain limitations, mainly the potentials for low specificity and variable efficacy. The low specificity could arise from direct, unintended effects on alternative pathways for endocytosis other than the ones targeted in this study. It could also arise from disruption of other cellular processes and/or activities that have indirect effects on endocytosis. An example of the latter case is the disruption of the actin cytoskeleton which is required for several endocytic pathways, involving invagination and formation of vesicles as well as changes in functions of various proteins and lipid components in the cell membrane.26
Efficacy of pharmacological inhibitors has also been shown to vary in a cell-dependent manner, even with optimization for each cell line.18 CPZ, used in our study to target clathrin-mediated endocytosis, has been shown to block phagocytosis in neutrophils and macrophages27,28 and potentially, macropinocytosis.26 These effects of CPZ on alternate pathways may result from the amphipathic property that allows this molecule to embed readily into the phospholipid bilayer to alter membrane fluidity and consequently, membrane invagination behavior. The side effects of CPZ may also stem from the drug’s purported inhibitory effect on phospholipase C, a regulatory molecule of actin polymerization and macropinocytosis.26,29–31
A systematic study of the efficacy and specificity of various endocytic inhibitors in several cell lines shows that CPZ can inhibit uptake of human transferrin, a specific marker of clathrin-mediated endocytosis, in a cell line-specific manner, with some cell lines unaffected by the treatment.18 The same study also investigated genistein, which was used in our study to target caveolae-dependent endocytosis, in inhibiting the uptake of LacCer, a specific marker for lipid raft dependent endocytosis. While genistein exhibits high efficacy in reducing LacCer uptake across all cell lines, it also causes a dramatic, 80% diminishment in the clathrin-mediated uptake of transferrin in one particular cell line.18 The observation is not surprising, considering that genistein, although a commonly used inhibitor of caveolae-mediated endocytosis,32–34 is a tyrosine kinase inhibitor that may have additional effects on molecules involved in or shared with other endocytic pathways. Amiloride, used in our study to target macropinocytosis, has been shown to have alternate inhibitory effects on clathrin-mediated uptake of junctional protein complexes in epithelial cells35 and of receptor-bound albumin in opossum kidney cells.36 An amiloride analogue has been shown to cause disruption of actin cytoskeleton dynamics in fibroblasts, leading to inhibited cell motility, pseudopodial retraction, and cell detachment from substrate.37 As a result, the drug-induced disassembly of actin filaments may affect multiple actin-dependent endocytic pathways. To overcome the low specificity and variable efficacy problems of pharmacological inhibitors, we also used RNAi technology to block endocytosis prior to electrotransfection of cells.
Molecular biological methods, such as RNAi induced gene silencing, provide the advantages of high specificity, superior efficacy, minimal toxicity, and sustained inhibition of the target proteins or mechanisms. The RNAi strategy was employed in this study to impair the same three endocytic pathways by targeting crucial proteins exclusively implicated for each pathway. For inhibition of clathrin-mediated endocytosis, siRNA was directed against the CLTC, an important component of the clathrin protein that dictates its triskelion structure and permits its assembly into polygonal lattices at the clathrin-coated pits along the membrane.38 For inhibition of caveolae-dependent endocytosis, siRNA was directed against CAV-1, the main protein found within caveolae in most cells that is responsible for the formation and structural stability of caveolae.39 Macropinocytosis was inhibited with siRNA directed against Rab34, a member of the Rab family of small GTPases that has been found to localize in regions of membrane ruffling, the predominant site for macropinocytosis, and be deemed necessary for the formation of macropinosomes.40 Our experimental data shown in Figure 5a revealed that the knockdown of CLTC could significantly reduce eTE in all three cell lines (P < 0.05), indicating that pDNA entered cells through clathrin-mediated endocytosis during electrotransfection. This finding was consistent with the pharmacological results shown in Figure 3a, where cells were pretreated with CPZ, an inhibitor for clathrin-mediated endocytosis.
An interesting point to note is the modest reductions in eTE in all three cell lines resulting from siRNA treatment, compared to those achieved with pharmacological inhibitors. CLTC knockdown yielded 21–25% decrease in eTE relative to the matched controls, whereas the eTE decrease was 38–46% for CPZ-treated cells. The disparity between the two different approaches might be attributable to the duration of inhibition. siRNA knockdown caused a gradual and sustained reduction in protein expression, whereas the effects of pharmacological agents were immediate and short-lived. Since endocytosis is crucial for cellular homeostasis and viability, cells in the former approach may develop compensatory mechanisms to rectify the deficiencies caused by siRNA-targeted inhibition of endocytosis. This adaptation often manifests as upregulation of alternative endocytic pathways, which seems reasonable given that the surface area of cells does not change drastically over the course of siRNA-induced inhibition of endocytosis. Evidence of this so-called “cross-regulation”,41 between different endocytic pathways can be gathered from the studies of Damke et al.42, which showed upregulation of fluid-phase pinocytosis in dynamin mutants subjected to temperature dependent inhibition of clathrin-mediated endocytosis. The existence of the “cross-regulation” mechanism implies that siRNA-induced inhibition of one endocytic mechanism may induce upregulation of alternate pathways that can facilitate pDNA uptake during electrotransfection. Pharmacological inhibitors, on the other hand, only cause immediate, transient effects on endocytosis, which might not provide sufficient amount of time for cells to adapt or develop the same compensatory mechanisms, thereby resulting in the higher eTE reduction. Furthermore, pharmacological inhibitors are relatively less specific as discussed earlier, which may cause unintended effects on alternate endocytic pathways or nonendocytic mechanisms that can reduce eTE.
Electrically-induced endocytosis of non-DNA macromolecules has been observed in previous studies. Antov et al.43 observed enhancement in membrane absorption of bovine serum albumin and dextran after application of nonpermeabilizing, low electric field pulses. The enhancement in absorption was followed by increased cellular uptake of these macromolecules. β-galactosidase and large dextran molecules, added into the medium up to 60 minutes after the application of electric pulses, were observed within large vesicles in cells. The postpulsing uptake of the macromolecules diminished when cells were treated with an actin-depolymerizing drug, colchicine,44 implying that the macromolecules were internalized by cells via macropinocytosis. Similar results have also been observed for fluorescently labeled bovine serum albumin.45 These experimental observations indicate that exposure of cells to electric pulses can enhance endocytosis of non-DNA macromolecules over a period that is several orders of magnitude longer than the lifetime of transient pores in the membrane induced by pulsed electric fields.46
In summary, the present study provided new insights into the mechanisms of electrotransfection. It supported the notion that endocytosis played an important role in electric pulse-induced cellular uptake of pDNA, and identified clathrin-mediated endocytosis as a mechanism of pDNA internalization after the cells were exposed to pulsed electric fields. Although the new findings cannot exclude the possibility that some fraction of DNA molecules may enter cells through electric pulse-induced defects in the membrane, they will stimulate further investigations of electrotransfection mechanisms, and potentially lead to development of new directions for improving electrogene therapy.
Materials and Methods
Cells culture
Three types of cells were used in the study: HEK293, a human embryonic kidney cell line, HCT116, a human colorectal carcinoma cell line, and HT29, a human colon adenocarcinoma cell line (ATCC, Manassas, VA). HEK293 was cultured in high glucose Dulbecco’s modified Eagle’s medium (Gibco, Grand Island, NY), supplemented with 10% (vol/vol) fetal bovine serum and 1× antibiotic-antimycotic solutions (Gibco). HCT116 and HT29 were cultured in McCoy’s 5A medium with 10% (vol/vol) fetal bovine serum and 1× antibiotic-antimycotic solutions (Gibco). All cells were incubated in 100 × 20 mm dishes or six-well plates (Clontech, Mountain View, CA) at 37 °C in 5% CO2 and 95% air, and passaged every 2–3 days. Cells were electrotransfected with GFP-encoding plasmid (pEGFP-N1).
Plasmid labeling
For visualization studies, pEGFP-N1was labeled with LabelIT Tetramethyl-rhodamine (Mirus, Madison, WI), conducted as per manufacturer’s protocol. The fluorescent dye was covalently linked with DNA; the labeled plasmid was purified through ethanol precipitation and resuspension in water before use.
Electrotransfection
1 × 106 cells were suspended in OptiMEM and mixed with 6 µg pEGFP-N1 to achieve a final sample volume of 100 µl. The samples were incubated on ice for 10 minutes (with occasional flicking to resuspend cells) before being transferred into BTX disposable 4-mm gap aluminum cuvettes (Harvard Apparatus, Holliston, MA). Cells were then subjected to electric field pulse treatments using a BTX ECM 830 Square Wave Electroporation System (Harvard Apparatus). To achieve high transfection efficiencies and cell viabilities, the optimal parameters for electric pulse sequences were: eight pulses at 400 V/cm, 5 msec duration, and 1 Hz frequency for HEK293 and HCT116 cells, and six pulses at 600V/cm, 5 msec duration, and 1 Hz frequency for HT29 cells. After exposure to electric pulses, 500 µl of complete culture media (described above) at room temperature were added into the cuvettes with the pulsed cells, and samples were left undisrupted for 10 minutes at room temperature postelectrotransfection to promote cell recovery and resealing of pores in the membrane. The samples were then seeded in fresh media in six-well plates; and the GFP expression was quantified using flow cytometry after incubation for 24 hours.
Changing temperature of pulsed cells
The experimental protocol was similar to that described above for electrotransfection. Immediately after the pulse applications, 500 µl of complete culture media, which were either ice-cold or at the room temperature, were added into the cuvettes with the pulsed cells, and samples were left undisrupted for 30 minutes on ice for cells in the cold medium or at room temperature for cells in the other group. All cells were then incubated at 37 °C for 1 hour. Then, the cells in each group were divided into two subgroups. One subgroup was used immediately for visualization of internalized pDNA molecules with confocal microscopy. Another subgroup was cultured for another 24 hours for the GFP expression analysis with flow cytometry, or cell viability analysis.
Confocal microscopy
Electrotransfected cells were imaged using an EMCCD camera (Andor Technology, Belfast, UK) coupled to an Andor XD revolution spinning disk confocal microscope (Andor Technology) at the Duke University Light Microscopy Core Facility. Before confocal imaging, all cells were stained with CellTracker Green CMFDA. The lasers provided excitation light at wavelengths of 488 and 561 nm for imaging CMFDA and rhodamine, respectively. Intensities of rhodamine and CMFDA in confocal images were then adjusted with Zen Software (Carl Zeiss, Thornwood, NY) for visualization of pDNA localization in cells.
Flow cytometry
Culture medium in six-well plates was aspirated, and adherent cells were washed twice with phosphate-buffered saline without Ca2+ and Mg2+. The cells were collected by trypsinization, and resuspended in 350–400 μl phosphate-buffered saline containing propidium iodide (PI) (5 μg/ml). Flow cytometry analysis was performed with a BD FACSCanto II flow cytometer (Becton Dickinson, Franklin Lakes, NJ) at a Duke Cancer Institute Core Research Facility, which was equipped with 488 and 633 nm lasers for simultaneous detection of GFP and PI fluorescence. Forward and side light scattering were used as independent variables to exclude debris and isolate the cell population of interest. Compensation was set between 20 and 25% to resolve spectral emission overlap between the two detection channels; 10,000 events were collected for each sample; and autofluorescence was corrected in each experimental group by using control cells treated with the same experimental protocols and subjected to the same electric fields but without GFP-encoding pDNA. The BD FACSDiva software was used for data acquisition and analysis. The eTE was defined as the percentage of total viable cells (PI negative) that expressed GFP (PI negative, GFP positive).
Cell viability assay
Cell suspension was placed in 96-well plates at a density of 1 × 104/well in 200 μl medium. 20 μl 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution (5 mg/ml, Molecular Probes, Grand Island, NY) was added to each well and incubated for 3.5 hours. MTT solvent (4 mmol/l HCl, 0.1% Nondet P-40 (NP40) in isopropanol) was used to dissolve the pellets. Plates were illuminated at 595 nm with a Victor X Multilabel Plate Reader (Perkin Elmer, Waltham, MA) and cell viability was defined as the absorbance of each experimental sample normalized by the absorbance of the sample in the corresponding control group. A total of three trials were conducted and averaged to yield the final viability value for each experiment. Results from eight independent experiments are reported in the paper.
Treatment of cells with pharmacological inhibitors of endocytosis
Cells were seeded in six-well plates to achieve 80–90% confluency within 24 hours. The media was aspirated, and adherent cells were washed twice with phosphate-buffered saline without Ca2+ and Mg2+. Two milliliters of OptiMEM (Gibco) were added to each well, and appropriate volumes of each drug stock prepared with dimethyl sulfoxide (DMSO) were added to achieve final drug concentrations of: 10 μmol/l for chlorpromazine; 100 or 300 μmol/l for genistein; and 300 μmol/l, 1 mmol/l, or 2.5 mmol/l for amiloride (Sigma-Aldrich, St Louis, MO). Cells in the control groups were treated with the equivalent volumes of DMSO without the addition of drugs. Cells were incubated at 37 °C in 5% CO2 and 95% air for 1 hour, and subsequently washed with phosphate-buffered saline without Ca2+ and Mg2+. Cells were then trypsinized, washed again with serum-containing media, and resuspended in OptiMEM at a density of 1 × 107 cell/ml. They were electrotransfected with pEGFP-N1 using the protocol described above to investigate effects of the drug treatments on eTE.
Small interfering RNA treatment
Cells were transfected with Stealth small interfering RNA (siRNA) oligos (Life Technologies, Grand Island, NY) directed against clathrin heavy chain, caveolin-1, and Rab34 or the Stealth RNAi negative control duplex with similar GC content (Life Technologies). For each gene target, two siRNA oligos targeting different sequences within its gene were used (see Table 1). After the transfection, the cells were incubated for 48 to 72 hours before they were harvested for western blot analysis, RT-PCR analysis, or electrotransfection experiments. The experimental details are as follows.
Lipofectamine RNAiMax Transfection Reagent (Life Technologies) was used to transfect HEK293 and HCT116 cells with the siRNA sequences directed against all three genes or negative control duplex. The transfection reagent was used according to the manufacturer’s forward transfection protocol. Before performing experiments, cells were allowed to grow to 60–70% confluency in six-well plates and were treated with 30 pmol of siRNA mixed with 9 µl of Lipofectamine RNAiMax per well. After treatment, the cells were incubated for 72 hours.
For HT29 cells, Lipofectamine RNAiMax failed to silence gene expressions with the same siRNA. Therefore, siRNA transfections were carried out using the Nucleofector Kit R (Amaxa, Cologne, Germany) and the W-017 program on the Nucleofector II System, according to the manufacturer’s protocol. Before performing experiments, cells were allowed to grow to 60–70% confluency. For transfection of HT29 cells with siRNA sequences directed against CLTC and CAV-1, 2 × 106 cells were treated with 300 nmol/l of siRNA or negative control duplex in a sample volume of 100 µl, seeded in six-well plates, and incubated for 48 hours. It should be noted that the difference in the methods used for siRNA transfection among different cell lines had little influence on the conclusions of this study.
Western blot analysis
To determine protein expression levels after siRNA treatments, total proteins were extracted from siRNA treated cells using M-PER Mammalian Protein Extraction Reagent (Thermo Scientific, Waltham, MA). Cells were gently scraped from the culture plates, resuspended in 100 µl M-PER buffer with protease inhibitor cocktail (Sigma Aldrich), and gently shaken for 15 minutes. Samples were then centrifuged at ~14,000 g for 5–10 min. Clear protein extract supernatants were collected and protein concentration was quantified using a Bradford Assay (Bio-Rad, Hercules, CA). The extract was resolved on 4–12% sodium dodecyl sulfate/polyacrylamide gel electrophoresis (Life Technologies) and transferred onto PVDF membrane (Bio-Rad), blocked for 1 hour at room temperature in 5% milk, 20 mmol/l Tris, 500 mmol/l NaCl, 0.1% Tween20 (pH 7.5), and incubated overnight at 4 °C with primary antibodies: rabbit polyclonal anti-caveolin-1 (Santa Cruz Biotech, Santa Cruz, CA), rabbit polyclonal anti-Rab34 (Abcam, Cambridge, MA), and rabbit polyclonal anti-clathrin heavy chain (Abcam). Rabbit polyclonal anti-actin (Santa Cruz) was used as a loading control. The membrane was then probed with goat anti-rabbit secondary antibody conjugated to horseradish peroxidase (GE Healthcare Bio-Sciences, Pittsburgh, PA) for 30 minutes at room temperature, developed with SuperSignal West Femto Chemiluminescent Substrate (Thermo Scientific), and imaged with the IS2000MM Image System (Carestream, Rochester, NY).
RT-PCR analysis
RT-PCR was performed to determine the level of Rab34 mRNA in cells. The total RNA extracted from cells was first converted into single-strand cDNA, using the RNA to cDNA EcoDry Premix (Clontech) before PCR. SYBR label-based PCR was performed, using Chromo 4 real-time PCR system (Bio-Rad). The PCR products were analyzed with the agarose gel electrophoresis method, and imaged with the IS2000MM Image System (Carestream).
Statistical analysis
Unpaired groups were compared with the Mann–Whitney U-test; paired groups were compared with the Wilcoxon Signed-Rank test. The difference was considered to be statistically significant if the P value was < 0.05.
Acknowledgments
The work was supported partly by grants from National Institutes of Health (GM098520) and National Science Foundation (BES-0828630). This work is dedicated to Feng Liu, a professor at The University of North Carolina Eshelman School of Pharmacy, who passed away tragically on July 24th, 2014.
The authors declare no conflict of interest.
References
- Heller LC, Ugen K, Heller R. Electroporation for targeted gene transfer. Expert Opin Drug Deliv. 2005;2:255–268. doi: 10.1517/17425247.2.2.255. [DOI] [PubMed] [Google Scholar]
- Heller LC, Heller R. In vivo electroporation for gene therapy. Hum Gene Ther. 2006;17:890–897. doi: 10.1089/hum.2006.17.890. [DOI] [PubMed] [Google Scholar]
- Mir LM, Moller PH, André F, Gehl J. Electric pulse-mediated gene delivery to various animal tissues. Adv Genet. 2005;54:83–114. doi: 10.1016/S0065-2660(05)54005-7. [DOI] [PubMed] [Google Scholar]
- Henshaw JW, Yuan F. Field distribution and DNA transport in solid tumors during electric field-mediated gene delivery. J Pharm Sci. 2008;97:691–711. doi: 10.1002/jps.21000. [DOI] [PubMed] [Google Scholar]
- Dimitrov DS, Sowers AE. Membrane electroporation–fast molecular exchange by electroosmosis. Biochim Biophys Acta. 1990;1022:381–392. doi: 10.1016/0005-2736(90)90289-z. [DOI] [PubMed] [Google Scholar]
- Michel MR, Elgizoli M, Koblet H, Kempf C. Diffusion loading conditions determine recovery of protein synthesis in electroporated P3X63Ag8 cells. Experientia. 1988;44:199–203. doi: 10.1007/BF01941705. [DOI] [PubMed] [Google Scholar]
- Klenchin VA, Sukharev SI, Serov SM, Chernomordik LV, Chizmadzhev Y. Electrically induced DNA uptake by cells is a fast process involving DNA electrophoresis. Biophys J. 1991;60:804–811. doi: 10.1016/S0006-3495(91)82115-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau MF, Gehl J, Deleuze V, Mir LM, Scherman D. Importance of association between permeabilization and electrophoretic forces for intramuscular DNA electrotransfer. Biochim Biophys Acta. 2000;1474:353–359. doi: 10.1016/s0304-4165(00)00028-3. [DOI] [PubMed] [Google Scholar]
- Liu F, Heston S, Shollenberger LM, Sun B, Mickle M, Lovell M. Mechanism of in vivo DNA transport into cells by electroporation: electrophoresis across the plasma membrane may not be involved. J Gene Med. 2006;8:353–361. doi: 10.1002/jgm.851. [DOI] [PubMed] [Google Scholar]
- Golzio M, Teissie J, Rols MP. Direct visualization at the single-cell level of electrically mediated gene delivery. Proc Natl Acad Sci USA. 2002;99:1292–1297. doi: 10.1073/pnas.022646499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phez E, Faurie C, Golzio M, Teissié J, Rols MP. New insights in the visualization of membrane permeabilization and DNA/membrane interaction of cells submitted to electric pulses. Biochim Biophys Acta. 2005;1724:248–254. doi: 10.1016/j.bbagen.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Faurie C, Rebersek M, Golzio M, Kanduser M, Escoffre JM, Pavlin M. Electro-mediated gene transfer and expression are controlled by the life-time of DNA/membrane complex formation. J Gene Med. 2010;12:117–125. doi: 10.1002/jgm.1414. [DOI] [PubMed] [Google Scholar]
- Escoffre JM, Portet T, Wasungu L, Teissié J, Dean D, Rols MP. What is (still not) known of the mechanism by which electroporation mediates gene transfer and expression in cells and tissues. Mol Biotechnol. 2009;41:286–295. doi: 10.1007/s12033-008-9121-0. [DOI] [PubMed] [Google Scholar]
- Wu M, Yuan F. Membrane binding of plasmid DNA and endocytic pathways are involved in electrotransfection of mammalian cells. PLoS One. 2011;6:e20923. doi: 10.1371/journal.pone.0020923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LH, Rothberg KG, Anderson RG. Mis-assembly of clathrin lattices on endosomes reveals a regulatory switch for coated pit formation. J Cell Biol. 1993;123:1107–1117. doi: 10.1083/jcb.123.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelkmans L, Püntener D, Helenius A. Local actin polymerization and dynamin recruitment in SV40-induced internalization of caveolae. Science. 2002;296:535–539. doi: 10.1126/science.1069784. [DOI] [PubMed] [Google Scholar]
- Koivusalo M, Welch C, Hayashi H, Scott CC, Kim M, Alexander T. Amiloride inhibits macropinocytosis by lowering submembranous pH and preventing Rac1 and Cdc42 signaling. J Cell Biol. 2010;188:547–563. doi: 10.1083/jcb.200908086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercauteren D, Vandenbroucke RE, Jones AT, Rejman J, Demeester J, De Smedt SC. The use of inhibitors to study endocytic pathways of gene carriers: optimization and pitfalls. Mol Ther. 2010;18:561–569. doi: 10.1038/mt.2009.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman RM, Mellman IS, Muller WA, Cohn ZA. Endocytosis and the recycling of plasma membrane. J Cell Biol. 1983;96:1–27. doi: 10.1083/jcb.96.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlin M, Pucihar G, Kandušer M. The role of electrically stimulated endocytosis in gene electrotransfer. Bioelectrochemistry. 2012;83:38–45. doi: 10.1016/j.bioelechem.2011.08.005. [DOI] [PubMed] [Google Scholar]
- Rols MP, Delteil C, Serin G, Teissié J. Temperature effects on electrotransfection of mammalian cells. Nucleic Acids Res. 1994;22:540. doi: 10.1093/nar/22.3.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Saltzman WM. Synthetic DNA delivery systems. Nat Biotechnol. 2000;18:33–37. doi: 10.1038/71889. [DOI] [PubMed] [Google Scholar]
- Rejman J, Bragonzi A, Conese M. Role of clathrin- and caveolae-mediated endocytosis in gene transfer mediated by lipo- and polyplexes. Mol Ther. 2005;12:468–474. doi: 10.1016/j.ymthe.2005.03.038. [DOI] [PubMed] [Google Scholar]
- Rejman J, Conese M, Hoekstra D. Gene transfer by means of lipo- and polyplexes: role of clathrin and caveolae-mediated endocytosis. J Liposome Res. 2006;16:237–247. doi: 10.1080/08982100600848819. [DOI] [PubMed] [Google Scholar]
- Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- Ivanov AI. Pharmacological inhibition of endocytic pathways: is it specific enough to be useful? Methods Mol Biol. 2008;440:15–33. doi: 10.1007/978-1-59745-178-9_2. [DOI] [PubMed] [Google Scholar]
- Elferink JG. Chlorpromazine inhibits phagocytosis and exocytosis in rabbit polymorphonuclear leukocytes. Biochem Pharmacol. 1979;28:965–968. doi: 10.1016/0006-2952(79)90287-9. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Hirose M, Miyazaki A, Tomono M, Takeuchi M, Kitamura T. Calmodulin antagonists inhibit the phagocytic activity of cultured Kupffer cells. Lab Invest. 1988;59:214–218. [PubMed] [Google Scholar]
- Amyere M, Payrastre B, Krause U, Van Der Smissen P, Veithen A, Courtoy PJ. Constitutive macropinocytosis in oncogene-transformed fibroblasts depends on sequential permanent activation of phosphoinositide 3-kinase and phospholipase C. Mol Biol Cell. 2000;11:3453–3467. doi: 10.1091/mbc.11.10.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiso T, Iwaki M, Mori K. Fluidity of human erythrocyte membrane and effect of chlorpromazine on fluidity and phase separation of membrane. Biochim Biophys Acta. 1981;649:325–335. doi: 10.1016/0005-2736(81)90422-3. [DOI] [PubMed] [Google Scholar]
- Wells A, Ware MF, Allen FD, Lauffenburger DA. Shaping up for shipping out: PLCgamma signaling of morphology changes in EGF-stimulated fibroblast migration. Cell Motil Cytoskeleton. 1999;44:227–233. doi: 10.1002/(SICI)1097-0169(199912)44:4<227::AID-CM1>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Gabrielson NP, Pack DW. Efficient polyethylenimine-mediated gene delivery proceeds via a caveolar pathway in HeLa cells. J Control Release. 2009;136:54–61. doi: 10.1016/j.jconrel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Rea JC, Gibly RF, Davis NE, Barron AE, Shea LD. Engineering surfaces for substrate-mediated gene delivery using recombinant proteins. Biomacromolecules. 2009;10:2779–2786. doi: 10.1021/bm900628e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieczkarski SB, Whittaker GR. Influenza virus can enter and infect cells in the absence of clathrin-mediated endocytosis. J Virol. 2002;76:10455–10464. doi: 10.1128/JVI.76.20.10455-10464.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov AI, Nusrat A, Parkos CA. Endocytosis of epithelial apical junctional proteins by a clathrin-mediated pathway into a unique storage compartment. Mol Biol Cell. 2004;15:176–188. doi: 10.1091/mbc.E03-05-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gekle M, Freudinger R, Mildenberger S. Inhibition of Na+-H+ exchanger-3 interferes with apical receptor-mediated endocytosis via vesicle fusion. J Physiol. 2001;531 Pt 3:619–629. doi: 10.1111/j.1469-7793.2001.0619h.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagana A, Vadnais J, Le PU, Nguyen TN, Laprade R, Nabi IR. Regulation of the formation of tumor cell pseudopodia by the Na(+)/H(+) exchanger NHE1. J Cell Sci. 2000;113 (Pt 20):3649–3662. doi: 10.1242/jcs.113.20.3649. [DOI] [PubMed] [Google Scholar]
- Conner SD, Schmid SL. Regulated portals of entry into the cell. Nature. 2003;422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- Nabi IR, Le PU. Caveolae/raft-dependent endocytosis. J Cell Biol. 2003;161:673–677. doi: 10.1083/jcb.200302028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P, Yamamoto H, Suetsugu S, Miki H, Takenawa T, Endo T. Small GTPase Rah/Rab34 is associated with membrane ruffles and macropinosomes and promotes macropinosome formation. J Biol Chem. 2003;278:4063–4071. doi: 10.1074/jbc.M208699200. [DOI] [PubMed] [Google Scholar]
- Mayor S, Pagano RE. Pathways of clathrin-independent endocytosis. Nat Rev Mol Cell Biol. 2007;8:603–612. doi: 10.1038/nrm2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damke H, Baba T, van der Bliek AM, Schmid SL. Clathrin-independent pinocytosis is induced in cells overexpressing a temperature-sensitive mutant of dynamin. J Cell Biol. 1995;131:69–80. doi: 10.1083/jcb.131.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antov Y, Barbul A, Mantsur H, Korenstein R. Electroendocytosis: exposure of cells to pulsed low electric fields enhances adsorption and uptake of macromolecules. Biophys J. 2005;88:2206–2223. doi: 10.1529/biophysj.104.051268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rols MP, Femenia P, Teissie J. Long-lived macropinocytosis takes place in electropermeabilized mammalian cells. Biochem Biophys Res Commun. 1995;208:26–35. doi: 10.1006/bbrc.1995.1300. [DOI] [PubMed] [Google Scholar]
- Glogauer M, Lee W, McCulloch CA. Induced endocytosis in human fibroblasts by electrical fields. Exp Cell Res. 1993;208:232–240. doi: 10.1006/excr.1993.1242. [DOI] [PubMed] [Google Scholar]
- Krassowska W, Filev PD. Modeling electroporation in a single cell. Biophys J. 2007;92:404–417. doi: 10.1529/biophysj.106.094235. [DOI] [PMC free article] [PubMed] [Google Scholar]


