SUMMARY
Ddi1 belongs to a family of shuttle proteins targeting polyubiquitinated substrates for proteasomal degradation. Unlike the other proteasomal shuttles, Rad23 and Dsk2, Ddi1 remains an enigma: its function is not fully understood and structural properties are poorly characterized. We determined the structure and binding properties of the ubiquitin-like (UBL) and ubiquitin-associated (UBA) domains of Ddi1 from Saccharomyces cerevisiae. We found that, while Ddi1UBA forms a characteristic UBA:ubiquitin complex, Ddi1UBL has entirely uncharacteristic binding preferences. Despite having a ubiquitin-like fold, Ddi1UBL does not interact with typical UBL-receptors but, unexpectedly, binds ubiquitin, forming a unique interface mediated by hydrophobic contacts and by salt-bridges between oppositely-charged residues of Ddi1UBL and ubiquitin. In stark contrast with ubiquitin and other UBLs, the β-sheet surface of Ddi1UBL is negatively charged and, therefore, is recognized in a completely different way. The dual functionality of Ddi1UBL, capable of binding both ubiquitin and proteasome, suggests a novel mechanism for Ddi1 as a proteasomal shuttle.
INTRODUCTION
The ubiquitin-proteasome system plays an essential role in the biology of eukaryotes. Through turnover of short-lived proteins, it regulates such vital processes as cell cycle progression, transcription, misfolded-protein degradation, and immune response (Ciechanover, 1994a, b; Conaway et al., 2002; Hicke, 2001a, b; Hoege et al., 2002; Muratani and Tansey, 2003; Rock and Goldberg, 1999; Schubert et al., 2000; Spence et al., 1995; Yamaguchi and Dutta, 2000). Ubiquitinated proteins are either directly recognized by the proteasomal ubiquitin (Ub) receptors (Rpn10, Rpn13) or first bound by the so-called shuttle proteins (Rad23, Dsk2, or Ddi1) which then drive the ubiquitinated proteins to the 26S proteasome for degradation (Clarke et al., 2001; Elsasser et al., 2004; Elsasser and Finley, 2005; Gomez et al., 2011; Hartmann-Petersen and Gordon, 2004; Hicke et al., 2005; Husnjak et al., 2008; Kaplun et al., 2005; Kleijnen et al., 2000; Lambertson et al., 1999; Saeki et al., 2002). Recognition of ubiquitinated substrates by either or both pathways is supported by the discovery that the function of Ub-receptors is redundant, and only concurrent deletion of two or more of the receptors causes accumulation of ubiquitinated proteins (Díaz-Martínez et al., 2006; Kang et al., 2006; Rao and Sastry, 2002; Saeki et al., 2002; Verma et al., 2004). The proteasomal shuttle proteins contain an N-terminal ubiquitin-like domain (UBL) and a C-terminal ubiquitin-associated domain (UBA) (Fig 1A). Rad23 has an additional UBA domain that follows its UBL domain in the gene structure. Unlike the proteasomal Ub-receptors (e.g., Rpn10, Rpn13), the UBL-UBA shuttles are not integral subunits of the proteasomal complex. It has been hypothesized that the UBL-UBA shuttle proteins utilize their UBL domain to interact with the proteasome (primarily through the Rpn1 subunit), and their UBA domain(s) to interact with Ub moieties on ubiquitinated proteins (Bertolaet et al., 2001b; Chen et al., 2001; Elsasser and Finley, 2005; Funakoshi et al., 2002; Kang et al., 2006; Wilkinson et al., 2001; Zhang et al., 2009).
Figure 1.
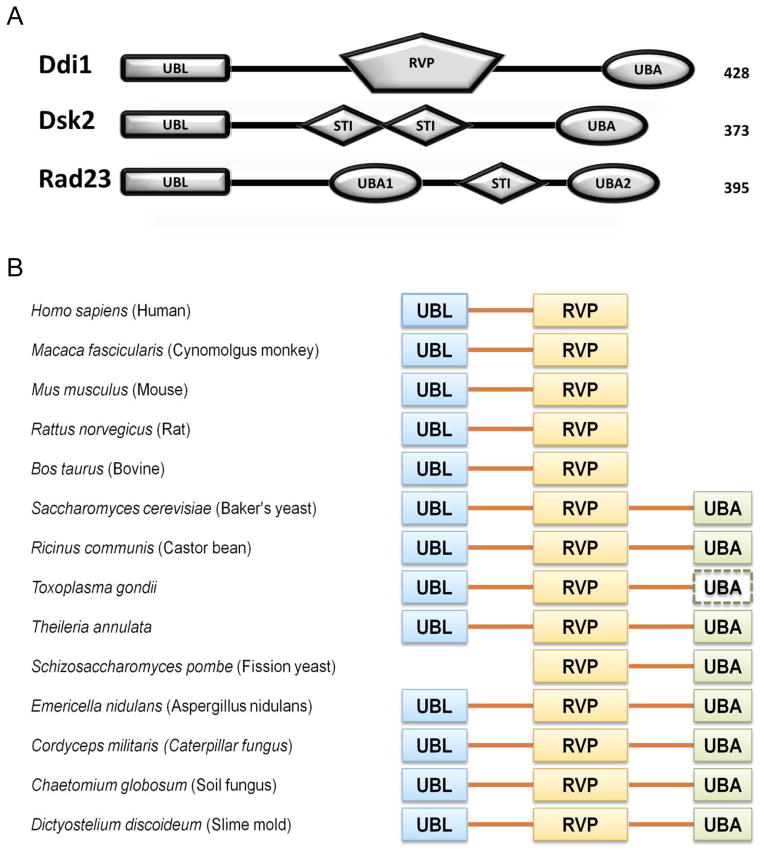
Ddi1 gene structure and domain conservation among eukaryotes. (A) Domain composition of Ddi1, Dsk2, and Rad23 from Saccharomyces cerevisiae. (B) Domain composition of Ddi1 from selected organisms; UBL – ubiquitin like domain, RVP – retroviral protease-like domain, UBA – ubiquitin associated domain are shown as solid blocks; the dashed block indicates the potential presence of a C-terminal UBA, that was identified by the domain prediction software but did not pass the threshold criteria. The UBL and RVP domains are present in almost all species whereas the UBA domain was lost in mammals upon evolution. See also Supplemental Information. (See also Fig S1)
Ddi1 (DNA Damage-Inducible 1) protein has an unusual composition for a UBL-UBA shuttle protein as it contains a conserved retroviral protease fold domain (RVP), followed by a putative PEST region and Sso-binding domains (Sso-BD) (Gabriely et al., 2008; Sirkis et al., 2006). The detailed substrate specificity of Ddi1 as a shuttle protein is not known, however, it was found that Ddi1 is required for degradation of Ho endonuclease and F-box protein Ufo1, two proteins involved in cell cycle progression and regulation (Ivantsiv et al., 2006; Kaplun et al., 2005). Ho endonuclease is recruited by Ufo1 to the SCF ubiquitin ligase complex for ubiquitination and subsequently transferred by Ddi1 to the 26S proteasome for degradation (Kaplun et al., 2003; Kaplun et al., 2000). Ufo1 is a unique F-box protein, which in addition to the F-box and a protein-protein interaction domain contains three ubiquitin-interacting motifs (UIM) that are important for Ufo1 autoubiquitination (Baranes-Bachar et al., 2008). Interestingly, it was suggested that turnover of Ufo1 depends on the presence of Ufo1UIMs and the UBL domain of Ddi1 (Ivantsiv et al., 2006). It is not fully understood whether Ddi1 interacts with Ufo1 directly, but importantly, in the absence of Ddi1, Ufo1 and ubiquitinated Ho endonuclease accumulate in the cell (Ivantsiv et al., 2006; Kaplun et al., 2005).
Although yeast Ddi1 participates in many cellular processes (Bertolaet et al., 2001b; Gabriely et al., 2008; Ivantsiv et al., 2006; Kaplun et al., 2000), its function is not fully understood, and its structure and ligand-binding preferences are poorly characterized. The evidence that Ddi1 transfers ubiquitinated substrates to the proteasome for degradation is sporadic (Ivantsiv et al., 2006; Kaplun et al., 2005; Saeki et al., 2002; Voloshin et al., 2012), and the role of Ddi1 in proteasomal targeting is the least studied of all shuttle proteins (Gomez et al., 2011; Rosenzweig et al., 2012). It has been reported that, unlike Rad23 and Dsk2, Ddi1 is not involved in the UFD proteolytic pathway (Kim et al., 2004). Moreover, interaction with Ufo1 appears unique to Ddi1 (Ivantsiv et al., 2006) and beyond the classical role of a shuttle protein. It is believed that yeast Ddi1 contains an N-terminal UBL domain and a C-terminal UBA domain, which enable Ddi1’s function as a shuttle. Indeed, the putative UBA domain of yeast Ddi1 has been shown to directly interact with Ub (Bertolaet et al., 2001b), although its three-dimensional structure has not been determined. However, the putative UBL domain of Ddi1 remains an enigma. In contrast to Ub and the UBLs of Rad23 and Dsk2, it does not interact with the UIM of Rpn10 (Zhang et al., 2009). Moreover, unlike the UBLs of Rad23, Dsk2, and Ubp6, the UBL of Ddi1 associates very weakly, if at all, with the proteasomal subunit Rpn1 (Rosenzweig et al., 2012). These observations raise the question whether the N-terminal region of Ddi1 is indeed a UBL domain, and if it is, what is its function and receptor-binding preferences. Furthermore, the RVP-domain’s activity as a protease has not been tested in the context of the ubiquitin-proteasome system. In order to shed light on the complex role of Ddi1 in protein degradation and to identify the source of its functional difference from other UBL-UBA proteins, we performed structural and functional characterization of this multidomain protein, described in this paper. These studies unveiled completely novel, unexpected binding preferences of the UBL domain of Ddi1 and provide new insights into Ddi1 function in cells.
RESULTS
Is Ddi1 a member of the UBL-UBA protein family?
As mentioned above, it is believed that Ddi1 from S. cerevisiae belongs to the UBL-UBA shuttle proteins family. Such classification is based largely on bioinformatic analysis suggesting that the N- and C-terminal regions of the Ddi1 gene contain the UBL and UBA domains, respectively (Bertolaet et al., 2001a; Clarke et al., 2001). The putative UBA domain of Ddi1 shares 30% (or higher) of its sequence with other UBA domains, whereas the low sequence identity of the putative UBL domain with Ub and other UBLs (Fig S1) questions its classification as a UBL domain. Moreover, standard analysis of Ddi1 sequence using databases of conserved domains (CDD, SMART) or protein families (Pfam) classified its C-terminal region as a UBA domain but did not predict that the protein contains a UBL domain. Interestingly, our analysis of Ddi1 domain conservation among different species indicates that, with just one exception, the UBL domain is always present in Ddi1 sequence, whereas the UBA domain is not present in mammals (Fig 1B). This loss of the UBA domain during evolution might be a result of its lack of functional importance in higher organisms, which then raises the question, whether and how can Ddi1 act as a shuttle protein without a Ub-binding component? These evolutionary changes in the Ddi1 gene domain composition combined with the low sequence identity to other UBL and UBA domains necessitate a thorough characterization of the structure and function of the putative UBL and UBA domains of Ddi1.
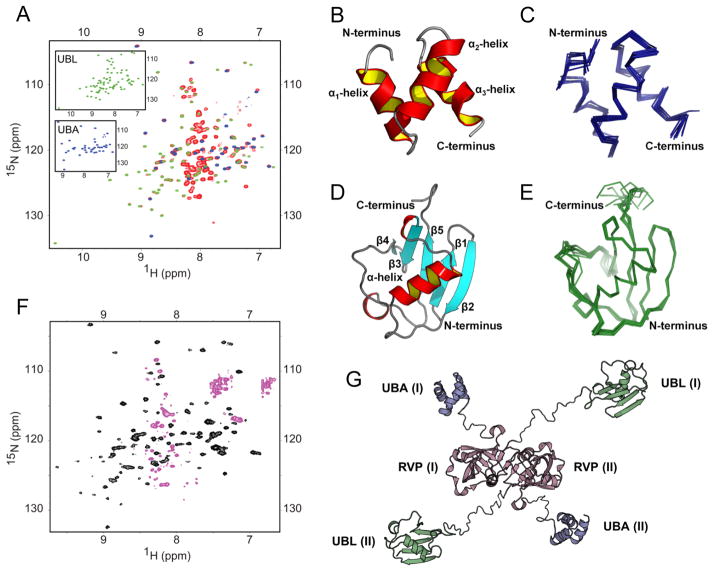
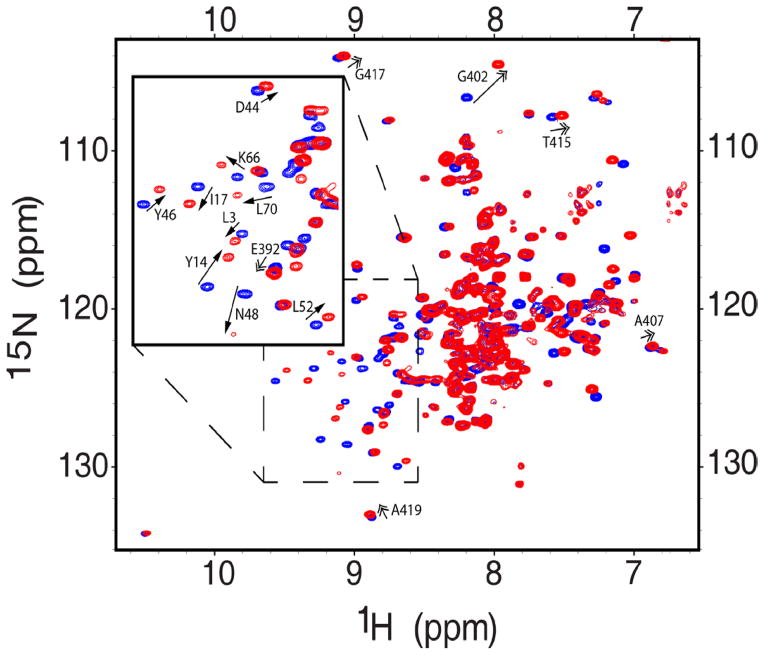
Due to the large size (47 kDa) of Ddi1, we decided to focus our NMR studies on the isolated N- and C-terminal Ddi1 fragments, containing the putative UBL and UBA domains, respectively. Prior to characterizing these fragments in isolation, we established that their structural/spectral properties are the same as in the context of the full-length (FL) Ddi1 protein. For this purpose, we recorded and compared NMR spectra of FL Ddi1 with the corresponding spectra of the isolated fragments. The majority of 1H-15N NMR signals overlaid almost perfectly, indicating that the chemical environment of each amide group is essentially the same in the FL protein and in the isolated fragments (Fig 2A). The small discrepancies observed only for the few C-terminal residues in Ddi1UBL and the few N-terminal residues in Ddi1UBA are not unexpected, and reflect a change in the chemical environment caused by the truncation. These results justified the reductionist approach, focusing on the relevant Ddi1 fragments in isolation.
Figure 2.
NMR spectra and structural models of Ddi1UBA and Ddi1UBL as individual domains and in full-length Ddi1. (A) Overlay of the 1H-15N TROSY spectra of the full-length Ddi1 (red) and of the isolated fragments containing only UBA (blue) or UBL (green) domains. Shown in insets are spectra of the individual domains with the same color coding. (B–E) 3D structures of the UBA (B, C) and UBL (D, E) domains of Ddi1, shown as cartoon representation (B and D) and as overlay of backbone traces of 10 lowest-energy structures (C and E, respectively). (F) Steady-state heteronuclear 15N{1H} NOE spectrum of full-length Ddi1; the black contours represent positive signal intensities (signals of the UBL and UBA) while the magenta contours correspond to negative intensities, indicating highly flexible amide groups. (G) A putative structural model of full-length Ddi1 protein (shown as a dimer) based on our NMR data indicating that the well-folded domains (UBL, UBA) are connected by flexible linkers. The Ddi1UBA and Ddi1UBL structures are from panels (B) and (D), the Ddi1RVP homodimer structure is from PDB ID 3S8I (it has Cα RMSD of 0.6 Å to that of yeast Ddi1RVP, PDB ID 2IIA). (See also Fig S2)
It is worth pointing out that the UBA and UBL signals in the spectrum of the FL Ddi1 are relatively sharp despite the large size of the protein; this indicates that both domains tumble somewhat independently from the rest of the protein and from each other (see also below). This is further supported by the measured values of 15N longitudinal relaxation time (T1), which depend on the overall tumbling rate, and hence reflect the apparent size of the molecule (Varadan et al., 2005). Specifically, in the case of Ddi1UBL the general pattern of 15N T1 values is retained, and the overall level of T1 in the context of FL Ddi1 is only slightly higher than for the isolated domain (Fig S2). The similarity of the tumbling rates for the putative UBL domain as part of FL Ddi1 and in isolation indicates that Ddi1UBL behaves almost independently from the rest of the protein, thus further supporting the above conclusion that the putative UBL domain is not significantly affected by the rest of the FL Ddi1 protein.
Three-dimensional structure of the UBA and UBL domains of Ddi1
Having confirmed the relevance of studying the isolated putative UBA and UBL domains of Ddi1, we set to determine their three-dimensional structures. Using multidimensional NMR methods we obtained a complete 1H, 13C, and 15N assignment of both Ddi1UBA and Ddi1UBL constructs. We then determined high-resolution structures of the UBA and UBL domains in solution using inter-proton NOEs as the main distance constraints for structure calculations (Table 1) together with the hydrogen bonding constraints confirmed by H/D exchange experiments, backbone torsion angles predicted by TALOS+ (Cornilescu et al., 1999; Shen et al., 2009) based on the chemical shifts, and 15N-1H residual dipolar couplings (RDCs) measured in weak liquid-crystalline medium.
Table 1.
NMR constraints and refinement statistics for Ddi1 UBL and UBA structures.
| Ddi1UBA | Ddi1UBL | |
|---|---|---|
|
| ||
| NMR distance and dihedral constraints | ||
| Distance constraints | ||
| Total NOE | 935 | 1101 |
| Intra-residue | 544 | 431 |
| Inter-residue | 391 | 670 |
| Sequential (|i − j| = 1) | 167 | 296 |
| Medium-range (|i − j| < 4) | 113 | 141 |
| Long-range (|i − j| > 5) | 111 | 233 |
| Intermolecular | N/A | N/A |
| Hydrogen bonds | 27 | 25 |
| Total dihedral angle restraints | ||
| ϕ | 27 | 63 |
| ψ | 27 | 63 |
| Total RDCs | 28 | 55 |
| Structure statistics | ||
| Violations (mean and s.d.) | ||
| Distance constraints (Å) | 0.07 ± 0.01 | 0.05 ± 0.05 |
| Dihedral angle constraints (°) | 1.47 ± 0.32 | 3.57 ± 0.38 |
| Max. dihedral angle violation (°) | 2.27 | 6.70 |
| Max. distance constraint violation (Å) | 0.98 | 0.22 |
| Deviations from idealized geometry | ||
| Bond lengths (Å) | 0.012 ± 0.001 | 0.162 ± 0.004 |
| Bond angles (°) | 1.02 ± 0.03 | 1.26 ± 0.04 |
| Impropers (°) | 2.07 ± 0.17 | 2.79 ± 0.17 |
| Average pairwise r.m.s. deviationa (Å) | ||
| All residues | Residues 389–428 | Residues 2–80 |
| Heavy | 1.04 ± 0.12 | 1.90 ± 0.35 |
| Backbone | 0.46 ± 0.10 | 1.34 ± 0.32 |
| Secondary structure | ||
| Heavy | 1.11 ± 0.12 | 0.89 ± 0.11 |
| Backbone | 0.40 ± 0.10 | 0.39 ± 0.05 |
Pairwise r.m.s. deviation was calculated among 10 refined structures using MolMol (Koradi et al., 1996)
The structure of the nominal UBA domain of Ddi1 shows the characteristic UBA fold, where three α-helices connected through two loops form a compact bundle (Fig 2B). The helices comprise residues 392–399, 405–414, and 419–426. The ensemble of ten Ddi1UBA structures derived from our NMR data has the backbone RMSD of 0.46 ± 0.10 Å (Fig 2C). The back-calculated RDCs for the lowest-energy Ddi1UBA structure have a correlation coefficient of 0.993 and the quality factor (Clore and Garrett, 1999) of 0.078 to the experimental RDC data (Fig S2C). The backbone RMSD values between Ddi1UBA and other UBAs range from 0.87 Å (Mud1UBA and hHR23A-UBA1) to 1.62 Å (Ubiquilin-1 UBA) (Fig S2D), similar to other UBAs (Zhang et al., 2008).
The structure of the nominal UBL domain of Ddi1 is shown in Figure 2D. The ten lowest-energy structures (Fig 2E) have a backbone RMSD of 0.80 ± 0.16 Å for residues 2–76 and 0.39 ± 0.05 Å for the secondary structure elements. This difference between the RMSD values reflects the presence of a long unstructured loop (residues 52–69) connecting strands β4 and β5 in Ddi1UBL. The C-terminal residues 77–80, not included in this comparison, had no inter-residue NOE constraints and therefore are disordered in the calculated structures. Note the excellent agreement between the experimental and back-calculated RDC values (correlation coefficient 0.987, quality factor 0.104, Fig S2C). Finally, our NMR-derived structure of Ddi1UBL is independently supported by the small-angle neutron scattering (SANS) data (Fig S7F).
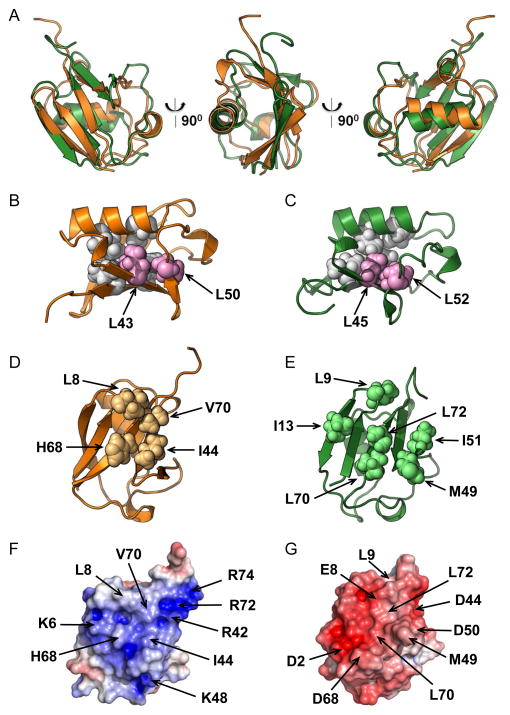
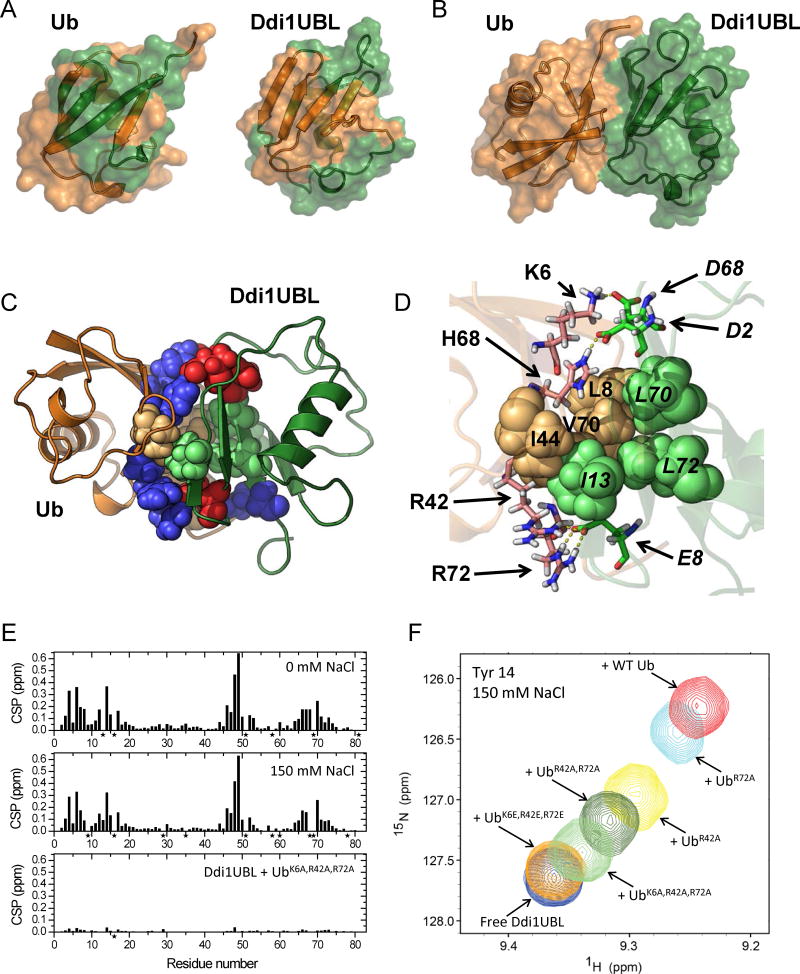
Our structural data clearly show that despite the low sequence identity to Ub, the N-terminal segment of Ddi1 indeed contains a domain with a Ub-like fold, where a 5-strand β-sheet is packed against the α-helix, forming the hydrophobic core of the protein (Fig 2D). Superimposition of the atom coordinates of Ddi1UBL and Ub revealed significant overall structural similarity between the two proteins (Fig 3A), although Ddi1UBL’s α-helix is slightly tilted compared to Ub, and the β-sheet in Ddi1UBL appears somewhat flatter and not wrapped around the helix to the same extent as in Ub. The backbone RMSD between Ddi1UBL and Ub is 1.92 Å (secondary structure).
Figure 3.
Structure comparison of Ddi1UBL and Ub. (A) Cartoon representation of the overlay of 3D structures of Ub (orange) and Ddi1UBL (green). (B–C) Cartoon representation of the structures of Ub (B) and Ddi1UBL (C) with the side chains of the hydrophobic core residues conserved among all the UBLs (see Fig S1): L43, L50 in Ub (B) and L45, L52 in Ddi1UBL (C) shown as spheres, colored pink. Also shown in light grey spheres are several other residues that, together with the abovementioned residues form the hydrophobic core in Ub (V26, I30, L67, L69) and Ddi1 UBL (L28, L32, L71, I73). (D) Sphere representation of the hydrophobic patch residues on Ub surface. (E) Sphere representation of the residues on the surface of Ddi1UBL that form the hydrophobic surface patch. (F–G) Surface electrostatic potential (positive is blue, negative is red) of Ub (F) and Ddi1UBL (G), calculated using Adaptive Poisson Boltzmann Solver (APBS) (Dolinsky et al., 2004). The coloring range is ±4 kT/e for Ub and ±8 kT/e for Ddi1UBL. Both proteins are oriented similarly, with the β-sheet surface facing the reader. Location of charged side chains and major hydrophobic residues on the surface of each protein is indicated with arrows. (See also Fig S3)
Most of side chain contacts forming and stabilizing the hydrophobic core of Ub are preserved in Ddi1UBL (Fig 3B, C). These include Leu28 and Leu32 (Val26 and Ile30 in Ub) that “anchor” the α-helix, Leu71 and Ile73 (Leu67 and Leu69 in Ub) that “anchor” strand β5 to the core of the protein (Haririnia et al., 2008), and Leu45 and Leu52 (Leu43 and Leu50 in Ub) that are conserved among Ub and several UBLs (see Fig S1).
It is clear from the sequence and structure alignment with Ub that Ddi1UBL does not have the “canonical” Leu8-Ile44-His68-Val70 hydrophobic patch (Beal et al., 1996) characteristic for Ub (Fig 3E) as well as for the UBLs of Dsk2 and Rad23 (Fig S3). However, several hydrophobic side chains form a similar solvent-exposed hydrophobic surface on the β-sheet face of Ddi1UBL that could mediate the UBL’s interactions with potential binding partners (Fig 3F). These hydrophobic side chains are spread over the entire β-sheet surface of Ddi1UBL compared to a more compact arrangement seen in Ub and the UBLs of Dsk2 and Rad23 (Fig S3).
A detailed inspection of the surface charges on the β-sheet face of Ddi1UBL revealed a striking contrast with Ub, as well as with the UBLs of Rad23 and Dsk2. In Ub the hydrophobic patch is surrounded by basic side chains. However, in Ddi1UBL most of them are replaced with acidic side chains, plus some additional acidic residues. Most notably, Asp44 and Asp50 in Ddi1UBL are located where Arg42 and Lys48 are in Ub, and Glu8 is positioned close to where Lys6 is in Ub. This results in the opposite sign of the electrostatic potential of the β-sheet surface of Ddi1UBL compared to Ub and other UBLs (Fig 3F, G and S3). This drastic difference in the distribution of surface charges suggests that although Ddi1UBL has a Ub-like fold, its binding partners (and recognition preferences) should be different from those of Ub and the other UBLs (see also below).
In order to further explore the similarities and differences between Ddi1UBL and Ub, we measured steady-state heteronuclear 15N{1H} NOEs (hetNOE) which are sensitive indicators of the backbone flexibility in a protein. Specifically, we wanted to independently verify some salient features of the derived Ddi1UBL structures, namely, the rigidity of the secondary structure elements, the less structured (compared to Ub) β4/β5 loop, and the unstructured C-terminus. Indeed, our results show that the amide bonds in the elements of secondary structure of Ddi1UBL have hetNOE values at or above 0.75, typical for well-defined regions in a protein (Fig S2E). Similarly to Ub, Ddi1UBL has a highly flexible C-terminus, as obvious from the extremely low hetNOE values for residues 76–80. Unlike Ub, Ddi1 residues 52–60 exhibited noticeably lower hetNOEs compared to the rest of the backbone (excluding the C-terminus), which indicates that the long β4/β5 loop in Ddi1UBL is more dynamic than a similar loop in Ub. This explains why that loop in Ddi1UBL appears less structured than in Ub.
Ddi1 as a multidomain protein
Previously it was shown that UBL-UBA proteins can homodimerize through intermolecular interaction between the UBL and UBA domains (Bertolaet et al., 2001a; Sasaki et al., 2005). In the case of Ddi1, the abovementioned similarity between the NMR spectra of these domains in isolation and in the context of FL Ddi1 (Fig 2A) rules out both intramolecular (in cis) and intermolecular (in trans, at least at the Ddi1 concentration of 250 μM) interaction between the UBL and UBA domains. Additionally, no UBL-UBA interaction was detected upon titration of 15N-labeled Ddi1UBL with Ddi1UBA (Fig S2B). These results are consistent with the published reports that only the RVP domain is important for Ddi1 homodimerization (Bertolaet et al., 2001a; Sirkis et al., 2006). Note that signals from the structured RVP domain are not easily discernible in the NMR spectrum of FL Ddi1 (Fig 2A), likely due to line broadening caused by slow tumbling of this internal dimerization domain (Kang et al., 2006). The resonances detected beside the UBA and UBL signals are mostly positioned in the middle of the spectrum. Unlike the UBA and UBL signals that have positive intensities in hetNOE spectra, these signals show negative or near zero intensities (Fig 2F) indicating that they belong to some unstructured and highly flexible regions of Ddi1.
Except for being responsible for Ddi1 homodimerization and being involved in pds1-128 rescue (Bertolaet et al., 2001a; Gabriely et al., 2008), no other function of the RVP domain especially in the UPS system has been reported. To examine whether this retroviral protease fold domain has any deubiquitinase activity, we incubated FL Ddi1 with di-Ubs of all seven possible lysine linkages. No cleavage was detected for any of the di-Ubs even after 24h (Fig S4). Although we found no activity against unanchored di-Ubs, we cannot exclude the possibility that Ddi1 functions as deubiquitinase with a different substrate specificity.
Combining the above observations that the UBA and UBL domains in Ddi1 tumble/behave independently with the role of RVP as the homodimerization domain, we propose a putative model of the FL Ddi1 structure, in which the three structured domains are connected through flexible linkers (Fig 2G) making the UBL and UBA domains available for independent interactions with their respective binding partners.
UBA domain of Ddi1 binds ubiquitin
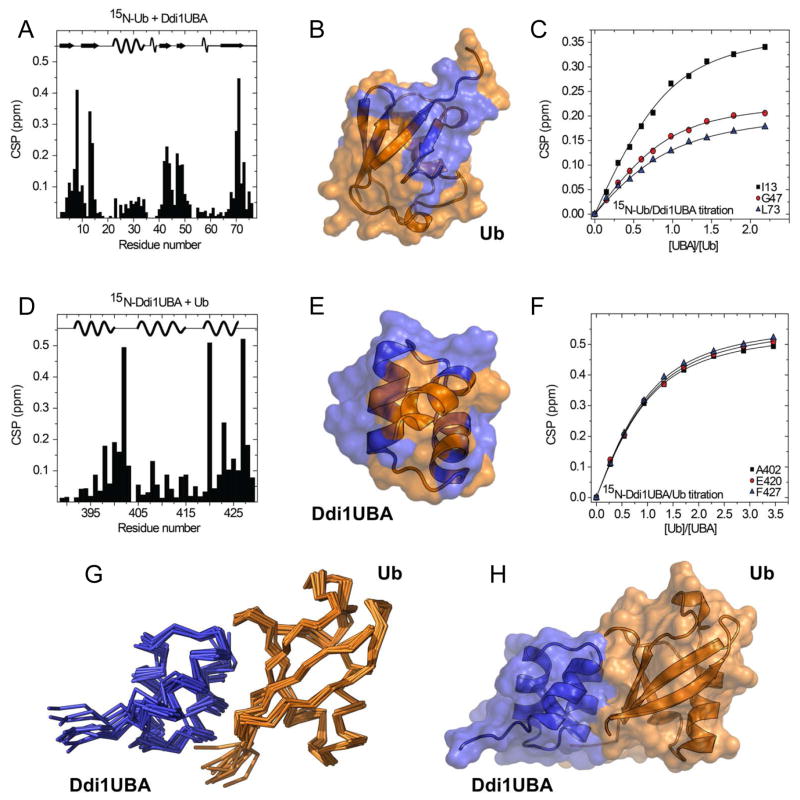
Our structural studies confirmed the bioinformatic predictions that Ddi1 contains an N-terminal domain with the UBL fold and a C-terminal domain with the UBA fold. It is critical, however, to verify that these domains not only have the expected folds but also the functions characteristic for a UBL and a UBA domain. It was shown in previous studies that Ddi1 interacts with Ub and polyUb through its UBA domain (Bertolaet et al., 2001b; Kang et al., 2006); however, these interactions were never studied in detail. In order to characterize Ub binding to the UBA domain of Ddi1, we titrated Ddi1UBA into Ub and vice versa. The results show strong site-specific chemical shift perturbations (CSPs) in the hydrophobic-patch residues of Ub, suggesting their involvement in the interaction with Ddi1UBA (Fig 4A–B). In Ddi1UBA, the most perturbed residues are clustered around Gly402, located in the loop between helices 1 and 2, Glu420 and Ala423 in helix 3, and Phe427 at the C terminus (Fig 4D–E). The analysis of NMR titration curves from this titration yielded the dissociation constant Kd of 150 ± 16 μM (Fig 4C, F), which is in the typical Kd range (4–500 μM) reported for interactions between monomeric Ub and other UBA domains (Ohno et al., 2005; Raasi et al., 2005; Trempe et al., 2005; Varadan et al., 2005; Walinda et al., 2014; Zhang et al., 2008). Based on the CSP data from Ub:Ddi1UBA titrations, complemented with intermolecular distance constraints derived using site-directed paramagnetic spin-labeling (Fig S5), we generated a structural model of the Ub:Ddi1UBA complex using biomolecular docking program HADDOCK (de Vries et al., 2010). The 10 lowest-energy structures showed good convergence, with the backbone RMSD of 1.0 ± 0.2 Å for the secondary structure elements (Fig 4G–H). In this complex, the interface is formed by the hydrophobic patch of Ub and, on the Ddi1UBA side, by the LGF motif in the loop connecting helices 1 and 2 as well as by Phe427 C-terminal to helix 3. This structure is in good agreement with those reported for other Ub:UBA complexes (Zhang et al., 2008).
Figure 4.
Characterization of Ub-Ddi1UBA binding by NMR. (A–C) Titration of 15N-labeled Ub with Ddi1UBA. (A) Amide chemical shift perturbations (CSPs) in Ub at the titration endpoint. (B) CSP-based mapping of Ddi1UBA-binding site (colored blue) on the surface of Ub (orange). (C) Titration curves for selected Ub residues in close proximity to the hydrophobic patch. (D–F) Titration of 15N-labeled Ddi1UBA with Ub. (D) CSPs in Ddi1UBA at the titration endpoint. (E) Map of the Ub-binding site (orange) on the surface of Ddi1UBA (blue) based on the CSPs from panel (D). The CSP threshold in (B) and (E) was set to 0.1 ppm. (F) Representative titration curves for selected Ddi1UBA residues. (G–H) Structure of the complex of Ub (orange) and Ddi1UBA (blue): (G) backbone traces of 10 lowest-energy HADDOC-derived structures of Ub:Ddi1UBA complex, and (H) cartoon representation of the best structure of the complex. The location of the secondary structure elements in Ub and Ddi1UBA is shown in the top portions of the graphs in (A) and (D), respectively. (See also Fig S5)
Unexpectedly, the UBL domain of Ddi1 binds ubiquitin
Because the Ub-binding studies presented above were performed for the isolated UBA domain of Ddi1 we wanted to test whether our observations hold in the context of FL Ddi1. For this, we added, in two-fold molar excess, unlabeled Ub to 15N-labeled FL Ddi1 (Fig 5). As expected, NMR signals corresponding to Ddi1UBA showed similar shifts as for the isolated UBA, thus confirming the relevance of our binding studies described above. Surprisingly, however, in addition to the shifts in Ddi1UBA signals, we observed even a bigger change in the positions and intensities of the signals corresponding to Ddi1UBL. This entirely unexpected result indicates that Ddi1 interacts with Ub not only through the UBA domain but also through the UBL domain.
Figure 5.
Both UBA and UBL domains of Ddi1 bind ubiquitin. Overlay of the 1H-15N TROSY spectra of FL Ddi1 alone (blue) and in the presence of Ub in 2-fold molar excess (red). Single-headed arrows indicate shifts of the UBL domain signals, double-headed arrows mark shifts of the UBA domain signals. The inset shows zoom on the spectral area indicated by the dashed box. (See also Fig S6)
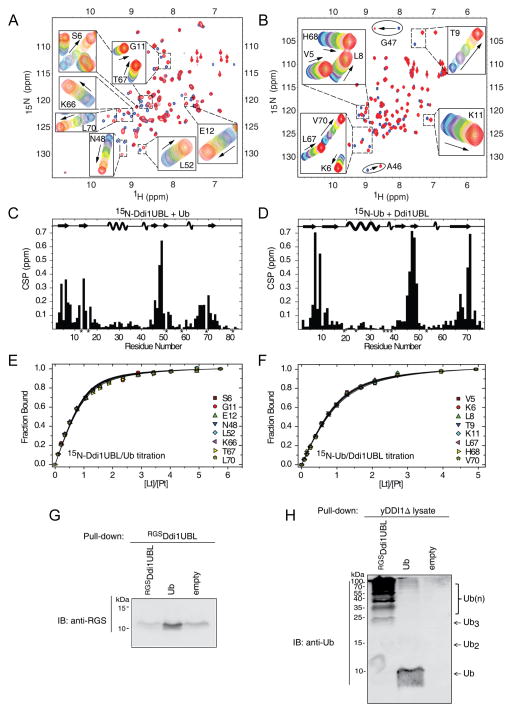
Since, to the best of our knowledge, no interaction between a UBL domain and Ub has ever been reported, we examined Ub binding to an isolated Ddi1UBL domain. When Ub was added to 15N-labeled Ddi1UBL, a number of UBL residues showed strong CSPs (Fig 6A, C). Mapping residues with CSPs > 0.075 ppm at the endpoint of titration on the Ddi1UBL structure allowed us to identify the putative binding site for Ub (Fig 7A). Our results clearly show that residues mostly affected by Ub binding are located on all five β-strands of Ddi1UBL and form a contiguous surface on the β-sheet side of the protein. By contrast, the opposite (α-helix) side of Ddi1UBL appears not to be involved in Ub binding, as inferred from the almost negligible CSPs observed there. It should be emphasized here that the location of the binding site on the β-sheet side of Ddi1UBL is consistent with the location of the ligand-binding site on Ub and other UBL proteins (Beal et al., 1996; Ryu et al., 2003; Zhang et al., 2009). Fitting the titration curves to a 1:1 binding model (Fig 6E) gave the Kd value of 45 ± 7 μM (averaged over 15 residues). The 1:1 stoichiometry of binding is supported by the 15N T1 data (the average T1 at 14.1 T was 601 ± 37 ms for Ddi1UBL alone and 951 ± 52 ms at the titration endpoint) as well as by the fact that this binding model gave the best fit to the titration curves.
Figure 6.
NMR characterization of Ub binding to Ddi1UBL. (A) Overlay of 1H-15N SOFAST-HMQC spectra of 15N-labeled Ddi1UBL free in solution (blue) and in Ub-bound state (at Ub:Ddi1UBL molar ratio 5:1). Insets show zoom on selected regions to illustrate gradual shifts in the peak positions upon titration. (B) Overlay of 1H-15N SOFAST-HMQC spectra of 15N-labeled Ub alone (blue) and saturated (red) with Ddi1UBL (at 5-fold molar excess). Gradual signal shifts of the residues used for Kd estimation are shown in insets. A46 and G47 exhibiting slow exchange upon Ub binding are marked with ovals. (C) Amide CSPs in Ddi1UBL at the endpoint of titration with Ub (molar ratio 5.7:1) as a function of residue number. Asterisks indicate P16 and those residues where N-H resonances cannot be followed due to signal overlap. (D) CSPs in Ub at the endpoint of titration with Ddi1UBL (5:1 molar ratio). P19, P37, P38, as well as several amides that could not be reliably observed in the NMR spectra are marked with asterisks. The location of the secondary structure elements in Ddi1UBL and Ub is shown in the top portions of the graphs in (C) and (D), respectively. (E–F) Representative normalized titration curves for selected β-sheet residues of Ddi1UBL (E) or Ub (F). The curves show fits of the data to a 1:1 binding model. (G) Ub pulls down Ddi1UBL. Purified RGS-His6-Ddi1UBL or His6-Ub was immobilized to activated CH-sepharose beads and was incubated with RGS-His6-Ddi1UBL. Eluted analyte proteins were visualized by immunoblotting against RGS tag (see Supplemental Experimental Procedures for further details). (H) Ddi1UBL pulls out Ub conjugates from yeast cell extract. As in (G), purified RGS-His6-DDI1UBL or His6-Ub was attached to activated CH-sepharose beads. Total protein extract was obtained from growing DDI1Δ yeast cells (strain lacking Ddi1 gene). Beads loaded with proteins were incubated with cell extract. Eluted proteins were detected by immunoblotting with anti-Ub. The monomeric Ub seen in the elution from Ub-loaded beads likely reflects a small amount of the protein that came off the beads during elution with 8M urea. (See also Fig S7)
Figure 7.
Structure of the Ub:Ddi1UBL complex. (A) NMR-based mapping of the Ub:Ddi1UBL interface. Left: Ddi1UBL-binding site (colored green) on the surface of Ub (orange). Right: Ub-binding site (orange) on the surface of Ddi1UBL (green). CSPs > 0.075 ppm were used for mapping in both cases. (B) HADDOCK-derived structure of the complex of Ub (orange) and Ddi1UBL (green). (C) The Ub:Ddi1UBL interface is formed by both hydrophobic and polar/charged amino acids (shown as spheres colored orange and blue, respectively, for Ub, green and red for Ddi1UBL). Ribbon colors are as in (A–B). (D) Specific side-chain contacts at the interface of the Ub:Ddi1UBL complex. Hydrophobic amino acids of Ub: L8, I44, V70 (orange) and Ddi1: I13, L70, L72 (green) are shown as spheres. Shown in stick representation are K6, R42, H68, R72 of Ub (pink) and D2, E8, D68 of Ddi1UBL (green) that form polar/electrostatic contacts (shown as yellow dash lines). To distinguish between Ub and Ddi1UBL residues, the latter are indicated in italic. (E, F) Validation of the Ub:Ddi1UBL complex by measurements at physiological ionic strength and by site-directed mutagenesis. (E) Amide CSPs in Ddi1UBL at the endpoint of titration with WT Ub at 0 mM NaCl (top) and at 150 mM NaCl (center), and in the presence of 4-fold molar excess of UbK6A,R42A,R72A, also at 150 mM NaCl (bottom). (F) Overlay of the NMR signals of Tyr14 of Ddi1UBL free in solution and upon addition of WT Ub or indicated Ub mutants, at [Ub]:[UBL]=4 and 150 mM NaCl. (See also Fig S7)
Ubiquitin interacts with the UBL domain of Ddi1 via the hydrophobic surface patch
This totally unexpected interaction between a UBL domain and Ub raised the question whether Ub interacts with Ddi1UBL through the same (hydrophobic-patch) surface as with other known Ub-ligands or perhaps it utilizes a different binding site. To answer this question, we titrated 15N-labeled Ub with unlabeled Ddi1UBL and monitored their interaction by NMR. Upon addition of Ddi1UBL, a number of amide signals of Ub changed their resonance positions significantly (Fig 6B). Moreover, signals of Ala46 and Gly47 disappeared and then reappeared in the course of titration, which is consistent with slow exchange on the NMR chemical shift time scale, indicative of tight binding. Interestingly, the overall magnitude of the observed CSPs is similar to that in 15N Ddi1UBL titrated with Ub (Fig 6C, D). Mapping the CSPs on the structure of Ub revealed that Ub interacts with Ddi1UBL through the same hydrophobic patch (centered at residues Leu8-Ile44-Val70) as when binding to other known Ub-ligands (Fig 7A). Finally, the analysis of titration curves (Fig 6F) gave the dissociation constant of 71 ± 6 μM (averaged over 13 residues with CSP > 0.075 ppm), in good agreement with the results obtained above from the CSPs in 15N Ddi1UBL. The averaged 15N T1 value (956 ± 53ms) for Ub in the complex with Ddi1UBL suggested the 1:1 stoichiometry of binding, and indeed the titration curves were best fit with the 1:1 binding model.
Ddi1UBL pulls out ubiquitin conjugates from cell extract
In support of our NMR results, the interaction between Ddi1UBL and Ub was also independently observed in a pull-down experiment (Fig 6G), where immobilized His6-Ub was able to pull out purified Ddi1UBL. Furthermore, His6-Ddi1UBL was capable of pulling out Ub-conjugates from the yeast cell extract lacking Ddi1, DDI1Δ (Fig 6H), thus indicating that Ddi1UBL and Ub interact at close to physiological conditions. The latter result suggests that the unusual capability of the UBL domain of Ddi1 to bind Ub could be utilized in the cell.
Ubiquitin interacts with UBL domain of Ddi1 but not with the UBL domains of Dsk2 or Rad23
Encouraged by the above finding, we examined whether the UBL domains from other UBL-UBA shuttle proteins can interact with Ub. For this, the UBL domains of Rad23 and Dsk2 were separately titrated into 15N-labeled Ub up to 2:1 and 5:1 molar ratio, respectively. No changes in the NMR spectra of Ub were observed (data not shown), indicating that neither Rad23UBL nor Dsk2UBL interact with Ub. This then makes the Ddi1UBL-Ub interaction entirely unique (at least) among the shuttle proteins family.
Structure of the ubiquitin:Ddi1UBL complex
Given that the interaction between Ub and Ddi1UBL is entirely novel and never studied before, we set out to characterize the structure of their complex. We obtained a structural model of the complex using HADDOCK (de Vries et al., 2010), and two sets of experimental intermolecular NMR constraints: (1) ambiguous constraints based on the observed CSPs (Table S1) and (2) unambiguous long-distance constraints (Table S2) derived from paramagnetic relaxation enhancement (PRE) detected in 15N-labeled Ddi1UBL upon addition of an equimolar amount of Ub carrying a nitroxide spin-label (MTSL) attached at residue 12 (Ub T12C). The derived structure of the Ub:Ddi1UBL complex is shown in Figures 7 and S7A. To verify the resulting structure, the atom coordinates of Ddi1UBL in complex with Ub were used to reconstruct the position of the spin label on Ub. The calculated position matched almost exactly the position of the Ub’s cysteine residue (Cys12) to which MTSL was attached (Fig S7B).
As an independent validation of our structure of the Ub:Ddi1UBL complex, the PREs in 15N-labeled Ddi1UBL caused by MTSL attached to residue 75 in Ub (Ub G75C) agreed nicely with their back-calculated values, and the reconstructed position of the MTSL’s unpaired electron was in close proximity to Cys75 (Fig S7C). Note that these data were not used for the docking. As a negative control, no major/systematic PREs were detected in Ddi1UBL when MTSL was attached to Cys63 in Ub K63C. In the Ub:Ddi1UBL complex, residue 63 of Ub is located at or above the MTSL’s PRE range (≈25 Å) from the amides in Ddi1UBL, hence too far to produce significant PREs in Ddi1UBL (Fig S7D). Finally, as a separate independent support for our findings, the NMR-derived structure of the Ub:Ddi1UBL complex and the Kd value agree well with the results of SANS measurements (Fig S7G).
A detailed analysis of the Ub:Ddi1UBL interface in the complex revealed that the interprotein interaction is mediated primarily through side chains of hydrophobic amino acids (Fig 7C, D). Ub utilizes its hydrophobic patch: Leu8, Ile44, Val70, whereas Ddi1UBL employs Ile13, Leu70, and Leu72 (Fig 7D) that form an analogous patch on the UBL surface (Fig 3E), resulting in the total buried surface area of ~1500 Å2. Interestingly, the interface contacts also include electrostatic/polar interactions, most notably, between Lys6 (Ub) and Asp68 (Ddi1), Arg42 (Ub) and Arg72 (Ub) and Glu8 (Ddi1), as well as His68 (Ub) and Asp2 (Ddi1) (Fig 7D). These interactions, involving the opposite charges on the β-sheet surfaces of Ub and Ddi1UBL (Fig 3F, G), could be responsible for strengthening the binding between Ub and Ddi1UBL.
The mode of Ub:Ddi1UBL and Ub:Ddi1UBA interactions is preserved at physiological salt concentrations
The pulldowns (Fig 6G, H) clearly show that Ub binds Ddi1UBL at physiological salt concentrations. To verify that the mode of Ub:Ddi1UBL binding observed in our NMR studies at low ionic strength (20 mM phosphate, no NaCl) is preserved at physiological conditions, the NMR titration studies were repeated in the presence of 150 mM NaCl. The CSPs in Ddi1UBL at the endpoint of titration ([Ub]:[UBL] = 6:1) were essentially the same as in the absence of NaCl (Fig 7E and S7), indicating that the binding interface and the physical nature of the Ub:Ddi1UBL interactions are fully preserved at physiological ionic strength. The Kd value increased to 175 ± 25 μM (averaged over 6 residues), consistent with weakening of charge-charge interactions at the interface. These results support the relevance of our NMR data obtained at low ionic strength and the important role of electrostatic/polar contacts revealed by the structure of the Ub:Ddi1UBL complex.
Similar conclusions were obtained for Ub:Ddi1UBA interaction, where the CSPs in Ddi1UBA at saturation ([Ub]:[UBA] = 8:1) in the presence of 150 mM NaCl were almost identical to those at low ionic strength (Fig S5), while the Kd increased to 291 ± 13 μM reflecting the weakening of electrostatic contacts at the Ub:Ddi1UBA interface.
Validation of the ubiquitin:Ddi1UBL structure by site-directed mutagenesis
To further validate the structure of the Ub:Ddi1UBL complex and the critical role of the electrostatic contacts in this binding, we performed a series of mutations, where the positively charged residues (K6, R42, and R72 of Ub) forming contacts with the negatively charged residues of Ddi1UBL were substituted with alanines or glutamates. The results (Figs 7E, F, and S7J) show that substitutions of these residues one by one with an Ala gradually decreased the strength of Ub:Ddi1UBL interaction, such that a triple-Ala mutation (UbK6A,R42A,R72A) reduced the binding drastically, and the triple-Glu mutation (UbK6E,R42E,R72E) essentially abolished binding. In agreement with the CSPs, no increase in 15N T1 of Ddi1UBL was detected upon addition of UbK6E,R42E,R72E and only marginal (5%, within the standard deviation) increase was observed in the presence of UbK6A,R42A,R72A. Thus, these results corroborate our structure of the Ub:Ddi1UBA complex.
UIMs of Ufo1 interact with ubiquitin but not with Ddi1
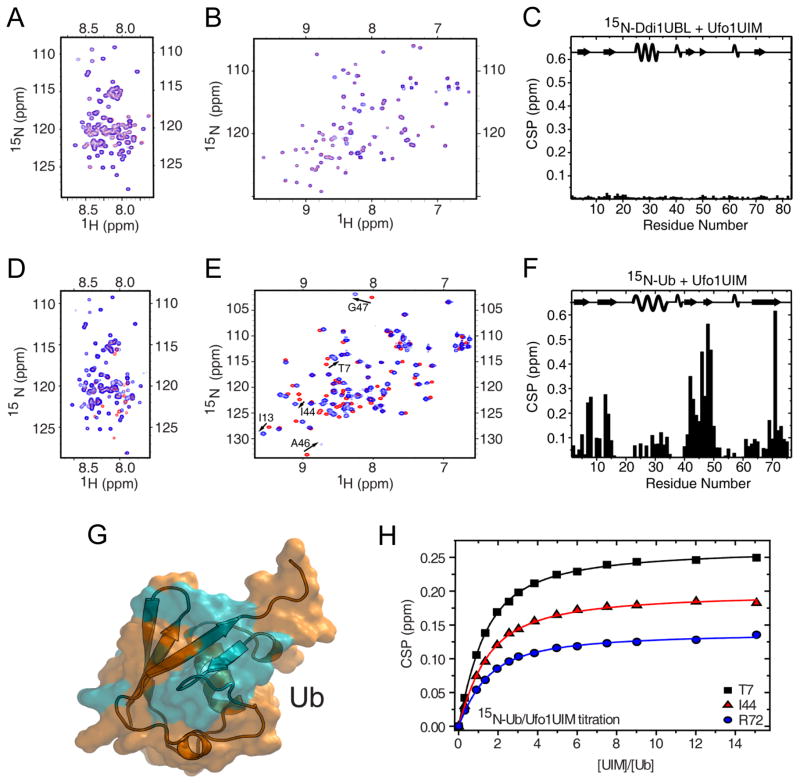
We found earlier that, unlike Ub and the UBLs of Rad23 and Dsk2, Ddi1UBL does not interact with the UIM of Rpn10 (Zhang et al., 2009). In order to determine if the lack of UIM binding is a general feature of Ddi1UBL or merely Rpn10-specific, we examined Ddi1 interaction with the UIM domains of Ufo1, the only ligand of Ddi1UBL proposed in the literature (Ivantsiv et al., 2006). For this purpose, 15N-Ddi1UBL was titrated with unlabeled Ufo1 construct containing all three UIMs (Ufo1UIMs) (Fig 8B). Surprisingly, despite overloading Ddi1UBL with Ufo1UIMs, up to Ufo1UIMs:Ddi1UBL molar ratio of 10:1, we did not detect any significant changes in the Ddi1UBL spectrum (Fig 8C). To further verify the lack of binding between the two proteins, we performed a reverse titration, this time adding unlabeled Ddi1UBL to 15N-labeled Ufo1UIMs; the Ufo1UIMs signals were not perturbed even at a 10-fold molar excess of Ddi1UBL (Fig 8A). Furthermore, no binding was detected when 15N-labeled FL Ddi1 was titrated with Ufo1UIMs also up to 10-fold molar excess (Fig S8). All these results indicate that Ddi1 does not interact directly with Ufo1UIMs. Note that, based on both NMR and circular dichroism data (Fig S8), Ufo1UIMs were in the folded state after expression and purification, and therefore capable of binding.
Figure 8.
NMR characterization of the interactions between Ufo1UIMs and Ddi1UBL or Ub. (A) Overlay of 1H-15N SOFAST-HMQC spectra of Ufo1UIMs alone (red) and upon saturation (blue) with 10-fold molar excess of Ddi1UBL. (B) Overlay of 1H-15N SOFAST-HMQC spectra of Ddi1UBL alone (red) and in the presence of Ufo1UIMs (blue) at 10:1 molar ratio. (C) CSPs in Ddi1UBL at saturation with Ufo1UIMs. (10:1 molar ratio, as in B) (D) Overlay of 1H-15N SOFAST-HMQC spectra of Ufo1UIMs alone (red) and upon addition of Ub (blue) at equimolar ratio. (E) Overlay of 1H-15N SOFAST-HMQC spectra of Ub alone (red) and in the presence of Ufo1UIMs (blue) at equimolar ratio. (F) CSPs in Ub at the titration endpoint with Ufo1UIMs (5:1 molar ratio). The location of the secondary structure elements in Ddi1UBL and Ub is shown in the top portions of the graphs in (c) and (f), respectively. (G) CSP-based mapping of the Ufo1-binding site (colored cyan) on the surface of Ub (orange). The CSP threshold was set to 0.1 ppm. (H) Representative titration curves for Ufo1UIMs binding to Ub. The lines show fits of the data to a model where all three UIMs in Ufo1UIMs bind Ub independently and with equal affinities. (See also Fig S8)
We then used Ub as a positive control, anticipating binding between Ub and the UIMs. Indeed, after mixing 15N-labeled Ufo1UIMs with Ub at a 1:1 molar ratio, we observed shifts and signal attenuations in several Ufo1UIM residues (Fig 8D). We then performed a full titration of 15N-labeled Ub with Ufo1UIMs until saturation was reached (at 5:1 molar ratio) and used the CSPs observed in Ub to map the Ufo1UIM-binding surface and quantify the strength of interaction (Fig 8E, F). Residues with significant CSPs pointed to the hydrophobic patch of Ub as the site for Ufo1UIMs binding (Fig 8G). Taking into account the presence of three UIMs in Ufo1UIMs and assuming that they bind Ub independently and with similar affinities, the average dissociation constant for UIM:Ub interaction was estimated to be 237 ± 52 μM (Fig 8H).
DISCUSSION
Ddi1 is a multi-domain protein in which each domain performs a different function. We found that the N- and C-terminal regions of Ddi1 are structured, and despite low sequence identity to other UBL or UBA domains, they do adopt the characteristic UBL and UBA folds, respectively. Moreover, all three main domains of Ddi1: UBL, RVP, and UBA do not interact with each other and are connected via flexible linkers that allow them to behave and interact with their targets independently.
As mentioned above, Ddi1 differs from the other shuttle proteins in its proteolytic roles and interacting partners. Our binding studies revealed the source of Ddi1’s unique status in the family of UBL-UBA proteins. As expected, the UBA domain of Ddi1 binds Ub and forms a typical UBA:Ub complex. The affinity of Ddi1UBA for Ub is in-between those of Rad23 UBA2 and Dsk2 UBA. Surprisingly, however, the UBL domain of Ddi1 exhibited completely unexpected binding preferences: it binds Ub quite strongly (Kd ≈ 45–70 μM) and is capable of pulling out Ub conjugates from yeast cell extract. This interaction appears specific to Ddi1UBL, as none of the other UBLs that we tested binds Ub.
The ability of Ddi1UBL to bind Ub is unusual and has not been reported for any other UBL domain. It should be mentioned here that monomeric Ub does bind Ub albeit with the Kd (~5 mM) much greater than the physiologically relevant concentrations of Ub (up to ca. 85 μM (Kaiser et al., 2011)). A non-covalent interaction between Ub molecules has been observed within some polyUb chains (Fushman and Wilkinson, 2011), as a result of either crystal packing forces or of tethering Ubs to each other, which facilitates Ub:Ub binding through a drastic increase, up to ~80 mM (Liu et al., 2012), in the effective local concentration of Ub.
Furthermore, Ddi1UBL is strikingly different from Ub and other UBLs in that it does not interact with the typical Ub/UBL-binding domains such as UIM and UBA (at least those tested so far). Unlike the other two yeast UBL-UBA shuttle proteins (Rad23, Dsk2) where the UBL and UBA domains interact with each other (Díaz-Martínez et al., 2006; Heessen et al., 2005; Kang et al., 2006; Zhang et al., 2009), there is no evidence for any in cis or in trans interactions between the UBL and UBA domains of Ddi1 (this work and (Bertolaet et al., 2001a; Gabriely et al., 2008)). To the best of our knowledge, this is the first protein with a Ub-like fold that does not behave like Ub with respect to its binding preferences.
What makes the UBL of Ddi1 unique? Like Ub and other UBLs (Dsk2, Rad23, Rub1/Nedd8), Ddi1UBL utilizes residues located on the β-sheet side of its surface for binding. The interface between Ub and Ddi1UBL is formed by hydrophobic contacts and stabilized by neighboring polar and charged residues. In Ub (as well as UBLs of other shuttle proteins, Dsk2, Rad23) the hydrophobic patch is surrounded by positively charged side chains (Figs 3 and S3). By contrast, the hydrophobic residues on the β-sheet surface of Ddi1UBL are surrounded by negatively charged side chains, making this surface complementary to that on Ub. The interactions between the opposite charges on Ub and Ddi1UBL are likely an important contributing factor to the observed strong binding between the two proteins. Thus, in addition to Ddi1UBL being a novel Ub-binding domain, the interface formed by the β-sheet surfaces of Ub and Ddi1UBL presents a novel mode of interaction between Ub and its binding partner, in which hydrophobic interface contacts are further strengthened by the complementarity of the surface charges.
A striking feature of Ddi1UBL is its negatively charged β-sheet surface. Interestingly, we found a similar arrangement of the hydrophobic side chains and the negative electrostatic potential on the β-sheet surface of the UBL domain from a mouse protein homologous to Ddi1 (Fig S9). By contrast, and similarly to their yeast variants, the UBLs of the human homologues of Rad23 and Dsk2 (Fig S9) feature a positively charged surface on their β-sheet side; the same holds for Ub’s closest kin Rub1 and Nedd8 (in mammals) (Singh et al., 2012). Note that the β-sheet surface is partially negatively charged in SUMO (Walters et al., 2004), but not to the same extent as in Ddi1UBL. These observations suggest that the unusual and unique binding preferences of yeast Ddi1UBL discovered in this study might be preserved in other organisms.
By finding that Ddi1UBL has binding preferences opposite to other UBLs, we identified it as the source of the functional difference between Ddi1 and other UBL-UBA proteins. The UBL domain of Ddi1 stands out in the UBL family because it contains a novel surface for a Ub-fold protein, which is recognized in a completely different way. While the other UBLs are competing with each other and with Ub for binding to their shared receptors (such as Rpn1, Rpn10 UIM, UBAs) (Rosenzweig et al., 2012; Zhang et al., 2009), Ddi1 UBL is suited for Ub binding and likely has its own receptors. In fact, a recent study (Gomez et al., 2011) identified a Ddi1-docking region on Rpn1 that differs from those for Rad23 and Dsk2. This suggests that Ddi1 might supplement Rad23 and Dsk2 rather than compete with them for targeting ubiquitinated substrates to the proteasome. Thus, our findings reveal the structural basis for the suggested role (Kim et al., 2004) of the UBL domain as the critical element that differentiates the proteolytic functions of Ddi1 and other yeast UBL-UBA proteins, Rad23 and Dsk2.
That Ddi1 can bind Ub through both the UBL and UBA domains, with the latter being a stronger Ub-binder (e.g. Fig S6), suggests that some of the published data on Ddi1 binding to Ub or ubiquitinated proteins might have to be revisited. For example, our results presented here suggest that the strong binding between full-length Ddi1 and Ub observed before (Bertolaet et al., 2001b) could primarily reflect Ub’s interaction with the UBL domain rather than the UBA domain. It was shown in previous studies that turnover of the Ufo1 protein depends upon Ddi1 (Ivantsiv et al., 2006). Surprisingly, we found no evidence for direct interaction between Ufo1UIMs and FL Ddi1 (Fig S8) or Ddi1UBL domain (Figs 8, S8). Instead, we discovered and characterized Ufo1UIMs binding to Ub, suggesting that the previously reported interaction between Ufo1 and Ddi1 might be indirect and mediated through binding to a Ub chain.
The finding that the UIMs of Ufo1 bind Ub but not Ddi1 suggests a possible interplay between Ufo1 and Ddi1 in the degradation of Ho endonuclease. Since Ho endonuclease is recruited by Ufo1 to the SCF complex for ubiquitination, the UIMs of Ufo1 could interact with the Ub chain being built on Ho endonuclease and possibly stabilize the chain’s buildup. That ubiquitination of Ho endonuclease is required for its interaction with Ddi1 (Kaplun et al., 2005; Tzirkin et al., 2003) implies Ddi1 binding (through the UBL or UBA domains, or both) to the Ub chain on Ho endonuclease. Thus, both Ufo1 and Ddi1 could compete for binding to the same Ub chain. Eventually, Ddi1 would bind to this chain and trigger release of ubiquitinated Ho endonuclease from the SCF either by outcompeting Ufo1UIMs or, as the chain becomes long enough, by interacting with the “overhanging” portion of the chain. The latter could provide a mechanism for controlling the appropriate chain length to signal degradation of Ho endonuclease.
As a shuttle protein, Ddi1 is expected to recognize ubiquitinated substrates and deliver them to the proteasome for degradation. “Classical” UBL-UBA shuttles (Rad23, Dsk2) utilize their UBA domain(s) to bind to the (poly)Ub tag on a substrate, and their UBL domain to bind to the 26S proteasome (Chen and Madura, 2002; Saeki et al., 2002), primarily through the Rpn1 subunit in the 19S regulatory particle (Elsasser and Finley, 2005; Gomez et al., 2011; Rosenzweig et al., 2012). The unusual dual functionality of Ddi1 UBL, which is capable of binding both Ub and 19S, suggests that Ddi1 might not act as a classical shuttle protein. One can envision that both UBL and UBA domains of Ddi1 could be bound to a (long) polyUb tag (Fig 9), with subsequent dissociation of the UBL to bind to Rpn1 when in close proximity to 19S. Another scenario could involve Ddi1 heterodimerization with Rad23 through Ddi1 UBA domain (Bertolaet et al., 2001a) and binding to polyUb through its UBL domain. In this case, Ddi1 could act together with Rad23 as a tandem shuttle, and Ddi1UBL could also contribute, together with UBA domains of Rad23, to the protection of polyUb chain (Kang et al., 2006) by “decorating” it with Ub-binding domains. Puzzlingly, unlike other UBL-UBA shuttle proteins having both UBA and UBL domains conserved among eukaryotes, Ddi1 has kept its UBL domain in higher eukaryotes but lost its UBA domain. This evolutionary change might be another indication that Ddi1 performs its shuttle-protein role differently from Rad23 and Dsk2, if at all. We can speculate that Ddi1 dimerization through the RVP domain might make it possible for a Ddi1 homodimer to utilize two UBL domains for both binding to polyUb tag and targeting it (together with the substrate) to the proteasome (see Fig 9). Further structural and functional studies are required in order to fully characterize and understand the role of Ddi1 in cells. Nevertheless, the uncharacteristic properties of the Ddi1 UBL domain revealed by our studies exemplify remodeling of protein function while preserving the 3-D fold, and emphasize the importance of understanding what it means for a domain to be ubiquitin-like and for a shuttle protein to be a shuttle.
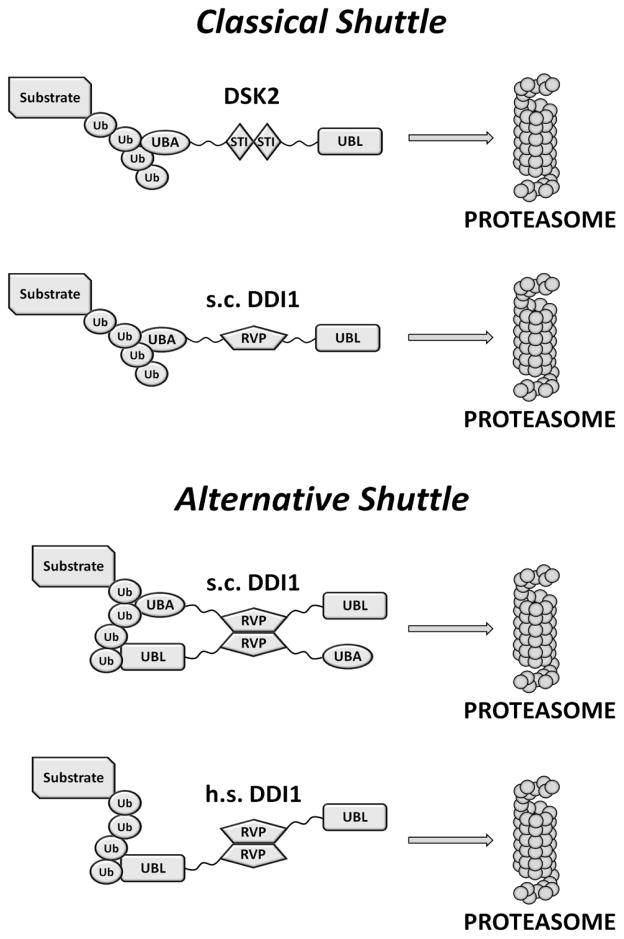
Figure 9.
Schematic representation of a possible function of Ddi1 as a proteasomal shuttle in yeast (s.c.) and humans (h.s.). A “classical” shuttle protein (e.g. Dsk2) employs a UBA domain to recognize and bind (poly)Ub tag on a substrate protein and a UBL domain to target it to the proteasome (e.g.,(Zhang et al., 2009)). In yeast Ddi1, both the UBA and UBL domains can recognize polyubiquitinated substrates. Human Ddi1 lost its UBA domain during the evolution but still contains the UBL domain; the dual functionality of the UBL domain should allow Ddi1 to both bind polyUb tag and deliver polyubiquitinated substrates to the 26S proteasome for degradation.
EXPERIMENTAL PROCEDURES
Protein constructs and purification procedures
The Ddi1UBL and Ddi1UBA constructs used in these studies contained residues 2–80 and 389–428, respectively, of Ddi1 from S. cerevisiae. The Ufo1UIMs fragment included Ufo1 residues 547–668 comprising three UIM motifs. Complete details on all protein constructs and purification procedures are in Supplemental Experimental Procedures.
NMR experiments
All NMR measurements were performed at 23°C on Avance III 600 MHz and 800 MHz Bruker Biospin spectrometers equipped with cryoprobes. Protein samples were prepared in 20 mM sodium phosphate buffer, pH 6.8, containing, when indicated, 150 mM NaCl. Complete details on all the NMR measurements and analyses and the structure calculations are in Supplemental Experimental Procedures.
Supplementary Material
Highlights.
The N- and C-terminal regions of Ddi1 adopt the characteristic UBL and UBA 3D folds
Unexpectedly, Ddi1UBL binds Ub and pulls out Ub conjugates from yeast cell extract
Unique Ddi1UBL:Ub interface is formed by hydrophobic contacts and salt bridges
Dual functionality of Ddi1UBL suggests novel proteasomal shuttle mechanism for Ddi1
Acknowledgments
We thank Emma Dixon for providing some di-Ub chains, Yan Kazansky and Christina Camara for Ub mutants, Hongpeng Wang for help with making Ddi1UBA, Dina Raveh for the original Ufo1 plasmid, Susan Krueger for help with SANS measurements, and Dina Schneidman-Duhovny for advice with SANS data analysis. This research was funded by NIH grant GM095755 to D.F. and M.H.G. and in part by a joint grant from the US-Israel Binational Foundation (BSF) to D.F. and M.H.G., and utilized NMR instrumentation supported in part by NSF grant DBI1040158. We acknowledge the support of NIST, U.S. Department of Commerce, in providing the neutron research facilities used in this work.
Atom coordinates deposited with the Protein Data Bank: Ddi1UBA (PDB ID 2mr9), Ddi1UBL (2mrp), Ddi1UBA:Ub (2mro), Ddi1UBL:Ub (2mws).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baranes-Bachar K, Baranes-Bacher K, Khalaila I, Ivantsiv Y, Lavut A, Voloshin O, Raveh D. New interacting partners of the F-box protein Ufo1 of yeast. Yeast. 2008;25:733– 743. doi: 10.1002/yea.1615. [DOI] [PubMed] [Google Scholar]
- Beal R, Deveraux Q, Xia G, Rechsteiner M, Pickart C. Surface hydrophobic residues of multiubiquitin chains essential for proteolytic targeting. Proc Natl Acad Sci USA. 1996;93:861–866. doi: 10.1073/pnas.93.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolaet BL, Clarke DJ, Wolff M, Watson MH, Henze M, Divita G, Reed SI. UBA domains mediate protein-protein interactions between two DNA damage-inducible proteins. J Mol Biol. 2001a;313:955–963. doi: 10.1006/jmbi.2001.5105. [DOI] [PubMed] [Google Scholar]
- Bertolaet BL, Clarke DJ, Wolff M, Watson MH, Henze M, Divita G, Reed SI. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nature Structural Biology. 2001b;8:417–422. doi: 10.1038/87575. [DOI] [PubMed] [Google Scholar]
- Chen L, Madura K. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol Cell Biol. 2002;22:4902–4913. doi: 10.1128/MCB.22.13.4902-4913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Shinde U, Ortolan TG, Madura K. Ubiquitin-associated (UBA) domains in Rad23 bind ubiquitin and promote inhibition of multi-ubiquitin chain assembly. EMBO Rep. 2001;2:933–938. doi: 10.1093/embo-reports/kve203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A. The ubiquitin-mediated proteolytic pathway: mechanisms of action and cellular physiology. Biol Chem Hoppe Seyler. 1994a;375:565–581. doi: 10.1515/bchm3.1994.375.9.565. [DOI] [PubMed] [Google Scholar]
- Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994b;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- Clarke DJ, Mondesert G, Segal M, Bertolaet BL, Jensen S, Wolff M, Henze M, Reed SI. Dosage suppressors of pds1 implicate ubiquitin-associated domains in checkpoint control. Molecular and Cellular Biology. 2001;21:1997–2007. doi: 10.1128/MCB.21.6.1997-2007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore G, Garrett D. R-factor, free R, and complete cross-validation for dipolar coupling refinement of NMR structures. J Am Chem Soc. 1999;121:9008–9012. [Google Scholar]
- Conaway RC, Brower CS, Conaway JW. Emerging roles of ubiquitin in transcription regulation. Science. 2002;296:1254–1258. doi: 10.1126/science.1067466. [DOI] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- de Vries SJ, van Dijk M, Bonvin AM. The HADDOCK web server for data-driven biomolecular docking. Nat Protoc. 2010;5:883–897. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- Díaz-Martínez LA, Kang Y, Walters KJ, Clarke DJ. Yeast UBL-UBA proteins have partially redundant functions in cell cycle control. Cell Div. 2006;1:28. doi: 10.1186/1747-1028-1-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsasser S, Chandler-Militello D, Müller B, Hanna J, Finley D. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J Biol Chem. 2004;279:26817–26822. doi: 10.1074/jbc.M404020200. [DOI] [PubMed] [Google Scholar]
- Elsasser S, Finley D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat Cell Biol. 2005;7:742–749. doi: 10.1038/ncb0805-742. [DOI] [PubMed] [Google Scholar]
- Funakoshi M, Sasaki T, Nishimoto T, Kobayashi H. Budding yeast Dsk2p is a polyubiquitin-binding protein that can interact with the proteasome. Proc Natl Acad Sci USA. 2002;99:745–750. doi: 10.1073/pnas.012585199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushman D, Wilkinson KD. Structure and recognition of polyubiquitin chains of different lengths and linkage. F1000 Biol Rep. 2011;3:26. doi: 10.3410/B3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriely G, Kama R, Gelin-Licht R, Gerst JE. Different domains of the UBL-UBA ubiquitin receptor, Ddi1/Vsm1, are involved in its multiple cellular roles. Mol Biol Cell. 2008;19:3625– 3637. doi: 10.1091/mbc.E07-05-0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez TA, Kolawa N, Gee M, Sweredoski MJ, Deshaies RJ. Identification of a functional docking site in the Rpn1 LRR domain for the UBA-UBL domain protein Ddi1. BMC Biol. 2011;9:33. doi: 10.1186/1741-7007-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haririnia A, Verma R, Purohit N, Twarog MZ, Deshaies RJ, Bolon D, Fushman D. Mutations in the hydrophobic core of ubiquitin differentially affect its recognition by receptor proteins. J Mol Biol. 2008;375:979–996. doi: 10.1016/j.jmb.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann-Petersen R, Gordon C. Integral UBL domain proteins: a family of proteasome interacting proteins. Semin Cell Dev Biol. 2004;15:247–259. doi: 10.1016/j.semcdb.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Heessen S, Masucci MG, Dantuma NP. The UBA2 domain functions as an intrinsic stabilization signal that protects Rad23 from proteasomal degradation. Mol Cell. 2005;18:225–235. doi: 10.1016/j.molcel.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Hicke L. A new ticket for entry into budding vesicles-ubiquitin. Cell. 2001a;106:527–530. doi: 10.1016/s0092-8674(01)00485-8. [DOI] [PubMed] [Google Scholar]
- Hicke L. Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol. 2001b;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivantsiv Y, Kaplun L, Tzirkin-Goldin R, Shabek N, Raveh D. Unique role for the UbL-UbA protein Ddi1 in turnover of SCFUfo1 complexes. Mol Cell Biol. 2006;26:1579–1588. doi: 10.1128/MCB.26.5.1579-1588.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser SE, Riley BE, Shaler TA, Trevino RS, Becker CH, Schulman H, Kopito RR. Protein standard absolute quantification (PSAQ) method for the measurement of cellular ubiquitin pools. Nat Methods. 2011;8:691–696. doi: 10.1038/nmeth.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Vossler RA, Diaz-Martinez LA, Winter NS, Clarke DJ, Walters KJ. UBL/UBA ubiquitin receptor proteins bind a common tetraubiquitin chain. J Mol Biol. 2006;356:1027– 1035. doi: 10.1016/j.jmb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Kaplun L, Ivantsiv Y, Bakhrat A, Raveh D. DNA damage response-mediated degradation of Ho endonuclease via the ubiquitin system involves its nuclear export. J Biol Chem. 2003;278:48727–48734. doi: 10.1074/jbc.M308671200. [DOI] [PubMed] [Google Scholar]
- Kaplun L, Ivantsiv Y, Kornitzer D, Raveh D. Functions of the DNA damage response pathway target Ho endonuclease of yeast for degradation via the ubiquitin-26S proteasome system. Proc Natl Acad Sci USA. 2000;97:10077–10082. doi: 10.1073/pnas.97.18.10077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplun L, Tzirkin R, Bakhrat A, Shabek N, Ivantsiv Y, Raveh D. The DNA damage-inducible UbL-UbA protein Ddi1 participates in Mec1-mediated degradation of Ho endonuclease. Mol Cell Biol. 2005;25:5355–5362. doi: 10.1128/MCB.25.13.5355-5362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Mi K, Rao H. Multiple interactions of rad23 suggest a mechanism for ubiquitylated substrate delivery important in proteolysis. Mol Biol Cell. 2004;15:3357–3365. doi: 10.1091/mbc.E03-11-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleijnen MF, Shih AH, Zhou P, Kumar S, Soccio RE, Kedersha NL, Gill G, Howley PM. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol Cell. 2000;6:409–419. doi: 10.1016/s1097-2765(00)00040-x. [DOI] [PubMed] [Google Scholar]
- Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- Lambertson D, Chen L, Madura K. Pleiotropic defects caused by loss of the proteasome-interacting factors Rad23 and Rpn10 of Saccharomyces cerevisiae. Genetics. 1999;153:69–79. doi: 10.1093/genetics/153.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Zhang WP, Xing Q, Ren X, Liu M, Tang C. Noncovalent dimerization of ubiquitin. Angew Chem Int Ed Engl. 2012;51:469–472. doi: 10.1002/anie.201106190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratani M, Tansey WP. How the ubiquitin-proteasome system controls transcription. Nat Rev Mol Cell Biol. 2003;4:192–201. doi: 10.1038/nrm1049. [DOI] [PubMed] [Google Scholar]
- Ohno A, Jee J, Fujiwara K, Tenno T, Goda N, Tochio H, Kobayashi H, Hiroaki H, Shirakawa M. Structure of the UBA domain of Dsk2p in complex with ubiquitin molecular determinants for ubiquitin recognition. Structure. 2005;13:521–532. doi: 10.1016/j.str.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Raasi S, Varadan R, Fushman D, Pickart CM. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol. 2005;12:708–714. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- Rao H, Sastry A. Recognition of specific ubiquitin conjugates is important for the proteolytic functions of the ubiquitin-associated domain proteins Dsk2 and Rad23. J Biol Chem. 2002;277:11691–11695. doi: 10.1074/jbc.M200245200. [DOI] [PubMed] [Google Scholar]
- Rock KL, Goldberg AL. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu Rev Immunol. 1999;17:739–779. doi: 10.1146/annurev.immunol.17.1.739. [DOI] [PubMed] [Google Scholar]
- Rosenzweig R, Bronner V, Zhang D, Fushman D, Glickman MH. Rpn1 and Rpn2 coordinate ubiquitin processing factors at proteasome. J Biol Chem. 2012;287:14659–14671. doi: 10.1074/jbc.M111.316323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu KS, Lee KJ, Bae SH, Kim BK, Kim KA, Choi BS. Binding surface mapping of intra- and interdomain interactions among hHR23B, ubiquitin, and polyubiquitin binding site 2 of S5a. J Biol Chem. 2003;278:36621–36627. doi: 10.1074/jbc.M304628200. [DOI] [PubMed] [Google Scholar]
- Saeki Y, Saitoh A, Toh-e A, Yokosawa H. Ubiquitin-like proteins and Rpn10 play cooperative roles in ubiquitin-dependent proteolysis. Biochem Biophys Res Commun. 2002;293:986– 992. doi: 10.1016/S0006-291X(02)00340-6. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Funakoshi M, Endicott JA, Kobayashi H. Budding yeast Dsk2 protein forms a homodimer via its C-terminal UBA domain. Biochem Biophys Res Commun. 2005;336:530– 535. doi: 10.1016/j.bbrc.2005.08.126. [DOI] [PubMed] [Google Scholar]
- Schubert U, Antón LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770– 774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Zerath S, Kleifeld O, Scheffner M, Glickman MH, Fushman D. Recognition and cleavage of related to ubiquitin 1 (Rub1) and Rub1-ubiquitin chains by components of the ubiquitin-proteasome system. Mol Cell Proteomics. 2012;11:1595–1611. doi: 10.1074/mcp.M112.022467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirkis R, Gerst JE, Fass D. Ddi1, a eukaryotic protein with the retroviral protease fold. J Mol Biol. 2006;364:376–387. doi: 10.1016/j.jmb.2006.08.086. [DOI] [PubMed] [Google Scholar]
- Spence J, Sadis S, Haas AL, Finley D. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol Cell Biol. 1995;15:1265–1273. doi: 10.1128/mcb.15.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempe JF, Brown NR, Lowe ED, Gordon C, Campbell ID, Noble ME, Endicott JA. Mechanism of Lys48-linked polyubiquitin chain recognition by the Mud1 UBA domain. EMBO J. 2005;24:3178–3189. doi: 10.1038/sj.emboj.7600797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzirkin R, Ivantsiv Y, Klyman E, Raveh D. The ubiquitin domain protein Ddi1 functions in DNA damage response mediated degradation of Ho endonuclease of yeast. Yeast. 2003;20:S144–S144. [Google Scholar]
- Varadan R, Assfalg M, Raasi S, Pickart C, Fushman D. Structural determinants for selective recognition of a Lys48-linked polyubiquitin chain by a UBA domain. Mol Cell. 2005;18:687–698. doi: 10.1016/j.molcel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Voloshin O, Bakhrat A, Herrmann S, Raveh D. Transfer of Ho endonuclease and Ufo1 to the proteasome by the UbL-UbA shuttle protein, Ddi1, analysed by complex formation in vitro. PLoS One. 2012;7:e39210. doi: 10.1371/journal.pone.0039210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walinda E, Morimoto D, Sugase K, Konuma T, Tochio H, Shirakawa M. Solution structure of the ubiquitin-associated (UBA) domain of human autophagy receptor NBR1 and its interaction with ubiquitin and polyubiquitin. J Biol Chem. 2014;289:13890–13902. doi: 10.1074/jbc.M114.555441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters KJ, Goh AM, Wang Q, Wagner G, Howley PM. Ubiquitin family proteins and their relationship to the proteasome: a structural perspective. Biochim Biophys Acta. 2004;1695:73–87. doi: 10.1016/j.bbamcr.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Wilkinson CR, Seeger M, Hartmann-Petersen R, Stone M, Wallace M, Semple C, Gordon C. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat Cell Biol. 2001;3:939–943. doi: 10.1038/ncb1001-939. [DOI] [PubMed] [Google Scholar]
- Yamaguchi R, Dutta A. Proteasome inhibitors alter the orderly progression of DNA synthesis during S-phase in HeLa cells and lead to rereplication of DNA. Exp Cell Res. 2000;261:271– 283. doi: 10.1006/excr.2000.5053. [DOI] [PubMed] [Google Scholar]
- Zhang D, Chen T, Ziv I, Rosenzweig R, Matiuhin Y, Bronner V, Glickman MH, Fushman D. Together, Rpn10 and Dsk2 can serve as a polyubiquitin chain-length sensor. Mol Cell. 2009;36:1018–1033. doi: 10.1016/j.molcel.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Raasi S, Fushman D. Affinity makes the difference: nonselective interaction of the UBA domain of Ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains. J Mol Biol. 2008;377:162–180. doi: 10.1016/j.jmb.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.