Research during the past 2 decades has established that the diverse biologic actions of 1,25-dihydroyxyvitamin D3 (1,25(OH)2D3) are initiated through precise changes in gene expression that are mediated by an intracellular vitamin D receptor (VDR).1 Activation of the VDR through direct interaction with 1,25(OH)2D3 prompts the receptor’s rapid binding to regulatory regions of target genes, where it acts to nucleate the formation of large protein complexes whose functional activities are essential for directed changes in transcription.2 In most target cells, these actions trigger the expression of networks of target genes whose functional activities combine to orchestrate specific biologic responses. These responses are tissue-specific and range from highly complex actions essential for homeostatic control of mineral metabolism to focal actions that control the growth, differentiation, and functional activity of numerous cell types including those of the immune system, skin, the pancreas and bone, as well as many other targets that are described in this issue devoted to vitamin D.3 In these tissues, gene targets are numerous. New studies combined with new techniques are now revealing a surprising increase in mechanistic complexity wherein multiple regulatory regions, frequently located many kilobases upstream, within, or downstream of a target gene’s transcription unit, seem to participate in transcriptional modulation.4–7
VDR STRUCTURE AND FUNCTION
The VDR is Structurally Organized to Mediate Changes in Transcription in Response to 1,25(OH)2D3
Despite nearly 2 decades of extensive biochemical characterization of the VDR after its discovery in 1974,8,9 it was the cloning of this receptor’s gene and the subsequent analysis of recombinant protein that led to key insights into its structure and its function.10,11 As depicted in Fig. 1A, the VDR protein is comprised of 3 distinct regions, an N-terminal dual zinc finger DNA-binding domain, a C-terminal ligand-binding activity domain, and an extensive and unstructured region that links the 2 functional domains of this protein together. The C-terminal region of the molecule, whose three-dimensional structure has been solved by X-ray crystallography,12,13 is the most complex and is comprised of 12 α-helices as illustrated in Fig. 1B. Amino acid contacts within a subset of these α-helices form a dynamic ligand-binding pocket, as shown in Fig. 1C. Selective occupancy by 1,25(OH)2D3 leads to the formation of 2 independent protein interaction surfaces on the VDR protein: 1 that facilitates interaction with a heterodimer partner required for specific DNA binding and 1 that is essential for the recruitment of large coregulatory complexes required for gene modulation.14 Additional studies suggest that the VDR can also be posttranslationally modified through phosphorylation, an alteration in the protein that may be capable of modulating and fine-tuning its transcriptional activity.15–17 Collectively, these domains within the VDR create a macromolecule receptive to physiologically relevant levels of circulating 1,25(OH)2D3 and capable of directing cellular regulatory machinery to specific subsets of genes whose protein products are key to 1,25(OH)2D3 response.
Fig. 1.
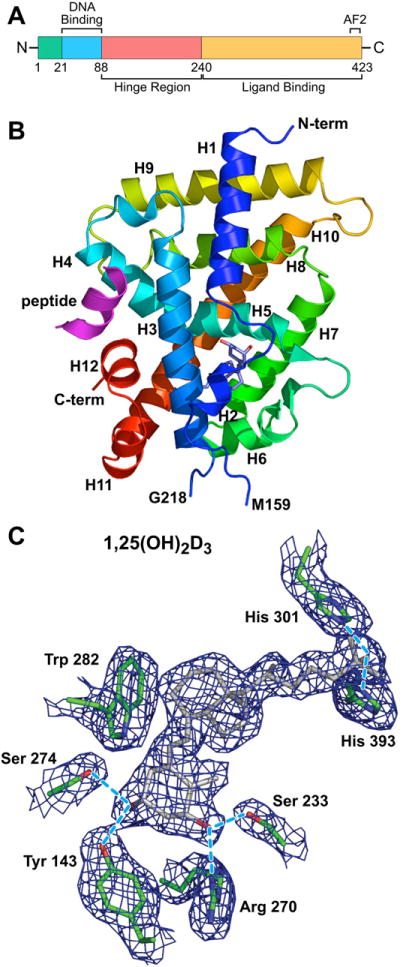
Structure and key features of the VDR. (A) The VDR protein comprised of a DNA-binding domain, a large ligand-binding domain, and a hinge region that links the 2 functional domains of the protein together. N, amino terminal end; C, carboxy terminal end; AF2, activation function 2. Amino acid numbers are shown. (B) Crystal structure of the VDR ligand-binding domain comprised of 12 α-helices (H1–H12). The N-terminal and C-terminal portions of the molecule are shown. A deletion in the molecular from G218 to M159 was required to achieve the formation of crystals. The position of 1,25(OH)2D3 is shown in the ligand-binding pocket as a stick figure. The ligand-binding domain was crystallized in the presence of a short peptide (indicated) representing a key LxxLL motif located in all coregulatory proteins that interact directly with the VDR. The repositioning of H12 as a consequence of 1,25(OH)2D3 binding provides the structural change necessary for interaction of the VDR with the LxxLL motif. (C) An electron density map of 1,25(OH)2D3 and adjacent amino acids within the VDR protein that make direct contact with the ligand. (Data from Vanhooke JL, Benning MM, Bauer CB, et al. Molecular structure of the rat vitamin D receptor ligand-binding domain complexed with 2-carbon-substituted vitamin D3 hormone analogues and a LXXLL-containing coactivator peptide. Biochemistry 2004;43(14):4101–10.)
The VDR Specifies Target Genes Through its DNA-binding Properties
The zinc finger containing the DNA-binding domain of the VDR is typical of that found in all members of the steroid receptor gene family including those for estrogens, androgens, and glucocorticoids, as well as for thyroid hormone, retinoid acid, and other lipophilic regulators.18,19 The VDR is now known to recognize a specific DNA sequence or vitamin D response element (VDRE) comprised of 2 hexameric nucleotide half-sites separated by 3 base pairs (bp).1,20 Other response element structures also occur, although these appear much less frequently.21 The 2 DNA half-sites accommodate the binding of a heterodimer comprised of a VDR molecule and a retinoid X receptor (RXR) molecule.19 The latter forms a heterodimer with other members of the steroid receptor family as well, including receptors for retinoic acid and thyroid hormone, thus linking the activities of several different endocrine systems. Recent studies, described later, suggest that RXR is independently bound to many sites on the genome in the absence of an activating ligand, thereby marking potential regulatory sites for subsequent activation by 1,25(OH)2D3. 1,25(OH)2D3 via its receptor also suppresses the transcriptional expression of numerous genes.1,22 The requirements for direct VDR DNA binding and for heterodimer formation with RXR in the suppression of gene activity are currently unclear.
The VDR Regulates Transcription Through its Ability to Recruit Coregulatory Complexes
Selective VDR DNA binding in a cell serves to highlight that subset of genes within a genome whose transcriptional activities are targeted under a specific set of conditions for modification by 1,25(OH)2D3. Changes in gene expression are not mediated directly via the VDR, however, but rather indirectly through the protein’s ability to facilitate through its transactivation domain the recruitment of large and diverse coregulatory machines that directly mediate such changes.2,23 This recruitment is often gene specific, suggesting a role for additional and as yet unidentified components. Coregulatory complexes generally contain 1 VDR-interacting component as well as many additional subunits, several of which can contain inherent enzymatic activity. These complexes include machines with ATPase-containing nucleosomal remodeling ability, enzymes such as acetyl- and deacetyltransferases and methyl- and demethyltransferases containing selective chromatin histone modifying capabilities, and complexes that play a role in RNA polymerase II (RNA pol II) recruitment and initiation such as Mediator, as documented in Fig. 2. Each of these groups of proteins identifies a key step in the process of transcription regulation and many more are likely to be identified in the future. The details of how these machines operate to enhance or suppress the expression of these gene targets are only now beginning to emerge.
Fig. 2.
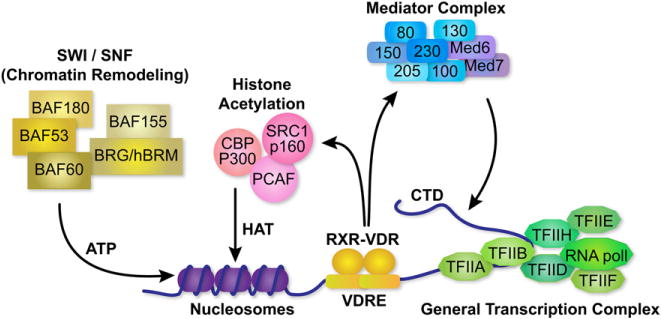
Coregulatory complexes that are involved in mediating the actions of 1,25(OH)2D3 and the VDR. The general transcriptional apparatus is shown at the TSS and the VDR/RXR heterodimer is shown bound to its regulatory vitamin D response element or VDRE. Three regulatory complexes are shown that interact with the VDR: an ATPase-containing, chromatin remodeling complex termed SWI/SNF, a histone acetylation complex containing histone acetyltransferases (HAT) and Mediator complex. The latter facilitates the activation of RNA pol II through its C-terminal domain (CTD). Nucleosomes as well as individual proteins that comprise the individual coregulatory complexes are indicated.
VITAMIN D TARGET GENES
1,25(OH)2D3 Regulates Networks of Genes in a Tissue/Cell-specific Fashion
As described earlier, the role of ligand-activated VDR is to direct cellular transcription machinery to specific sites on the genome where these complexes can influence the production of RNA, which encodes proteins that are integral to specific biologic activities. It is in this manner that 1,25(OH)2D3 plays a central role in regulating mineral metabolism via its actions in intestinal and kidney epithelial cells and in specific bone cells. Although many target genes that play important roles in calcium and phosphorus homeostatic have been identified, additional targets important to these processes continue to be discovered. These include the calcium and phosphate transporters and their associated basolaterally located, energy-driven ion pumps in the intestine and kidney,24–26 and the osteoblast-synthesized osteoclastogenic differentiation factor receptor activator of NF-κB ligand (RANKL),27 which stimulates the activity of existing bone-resorbing osteoclasts, prolongs their lifespan, and induces the formation of new replacements.28 Vitamin D also regulates gene networks involved in bile acid metabolism in the colon,29 the degradation of xenobiotic compounds in several tissues,24 the differentiation of keratinocytes in skin,30 the development and cycling of dermal hair follicles,31 and the functions of key cell types involved in innate and adaptive immunity.32 The genes and gene networks that have been identified as responsible for these biologic actions of 1,25(OH)2D3 are extensive. Indeed, many have emerged as a consequence of contemporary genome-wide analyses that are almost routinely conducted by investigators currently, and which are capable of measuring the effects of the hormone on entire cellular or tissue transcriptomes. Many of these gene networks are regulated by the hormone in a tissue-specific fashion. Perhaps most interesting is the intricate regulatory controls exerted directlyby1,25(OH)2D3 and its receptor at genes involved in the vitamin D ligand’s production and degradation, actions that contribute to the maintenance of biologically active levels of intracellular 1,25(OH)2D3. Thus, as outlined in Fig. 3, 1,25(OH)2D3 suppresses the renal expression of Cyp27b1,33 whose protein product is responsible for its synthesis, and induces Cyp24a1,33,34 whose product is responsible for its degradation to calcitroic acid. In addition to these activities, 1,25(OH)2D3 also autoregulates the expression of its own receptor gene (see Fig. 3), thus modulating not only levels of the ligand but also of the VDR.5,35 Some of the mechanistic details of this regulation are discussed later. Thus, 1,25(OH)2D3 also contributes directly to the maintenance of the key signaling components essential for generating and mediating hormonal response.
Fig. 3.
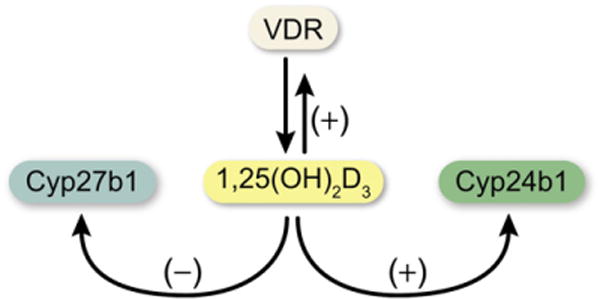
Regulatory control of the synthesis (Cyp27b1), degradation (Cyp24a1), and mediation of activity (Vdr) of 1,25(OH)2D3 The concentration of 1,25(OH)2D3 in cells is determined through its synthesis and its degradation. Its functional activity is determined by the presence and intracellular concentration of the VDR.
Traditional Studies were Initiated by Identifying Target Genes and Defining Regions that Mediate Regulation by the Vitamin D Hormone
Identifying the site(s) of action of 1,25(OH)2D3 at a target gene locus represents the first step in defining the molecular processes that are essential for altering a gene’s transcriptional output. This step is also important because it often leads to the identification of a region that is likely to provide important regulatory control after activation through other signaling pathways as well. Early studies of the osteocalcin gene and its regulation by 1,25(OH)2D3 in bone cells provide an excellent example of this principle. Based on the ability of 1,25(OH)2D3 to induce osteocalcin in bone cells, our early molecular studies, using a traditional human osteocalcin promoter-reporter plasmid approach coupled to classic protein-DNA interaction analyses, revealed the first DNA-binding site for the VDR.20,36 This site was located approximately 485 bp upstream of the human gene’s transcriptional start site (TSS) and was comprised of 2 directly repeated 6-bp sequences separated by 3 bp. Follow-on studies confirmed the general location and highly conserved nature of this vitamin D responsive region in the rat37 and mouse genes.38 The latter was functionally suppressed by 1,25(OH)2D3 as a result of a strategic change in the regulatory element’s base structure thereby highlighting an important species-specific difference in vitamin D response. An extensive series of studies conducted more than a decade after these initial discoveries firmly established that this general region was a direct target for many different transcription factors some of which were activated by either separate or overlapping signal transduction pathways.39 The ability of these proteins to influence response to 1,25(OH)2D3 and for the vitamin D hormone and its receptor to influence their actions was characterized. Perhaps the most important transcription factor to be discovered at the osteocalcin promoter was RUNX2, a regulatory protein now known to be essential to the formation and bone-forming activity of osteoblasts.40,41 During the ensuing years, many genes have been explored for the location of regulatory sites that are capable of mediating 1,25(OH)2D3 action, binding the VDR and its heterodimer partner, and recruiting coregulatory complexes necessary for changes in transcriptional output. These include the genes for osteocalcin, osteopontin, bone sialoprotein, TRPV6, PTH, PTHrp, Cyp24a1, and Cyp27b1 as well as many others. In the case of Cyp24a1, 2 sites located within 300 bp of the TSS were identified as significant mediators of the actions of 1,25(OH)2D3.34
NEW APPROACHES REVEAL NEW INSIGHTS INTO VITAMIN D3–MEDIATED GENE REGULATION
Development of New Approaches to the Study of Transcription Research
The study of gene regulation in the past several decades has relied heavily on the analysis of transcriptional activity generated from gene promoter/reporter plasmids transfected into host cells to identify key components of regulatory processes. These analyses, together with biochemical assays that assess direct protein-protein and protein-DNA interactions have provided considerable insight into how genes are regulated. These approaches are inherently biased, however, because they rely on cellular transfection, involve the analysis of short segments of target genes that are not in context with the gene’s normal chromatin environment, are often dependent on co-expression and/or over-expression of DNA-binding proteins and/or specific coregulators for measureable activity and, in many cases, use concentrations of reactants that are manyfold higher than that normally found in cells. These deficiencies as well as many others prompted the development and application of new techniques to assess the molecular details of transcriptional regulation. Perhaps the most important has been the development of chromatin immunoprecipitation (ChIP) analysis, a technique summarized in Fig. 4 that permits the detection of regulatory proteins and/or the appearance of covalent activity at specific DNA targets in unmodified cells or tissues.42,43 However, it has been the analysis of amplified ChIP products using tiled microarrays (ChIP-chip)44 or massively parallel sequencing techniques (ChIP-seq),45–47 as also shown in the figure, that has provided the most important new insights yet into how genes are regulated. These latter extensions to ChIP analysis impose no restrictions on the regions within the genome that can be evaluated. Thus, they can be used to examine at an equally high level of resolution a single gene locus of several hundreds of kilobases or an entire genome comprised of several billion bases. These techniques are currently being used to re-examine the mechanisms whereby 1,25(OH)2D3 regulates known targets of vitamin D action, to explore regulated genes for which the underlying mechanisms have yet to be discovered, to identify and assess new gene targets, and to establish overarching principles of VDR gene regulation at the genome-wide level. One principle that seems to be emerging from these studies is that the regulation of most genes, including those that are targets of vitamin D action, is mediated by multiple regulatory regions often located many kilobases from their respective gene’s start site. Specific examples of how ChIP-chip and ChIP-seq analyses are being used to illuminate the transcriptional actions of 1,25(OH)2D3 are provided in the following sections. As mentioned earlier, many of our preconceived notions of how this hormone regulates transcription were only partially correct.
Fig. 4.
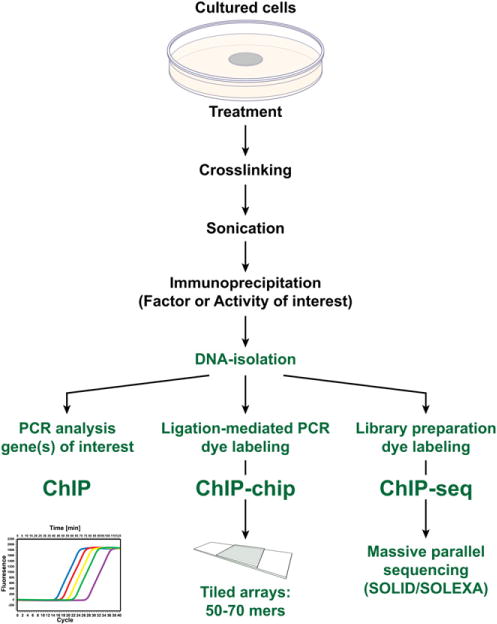
Methodology associated with chromatin immunoprecipitation (ChIP) analysis and subsequent ChIP-DNA microarray (ChIP-chip) or massive parallel sequencing (ChIP-seq) analyses. Biologic samples are cross-linked, sonicated to prepare discrete size chromatin fragments, and then subjected to immunoprecipitation using selected antibodies. The precipitated DNA is then isolated and evaluated by polymerase chain reaction analysis or amplified and then subjected to either ChIP-chip or ChIP-seq analyses.
Regulation of Cyp24a1 Expression by 1,25(OH)2D3 Before and After ChIP-Chip Analysis
As indicated earlier, 1,25(OH)2D3 regulates the expression of Cyp27b1 and Cyp24a1. Because the expression of these genes is central to the maintenance of an effective vitamin D endocrine system, the diverging mechanisms whereby 1,25(OH)2D3 suppresses the expression of Cyp27b1 while inducing the expression of Cyp24a1 have receive considerable attention. Although many details remain to be worked out, it seems that 1,25(OH)2D3 prompts the displacement of a key transcription factor at the Cyp27b1 proximal promoter that is responsible for basal expression.48 This displacement suppresses the expression of Cyp27b1. In the case of Cyp24a1, numerous studies have shown the presence of 2 regulatory elements (VDREs) located approximately 150 and 250 bp upstream of the TSS that mediate the inducing capability of 1,25(OH)2D3 via the VDR and its partner RXR.34,49 Several additional regulatory sites are also present in this proximal region that contribute to the up-regulation of Cyp24a1, including sites for the transcription factor C/EBPβ and for Ets-1.50 Parathyroid hormone (PTH) also regulates Cyp24a1, although this action is indirect and mediated via either a modification of 1,25(OH)2D3 response and/or through posttranslational events.51,52 Recent ChIP studies of 1,25(OH)2D3-induced activation of Cyp24a1 reveal that the hormone induces rapid binding of VDR and RXR to the proximal promoter elements and that this binding leads to the recruitment of coregulators such as the p160 family members, the integrators CBP and p300, the Med1 cofactor TRAP220, and RNA polymerase II (RNA pol II).53 This region also undergoes rapid histone H4 acetylation, likely the result of the appearance of the p160 family members. The appearance of these factors at the Cyp24a1 proximal promoter is cyclic within the first 3 hours, with a periodicity of approximately 45 minutes.53 This periodicity has been observed for other nuclear receptors and its mechanism recently modeled for PPARγ in HEK293 cells.54 These and other studies provide excellent overviews of Cyp27b1 and Cyp24a1 regulation by 1,25(OH)2D3.
In recent studies, the authors used ChIP-chip and ChIP-seq analyses to examine the ability of 1,25(OH)2D3 to induce not only VDR and RXR binding to the human CYP24A1 promoter but also to stimulate the recruitment of RNA pol II to the gene’s TSS and to promote changes in histone H4 acetylation.55 These studies confirmed the earlier findings of a region located immediately proximal to the CYP24A1 promoter to which the VDR/RXR heterodimer binds on induction by 1,25(OH)2D3. The hormone also induced an increase in H4 acetylation and the recruitment of RNA pol II at this region, and at sites within the transcription unit. Surprisingly, ChIP-chip analysis also revealed that 1,25(OH)2D3 induced VDR/RXR heterodimer binding to a robust cluster of intergenic sites located 50 to 70 kb downstream of the human CYP24A1 gene. H4 acetylation and RNA pol II recruitment were increased across these sites in a fashion similar to that identified at the proximal promoter. This cluster of 1,25(OH)2D3-regulated enhancers was also conserved, in position and function, in the mouse Cyp24a1 gene locus. Functional analysis of these regions using large recombineered bacterial artificial chromosome (BAC) clones containing the entire mouse and human CYP24A1 gene loci confirmed the contribution of these downstream clusters of enhancers. Thus, ChIP-chip analysis has revealed unexpectedly that CYP24A1, a quintessential target of 1,25(OH)2D3 action, is regulated by multiple enhancers located not only proximal but also downstream of and distal to the promoter. This characteristic of the CYP24A1 gene is emerging as typical of most highly regulated genes, and highlights an important new feature of gene regulation, as revealed by ChIP-chip analysis.
1,25(OH)2D3 Autoregulates the Expression of the VDR Gene Through Intronic and Upstream Enhancers
The VDR is an absolute determinant of the biologic activity of 1,25(OH)2D3.1 Thus, the receptor’s expression in cells is a requirement for response, and the receptor’s concentration itself a key component of sensitivity to the hormone. Although little is known of the molecular determinants of basal expression of the VDR in cells, the VDR gene is known to be regulated by a variety of hormones including PTH, retinoic acid, and the glucocorticoids.56 Perhaps most interesting is the ability of 1,25(OH)2D3 to increase the level of VDR gene expression itself. Despite the discovery of this autoregulatory feature of the VDR gene several decades ago,10,35,57 a general lack of a regulatory response to 1,25(OH)2D3 at the promoter for the VDR gene left the mechanism unresolved. To elucidate this mechanism, however, the authors turned to ChIP-chip analysis and explored the entire mouse Vdr gene locus for the presence of regions that might mediate the inducing actions of 1,25(OH)2D3. This analysis revealed the presence of several enhancers that bound the VDR and its heterodimer partner RXR that were located in 2 separate introns approximately 20 and 30 kb downstream of the gene’s TSS.5 No activity was observed at the Vdr gene’s proximal promoter thus confirming the lack of activity observed in earlier studies. At least 1 of these regions contained a functional VDRE capable of mediating vitamin D hormone action when analyzed independently in host cells. More recent studies have now identified additional sites of regulation, at least 1 of which is located many kilobases upstream of the Vdr gene’s TSS.58 Subsets of these enhancers also mediate the actions of PTH, retinoic acid, and the glucocorticoids, through the binding of the transcription factors CREB, RAR, and GR, respectively, thus underscoring a previously known characteristic of enhancers, that of modularity. Further examination resulted in the identification of additional transcription factors such a C/EBPβ, which likely participate in the basal expression of the VDR in selected cell types. Subsequent BAC clone analysis, as described earlier, has confirmed the roles of these enhancers in the regulation of Vdr gene expression. Current studies are focused on the use of these large DNA constructs to recapitulate Vdr gene expression in vivo in transgenic mice.
1,25(OH)2D3 and PTH Regulates the Expression of the Mouse Rankl Gene Through Multiple Upstream Distal Enhancers
Rankl is a TNFα-like factor that is produced by stromal cells and osteoblasts and which regulates the differentiation, activation, and survival of osteoclasts, cells responsible for bone resorption.28,59,60 The expression of this factor in osteoblast lineage cells is regulated by the 2 primary calciotropic hormones, 1,25(OH)2D3 and PTH, as well as several of the inflammatory cytokines including IL-1, TNFα, and IL-6. These actions on Rankl expression facilitate the normal bone remodeling function of 1,25(OH)2D3 and PTH in particular but also highlight the bone loss that is associated with increased levels of these hormones. As with the genes discussed earlier, early studies aimed at understanding the regulation of Rankl gene expression focused on the proximal promoter and regions immediately upstream. Although 1,25(OH)2D3 was shown to manifest activity at the proximal promoter, this activity was modest and difficult to interpret.61–63 Activity as a consequence of PTH treatment was not detected. These features of the mouse and human RANKL proximal promoters suggested the possibility that the genes might be regulated through additional unidentified control regions. To explore this possibility, the authors conducted a ChIP-chip analysis and explored the ability of 1,25(OH)2D3 to induce VDR binding across the mouse Rankl gene locus. This analysis revealed the presence of 5 regions capable of mediating the regulatory activity of the vitamin D hormone.4 Surprisingly, these regions were located 16, 22, 60, 69, and 75 kilobases (kb) upstream of the Rankl TSS. The region at 75 kb was shown to contain several VDREs and was particularly active. Studies in parallel by Fu and colleagues64 revealed that a region immediately upstream of the enhancer at −75 kb mediated the actions of PTH through CREB as well. This combined enhancer was thus termed the distal control region or DCR. Subsequent studies suggested that the actions of PTH were not limited to the DCR, but were also observed at several of the more proximal enhancers identified for the VDR.65 Although basal levels of H4 acetylation were noted at many of these enhancers, 1,25(OH)2D3 and PTH induced a striking increase in this epigenetic activity. The vitamin D hormone also induced an increase in RNA pol II at these sites.4,66 These studies suggested that the binding of VDR and CREB to these sites initiated changes in chromatin structure and function, thus supporting the hypothesis that they represent true regulatory enhancers. The central role of the enhancer located at −75/76 prompted Galli and colleagues66 to delete this region in the mouse genome. Surprisingly, this deletion resulted in a significant suppression of the basal expression of Rankl in osteoblasts and limited responsiveness to exogenous 1,25(OH)2D3 and PTH. In addition, these mice displayed a modest increase in bone mineral density in adults that was similar to that observed in PTH-null mice. These studies support a distinct biologic role for a unique Rankl enhancer in basal and inducible Rankl gene expression and highlight the usefulness of ChIP-chip analysis in identifying this and additional regulatory regions. These results reinforce the emerging concept that many if not most genes are regulated through the actions of multiple enhancers that can be located in often remote regions surrounding a gene’s transcription unit. More recent studies have now identified an even more distal region, located 88 kb upstream of the mouse Rankl TSS that mediates the actions of the gp130-activating cytokines such as IL-6 through the STAT3 transcription factor.67
GENOME-WIDE STUDIES REVEAL OVERARCHING PRINCIPLES OF GENE REGULATION BY STEROID HORMONES AND BY 1,25(OH)2D3
ChIP-chip analyses on a genome-wide scale have been conducted recently for several steroid hormones and their respective receptors.42–44,46,47,68 These studies include an examination of binding sites for the estrogen, androgen, and peroxisome proliferator-activated receptors. These studies have revealed new insights into the sites of action of these transcription factors and are currently establishing not only new gene targets but new principles through which hormones activate genomic targets. In several cases, investigators have identified the effect of transcription factor binding on RNA pol II recruitment and changes in epigenetic marks. Genome-wide studies of VDR binding sites in tissues and cells are currently in progress and have yet to be published. However, an extensive analysis of subsets of known 1,25(OH)2D3 target genes has been examined, and these studies together with the earlier observations on CYP24a1, Vdr, Rankl, and Lrp56 indicate several common features. These features confirm those reported through the genomewide studies conducted for other endocrine systems. First, it is now clear that the expression of target genes is commonly regulated by multiple control regions. Although many of these regulatory regions are located proximal to promoters, most are situated many kilobases from their respective promoters upstream and downstream, as well as at intronic and exonic sites within the transcription unit itself. Second, although the binding of the VDR to these regulatory regions is largely, although not exclusively, dependent on activation by 1,25(OH)2D3, RXR, the VDR’s heterodimer partner, can be found frequently at these regulatory sites before activation. Thus, as indicated earlier in this article, RXR may mark certain regulatory sites for subsequent activation by 1,25(OH)2D3. RXR also forms homodimers with itself as well as heterodimers with other members of the steroid receptor family. Accordingly, the presence of RXR at a specific site could alternatively represent the means for gene activation by other endocrine factors. Third, bioinformatic analysis of these regulatory sites of VDR/RXR activity has revealed that they are almost always associated with a recognizable regulatory element (VDREs) to which the heterodimer complex can bind directly. Functional studies of these elements have generally confirmed the validity of these projected binding sites. Fourth, the binding of the VDR/RXR heterodimer to regulatory sites within genes can be demonstrated by ChIP-chip analysis to be associated with subsequent genetic activity and frequently with a change in gene expression. Thus, VDR/RXR binding at enhancers correlates with the recruitment of many of the coregulators described earlier, including acetyltransferases, cointegrators such as CBP, corepressor such as SMRT or NCoR, and members of the Mediator complex. The appearance of regulatory complexes at these sites of VDR action are likely responsible for striking increases in histone H4 acetylation or methylation that are observed at these sites and for the increase in RNA pol II that is recruited to these sites and to transcriptional start sites. Thus, the binding of the VDR facilitates downstream molecular activities that are integral to changes in the transcriptional output of target genes. An investigation of the regulation of these same genes by other hormones and signaling pathways demonstrates that these regulatory regions also bind other transcription factors, thereby supporting the idea that regulatory regions are modular in nature and mediate the activity of multiple signaling inputs at target genes. These and additional features of gene regulation that have emerged as a result of ChIP-chip analyses provide new perspectives on the underlying mechanisms through which the expression of target genes is controlled.
SUMMARY
This article represents a summary of what is known of the VDR protein and its molecular mechanism of action at target genes. New methodologies now used, such as ChIP-chip and ChIP-seq, as well as novel reporter studies using large BAC clones stably transfected into culture cells or introduced as transgenes in mice, are providing new insights into how 1,25(OH)2D3-activated VDR modulates the expression of genes at single gene loci and at the level of gene networks. Many of these insights are unexpected and suggest that gene regulation is even more complex than previously appreciated. These studies also highlight new technologies and their central role in establishing fundamental biologic principles.
Acknowledgments
The author thanks the members of the Pike laboratory for helpful discussions related to the work described and Laura Vanderploeg for preparing the figures.
This work was supported by National Institutes of Health Grants DK-072281, DK-073995, DK-074993 and AR-045173.
References
- 1.Haussler MR, Whitfield GK, Haussler CA, et al. The nuclear vitamin D receptor: biological and molecular regulatory properties revealed. J Bone Miner Res. 1998;13(3):325–49. doi: 10.1359/jbmr.1998.13.3.325. [DOI] [PubMed] [Google Scholar]
- 2.Sutton AL, MacDonald PN. Vitamin D: more than a “bone-a-fide” hormone. Mol Endocrinol. 2003;17(5):777–91. doi: 10.1210/me.2002-0363. [DOI] [PubMed] [Google Scholar]
- 3.Bouillon R, Carmeliet G, Verlinden L, et al. Vitamin D and human health: lessons from vitamin D receptor null mice. Endocr Rev. 2008;29(6):726–76. doi: 10.1210/er.2008-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim S, Yamazaki M, Zella LA, et al. Activation of receptor activator of NF-kappaB ligand gene expression by 1,25-dihydroxyvitamin D3 is mediated through multiple long-range enhancers. Mol Cell Biol. 2006;26(17):6469–86. doi: 10.1128/MCB.00353-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zella LA, Kim S, Shevde NK, et al. Enhancers located within two introns of the vitamin D receptor gene mediate transcriptional autoregulation by 1,25-dihydrox-yvitamin D3. Mol Endocrinol. 2006;20(6):1231–47. doi: 10.1210/me.2006-0015. [DOI] [PubMed] [Google Scholar]
- 6.Fretz J, Zella L, Kim S, et al. 1,25-Dihydroxyvitamin D3 regulates the expression of low-density lipoprotein receptor-related protein 5 via deoxyribonucleic acid sequence elements located downstream of the start site of transcription. Mol Endocrinol. 2006;20(9):2215–30. doi: 10.1210/me.2006-0102. [DOI] [PubMed] [Google Scholar]
- 7.Meyer M, Watanuki M, Kim S, et al. The human transient receptor potential vanilloid type 6 distal promoter contains multiple vitamin D receptor binding sites that mediate activation by 1,25-dihydroxyvitamin D3 in intestinal cells. Mol Endocrinol. 2006;20(6):1447–61. doi: 10.1210/me.2006-0031. [DOI] [PubMed] [Google Scholar]
- 8.Brumbaugh PF, Haussler MR. 1α,25-Dihydroxycholecalciferol receptors in intestine. I. Association of 1α,25-dihydroxycholecalciferol with intestinal mucosa chromatin. J Biol Chem. 1974;249(4):1251–7. [PubMed] [Google Scholar]
- 9.Brumbaugh PF, Haussler MR. 1α,25-Dihydroxycholecalciferol receptors in intestine. II. Temperature-dependent transfer of the hormone to chromatin via a specific cytosol receptor. J Biol Chem. 1974;249(4):1258–62. [PubMed] [Google Scholar]
- 10.McDonnell DP, Mangelsdorf DJ, Pike JW, et al. Molecular cloning of complementary DNA encoding the avian receptor for vitamin D. Science. 1987;235(4793):1214–7. doi: 10.1126/science.3029866. [DOI] [PubMed] [Google Scholar]
- 11.Baker AR, McDonnell DP, Hughes M, et al. Cloning and expression of full-length cDNA encoding human vitamin D receptor. Proc Natl Acad Sci U S A. 1988;85(10):3294–8. doi: 10.1073/pnas.85.10.3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rochel N, Wurtz JM, Mitschler A, et al. The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand. Mol Cell. 2000;5(1):173–9. doi: 10.1016/s1097-2765(00)80413-x. [DOI] [PubMed] [Google Scholar]
- 13.Vanhooke JL, Benning MM, Bauer CB, et al. Molecular structure of the rat vitamin D receptor ligand binding domain complexed with 2-carbon-substituted vitamin D3 hormone analogues and a LXXLL-containing coactivator peptide. Biochemistry. 2004;43(14):4101–10. doi: 10.1021/bi036056y. [DOI] [PubMed] [Google Scholar]
- 14.Smith CL, O’Malley BW. Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr Rev. 2004;25(1):45–71. doi: 10.1210/er.2003-0023. [DOI] [PubMed] [Google Scholar]
- 15.Jurutka P, Hsieh J, Nakajima S, et al. Human vitamin D receptor phosphorylation by casein kinase II at Ser-208 potentiates transcriptional activation. Proc Natl Acad Sci U S A. 1996;93(8):3519–24. doi: 10.1073/pnas.93.8.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hilliard GT, Cook R, Weigel N, et al. 1,25-Dihydroxyvitamin D3 modulates phosphorylation of serine 205 in the human vitamin D receptor: site-directed mutagenesis of this residue promotes alternative phosphorylation. Biochemistry. 1994;33(14):4300–11. doi: 10.1021/bi00180a026. [DOI] [PubMed] [Google Scholar]
- 17.Jurutka P, Hsieh J, MacDonald P, et al. Phosphorylation of serine 208 in the human vitamin D receptor. The predominant amino acid phosphorylated by casein kinase II, in vitro, and identification as a significant phosphorylation site in intact cells. J Biol Chem. 1993;268(9):6791–9. [PubMed] [Google Scholar]
- 18.Mangelsdorf DJ, Thummel C, Beato M, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83(6):835–9. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83(6):841–50. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 20.Ozono K, Liao J, Kerner SA, et al. The vitamin D-responsive element in the human osteocalcin gene. Association with a nuclear proto-oncogene enhancer. J Biol Chem. 1990;265(35):21881–8. [PubMed] [Google Scholar]
- 21.Carlberg C. Molecular basis of the selective activity of vitamin D analogues. J Cell Biochem. 2003;88(2):274–81. doi: 10.1002/jcb.10337. [DOI] [PubMed] [Google Scholar]
- 22.Demay MB, Kiernan MS, DeLuca HF, et al. Sequences in the human parathyroid hormone gene that bind the 1,25-dihydroxyvitamin D3 receptor and mediate transcriptional repression in response to 1,25-dihydroxyvitamin D3. Proc Natl Acad Sci U S A. 1992;89(17):8097–101. doi: 10.1073/pnas.89.17.8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dowd DR, Sutton AL, Zhang C, et al. Comodulators of vitamin D receptor-mediated gene expression. In: Feldman D, Pike JW, Glorieux FH, editors. Vitamin D. 2. New York: Elsevier/Academic Press; 2005. pp. 291–304. [Google Scholar]
- 24.Bouillon R, Bischoff-Ferrari H, Willett W. Vitamin D and health: perspectives from mice and man. J Bone Miner Res. 2008;23(7):974–9. doi: 10.1359/jbmr.080420. [DOI] [PubMed] [Google Scholar]
- 25.Bouillon R, Carmeliet G, Boonen S. Ageing and calcium metabolism. Baillieres Clin Endocrinol Metab. 1997;11(2):341–65. doi: 10.1016/s0950-351x(97)80332-1. [DOI] [PubMed] [Google Scholar]
- 26.Benn BS, Ajibade D, Porta A, et al. Active intestinal calcium transport in the absence of transient receptor potential vanilloid type 6 and calbindin-D9k. Endocrinology. 2008;149(6):3196–205. doi: 10.1210/en.2007-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jimi E, Akiyama S, Tsurukai T, et al. Osteoclast differentiation factor acts as a multifunctional regulator in murine osteoclast differentiation and function. J Immunol. 1999;163(1):434–42. [PubMed] [Google Scholar]
- 28.Leibbrandt A, Penninger J. RANK/RANKL: regulators of immune responses and bone physiology. Ann N Y Acad Sci. 2008;1143:123–50. doi: 10.1196/annals.1443.016. [DOI] [PubMed] [Google Scholar]
- 29.Makishima M, Lu TT, Xie W, et al. Vitamin D receptor as an intestinal bile acid sensor. Science. 2002;296(5571):1313–6. doi: 10.1126/science.1070477. [DOI] [PubMed] [Google Scholar]
- 30.Bikle DD. Vitamin D regulated keratinocyte differentiation. J Cell Biochem. 2004;92(3):436–44. doi: 10.1002/jcb.20095. [DOI] [PubMed] [Google Scholar]
- 31.Demay M, MacDonald P, Skorija K, et al. Role of the vitamin D receptor in hair follicle biology. J Steroid Biochem Mol Biol. 2007;103(3–5):344–6. doi: 10.1016/j.jsbmb.2006.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adorini L. Regulation of immune responses by vitamin D receptor ligands. In: Feldman D, Pike JW, Glorieux FH, editors. Vitamin D. 2. New York: Elsevier/Academic Press; 2005. pp. 631–48. [Google Scholar]
- 33.Prosser DE, Jones G. Enzymes involved in the activation and inactivation of vitamin D. Trends Biochem Sci. 2004;29(12):664–73. doi: 10.1016/j.tibs.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 34.Zierold C, Darwish HM, DeLuca HF. Two vitamin D response elements function in the rat 1,25-dihydroxyvitamin D 24-hydroxylase promoter. J Biol Chem. 1995;270(4):1675–8. doi: 10.1074/jbc.270.4.1675. [DOI] [PubMed] [Google Scholar]
- 35.Costa EM, Hirst MA, Feldman D. Regulation of 1,25-dihydroxyvitamin D3 receptors by vitamin D analogs in cultured mammalian cells. Endocrinology. 1985;117(5):2203–10. doi: 10.1210/endo-117-5-2203. [DOI] [PubMed] [Google Scholar]
- 36.Kerner SA, Scott RA, Pike JW. Sequence elements in the human osteocalcin gene confer basal activation and inducible response to hormonal vitamin D3. Proc Natl Acad Sci U S A. 1989;86(12):4455–9. doi: 10.1073/pnas.86.12.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terpening C, Haussler C, Jurutka P, et al. The vitamin D-responsive element in the rat bone Gla protein gene is an imperfect direct repeat that cooperates with other cis-elements in 1,25-dihydroxyvitamin D3-mediated transcriptional activation. Mol Endocrinol. 1991;5(3):373–85. doi: 10.1210/mend-5-3-373. [DOI] [PubMed] [Google Scholar]
- 38.Clemens T, Tang H, Maeda S, et al. Analysis of osteocalcin expression in transgenic mice reveals a species difference in vitamin D regulation of mouse and human osteocalcin genes. J Bone Miner Res. 1997;12(10):1570–6. doi: 10.1359/jbmr.1997.12.10.1570. [DOI] [PubMed] [Google Scholar]
- 39.Lian JB, Stein GS, Javed A, et al. Networks and hubs for the transcriptional control of osteoblastogenesis. Rev Endocr Metab Disord. 2006;7(1–2):1–16. doi: 10.1007/s11154-006-9001-5. [DOI] [PubMed] [Google Scholar]
- 40.Stein GS, Lian JB, Stein JL, et al. Combinatorial organization of the transcriptional regulatory machinery in biological control and cancer. Adv Enzyme Regul. 2005;45:136–54. doi: 10.1016/j.advenzreg.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 41.Ducy P, Zhang R, Geoffroy V, et al. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89(5):747–54. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 42.Shang Y, Hu X, DiRenzo J, et al. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103(6):843–52. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 43.Shang Y, Myers M, Brown M. Formation of the androgen receptor transcription complex. Mol Cell. 2002;9(3):601–10. doi: 10.1016/s1097-2765(02)00471-9. [DOI] [PubMed] [Google Scholar]
- 44.Carroll J, Meyer C, Song J, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38(11):1289–97. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 45.Lin C, Vega V, Thomsen J, et al. Whole-genome cartography of estrogen receptor alpha binding sites. PLoS Genet. 2007;3(6):e87. doi: 10.1371/journal.pgen.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Welboren W, van Driel M, Janssen-Megens E, et al. ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 2009;28(10):1418–28. doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lefterova M, Zhang Y, Steger D, et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22(21):2941–52. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murayama A, Takeyama K, Kitanaka S, et al. Positive and negative regulations of the renal 25-hydroxyvitamin D3 1α-hydroxylase gene by parathyroid hormone, calcitonin, and 1α,25(OH)2D3 in intact animals. Endocrinology. 1999;140(5):2224–31. doi: 10.1210/endo.140.5.6691. [DOI] [PubMed] [Google Scholar]
- 49.Ohyama Y, Ozono K, Uchida M, et al. Functional assessment of two vitamin D-responsive elements in the rat 25-hydroxyvitamin D3 24-hydroxylase gene. J Biol Chem. 1996;271(48):30381–5. doi: 10.1074/jbc.271.48.30381. [DOI] [PubMed] [Google Scholar]
- 50.Dwivedi P, Omdahl J, Kola I, et al. Regulation of rat cytochrome P450C24 (CYP24) gene expression. Evidence for functional cooperation of Ras-activated Ets transcription factors with the vitamin D receptor in 1,25-dihydroxyvitamin D(3)-mediated induction. J Biol Chem. 2000;275(1):47–55. doi: 10.1074/jbc.275.1.47. [DOI] [PubMed] [Google Scholar]
- 51.Yang W, Friedman P, Kumar R, et al. Expression of 25(OH)D3 24-hydroxylase in distal nephron: coordinate regulation by 1,25(OH)2D3 and cAMP or PTH. Am J Physiol. 1999;276(4 Pt 1):E793–805. doi: 10.1152/ajpendo.1999.276.4.E793. [DOI] [PubMed] [Google Scholar]
- 52.Zierold C, Mings J, DeLuca H. Regulation of 25-hydroxyvitamin D3-24-hydroxylase mRNA by 1,25-dihydroxyvitamin D3 and parathyroid hormone. J Cell Biochem. 2003;88(2):234–7. doi: 10.1002/jcb.10341. [DOI] [PubMed] [Google Scholar]
- 53.Kim S, Shevde NK, Pike JW. 1,25-Dihydroxyvitamin D3 stimulates cyclic vitamin D receptor/retinoid X receptor DNA-binding, co-activator recruitment, and histone acetylation in intact osteoblasts. J Bone Miner Res. 2005;20(2):305–17. doi: 10.1359/JBMR.041112. [DOI] [PubMed] [Google Scholar]
- 54.Degenhardt T, Rybakova K, Tomaszewska A, et al. Population-level transcription cycles derive from stochastic timing of single-cell transcription. Cell. 2009;138(3):489–501. doi: 10.1016/j.cell.2009.05.029. [DOI] [PubMed] [Google Scholar]
- 55.Meyer MB, Goetsch PD, Pike JW. A downstream intergenic cluster of regulatory enhancers contributes to the induction of CYP24A1 expression by 1α,25-dihydroxyvitamin D3. J Biol Chem. 2010;285(20):15599–610. doi: 10.1074/jbc.M110.119958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Esteban L, Eisman J, Gardiner E. Vitamin D receptor promoter and regulation of receptor expression. In: Feldman D, Pike JW, Glorieux FH, editors. Vitamin D. 2. New York: Elsevier/Academic Press; 2005. pp. 193–217. [Google Scholar]
- 57.Santiso-Mere D, Sone T, Hilliard GM, 4th, et al. Positive regulation of the vitamin D receptor by its cognate ligand in heterologous expression systems. Mol Endocrinol. 1993;7(7):833–9. doi: 10.1210/mend.7.7.8413308. [DOI] [PubMed] [Google Scholar]
- 58.Zella LA, Meyer MB, Nerenz RD, et al. Multifunctional enhancers regulate mouse and human vitamin D receptor gene transcription. Mol Endocrinol. 2010;24(1):128–47. doi: 10.1210/me.2009-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kong YY, Feige U, Sarosi I, et al. Activated T cells regulate bone loss and joint destruction in adjuvant arthritis through osteoprotegerin ligand. Nature. 1999;402(6759):304–9. doi: 10.1038/46303. [DOI] [PubMed] [Google Scholar]
- 60.Kong YY, Yoshida H, Sarosi I, et al. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature. 1999;397(6717):315–23. doi: 10.1038/16852. [DOI] [PubMed] [Google Scholar]
- 61.Kitazawa S, Kajimoto K, Kondo T, et al. Vitamin D3 supports osteoclastogenesis via functional vitamin D response element of human RANKL gene promoter. J Cell Biochem. 2003;89(4):771–7. doi: 10.1002/jcb.10567. [DOI] [PubMed] [Google Scholar]
- 62.Kitazawa R, Mori K, Yamaguchi A, et al. Modulation of mouse RANKL gene expression by Runx2 and vitamin D3. J Cell Biochem. 2008;105(5):1289–97. doi: 10.1002/jcb.21929. [DOI] [PubMed] [Google Scholar]
- 63.Mori K, Kitazawa R, Kondo T, et al. Modulation of mouse RANKL gene expression by Runx2 and PKA pathway. J Cell Biochem. 2006;98(6):1629–44. doi: 10.1002/jcb.20891. [DOI] [PubMed] [Google Scholar]
- 64.Fu Q, Manolagas SC, O’Brien CA. Parathyroid hormone controls receptor activator of NF-kappaB ligand gene expression via a distant transcriptional enhancer. Mol Cell Biol. 2006;26(17):6453–68. doi: 10.1128/MCB.00356-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim S, Yamazaki M, Shevde NK, et al. Transcriptional control of receptor activator of nuclear factor-kappaB ligand by the protein kinase A activator forskolin and the transmembrane glycoprotein 130-activating cytokine, oncostatin M, is exerted through multiple distal enhancers. Mol Endocrinol. 2007;21(1):197–214. doi: 10.1210/me.2006-0315. [DOI] [PubMed] [Google Scholar]
- 66.Galli C, Zella LA, Fretz JA, et al. Targeted deletion of a distant transcriptional enhancer of the receptor activator of nuclear factor-kappaB ligand gene reduces bone remodeling and increases bone mass. Endocrinology. 2008;149(1):146–53. doi: 10.1210/en.2007-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bishop KA, Meyer MB, Pike JW. A novel distal enhancer mediates cytokine induction of mouse RANKL gene expression. Mol Endocrinol. 2009;23(12):2095–110. doi: 10.1210/me.2009-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Welboren W, Stunnenberg H, Sweep F, et al. Identifying estrogen receptor target genes. Mol Oncol. 2007;1(2):138–43. doi: 10.1016/j.molonc.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]


