Abstract
Intraspecific chemical communication is mediated by signals called pheromones. C. elegans secretes a mixture of small molecules (collectively termed dauer pheromone) that regulates entry into the alternate dauer larval stage and also modulates adult behavior via as yet unknown receptors. Here, we identify two G protein-coupled receptors (GPCRs) that mediate dauer formation in response to a subset of dauer pheromone components. The SRBC-64 and SRBC-66 GPCRs are members of the large Caenorhabditis-specific SRBC subfamily, and are expressed in the ASK chemosensory neurons, which are required for pheromone-induced dauer formation. Expression of both, but not each receptor alone, confers pheromone-mediated effects on heterologous cells. Identification of dauer pheromone receptors will allow a better understanding of the signaling cascades that transduce the context-dependent effects of ecologically important chemical signals.
Pheromones are molecules secreted by an individual that induce behavioral or developmental changes in other animals of the same species (1). Pheromone signals can trigger long-term changes in development or physiology (primer effects) by modulating endocrine signaling and gene expression, or short-term changes in behaviors (releaser effects) via acute effects on neuronal responses (2). The mechanisms by which the same molecule(s) can elicit these markedly distinct effects are not well understood. The nematode C. elegans secretes a complex mixture of chemicals collectively termed dauer pheromone. Dauer pheromone concentrations are assessed prior to the first larval molt to regulate the decision either to proceed through the reproductive cycle, or to enter into the alternate, developmentally arrested dauer developmental stage (3, 4). Dauer pheromone also modulates adult behaviors (5-7), indicating that it can evoke both primer and releaser effects in C. elegans. The active components of dauer pheromone have recently been shown to be structurally related derivatives of the dideoxysugar ascarylose (7-9), including the ascarosides C6, C9, C7 and C3 (these molecules are referred to based on the numbers of carbons in their side chains; fig. s1). However, pheromone receptors are as yet unidentified.
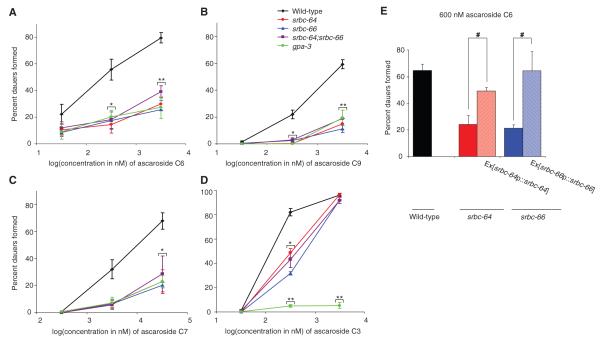
In the course of searching for mutants exhibiting altered responses to dauer pheromone (10), we found that mutations in the predicted GPCR gene srbc-64 resulted in defects in dauer formation. Loss-of-function mutations in both srbc-64, and the closely related srbc-66 gene (fig. s2) (11) resulted in similar strong defects in the ability to form dauers in response to C6, C9, and C7, with weaker defects in response to C3 (Fig. 1A-D). These defects could be rescued upon expression of wild-type srbc-64 and srbc-66 cDNAs driven under their respective upstream regulatory sequences (Fig. 1E), but not by the related srbc-65 gene (fig. s3). srbc-64; srbc-66 double mutants exhibited phenotypes similar to those of either single mutant alone (Fig. 1A-D). Single and double mutants retained the ability to form dauers at high concentrations of C6, C7, and C9, and showed wild-type responses to high concentrations of C3 (Fig. 1A-D), suggesting that additional receptors, perhaps of the SRBC subfamily, are also likely to play a role. These findings are consistent with previous observations which suggested that C6, C9, and C7 may act in the same pathway to promote dauer formation (8), whereas C3 may target a distinct pathway (8).
Fig. 1.
srbc-64, srbc-66 and gpa-3 mutants exhibit defects in dauer formation in response to pheromone components. (A-D) Percent dauers formed by animals of the indicated genotypes when grown in the presence of different concentrations of ascarosides. Error bars are the s.e.m. * and ** indicate values that are different from the corresponding wild-type values at P<0.01 and 0.001, respectively. (E) Percent dauers formed in response to 600 nM C6. Ex refers to animals carrying the indicated expression constructs as extrachromosomal arrays. # indicates values that are different at P<0.01 between the values indicated by brackets. Shown are the averages from at least 4 independent experiments with 50-100 animals each. For each experiment, all strains were assayed in parallel.
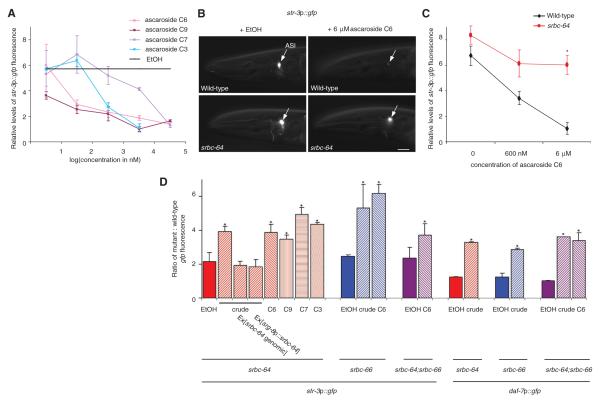
Pheromone regulates dauer formation in part via repression of daf-7 TGF-β expression in the bilateral ASI chemosensory neuron pair (12, 13), and also represses expression of the candidate str-3 chemoreceptor gene in the ASI neurons (14, 15). The four examined ascarosides also downregulated expression of the gfp reporter gene driven under str-3 or daf-7 regulatory sequences (Fig. 2A, B, fig. s4), with C3, C6, and C9 being more potent than C7. Repression of pheromone-mediated gene expression was affected in srbc-64, srbc-66, and srbc-64; srbc-66 mutants (Fig. 2B-D, fig. s4), and could be rescued upon expression of genomic receptor sequences (Fig. 2D). These observations were further validated by examining the effects of pheromone on endogenous str-3 message levels via qRT-PCR (fig. s4). The placement of SRBC-66 upstream of daf-7 TGF-β signaling in dauer formation was also verified by epistasis analysis (table s1). These results indicate that SRBC-64 and SRBC-66 partly mediate the effects of all examined ascarosides on both dauer formation and gene expression.
Fig. 2.
Mutations in srbc-64 and -66 decrease pheromone-mediated downregulation of gene expression. (A) Relative levels of str-3p::gfp expression in adult wild-type animals grown in the presence of the indicated concentrations of each ascaroside. str-3p::gfp expression levels in ethanol were assigned an arbitrary value (black line), and fluorescence levels in experimental conditions were quantified relative to this value. n > 30 for each; 2 independent experiments. (B) Expression of str-3p::gfp in wild-type and srbc-64 mutant adult animals grown in the presence of 6 μM C6. Lateral view; scale – 10 μm. Adult animals were examined using the same exposure time. (C) Relative levels of str-3p::gfp fluorescence in adult wild-type and srbc-64 mutant animals, grown in the presence of C6. n > 50 for each; 2 independent experiments. (D) Shown is the ratio of gfp fluorescence in animals of the indicated genotypes to gfp levels in wild-type animals grown on the same plates in the presence of ethanol, crude pheromone (1 μl/ml), or ascarosides (C3, C6, C9 - 6 μM; C7 - 60 μM). Adult animals were examined in all cases. n > 30 animals each; 2 independent experiments. Error bars are the s.e.m. * indicate values that are different from wild-type at P<0.05 in all panels (with the exception of (D), where values are compared to those of animals grown in ethanol).
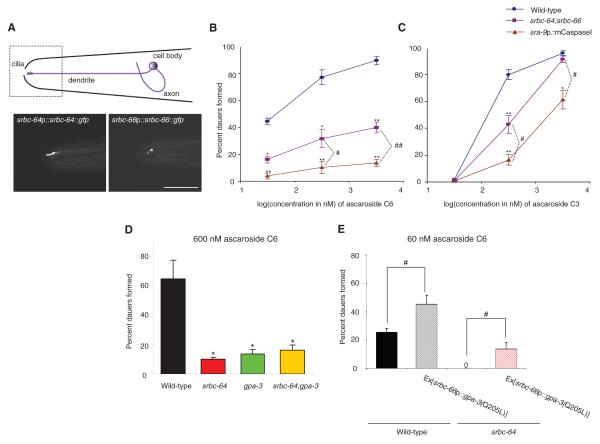
We next determined the site(s) of action of SRBC-64/66 in the regulation of dauer formation and gene expression. We found that full-length GFP-tagged SRBC-64 and SRBC-66 proteins were expressed in, and localized to the sensory cilia of the ASK chemosensory neurons (Fig. 3A; fig. s5). The expression levels and subcellular localization of each chemoreceptor were not altered in the absence of the other chemoreceptor (fig. s5), suggesting that these proteins are not dependent on each other for correct expression or localization. Expression of srbc-64 or srbc-66 specifically in the ASK neurons, but not in the ASI neurons, was sufficient to rescue the dauer formation defects (Fig. 1E; fig.s6). Expression of srbc-64 under the ASK-specific srg-8 promoter (16) also rescued the gene expression defects of srbc-64 mutants (Fig. 2D), further indicating that SRBC-64 and -66 act in the ASK neurons.
Fig. 3.
srbc-64 and srbc-66 are expressed in, and act via the GPA-3 Gα protein in the ASK chemosensory neurons. (A) Full-length GFP-tagged SRBC-64 and -66 proteins are localized to the cilia of the ASK neurons. Lateral view; scale - 10 μm. Images were acquired using confocal microscopy. (B-C) Percent dauers formed by animals of the indicated genotypes in response to C6 (C) and C3 (D). sra-9 regulatory sequences drive expression specifically in the ASK neurons (16). Shown are the averages from at least 4 independent experiments with 50-100 animals each. For each experiment, all strains were assayed in parallel. Error bars are the s.e.m. (D-E) Percent dauers formed when animals of the indicated genotypes were grown in the presence of C6. Shown are the averages from at least 4 independent experiments with 50-100 animals each. # and ## indicate values that are different at P<0.01 and <0.001 respectively, between the values indicated by brackets. * and ** indicate values that are different at P<0.01 and 0.001 from corresponding values of wild-type animals.
Previous work showed that ablation of the ASK neurons, particularly in combination with ablation of other chemosensory neuron types, decreases the efficacy of dauer formation by crude pheromone extracts (12). To further examine the contribution of the ASK neurons to dauer formation, we examined dauer formation in animals in which the ASK neurons have been genetically ablated via expression of the mouse caspase-1 gene expressed under an ASK-specific promoter (gift of Dr. R. Shingai; fig. s7). Animals lacking the ASK neurons exhibited strong defects in dauer formation in response to C6, and weaker defects in response to C3 (Fig. 3B-C). The dauer formation defect of ASK-ablated animals was significantly stronger than that of srbc-64; srbc-66 double mutants (Fig. 3B-C), suggesting that additional ASK-expressed chemoreceptors likely contribute to the response.
We next investigated the intracellular mechanisms mediating the dauer pheromone response in the ASK neurons. Downregulation of intracellular cGMP levels has previously been implicated in pheromone-regulated dauer formation. Addition of 8-Br-cGMP suppresses pheromone-induced dauer formation (17), and loss-of-function mutations in the daf-11 receptor guanylyl cyclase result in constitutive dauer formation, as well as downregulation of daf-7p::gfp expression (17, 18). DAF-11 appears to act in multiple cell types, including the ASK neurons to regulate dauer formation (12, 18). We confirmed that mutations in daf-11 also strongly downregulate str-3p::gfp expression (fig. s8), suggesting that addition of pheromone decreases intracellular cyclic nucleotide levels to both promote dauer formation and downregulate gene expression.
The GPA-2 and GPA-3 Gα proteins have been proposed to mediate pheromone signal transduction, although their sites of action are unknown (19). Loss-of-function mutations in gpa-3 resulted in defects in dauer formation and str-3p::gfp repression in response to ascarosides (Fig. 1A-D, Fig. 3D, fig. s9). srbc-64; gpa-3 and srbc-66 gpa-3 double mutants exhibited dauer formation defects similar to those of either single mutant alone (Fig. 3D, fig. s9), and expression of constitutively activated GPA-3[Q205L] specifically in the ASK neurons resulted in increased dauer formation, and partly bypassed the dauer formation defects of srbc-64 and srbc-66 mutants (Fig. 3E, fig. s9). Taken together, these observations are consistent with a model in which pheromone signals via SRBC-64 and -66 chemoreceptors and the GPA-3 Gα protein in larval ASK neurons to inhibit DAF-11 guanylyl cyclase activity, and promote dauer formation (fig. s10).
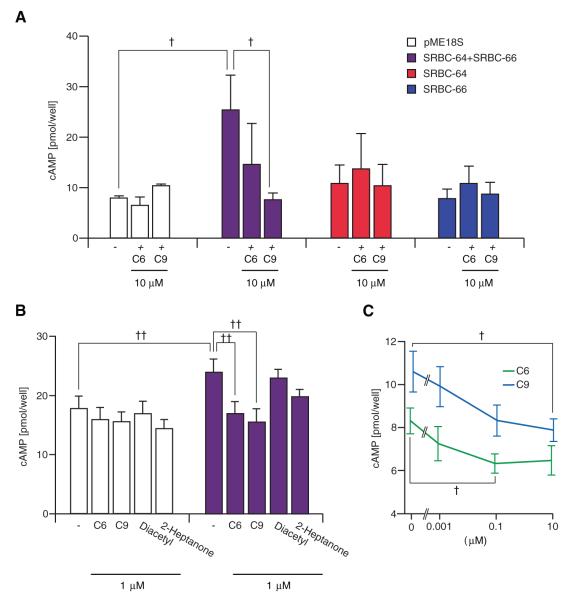
To investigate whether expression of SRBC-64/66 was sufficient to confer pheromone responses in a heterologous context, we expressed one or both receptors in HEK293 cells and monitored intracellular second messenger levels upon the addition of ascarosides. Co-expression of SRBC-64 and -66, but not each receptor alone, enhanced forskolin-mediated increases in cAMP levels (Fig. 4A, B), and this increase was significantly inhibited upon addition of C6 and C9 at different concentrations (Fig. 4A-C). Similar inhibition was not observed in cells transfected with the control construct, SRBC-64 or SRBC-66 alone, or upon addition of the structurally unrelated odorants diacetyl that is attractive to C. elegans, or a mouse pheromone 2-heptanone (Fig. 4A, B). Moreover, ascarosides did not affect the increase in cAMP levels resulting from activation of adenylyl cyclase by the mouse eugenol olfactory receptor in the presence of eugenol (fig. s11). Receptors that exhibit constitutive basal activity such as histamine H2 receptors similarly enhance forskolin-mediated cAMP production in transfected cells, and this increased response is inhibited by inverse agonists (20). Thus, it is conceivable that ascarosides act as inverse agonists of SRBC-64 and -66 receptors. These results indicate that co-expression of SRBC-64 and -66 is sufficient to confer ascaroside-dependent responses in HEK293 cells. Moreover, pheromone acts via SRBC-64 and -66 to decrease intracellular cyclic nucleotide levels both in vivo and in vitro.
Fig. 4.
Ascarosides inhibit forskolin-induced cAMP production in HEK293 cells co-expressing SRBC-64 and SRBC-66. (A-C) HEK293 cells were transiently transfected with the indicated expression constructs, and stimulated with 5 μM forskolin in the absence or presence of the indicated compounds. pME18S is a control vector. (C) Cells were co-transfected with SRBC-64 and SRBC-66. Representative data from one of four independent experiments are shown. Each data point is the average of three to eleven trials. † and †† indicate values that are different at P<0.05 and P<0.01, respectively, between the values indicated by brackets.
We next asked whether in addition to mediating the developmental effects, SRBC-64 and -66 also mediate the adult acute effects of dauer pheromone (5-7). Recently, ASK neurons in adult hermaphrodites have been shown to respond to a mixture of C3, C6, and C9 with a decrease in intracellular calcium levels, and this calcium response has been associated with a behavioral response of aggregation and related behaviors (5). We found that calcium dynamics in adult ASK neurons in response to C3 and C6 were unaffected in srbc-64; srbc-66 or gpa-3 mutant animals (fig. s12), despite the fact that mutations in each strongly affected pheromone-induced dauer formation (Fig. 1A-D). We were unable to monitor intracellular calcium levels in larval ASK neurons due to low calcium sensor expression levels. These results indicate that the long-term and short-term effects of dauer pheromone may be mediated via distinct receptors and signaling cascades at different developmental stages.
Taken together, our findings are consistent with the hypothesis that SRBC-64 and -66 are receptors for subsets of dauer pheromone components in the dauer formation pathway, and that alternate receptors and signaling cascades mediate adult pheromone responses. Whereas short-term releaser effects of pheromones appear to be mediated by fast neuronal and synaptic responses (5, 21), long-term primer effects of pheromones occur on a slower developmental timescale involving long-term changes in gene expression and modulation of neurohormonal signaling (22, 23). The C. elegans genome encodes 86 predicted members of the srbc subfamily (11), which has undergone extensive expansion in C. elegans. Large lineage-specific gains and losses in gene numbers have been noted in other chemoreceptor families that are proposed to mediate responses to niche- and species-specific signals such as pheromones (24). A unique characteristic of the SRBC subfamily is the presence of four invariant Cys residues in the third extracellular loop (11), which may bind structurally related ligands such as ascarosides. Expression of additional SRBC chemoreceptors, coupled to distinct downstream signaling proteins may account for the distinct effects of pheromone at different developmental stages or in different sexes (6, 7). Expansion of the SRBC subfamily may, therefore, contribute to the fidelity and diversity of the pheromone response in C. elegans.
Our results imply that following pheromone binding, the ASK neurons communicate with the ASI neurons via as yet unknown signaling pathways to regulate critical developmental decisions. Integration of multiple environmental cues such as pheromone, food and temperature signals, as well as information regarding internal state may allow for the precise regulation of neurohormone signaling and dauer entry by the ASI neurons (12, 13, 25, 26). This integration resembles the monitoring of external and internal signals such as nutritional status and temperature cues by the hypothalamus to regulate metabolism and behavior via neuroendocrine and autonomic mechanisms (27). Identification of members of the SRBC subfamily as receptors for a subset of pheromone components will now allow us to begin to explore the mechanisms by which complex species-specific chemical cues elicit the appropriate stage- and sex-specific developmental and behavioral responses, and may provide insights into related processes in other animals.
Supplementary Material
Acknowledgments
We are grateful to E. Macosko, C. Bargmann, J. Thomas, and R. Shingai for advice, assistance and reagents. We thank the Caenorhabditis Genetics Center and S. Mitani for strains, members of the Sengupta lab and P. Garrity for discussion, and C. Li for critical comments. This work was supported by the NSF (IOS 0542372 – P.S.) and the NIH (RO1 GM56223 – P.S.; P30 NS45713; RO1 CA24487 – J.C., F32 GM077943 – R.A.B.), and the Ministry of Education, Science, Sports, and Culture, Japan (K.T.)
References and Notes
- 1.Karlson P, Lüscher M. Nature. 1959;183:155. doi: 10.1038/183055a0. [DOI] [PubMed] [Google Scholar]
- 2.Wyatt TS. Pheromones and Animal Behavior:Communication by Smell and Taste. Cambridge University Press; Cambridge: 2003. [Google Scholar]
- 3.Golden JW, Riddle DL. Dev. Biol. 1984;102:368. doi: 10.1016/0012-1606(84)90201-x. [DOI] [PubMed] [Google Scholar]
- 4.Golden JW, Riddle DL. Science. 1982;218:578. doi: 10.1126/science.6896933. [DOI] [PubMed] [Google Scholar]
- 5.Macosko EZ, et al. Nature. 2009;458:1171. doi: 10.1038/nature07886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Srinivasan J, et al. Nature. 2008;454:1115. doi: 10.1038/nature07168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pungaliya C, et al. Proc. Natl. Acad. Sci. USA. 2009 [Google Scholar]
- 8.Butcher RA, Ragains JR, Kim E, Clardy J. Proc. Natl. Acad. Sci. USA. 2008;105:14288. doi: 10.1073/pnas.0806676105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Butcher RA, Fujita M, Schroeder FC, Clardy J. Nat. Chem. Biol. 2007;3:420. doi: 10.1038/nchembio.2007.3. [DOI] [PubMed] [Google Scholar]
- 10.Materials and Methods are provided in Supporting Material Online.
- 11.Thomas JH, Robertson HR. BMC Biol. 2008;6:42. doi: 10.1186/1741-7007-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schackwitz WS, Inoue T, Thomas JH. Neuron. 1996;17:719. doi: 10.1016/s0896-6273(00)80203-2. [DOI] [PubMed] [Google Scholar]
- 13.Ren P, et al. Science. 1996;274:1389. doi: 10.1126/science.274.5291.1389. [DOI] [PubMed] [Google Scholar]
- 14.Peckol EL, Troemel ER, Bargmann CI. Proc. Natl. Acad. Sci. USA. 2001;98:11032. doi: 10.1073/pnas.191352498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nolan KM, Sarafi-Reinach TR, Horne JG, Saffer AM, Sengupta P. Genes Dev. 2002;16:3061. doi: 10.1101/gad.1027702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Troemel ER, Chou JH, Dwyer ND, Colbert HA, Bargmann CI. Cell. 1995;83:207. doi: 10.1016/0092-8674(95)90162-0. [DOI] [PubMed] [Google Scholar]
- 17.Birnby DA, et al. Genetics. 2000;155:85. doi: 10.1093/genetics/155.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murakami M, Koga M, Ohshima Y. Mech. Dev. 2001;109:27. doi: 10.1016/s0925-4773(01)00507-x. [DOI] [PubMed] [Google Scholar]
- 19.Zwaal RR, Mendel JE, Sternberg PW, Plasterk RH. Genetics. 1997;145:715. doi: 10.1093/genetics/145.3.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alewijnse AE, et al. FEBS Lett. 1997;419:171. doi: 10.1016/s0014-5793(97)01440-3. [DOI] [PubMed] [Google Scholar]
- 21.Kurtovic A, Widmer A, Dickson BJ. Nature. 2007;446:542. doi: 10.1038/nature05672. [DOI] [PubMed] [Google Scholar]
- 22.Grozinger CM, Sharabash NM, Whitfield CW, Robinson GE. Proc. Natl. Acad. Sci. USA. 2003;100(Suppl 2):14519. doi: 10.1073/pnas.2335884100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alaux C, et al. Genes Brain Behav. 2009;8:309. doi: 10.1111/j.1601-183X.2009.00480.x. [DOI] [PubMed] [Google Scholar]
- 24.Grus WE, Zhang J. Mol. Biol. Evol. 2008;25:1593. doi: 10.1093/molbev/msn107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li W, Kennedy SG, Ruvkun G. Genes Dev. 2003;17:844. doi: 10.1101/gad.1066503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bishop NA, Guarente L. Nature. 2007;447:545. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- 27.Dowell P, Hu Z, Lane MD. Annu. Rev. Biochem. 2005;74:515. doi: 10.1146/annurev.biochem.73.011303.074027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.