Abstract
Until recently, Toxoplasma gondii was considered clonal with very little genetic variability. Recent studies indicate that T. gondii isolates from Brazil are genetically and biologically different from T. gondii isolates from USA and Europe. In the present study, we retyped 151 free range chicken isolates from Brazil including 117 newly isolated samples from 11 geographically areas (Alagoas, Bahia, Ceará, Maranhão, Paraná, Pernambuco, Rio de Janeiro, Rio Grande do Norte, São Paulo, Sergipe, and Rondonia) and 34 previously reported isolates from the very north (Pará) and the very south (Rio Grande do Sul). Ten PCR-RFLP markers including SAG1, SAG2, SAG3, BTUB, GRA6, c22-8, c29-2, L358, PK1, and Apico were used to genotype all isolates. Overall analysis of 151 T. gondii isolates revealed 58 genotypes. Half (29/58) of these genotypes had single isolate and the other half of the genotypes were characterized with two or more isolates. Only 1 of 151 isolates was clonal Type I strain and 5 were clonal Type III strains. Two isolates had mixed infections. Clonal Type II strain was absent. One strain was Type II at all loci, except BTUB. The results confirm high genetic diversity of T. gondii isolates from Brazil.
Keywords: Toxoplasma gondii, Chickens, Genotype, PCR-RFLP, Brazil
1. Introduction
Toxoplasma gondii infections are widely prevalent in human beings and other animals worldwide (Dubey and Beattie, 1988). Humans become infected post-natally by ingesting tissue cysts from undercooked meat, consuming food or drink contaminated with oocysts, or by accidentally ingesting oocysts from the environment. However, only a small percentage of exposed adult humans or other animals develop clinical signs of disease. Whether the severity of toxoplasmosis in immunocompetent hosts is due to the parasite strain, host variability, or to other factors is largely unknown. Recently, attention has been focused on the genetic variability among T. gondii isolates from apparently healthy and sick hosts. In humans in French Guiana and Suriname, severe cases of toxoplasmosis in immuno-competent patients have been related to T. gondii strains with unusual genetic characteristics (Ajzenberg et al., 2004; Demar et al., 2007). An unusually high prevalence of clinical ocular toxoplasmosis in Erechim, Brazil is thought to be related to characteristics of prevailing T. gondii isolates (Glasner et al., 1992; Khan et al., 2006, 2007).
Most T. gondii isolates from human and animal sources have been grouped into one of three clonal lineages (Type I, II and III) by multilocus enzyme electrophoresis, polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP), and microsatellite typing (Dardé et al., 1992; Howe and Sibley, 1995; Ajzenberg et al., 2002a,b). We have started a worldwide evaluation of genetic diversity of T. gondii isolates based on DNA derived from live parasites. Our attention has been focused on South America, especially Brazil, because of the collaborative success in obtaining tissues from animals for the isolation of T. gondii. Initial results indicate that the isolates of T. gondii from Brazil are biologically and genetically different from those in North America and Europe (Dubey et al., 2002; Lehmann et al., 2006; Dubey et al., 2007a,b,c). T. gondii isolates from asymptomatic chickens from Brazil were more pathogenic to mice than isolates from Europe or North America, irrespective of the genotype (Dubey et al., 2006). Additionally, most isolates from chickens from Brazil were not clonal, and Type II lineage was absent (Dubey et al., 2007a,c).
Our initial studies were based on one marker (SAG2) (Dubey et al., 2002) and six microsatellites (Lehmann et al., 2006). In the present paper we genotyped 94 T. gondii isolates (previously analyzed with only SAG2) using 10 PCR-RFLP markers to achieve a high resolution in identification and evaluated the entire PCR-RFLP data set on chickens from Brazil (Table 1). Results indicate a very high genetic diversity among isolates of T. gondii, irrespective of the geographic location.
Table 1.
Genetic diversity of T. gondii isolates from different regions of Brazil
| T. gondii isolate | No. of isolates | States in Brazil | Reference for isolates | Genotype |
|---|---|---|---|---|
| TgCkBr 117, 119, 120, 122, 123, 124, 126, 127, 129, 130, 131, 132, 133, 134, 135, 136, 137, 138, 139, 140 | 20 | Rond?nia | Dubey et al. (2006) | 7 genotypes – this study |
| TgCkBr 7, 8, 10, 11, 13, 16, 17, 19, 23, 24 | 10 | São Paulo | Dubey et al. (2002, 2006) | 6 genotypes – this study |
| TgCkBr 26, 27, 28, 30, 31, 32, 33, 34, 36, 37, 38, 40, 41, 42, 44, 45, 46, 47, 48, 49, 50, 51, 52, 54, 55, 56, 57, 58, 59, 60, 61, 62, 64, 65, 66, 67, 69, 74, 75, 76, 78, 79, 80, 81, 82, 84, 85, 86, 87, 88, 89, 90, 92. | 53 | Rio de Janeiro | Dubey et al. (2003a,b, 2006) | 29 genotypes – this study |
| TgCkBr 93, 94, 95, 96, 97, 98, 99, 100, 101, 102, 104 | 11 | Paraná | Dubey et al. (2003a,b, 2006) | 7 genotypes – this study |
| TgCkBr 107–116, 141–145 | 15 | Pará | Dubey et al. (2007a) | 11 genotypes – Dubey et al. (2007a) |
| TgCkBr 146–164 | 19 | Rio Grande do Sul | Dubey et al. (2007a) | 9 genotypes – Dubey et al. (2007a) |
| TgCkBr 165, 166 | 2 | Pernambuco | de Oliveira et al. (in press) | 2 genotypes – this study |
| TgCkBr 167–170 | 4 | Rio Grande do Norte | de Oliveira et al. (in press) | 3 genotypes – this study |
| TgCkBr 171, 172 | 2 | Maranhão | de Oliveira et al. (in press) | 1 genotype – this study |
| TgCkBr 173–176 | 4 | Bahia | de Oliveira et al. (in press) | 3 genotypes – this study |
| TgCkBr 177–182 | 6 | Ceará | de Oliveira et al. (in press) | 5 genotypes – this study |
| TgCkBr 183 | 1 | Sergipe | de Oliveira et al. (in press) | 1 genotype – this study |
| TgCkBr 184–187 | 4 | Alagoas | de Oliveira et al. (in press) | 3 genotypes – this study |
2. Materials and methods
For the present study, in total 151 T. gondii isolates from 11 different regions of Brazil (Fig. 1) were genotyped (Table 1). Of these SAG2 data on 94 isolates were previously reported (Dubey et al., 2002, 2003a,b, 2006). For the present study, these cryopreserved 94 T. gondii isolates were revived in mice at the Animal Parasitic Diseases Laboratory, Beltsville, MD (Dubey et al., 2002). Viable T. gondii parasites were collected from lung tissue of mice that died, and from the brains of mice that survived for more than 30 days after inoculation with cryopreserved material were processed for the present study, and were cryopreserved in liquid nitrogen with DMSO (Dubey and Beattie, 1988) for future studies. Additionally, 23 isolates were recently obtained at the Universidade de São Paulo, São Paulo, SP, Brazil (de Oliveira et al., in press); these isolates were genetically characterized for the first time. Genetic data on the remaining 34 isolates had been published previously (Dubey et al., 2007a) and incorporated in the present study. Details of T. gondii isolation in mice have been published in detail in the papers listed in Table 1 and not repeated here.
Fig. 1.
Map of Brazil with sources of chickens sampled.
T. gondii DNA was extracted from tissues of positive mice using DNeasy kit (Qiagen) and genotyped using the genetic markers SAG1, SAG2, SAG3, BTUB, GRA6, c22-8, c29-2, L358, PK1, and Apico (Dubey et al., 2006; Su et al., 2006). Data on 34 T. gondii isolates recently published (Dubey et al., 2007a) from chickens from Pará and Rio Grande do Sul (Table 1) were combined with data on newly genotyped 117 isolates and analyzed by SplitsTree4 (Huson, 1998; Huson and Bryant, 2006) in the present study.
3. Results
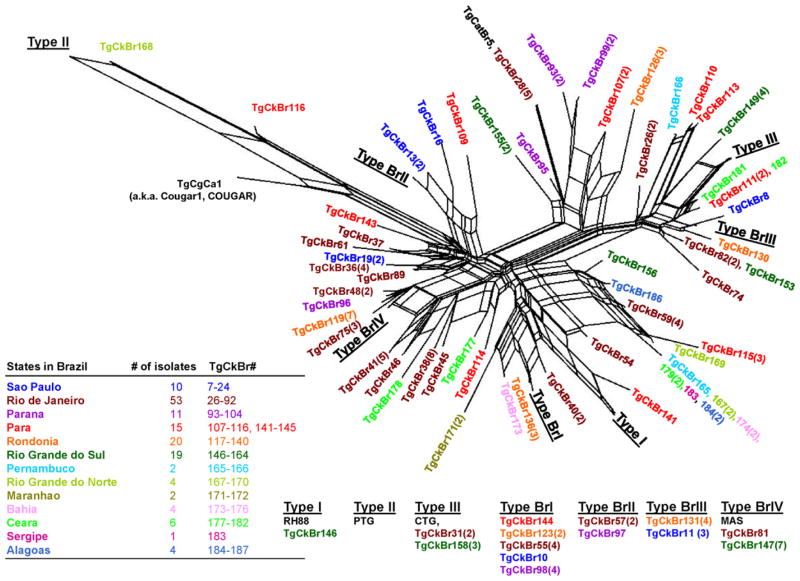
Multilocus PCR-RFLP genotyping of a total of 151 T. gondii isolates from free range chickens in 13 geographically areas including the states of Alagoas, Bahia, Ceará, Maranhão, Paraná, Pernambuco, Rio de Janeiro, Rio Grande do Norte, São Paulo, Sergipe, Rondönia, Pará and Rio Grande do Sul identified 58 genotype groups (Genotype #1–58) and two isolates with mixed infection (Table 2). Twenty-nine of the 58 multilocus genotypes identified are characterized with two or more isolates, while the other 29 genotypes have single isolate each. Only 1 (TgCkBr 146, now lost) of 151 isolates was clonal Type I strain, and 5 were clonal Type III strains. No clonal Type II lineage was found. One strain (TgCkBr 168) was Type II at all loci, except BTUB. The four major T. gondii lineages (Type BrI, BrII, BrIII and BrIV) previously identified from different animal hosts in Brazil (Pena et al., 2008) were also identified from chickens from much wider geographical regions in this study. In addition, the genotype group #21 was also identified in several states in Brazil (Table 2). Phylogenetic relationships of these 149 chicken isolates (exclude the two with mixed infection) were analyzed by SplitsTree4 (Huson, 1998; Huson and Bryant, 2006). The result is presented as reticulated network in Fig. 2. This results show that there is no clear clustering of genotypes with different geographical regions, and there is lack of type II alleles in the parasite population in Brazil. In addition, most genotypes are found only in one of the 13 states analyzed in this study, with exception of the lineages Type III, BrI, BrII, BrIII, BrIVand Genotype groups #21 (with 10 isolates from six different states in Brazil (Table 1, Table 2 and Fig. 2).
Table 2.
Summary of T. gondii PCR-RFLP genotypes from chickens from Brazil
| Genotypes (clonal types) | T. gondii isolate | Genetic markers
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SAG1* | (5′ + 3′) SAG2a | SAG2b | SAG3 | BTUB | GRA6 | c22-8 | c29-2 | L358 | PK1 | Apico | ||
| Reference (I) | RH88 | I | I | I | I | I | I | I | I | I | I | I |
| Reference (II) | PTG | II or III | II | II | II | II | II | II | II | II | II | II |
| Reference (III) | CTG | II or III | III | III | III | III | III | III | III | III | III | III |
| Reference | COUGAR | I | II | II | III | II | II | II | u-1 | I | u-2 | I |
| Reference | MAS | u-1 | I | II | III | III | III | u-1 | I | I | III | I |
| Reference | TgCatBr5 | I | III | III | III | III | III | I | I | I | u-1 | I |
| #1 | 7 (TgCkBr 119, 120, 122, 129, 135, 137, 140) | u-1 | I | II | III | III | III | III | I | I | III | I |
| #2 | 2 (TgCkBr 48, 88) | u-1 | I | II | III | III | III | II | I | I | III | III |
| #3 | 1 (TgCkBr 96) | u-1 | I | II | III | III | III | II | I | I | III | I |
| #4 | 3 (TgCkBr 75, 76, 92) | u-1 | I | II | III | III | III | III | III | I | III | I |
| #5 (BrIV) | 8 (TgCkBr 81, 147, 148, 151, 154, 160, 162, 163) | u-1 | I | II | III | III | III | u-1 | I | I | III | I |
| #6 | 5 (TgCkBr 41, 42, 49, 60, 62) | u-1 | I | II | III | I | III | u-1 | I | I | I | I |
| #7 | 1 (TgCkBr 46) | u-1 | I | II | III | I | III | II | I | I | I | I |
| #8 | 1 (TgCkBr 178) | u-1 | I | II | III | I | III | II | III | III | I | III |
| #9 | 8 (TgCkBr 38, 27, 44, 51, 65, 66, 78, 80) | u-1 | I | II | III | III | III | u-1 | I | III | III | III |
| #10 | 1 (TgCkBr 45) | u-1 | I | II | III | III | III | II | I | III | III | I |
| #11 | 1 (TgCkBr 177) | I | I | II | III | III | III | III | I | III | III | III |
| #12 | 1 (TgCkBr 114) | I | I | II | III | I | III | II | I | III | III | I |
| #13 | 2 (TgCkBr 171, 172) | I | I | I | III | I | II | u-1 | I | III | II | III |
| #14 | 1 (TgCkBr 173) | I | I | I | III | I | II | u-1 | I | I | III | III |
| #15 | 3 (TgCkBr 136, 138, 139) | I | I | I | III | I | II | I | I | I | I | I |
| #16 (BrI) | 12 (TgCkBr 123, 124, 55, 79, 86, 87, 10, 98, 101, 102, 104, 144) | I | I | I | III | I | II | u-1 | I | I | I | I |
| #17 | 2 (TgCkBr 40, 47) | I | I | I | III | III | II | u-1 | I | I | I | I |
| #18 | 1 (TgCkBr 146) | I | I | I | I | I | I | I | I | I | I | I |
| #19 | 1 (TgCkBr 141) | I | I | I | I | I | I | u-1 | I | I | III | III |
| #20 | 1 (TgCkBr 54) | I | I | I | III | I | III | II | I | I | I | III |
| #21 | 10 (TgCkBr 165, 167, 170, 174, 176, 179, 180, 183, 184, 185 | I | I | I | I | I | III | II | III | III | I | III |
| #22 | 1 (TgCkBr 169) | I | I | I | I | I | III | II | I | III | I | III |
| #23 | 3 (TgCkBr 115, 142, 145) | I | I | I | I | I | I | II | I | III | I | III |
| #24 | 4 (TgCkBr 59, 30, 34, 67) | I | I | I | III | I | III | II | I | III | I | III |
| #25 | 1 (TgCkBr 186) | I | I | I | III | III | III | II | I | III | I | III |
| #26 | 1 (TgCkBr 156) | I | I | I | III | III | III | I | I | III | I | III |
| #27 | 1 (TgCkBr 74) | u-1 | III | III | III | III | III | III | I | III | III | III |
| #28 | 3 (TgCkBr 82, 90, 153) | I | III | III | III | III | III | III | I | III | III | III |
| #29 | 1 (TgCkBr 130) | I | III | III | III | I | III | II | III | III | III | III |
| #30 (BrIII) | 7 (TgCkBr 11, 7, 17, 131, 132, 133, 134) | I | III | III | III | III | III | II | III | III | III | III |
| #31 | 1 (TgCkBr 8) | I | III | III | III | III | III | II | III | III | u-2 | III |
| #32 | 3 (TgCkBr 111, 112, 182) | I | III | III | III | III | III | III | III | III | III | I |
| #33 | 1 (TgCkBr 181) | I | III | III | III | III | III | III | III | III | III | III |
| #34 | 5 (TgCkBr 31, 56, 158, 161, 164) | II or III | III | III | III | III | III | III | III | III | III | III |
| #35 | 4 (TgCkBr 149, 150, 152, 157) | II or III | III | III | III | III | III | I | III | III | I | III |
| #36 | 1 (TgCkBr 113) | I | III | III | I | III | III | III | III | III | I | III |
| #37 | 1 (TgCkBr 110) | I | III | III | I | III | III | III | III | III | I | I |
| #38 | 1 (TgCkBr 166) | I | III | III | I | III | III | III | I | III | I | I |
| #39 | 2 (TgCkBr 26, 69) | I | III | III | III | III | III | II | I | III | III | I |
| #40 | 3 (TgCkBr 126, 127, 117) | I | III | III | III | I | II | II | III | I | I | III |
| #41 | 2 (TgCkBr 107, 108) | I | III | III | III | III | II | u-1 | I | I | I | III |
| #42 | 2 (TgCkBr 99, 100) | I | III | III | III | III | II | u-1 | I | I | II | I |
| #43 | 2 (TgCkBr 93, 94) | I | III | III | III | III | II | I | III | I | II | I |
| #44 | 5 (TgCkBr 28, 33, 50, 52, 58) | I | III | III | III | III | III | I | I | I | u-1 | I |
| #45 | 1 (TgCkBr 95) | I | III | III | III | III | III | I | I | I | III | III |
| #46 | 2 (TgCkBr 155, 159) | u-1 | III | III | III | III | III | u-1 | I | I | III | I |
| #47 | 1 (TgCkBr 109) | I | I | II | I | III | III | II | III | I | III | III |
| #48 | 1 (TgCkBr 16) | I | I | II | I | III | III | I | I | I | II | I |
| #49 | 2 (TgCkBr 13, 23) | I | I | II | III | III | III | I | III | I | II | I |
| #50 (BrII) | 3 (TgCkBr 57, 64, 97) | I | I | II | III | III | III | I | III | I | II | III |
| #51 | 1 (TgCkBr 116) | u-1 | II | II | III | III | II | II | nd | II | II | I |
| #52 | 1 (TgCkBr 168) | II or III | II | II | II | III | II | II | II | II | II | II |
| #53 | 1 (TgCkBr 143) | I | I | II | III | III | II | u-1 | III | III | III | I |
| #54 | 1 (TgCkBr 37) | I | I | II | III | III | II | u-1 | I | I | III | I |
| #55 | 1 (TgCkBr 61) | I | I | II | I | III | II | u-1 | I | I | III | I |
| #56 | 2 (TgCkBr 19, 24) | I | I | II | III | III | III | u-1 | I | I | u-2 | I |
| #57 | 4 (TgCkBr 36, 32, 84, 85) | I | II | II | III | III | III | u-1 | I | I | III | I |
| #58 | 1 (TgCkBr 89) | I | I | II | III | III | III | u-1 | I | I | III | I |
| Mixed infections | 1 (TgCkBr 175) | I | I + III | I + III | I + III | I | II + III | II | I + III | I + III | I | I + III |
| 1 (TgCkBr 187) | I | I | I | I + III | I + III | III | II | I + III | III | I | III | |
u-1 and u-2 are new alleles that are different from the clonal Type I, II and III alleles.
SAG2 marker based on 5′- and 3′-ends of the gene sequence (Howe et al., 1997).
A new SAG2 marker based on the 5′-end of the gene sequence (Su et al., 2006).
Fig. 2.
NeighborNet phylogenetic network of T. gondii isolates in chickens from Brazil. Isolates from different states of Brazil are color-coded for clarity. RH88, PTG, CTG, TgCgCa1 (a.k.a. Cougar1, COUGAR), MAS and TgCatBr5 are used as reference strains. Numeric number in parenthesis following each isolate’s identification number is the number of strains with the identical genotype from the same states.
4. Discussion
Multilocus genotyping data on these 151 chicken isolates (including two mixed infections) showed that half (29/58) of the identified genotype groups have a single isolate, indicating high diversity of T. gondii isolates in Brazil. This is in supporting of recent findings from cat and dog isolates in Brazil (Pena et al., 2008). The current study of chicken isolates confirms the previous finding in that the Type I strain is rare and there is lack of the clonal Type II strain in Brazil, which is in sharp contrast in that these two lineages are highly prevalent in North America and Europe (Dardé et al., 1992; Howe and Sibley, 1995; Ajzenberg et al., 2002a,b). Identification of the Type III, BrI, BrII, BrIII, BrIV and the genotype group #21 strains from different states of Brazil do suggest that these lineages are wide spread in different regions. The high proportion of genotypes with single isolate indicates high diversity of parasite strains in Brazil. This would suggest there could be many more unique genotypes circulating in the environment. To better understand molecular epidemiology and population structure of T. gondii in Brazil, a much deeper sampling of a variety of animal hosts is necessary.
References
- Ajzenberg D, Bañuls AL, Tibayrenc M, Dardé ML. Microsatellite analysis of Toxoplasma gondii shows considerable polymorphism structured into two main clonal groups. Int J Parasitol. 2002a;32:27–38. doi: 10.1016/s0020-7519(01)00301-0. [DOI] [PubMed] [Google Scholar]
- Ajzenberg D, Cogné N, Paris L, Bessières MH, Thulliez P, Filisetti D, Pelloux H, Marty P, Dardé ML. Genotype of 86 Toxoplasma gondii isolates associated with human congenital toxoplasmosis, and correlation with clinical findings. J Infect Dis. 2002b;186:684–689. doi: 10.1086/342663. [DOI] [PubMed] [Google Scholar]
- Ajzenberg D, Bañuls AL, Su C, Dumètre A, Demar M, Carme B, Dardé ML. Genetic diversity, clonality and sexuality in Toxoplasma gondii. Int J Parasitol. 2004;34:1185–1196. doi: 10.1016/j.ijpara.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Dardé ML, Bouteille B, Perstreal M. Isoenzyme analysis of 35 Toxoplasma gondii isolates and the biological and epidemiologic implications. J Parasitol. 1992;78:909–912. [PubMed] [Google Scholar]
- Demar M, Ajzenberg D, Maubon D, Djossou F, Panchoe D, Punwasi W, Valery N, Peneau C, Daigre JL, Aznar C, Cottrelle B, Terzan L, Dardé ML, Carme B. Fatal outbreak of human toxoplasmosis along the Maroni River: epidemiological, clinical, and parasitological aspects. Clin Infect Dis. 2007;45:e88–e95. doi: 10.1086/521246. [DOI] [PubMed] [Google Scholar]
- de Oliveira LN, Costa LM, de Melo CB, Silva JCR, Bevilaqua CML, Azevedo SS, Muradian V, Araújo DAFV, Dubey JP, Gennari SM. Toxoplasma gondii isolates from free-range chickens from the northeast region of Brazil. J Parasitol. doi: 10.1645/GE-1730.1. in press. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Beattie CP. Toxoplasmosis of animals and man. CRC Press; Boca Raton, FL: 1988. pp. 1–220. [Google Scholar]
- Dubey JP, Graham DH, Blackston CR, Lehmann T, Gennari SM, Ragozo AMA, Nishi SM, Shen SK, Kwok OCH, Hill DE, Thulliez P. Biological and genetic characterisation of Toxoplasma gondii isolates from chickens (Gallus domesticus) from São Paulo Brazil: unexpected findings. Int J Parasitol. 2002;32:99–105. doi: 10.1016/s0020-7519(01)00364-2. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Graham DH, Silva DS, Lehmann T, Bahia-Oliveira LMG. Toxoplasma gondii isolates of free-ranging chickens from Rio de Janeiro, Brazil: mouse mortality, genotype, and oocyst shedding by cats. J Parasitol. 2003a;89:851–853. doi: 10.1645/GE-60R. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Navarro IT, Graham DH, Dahl E, Freire RL, Prudencio LB, Sreekumar C, Vianna MC, Lehmann T. Characterization of Toxoplasma gondii isolates from free range chickens from Paraná. Brazil Vet Parasitol. 2003b;117:229–234. doi: 10.1016/j.vetpar.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Gennari SM, Labruna MB, Camargo LMA, Vianna MCB, Marcet PL, Lehmann T. Characterization of Toxoplasma gondii isolates in free-range chickens from Amazon. Brazil J Parasitol. 2006;92:36–40. doi: 10.1645/GE-655R.1. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Sundar N, Gennari SM, Minervino AHH, Farias NAR, Ruas JL, dos Santos TRB, Cavalcante GT, Kwok OCH, Su C. Biologic and genetic comparison of Toxoplasma gondii isolates in free-range chickens from the northern Pará state and the southern state Rio Grande do Sul. Brazil revealed highly diverse and distinct parasite populations. Vet Parasitol. 2007a;143:182–188. doi: 10.1016/j.vetpar.2006.08.024. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Applewhaite L, Sundar N, Velmurugan GV, Bandini LA, Kwok OCH, Hill R, Su C. Molecular and biological characterization of Toxoplasma gondii isolates from free-range chickens from Guyana. South America identified several unique and common parasite genotypes. Parasitology. 2007b;134:1559–1566. doi: 10.1017/S0031182007003083. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Gennari SM, Sundar N, Vianna MCB, Bandini LM, Yai LEO, Kwok OCH, Su C. Diverse and atypical genotypes identified in Toxoplasma gondii from dogs in São Paulo. Brazil J Parasitol. 2007c;93:60–64. doi: 10.1645/GE-972R.1. [DOI] [PubMed] [Google Scholar]
- Glasner PD, Silveira C, Kruszon-Moran D, Martins MC, Burnier M, Silveira S, Camargo ME, Nussenblatt RB, Kaslow RA, Belfort R. An unusually high prevalence of ocular toxoplasmosis in southern Brazil. Am J Ophthalmol. 1992;114:136–144. doi: 10.1016/s0002-9394(14)73976-5. [DOI] [PubMed] [Google Scholar]
- Howe DK, Sibley LD. Toxoplasma gondii comprises three clonal lineages: correlation of parasite genotype with human disease. J Infect Dis. 1995;172:1561–1566. doi: 10.1093/infdis/172.6.1561. [DOI] [PubMed] [Google Scholar]
- Howe DK, Honoré S, Derouin F, Sibley LD. Determination of genotypes of Toxoplasma gondii strains isolated from patients with toxoplasmosis. J Clin Microbiol. 1997;35:1411–1414. doi: 10.1128/jcm.35.6.1411-1414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH. SplitsTree: a program for analyzing and visualizing evolutionary data. Bioinformatics. 1998;14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Khan A, Jordan C, Muccioli C, Vallochi AL, Rizzo LV, Belfort R, Jr, Vitor RWA, Silveira C, Sibley LD. Genetic divergence of Toxoplasma gondii strains associated with ocular toxoplasmosis. Brazil Emerg Infect Dis. 2006;12:942–949. doi: 10.3201/eid1206.060025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, Fux B, Su C, Dubey JP, Darde ML, Ajioka JW, Rosenthal BM, Sibley LD. Recent transcontinental sweep of Toxoplasma gondii driven by a single monomorphic chromosome. Proc Natl Acad Sci (Washington, DC) 2007;104:14872–14877. doi: 10.1073/pnas.0702356104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann T, Marcet PL, Graham DH, Dahl ER, Dubey JP. Globalization and the population structure of Toxoplasma gondii. Proc Natl Acad Sci. 2006;103:11423–11428. doi: 10.1073/pnas.0601438103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena HFG, Gennari SM, Dubey JP, Su C. Population structure and mouse-virulence of Toxoplasma gondii in Brazil. Int J Parasitol. 2008;38:561–569. doi: 10.1016/j.ijpara.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Su C, Zhang X, Dubey JP. Genotyping of Toxoplasma gondii by multilocus PCR-RFLP markers: a high resolution and simple method for identification of parasites. Int J Parasitol. 2006;36:841–848. doi: 10.1016/j.ijpara.2006.03.003. [DOI] [PubMed] [Google Scholar]