Abstract
Cordycepin, which is an analogue of a nucleoside adenosine, exhibits a wide variety of pharmacological activities including anticancer effects. In this study, ADA1- and ADA2-expressing HEK293 cells were established to determine the major ADA isoform responsible for the deamination of cordycepin. While the metabolic rate of cordycepin deamination was similar between ADA2-expressing and Mock cells, extensive metabolism of cordycepin was observed in the ADA1-expressing cells with Km and Vmax values of 54.9 μmol/L and 45.8 nmole/min/mg protein. Among five natural substances tested in this study (kaempferol, quercetin, myricetin, naringenin, and naringin), naringin strongly inhibited the deamination of cordycepin with Ki values of 58.8 μmol/L in mouse erythrocytes and 168.3 μmol/L in human erythrocytes. A treatment of Jurkat cells with a combination of cordycepin and naringin showed significant cytotoxicity. Our in silico study suggests that not only small molecules such as adenosine derivatives but also bulky molecules like naringin can be a potent ADA1 inhibitor for the clinical usage.
Keywords: ADA, adenosine deaminase, docking study, enzyme inhibition, naringin
Introduction
Cordycepin (3′-deoxyadenosine), which is an analogue of the nucleoside adenosine, was originally extracted from the fungi of the genus Cordyceps (Cunningham et al. 1950). Cordyceps has been used as a Chinese medicine as it exhibits a wide range of biological functions such as aphrodisiac (Bhattarai 1993), analgesic (Koyama et al. 1997), immune modulation (Gong et al. 2000), and free radical scavenging activity (Yamaguchi et al. 2000). Recent in vivo and in vitro studies have further demonstrated that cordycepin exhibits anticancer effects (Yoshikawa et al. 2004; Lee et al. 2012). Treatment of various leukemia and cancer cells, such as L1210, P388, CEM, and OEC-M1, with cordycepin resulted in decreased cell proliferation (Johns and Adamson 1976; Glazer et al. 1978; Wu et al. 2007). In vivo studies have also supported the effectiveness of cordycepin, as a treatment by cordycepin had a curative effect on mice infected with Trypanosoma evansi, Candida albicans, and Candida krusei (Sugar and McCaffrey 1998; Dalla et al. 2013). It was proposed that those pharmacological effects of cordycepin were largely attributed to its inhibitory effect of poly (A) synthesis and thus it interferes with the processing and maturation of both cellular and viral mRNA (Johns and Adamson 1976). However, the half-life of cordycepin elimination is significantly short in vivo in rats (t1/2 = 1.6 min) (Tsai et al. 2010).
Adenosine deaminase (ADA, EC 3.5.4.4), which has been found in plants, bacteria, and mammals including humans and others, catalyzes the hydrolysis of adenosine to yield inosine (Cristalli et al. 2001). It was demonstrated that ADA also catalyzes the hydrolysis of the derivatives of nucleoside adenosine such as 2′-deoxyadenosine (Cristalli et al. 2001). ADA is expressed on the surface of T and B cells and plays an important role in the development of immune system. Therefore, deficiency of ADA activity is associated with the defect in immunological development, which can result in the development of hematologic malignancies (Dong et al. 1997). Because cordycepin has an extremely high structural similarity to adenosine (Fig.1), it was suggested that cordycepin, too, could be a substrate of ADA. Indeed, Frederiksen et al. (1965) identified a metabolite of cordycepin, 3′-deoxyinosine. Further study revealed that the conversion from cordycepin to 3′-deoxyinosine was catalyzed by ADA (Cristalli et al. 2001). Human ADA has two distinct isozymes, ADA1 and ADA2. ADA1 is ubiquitous in all cells including lymphocytes and monocytes (Gakis 1996). While ADA2 is not ubiquitous and is only found in monocytes (Gakis 1996), its expression can be increased in certain diseases such as psoriasis (Bukulmez et al. 2000). As several inhibitors isoform-specifically inhibit ADA, there is a ligand specificity in ADA-catalyzed deamination of substrates.
Figure 1.
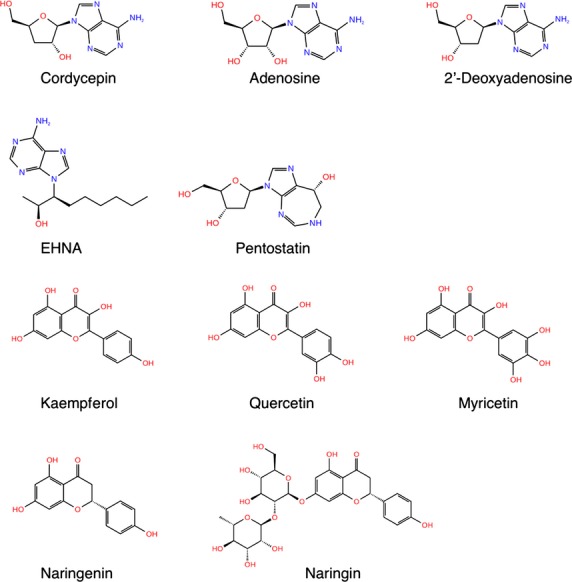
Secondary structures of ADA substrates and inhibitors. Secondary structure of ADA substrates (cordycepin, adenosine, 2′-deoxyadenosine) and inhibitors (EHNA, pentostatin, and flavonoids) are shown.
Extended half-life of cordycepin leads to successful treatment by cordycepin in vivo. Since cordycepin is metabolized by ADA, a potent inhibitor of ADA, such as pentostatin and erythro-9-(2-hydroxy-3-nonyl) adenine (EHNA), can be coadministered to extend the half-life of cordycepin (Adamson et al. 1977; North and Cohen 1978). However, due to the potency of the inhibitory effect of pentostatin and EHNA on ADA, administration of the potent ADA inhibitors can cause hemolytic uremic syndrome (HUS), leukopenia, thrombocytopenia, and anemia in clinical applications (Spiers et al. 1984; Leach et al. 1999).
Cordycepin is a valuable phytochemical that can be used for the treatment of various diseases including cancer. To extend the half-life of cordycepin without developing coadministered substance-induced adverse reactions, effective but less toxic inhibitors need to be identified. In this study, first, the ADA isozyme responsible for the deamination of cordycepin was determined. Second, inhibitory effects of natural substances, such as kaempferol, quercetin, myricetin, naringenin, and naringin, on the ADA-catalyzed deamination of cordycepin were examined. Finally, human T lymphocytes (Jurkat cells) were cotreated with cordycepin and natural ADA inhibitors to investigate the effects of the natural substance on cordycepin-induced cell death.
Materials and Methods
Chemicals and reagents
Cordycepin, adenosine, and 2′-deoxyadenosine were purchased from Wako Pure Chemical (Osaka, Japan). Pentostatin, 1-deazaadenosine, and EHNA were obtained from Tocris Bioscience (Bristol, UK). Kaempferol, myricetin, naringin, and naringenin were purchased from TCI Chemical Industries (Tokyo, Japan). Quercetin was purchased from Sigma–Aldrich (St Louis, MO). Anti-ADA1 antibody (H-300) and anti-ADA2 (anti-CECR1) antibody were purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and Abcam (Cambridge, MA), respectively. The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay kit was purchased from Nacalai Tesque (Kyoto, Japan). Human erythrocytes were purchased from Lee Biosolutions (St Louis, MO). All other chemicals and solvents were of analytical grade or the highest grade commercially available.
Establishment of stable expression systems of human ADA1 and ADA2 in HEK293 cells
Complementary DNA (cDNA) of human ADA1 and ADA2 was prepared by a reverse transcription-PCR method from human liver total RNA with sense and antisense oligonucleotide primers (ADA1 sense, 5′-GGC CCG TTA AGA AGA GCG T-3′; ADA1 antisense, 5′-GTG ACT CCA CAG GGT GAA GG-3′; ADA2 sense, 5′-ATC CCG ATG TTG GTG GAT GG-3′; and ADA2 antisense, 5′-AGA GGA AGT GAC AGC GTG TG-3′). The PCR products were subcloned into pTARGET Mammalian Expression Vector (Promega, Madison, WI) and the DNA sequences of the inserts were determined using a BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, CA) with a sequence analyzer (3130 Genetic Analyzer, Applied Biosystems). HEK293 cells were obtained from the Cell Resource Center for Biomedical Research, Tohoku University (Sendai, Japan) and were maintained at 37°C in a humidified atmosphere containing 5% of CO2. Culture medium consisted of Dulbecco's modified Eagle's medium, 100 U/mL penicillin, 100 mg/L streptomycin, and 10% fetal bovine serum. Ten micrograms of the ADA1 and ADA2 expression vectors was transfected into HEK293 cells in six-well plates with Lipofectamine (Invitrogen, Carlsbad, CA). To establish the stable expression systems, the cells transfected with the vectors were grown in Dulbecco's modified Eagle's medium containing 100 U/mL penicillin, 100 mg/L streptomycin, 600 mg/L antibiotic G418 sulfate, and 10% fetal bovine serum. Several clones were isolated and the clone highly expressing ADA was selected for the following study.
Western blot analysis
To determine the protein expression of ADA1 and ADA2 in the ADA1- and ADA2-transfected cells, sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) followed by immunoblotting was carried out using anti-ADA1 and anti-ADA2 antibodies. The total cell homogenates (50 μg of protein) were separated on 4–12% NuPAGE Bis-Tris polyacrylamide gels (Invitrogen). Separated proteins were transferred to a PVDF membrane. The membrane was washed with phosphate-buffered saline (PBS) three times and blocked with skim milk for 1 h. The membrane was probed overnight with the anti-ADA1 (1:2000) or anti-ADA2 (1:1000) primary antibodies diluted with PBS. The membrane was then washed with PBS three times and incubated with HRP-conjugated antibody for 1 h. After the membrane was washed three times, the immunoreactive proteins were visualized with Chemi-Lumi One L (Nacalai).
Enzyme assays
Blood was collected from C57BL/6 mice (SLC, Shizuoka, Japan). Mouse erythrocytes were prepared to determine the ADA activity in mouse blood according to the method described previously (Meyskens and Williams 1971). To determine the ADA activity in the stable expression systems, cells were harvested by centrifugation. The protein concentrations were determined according to Bradford (1976). A typical incubation mixture (200 μL total volume) contained 400 mmol/L sodium acetate buffer (pH 5.8), substrate, and enzyme sources (erythrocytes or ADA-expressing cells). After a 3 min-preincubation at 37°C, the reaction was initiated by adding substrates (adenosine, cordycepin, or 2′-deoxyadenosine; Fig.1). The incubation mixture was incubated for 2 min at 37°C and then the reaction was terminated by adding 400 μL of acetonitrile. It was confirmed that the deamination activities against adenosine, 2′-deoxyadenosine, and cordycepin were linear over 10 min. The reaction mixture was centrifuged at 10,000g for 3 min and 50 μL aliquot of the supernatant was subjected to a high-performance liquid chromatography (HPLC). The analytical column was a Mightysil RP-18 GP 150-4.6 (4.6 × 250 mm; 5 μm) (Kanto Chemical, Tokyo, Japan). The mobile phase was 6% acetonitrile and the flow rate was 1.0 mL/min. The column elute was monitored at 260 nm (Ikeda et al. 2008). The retention times of 3′-deoxyinosine, inosine, and 2′-deoxyinosine were 4.1, 3.3, and 3.7 min, respectively.
The metabolic data were analyzed by the Michaelis–Menten Equation (1):
| 1 |
where V is the metabolic rate, S is the substrate concentration, Vmax is the maximum metabolic rate, and Km is the Michaelis constant.
To estimate the inhibitory effects of ADA inhibitors (Fig.1), analyses were conducted using varied concentrations of cordycepin. The type of inhibition and the Ki values were determined by a Dixon plot analysis. Enzyme assays were also conducted in the presence of heparin (40 μmol/L) and bovine serum albumin (BSA) (6 μmol/L).
Cell culture and MTT assay
The human T-cell leukemia (Jurkat) cells were maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum, 1% penicillin–streptomycin-mixed solution (Nacalai), 2.0 g/L NaHCO3, and 0.3 g/L glutamine. The cells were grown in a humidified atmosphere with 5% CO2 at 37°C. Jurkat cells were seeded into 96-well plates at the density of 1 × 104 cells/well. After 24 h, the cells were treated with cordycepin (100 mmol/L or 1 mmol/L) with or without pentostatin (50 μmol/L) or naringin (50 μmol/L) for 24 h. MTT assay was carried out according to the manufacturers' instructions.
MD simulation and docking studies
The three-dimensional (3D) structure of human ADA1 (PDB code: 3IAR) was used in the in silico analysis. Molecular dynamics (MD) simulations of ADA1 and docking studies were carried out according to a method described previously (Handa et al. 2013). Briefly, MD simulations of the 3D structure were carried out with a Maestro program from the Schrödinger Suite 2010 (Schrödinger K.K., Tokyo, Japan). Minimizations of the protein structure were carried out using force field OPLS_2005 until the average root-mean-square deviation (RMSD) of the heavy atoms reached 0.3 Å. The two-dimensional (2D) structure of naringin was converted into the 3D structure using the LigPrep program from the Schrödinger Suite 2010. The final step of a LigPrep preparation was an energy minimization of the 3D conformers using OPLS_2005. For conformational search of the compound, the CongGen program from the Schrödinger Suite 2010 was used. The computational docking was carried out using the Glide SP docking program from the Schrödinger Suite 2008 (Schrödinger K.K.).
Statistical analysis
Data were presented as means ± SD and were analyzed by analysis of variance (ANOVA) and Dunnett's procedure for multiple comparisons. P < 0.05 was considered significant.
Results
Stable expression systems of human ADA1 and ADA2
Several clones were isolated from the ADA1- and ADA2-transfected HEK293 cells. By conducting an immunoblotting analysis of the clones with the anti-ADA1 and anti-ADA2 antibodies, the clones with the highest ADA protein levels were selected for the subsequent analyses (Fig.2). In Mock cells, faint bands of ADA1 and ADA2 were observed, indicating that HEK293 cells endogenously express ADAs.
Figure 2.
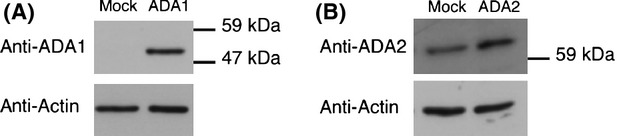
Western blot analysis of human ADA1 and ADA2. The total cell homogenates (50 μg protein) from the ADA-expressing HEK293 cells were subjected to western blot analysis with anti-ADA1 (A) or anti-ADA2 (B) antibodies. As a loading control, bands obtained with antiactin antibody are also shown.
Kinetic analysis of ADA-catalyzed deamination of substrates
To determine the ADA isoform responsible for the metabolism of cordycepin, ADA1- and ADA2-expressing HEK293 cells and Mock cells were subjected to the enzyme assay with cordycepin. While the ADA1-expressing HEK293 cells exhibit high enzyme activity toward cordycepin, the cordycepin-deamination activity in the ADA2-expressing HEK293 cells was low and comparable to that in the Mock cells (Fig.3). The cordycepin deamination by the ADA1-expressing HEK293 cells followed the Michaelis–Menten Equation (1), yielding Km = 54.9 μmol/L and Vmax = 45.8 nmol/min/mg (Fig.3A and Table1).
Figure 3.
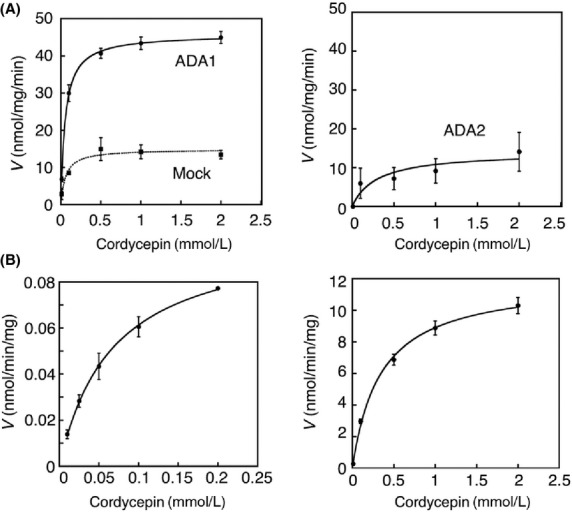
The deamination of cordycepin in ADA1- and ADA2-expressing cells, Mock cells, human erythrocytes, and mouse erythrocytes. Cordycepin was incubated with total cell homogenates of ADA1- and ADA2-expressing cells, and Mock cells (0.05 mg protein/mL) at 37°C for 2 min (A). Cordycepin was also incubated with human (left) and mouse (right) erythrocytes (0.5 mg protein/mL) at 37°C for 30 min (B). The metabolite was quantified with HPLC analysis. Data are the means ± SD of three independent determinations.
Table 1.
Kinetic parameters of deamination of cordycepin in ADA1- and ADA2-expressing cells and mouse and human erythrocytes
| Km (μmol/L) | Vmax (nmol/min/mg) | |
|---|---|---|
| Cordycepin | ||
| ADA1 | 54.9 ± 2.4 | 45.8 ± 0.6 |
| ADA2 | 165.2 ± 126.4 | 17.2 ± 8.8 |
| Mouse erythrocytes | 350.3 ± 78.7 | 12.1 ± 0.4 |
| Human erythrocytes | 70.4 ± 17.9 | 0.1 ± 0.005 |
The established ADA1- and ADA2-expressing HEK293 cells were further subjected to the enzyme assays using known ADA substrates, adenosine, and 2′-deoxyadenosine. It has been demonstrated that adenosine and 2′-deoxyadenosine are mainly metabolized by ADA1 (Cristalli et al. 2001). The ADA1-expressing HEK293 cells exhibited much higher deamination activities toward adenosine and 2′-deoxyadenosine with the Km = 125.8 μmol/L and Vmax = 621.9 nmol/min/mg protein (adenosine) and the Km = 51.3 μmol/L and Vmax = 467.8 nmol/min/mg protein (2′-deoxyadenosine). In the ADA2-expressing HEK293 cells, the enzyme activities toward adenosine and 2′-deoxyadenosine were comparable to those in Mock cells. These findings indicate that cordycepin is mainly metabolized by ADA1, which is similar to adenosine and 2′-deoxyadenosine.
Substrates of ADA are extensively metabolized in erythrocytes in vivo (Meyskens and Williams 1971; Agarwal et al. 1977). To obtain the kinetic parameters for deamination of cordycepin, we carried out the enzyme assays in mouse and human erythrocytes (Fig.3B). The enzyme activities toward cordycepin were relatively low in the erythrocytes compared to those in the ADA1-expressing HEK293 cells. In the mouse erythrocytes, the estimated kinetic parameters were Km = 350.3 μmol/L and Vmax =12.1 nmol/min/mg protein (cordycepin) (Table1). In the human erythrocytes, the estimated kinetic parameters were Km = 70.4 μmol/L and Vmax = 0.1 nmol/min/mg protein (cordycepin) (Table1). The Km value obtained in human erythrocytes was similar to that in ADA1-expressing cells.
Inhibitory effects of ADA inhibitors on ADA1-catalyzed cordycepin deamination
Pentostatin, EHNA, and 1-deazaadenosine are known ADA inhibitors. It has been demonstrated that EHNA specifically inhibits ADA1, while pentostatin and 1-deazaadenosine can inhibit both ADA1 and ADA2 (Ratech et al. 1981; Cristalli et al. 1993, 2001; Dalla et al. 2013). The cordycepin deamination in the erythrocytes was dramatically inhibited by pentostatin and 1-deazaadenosine, while EHNA partially inhibited the deamination of cordycepin (Fig.4). This indicates that not only ADA1 but also ADA2 might contribute to the metabolism of cordycepin in erythrocytes (Fig.4). The inhibition pattern of pentostatin and EHNA against the cordycepin deamination in mouse erythrocytes was competitive and noncompetitive with Ki values of 4.9 nmol/L and 139 nmol/L, respectively (Table2).
Figure 4.
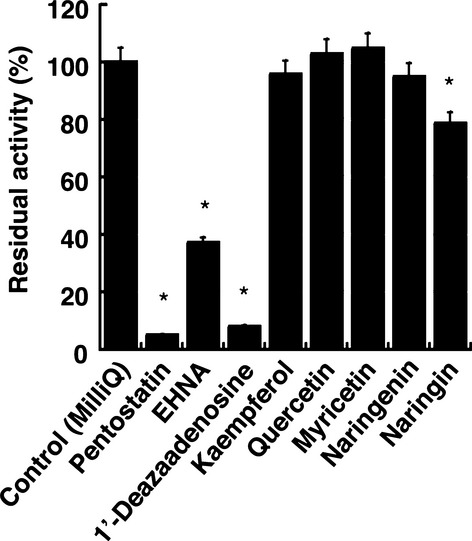
Inhibition of deamination of cordycepin. Cordycepin (100 μmol/L) was incubated at 37°C with erythrocytes in the presence of inhibitors (100 μmol/L). Data are the means ± SD of three independent determinations. *P < 0.05 compared to control.
Table 2.
Inhibition constants of pentostatin, EHNA, and naringin for deamination of cordycepin, adenosine, and 2′-deoxyadenosine in mouse and human erythrocytes
| Substrates | Inhibitors | Ki (μmol/L) | |
|---|---|---|---|
| Mouse | Cordycepin | Pentostatin | 0.0049 ± 0.00085 |
| EHNA | 0.139 ± 0.020 | ||
| Naringin | 58.8 ± 0.7 | ||
| Adenosine | Naringin | 371.0 ± 10.1 | |
| 2′-Deoxyadenosine | Naringin | 197.2 ± 49.3 | |
| Human | Cordycepin | Naringin | 168.3 ± 22.4 |
Pentostatin has been used to extend the half-life of cordycepin in vivo (Dalla et al. 2013). However, it has been shown that pentostatin can cause severe adverse reactions due to their extreme potency as ADA inhibitors. Therefore, mild but effective ADA inhibitors need to be identified. In this study, inhibitory effects of natural substances, kaempferol, quercetin, myricetin, naringenin, and naringin, on the ADA-catalyzed deamination of cordycepin were examined. The inhibitory effects of kaempferol, quercetin, myricetin, and naringenin on ADA-catalyzed deamination of cordycepin were slight, as they inhibited only 0–10% of the enzyme activity (Fig.4). In contrast, naringin moderately inhibited the deamination of cordycepin. The ADA1-catalyzed deamination of cordycepin at 100 μmol/L cordycepin was inhibited by 20% by 100 μmol/L naringin (Fig.4). The inhibition pattern of naringin against cordycepin was competitive and the Ki value was 58.8 μmol/L (Fig.5A and Table2). It was further demonstrated that naringin was able to inhibit the deamination of adenosine and 2′-deoxyadenosine with Ki values of 371 μmol/L and 197 μmol/L, respectively (Table2). This indicates that naringin is more potent as an inhibitor of the deamination of cordycepin vs. the other substrates. To further add the significance of this study, we conducted the inhibition assay using human erythrocytes. Naringin competitively inhibited the deamination of cordycepin with Ki values of 168.3 μmol/L (Fig.5B and Table2).
Figure 5.
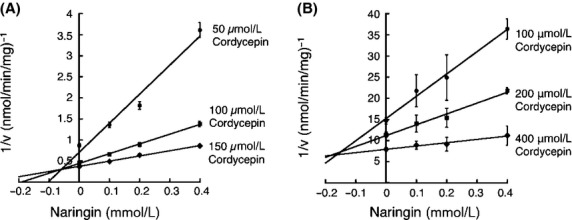
Analyses of inhibitory pattern. Inhibitory effects of naringin on deamination of cordycepin was examined in mouse (A) and human (B) erythrocytes (0.5 mg protein/mL). Data are the means ± SD of three independent determinations.
ADA-catalyzed cordycepin deamination in mouse tissues
To determine the tissue important for the metabolism of cordycepin in vivo, cordycepin deamination was examined in mouse erythrocytes as well as in mouse liver, kidney, heart, spleen, lung, stomach, and small and large intestines. While it has been widely recognized that blood is responsible for deamination of compounds in vivo, our results demonstrated that cordycepin deamination in tissues such as liver, lung, and intestines were comparable to that in erythrocytes (Fig.6). This finding indicates that not only blood and liver, which are important tissues for metabolism but also other tissues such as lung and intestines would contribute to the metabolism of cordycepin in vivo.
Figure 6.
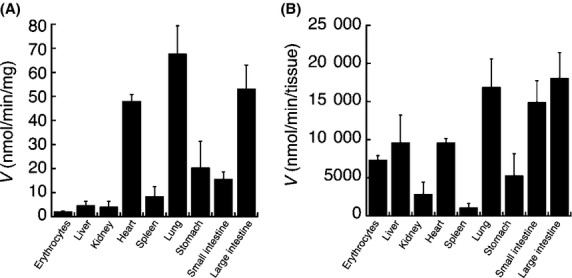
Cordycepin deamination in mouse tissues. Cordycepin (100 μmol/L) were incubated with erythrocytes or tissue homogenates of liver, kidney, heart, spleen, lung, stomach, small and large intestines. Metabolic rates of the deamination of cordycepin were shown as nmol/min per mg protein (A) or nmol/min per tissue (B). Data are the means ± SD of three independent determinations.
Naringin increases the cytotoxic effect of cordycepin on Jurkat cells
Our study indicated that naringin could be an inhibitor for the deamination of cordycepin. To investigate whether naringin can increase the cytotoxicity of cordycepin, cell-based assays were conducted using human T lymphocytes (Jurkat cells). Cordycepin concentration-dependently decreased the cell viability of the Jurkat cells (Fig.7), which agrees with a previous report that cordycepin had a cytotoxic effect toward L1210 or P388 (Johns and Adamson 1976). By cotreating the cells with pentostatin, cordycepin exhibited stronger cytotoxicity, possibly due to the protective effect of pentostatin on the cordycepin metabolism. Although the effect of naringin on the cytotoxic effect of cordycepin was weaker than that of pentostatin, naringin significantly increased the cytotoxic effect of cordycepin (Fig.7).
Figure 7.
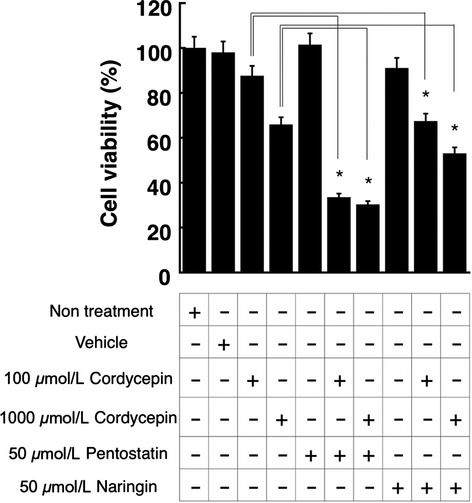
Cell viability assays. Jurkat cells were treated with cordycepin, pentostatin, cordycepin, and naringin as indicated. Twenty-four hours after the treatment, cell viability was determined by MTT assays. Data are the means ± SD of three independent determinations. *P < 0.05.
As shown in Figure8A, ADA1 was highly expressed in Jurkat cells. This is in agreement with a previous publication that demonstrated that Jurkat cells primarily express ADA1. As shown in Figure8B, pentostatin and naringin strongly and moderately inhibited deamination of cordycepin in Jurkat cell homogenates. Therefore, it is likely that the decreases of cell viability by pentostatin and naringin were attributed to inhibition of ADA1-catalyzed deamination of cordycepin in the cells.
Figure 8.
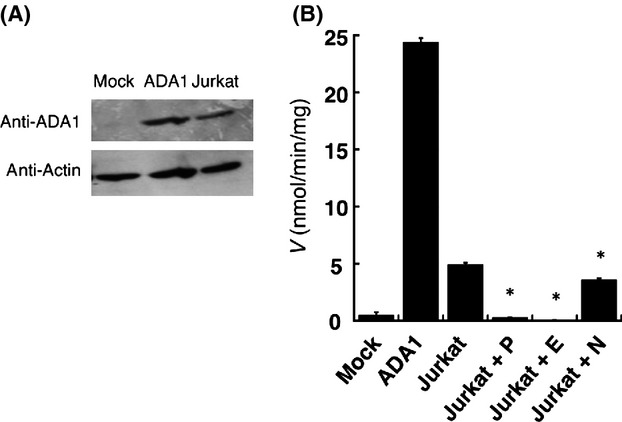
ADA1 expression and activity in Jurkat cells. The total cell homogenates (2 μg protein) of the ADA1-expressing HEK293 cells, Mock cells, and Jurkat cells were subjected to Western blot analysis with anti-ADA1 or antiactin antibodies (A). Cordycepin deamination activities were determined using the cell homogenates in the absence or presence of pentostatin (P), EHNA (E), and naringin (N) (B). Concentrations of substrate and inhibitors were 100 μmol/L and 50 μmol/L, respectively. *P < 0.05.
In silico docking study
ADA1 metabolizes adenosine and its derivatives such as 2′-deoxyadenosine. However, the secondary structure of naringin, which was found to inhibit ADA1 in this study, is relatively bulky (Fig.1). Our inhibition study revealed that naringin competitively inhibited deamination of adenosine and cordycepin. To investigate whether naringin binds to the substrate recognition site of ADA1, an in silico docking study was carried out. The 3D structure of human ADA1 was obtained from Protein Data Bank (http://www.rcsb.org/pdb). As shown in Figure9, our docking study revealed that naringin bound to the active site of ADA1 with six hydrogen bonds (Fig.9B), which is comparable to the number of hydrogen bonds observed in the docking model of ADA1 with adenosine (Fig.9A). In the docking model of ADA1 with naringenin, which did not inhibit ADA1 in this study, there were only two hydrogen bonds between the ligand and the active site (Fig.9C). Therefore, it is notable to mention that the extra hydrogen bonds around the active site of ADA1 make a bulky ligand possible to bind the binding pocket of ADA1.
Figure 9.
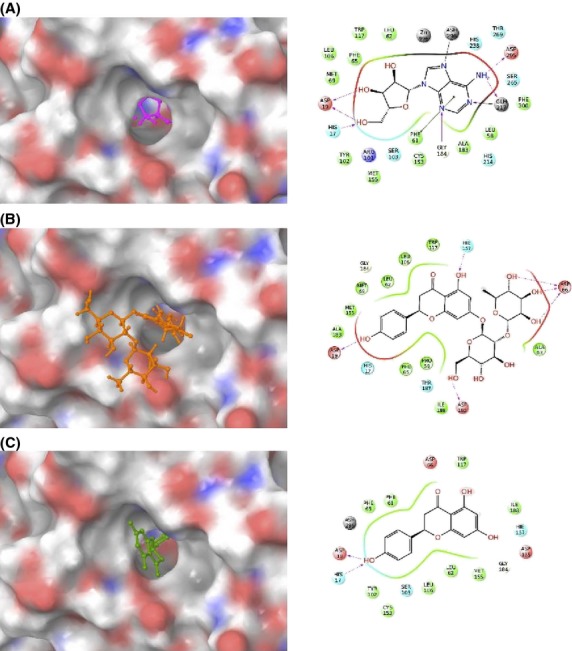
In silico docking study of ADA1 with adenosine (A), naringin (B), and naringenin (C). The ligand-binding pocket of ADA1 is shown as a transparent surface (left panels). Interactions between amino acid residues of ADA1 and ligands, such as hydrogen bonds (pink) and pi-pi stacking interactions (green), are shown (right panels).
Discussion
Human genome encodes two different ADA isoforms, ADA1 and ADA2. Specific inhibitors for certain ADA isoform have been used to determine the ADA isoform responsible for the deamination of substrates. EHNA has been known as an ADA1-specific inhibitor, while pentostatin nonspecifically inhibits both ADA1 and ADA2 (Ratech et al. 1981). However, accumulated evidence indicates that EHNA is able to inhibit ADA2 at a high concentration (Ratech et al. 1981). To fully understand the substrate specificity of ADA1 and ADA2, therefore, an enzyme assay utilizing recombinant systems of human ADAs was conducted in this study. The ADA isoform responsible for the metabolism of cordycepin was unclear. To date, the nonspecific ADA inhibitor, pentostatin, has been widely used to protect cordycepin from its degradation in order to increase the efficacy of the cordycepin therapy (Ratech et al. 1981). Our data indicate that cordycepin is mainly metabolized by ADA1, since not ADA2-expressing cells but ADA1-expressing HEK293 cells extensively generated a metabolite of cordycepin. This finding leads to a speculation that a combination of cordycepin with a specific ADA1 inhibitor would further make it possible to increase the efficacy of the cordycepin therapy without inducing adverse reactions associated with an inhibition of ADA2.
Importantly, evidence that functional ADA2 was expressed in the stable expression system was not obtained in this study, since the adenosine deamination activity in the ADA2-expressing cells was comparable to that in the Mock-transfected cells (Fig.3), even though adenosine has been suggested as a substrate of ADA2. It was reported that freeze–thaw cycles decreased ADA2 activities (Andreasyan et al. 2005). It was also reported that heparin and BSA increased ADA2 activities (Sargisova et al. 2012). Under these experimental conditions, however, the enzyme activity of ADA2 was still indistinguishable from mock-transfected cells (Fig. S1). While our data strongly suggest that ADA1 mainly contributes to the metabolism of cordycepin, a possibility that ADA2 might also contribute to the metabolism cannot be excluded. Indeed, coincubation of mouse erythrocytes with EHNA resulted in a partial inhibition of the deamination of cordycepin (Fig.4), indicating that the other enzyme, probably ADA2, partially contribute to the metabolism of cordycepin in erythrocytes.
Cordycepin therapy requires coadministration of an ADA inhibitor to protect cordycepin from its rapid metabolism. However, potent ADA inhibitors could induce severe adverse reactions, which is because ADA is involved in the development of immune system in the body. Previous studies demonstrated that natural substances such as quercetin, myricetin, and kaempferol were able to inhibit deamination of adenosine (Koch et al. 1992; Melzig 1996). In this study, we investigated the inhibitory effects of natural substances on deamination of cordycepin, finding that while quercetin, myricetin, and kaempferol did not, but naringin showed the inhibitory effect on the deamination of cordycepin (Fig.4). We further demonstrated that a treatment of Jurkat cells with a combination of cordycepin and naringin exhibited stronger cytotoxic effect compared to a single treatment of the cells with cordycepin (Fig.8), indicating that naringin is promising as an in vivo ADA inhibitor.
To date, several research groups carried out in silico docking study to discover new molecules that bind strongly to the active site of human ADA1. Because small molecules such as adenosine and pentostatin have been known to bind to the active site of ADA1, only these and their derivatives compounds have been used in pharmacophore modeling parameters (Terasaka et al. 2003, 2005). It should be noted that the secondary structure of naringin is quite bulky compared to that of ADA substrates (adenosine and 2′-deoxyadenosine) and known ADA inhibitors (EHNA and pentostatin) (Fig.1). Our in silico study indicated that naringin forms hydrogen bonds not only with the active site of ADA1 but also with the surface of the ADA protein around the substrate pocket (Fig.9). This suggests that bulky molecules like naringin might have a potency to be a potent ADA1 inhibitor. As cordycepin exhibits a wide range of biological functions including anticancer effects, derivatives of adenosine could be a promising candidate for a new drug. However, to protect the molecules from the rapid degradation by ADA, noncytotoxic but effective ADA inhibitors need to be coadministered, except for a case like vidarabin that a metabolite of an ADA substrate still exhibits the pharmacological activity (Shipman et al. 1976). Further in silico investigation is required to discover more potent bulky ADA inhibitors.
In conclusion, this study identified the ADA isoform responsible for the deamination of cordycepin by conducting enzyme assays in the developed ADA1- and ADA2-expressing HEK293 cells. Inhibition assays revealed that naringin exhibited an inhibitory effect against the deamination of cordycepin. Strong inhibition of ADA activities by potent ADA inhibitors such as pentostatin or deficiency of ADA1 gene can cause immune deficiency (Kraut et al. 1990; Ozsahin et al. 1997). Meanwhile, the inhibitory effect of naringin was moderate, suggesting that severe adverse reactions such as immune deficiency would not occur with naringin. We further demonstrated that naringin forms extra hydrogen bonds with the active site and the surface of the ADA1 protein, suggesting that not only small molecules such as known ADA substrates and inhibitors but also bulky molecules can be a potent ADA inhibitor.
Author Contributions
Li, Hirono, Itoh, and Fujiwara participated in research design. Li, Nakagome, and Fujiwara conducted experiments. Li, Nakagome, Hirono, and Fujiwara performed data analysis. Itoh and Fujiwara wrote or contributed to the writing of the manuscript.
Disclosures
None declared.
Glossary
- 3D
three-dimensional
- ADA
adenosine deaminase
- Cordycepin
3′-deoxyadenosine
- EHNA
erythro-9-(2-hydroxy-3-nonyl) adenine
- HPLC
high-performance liquid chromatography
- HUS
hemolytic uremic syndrome
- MD
molecular dynamics
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1. Adenosine deamination activities in Mock cells and ADA2-expressing cells. Adenosine deamination activities were determined using the cells that were frozen or nonfrozen in the presence or absence of heparin (H) or BSA.
References
- Adamson RH, Zaharevitz DW, Johns DG. Enhancement of the biological activity of adenosine analogs by the adenosine deaminase inhibitor 2'-deoxycoformycin. Pharmacology. 1977;15:84–89. doi: 10.1159/000136666. [DOI] [PubMed] [Google Scholar]
- Agarwal RP, Spector T, Parks RE., Jr Tight-binding inhibitors IV. Inhibition of adenosine deaminase by various inhibitors. Biochem Pharmacol. 1977;26:359–367. doi: 10.1016/0006-2952(77)90192-7. [DOI] [PubMed] [Google Scholar]
- Andreasyan NA, Hairapetyan HL, Sargisova YG, Mardanyan SS. ADA2 isoform of adenosine deaminase from pleural fluid. FEBS Lett. 2005;579:643–647. doi: 10.1016/j.febslet.2004.11.109. [DOI] [PubMed] [Google Scholar]
- Bhattarai NK. Folk herbal medicines of Dolakha district, Nepal. FITOTERAPIA-MILANO- 1993;64:387. [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bukulmez G, Akan T, Ciliv G. Serum adenosine deaminase levels in patients with psoriasis: a prospective case-control study. European Journal of Dermatology. 2000;10:274–276. [PubMed] [Google Scholar]
- Cristalli G, Eleuteri A, Vittori S, Volpini R, Camaioni E, Lupidi G. Adenosine deaminase inhibitors structure activity relationships in 1-deazaadenosine and erythro-9-(2-hydroxy-3-nonyl)adenine analogs. Drug Dev Res. 1993;28:253–258. [Google Scholar]
- Cristalli G, Costanzi S, Lambertucci C, Lupidi G, Vittori S, Volpini R, et al. Adenosine deaminase: functional implications and different classes of inhibitors. Med Res Rev. 2001;21:105–128. doi: 10.1002/1098-1128(200103)21:2<105::aid-med1002>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Cunningham KG, Manson W, Spring FS, Hutchinson SA. Cordycepin, a metabolic product isolated from cultures of Cordyceps militaris (Linn.) Link. Nature. 1950;166:949. doi: 10.1038/166949a0. [DOI] [PubMed] [Google Scholar]
- Dalla RL, da Silva AS, Gressler LT, Oliveira CB, Dambrós MG, Miletti LC, et al. Cordycepin (3'-deoxyadenosine) pentostatin (deoxycoformycin) combination treatment of mice experimentally infected with Trypanosoma evansi. Parasitology. 2013;140:663–671. doi: 10.1017/S0031182012001990. [DOI] [PubMed] [Google Scholar]
- Dong RP, Tachibana K, Hegen M, Munakata Y, Cho D, Schlossman SF, et al. Determination of adenosine deaminase binding domain on CD26 and its immunoregulatory effect on T cell activation. J Immunol. 1997;159:6070–6076. [PubMed] [Google Scholar]
- Frederiksen S, Malling H, Klenow H. Isolation of 3'-deoxyadenosine (cordycepin) from the liquid medium of Cordyceps militarisBiochimica et Biophysica Acta (BBA)-Nucleic Acids and Protein. Synthesis. 1965;95:189–193. doi: 10.1016/0005-2787(65)90483-1. [DOI] [PubMed] [Google Scholar]
- Gakis C. Adenosine deaminase (ADA) isoenzymes ADA1 and ADA2: diagnostic and biological role. Eur Respir J. 1996;9:632–633. doi: 10.1183/09031936.96.09040632. [DOI] [PubMed] [Google Scholar]
- Glazer RI, Lott TJ, Peale AL. Potentiation by 2'-deoxycoformycin of the inhibitory effect by 3'-deoxyadenosine (Cordycepin) on nuclear RNA synthesis in L1210 cells in vitro. Cancer Res. 1978;38:2233–2238. [PubMed] [Google Scholar]
- Gong XJ, Ji H, Lu SG, Li SP, Li P. Effects of polysaccharides from cultured cordyceps sinensis on murine immunologic function. Zhongguo Yaoke Daxue Xuebao. 2000;31:53–55. [Google Scholar]
- Handa K, Nakagome I, Yamaotsu N, Gouda H, Hirono S. Three-dimensional quantitative structure-activity relationship analysis of inhibitors of human and rat cytochrome P4503A enzymes. Drug Metab Pharmacokinet. 2013;28:345–355. doi: 10.2133/dmpk.dmpk-12-rg-133. [DOI] [PubMed] [Google Scholar]
- Ikeda R, Nishimura M, Sun Y, Wada M, Nakashima K. Simple HPLC-UV determination of nucleosides and its application to the authentication of Cordyceps and its allies. Biomed Chromatogra. 2008;22:630–636. doi: 10.1002/bmc.980. [DOI] [PubMed] [Google Scholar]
- Johns DG, Adamson RH. Enhancement of the biological activity of cordycepin (3'-deoxyadenosine) by the adenosine deaminase inhibitor 2'-deoxycoformycin. BiochemPharmacol. 1976;25:1441–1444. doi: 10.1016/0006-2952(76)90121-0. [DOI] [PubMed] [Google Scholar]
- Koch HP, Jäger W, Groh U, Plank G. In vitro inhibition of adenosine deaminase by flavonoids and related compounds. New insight into the mechanism of action of plant phenolics. Methods Find Exp Clin Pharmacol. 1992;14:413–417. [PubMed] [Google Scholar]
- Koyama K, Imaizumi T, Akiba M, Kinoshita K, Takahashi K, Suzuki A, et al. Antinociceptive components of Ganoderma lucidum. Planta Med. 1997;63:224–227. doi: 10.1055/s-2006-957658. [DOI] [PubMed] [Google Scholar]
- Kraut EH, Neff JC, Bouroncle BA, Gochnour D, Grever MR. Immunosuppressive effects of pentostatin. J Clin Oncol. 1990;8:848–855. doi: 10.1200/JCO.1990.8.5.848. [DOI] [PubMed] [Google Scholar]
- Leach JW, Pham T, Diamandidis D, George JN. Thrombotic thrombocytopenic purpura-hemolytic uremic syndrome (TTP-HUS) following treatment with deoxycoformycin in a patient with cutaneous T-cell lymphoma (Sezary syndrome): a case report. Am J Hematol. 1999;61:268–270. doi: 10.1002/(sici)1096-8652(199908)61:4<268::aid-ajh9>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Burger P, Vogel M, Friese K, Brüning A. The nucleoside antagonist cordycepin causes DNA double strand breaks in breast cancer cells. Invest New Drugs. 2012;30:1917–1925. doi: 10.1007/s10637-012-9859-x. [DOI] [PubMed] [Google Scholar]
- Melzig MF. Inhibition of adenosine deaminase activity of aortic endothelial cells by selected flavonoids. Planta Med. 1996;62:20–21. doi: 10.1055/s-2006-957788. [DOI] [PubMed] [Google Scholar]
- Meyskens FL, Williams HE. Adenosine metabolism in human erythrocytes. Biochimica et Biophysica Acta (BBA)-Nucleic Acids and Protein. Synthesis. 1971;240:170–179. doi: 10.1016/0005-2787(71)90654-x. [DOI] [PubMed] [Google Scholar]
- North TW, Cohen SS. Erythro-9-(2-hydroxy-3-nonyl) adenine as a specific inhibitor of herpes simplex virus replication in the presence and absence of adenosine analogues. Proc Natl Acad Sci. 1978;75:4684–4688. doi: 10.1073/pnas.75.10.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsahin H, Arredondo-Vega FX, Santisteban I, Fuhrer H, Tuchschmid P, Jochum W, et al. Adenosine deaminase deficiency in adults. Blood. 1997;89:2849–2855. [PubMed] [Google Scholar]
- Ratech H, Thorbecke GJ, Meredith G, Hirschhorn R. Comparison and possible homology of isozymes of adenosine deaminase in Aves and Humans. Enzyme. 1981;26:74–84. doi: 10.1159/000459153. [DOI] [PubMed] [Google Scholar]
- Sargisova YG, Andreasyan NA, Hayrapetyan HL, Harutyunyan HA. Nitric oxide - an activating factor of adenosine deaminase 2 in vitro. Biochemistry (Mosc) 2012;77:92–97. doi: 10.1134/S0006297912010117. [DOI] [PubMed] [Google Scholar]
- Shipman C, Smith SH, Carlson RH, Drach JC. Antiviral activity of arabinosyladenine and arabinosylhypoxanthine in herpes simplex virus-infected KB cells: selective inhibition of viral deoxyribonucleic acid synthesis in synchronized suspension cultures. Antimicrob Agents Chemother. 1976;9:120–127. doi: 10.1128/aac.9.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiers AS, Parekh SJ, Bishop MB. Hairy-cell leukemia: induction of complete remission with pentostatin (2'-deoxycoformycin) J Clin Oncol. 1984;2:1336–1342. doi: 10.1200/JCO.1984.2.12.1336. [DOI] [PubMed] [Google Scholar]
- Sugar AM, McCaffrey RP. Antifungal Activity of 3'-Deoxyadenosine (Cordycepin) Antimicrob Agents Chemother. 1998;42:1424–1427. doi: 10.1128/aac.42.6.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaka T, Nakanishi I, Nakamura K, Eikyu Y, Kinoshita T, Nishio N, et al. Structure-based de novo design of non-nucleoside adenosine deaminase inhibitors. Bioorg Med Chem Lett. 2003;13:1115–1118. doi: 10.1016/s0960-894x(03)00026-x. [DOI] [PubMed] [Google Scholar]
- Terasaka T, Tsuji K, Kato T, Nakanishi I, Kinoshita T, Kato Y, et al. Rational design of non-nucleoside, potent, and orally bioavailable adenosine deaminase inhibitors: predicting enzyme conformational change and metabolism. J Med Chem. 2005;48:4750–4753. doi: 10.1021/jm050413g. [DOI] [PubMed] [Google Scholar]
- Tsai YJ, Lin LC, Tsai TH. Pharmacokinetics of adenosine and cordycepin, a bioactive constituent of Cordyceps sinensis in Rat. J Agric Food Chem. 2010;58:4638–4643. doi: 10.1021/jf100269g. [DOI] [PubMed] [Google Scholar]
- Wu WC, Hsiao JR, Lian YY, Lin CY, Huang BM. The apoptotic effect of cordycepin on human OEC-M1 oral cancer cell line. Cancer Chemother Pharmacol. 2007;60:103–111. doi: 10.1007/s00280-006-0354-y. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Kagota S, Nakamura K, Shinozuka K, Kunitomo M. Antioxidant activity of the extracts from fruiting bodies of cultured Cordyceps sinensis. Phytother Res. 2000;14:647–649. doi: 10.1002/1099-1573(200012)14:8<647::aid-ptr670>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Yoshikawa N, Nakamura K, Yamaguchi Y, Kagota S, Shinozuka K, Kunitomo M. Antitumour activity of cordycepin in mice. Clin Exp Pharmacol Physiol Suppl. 2004;2:S51–S53. doi: 10.1111/j.1440-1681.2004.04108.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Adenosine deamination activities in Mock cells and ADA2-expressing cells. Adenosine deamination activities were determined using the cells that were frozen or nonfrozen in the presence or absence of heparin (H) or BSA.


