Abstract
The f subunit of the eukaryotic initiation factor 3 (eIF3f) is downregulated in several cancers and in particular in melanoma and pancreatic cancer cells. Its enforced expression by transient gene transfection negatively regulates cancer cell growth by activating apoptosis. With the aim to increase the intracellular level of eIF3f proteins and activate apoptosis in cancer cell lines, we developed a protein transfer system composed of a cell-penetrating peptide sequence fused to eIF3f protein sequence (MD11-eIF3f). To determine whether exogenously administered eIF3f proteins were able to compensate the loss of endogenous eIF3f and induce cancer cell death, we analyzed the therapeutic action of MD11-eIF3f in several tumor cells. We identified four cell lines respondent to eIF3f-treatment and we evaluated the antitumor properties of the recombinant proteins using dose- and time-dependent studies. Our results demonstrate that this protein delivery approach represents an innovative and powerful strategy for cancer treatment.
Introduction
Protein therapy is one of the most direct and safe approach for treating some of the most difficult-to-treat diseases, by which the missing or defective protein is produced by recombinant methods and delivered directly into human cells.1 As only compounds within a narrow range of molecular size and polarity passively penetrate into cells, the plasma membrane represents a major limit for the delivery of peptides or proteins. In recent years, substantial progresses have been made to find and design novel therapeutic technologies for improving the cellular entry of hydrophilic macromolecules with cytoplasmic or nucleic targets. Cell-penetrating peptides (CPPs) or Protein Transduction Domains (PTDs) are short peptides able to gain access to cytoplasm and subcellular compartments by different mechanisms, including endocytosis, and to promote the intracellular delivery of different cargoes.2 CPP-mediated delivery of functional proteins or peptides has been extensively employed both for studying cellular processes and for developing novel therapeutic macromolecules.3 A novel cell-penetrating peptide deriving from the Epstein-Barr virus (EBV) ZEBRA transcription factor has been recently described by our group.4 The minimal amino acid region implicated in cellular uptake spans residues 178–220 of full-length ZEBRA protein (MD11), and is able to translocate high molecular weight proteins in an endocytosis-independent mechanism, allowing the internalization of cargo proteins in fully biologically active form.
Eukaryotic gene expression is a process mainly regulated at the levels of gene transcription and protein synthesis. Minimal deregulation of protein synthesis can lead to uncontrolled cell growth and cancer formation. The translation process is largely regulated at initiation level by 12 different eukaryotic initiation factors (eIFs). The involvement of several eIFs in pathogenesis and cancer development has been already reported.5 In particular, several lines of evidence suggest that the eukaryotic initiation factor 3 (eIF3) contributes to tumorigenesis. This complex is composed of 13 subunits (designated eIF3 a-m). Amongst the 13 subunits, overexpression of subunits eIF3-a, -b, -c, -h, -i, and -m has been detected in several different solid tumors and in different cancer cell lines.6–9 Two other subunits eIF3e and eIF3f are downregulated in many human tumors, and in particular the f subunit expression is significantly decreased in 100% of pancreas and vulva tumors, 90% breast tumors, 71% melanomas, and 70% of ovary and small intestine tumors.10
eIF3f belongs to the Mov34 protein family containing the MPN motif11 and acts as a negative regulator of translation inhibiting both cap-dependent and cap-independent translation.12 The exact molecular mechanisms, by which eIF3f expression decreases contributing to cancer development, are still unclear. It is likely that the loss of eIF3f in cancer cells induces increased eIF3 activity, which in turn stimulates translation of specific mRNAs encoding proteins involved in cell proliferation.13,14 In melanoma and pancreas cancer cells, eIF3f exhibits a tumor-suppressive role as its enforced expression by gene transfection negatively regulates cancer cell growth by activating apoptosis.10,12–14
However, in proliferating myoblasts eIF3f protein is barely detectable, but is dramatically upregulated during terminal differentiation and maintained in adult skeletal muscle.15 Interestingly, the genetic repression of eIF3f in normal myotubes induces atrophy while genetic activation is sufficient to induce hypertrophy through modulation of protein synthesis via the mTORC1 pathway.16,17 Therefore, eIF3f plays a central role in the control of muscle mass and size and a cell-type-specific role is believed to exist for eIF3f.18
In this study, we hypothesize that increasing of eIF3f protein intracellular level by protein transduction could represent a powerful approach to cancer treatment. To test this hypothesis, we produced fusion recombinant proteins (MD11-eIF3f) in which the eIF3f sequence is directly fused to the MD11 cell-penetrating peptide, previously isolated and characterized in our laboratory.4 We performed experiments to determine whether exogenously administered MD11-eIF3f recombinant proteins were able to compensate the loss of endogenous eIF3f and induce a cellular death by inhibiting the protein synthesis process. We engineered two eIF3f truncations (MD11-AN and MD11-NF) and an ubiquitination-resistant mutant (MD11-K510R).19 The therapeutic efficiency of these proteins was screened in several tumor cells. We identified four eIF3f-treatment respondent cell lines (B16 and Colo857 melanoma cells; HCT116 p53+/+ and HCT116 p53−/− colorectal carcinoma) and we evaluated the antitumor properties of the recombinant proteins using dose- and time-dependent studies. Our results demonstrate that the MD11 peptide can efficiently deliver functional eIF3f in tumor cells, and that this protein delivery system represents a potentially powerful tool for treating melanoma and human colorectal carcinoma.
Results
Expression and purification of recombinant fusion proteins
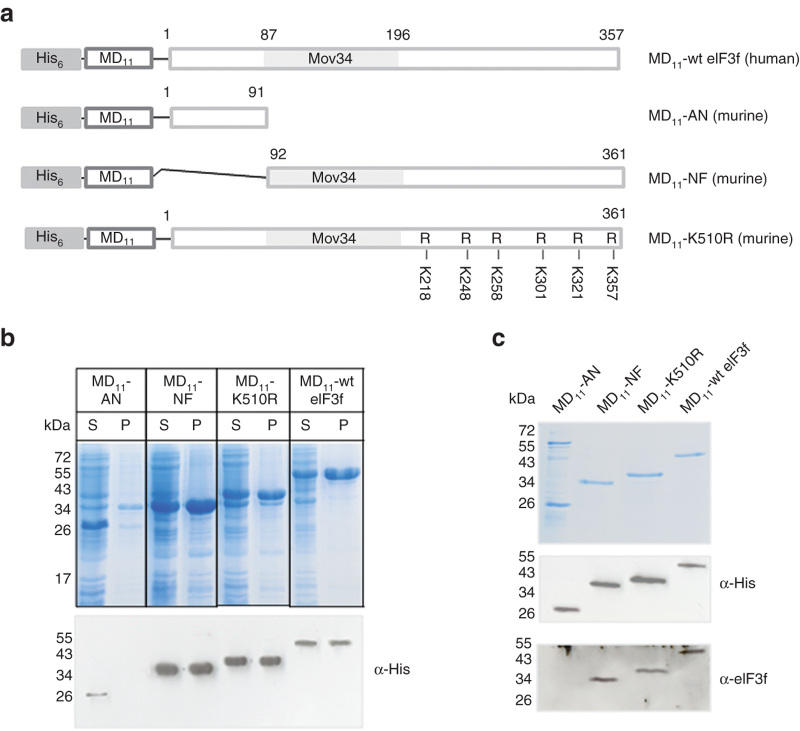
Cell-penetrating proteins are designed and developed as recombinant fusion proteins containing MD11 cell-penetrating peptide attached to the N-terminus of eIF3f sequence20 (Figure 1a). We used the full length human protein (MD11-wt eIF3f) and three mutants (Figure 1a): the N-terminal protein truncation lacking of Mov34 domain (MD11-AN, residues 1–91), the C-terminal truncation containing the Mov34 domain (MD11-NF, residues 92–362) and the ubiquitination-resistant mutant19 in which six C-terminal lysines (from K5 to K10) are replaced by arginine residues (MD11-K510R). The murine eIF3f amino acid sequence shows extensive homology with its human counterpart (92%), particularly in the region that contains the Mov34 domain.
Figure 1.
Design, expression, and purification of MD11-eIF3f recombinant fusion proteins. (a) Scheme of recombinant MD11-eIF3 constructs containing His tag for affinity purification. Mutations in which C-terminal lysines were replaced to arginines are indicated in MD11-K510R mutant. (b) Protein expression level in E. coli. After bacterial cell lysis, the soluble (S) and the insoluble (P) fractions were analyzed by SDS-PAGE, stained with Coomassie blue and identified by western blotting using an anti-His antibody. (c) After purifications, soluble recombinant fusion proteins were analyzed by SDS-PAGE, stained with Coomassie blue and detected by western blotting with an anti-His antibody and an anti-eIF3f antibody.
The his-tagged recombinant fusion proteins were produced using an E. coli expression system (Figure 1b) and purified by nickel affinity chromatography. With the exception of MD11-AN, MD11-NF, MD11-wt eIF3f, and MD11-K510R were expressed in both soluble and insoluble fractions. The yields of purified soluble proteins were negligible when compared to the total production, presumably due to the lack of tag exposition in native structure. For this reason, MD11-NF, MD11-wt eIF3f, and MD11-K510R were purified from inclusion bodies under denaturing conditions and refolded by dialysis. The purity of the MD11-eIF3f proteins was analyzed by Coomassie blue staining and by western-blotting using anti-His tag and anti-eIF3f antibodies (Figure 1c). MD11-AN was pure at around 75% and MD11-NF, MD11-wt eIF3f, and MD11-K510R at around 95%.
51Cr release cytotoxicity assay
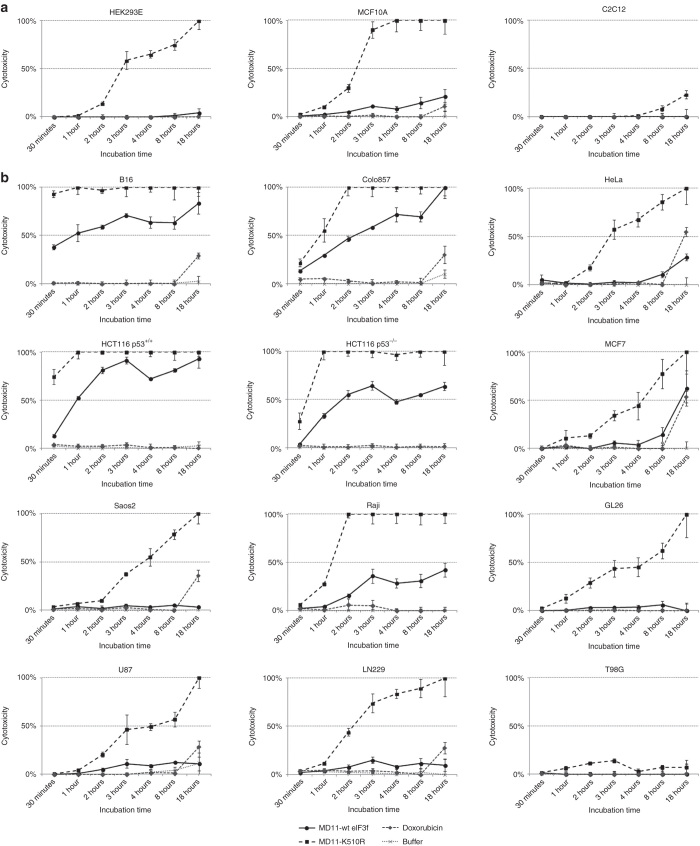
Cytotoxic activities of MD11-wt eIF3f and MD11-K510R were measured on different cell lines using a time course 51Cr release cytotoxicity assay (Figure 2). As control of toxicity, cells were incubated with 1 µmol/l doxorubicin which becomes toxic after 18 hours incubation. To discard any toxicity linked to buffer, cells were also incubated with protein-free buffer (50 mmol/l Na citrate pH 6.1, 50 mmol/l NaCl, 10% glycerol) in same volumes used for protein treatments. Recombinant MD11-wt eIF3f and MD11-K510R cytotoxic effects were screened on both tumor cells, such as human (Colo857) and mouse (B16) melanoma, human (LN229, T98G, U87) and mouse (GL26) glioblastoma, human osteosarcoma (Saos2), human cervix adenocarcinoma (HeLa), human breast adenocarcinoma (MCF7), Burkitt’s lymphoma (Raji), human colorectal carcinoma (HCT116), and on immortal nontumorigenic cell lines, such as mouse skeletal myoblasts (C2C12), human embryonic kidney cells (HEK293E), and human breast cells (MCF10A).
Figure 2.
Screening of MD11-wt eIF3f and MD11-K510R cytotoxic effect by 51Cr-release assay on human and murine cell lines. 10 nmol/l of MD11-wt eIF3f and MD11-K510R were incubated with (a) nontumorigenic and (b) tumor cells and 51Cr release measurements were performed 0.5, 1, 2, 3, 4, 8, and 18 hours after protein incubation. Doxorubicin (1 µmol/l) was used as toxicity control and protein-free buffer was tested to discard any buffer-linked toxicity. Each data point is the mean of three values ± SD.
The cell-death values detected after MD11-wt eIF3f and MD11-K510R treatment are used as parameter to discriminate their cell-type dependency (Figure 2). Generally, MD11-K510R induced a stronger cytotoxic effect than wild-type eIF3f. As matter of facts 18 hours incubation with MD11-K510R induced 100% death in HEK293E, MCF10A, MCF7, HeLa, Saos2, Raji and LN229, U87, GL26 cells, while at the same time point the wt protein mildly affect viability (from 0 to 50% in MCF7) (Figure 2). Interestingly, T98G glioblastoma cells were resistant to both treatments (Figure 2). C2C12 myoblast cell line viability is not affected by the treatment with recombinant eIF3f fusion proteins. This result is in line with previous observations demonstrating that eIF3f upregulation in muscle cells does not affect cell viability.17 A massive and rapid cytotoxic effect was detected in melanoma (B16 and Colo857) and colorectal carcinoma (HCT116 p53+/+ and HCT116 p53−/−) cells (Figure 2). For instance, one hour treatment of MD11-wt eIF3f induced 30% death in Colo857 and HCT116 p53−/− and 50% in B16 and HCT116 p53+/+, while MD11-K510R killed as expected 50% of Colo857 cells and 100% of the others. In particular, the HCT116 cell model (p53+/+ and p53−/−) was used to evaluate an eventual contribution of p53 in death induction. We found that at low dosages (2.5–10 nmol/l) HCT116 p53−/− cells were more resistant to the protein treatment when compared to the wild-type cell line. On the basis of these screening results B16, Colo857, HCT116 p53+/+, and HCT116 p53−/− cells were chosen as target tumor models for following experiments.
Protein labeling and intracellular detection
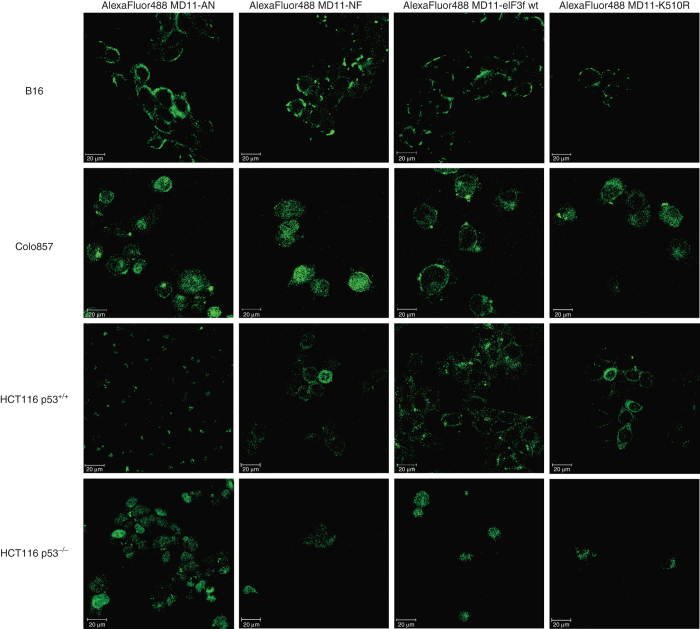
To assess the ability of fusion recombinant proteins to transduce into cells, MD11-AN, MD11-NF, MD11-wt eIF3f, and MD11-K510R were labeled with AlexaFluor488 dye and incubated with B16 cells for 30 minutes, with Colo857 for 1 hour, and with HCT116 p53+/+ and HCT116 p53−/− for 2 hours. Treatment times were chosen according to 51Cr kinetics results (Figure 2). The intracellular delivery of recombinant eIF3f was detected in living target cells by confocal laser scanning microscopy. Cellular Z-stack analysis (Figure 3) revealed that all four recombinant proteins were detectable in cell cytoplasm, confirming that MD11-bearing eIF3f proteins were able to penetrate cells. We also noticed that exogenous recombinant proteins were able to localize into cell nuclei, effect that has been already observed and reported for endogenous proteins.21,22
Figure 3.
Confocal laser scanning microscopy images of living B16 (0.5 hour), Colo857 (1 hour), HCT116 p53+/+, (2 hours) and HCT116 p53−/− (2 hours) cells incubated with AlexaFluor 488-labeled MD11-AN, MD11-NF, MD11-wt eIF3f, and MD11-K510R fusion protein (10 nmol/l). 16 successive optical slices were captured along the cellular z-axis with a step of 1 µm. The presented images correspond to the middle plan of cellular z-axis sectioning.
Cellular uptake and Annexin V FACS analysis
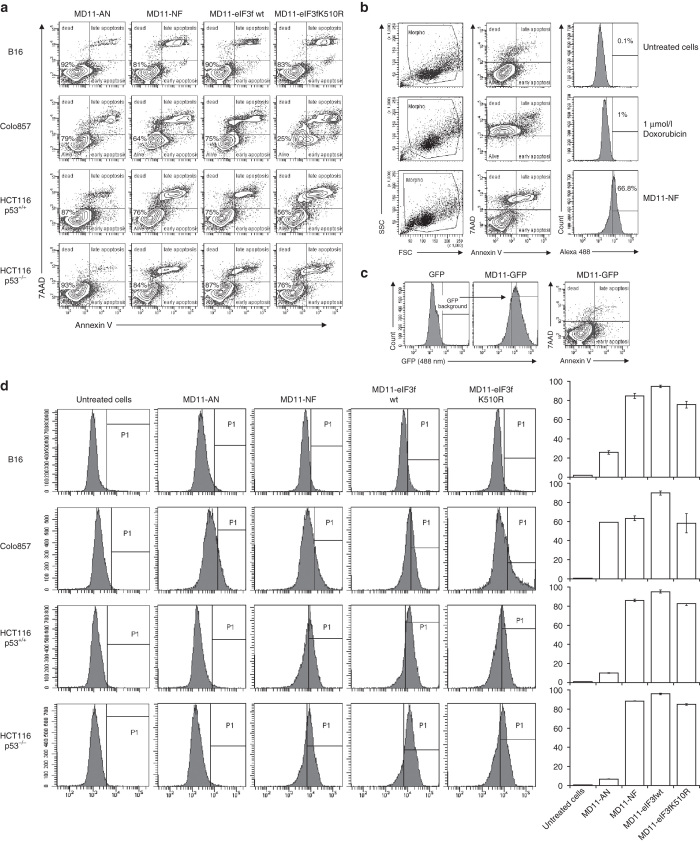
To estimate the transduction efficiency and the cell death induction, a FACS analysis using AlexaFluor488 labeled fusion proteins was performed (Figure 4). Recombinant eIF3f proteins were efficiently transduced into selected cell lines (Figure 4d). A simultaneous Annexin V-7AAD staining was used to quantitatively determine the percentage of cells undergoing apoptosis. Annexin V has a high affinity for membrane phospholipid phosphatidylserine (PS) which is translocated from the inner to the outer leaflet of the plasma membrane in apoptotic cells. 7-AAD is a viability probe that is excluded from viable cells and is internalized in dead cells. The FACS analysis (Figure 4a) on B16, Colo857, HCT116 p53+/+, and HCT116 p53−/− cells showed that the incubation with MD11-NF, MD11-wt eIF3f, and MD11-K510R increases the percentage of apoptosis compared to the negative controls (untreated cells, Figure 4b). These results indicate that the cell death quantified by 51Cr release assay can be ascribed as apoptotic-like process linked to PS flip-flop. To exclude that the toxicity of fusion proteins was not induced by the MD11 penetrating sequence, cells were also incubated with MD11-eGFP reporter protein (at 30-fold higher dose than therapeutic proteins) and no effect on viability was detected (Figure 4c). The effect of the four recombinant proteins was also investigated in normal BJ fibroblasts using Annexin V-7AAD staining (Supplementary Figure S3). Except for MD11-K510R, the recombinant proteins do not affect the viability of normal cells, even after 8 hours treatment.
Figure 4.
Annexin V-7AAD apoptosis assay and protein delivery efficiency analysis by flow cytometry. (a) B16, Colo857, HCT116 p53+/+, and HCT116 p53−/− cells were incubated with AlexaFluor 488 labeled proteins (10 nmol/l) and stained with Annexin V-APC and 7-AAD. Alive cells show negative staining for Annexin V and 7-AAD. Cells undergoing apoptosis show positive staining for Annexin V and negative for 7-AAD, while cells positive for both Annexin V and 7-AAD are either in the end stage of apoptosis, undergoing necrosis or already dead. (b) Examples of FACS profiles of untreated cells and doxorubicin-treated cells, used as controls for viability/toxicity setting gates. (c) Representative plot of HCT116 p53−/− cells incubated with 0.3 µmol/l MD11-eGFP, showing no toxic effect associated to the MD11-mediated protein internalization. (d) Evaluation of delivery efficiency of AlexaFluor 488-labeled recombinant proteins in selected cells. Histograms represent percentages of internalized MD11-AN, MD11-NF, MD11-wt, and MD11-K510R proteins in target cells.
In addition, the apoptotic action is specific to full-length eIF3f or its C-terminal truncation, as demonstrated by the lack of significant death in cells incubated with MD11-AN (Figure 4a). These findings confirm that exogenous wt eIF3f, NF truncation or ubiquitination-resistant mutant induce apoptosis in melanoma and colorectal carcinoma cells.
51Cr release assay on selected tumor cell lines
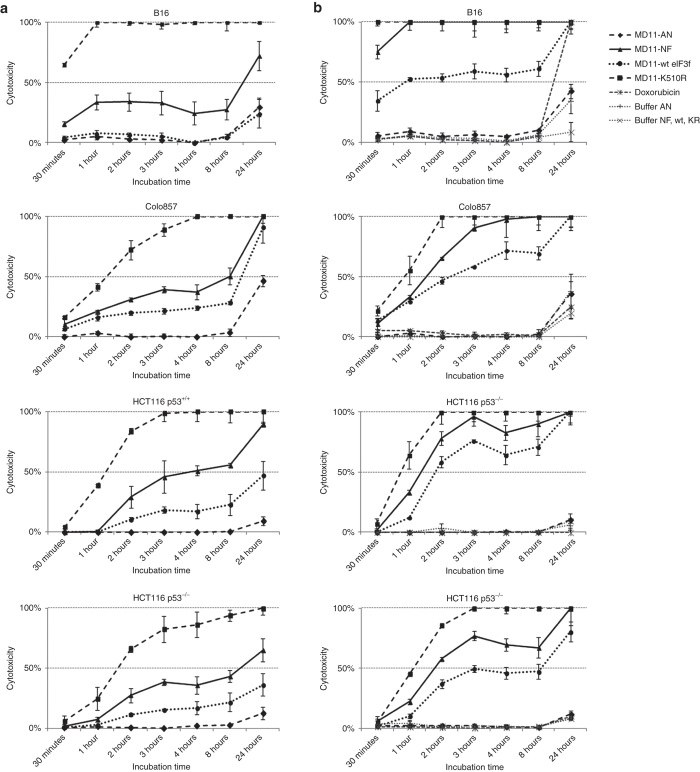
Once determined that the four recombinant proteins are internalized in target cells and that retain their therapeutic anti-cancer activity, we conducted a dose- and a time-course study using 51Cr release assay (Figure 5). Cells were incubated with 2.5, 5, 10, and 50 nmol/l of each recombinant protein and measurements were taken at various times after protein incubation. As controls, cells were incubated with doxorubicin and with protein-free buffer (50 mmol/l Na citrate pH 6.1, 50 mmol/l NaCl, 10% glycerol for MD11-NF, MD11-wt eIF3f and MD11-K510R and 25 mmol/l Hepes/KOH pH 7.0, 50 mmol/l NaCl and 10% glycerol for MD11-AN) in same volumes used for protein treatments.
Figure 5.
Time-dependent kinetics of MD11-AN, MD11-NF, MD11-wt eIF3f, and MD11-K510R effect on B16, Colo857, HCT116 p53+/+, and HCT116 p53−/− cells. Treatment with (a) 5 and (b) 10 nmol/l of recombinant proteins were performed and 51Cr release measurements were collected 0.5, 1, 2, 3, 4, 8, and 18 hours after protein incubations. Doxorubicin (1 µmol/l) was used as toxicity control and protein-free buffers were tested to discard any buffer-linked toxicity. Each data point is the mean of three values ± SD.
MD11-NF, MD11-wt eIF3f, and MD11-K510R induced inhibition of cellular proliferation in all target cell lines in a dose- and a time-dependent manner. At lower tested dose (2.5 nmol/l), the time required to obtain a maximum of 50% cellular death was 24 hours in both melanoma and colorectal carcinoma cell lines (Supplementary Figure S1). In murine melanoma cells, at 10 nmol/l concentration the 50% cellular death was achieved in 30 minutes after MD11-K510R treatment and in 1 hour after MD11-wt eIF3f treatment (Figure 5b). At the same dose, in human melanoma cells (Colo857) and in colorectal carcinoma cells (HCT116 p53+/+) the time necessary for 50% inhibition of cellular proliferation was less than 1 hour for MD11-K510R and 1.5 hours for MD11-NF and 2 hours for MD11-wt eIF3f (Figure 5b). The same effect was reached in colorectal carcinoma cells lacking of p53 (HCT116 p53−/−) in about 1 hour for MD11-K510R, 1.5 hours for MD11-NF and 3 hours for MD11-wt eIF3f, confirming that this cell line is more resistant than wild-type to the death stimulus and that p53 may play a role in eIF3f-induced death (Figure 5b). Apoptotic effects were detected also at lower doses (5 nmol/l) of recombinant proteins, but at longer times (24 hours, Figure 5a). At 50 nmol/l doses, the apoptotic response curves reached the plateau within 30 minutes (Supplementary Figure S2). Treatments with the nonfunctional MD11-AN did not affect cell viability when compared to control cells incubated with protein-free buffer (Figure 5a,b).
Discussion
The f subunit is a negative regulator of translation initiation, and the decrease of its expression contributes to tumor cells’ evading apoptosis via upregulation of protein synthesis.12 The endogenous level of eIF3f protein is downregulated in several cancers and in particular in melanoma and pancreatic cancer cells.10,13,14 Advances in the understanding of the molecular mechanism behind cellular carcinogenesis have shown that ectopic expression of eIF3f by transient gene transfection inhibits translation, cellular growth and proliferation and induces apoptosis.10,13,14 Increasing the intracellular level of eIF3f protein seems to be a crucial element for treating cancer. Due to the difficulties in developing safe and efficient gene-delivery methods and in controlling the levels of transient gene expression, the protein therapy represents a valid alternative to gene therapy. To this aim, we developed an approach based on the direct delivery of functional exogenous eIF3f proteins into cells. Thus, the MD11 cell-penetrating sequence4 was fused to eIF3f therapeutic proteins, and the effect of their cellular internalization was monitored.
The first evidence on the efficient cellular uptake and consequent protein therapeutic effect arises from the results of 51Cr screening assay on different cell lines. MD11 fused with wild-type eIF3f and its ubiquitination-resistant mutant (K510R) showed a cell-type dependent effect and, in the same cell line, they behaved differently. This difference confirms both the functional state and the expected intracellular processing of the delivered molecules. In particular, we observed a stronger MD11-K510R-mediated death induction when compared to the wt molecules. Indeed, the C-terminal point mutations (K510R) prevent polyubiquitination of eIF3f and proteasome degradation, leading to increased in cellulo protein stabilization. Interestingly, in T98G and C2C12 cells any effect on the viability is observed. While the lacking of literature does not allow formulate any hypothesis on glioblastoma cells and eIF3f correlation, our result on C2C12 confirms that overexpression of eIF3f does not induce cell death but it acts as “cell growth enhancer” instead of “death executer.”17 Among the tested cell lines, melanoma and colorectal carcinoma cells showed the most interesting death profiles and we decided to pursuit our investigation on B16, Colo857, HCT116 p53+/+, and HCT116 p53−/−.
Knowing that the Mov34 domain plays a crucial role in regulating the activity of eIF3f protein and its interaction with most identified partners,18 we engineered two eIF3f truncations: an N-terminal truncation (MD11-AN) lacking of the Mov34 domain and a C-terminal truncation containing the Mov34 domain (MD11-NF). Once incubated with target cells, MD11-NF effectively suppresses target cancer cells proliferation with a higher efficiency than the wild-type protein. In contrast, cells incubated with MD11-AN show unchanged cell proliferation. These results demonstrate that the N-terminal part of eIF3f is not implicated in apoptosis-inducing activity, and that the Mov34 domain plays a key role in regulating the activity of this subunit in cancer cells.
When transduced by MD11, fluorescently labeled wt, AN, NF, and K510R eIF3f proteins localize into cytoplasm or nucleus of selected cell lines. Our findings are in line with previous observations reporting that endogenous eIF3f can localize at both cytoplasm and nuclear levels.21 During apoptosis, endogenous eIF3f is phosphorylated by CDK11 which is mainly a nuclear protein21,22 and can assembly with other eIF3 subunits to form nuclear complexes which are involved in functions other than translation initiation, such as ribosome biogenesis that takes place into nucleus.22 The observation of nuclear localization together with the apoptotic effect observed by Annexin V assay strongly suggests that the exogenous and endogenous eIF3f are subjected to similar intracellular processing. It has been shown that in melanoma and pancreatic cancer cells eIF3f can induce apoptosis in caspase 3/7 and in a Bcl-2, Bax, or Bcl-XL–independent manner.10,13,14
An ideal protein-derived therapeutic drug should ideally be able to trigger apoptosis in cancer cells without inducing side effects in normal cells. Analyzing the results obtained with the recombinant eIF3f proteins, we can consider that MD11-NF, MD11-wt eIF3f, and MD11-K510R possess an interesting therapeutic potential for tumor treatment. As matter of fact MD11-K510R shows a high toxic effect on both normal and tumor cells, while MD11-wt eIF3f has lesser toxicity on tumor cells and no effect on normal cell viability.
Considering this MD11-wt eIF3f peculiar property, we can claim that this molecule is a possible and suitable candidate for melanoma and human colorectal carcinoma treatment. Some additional investigations need to be conducted for deciphering the detailed molecular mechanisms behind eIF3f action and its effects on cell viability before validating its therapeutic efficiency in in vivo models. Optimization of this protein-derived drug, including the fusion to tumor homing peptide sequences, will also be considered.
In conclusion, the work presented here establishes the first proof of concept on the use of MD11-based delivery system as an efficient tool for transporting functional protein cargos in living cells, and for developing protein therapy strategies for melanoma or colorectal carcinoma. This technology opens a broad range of possible applications; in fact the controlled intracellular protein delivery can be exploited for treating other malignancies or diseases by fusing the MD11 peptide sequence to other therapeutic target proteins.
Materials and Methods
Expression vectors of MD11-eIF3f fusion proteins
The coding sequence of full-length human eIF3-f was generated by PCR using the forward primer 5′-TAACGCCTCGAGGCCACACCGGCGGTACCAGTAAGTGCTCCTCCG-3′ and the reverse primer 5′-ATTCTTATGGATCCTTACAGGTTTACAAGTTTTTCATTG-3′, which contain at 5′ ends XhoI and BamHI restriction sites respectively. Two truncation mutants of murine eIF3f were generated: the mutant AN extending from nucleotide 1 to 91 and the mutant NF extending from nucleotide 91 to 361 using the following primers:
eIF3f 1–91 forward: 5′-TAACGCCTCGAGGCTTCTCCGGCCGTACCG-3′;
eIF3f 1–91 reverse: 5′-CGCGGATCCTCAGAAAGGCCCCGGGAGG-3′;
eIF3f 92–361 forward: 5′-TAACGCCTCGAGCCGGGCGGCCGCGTGG-3′;
eIF3f 92–391 reverse: 5′-CGCGGATCCTCACAGGTTTACAAGTTTCTCGT-3′.
The murine ubiquitination resistant mutant K5-10R sequence, previously described in ref. 19, was amplified using eIF3f1-91 forward primer and the reverse primer 5′-CGCGGATCCTCACAGGTTTACAAGTCTCTC-3′. Downstream fusion of the amplified fragments to ZEBRA-MD11 sequence was performed as described in ref. 20. After amplification, the DNAs of interest were digested with appropriate restriction enzymes and ligated into the optimized pET15b vector containing the sequence of ZEBRA MD11. All cloned plasmid sequences were verified by sequencing.
Expression and purification of recombinant fusion proteins
For optimum expression yields, the MD11-AN, MD11-NF, MD11-wt eIF3f, and MD11-K510R recombinant fusion proteins were expressed in E. coli after induction at OD600 nm = 0.8 with IPTG for 18 hours at 16 °C in the conditions given in Table 1.
Table 1. Optimized expression conditions used for recombinant protein production in E. coli system.
| Protein | E. coli bacterial strain | (IPTG) mmol/l | Additive |
|---|---|---|---|
| MD11-AN | BL21-CodonPlus (DE3)-RIPL (Stratagene) | 0.2 | 0.4 mol/l sucrose |
| MD11-NF | E. coli BL21 (DE3) (Novagen) | 0.5 | None |
| MD11-wt | BL21-CodonPlus (DE3)-RIPL | 0.5 | None |
| MD11-K510R | BL21-CodonPlus (DE3)-RIPL | 0.2 | 10 g/l glycerol |
Bacteria were then harvested by centrifugation at 10,000g for 10 minutes at 4 °C. After washing with PBS buffer, cell pellets were resuspended in 5 ml/g cell pellet of 20 mmol/l MOPS pH 7.0, 250 mmol/l NaCl, 5% glycerol, 5 mmol/l imidazole (lysis buffer) supplemented with complete protease inhibitor cocktail (Roche) and sonicated. The soluble and insoluble fractions (inclusion bodies, IBs) were separated by centrifugation at 13,000g for 30 minutes at 4 °C.
MD11-AN was purified from the soluble fraction onto a Ni-NTA column (Qiagen) using FPLC liquid chromatography system. Protein of interest was eluted in lysis buffer by a stepwise increase (at 100, 250, and 500 mmol/l) of imidazole content.
MD11-NF, MD11-wt eIF3f, and MD11-K510R were purified from inclusion bodies under denaturing conditions (insoluble fraction). As a first step of the purification procedure, IBs were washed in 20 mmol/l MOPS pH 7.0, 15 mmol/l cysteine, 2 mol/l NaCl for 1 hour at 4 °C and centrifuged at 14,000g for 30 minutes at 4 °C. Two additional washing steps were performed in 20 mmol/l MOPS, 15 mmol/l cysteine, 250 mmol/l NaCl, 2% Triton X-100, and in 20 mmol/l MOPS pH 7.0, 15 mmol/l cysteine, 250 mmol/l NaCl for 1 hour at 4 °C respectively. IBs were then solubilized for 18 hours at 4 °C in 20 mmol/l MOPS pH 7.0, 15 mmol/l cysteine, 250 mmol/l NaCl, 8 mol/l Urea (solubilization buffer). The samples were centrifuged at 14,000g for 30 minutes at 4 °C and the supernatants were applied to HisGraviTrap columns (GE Healthcare). Proteins of interest were eluted at room temperature in solubilization buffer at pH 6.5 by a stepwise increase (at 250 mmol/l, 500 mmol/l, and 1 mol/l) of imidazole content.
The following refolding steps were performed at 4 °C. Elution fractions containing purified proteins were dropwise diluted at concentrations ranging from 0.1–0.05 mg/ml in 50 mmol/l Na citrate pH 6.1, 2 mmol/l MgCl2, 4 mmol/l DTT, 50 mmol/l NaCl, 0.5 mol/l L-arginine, 0.5% Tween 20 and degassed with N2 for 20 minutes. After 24 hours of gentle stirring, the samples were concentrated to 5 ml using MWCO 10 kDa Amicon centrifugal filter (Millipore) and dialyzed twice against 50 mmol/l Na citrate pH 6.1, 2 mmol/l MgCl2, 4 mmol/l DTT, 50 mmol/l NaCl, 0.05 mol/l L-arginine, 0.01% N-lauroylsarcosine. Soluble refolded proteins were recovered by centrifugation at 17,000g for 10 minutes. Prior to their use for cellular uptake experiments, MD11-NF, MD11-wt eIF3f, and MD11-K510R purified proteins were dialyzed twice against 50 mmol/l Na citrate pH 6.1, 50 mmol/l NaCl, 10% glycerol and MD11-AN against 25 mmol/l Hepes/KOH pH 7.0, 50 mmol/l NaCl and 10% glycerol.
Proteins were separated onto a 12–15% SDS-polyacrylamide gel electrophoresis and analyzed by Coomassie blue staining or by Western-blotting using an anti-His tag antibody HRP-coupled (Sigma, 1:10,000 dilution) or using anti-eIF3f antibody (Rockland, 1:1,000 dilution) and an anti-rabbit HRP-labeled secondary (GE Healthcare). The yields of purified his-tagged proteins were quantified by BCA Protein Assay Kit according to the manufacturer’s instructions (Pierce).
Cell culture and treatments
Cells were cultured at 37 °C in a humidified 5% CO2 atmosphere incubator in media given in Table 2.
Table 2. List of used cell lines and corresponding growth media.
| Cell line | Medium |
|---|---|
| B16, GL26, HEK293E, HeLa | DMEM with stable glutamine and Na pyruvate (PAA, GE Healthcare), 10% FBS (PAA, GE Healthcare), 1% Penicillin/Streptomycin (Gibco) |
| C2C12 | DMEM with stable glutamine and Na pyruvate, 15% FBS, 1% Penicillin/Streptomycin |
| MCF7 | DMEM with stable glutamine and Na pyruvate, 10% FBS, 1% Penicillin/Streptomycin, 10 µg/ml insulin (Sigma), 0.1 mmol/l nonessential amino acids (Gibco) |
| MCF10-2A | F12 DMEM with stable glutamine and Na pyruvate (Gibco), 10% FBS, 10 µg/ml insulin, 0.5 µg/ml hydrocortisone (Gibco), 100 ng/ml cholera toxin (Sigma), 20 ng/ml epidermal growth factor (Sigma) and 1% Penicillin/Streptomycin |
| HCT116 p53+/+ and HCT116 p53−/− | McCoy’s with stable glutamine (PAA, GE Healthcare), 10% FBS, 1% Penicillin/Streptomycin (supplemented with 50 µg/ml G418 (Gibco) for p53−/− cells) |
| Saos-2 | McCoy’s with stable glutamine, 15% FBS, 1% Penicillin/Streptomycin |
| LN-229, T98G, U87, Colo857 | RPMI-glutamax (Gibco), 10% FBS, 1% Penicillin/Streptomycin |
| Raji | RPMI-glutamax, 10% FBS, 1 mmol/l Na pyruvate (Gibco), 0.1 mmol/l nonessential aminoacids, 10 µg/ml gentamicin (Gibco) |
| BJ | DMEM with stable glutamine and Na pyruvate, 10% FBS, 0.1 mmol/l nonessential aminoacids |
51Cr-release cytotoxicity assays
For cytotoxicity activity, 51Cr-release assay was used to screening protein-respondent cell lines and to determine time- and dose- dependent kinetics on selected cell lines. From 1 to 3 × 106 cells were harvested in complete medium (described above) and then incubated with 51Cr (Perkin Elmer) at 37 °C in 5% CO2 humidified air for 1 hour. Cells were washed with complete medium twice and reincubated at 37 °C for 1 hour. Cells were washed with serum-free medium and resuspended to a concentration of 1 × 105 cells per ml. Aliquots of 100 µl of labeled cells were loaded into 96-well plates in triplicate. Saponin (0.1%, Sigma) was added to determine cellular maximum 51Cr release and serum-free medium was used to determine the spontaneous release. Doxorubicin (1 µmol/l, Sigma) was used as toxicity control and protein-free buffers were tested to discard any buffer-linked toxicity.
Recombinant purified proteins were diluted to different concentrations in appropriate serum-free media and aliquots of 100 µl were added to cells. After 4 hours protein incubation, 10% FBS was added to cells. The 51Cr incorporated into the cell cytosol by diffusion is released into cell medium in case of protein induced membrane damage. For measurements of 51Cr release, 20 µl of cell-free supernatant were read in a gamma counter (Perkin Elmer).
For cell/line screening assay, 10 nmol/l MD11-wt eIF3f and MD11-K510R were tested and measurements were performed 0.5, 1, 2, 3, 4, 8, and 18 hours after protein incubation. For dose- and time- dependent study, 2.5, 5, 10, and 50 nmol/l of MD11-AN, MD11-NF, MD11-wt eIF3f, and MD11-K510R were used and measurements were collected at 0.5, 1, 2, 3, 4, 8, and 18 hours after protein incubations.
Percent specific cell death was calculated using the following formula: (x − SR)/(TR − SR) × 100, where x represents the amount of radioactivity released from target cells incubated with proteins, SR represents the amount of spontaneous release of radioactivity from the target cells (target cells incubated with serum-free medium), and TR represents the maximum amount of radioactivity released when target cells are lysed with saponin.
Protein labeling and intracellular detection
To track the cellular internalization of MD11-AN, MD11-NF, MD11-wt eIF3f, and MD11-K510R, fusion proteins were fluorescently labeled using Alexa Fluor 488 Protein Labeling Kit (Molecular Probes, Invitrogen) according to manufacturer’s instructions. For microscopy analysis, B16, Colo857, HCT116 p53+/+, and HCT116 p53−/− cells (7 × 104 cells/well) were seeded onto an eight-well Lab-Tek chambered coverglass (Nunc) in complete cell culture media 24 hours before treatment. After removal of the medium, the cell layers were rinsed twice with DPBS and subsequently exposed to 10 nmol/l of AlexaFluor 488 labeled-recombinant proteins in fresh serum-free media at 37 °C. Proteins were incubated with B16 cells for 0.5 hour, with Colo857 for 1 hour, and with HCT116 p53+/+ and HCT116 p53−/− for 2 hours. Thereafter, the incubation solutions were removed and the cells were washed three times with DPBS. Living cell preparations were observed with a LSM 710 confocal scanning laser microscope (Carl Zeiss, Jena, Germany), using a 63×, NA 1.2, C-apochromat water-immersion objective (Carl Zeiss). The experiment was carried out at 488 nm excitation and fluorescence was collected with a 510–560 nm filter. 16 successive optical slices were captured along the cell z-axis, with a step of 1 μm.
Cellular uptake FACS analysis
To analyze the transduction efficiency and phosphatidyl serine (PS) exposure in response to recombinant protein treatment, a FACS analysis was performed. B16, Colo857, HCT116 p53+/+, and HCT116 p53−/− cells (1 × 105 cells/well) were cultured on 12-well plates until 70% of confluence. Cells were washed twice with DPBS and incubated with 10 nmol/l of AlexaFluor 488 labeled-recombinant proteins in fresh serum-free culture medium for 2 hours at 37 °C. As 488 nm fluorescence positive control, recombinant proteins containing the MD11 peptide fused to a non toxic eGFP reporter protein (MD11-eGFP, 0.3 µmol/l) were incubated for 2 hours with cells. As toxicity control, doxorubicin (1 µmol/l) was incubated with cells 24 hours before analysis. After incubation, cells were trypsinized with 0.5% trypsin/EDTA solution (PAA, GE Healthcare) for 10 minutes at 37 °C. The trypsin was then neutralized by adding complete medium, cells were washed twice with DPBS and then stained with Annexin V-APC and the vital dye 7-Amino-actinomycin (7–AAD, BD Biosciences). Cell-associated fluorescence was detected using a FACS Canto II BD Biosciences flow cytometer (equipped with three lasers). The 488 nm and 633 nm excitation and the 647 nm emission filters (for 7AAD) and the 660/20 (for Annexin V-APC) were used. The cytometer parameters were performed using the seven color set-up beads (BD) and the compensations were calculated with the Set-up compbeads kit from BD using antibodies for conjugates and cells for viability dye. The results are reported as the percent of fluorescent cells from living cells gate of 30,000 events recorded and analyzed with the FACSDiva software.
Acknowledgments
The authors thank Antimo Di Maro for his help in protein purification; Caroline Aspord for generously providing Colo857 cells, Joel Eyer for U87 and T98G cells and Pierre Champelovierre for LN229 cells.
References
- Yan M, Du J, Gu Z, Liang M, Hu Y, Zhang W. A novel intracellular protein delivery platform based on single-protein nanocapsules. Nat Nanotechnol. 2010;5:48–53. doi: 10.1038/nnano.2009.341. [DOI] [PubMed] [Google Scholar]
- Langel U.2006Handbook of Cell-Penetrating Peptides, 2nd edition CRC Press/Taylor & Francis Ed.Boca Raton, FL. [Google Scholar]
- Dietz GP, Bähr M. Delivery of bioactive molecules into the cell: the Trojan horse approach. Mol Cell Neurosci. 2004;27:85–131. doi: 10.1016/j.mcn.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Rothe R, Liguori L, Villegas-Mendez A, Marques B, Grunwald D, Drouet E. Characterization of the cell-penetrating properties of the Epstein-Barr virus ZEBRA trans-activator. J Biol Chem. 2010;285:20224–20233. doi: 10.1074/jbc.M110.101550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilka R, Ernst C, Mehta AK, Haybaeck J. Eukaryotic translation initiation factors in cancer development and progression. Cancer Lett. 2013;340:9–21. doi: 10.1016/j.canlet.2013.06.019. [DOI] [PubMed] [Google Scholar]
- Dong Z, Zhang JT. Initiation factor eIF3 and regulation of mRNA translation, cell growth, and cancer. Crit Rev Oncol Hematol. 2006;59:169–180. doi: 10.1016/j.critrevonc.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Zhang L, Pan X, Hershey JW. Individual overexpression of five subunits of human translation initiation factor eIF3 promotes malignant transformation of immortal fibroblast cells. J Biol Chem. 2007;282:5790–5800. doi: 10.1074/jbc.M606284200. [DOI] [PubMed] [Google Scholar]
- Spilka R, Ernst C, Bergler H, Rainer J, Flechsig S, Vogetseder A. eIF3a is over-expressed in urinary bladder cancer and influences its phenotype independent of translation initiation. Cell Oncol (Dordr) 2014;37:253–267. doi: 10.1007/s13402-014-0181-9. [DOI] [PubMed] [Google Scholar]
- Haybaeck J, O’Connor T, Spilka R, Spizzo G, Ensinger Ch, Mikuz G. Overexpression of p150, a part of the large subunit of the eukaryotic translation initiation factor 3, in colon cancer. Anticancer Res. 2010;30:1047–1055. [PubMed] [Google Scholar]
- Shi J, Kahle A, Hershey JW, Honchak BM, Warneke JA, Leong SP. Decreased expression of eukaryotic initiation factor 3f deregulates translation and apoptosis in tumor cells. Oncogene. 2006;25:4923–4936. doi: 10.1038/sj.onc.1209495. [DOI] [PubMed] [Google Scholar]
- Hofmann K, Bucher P. The PCI domain: a common theme in three multiprotein complexes. Trends Biochem Sci. 1998;23:204–205. doi: 10.1016/s0968-0004(98)01217-1. [DOI] [PubMed] [Google Scholar]
- Wen F, Zhou R, Shen A, Choi A, Uribe D, Shi J. The tumor suppressive role of eIF3f and its function in translation inhibition and rRNA degradation. PLoS ONE. 2012;7:e34194. doi: 10.1371/journal.pone.0034194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doldan A, Chandramouli A, Shanas R, Bhattacharyya A, Cunningham JT, Nelson MA. Loss of the eukaryotic initiation factor 3f in pancreatic cancer. Mol Carcinog. 2008;47:235–244. doi: 10.1002/mc.20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doldan A, Chandramouli A, Shanas R, Bhattacharyya A, Leong SP, Nelson MA. Loss of the eukaryotic initiation factor 3f in melanoma. Mol Carcinog. 2008;47:806–813. doi: 10.1002/mc.20436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csibi A, Tintignac LA, Leibovitch MP, Leibovitch SA. eIF3-f function in skeletal muscles: to stand at the crossroads of atrophy and hypertrophy. Cell Cycle. 2008;7:1698–1701. doi: 10.4161/cc.7.12.6090. [DOI] [PubMed] [Google Scholar]
- Lagirand-Cantaloube J, Offner N, Csibi A, Leibovitch MP, Batonnet-Pichon S, Tintignac LA. The initiation factor eIF3-f is a major target for atrogin1/MAFbx function in skeletal muscle atrophy. EMBO J. 2008;27:1266–1276. doi: 10.1038/emboj.2008.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csibi A, Cornille K, Leibovitch MP, Poupon A, Tintignac LA, Sanchez AM. The translation regulatory subunit eIF3f controls the kinase-dependent mTOR signaling required for muscle differentiation and hypertrophy in mouse. PLoS ONE. 2010;5:e8994. doi: 10.1371/journal.pone.0008994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchione R, Leibovitch SA, Lenormand JL. The translational factor eIF3f: the ambivalent eIF3 subunit. Cell Mol Life Sci. 2013;70:3603–3616. doi: 10.1007/s00018-013-1263-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csibi A, Leibovitch MP, Cornille K, Tintignac LA, Leibovitch SA. MAFbx/Atrogin-1 controls the activity of the initiation factor eIF3-f in skeletal muscle atrophy by targeting multiple C-terminal lysines. J Biol Chem. 2009;284:4413–4421. doi: 10.1074/jbc.M807641200. [DOI] [PubMed] [Google Scholar]
- Rothe R, Lenormand JL. Expression and purification of ZEBRA fusion proteins and applications for the delivery of macromolecules into mammalian cells. Curr Protoc Protein Sci. 2008;Chapter 18:Unit 18.11. doi: 10.1002/0471140864.ps1811s54. [DOI] [PubMed] [Google Scholar]
- Shi J, Feng Y, Goulet AC, Vaillancourt RR, Sachs NA, Hershey JW. The p34cdc2-related cyclin-dependent kinase 11 interacts with the p47 subunit of eukaryotic initiation factor 3 during apoptosis. J Biol Chem. 2003;278:5062–5071. doi: 10.1074/jbc.M206427200. [DOI] [PubMed] [Google Scholar]
- Shi J, Hershey JW, Nelson MA. Phosphorylation of the eukaryotic initiation factor 3f by cyclin-dependent kinase 11 during apoptosis. FEBS Lett. 2009;583:971–977. doi: 10.1016/j.febslet.2009.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.