Abstract
Human embryonic stem cells (hESC) and induced pluripotent stem cells (hiPSC) assert a great future for the cardiovascular diseases, both to study them and to explore therapies. However, a comprehensive assessment of the viral vectors used to modify these cells is lacking. In this study, we aimed to compare the transduction efficiency of recombinant adeno-associated vectors (AAV), adenoviruses and lentiviral vectors in hESC, hiPSC, and the derived cardiomyocytes. In undifferentiated cells, adenoviral and lentiviral vectors were superior, whereas in differentiated cells AAV surpassed at least lentiviral vectors. We also tested four AAV serotypes, 1, 2, 6, and 9, of which 2 and 6 were superior in their transduction efficiency. Interestingly, we observed that AAVs severely diminished the viability of undifferentiated cells, an effect mediated by induction of cell cycle arrest genes and apoptosis. Furthermore, we show that the transduction efficiency of the different viral vectors correlates with the abundance of their respective receptors. Finally, adenoviral delivery of the calcium-transporting ATPase SERCA2a to hESC and hiPSC-derived cardiomyocytes successfully resulted in faster calcium reuptake. In conclusion, adenoviral vectors prove to be efficient for both differentiated and undifferentiated lines, whereas lentiviral vectors are more applicable to undifferentiated cells and AAVs to differentiated cells.
Introduction
Pluripotent stem cells (PSCs) have the ability to self-renew and to differentiate to most cell types (pluripotency), including to cardiac myocytes. Human embryonic stem cells (hESC) are pluripotent cell lines derived from the inner cell mass of human blastocysts.1 A few years ago, the group of Dr. Yamanaka published ground-breaking studies in which they showed that pluripotency can be induced in somatic cells, such as dermal fibroblasts, from both mouse2 and most importantly human origin.3 The differentiation of these pluripotent stem cells to cardiac myocytes is of great importance, as it provides a patient-specific disease model system to test cardioactive compounds and may also provide an unlimited source of cells for cell replacement–based therapies for the injured heart. The differentiation of hESCs to cardiac myocytes (CM) was first reported more than 10 years ago and relied on the spontaneous differentiation of hESCs that were dissociated, developed into spheroids, termed embryoid bodies (EBs) and grown in suspension.4 Since the first reports of spontaneous differentiation, several protocols have been established for the directed differentiation of not only hESC, but also hiPSC, into the cardiac lineage. Most of these induced differentiation protocols rely on the timely addition of specific factors, including activators of the BMP and nodal/activin signaling pathways (BMP4, Activin A and FGF2 (ref. 5)) and inhibitors of the Wnt/b-catenin pathway (DKK1 and IWR1 (ref. 6)), to transition the cell fate from pluripotent to mesodermal, precardiac and then cardiac mesodermal, and finally to immature cardiac myocytes.4 These PSC-CMs are immature structurally and functionally, but offer a valuable tool nonetheless for pharmacological and genetic studies.
The potential for the use of hESC and hiPSC and thereof derived cardiac myocytes in research and therapy is well established in the literature. Identifying the viral vectors that can be used to genetically manipulate the hESC/hiPSC system is necessary because it will not only provide valuable information regarding therapeutic applications but also a level of consistency among different research groups in order to interpret data. Three types of viral vectors are widely used not only in in vitro but also in in vivo studies in the cardiovascular field: adeno-associated viral (AAV), adenoviral, and lentiviral vectors.
Of the three, AAV-based vectors are a new promising tool for gene therapy.7 AAV are members of the family of Parvoviridae and are nonenveloped, single-stranded DNA viruses that in their native form, at least AAV2, can site-specifically integrate into the genome. The recombinant AAVs however are predominately episomal. There are 13 reported serotypes and each one displays a distinct tropism profile. AAVs 1, 6, and 9 are the most cardiotropic, whereas AAV2 is the archetypical serotype, whose biology has been the most extensively studied. In AAV vectors, the 4.7 kb viral genome between the inverted terminal repeats (ITRs) is replaced by the therapeutic expression cassette.7,8 AAV-based vectors have arisen in the past decade as a potent gene therapy vector owing to their tropism for specific tissues, their low immunogenicity, their lack of pathogenicity, and their ability to sustain long-term expression. As a testament to their potential, several clinical trials are underway using AAVs for cardiac gene therapy7 and, recently, phase 2 of the CUPID clinical trial was successfully completed. Importantly, AAV1 is the vector used in the first approved gene therapy (Glybera) which is currently being made commercially available.9
Adenoviruses are nonenveloped, double-stranded DNA viruses whose genomic DNA remains episomal after infection and triggers transient expression of the transgene. They are mostly associated with infections of the respiratory system. Of the more than 50 identified serotypes, serotypes 2 and 5 are the most commonly used.10 The adenoviral genome is 36 kb in size. In adenoviral vectors, part of the viral genome (first and second generation), or all of it (third generation or gutless), is replaced by the therapeutic expression cassette. The gutless adenoviral vectors were generated, because the expression of adenoviral genes after in vivo transduction leads to strong cellular immune responses against the vector infected cells. However, so far, the most commonly used adenoviral vectors are the first-generation ones, owing to their simplicity. They are missing one adenoviral gene, E1, which is supplied by the packaging cell line. This renders them replication-deficient, and therefore safe.10 Adenoviral-based vectors are widely used in clinical trials, but their usage has seen a significant decrease due to safety issues stemming from strong immune reactions as opposed to the use of AAVs.7 In vitro, adenoviral vectors still achieve significant expression, albeit the expression of adenoviral genes present in the viral vectors could be a source of concern and complexity in interpreting data.10
Lentiviruses belong to the family of Retroviridae, are single-stranded RNA viruses and integrate into the host cell genome after reverse transcription of their viral genome, which is flanked by ITRs. They can infect nondividing cells, such as the terminally differentiated cardiac myocytes, efficiently, which makes them potent gene therapy vectors.7,8 Most lentiviral vectors are based on human immunodeficiency virus. For the recent generation of replication defective vectors, most of the lentiviral genome, which is 7.5 kb in size, is replaced by the gene expression cassette, which is flanked by the long terminal repeats. They have already been used in in vivo models for cardiac disease.8,11 In vitro, they are used predominately when stable expression is needed in dividing cells, such as hiPSC,3 because their integration to the host genome ascertains their long-term expression, which cannot be achieved with episomal vectors. Finally, because the natural tropism of lentiviruses is restricted to CD4+ cells, the vectors are pseudotyped through the incorporation of glycoproteins, which extends their host range.12
The use of viral vectors for the genetic manipulation of PSCs is quite extensive, with most prominent the use of lentiviral vectors carrying the pluripotency factors on skin fibroblasts for the generation of PSC.13 Independently, adeno-associated viral, adenoviral and lentiviral vectors have been used,13 but to the best of our knowledge, no systematic comparison of these three major groups has ever been conducted neither in PSCs, nor in thereof derived cardiac myocytes. Having all the aforementioned information in mind, we set out to compare the major viral vectors currently used in gene therapy applications, AAV, adenoviral and lentiviral vectors using different undifferentiated cells, both hES and hiPS (hES2, H9, hiPS31.3, hiPS24.1). We also included in our studies hES (hES2) and hiPS (hiPS31.3) cells differentiated into cardiomyocytes. As a first step, we decided to compare four different AAV serotypes, the cardiotropic AAV serotypes 1, 6, and 9 and the archetypical serotype 2. We found that AAV serotypes 2 and 6 were superior both in PSCs and in PSC-CMs. We next sought to compare these two AAV serotypes to adenoviral and lentiviral vectors. Overall, adenoviral vectors proved efficient in both cells types, AAVs in differentiated cells, that is PSC-CM, whereas lentiviral vectors transduced all cell types with low to moderate efficiency (Table 1). It is noteworthy that infection of PSC with AAVs results in cell death. Finally, adenoviraly expressed Serca2a in PSC-CM, improved functional parameters, thereby validating the use of adenoviruses at least, the most potent vectors in our study, for the genetic manipulation of PSC-CMs.
Table 1. Summary of transduction efficiencies.
|
Vectors |
|||||||
|---|---|---|---|---|---|---|---|
| AAV1 | AAV2 | AAV6 | AAV9 | Adenoviral | Lentiviral | ||
| hESC | hES2C | 0a | 0a | 0a | 0a | ++ | + |
| H9C | + | +++ | ++ | + | ++ | + | |
| hiPSC | iPS31.3C | 0a | 0a | 0a | 0a | + | + |
| iPS24.1C | 0a | 0a | +a | 0a | + | + | |
| hESC-CMs | hES2C-CMs | ++ | +++ | +++ | + | +++ | ++ |
| hiPSC-CMs | hiPSC-CMs | ++ | +++ | +++ | + | ++ | + |
Extensive cell death.
Results
Comparison of AAV serotypes 1, 2, 6, and 9 in PSCs and PSC-CMs
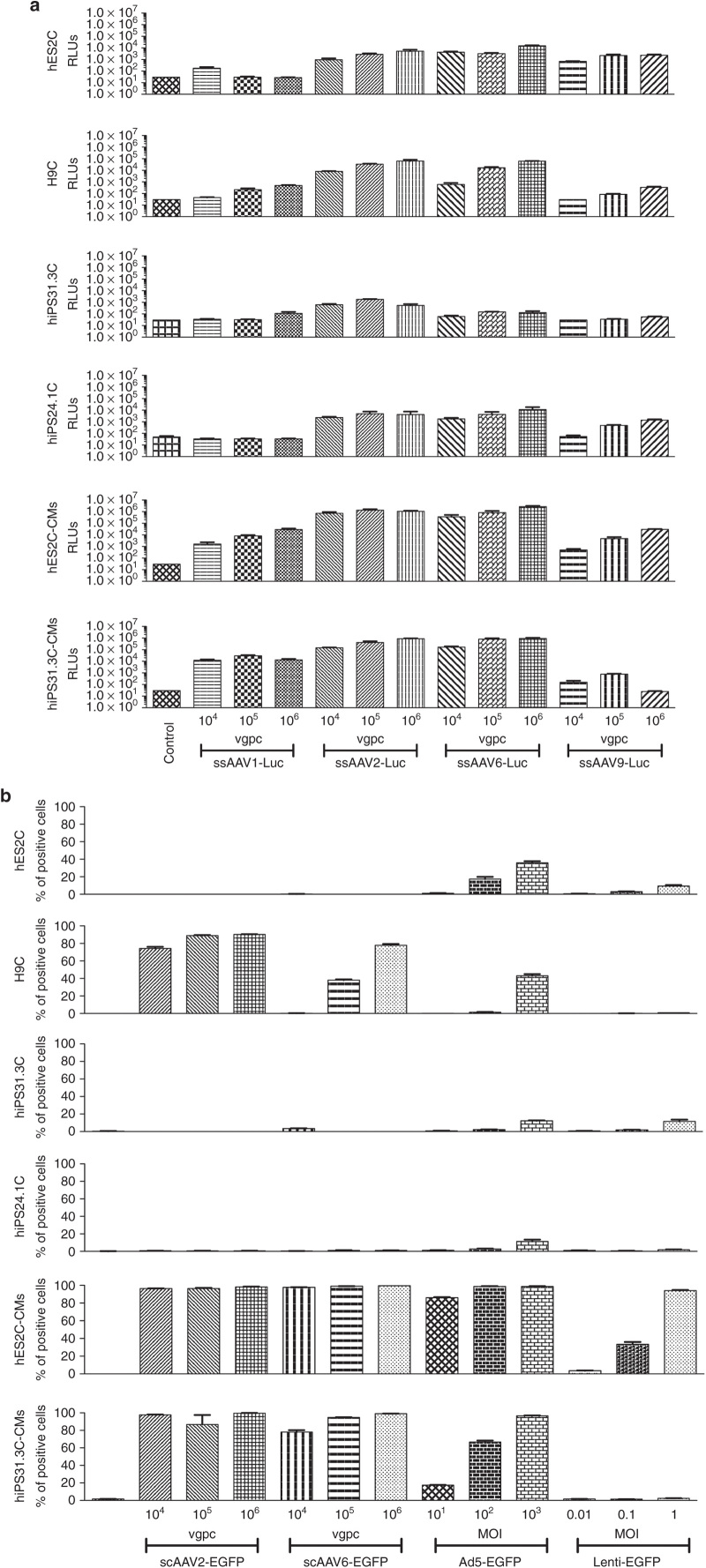
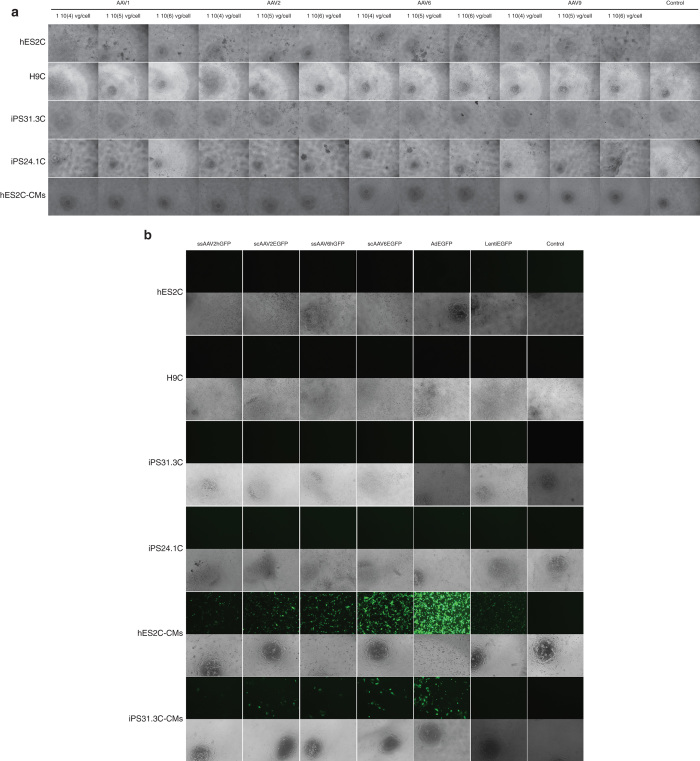
As a first step in our study, we set out to compare three known cardiotropic AAV serotypes 1, 6, and 9 and the archetypical serotype 2 (ref. 8), in PSCs and PSC-CMs. For this part of the study, we used luciferase as a reporter system, as it was anticipated that the serotypes would display several orders of magnitude of difference in efficiency and during this preliminary investigation, the purpose was to select the most efficient vector for further studies without focusing on the number of cells transduced or the individual cell transduction levels. We tested the AAV vectors in two hESC lines (hES2, H9), two hiPSC (hiPS31.3, hiPS24.1) lines and cardiomyocytes derived from one hESC (hES2), and one hiPSC (hiPS31.3) line. Three different amounts of viral genomes (vg) per cell were used, each 10-fold apart from the other based on our previous experience, to cover a broad range of infectivity and achieve at the higher dosages saturation. For all AAV serotypes, we used single-stranded vectors. Figure 1a shows that ssAAV2Luc and ssAAV6Luc exhibit the highest transduction efficiency compared to AAV1 (moderate transduction efficiency) and AAV9 (lowest transduction efficiency). It is noteworthy that all AAVs transduce differentiated cells more efficiently, approximately 100-fold, compared to undifferentiated cells. During these experiments, we observed extensive cell death in most undifferentiated cell lines (hES2, hiPS31.3, and hiPS24.1), with the exception of H9. No such effect was observed in the differentiated cells (Figure 2a).
Figure 1.
Transduction efficiency by adeno-associated virus (AAV), adenoviral, and lentiviral vectors of different human embryonic stem cell (hES2, H9), human induced pluripotent stem cell (hiPS31.3, hiPS24.1) lines, and hES2C- and hiPS31.3C-derived cardiomyocytes (hES2C-CMs and hiPS31.3C-CMs). (a) Luciferase assay was used to assess the transduction efficiency by four AAV serotypes (the cardiotropic serotypes 1, 6, and 9 and the archetypical serotype 2) of different pluripotent cell lines (hES2, H9, hiPS31.3, hiPS24.1) and cardiomyocytes derived from two pluripotent cell lines (hES2, hiPS31.3). AAV serotypes 2 and 6 appear to be the most efficient ones in transducing all cell types, with AAV2 showing higher efficiency in undifferentiated cells. In differentiated cells however, AAV6 is superior. In particular, in hES2C-derived cardiomyocytes, AAV2 is more efficient in lower concentrations (vg/cell), whereas at higher concentrations it saturates and AAV6 outperforms AAV2. Error bars depict ± standard error mean (SEM). (b) Flow cytometric analysis (% of green fluorescent protein (GFP)-positive cells) was used to assess the transduction efficiency by AAVs 2 and 6 (carrying self-complementary genomes), adenoviral (serotype 5) and lentiviral vectors. In undifferentiated cells, adenoviral and lentiviral vectors are superior to AAVs, whereas in differentiated cells (cardiomyocytes), the transduction efficiency of AAVs and in particular AAV6, is enhanced. Error bars depict ± SEM. Groups were analyzed by analysis of variance test. Please see Supplementary Table S1 (Luciferase, Figure 1a) and Supplementary Table S2 (Flow Cytometry, Figure 1b) for detailed statistical analysis. Error bars depict ± SEM.
Figure 2.
Representative images of different pluripotent (hES2, H7, hiPS31.3, hiPS24.1) and differentiated (hES2-CM and hiPS31.3-CM) cells transduced with adeno-associated virus (AAV), adenoviral, and lentiviral vectors. (a) Representative images of hES2, H7, hiPS31.3, hiPS24.1 cells, hES2-CM, and hiPS31.3-CM infected with ssAAV vectors carrying the luciferase gene. ssAAV serotypes 2 and 6, and highest concentrations of serotypes 1 and 9 induced cell death in hES2 and iPS31.3 and iPS24.1 cells. AAVs did not appear to affect the viability of differentiated cells. (b) Representative images of hES2, H7, hiPS31.3, hiPS24.1 cells, hES2C-CMs, and hiPS31.3C-CMs infected with scAAVs 2 and 6, adenoviral and lentiviral vectors carrying the GFP gene. Images of the infections using the highest concentration of virus per cell are shown. High levels of AAVs reduced the survival of hES2 and hiPS31.3 cells. No cell death was observed in hES2C- and hiPS31.3C-derived cardiomyocytes, which also exhibited high levels of transduction compared to their undifferentiated parental cell lines.
Comparison of AAV, adenoviral, and lentiviral vectors infectivity of PSCs and PSC-CMs
Once we determined that AAV2 and AAV6 vectors provide the strongest transduction efficiency, we sought to compare them to two other, commonly used, viral vectors: adenoviral and lentiviral. For this set of experiments, we also included AAV vectors of both serotypes containing self-complementary (sc) viral genomes, because they have been shown to exert higher transduction levels compared to single-stranded (ss) genomes.14,15 We also elected to determine the number of cells transduced in the populations, as well as the individual cell expression levels. For this reason, we chose to use the enhanced green fluorescent protein (EGFP) as a reporter gene and assess viral transduction efficiency by flow cytometry. The same cell lines and types as before were tested. A general observation stemming from the flow cytometry measurements (Figures 1b and 2b and Supplementary Figure S1a,b) is that none of the viral vectors tested transduces undifferentiated cells efficiently, compared to cells differentiated into cardiomyocytes. In general, undifferentiated cells, both hESC and hiPSC, were infected most efficiently by adenoviral vectors followed by lentiviral vectors. On the other hand, PSC-CM both hESC-CMs and hiPSC-CMs, were transduced most efficiently by AAV vectors in all the conditions tested, even when lower infection titers were used, compared to adenoviral and lentiviral vectors. In particular, AAV vectors of both serotypes were largely inefficient to infect PSCs, with the exception of the H9 line. It is of note that extensive cell death was observed in the PSC populations infected with AAVs. Contrastingly, hPSC-CMs and in particular hES2-CMs were infected by AAVs at 100% efficiency even at low viral genome (vg) per cell. Adenoviral vectors were efficient across all cell types and lines tested, exhibiting higher transduction efficiency in hPSC-CMs and in particular in hES2C-CMs, where infection at all multiplicity of infections (MOIs) showed almost complete transduction. Lentiviral vectors were moderately efficient in hPSC, compared to adenoviral vectors, but more efficient in hPSC-CMs, especially in hES2C-CMs, where they reached 90–95% transduction efficiency, but only at the higher MOIs.
To better understand the extent of the transduction efficiency of the different serotypes, we measured the expression index (% of positive cells × mean fluorescence of infected to noninfected cells)16 in the flow cytometry experiments. This index incorporates not only the factor of positive cell number, but also the transduction level for each cell through the expression levels of the reporter gene. In particular, infection of hES2-CMs with 1 × 106 vg per cell of scAAV6EGFP is equal to the higher adenoviral MOI. In hiPS31.3-CMs both self-complementary AAV serotypes tested, 2 and 6, outperformed adenoviral and lentiviral vectors (Figure 2b; Supplementary Figure S1a). It seems thus, that the transduction hindrance in PScs is higher for AAV vectors compared to others. We also sought to determine the transduction efficiency of single stranded AAV vectors (Supplementary Figure S1b). For these experiments, we used vectors carrying the humanized GFP (hGFP). The transduction efficiency pattern of ssAAVs was in line with that of scAAVs. They transduced differentiated cells efficiently, whereas they induced cell death in pluripotent cells, albeit to a lesser extent. The transduction efficiency and cell death induction efficiency was much lower for ssAAVs, compared to scAAVs. These vectors have not only different genomes (sc versus ss), but also different expression cassettes and transgenes (EGFP versus hGFP). To test for the contribution of the expression cassette, we performed transfection experiments using the respective plasmids in two cell lines (293T and HELA), taking into consideration the fact that hPSCs are almost refractory to transfection (data not shown). We did not observe any major differences that could fully account for the robust differences in transduction efficiency.
Characterization of cell death observed in PSCs after infection with AAV vectors
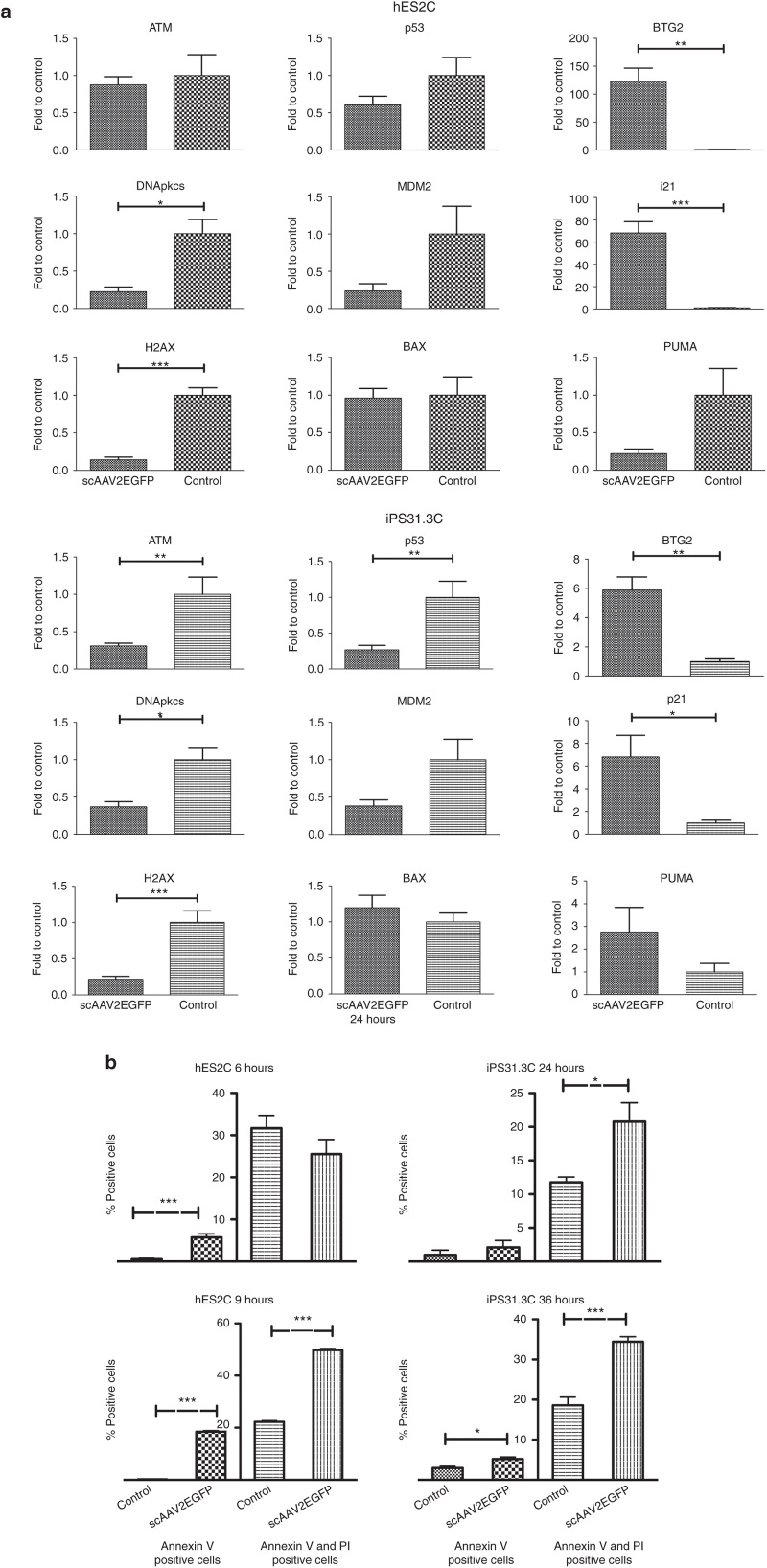
Adenoviral and lentiviral vectors, as mentioned, transduce pluripotent cells with moderate to high efficiency (Table 1). Infection of these cells by AAV vectors, on the other hand, resulted in pronounced cell death within one or 2 days postinfection (Figure 2a,b). As a first step to characterize the induction of cell death by AAV infection, we assessed the effect of all viral vectors on two cell lines, hES2 and hiPS31.3, at 12 hours postinfection (hpi) using the highest infectivity ratios for adenoviral and lentiviral and the lowest for AAV vectors (Supplementary Figure S2), because no cell death was evident in the former and pronounced cell death in the latter in our initial experiments. The time point of 12 hours was based on our observation that cell death was first evident 12 hours postinfection. The time point chosen for the transduction studies, at 3 days postinfection, was deemed too progressed for this type of study based on initial experiments (data not shown) and visual observation of the cell culture. We sought to test the expression of tumor protein p53 (TP53), which is known to be involved in viral DNA induced apoptosis in embryonic stem cells.17 We also assessed several TP53 target genes. B-cell translocation gene family member 2 (BTG2) and cyclin-dependent kinase inhibitor 1A (CDKN1A) (or p21) are involved in cell cycle arrest at G1, that are known to be upregulated upon AAV infection.17 We also tested expression of the mouse double minute 2, also known as E3 ubiquitin protein ligase (MDM2), a negative regulator of TP53. Furthermore, we examined genes upstream of TP53 that are involved in the DNA damage response (DDR) pathway, such as ataxia telangiectasia mutated (ATM), protein kinase, DNA-activated catalytic polypeptide (PRKDC) and H2A histone family member X (H2AFX), and its downstream transcriptional targets involved in apoptosis induction, such as BCL2-associated X protein (BAX), BCL2 binding component 3 (BBC3 or PUMA) and NADPH oxidase activator 1 (NOXA1).18 Treatment with hydroxyurea (HU), a known DDR pathway inducer, was used as a positive control.19
Of all viral vectors tested on hES2, only AAV vectors (serotypes 2 and 6) and in particular the self-complementary AAV2, were able to induce cell cycle arrest, as indicated by the significant upregulation of CDKN1A (p21), a known TP53 target. BTG2, another cell cycle arrest gene and TP53 target,20 was significantly upregulated upon scAAV2 infection only, although it was increased, albeit nonsignificantly, by scAAV6 (Supplementary Figure S2). MDM2 was significantly downregulated upon infection with scAAV2, ssAAV2 and scAAV6. The expression of several DDR pathway genes, such as ATM, PRKDC, and TP53 (ref. 18) remained stable. Two TP53 translational targets involved in apoptosis, BAX and PUMA, along with two other TP53 targets, NOXA1 and TP53AIP1, were also unaltered (data not shown). Somewhat similar results were observed in hiPS31.3C, although the effect was much more mild and never reached significance (Supplementary Figure S2).
To explore this pathway further, we decided to test the most efficient vector in inducing the DDR pathway, scAAV2, and increase the viral genomes used per cell to 5 × 104 vg/cell), using the same cell lines. We also chose two time-points (6 hours for hES2C and 24 hours for hiPS31.3C), which we presumed to have high DDR induction, because between those time point we observed the first indication of cell death, based on visual observation and initial experiments (data not shown). Later time points could not be tested, as cell death was extensive beyond that point. As shown in Figure 3a, CDKN1A (p21) and BTG2 were robustly induced in hES2 cells and significantly upregulated in hiPS31.3. We also observed a moderate downregulation of DDR pathway genes but no significant difference in apoptotic genes. AAV vectors consistently induced cell death, however we were not able to detect significant upregulation of known p53 targets (BAX, NOXA1, PUMA (BBC3)). Of note, PUMA appears to decrease in hES2 cells and increase in hiPS31.3 cells upon infection with scAAV2. However, neither of these changes reached statistical significance.
Figure 3.
Induction of DNA damage response pathways and apoptosis by adeno-associated virus (AAVs) in undifferentiated cell lines. (a) Induction of DDR, p53, and cell cycle arrest pathways by scAAV2 (5 × 104 vg per cell) assessment by quantitative polymerase chain reaction. For these experiments, the viral vector with the highest effect, scAAV2, was used at a dose of 5 × 104 vg per cell. The expression of key genes of the DDR, p53, and cell cycle arrest pathways was examined at the cDNA level. In particular, hES2C cells showed robust upregulation of BTG2 and p21 6 hours postinfection, whereas hiPS31.3C showed a significant increase in these two genes at 24 hours postinfection. Earlier time points for both cell lines showed no significant differences (data not shown). Some of the DDR and p53 pathway genes showed significant decrease in these time points (data not shown). Error bars depict ± standard error mean (SEM) (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001). (b) Annexin V staining and flow cytometric analysis of hES2C and hiPS31.3C infected with scAAV2. hES2C at 6 and 9 hours postinfection (hpi) and hiPS31.3C 36 hpi with scAAV2 (1 × 105 vg per cell) showed a significant increase, compared to no infection control, of Annexin V positive cells (early apoptosis), as well as of cells positive for both Annexin V and propidium iodide (PI) (late apoptosis). At later time points for hES2 (9 hpi) and at both time points for hiPS31.3 (24 hours and 36 hpi) cells were significantly positive for either Annexin V alone or Annexin V and PI. Error bars depict ± SEM (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001).
We next sought to determine if the observed cell death was apoptotic using Annexin V staining. scAAV2 induced apoptosis in both hES2 and hiPS31.3 cells (Figure 3b). In particular, at 6 hpi, a small population of hES2C underwent early apoptosis, whereas at 9 hpi both early and late apoptosis were significantly increased. Similar results were observed for hiPS31.3C, but at 24 hpi and 36 hpi respectively. It is noteworthy that hiPS31.3 cells have higher basal levels of early apoptosis (between 5 and 6%) compared to hES2 cells (less than 0.5%), as exhibited by the noninfected control at the time points tested. Overall, AAV vectors consistently upregulated cell cycle arrest genes (BTG2, CDKN1A (p21)) and induced cell death in both cell types. Importantly, only a small fraction of cells undergo apoptosis at a particular time as shown in Figure3b, an occurrence more pronounced in hiPSC.
Identification of the abundance of AAV and adenoviral receptors in PSCs and PSC-CMs
In order to investigate the cause of the different viral transduction efficiencies, the abundance of identified receptors for adenoviruses and some of the AAV serotypes was explored in hES2Cs, hES2-CMS, hiPS31.3Cs, and hiPS31.3-CMs. Sialic acids that are present on N-linked glycoproteins act as primary receptors for AAV serotypes 1 and 6 (ref. 21). N-terminal galactose acts as a primary receptor for AAV9 (refs. 22,23). Staining with wheat germ agglutinin lectin that binds to sialic acid shows that the AAV1/6 receptor is abundant in differentiated cells, hES2C-CMs and hiPS31.3C-CMs, compared to pluripotent cells, hES2C and hiPS31.1C (Supplementary Figure S3). Similarly, the AAV9 receptor, identified using another lectin, Erythrina cristagalli lectin (ECL), was abundant in differentiated cells, and undetectable in pluripotent cells (Supplementary Figure S3a). These observations are in accordance with the transduction efficiency differences observed between the cell types. The coxsackie virus and adenovirus receptor, CXADR, distribution was examined by quantitative polymerase chain reaction (qPCR) using primers in the conserved region of CXADR. As shown, it is highly expressed in pluripotent cells, and moderately less in differentiated cells (Supplementary Figure S4). Next, we examined the presence of the cell surface expression of CXADR, Integrin α1β3 (a different adenovirus receptor10), and Heparan Sulfate ProteoGlycan (HSPG) (an AAV2 primary receptor24) in hES2, iPS31.3, and HeLa cells, typically used in viral receptor binding and endocytosis studies.10,24 We found that hPScs express similar levels of all the receptors compared to HeLa cells (Supplementary Figure S5).
Adenoviral overexpression of Serca2a in hES and hiPS derived cardiomyocytes improves calcium cycling parameters
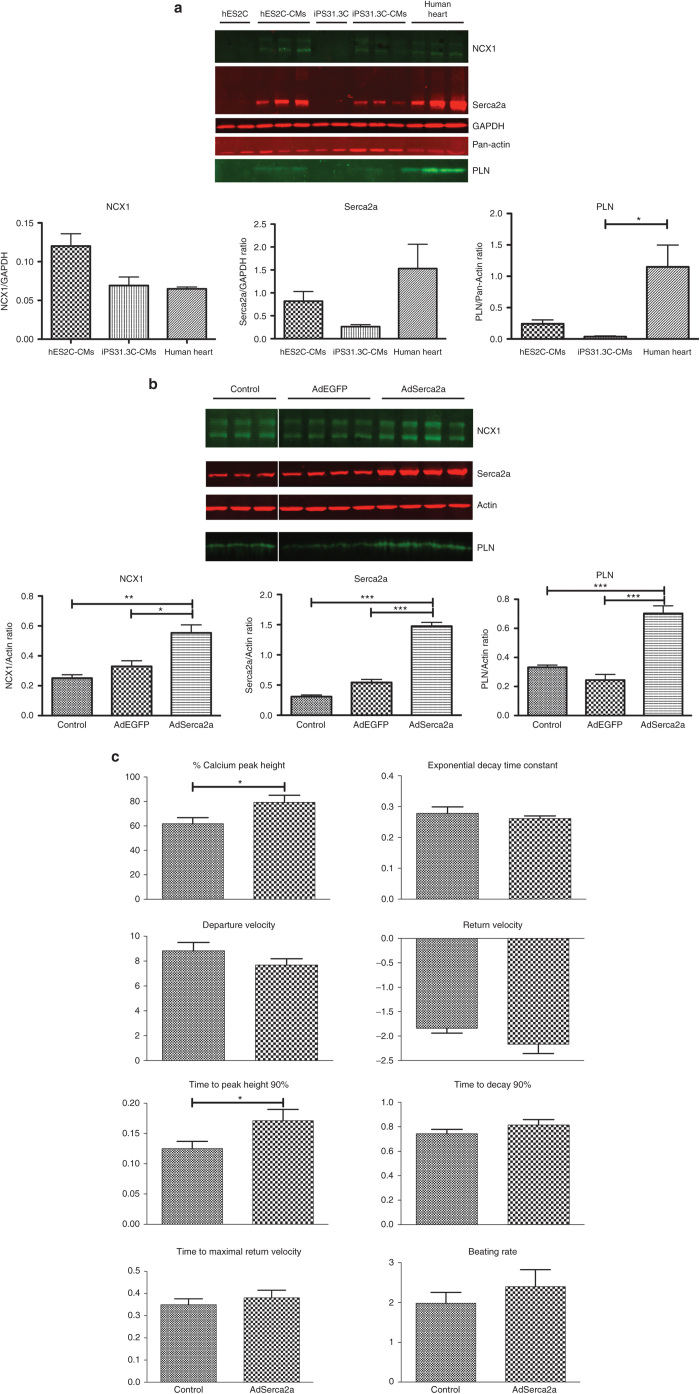
Adenoviral vectors were determined as the most efficient vectors for hESC- and hiPSC-derived cardiomyocytes. We then sought to examine the physiological effects of delivering a therapeutic gene by adenoviral transduction in hPSC-CM. Serca2a is an ATP-dependent Ca2+ pump responsible for the transfer of cytosolic calcium into the sarcoplasmic reticulum, during relaxation. As a first step, we sought to determine the levels of expression of three major cycling proteins in PSc derived cardiomyocytes. Serca2a , phospholamban (PLN), and the sodium-calcium exchanger 1 (NCX1) are expressed in hES2C-CMs and to a lesser extent in hiPS31.3C-CMs, both of which were lower compared to human hearts (Figure 4a). For the purpose of this experiment, hES2C-CMs and hiPS31.3C-CMs were infected with adenovirus carrying either the Serca2a (AdSerca2a) and/or the EGFP (AdEGFP) gene and the expression of cardiac markers and calcium cycling properties were examined (Figure 4b,c and Supplementary Figure S6a,b). The differences in calcium cycling between the cardiomyocytes derived from hESCs and hiPSCs, at baseline level are noteworthy. Overall, hES2C-CMs exhibit higher calcium peak height, higher departure and return velocities, shorter times to peak height and to decay (90%) (Figure 4c and Supplementary Figure S6b).
Figure 4.
Overexpression of Serca2a in hES2C-CMs increases the expression of calcium homeostasis genes and improves calcium cycling. (a) Protein expression of cardiac-specific markers by differentiated cells. Key calcium cycling proteins Serca2a, PLN and NCX1 are expressed highly in hES2C-CMs and at significant levels in hiPS31.3C-CMs, compared to human heart, whereas they are absent from the cell lines they were derived from. Error bars depict ± standard error mean (SEM) (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001). (b) Adenoviral overexpression of Serca2a in hES2C-CMs leads to increase in protein expression of cardiac specific markers. Serca2a and EGFP as a control were adenovirally overexpressed in hES2C-CMs. The AdSerca2a infected cells showed higher expression of not only Serca2a, but also PLN and NCX1. Error bars depict ± SEM (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001). (c) Effect of Serca2a overexpression on calcium cycling in hES2C-CMs. Adenoviral overexpression of Serca2a in hES2C-CM improved calcium cycling as shown by the significant increase in the peak height of the calcium transients as well as the time to reach the peak height. Error bars depict ± SEM (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001).
Interestingly, AdSerca2a transduced hES2C-CM expressed not only higher levels of Serca2a, but also PLN and NCX1 (Figure 4b), indicating a more efficient calcium ion homeostasis and higher differentiation state. Their calcium cycling properties, as indicated by the calcium transients, were also measured. As shown in Figure 4c, overexpression of Serca2a leads to a significant increase in cytosolic calcium during contraction, as indicated by the increase in calcium peak height. The time to reach peak cytosolic calcium (90%) was also increased. The change in both these parameters is in agreement with improved calcium handling. hiPS31.3C-CMs had similar but less pronounced effects (Supplementary Figure S6a,b).
Discussion
Cardiomyocytes derived from pluripotent stem cells, whether hESC or, more importantly, hiPSCs provide a valuable tool for the study of human diseases. Especially in the past few years, a vast wealth of information has been extracted through studies on hiPSC from patients.13,25 Using hiPSC derived cardiomyocytes as a model to study human disease implies also the evaluation of the therapeutic potential of different modalities, and importantly gene therapy. Gene therapy has emerged in the past few years as a promising alternative, when traditional therapeutic approaches prove to be insufficient, as recently shown by the success of clinical trials.7 It is therefore imperative to validate the potential of gene therapies, not only in animal models, but also in hiPS-derived cardiomyocytes. Nonviral gene therapy has proven safe, but relatively inefficient in delivering genes in vivo and, to lesser extent, in vitro. Viral vectors on the other hand are very efficient, albeit less safe, by reason of immunogenicity (mostly for adenoviruses) and insertion mutagenesis (lentiviruses). The attributes of three major viral vectors, adenoviral, AAV, and lentiviral vectors, in several animal models are well known.7,8 Despite their significance in gene therapy, the literature is lacking a systematic comparison in vitro of these viral vectors in PSCs and thereof derived cardiac myocytes. Consequently, we sought to compare these vectors in these two cell types. Table 1 summarizes our results. We found that adenoviruses are efficient in both cell types, lentiviruses in pluripotent cells and AAV vectors in differentiated cardiomyocytes. In order to further validate our study, based on our data, we overexpressed Serca2a, a sarcoplasmic reticulum calcium channel, found to be downregulated in a wide variety of pathological conditions involving heart failure, 7 in PSC-CM and studied its effects. As expected, Serca2a overexpression in PSC-CMs, results in improvement of various calcium cycling parameters, thereby validating the gene therapy therapeutic approach in this in vitro model.
Towards our goal, to assess the effectiveness of viral transduction systems in pluripotent cells and thereof derived cardiac myocytes, we compared four known AAV serotypes, three cardiotropic (AAV1, 6 and 9) and the archetypical AAV2. This study revealed the superiority of AAVs 2 and 6 in in vitro systems. It is also of interest that AAV2 transduction efficiency seems to saturate at higher viral genomes per cell concentrations, an occurrence that could be explained by limitations in receptor binding or intracellular trafficking.15,24,26 It is also of note that AAV9, which is the most cardiotropic and efficient serotype at least in small animals in vivo, was far less efficient than serotypes 2 and 6, in differentiated cells. This discrepancy between in vivo and in vitro data however has been observed by our (unpublished data) and other groups.27 For the next stage of our experiments, we chose to use EGFP as a reporter gene and flow cytometry, in order to measure level of expression as well as number of transduced cells. Our experiments show that adenoviral vectors are superior in both cell types. In differentiated cells they are very closely followed by AAVs and in particular the cardiotropic AAV6, whereas in undifferentiated cells they are followed in efficiency by lentiviral vectors. The AAV vectors used for these experiments were self-complementary (sc) as opposed to the single stranded (ss) used in the first experiments comparing AAV serotypes. Transduction by AAVs is partially limited by the need to convert the single stranded viral genome to double-stranded DNA. The scAAV vectors overcome this hurdle by packaging an inverted repeat genome that comes together as a double-stranded DNA in the cell.28 These vectors have proven valuable in vivo, and in vitro.14 In some of our experiments, we also used a single-stranded vector (Supplementary Figure S1b), which was less efficient than scAAVs, most importantly with regards to the number of transduced cells. The transduction, but also the cell death induction efficiency was much lower for ssAAVs, compared to scAAVs, which could be explained by the use of a different expression cassette and transgenes with different fluorescence intensities.29 However, transfection experiments in two cell lines (293T and HeLa) using the respective plasmids (data not shown) cannot account fully for the robust differences in transduction efficiency. Therefore, we speculate that the type of viral genomes used affects the viral efficiency, in accordance with the literature.14,15
Pluripotent cells, both hES and hiPS, are not easily transduced by AAVs in particular, which is in accordance with what we observed (Figure 1a,b) and the literature,30 and could be attributed to the Mre11 complex (MRN), which is involved in DNA damage sensing and inhibition of AAV transduction.30 hES2C-CMs and hiPS31.3C-CMs on the other hand, both showed higher transduction efficiencies, especially with AAV vectors. It is conspicuous that the self-complementary AAV vectors are almost as efficient as the adenoviral and much more efficient than the lentiviral vector in PSC-CM. Undifferentiated cells by AAVs could also be explained by the ensuing cell death observed in our experiments, verified via Annexin V staining, and those of others.17 We next set out to explore the DDR and apoptotic pathways. Undifferentiated/pluripotent cells have highly active DNA damage response (DDR) pathways, which interferes with the viral cell cycle,31 and they also lack the repair capacity.32 Notably, NOXA1 expression is significantly upregulated in hESCs.33 Additionally, the DDR genes are involved in the TP53-controlled balance between survival and cell death.20,33,34 The expression of several DDR pathway genes, such as ATM, PRKDC, and TP53 remained stable, which is expected, as regulation of these genes is exerted mostly at the posttranslational level.17,18
AAV transduction in our experiments induced cell death in both cell types, with the only deviation being that hiPSCs exhibit cell death at later time-points. In accordance to our observations, studies show no significant difference in the DNA damage response gene expression between hESCs and hiPSCs.35 The smaller percentage of hiPSCs undergoing apoptosis at a particular time point compared to hESCs could be due to the higher cell to cell variation observed in hiPSCs, which could affect transduction efficiency and DDR response at a particular time point.36,37 This small percentage of apoptosis-positive cells could explain the difficulty in detecting apoptotic gene upregulation in hiPSC, considering the gene profiling is from the whole population. However, apoptotic genes not tested in the present study20 or alternative pathways could be responsible for the observed cell death.38,39
To further establish the validity of our transduction efficiency patterns displayed by the viral vectors used in the two types of cells, pluripotent and thereof differentiated cardiac myocytes, we also sought to explore the expression of some known receptors, for AAV and adenoviral vectors in particular. We found that HSPG, the primary receptor for AAV2 (ref. 24), was abundantly expressed in hPSCs. We also found that the expression pattern of the sialic acid, receptor for AAV serotypes 1 and 6 (ref. 21), and galactose, receptor for AAV9 (refs. 22,23), follows a pattern similar to the transduction pattern. The expression of CXAD receptor, the known receptor for adenovirus serotype 5 (ref. 10), which was used in the present study, and also integrin α3β1, both known to mediate adenoviral transduction, was similar to that found in HeLa cells, known to be transduced efficiently by adenovirus.10 The CXAD receptor mRNA was also found to be more abundant in pluripotent cells rather than in differentiated cardiomyocytes. It is of note that adenoviral transduction was highly efficient in both cell types, and slightly better in differentiated cells. We speculate that the difference in transgene expression that does not correspond directly to receptor abundance is due to the capacity of undifferentiated cells to interfere with viral cell cycle.30,31 There is also the possibility, that the choice of a common promoter for our experiments, albeit necessary to have a baseline comparison, might drive transgene expression with different efficiency in the different cell types.40 Finally, the receptor for the pseudotyped lentiviral vectors used in this study,11 is phosphatidylserine, which is a ubiquitous membrane lipid.12
To decipher the elementary underlying calcium handling molecular pathway that drives contraction in PSC-CMs, we screened for the expression of two major calcium-handling proteins residing in the sarcoplasmic reticulum, Serca2a and PLN. Both hESC-CMs and hiPSC-CMs express lower levels of these proteins compared to human heart samples, in accordance with the literature reviewed in ref. 41). Additionally, as observed, hESC-CMs express higher levels of these proteins compared to hiPSC-CMs, which indicates the lower differentiation potential of the latter cells. Also, NCX1, as shown in this work and by others, exhibits relatively increased expression compared to the other calcium cycling proteins (reviewed in ref. 41). Potentially, the increased NCX1 expression could also be attributed to the presence of cardiac pacemaker cells,42 even though the differentiation protocol used in the study is optimized for ventricular cardiomyocyte production.43 High NCX1/Serca2a is characteristic of fetal and neonatal cardiac myocytes.41 PSC-derived cardiomyocytes also exhibit immature spontaneous beating, which is accompanied by calcium transients. As observed by others as well,44 hiPSC-derived cardiomyocytes exhibit lower calcium handling properties, compared to hESC-CM, which reflects the relative expression of the relevant proteins. Overall, our data indicate the presence of immature calcium handling properties in hESC, and even more so, in hiPSC-derived cardiomyocytes.
The purpose of this study, however, was to identify the most efficient vector to modulate the functional properties of PSC-CMs. Accordingly, as a final step toward identifying a potent transduction vector, we sought to investigate the potential of the adenoviral vectors in a functional assay in PSC-CM. PSC-derived CMs are promoted as a platform to evaluate different therapeutic applications in a patient specific manner and calcium cycling in these cells is critical to their potential. Overexpression of Serca2a, amongst others, ameliorates heart failure by improving calcium handling, an effect also observed in a phase 2 clinical trial.7,45,46 Accordingly, we chose the delivery of Serca2a using adenoviral vectors, which proved to be the most efficient in PSC-CMs. Adenoviral transduction with Serca2a resulted, as expected, in increased expression of not only Serca2a47 but also of PLN and NCX1. These results indicate that overexpression of Serca2a in PSC-CM results in overall increase in the calcium handling properties of these cells. Furthermore, and in accordance with the calcium handling protein expression pattern, hESC-CM showed better calcium cycling properties compared to hiPSC-CM. Spontaneous rhythmic Ca2+ transients reveal improved calcium cycling properties, more evidently for hESC-CM.
The novelty of our study lies in the systematic evaluation of known viral vectors for the infection of PSC and thereof derived cardiomyocytes and the validation in a functional manner. In conclusion, all viral vectors tested transduce differentiated cells more potently than pluripotent cells. This effect correlates with the distribution of their respective receptors, but also relates to inhibition of viral transduction. Adenoviral vectors transduce cells efficiently, across all cell lines tested. Importantly, AAV vectors induce cell cycle arrest and cell death in pluripotent cells, which would make them unsuitable not only for hES and hiPS cells, but also for the generation of hiPSC from fibroblasts. Finally, cardiac myocytes derived from hES and hiPS cells exhibit an immature cardiac phenotype, as shown by the lower levels of cardiac cycling proteins. Overexpression of Serca2a could be employed to improve calcium handling but most importantly to induce a more mature phenotype.
Materials and Methods
hESC and hiPSC culture
The hESC, HES-2 (ES02) and H9 (WA09) were obtained from the WiCell Research Institute (Madison, WI). The hiPS line (SKiPS-31.3 and 24.1) (described herein as hiPS31.3 and hiPS24.1) was derived from skin biopsies of two volunteers, 40- and 45-year old, with informed consent (Staten Island Hospital) as previously described.48 All lines were propagated under feeder-independent conditions as previously described. Briefly, the hESCs and hiPSC were maintained in an undifferentiated state using the mTeSR (Stem Cell Technologies, Vancouver, BC) media on hESC-qualified Matrigel (BD Biosciences, San Jose, CA) at 37 °C in 5% CO2/5% O2 and expanded by an enzyme-free treatment with Gentle Cell Dissociation Reagent (Stem Cell Technologies).
Cardiomyocyte differentiation
The protocol is described in detail elsewhere.49 Briefly, undifferentiated hESCs and hiPSCs were dissociated with Gentle Cell Dissociation Reagent (StemCell Technologies) for 4 minutes at room temperature. Small clusters (50–100 cells) were cultured in suspension on ultralow attachment cell culture dishes (Corning, Lowell, MA) with mTesR1 (days 0–1) and differentiation media (StemPro34, 50 μgml-1 ascorbic acid and 2 mmol/l GlutaMAX-I) (days 1-end of the experiment) supplemented with recombinant cytokines and small-molecules for the indicated time period and at the following concentrations; day 0: BMP4 (10 ng ml−1) and Blebbistatin (5 μmol/l); days 1–2: BMP4 (10 ng ml−1) and Activin-A (5 ng ml−1); days 4.5–8, IWR-1 (1 μmol/l). Beating EBs were typically identified at days 8–12.
rAAV vector production
AAV were produced as previously described.50 Briefly, all viral preparations were produced by Polyethylenimine (linear, MW ~25,000; cat. no.: 23966; Polysciences, Warrington, PA) transfection of 293T cells using a two-plasmid system: one plasmid to provide the ITR flanked transgene and another to provide the serotype-specific capsid proteins, the adenoviral helper functions and the AAV2 Rep proteins. The viruses were iodixanol purified (Optiprep; cat. no. D1556, Sigma-Aldrich, St Louis, MO), dialyzed in PBS (2×, overnight) (Spectra/Por 2 dialysis tubing, 12-14K MWCO; 132676, Thermo Fisher Scientific, Fremont, CA), and the titers were determined by qPCR using the SYBR Advantage qPCR Premix (cat. no.: 639676; Clontech Laboratories, Mountain View, CA) with an Applied Biosystems (Carlsbad, CA) 7500 real-time PCR system with primers against the Simian Virus 40 (SV40) polyA sequence (forward: AACCTCCCACACCTCCC, reverse: TTGGACAAACCACAACTAGAA) and the rAAV2 reference standard stock (RSS) (VR-1616; ATCC, Manassas, VA).
Helper plasmids.
AAV1: pXYZ1, AAV2: pDG plasmid (kindly provided by Jürgen Kleinschmidt, Deutsches Krebsforschungszentrum, Heidelberg, Germany), AAV6: pDP6rs (Plasmid Factory, Bielefeld, Germany) and AAV9: pDG9 plasmid (kindly provided by James M. Wilson, University of Pennsylvania School of Medicine, Philadelphia, PA).
Transgene plasmids.
pUF-CBA-Luc encodes for firefly luciferase under the control of the synthetic CMV (cytomegalovirus)/chicken β-actin promoter (CBA) and SV40 polyA (kindly provided by M. Nonnenmacher, Mount Sinai School of Medicine). pdsAAV-GFP (kindly provided by X. Xiao, University of North Carolina, Chapel Hill, NC) encodes for the EGFP under the control of a CMV promoter and the SV40 polyA. Deletion of the left-hand D-region in this construct results in the generation of self-complementary, i.e., double-stranded AAV vectors (Wang Z 2011). pTRUF11 encodes for the humanized GFP (hGFP), under the control of the CBA promoter/SV40polyA (kindly provided by Linden M.).
Adenoviral vector production
Adenoviral type 5 vectors were produced using the AdEasy System, as previously described. The EGFP gene (from the pEGFP-N1 vector) (Clontech Laboratories) was cloned in the pShuttleCMV plasmid and under the control of the CMV promoter and SV40 polyA to produce the AdEGFP vector. The Serca2a and GFP genes were placed under the control of separate CMV promoters and this cassete was used to produce the AdSerca2a vector. The adenoviruses were propagated in 293 cells and purified using two-step cesium chloride ultracentrifugation. Viral particle titers were determined by optical density measurement.
Lentiviral vector production
The lentiviral plasmids were previously described.11 The lentiviral vectors were prepared using third generation lentiviral vectors systems as previously described.11
Western blotting
Non failing (control) human heart samples were procured from donors with a median age of 62, whose echocardiography showed normal cardiac function and died of neurological diseases or trauma/motor vehicle accidents. Whole cell lysates and human heart samples were prepared with RIPA lysis buffer (10 mmol/l Tris-HCL pH7.4, 30 mmol/l NaCl, 1 mmol/l EDTA, 1% Nonidet P-40, supplemented with complete protease inhibitors cocktail (04693159001; Roche Applied Science, Indianapolis, IN)), analyzed in NuPAGE 4–12% Bis Tris Gel (NP0336BOX; Invitrogen, Grand Island, NY), transferred onto Immobilon PVDF membrane (IPFL00010; Thermo Fisher Scientific) and blocked for 1 hour at room temperature using 5% milk in Tris-buffered saline. Primary antibodies were incubated overnight in 5% bovine serum albumin. Membranes were washed in Tris-buffered saline-Tween 0.05% (TBST), incubated with secondary antibody for 1h at room temperature. The membranes were washed in TBST and the signal was visualized either by infra-red imaging at a LI-COR Odyssey IR imager (LI-COR Biosciences—Biotechnology, Lincoln, NE) or by chemiluminescence using Immobilon Western Chemiluminescent horseradish peroxidase Substrate (WBKLS0050; Millipore, Billerica, MA).
Antibodies
Primary: Rabbit anti-Serca2a (1:3,000) (custom made; Century 21st Biochemicals, Marlborough, MA), rabbit anti-CSQ (1:1,000) (PA1-913; ThermoScientific, Fremont, CA), mouse anti-NCX1 (1:1,000) (NB300-127; Novus Biologicals, Littleton, CO), rabbit anti-RYR2 (1:2,000) (AB9080; Millipore), mouse anti-PLN (1:5,000) (A010-30; Badrilla, Leeds, UK), rabbit anti-pan-actin (1:1,000) (#4968; Cell Signaling Technology, Beverly, MA), rabbit anti-beta actin (1:1,000) (#4970; Cell Signaling Technology), rabbit anti-GAPDH (1:10,000) (G9545; Sigma-Aldrich). Secondary (1:10,000): IRDye 800 anti-mouse (926–32212; Li-Cor Biosciences), IRDye 680RD anti-rabbit IgG (H+L) (926–68073; Li-Cor Biosciences), Horse Radish Peroxidase (HRP) anti-mouse IgG (H+L) (170–6516; BioRad, Hercules, CA), HRP anti-rabbit IgG (H+L) (#7074S, Cell Signaling Technology).
Luciferase assay
The assay was performed as previously described.50 Briefly, cells were infected with the viruses at the mentioned viral genome per cell and the assay was performed 3 days postinfection (dpi). For the assay, cells were lysed with 2× Lysis Buffer (10 mmol/l Tris–hydrochloride pH 8.0, 150 mmol/l NaCl, 1% (wt/vol) NP40, 10 mmol/l DTT), and 20 µl of cell lysate was mixed with 100 µl of luciferase reaction buffer (25 mmol/l Tricine–hydrochloride pH 7.8, 5 mmol/l magnesium sulfate, 0.5 mmol/l EDTA (pH 8.0), 3.3 mmol/l DTT, 0.5 mmol/l ATP pH ~7–8 (A26209; Sigma-Aldrich), 1 mg/ml bovine serum albumin, 0.05 mg/ml D-luciferin (cat. no.: LUCK-100; Gold Biotechnology, St Louis, MO), 0.05 mmol/l CoA (cat. no.: 13787, United States Biological, Swampscott, MA). The luminescence was read in a luminescence counter (Microbeta Trilux 1450 LSC and Luminescence Counter; Perkin Elmer, Carlsbad, CA).
Flow cytometric analysis
Cells were infected with the viruses at the mentioned viral genome per cell or MOI and the flow cytometric analysis was performed 3 days postinfection (dpi). For the analysis, cells were collected using the PromoCell DetachKit (C-41200; PromoCell GmbH, Heidelberg, Germany), washed with PBS without Mg++, Ca++, incubated with 7-AAD (7-aminoactinomycin D) (A1310; Invitrogen) and finally fixed with 4% paraformaldehyde. Because of extensive cell death in certain conditions, the total number of gated events was lower in these conditions (depicted with asterisk in Table 1). Cells were counted by flow cytometry by using the FACSCalibur flow cytometry system and analyzed using FlowJo Software (Tree Star, Ashland, OR). All fluorescence plots were in log scales. Cells were gated based on their forward-scatter and side-scatter properties. The threshold between GFP-positive and GFP-negative was set so that >99.5% of uninfected cells were considered fluorescence (FL1)-negative. Expression index (EI) is a parameter calculated by the following equation: EI = ((% GFP-positive cells) * (mean fluorescent intensity (MFI) of GFP-Positive cells))infected cells/((% GFP-positive cells) * (MFI of GFP-positive cells)noninfected control). Apoptosis and cell death was measured by using Annexin V/propidium iodide (PI) staining (Annexin V-FITC: 556570, APC-FITC: 561012; BD Biosciences). The cells were dissociated by incubation in PBS without Mg+2, Ca+2 for 5 minutes, followed by thorough pipetting for mechanical dissociation. The rest of the staining was performed according to manufacturer’s instructions. Cells were counted by using the LSRII flow cytometry system (BD Biosciences) and analyzed using FlowJo Software.
Quantitative PCR (qPCR) analysis of mRNA
Cells were seeded as previously described and infected with the viruses at the reported viral genome per cell and MOI. For the collection, at the reported time points, cells were washed once with PBS, RNA was isolated using TRIzol Reagent (15596-026; Invitrogen), DNase treated using the RQ1 RNase-Free DNase (M6101; Promega, Madison, WI) and reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (4368814; Invitrogen) according to manufacturer’s instructions. cDNA was analyzed using the SYBR Advantage qPCR Premix (639676; Clontech Laboratories) and 7500 real-time PCR system with the primers mentioned in Supplementary Table S3.
Immunocytochemistry
hESCs and iPSCs were cultured on matrigel-coated coverslips for 4–5 days and then fixed with 4% paraformaldehyde. Fixed cells were permeabilized in blocking/permeabilization buffer (2% bovine serum albumin/2% FBS/0.05 % NP-40 in PBS) for 45 minutes and incubated with primary antibodies overnight at 4 °C. The cells were then washed in PBS and incubated with Alexa-conjugated secondary antibodies (Invitrogen) diluted in blocking/permeabilization buffer (1:750). Finally, after washing in PBS, the cells were counterstained with DAPI. The following antibodies were used: rabbit polyclonal anti-Integrinα3β1 (orb10925; Biorbyt, San Francisco, CA), rabbit polyclonal anti-Coxsackie Adenovirus Receptor (orb4966; Biorbyt), and rabbit polyclonal anti-Syndecan-2 (sc-15348; Santa Cruz Biotechnology, Dallas, TX). Confocal imaging was performed using a Leica SP5 confocal system.
Lectin staining
hESCs and iPSCs cultured on matrigel-coated coverslips for 4–5 days were incubated with wheat germ agglutinin (Alexa Fluor 555 Conjugate) (W32464; Invitrogen) and ECL (Fluorescein labeled Erythrina Cristagalli Lectin (ECL, ECA)) (FL-1141; Vector) at 1:200, for 10 minutes at room temperature. The cells were then washed three times with PBS and fixed for 20 minutes with 4% paraformaldehyde. They were washed three times with PBS and counterstained with DAPI. Imaging was performed on a Zeiss LSM510 laser scanning microscope.
Calcium cycling measurement
Partially dissociated hES2-CM and hiPS31.3-CM embryoid bodies were plated onto Matrigel-coated, pretreated German glass coverslips (GG-25-PRE; NeuVitro, El Monte, CA). The cells were loaded with Fura2-AM (1 µmol/l) for 30 minutes at 37 °C in Tyrode solution (140 mmol/l NaCl, 5.4 mmol/l KCl, 1 mmol/l MgCl2, 10 mmol/l glucose, 1.8 mmol/l CaCl2, and 10 mmol/l Hepes (pH 7.4)) and then transferred to fresh Tyrode solution for 10 minutes. Calcium transients were recorded using a video-based edge detection system (Myocyte Calcium and Contractility Recording System) and analyzed using the IonWizard software (IonOptix LLT, Milton, MA).
Statistical significance
One-way analysis of variance followed by Bonferroni’s post hoc comparisons test (GraphPad Prism version 5.0b for MacIntosh; GraphPad Software, La Jolla, CA) was performed for the flow cytometry, luciferase assay, and western blot experiments. Student t-test was performed for the qPCR (samples were compared to control), Annexin V and calcium cycling experiments. Significance is defined as P < 0.05 (one symbol), 0.05 < P<0.01 (two symbols), P < 0.001 (three symbols).
Acknowledgments
This work was supported by National Institutes of Health grants RO1 HL083156, HL093183, HL119046, HL113497, and P20HL100396 and a National Heart, Lung, and Blood Institute Program of Excellence in Nanotechnology (18) Award, Contract HHSN268201000045C (R.J.H. and 1K99HL104002 (I.K)). Part of the work was funded by a Leducq Foundation grant. The authors thank Shihong Zhang, Dongtak Jeong, Erik Kohlbrenner, Lahouaria Hadri, and Patricia Rodriguez for support in tissue procurement, viral productions, and cardiac marker interpretations.
R.J.H. is the scientific cofounder of Celladon, which plans to commercialize AAV SERCA2a for the treatment of HF. All other authors declare no competing financial interests.
References
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Mummery CL, Zhang J, Ng ES, Elliott DA, Elefanty AG, Kamp TJ. Differentiation of human embryonic stem cells and induced pluripotent stem cells to cardiomyocytes: a methods overview. Circ Res. 2012;111:344–358. doi: 10.1161/CIRCRESAHA.110.227512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Willems E, Spiering S, Davidovics H, Lanier M, Xia Z, Dawson M. Small-molecule inhibitors of the Wnt pathway potently promote cardiomyocytes from human embryonic stem cell-derived mesoderm. Circ Res. 2011;109:360–364. doi: 10.1161/CIRCRESAHA.111.249540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjar RJ. Potential of gene therapy as a treatment for heart failure. J Clin Invest. 2013;123:53–61. doi: 10.1172/JCI62837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapti K, Chaanine AH, Hajjar RJ. Targeted gene therapy for the treatment of heart failure. Can J Cardiol. 2011;27:265–283. doi: 10.1016/j.cjca.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth T, Parker N, Ylä-Herttuala S. History of gene therapy. Gene. 2013;525:162–169. doi: 10.1016/j.gene.2013.03.137. [DOI] [PubMed] [Google Scholar]
- Douglas JT. Adenoviral vectors for gene therapy. Mol Biotechnol. 2007;36:71–80. doi: 10.1007/s12033-007-0021-5. [DOI] [PubMed] [Google Scholar]
- Kho C, Lee A, Jeong D, Oh JG, Chaanine AH, Kizana E. SUMO1-dependent modulation of SERCA2a in heart failure. Nature. 2011;477:601–605. doi: 10.1038/nature10407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin J, Zhang XY, Reiser J. Altering the tropism of lentiviral vectors through pseudotyping. Curr Gene Ther. 2005;5:387–398. doi: 10.2174/1566523054546224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellin M, Marchetto MC, Gage FH, Mummery CL. Induced pluripotent stem cells: the new patient? Nat Rev Mol Cell Biol. 2012;13:713–726. doi: 10.1038/nrm3448. [DOI] [PubMed] [Google Scholar]
- Wang Z, Ma H-I, Li J, Sun L, Zhang J, Xiao X. Rapid and highly efficient transduction by double-stranded adeno-associated virus vectors in vitro and in vivo. Gene therapy. 2003;10:2105–2111. doi: 10.1038/sj.gt.3302133. [DOI] [PubMed] [Google Scholar]
- Sanlioglu S, Monick MM, Luleci G, Hunninghake GW, Engelhardt JF. Rate limiting steps of AAV transduction and implications for human gene therapy. Curr Gene Ther. 2001;1:137–147. doi: 10.2174/1566523013348788. [DOI] [PubMed] [Google Scholar]
- Shaw JM, Al-Shamkhani A, Boxer LA, Buckley CD, Dodds AW, Klein N. Characterization of four CD18 mutants in leucocyte adhesion deficient (LAD) patients with differential capacities to support expression and function of the CD11/CD18 integrins LFA-1, Mac-1 and p150,95. Clin Exp Immunol. 2001;126:311–318. doi: 10.1046/j.1365-2249.2001.01661.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch ML, Fagan BM, Dumitru R, Bower JJ, Yadav S, Porteus MH. Viral single-strand DNA induces p53-dependent apoptosis in human embryonic stem cells. PLoS One. 2011;6:e27520. doi: 10.1371/journal.pone.0027520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal PK, Rossi DJ. DNA-damage-induced differentiation in hematopoietic stem cells. Cell. 2012;148:847–848. doi: 10.1016/j.cell.2012.02.011. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Huang X, Halicka HD, Zhao H, Traganos F, Albino AP. Cytometry of ATM activation and histone H2AX phosphorylation to estimate extent of DNA damage induced by exogenous agents. Cytometry A. 2007;71:648–661. doi: 10.1002/cyto.a.20426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- Wu Z, Miller E, Agbandje-McKenna M, Samulski RJ. Alpha2,3 and alpha2,6 N-linked sialic acids facilitate efficient binding and transduction by adeno-associated virus types 1 and 6. J Virol. 2006;80:9093–9103. doi: 10.1128/JVI.00895-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CL, Vandenberghe LH, Bell P, Limberis MP, Gao GP, Van Vliet K. The AAV9 receptor and its modification to improve in vivo lung gene transfer in mice. J Clin Invest. 2011;121:2427–2435. doi: 10.1172/JCI57367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Bryant KD, Brown SM, Randell SH, Asokan A. Terminal N-linked galactose is the primary receptor for adeno-associated virus 9. J Biol Chem. 2011;286:13532–13540. doi: 10.1074/jbc.M110.210922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonnenmacher M, Weber T. Intracellular transport of recombinant adeno-associated virus vectors. Gene Ther. 2012;19:649–658. doi: 10.1038/gt.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priori SG, Napolitano C, Di Pasquale E, Condorelli G. Induced pluripotent stem cell-derived cardiomyocytes in studies of inherited arrhythmias. J Clin Invest. 2013;123:84–91. doi: 10.1172/JCI62838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbert CL, Allen JM, Miller AD. Adeno-associated virus type 6 (AAV6) vectors mediate efficient transduction of airway epithelial cells in mouse lungs compared to that of AAV2 vectors. J Virol. 2001;75:6615–6624. doi: 10.1128/JVI.75.14.6615-6624.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suckau L, Fechner H, Chemaly E, Krohn S, Hadri L, Kockskämper J. Long-term cardiac-targeted RNA interference for the treatment of heart failure restores cardiac function and reduces pathological hypertrophy. Circulation. 2009;119:1241–1252. doi: 10.1161/CIRCULATIONAHA.108.783852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DM. Self-complementary AAV vectors; advances and applications. Mol Ther. 2008;16:1648–1656. doi: 10.1038/mt.2008.171. [DOI] [PubMed] [Google Scholar]
- Bierhuizen MF, Westerman Y, Visser TP, Wognum AW, Wagemaker G. Green fluorescent protein variants as markers of retroviral-mediated gene transfer in primary hematopoietic cells and cell lines. Biochem Biophys Res Commun. 1997;234:371–375. doi: 10.1006/bbrc.1997.6648. [DOI] [PubMed] [Google Scholar]
- Schwartz RA, Palacios JA, Cassell GD, Adam S, Giacca M, Weitzman MD. The Mre11/Rad50/Nbs1 complex limits adeno-associated virus transduction and replication. J Virol. 2007;81:12936–12945. doi: 10.1128/JVI.01523-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnell AS, Grand RJ. DNA viruses and the cellular DNA-damage response. J Gen Virol. 2012;93 Pt 10:2076–2097. doi: 10.1099/vir.0.044412-0. [DOI] [PubMed] [Google Scholar]
- Zhang M, Yang C, Liu H, Sun Y. Induced pluripotent stem cells are sensitive to DNA damage. Genomics Proteomics Bioinformatics. 2013;11:320–326. doi: 10.1016/j.gpb.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden DT, Davila-Kruger D, Melov S, Bredesen DE. Human embryonic stem cells express elevated levels of multiple pro-apoptotic BCL-2 family members. PLoS One. 2011;6:e28530. doi: 10.1371/journal.pone.0028530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvorova II, Katolikova NV, Pospelov VA. New insights into cell cycle regulation and DNA damage response in embryonic stem cells. Int Rev Cell Mol Biol. 2012;299:161–198. doi: 10.1016/B978-0-12-394310-1.00004-7. [DOI] [PubMed] [Google Scholar]
- Momcilovic O, Knobloch L, Fornsaglio J, Varum S, Easley C, Schatten G. DNA damage responses in human induced pluripotent stem cells and embryonic stem cells. PLoS One. 2010;5:e13410. doi: 10.1371/journal.pone.0013410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsinh KH, Sun N, Sanchez-Freire V, Lee AS, Almeida P, Hu S. Single cell transcriptional profiling reveals heterogeneity of human induced pluripotent stem cells. J Clin Invest. 2011;121:1217–1221. doi: 10.1172/JCI44635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder B, Sacher R, Rämö P, Damm EM, Liberali P, Pelkmans L. Population context determines cell-to-cell variability in endocytosis and virus infection. Nature. 2009;461:520–523. doi: 10.1038/nature08282. [DOI] [PubMed] [Google Scholar]
- Qin H, Yu T, Qing T, Liu Y, Zhao Y, Cai J. Regulation of apoptosis and differentiation by p53 in human embryonic stem cells. J Biol Chem. 2007;282:5842–5852. doi: 10.1074/jbc.M610464200. [DOI] [PubMed] [Google Scholar]
- Vaseva AV, Marchenko ND, Ji K, Tsirka SE, Holzmann S, Moll UM. p53 opens the mitochondrial permeability transition pore to trigger necrosis. Cell. 2012;149:1536–1548. doi: 10.1016/j.cell.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro K, Kawabata K, Inamura M, Takayama K, Furukawa N, Sakurai F. Adenovirus vector-mediated efficient transduction into human embryonic and induced pluripotent stem cells. Cell Reprogram. 2010;12:501–507. doi: 10.1089/cell.2010.0023. [DOI] [PubMed] [Google Scholar]
- Li S, Chen G, Li RA. Calcium signalling of human pluripotent stem cell-derived cardiomyocytes. J Physiol. 2013;591 Pt 21:5279–5290. doi: 10.1113/jphysiol.2013.256495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltsev AV, Yaniv Y, Stern MD, Lakatta EG, Maltsev VA. RyR-NCX-SERCA local cross-talk ensures pacemaker cell function at rest and during the fight-or-flight reflex. Circ Res. 2013;113:e94–e100. doi: 10.1161/CIRCRESAHA.113.302465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakikes I, Senyei GD, Hansen J, Kong CW, Azeloglu EU, Stillitano F. Small molecule-mediated directed differentiation of human embryonic stem cells toward ventricular cardiomyocytes. Stem Cells Transl Med. 2014;3:18–31. doi: 10.5966/sctm.2013-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Ng KM, Lai WH, Chan YC, Lau YM, Lian Q. Calcium homeostasis in human induced pluripotent stem cell-derived cardiomyocytes. Stem Cell Rev. 2011;7:976–986. doi: 10.1007/s12015-011-9273-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase Y, Ly HQ, Prunier F, Lebeche D, Shi Y, Jin H. Reversal of cardiac dysfunction after long-term expression of SERCA2a by gene transfer in a pre-clinical model of heart failure. J Am Coll Cardiol. 2008;51:1112–1119. doi: 10.1016/j.jacc.2007.12.014. [DOI] [PubMed] [Google Scholar]
- Zsebo K, Yaroshinsky A, Rudy JJ, Wagner K, Greenberg B, Jessup M. Long-term effects of AAV1/SERCA2a gene transfer in patients with severe heart failure: analysis of recurrent cardiovascular events and mortality. Circ Res. 2014;114:101–108. doi: 10.1161/CIRCRESAHA.113.302421. [DOI] [PubMed] [Google Scholar]
- Sun N, Yazawa M, Liu J, Han L, Sanchez-Freire V, Abilez OJ. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci Transl Med. 2012;4:130ra47. doi: 10.1126/scitranslmed.3003552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galende E, Karakikes I, Edelmann L, Desnick RJ, Kerenyi T, Khoueiry G. Amniotic fluid cells are more efficiently reprogrammed to pluripotency than adult cells. Cell Reprogram. 2010;12:117–125. doi: 10.1089/cell.2009.0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakikes I, Senyei GD, Hansen J, Kong CW, Azeloglu EU, Stillitano F. Small molecule-mediated directed differentiation of human embryonic stem cells toward ventricular cardiomyocytes. Stem Cells Transl Med. 2014;3:18–31. doi: 10.5966/sctm.2013-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapti K, Louis-Jeune V, Kohlbrenner E, Ishikawa K, Ladage D, Zolotukhin S. Neutralizing antibodies against AAV serotypes 1, 2, 6, and 9 in sera of commonly used animal models. Mol Ther. 2012;20:73–83. doi: 10.1038/mt.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.