Abstract
Cytoplasmic splicing represents a newly emerging level of transcriptional regulation adding to the molecular diversity of mammalian cells. As examples of this noncanonical form of transcript processing are discovered, the evidence of its importance to normal cellular function grows. Work from a number of groups using a variety of cell types is steadily identifying a large number of transcripts (and soon to be even larger as genome-wide analyses of retained introns across a number of cellular phenotypes are currently underway) that undergo some level of regulated endogenous extranuclear splicing as part of their normal biosynthetic pathway. Here, we review the existing data covering cytoplasmic retained intron sequences and suggest that such sequences may be a component of `sentinel RNA' that serves to generate transcript variants within the cytoplasm as well as a source for RNA-based secondary messages.
Intron retention and cytoplasmic splicing are emerging forms of transcriptional regulation that add to the molecular diversity of mammalian cells. Evidence of the importance of nonnuclear transcript processing for normal cellular function grows as more examples of this phenomenon are discovered. A number of groups, using a variety of cell types, are identifying a growing number of transcripts that feature some degree of intron retention. For a subset of these transcripts it appears as though they undergo some level of regulated endogenous extranuclear splicing as part of their normal biosynthetic pathway. Here, we review the existing data covering the identification of cytoplasmic intron-retaining transcripts (CIRTs), evidence for their modification and splicing outside of the nucleus, and their potential functional roles. We suggest that such retained intronic sequences may be a component of a `sentinel RNA' that serves to generate transcript variants within the cytoplasm as well as a source for RNA-based secondary messages.
Cytoplasmic splicing, by definition, requires the presence of introns or intronic sequences within transcripts that leave the nucleus. Intron retention, along with exon skipping, mutually exclusive exon utilization, and alternative donor or acceptor splice sites, is one of the primary modes of alternative splicing. Retained introns are a substrate for cytoplasmic splicing as well as a foundation for the identification of spliceosome constituents with cytoplasmic activity. As it is estimated that 84–92% of genes in the human genome undergo some form of alternative splicing,1 the presence of retained introns is not surprising; in fact, a number of retained introns have been reported across a number of cell types across many species.2 Examples of retained introns from individual transcripts include the BK channel (KCNMA1), FMRP, oxytocin, lamin B1 and IL1-b, as well as broad reports of several others from across the transcriptome (Table 1). As alternative splicing adds transcriptome and consequently proteome diversity, it is an important level of post-transcriptional regulation in eukaryotes.
TABLE 1.
Sampling of Reported Retained Introns Some with Known Biological Function in Mammals
| Gene Name | Description | Intron | Reference |
|---|---|---|---|
| APP | Amyloid B-protein | I1, i2,i5, i6,i8, i13, i151 | 3 |
| ATF4 | Activating transcription factor 4 | i1 | 4 |
| CACNA1H | Calcium channel, voltage-dependent, T type, α 1H subunit | i23 | 5 |
| CAMK2B | Calcium/calmodulin-dependent protein kinase II β | i3 | 3 |
| CTSD | Cathepsin D | i5 | 4 |
| CREB | cAMP response element binding protein | i1 | 3 |
| EIF1 | Eukaryotic translation initiation factor 1 | i1 | 4 |
| FMR1 | Fragile X mental retardation 1 | i1 | 3 |
| FANCA | Fanconi anemia, complementation group A | i21 | 4 |
| GABRG3 | γ-Aminobutyric acid (GABA) A receptor, γ | ||
| GRIK1 | Glutamate receptor, ionotropic, kainate 1 | i1, i7 | 3 |
| GRIN1 | NMDA 1 receptor | i8 | 3 |
| HP | Haptoglobin | i4 | 4 |
| ID3 | Inhibitor of DNA binding 3, dominant negative helix-loop-helix protein | i1 | 6 |
| IL1B | Interleukin 1, β | i1 | 7 |
| INS | Proinsulin | i1 | 8 |
| KCNMA1 | Potassium large conductance calcium-activated channel, subfamily M, α member 1 | i16 | 9 |
| KCNMA1 | Potassium large conductance calcium-activated channel, subfamily M, α member 1 | i17 | 10 |
| LBR | Lamin B receptor | i9 | 4 |
| LMNB1 | Lamin B1 | i7 | 4 |
| P2X2 | Purinergic receptor P2X, ligand-gated ion channel, 2 | i11 | 11 |
| Robo3 | Roundabout, axon guidance receptor, homolog 3 (Drosophila) | i26 | 12 |
| SRSF7 | Serine/arginine-rich splicing factor 7 (reported as 9G8) | i3 | 13 |
In instances where more than one retained intron is indicated for a given gene name, the retained introns need not all be present in a single CIRT. The retained introns may be found in different RNA molecules, each of which has been detected by sequencing.
Although a number of cytoplasmically retained introns have been identified across mammalian genomes, only sparse evidence exists for action upon these elements by cytosolic constituents of the spliceosome. A small number of well-characterized examples of cytoplasmic mRNA splicing can be found in yeast and plants, as well as in the biosynthetic pathways of tRNA and viral transcript processing, but few examples have been reported in mammals. Conventional splicing of introns from pre-mRNA or heteronuclear RNA (hnRNA) transcripts occurs in the nucleus of cells and involves the action and coordination of various nucleic acid and protein constituents of the spliceosome. The canonical spliceosomal unit is composed of five small nuclear RNAs (snRNA) and a number of protein factors. As many as 300 proteins have been implicated in spliceosomal function14 and the characterization of a minimal spliceosome is the subject of ongoing research.
The activity of the spliceosome in the cytoplasm is a highly controversial topic. The spliceosome is divided into major and minor complexes, with the major complex functioning in the nucleus and believed to assemble on each newly transcribed pre-mRNA molecule. The minor spliceosome acts upon a subset of introns which are found at much lower frequencies across eukaryotic genomes that are characterized by distinct and highly conserved 5′ splice sites and branch point sequences with respect to the vast majority of introns.15 The cellular site of action of the minor spliceosome has been the topic of much debate.
A paper from Konig et al.16 reports that the minor spliceosome functions predominantly in the cytoplasm and controls cell proliferation acting on U12-type introns. This finding challenges previous results showing that the minor spliceosome functions in the nucleus, and has since been refuted in a number of publications.17–19 A study by Steitz suggests that the control experiments and techniques used to argue in favor of the localization of the minor spliceosome to the cytoplasm are inconclusive. However, even with the dearth of direct evidence, those arguing the case for splicing exclusively in the nucleus do acknowledge the possibility of cytoplasmic spliceosome mediated pre-mRNA splicing. Indeed, the degree and rate of cytoplasmic splicing governed by the spliceosome is likely far lower than what is observed in the central processing body of the cell, and the phenomenon may occur only under very specific and specialized conditions, as in highly compartmentalized cells such as neurons or megakaryocytes, and will be discussed in greater detail later.
It is clear that cytoplasmic splicing is a real phenomenon, as it can occur in the enucleated platelet, and in the isolated neuronal dendrite, which is devoid of a nucleus. However, it should be noted that while extranuclear splicing can occur, the mechanism of cytoplasmic splicing is unknown and may differ from nuclear splicing. An example of such an alternative mechanism is cytoplasmic splicing of X-box binding protein 1 (XBP-1) pre-mRNA in the unfolded protein response (UPR) pathway.20 UPR initiates upon the continued accumulation of incorrectly folded proteins in mammalian cells via signaling through transmembrane receptors on the endoplasmic reticulum.21 Upon endoplasmic reticulum stress, the UPR-associated molecule activating transcription factor 6 (ATF6) leads to the downstream expression of XBP-1, which is processed by the UPR-associated molecule inositol requiring kinase 1 (IRE1). Activation of IRE1 during UPR catalyzes the removal of a short 26-nucleotide intron from XBP-1 mRNA via endoribonuclease activity of IRE1. The resulting XBP-1 protein is a frameshift variant that acts as a potent transcription factor that regulates the expression of chaperones and protein degradation factors.22,23 This mechanism of cytoplasmic XBP-1 splicing is more similar to pre-tRNA splicing than conventional pre-mRNA splicing. It is possible that a similar mechanism could be associated with other intron bearing transcripts identified in the cytoplasm, but at this time little data exists directly associating this mechanism with cytoplasmic splicing of other transcripts.
An early example of cytoplasmic mammalian splicing came from Denis et al.7 using platelets. They showed that IL1b transcripts harboring a retained intron accumulated in pro-platelet projections, and persisted after platelet budding and maturation. These cytoplasmic intron-retaining transcripts not only avoid conventional splicing and are exported from the nucleus, but they also escape nonsense mediated decay factors. Intron-retaining IL1b transcripts in platelets persist until platelet activation, when they are spliced in the enucleate platelet and give rise to canonical IL1b mature messenger RNA that is then translated into functional IL1b protein. It is important to note that mature platelets are enucleated cells, so by definition any splicing that may occur must be extranuclear.
This results demonstrate an endogenous extranuclear splicing system exists within a mammalian cell and has obvious biological relevance. Around the same time, both protein and nucleic acid constituents of the spliceosome were reported in the dendrites of primary rat neurons.24 While these molecules may have secondary function outside the nuclear compartment, the same study revealed that mechanically isolated dendrites, which had been physically separated from the soma and nucleus, were capable of splicing reporter RNA constructs. This result proved that the process of splicing could occur outside the nucleus of neurons. The identification of spliceosome molecules in neuronal cytoplasm suggests that some transcripts may be processed in a way that resembles conventional splicing. It should be noted that dendritic splicing of the reporter constructs in this study did not utilize canonical splice sites. While adherence to canonical donor and acceptor sequences was observed in some of the sequences resulting from splicing in dendrites, no single example adhered to both the canonical donor and acceptor sequences. It is possible that this is a consequence of introducing an artificial intron-retaining sequence or saturating the splicing machinery, an alternate explanation is that a cytoplasmic splicing code may exist.
These findings also suggested that endogenous cytoplasmic transcripts harboring retained intronic substrates are present in dendrites and could be subject to extranuclear splicing. These dendritic projections are located at great distances from the cell body, and a local splicing event would obviate the need for shuttling molecules to and from the nucleus for processing. Further, in a manner similar to the regulation of dendritic translation in response to synaptic stimulation, dendritic splicing could represent a novel method of post-transcriptional gene regulation similar to the case with IL1b platelets.
In continuation of this work, a large number of endogenous CIRTs were reported in neuronal dendrites as detected by a combination of microarray, in situ hybridization, and next-generation sequencing.3 While introns could be retained for a number of reasons, the identification of these molecules supports the speculation that functional dendritic splicing factors may have endogenous substrates. While the mechanism of cytoplasmic splicing is largely unknown, a working spliceosome within a subcellular compartment can be envisioned to provide the means for liberating retained introns. Spliceosome-independent cytoplasmic splicing, as part of the unfolded protein response pathway, has been described in yeast25 and mammalian cells.26 This splicing involves an unconventional mechanism involving tRNA ligase and an endonuclease instead of the spliceosome. However, this mechanism reportedly removes relatively small sized sequences and seems unlikely to be involved in processing the considerably larger general retained introns.
Evidence for intron retention in various classes of RNAs can be shown through functional changes associated with the perturbation of retained introns (Table 1). Retained introns have been reported for transcripts associated with proteins from a range of functional classes that include receptors, channels, enzymes and transcription factors. Several of these CIRTs have been shown to be functional although it is not clear for all identified CIRTs to date. Although common elements shared by all of these CIRTS have not been defined, two features include an imbedded sine ID retroviral element that is involved in subcellular targeting of some of the CIRT RNAs3 and a role for retained intron and nonsense mediated decay mechanisms that regulate developmentally important protein abundances in other CIRTs4 (these elements will be discussed later in this review). Based on these data, the functional consequences of cytoplasmic splicing could affect many levels of normal cellular activity. In addition to the splicing and translation of IL1B in platelets upon activation, a retained intron within the KCNMA1 gene has an important role in neuronal excitability. KCNMA1 is alternatively spliced to a high degree and it is an excellent candidate for containing CIRTs. Bell et al.9 found that siRNA knockdown of only that fraction of the KCNMA1 transcripts retaining intron 16 diminished the burst firing properties of treated cells and also altered the localization of channel protein in dendritic subcompartments. This functional effect may be explained in a number of ways: (1) a form of protein that is translated from the cytoplasmically spliced RNA that targets directly into the dendritic spine is lost; (2) the loss of this particular intron-retaining transcript impacts the fate (location or translation) of other forms of the KCNMA1 mRNA and therefore the localization of functional protein; or (3) the translation of the i16 containing transcript may give rise to a truncated protein that is critical to normal functioning of the full-length channel, and thereby acts as a dominant negative. It should be noted that it is unclear in this particular study whether the siRNA knockdown occurred in the cytoplasm of the cell27 or in the nucleus.28 However, since in situ hybridization shows that CIRTs are present in the cytoplasm, at a relatively high abundance as compared to the nucleus, it is likely that the siRNA is functional in the cytoplasm. Additionally, given the fact that RNA splicing occurs in isolated dendrites, it is highly likely that the dendritically localized KCNMA1 CIRT is also spliced in the dendrite. Further should the i16 siRNA function on hnRNA in the nucleus, one would expect all of the KCNMA1 RNA to be decreased in abundance, however this is not the case. Finally, a number of important points regarding the siRNA-mediated alteration of KCNMA1 protein distribution support the role of cytoplasmic splicing in normal cellular function. Termination codons are found within all reading frames for KCNMA1 intron 16, so its removal, or at the very least a partial removal that is in frame regardless of whether canonical donor and acceptor splice sites are utilized, is required for normal full-length protein synthesis. Further, the sequence coding for the epitope used for antibody detection of the protein is located 3′ to intron 16, therefore, only protein translated after the excision of intron 16 retaining transcripts have been detected. All of these considerations strongly suggest that cytoplasmic splicing of i16 bearing KCNMA1 CIRTs is necessary for properly functioning channels to be expressed in dendritic spines.
Another important aspect of this CIRT-related phenotypic change is the number of KCNMA1 transcripts retaining i16 that can be detected in cells. Approximately 90% of detectable KCNMA transcripts in the cytoplasm are fully spliced, and 10% of the total KCNMA1 transcript population is comprised of KCNMA1i16 CIRTs. This raises the question of the physiological relevance of a relatively small number of KCNMA1 transcripts. The fact that knockdown of only this small fraction of the KCNMA1 mRNA population has significant implications for the function of KCNMA1 proteins implies that low abundance transcript variants and their potential cytoplasmic splicing may be critically important to protein distribution and function in cells. As a result, a threshold for transcript abundance that is functionally relevant may be very low, making it difficult to rule out that other low abundance CIRTs may also have critical cellular functions.
The features that determine which introns are retained and subject to cytoplasmic splicing is unknown. The retained introns in CIRTS are not full length and not all introns for a particular gene are retained. For example, another CIRT variant of KCNMA1 has shed light on how transcript variants are organized among a variety of possible subtypes. Bell et al.10 found that cytoplasmic retention and splicing of intron 17 may be a requirement for inclusion of the alternative stress-axis regulated exon (STREX) in channel transcripts and proteins. An evaluation of transcript populations in this study found that the STREX exon was only found in mature transcripts and those containing i17. Once again, the retention of an intronic sequence that must be removed for proper translation of an important regulatory protein domain (STREX domain) has an impact on downstream cellular function by determining the makeup of channel protein populations.
These examples highlight intron retention, and cytoplasmic splicing, as potentially having an important role for the physiological function of the proteins encoded by these genes. A more generalized function associated with retained introns was reported by Buckley et al.3 along with a host of detectable retained intronic sequences. Retained introns within diverse genes were shown to be capable of harboring sequences that confer novel cellular function. In this study, short interspersed element (SINE) sequences called identifier (ID) elements were often found in retained introns in mRNA harvested from neuronal dendrites. These CIRT-embedded ID elements were found to be capable of interacting with endogenous transport mechanisms in localizing transcripts to the dendrite. This mechanism was hypothesized to allow for a common dendritic localization mechanism across transcripts that can be removed prior to translation. The retained sequences in the transcripts in this study also contained termination codons, as discussed previously, necessitating their removal prior to translation for proper protein formation. In the same study that identified the intronic targeting element, it was shown that competition with endogenous transport mechanisms affected the distribution of protein in the dendrite, showing that these CIRT transcripts contribute to canonical protein populations in neurons.
In addition to intronic targeting elements, early evidence suggests that retained introns in dendrites of rat neurons harbor other potentially functional intronic RNA elements (Table 2), including predicted microRNAs. Given their existence, it is intriguing to speculate that functional non-coding molecules like microRNAs may be liberated from CIRTs in the cytoplasm through extranuclear processing, and exert their effects on local transcript populations. Our work with CIRTs in neurons in addition to other evidence of functional retained introns suggests the presence of a novel post-transcriptional regulation mechanism. One could envision a system in which an incompletely processed transcript is exported from the nucleus, localized to a subcellular compartment by virtue of a retained RNA targeting element, spliced by components of the cytoplasmic spliceosome, perhaps in response to some external stimuli activating the process within the cell, and translated into functional protein. Meanwhile, the liberated retained intron sequence may contain other functional elements such as miRNA sequences that act upon local transcript populations to regulate the local microenvironment or act as a component of regulatory signal cascade back to the nucleus. With cytoplasmic splicing and RNA processing, this would be envisioned to occur without the need for activation of gene expression in the nucleus, and could add to the diversity of cellular phenotype without the need for the expression of an expanded population of genes. This would provide a more rapid regulation of the local environment allowing subcellular functional dependence upon the splicing and translation of the local RNA pool. As translation is a biological amplification, such a mechanism may act to produce a signal that communicates changes in the subcellular RNA and translation states back to the nucleus. While this is speculative, this is a testable hypothesis, as many of the CIRT candidates that would be involved in such communication are known.
TABLE 2.
Clusters of Shared Sequence Elements from Hierarchical Clustering of the Cytoplasmically Retained Intronic Contigs from Rat Neurons
| Sequence Element | Cluster Size |
|---|---|
| B2 | 65 |
| ID | 63 |
Many retained introns that have been reported in the literature contain termination codons that have consequences for the proteins encoded by transcripts bearing these intronic sequences. As discussed, there is evidence that suggests some of these non-coding sequences may be removed by cytoplasmic splicing events, but their presence in cytoplasmic transcripts raises questions about their interaction with the nonsense mediated decay (NMD) pathway within cells. NMD is a highly conserved process in eukaryotes that regulates the degradation of many mRNA transcripts in cells.29,30 It is primarily considered to function as a translation-coupled detector of transcripts with truncated open reading frames. This results from the existence of inappropriate termination codons that occur at a high frequency in intronic sequence that would lead to aberrant protein products upon translation. It is straightforward to see how mutations, transcription or splicing errors could give rise to transcripts destined for NMD. Interestingly, there a large number of `normal' transcripts have also been associated with an NMD regulated degradation pathway.
During conventional splicing, exon–junction complexes (EJC), which are composed of at least three proteins (MAGOH, Y14, and eIF4AIII), are deposited on transcripts to mark sites of intron removal by the spliceosome. These EJCs are also the points of contact for NMD surveillance factors. In particular, when an RNA contains a premature termination codon 5′ to an EJC, NMD is triggered through the activity of these surveillance factors. The inclusion of a retained intron in an mRNA, particularly one bearing an in-frame termination codon, would therefore traditionally be thought to define CIRTs as NMD substrates. NMD is selective in its degradation of RNA molecules, and despite direct evidence pertaining to their status as NMD substrates, the persistence and function of CIRTs suggests that these molecules are able to avoid this particular RNA surveillance pathway. This may occur though the camouflaging of the retained introns by protein association, the inhibition of translation prior to cytoplasmic splicing, or perhaps by a heretofore-unexpected complexity to the signal that targets a particular RNA to the NMD pathway. Interestingly, a recent study4 has linked intron retention and NMD as a mechanism of gene expression regulation. This study identified 86 genes that were involved in immune response, including nuclear lamina genes, with intron retention during differentiation of myeloid cells. They showed that the level of intron retention in these genes increases markedly during each stage of differentiation, resulting in greatly reduced protein levels due to NMD; however, there was no down-regulation of nascent transcripts of these genes. This suggests that NMD specifically destroys a particular `type of RNA,' and as such, may act analogously to the immune system in which particular antigens are targeted for elimination based upon the presence of appropriate antibodies.
In the same way that protein complexes are deposited on transcripts during splicing to indicate a premature stop codon and subsequent transcript degradation, a complex may exist to indicate that a retained intron bearing a stop codon is not to be degraded. This complex could be recruited to a CIRT early in its biogenesis in order to ensure its survival and delivery to the appropriate cellular region before exerting its intended function. Intronic sequence tags may serve to recruit this complex, or the nuclear spliceosome may recognize particular intronic sequences and deposit complexes analogous to the EJC in order to prevent these transcripts from entering the NMD pathway. Even in the absence of such a complex, the evasion of the NMD pathway by CIRTs suggests that particular introns may have sequence tags that promote their stability in the cellular environment. Such intronic sequence tags could also regulate their retention in specific transcript populations under different biological conditions through their interaction with the spliceosome.
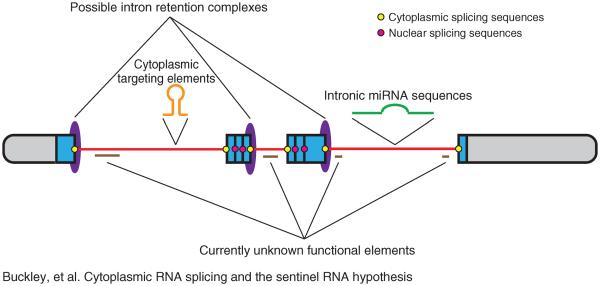
As examples of intron retention, their functional relevance, and the possibility of cytoplasmic splicing continues to emerge for transcripts across a range of cell types, it is not unreasonable to question the origin of mature transcripts in the cytoplasm. Most are certainty fully processed in the nucleus and exported, yet it is possible that a significant number could be the product of a RNA transcript that leaves the nucleus harboring all the features and elements needed to create multiple subpopulations of RNAs under specific cellular conditions, such as restricted to a subcellular compartment or following stimulation. Such an RNA, which we refer to as a `sentinel RNA' (Figure 1), could then serve as a master transcript from which a number of other variants could be derived via splicing in the cytoplasm in response to specific stimulation. This `sentinel RNA hypothesis' is likely to become an area of keen interest as cytoplasmic splicing and its functional consequences continue to emerge.
FIGURE 1.
Model of a sentinel RNA. Includes reported features (e.g., nuclear splicing sequences, ID targeting elements, microRNAs) as well as speculated features (e.g., intron–retention complexes, cytoplasmic splicing sequences, undescribed functional elements).
Footnotes
Conflict of interest: The authors ha•ve declared no conflicts of interest for this article.
REFERENCES
- 1.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmorre SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khaladkar M, Buckley P, Lee M, Francis C, Eghbal M, Chuong T, Suresh S, Kuhn B, Eberwine J, Kim J. Subcellular RNA sequencing reveals broad presence of cytoplasmic intron-sequence retaining transcripts in mouse and rat neurons. PLOS ONE. 2013;8:e76194. doi: 10.1371/journal.pone.0076194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buckley PT, Lee MT, Sul JY, Miyashiro KY, Bell TJ, Fisher SA, Kim J, Eberwine J. Cytoplasmic intron sequence-retaining transcripts can be dendritically targeted via ID element retrotransposons. Neuron. 2011;69:877–884. doi: 10.1016/j.neuron.2011.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong JJ, Ritchie W, Ebner OA, Selbach M, Wong JW, Huang Y, Gao D, Pinello N, Gonzalez M, Baidya K, et al. Orchestrated intron retention regulates normal granulocyte differentiation. Cell. 2013;154:583–595. doi: 10.1016/j.cell.2013.06.052. [DOI] [PubMed] [Google Scholar]
- 5.Zhong X, Liu JR, Kyle JW, Hanck DA, Agnew WS. A profile of alternative RNA splicing and transcript variation of CACNA1H, a human T-channel gene candidate for idiopathic generalized epilepsies. Hum Mol Genet. 2006;15:1497–1512. doi: 10.1093/hmg/ddl068. [DOI] [PubMed] [Google Scholar]
- 6.Forrest ST, Barringhaus KG, Perlegas D, Hammarskjold ML, McNamara CA. Intron retention generates a novel Id3 isoform that inhibits vascular lesion formation. J Biol Chem. 2004;279:32897–32903. doi: 10.1074/jbc.M404882200. [DOI] [PubMed] [Google Scholar]
- 7.Denis MM, Tolley ND, Bunting M, Schwertz H, Jiang H, Lindemann S, Yost CC, Rubner FJ, Albertine KH, Swoboda KJ, et al. Escaping the nuclear confines: signal-dependent pre-mRNA splicing in anucleate platelets. Cell. 2005;122:379–391. doi: 10.1016/j.cell.2005.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mansilla A, Lopez-Sanchez C, de la Rosa EJ, Garcia-Martinez V, Martinez-Salas E, de Pablo F, Hernandez-Sanchez C. Developmental regulation of a proinsulin messenger RNA generated by intron retention. EMBO Rep. 2005;6:1182–1187. doi: 10.1038/sj.embor.7400539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bell TJ, Miyashiro KY, Sul JY, McCullough R, Buckley PT, Jochems J, Meaney DF, Haydon P, Cantor CR, Parsons TD, et al. Cytoplasmic BK(Ca) channel intron-containing mRNAs contribute to the intrinsic excitability of hippocampal neurons. Proc Natl Acad Sci U S A. 2008;105:1901–1906. doi: 10.1073/pnas.0711796105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bell TJ, Miyashiro KY, Sul JY, Buckley PT, Lee MT, McCullough R, Jochems J, Kim J, Cantor CR, Parsons TD, et al. Intron retention facilitates splice variant diversity in calcium-activated big potassium channel populations. Proc Natl Acad Sci U S A. 2010;107:21152–21157. doi: 10.1073/pnas.1015264107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linan-Rico A, Jaramillo-Polanco J, Espinosa-Luna R, Jimenez-Bremont JF, Linan-Rico L, Montano LM, Barajas-Lopez C. Retention of a new-defined intron changes pharmacology and kinetics of the full-length P2X2 receptor found in myenteric neurons of the guinea pig. Neuropharmacology. 2012;63:394–404. doi: 10.1016/j.neuropharm.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 12.Chen Z, Gore BB, Long H, Ma L, Tessier-Lavigne M. Alternative splicing of the Robo3 axon guidance receptor governs the midline switch from attraction to repulsion. Neuron. 2008;58:325–332. doi: 10.1016/j.neuron.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 13.Lejeune F, Cavaloc Y, Stevenin J. Alternative splicing of intron 3 of the serine/arginine-rich protein 9G8 gene. Identification of flanking exonic splicing enhancers and involvement of 9G8 as a trans-acting factor. J Biol Chem. 2001;276:7850–7858. doi: 10.1074/jbc.M009510200. [DOI] [PubMed] [Google Scholar]
- 14.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 15.Turunen JJ, Niemela EH, Verma B, Frilander MJ. The significant other: splicing by the minor spliceosome. WIREs RNA. 2013;4:61–76. doi: 10.1002/wrna.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Konig H, Matter N, Bader R, Thiele W, Muller F. Splicing segregation: the minor spliceosome acts outside the nucleus and controls cell proliferation. Cell. 2007;131:718–729. doi: 10.1016/j.cell.2007.09.043. [DOI] [PubMed] [Google Scholar]
- 17.Friend K, Kolev NG, Shu MD, Steitz JA. Minor-class splicing occurs in the nucleus of the Xenopus oocyte. RNA. 2008;14:1459–1462. doi: 10.1261/rna.1119708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pessa HK, Will CL, Meng X, Schneider C, Watkins NJ, Perala N, Nymark M, Turunen JJ, Luhrmann R, Frilander MJ. Minor spliceosome components are predominantly localized in the nucleus. Proc Natl Acad Sci U S A. 2008;105:8655–8660. doi: 10.1073/pnas.0803646105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh J, Padgett RA. Rates of in situ transcription and splicing in large human genes. Nat Struct Mol Biol. 2009;16:1128–1133. doi: 10.1038/nsmb.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshida H. Unconventional splicing of XBP-1 mRNA in the unfolded protein response. Antioxid Red Signal. 2007;9:2323–2333. doi: 10.1089/ars.2007.1800. [DOI] [PubMed] [Google Scholar]
- 21.Chakrabarti A, Chen AW, Varner JD. A review of the mammalian unfolded protein response. Biotechnol Bioeng. 2011;108:2777–2793. doi: 10.1002/bit.23282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malhotra JD, Kaufman RJ. The endoplasmic reticulum and the unfolded protein response. Sem Cell Dev Biol. 2007;18:716–731. doi: 10.1016/j.semcdb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao RV, Bredesen DE. Misfolded proteins, endoplasmic reticulum stress and neurodegeneration. Curr Opin Cell Biol. 2004;16:653–662. doi: 10.1016/j.ceb.2004.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glanzer J, Miyashiro KY, Sul JY, Barrett L, Belt B, Haydon P, Eberwine J. RNA splicing capability of live neuronal dendrites. Proc Natl Acad Sci U S A. 2005;102:16859–16864. doi: 10.1073/pnas.0503783102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruegsegger U, Leber JH, Walter P. Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell. 2001;107:103–114. doi: 10.1016/s0092-8674(01)00505-0. [DOI] [PubMed] [Google Scholar]
- 26.Back SH, Lee K, Vink E, Kaufman RJ. Cytoplasmic IRE1α-mediated XBP1 mRNA splicing in the absence of nuclear processing and endoplasmic reticulum stress. J Biol Chem. 2006;281:18691–18706. doi: 10.1074/jbc.M602030200. [DOI] [PubMed] [Google Scholar]
- 27.Berezhna S, Supekova L, Supek F, Schultz P, Deniz A. siRNA in human cells selectively localizes to target RNA sites. Proc Natl Acad Sci. 2006;103:7682–87. doi: 10.1073/pnas.0600148103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allo M, Buggiano V, Fededa JP, Petrillo E, Schor I, de la Mata M, Agirre E, Plass M, Eyras E, Elela SA, et al. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat Struct Mol Biol. 2009;16:717–724. doi: 10.1038/nsmb.1620. [DOI] [PubMed] [Google Scholar]
- 29.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Ann Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 30.Schweingruber C, Rufener SC, Zund D, Yamashita A, Muhlemann O. Nonsense-mediated mRNA decay—mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim Biophys Acta. 2013;1829:612–623. doi: 10.1016/j.bbagrm.2013.02.005. [DOI] [PubMed] [Google Scholar]