Abstract
While much is known about human genetic variation, such information is typically ignored in assembling novel genomes. Instead, reads are mapped to a single reference, which can lead to poor characterization of regions of high sequence or structural diversity. We introduce a population reference graph, which combines multiple reference sequences and catalogues of variation. The genomes of novel samples are reconstructed as paths through the graph using an efficient hidden Markov model, allowing for recombination between different haplotypes and additional variants. By applying the method to the 4.5Mb extended MHC region on human chromosome 6, combining eight assembled haplotypes, sequences of known classical HLA alleles and 87,640 SNP variants from the 1000 Genomes Project, we demonstrate, using simulations, SNP genotyping, short-read and long-read data, how the method improves the accuracy of genome inference and reveals regions where the current set of reference sequences is substantially incomplete.
Introduction
The current paradigm for analyzing human genomes using high throughput sequence (HTS) data is to map to a single haploid reference sequence in which there is no representation of variation1-3. Across much of the genome, such exclusion has little effect on the accuracy of genome inference because of the relatively low genetic diversity of humans4. However, for some regions, such as the major histocompatibility complex (MHC) on chromosome 6, which contains the human leukocyte antigen (HLA) genes, there is very substantial sequence and structural variation5. Such diversity can result in poor genomic characterization in individuals who carry sequence that is either missing or highly divergent from the single reference. Other locations of high diversity include the KIR6 region, olfactory gene clusters7, ancient inversions such as that on 17q21.318-10 and regions of recurrent genomic rearrangement11, many of which have substantial influence on phenotype and disease risk. In many of these cases, multiple alternative haplotypes have been characterized and are available. For example, there are seven alternative, plus one primary (PGF), MHC haplotypes in the human reference (GRCh37). More generally, sequencing projects have greatly advanced our understanding of human genetic variation12-14 and using such information to help characterize human genomes represents an important and unsolved problem
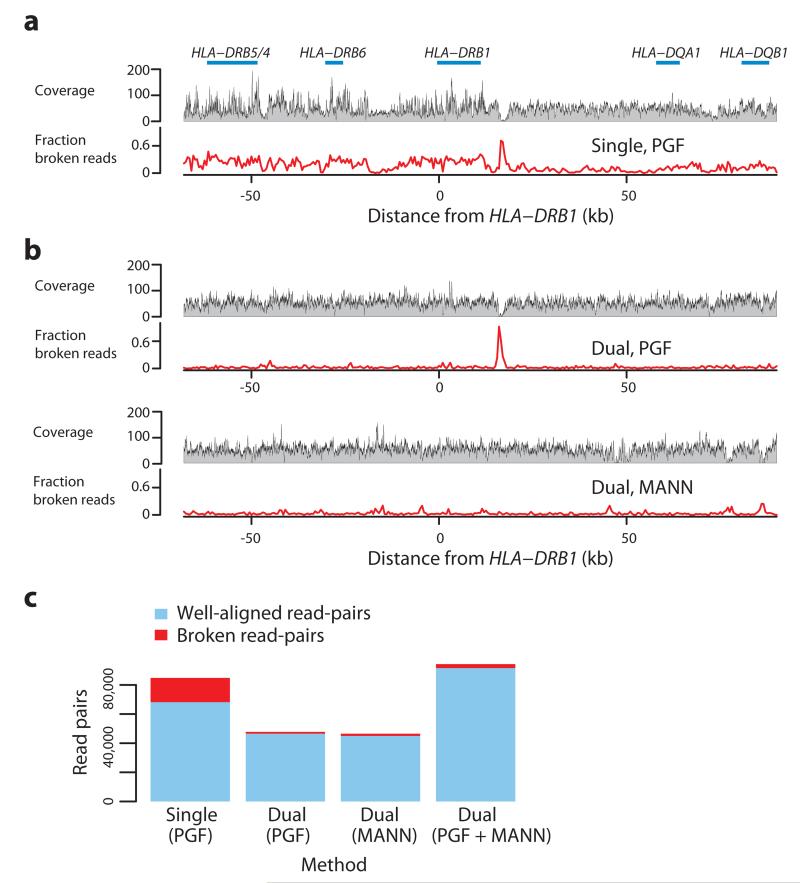
The problem of the single reference approach and the potential for using known MHC variation is demonstrated in Figure 1. When mapping to the standard reference (the PGF MHC haplotype) results in large fluctuations in coverage and many poorly-mapped reads (Fig. 1a). However, when the reference is augmented with an additional haplotype, identified by comparing the classical HLA genotypes of the sample with those of the eight reference haplotypes and noting that one of the eight haplotypes was a close match, read coverage and alignment is greatly improved (Fig. 1b, c).
Figure 1. Read-mapping in the MHC Class II region.
a. Summary of read alignment to a single reference (GRCh37 without alternative loci, containing the ‘PGF’ haplotype in the xMHC) for a single sample (CS1) in the MHC Class II region (around HLA-DRB1) showing coverage (grey profile) and the proportion of ‘broken’ read-pairs (red line; defined as mapping to different chromosomes, incompatible strands, or implausible insert size). b The same metrics as for part a, where mapping has been performed to a reference augmented with the MANN (ALT_REF_LOCI_4, Genbank ID GL000253.1) haplotype (i.e. in addition to PGF), chosen because the combined classical HLA genotypes from PGF and MANN match those of the sample. c. Number of mapped intact (blue) and broken (red) read pairs for experiments underlying panels a and b (results from the latter split according to which haplotype reads map to, and a combined metric), demonstrating that the augmented reference results in many more well-mapped and many fewer broken read-pairs.
Using prior information about variation raises five main challenges. First, a data structure for representing genomic variation must be defined, which can accommodate multiple sources of information, from assembled reference sequence (such as the ALT paths in GRCh37) to catalogues of small variants such as the 1000 Genomes Project12,14. Second, algorithms must exist for matching high-throughput sequencing (HTS) data to the variation aware reference structure. Third, and potentially simultaneously with step two, additional variation not yet represented in the reference data structure must be detected. Fourth, because most functional information (such as gene location and structure) uses the coordinates of a single linear reference, information from a variation-aware reference must be projected onto a primary sequence. Finally, benchmarks must be established to validate and compare the output from a variation aware reference tool-chain to that provided by existing approaches.
To date, these challenges have only been partially addressed. Traditional multiple sequence alignments, representing inter- and intra-species genetic variation, have been generalized to partial order alignment (POA) graphs15 to represent shared sequence and to represent mosaic sequences arising from recombination, and then further to A-Bruijn graphs16 and cactus graphs17 to support rearrangements and duplications. However, these graphs have not been used in the assembly of individual genomes from HTS data. Conversely, multiple approaches of mapping individual reads to variation-aware data structures have been proposed18-22. However, none of these are practical for representing a heterogeneous catalogue of population variation with large and small events and the additional mutation and recombination-driven differences found between reference material and the sample being studied.
Here, we present a solution to these challenges. We describe a structure for representing known variation called a population reference graph (PRG) and a series of algorithms that enable characterization of the genomes present in an individual from HTS data. We build on previous work for using coloured de Bruijn graphs for analyzing sequence variation23, but also take advantage of the existing tool chain for read mapping and variant calling3,19. To demonstrate the value of the method we develop a PRG for the MHC region and combine simulation with analysis of empirical data on SNP genotypes, classical HLA types, short-read and synthetic long-read Moleculo data.
Results
The population reference graph
A population reference graph (PRG) is a directed acyclic graphical model for genetic variation generated by using known allelic relationships between sequences (Figs. 2a, 2b; Supplementary Fig 1). The graph is constructed in three steps (see Supplementary Note for details). First, reference sequences are aligned using standard multiple sequence alignment (MSA) methods24,25. Second, a graph structure is generated from the MSA by collapsing aligned regions with sequence identity over a defined kmer size. This structure is related to the POA graph15, though differs in preserving more information about local haplotype structure, which is important for read alignment in regions of high sequence diversity. Third, small variants, defined (as in VCF) by a reference position and alternative alleles, are added to all valid paths (i.e., a SNP cannot be added to a path with a deletion). Here, we use the primary assembly and seven MHC ALT sequences from GRCh37, along with SNPs from Phase 1 of the 1000 Genomes Project and f classical HLA allele sequences from the International Immunogenetics Information System (IMGT26) at key HLA Class I and Class II loci (Supplementary Table 1). The resulting graph structure can be thought of as a generative model for genomes. From a limited set of input sequences, many different paths through the graph are possible, capturing the effect of recombination..
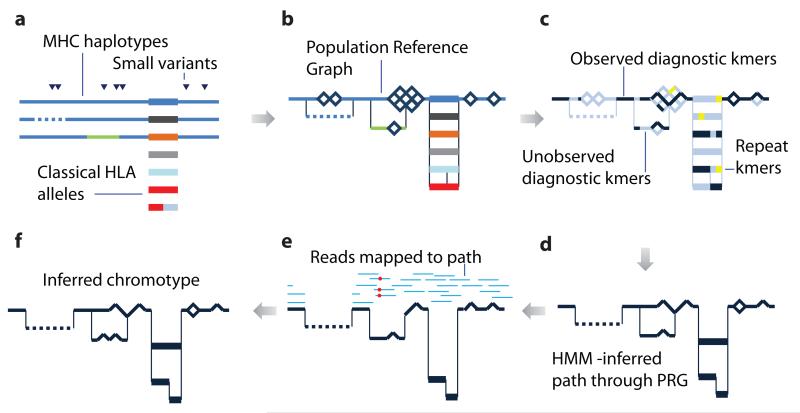
Figure 2. Schematic illustration showing the construction and application of a population reference graph.
a. Multiple sources of information about genetic variation, including alternative reference haplotypes (lines), classical HLA alleles (rectangles) and SNPs / short indels (triangles) are aligned. Colours indicate divergent sequence, dashes indicate gaps. b. A population reference graph (PRG) is constructed from the alignment, resulting in a generative model for variation within the region. SNPs, indicated by diamonds, are added as alternative paths to all valid backgrounds (i.e. excluding sequence with gaps or a third allele at the position). c. The PRG is compared to the de Bruijn graph constructed from reads obtained from a sample. Informative kmers (i.e. those that are found at only one level in the PRG) are identified (dark blue). Those found elsewhere in the genome (yellow) are ignored. d. A hidden Markov model is used to infer the most likely pair of paths through the PRG, allowing for read errors, resulting in an individualized reference chromotype for the sample. e. Two haploid genomes are constructed from the reference chromotype, with arbitrary phasing between adjacent bubbles, and reads (light blue lines) from the sample are aligned and assigned (on the basis of mapping quality) to a reference, thus identifying places where the sample contains novel variation (red circles; only one path through the chromotype is shown). f. Newly-discovered variants modify the reference chromotype, resulting in the inferred chromotype for the sample.
Using the PRG to infer individual genomes
The use of HTS in humans largely relies on genome(s) being closely related to the reference, thus enabling reads to be mapped accurately and with appropriate certainty. We extend this idea by inferring the (diploid) path through the PRG that most closely resembles the two haplotypes of the sample. Specifically, by comparing the HTS data from a sample to the PRG we construct a diploid personalized reference genome, here referred to as a chromotype (which could be generalized to higher ploidies or mixtures). To infer novel variation, we map reads to the chromotype and use existing variant calling software19. A chromotype for a diploid is best understood as a bifurcating/merging sub-graph of the PRG, analogous to paired homologous chromosomes with bubbles at regions of divergence.
To infer chromotypes we exploit the computational efficiency of hidden Markov models. Briefly, HTS data is summarized (using Cortex23) by the counts of each string of length k (kmer). Similarly, the set of kmers that can be emitted from the PRG is enumerated, eliminating those that occur at more than one level within the PRG (i.e. are paralogous), hence uninformative (Fig. 2c). Finally, by using a probabilistic model for the emission of kmers (Methods), the Viterbi-algorithm infers the maximum-likelihood (ML) chromotype (Fig. 2d). Note that this approach does not preserve long-range haplotype phase information and cannot detect variants absent from the PRG. In addition, the focus on diagnostic kmers limits our ability to analyze low-complexity regions, such as segregating segmental duplications, where read-depth information is required for genotyping.
To detect novel variation, the inferred chromotype is decomposed into two haplotypes (with arbitrary phasing between adjacent bubbles), which then replace the homologous region in the primary reference. Reads are mapped to the two resulting reference genomes and placed at their best position across the two reference genomes, as measured by mapping quality, or uniformly if mapping qualities are identical. A standard variant caller19 is used to discover new alleles independently in the two mappings and a heuristic algorithm modifies the chromotype, incorporating novel variants. We have also developed an algorithm for mapping reads directly to the chromotype, however this is currently too slow for analyzing millions of reads and hence only used for Moleculo validation, see below.
Validation and comparison to other methods
To assess the value of the PRG approach in characterizing variation within samples we used simulations and empirical data analysis. We compare four approaches to characterizing variation.
As a base-line we use a single reference (the PGF haplotype within the MHC region from GRCh37) and look at the effect of calling a sample as everywhere homozygous-reference (“PGF Reference”).
We use a read-mapping approach (Stampy3 followed by Platypus19) in which the components were designed explicitly for high sensitivity detection and genotyping of short INDELs and clustered variants (“Platypus”). The resulting VCF is converted into a chromotype (see Methods) for comparison.
From the PRG, we assess the Viterbi chromotype, representing a “best guess” diploid path through the PRG (“PRG-Viterbi”). These are also reported as VCF.
From the PRG, we assess the mapping-modified Viterbi chromotypes, containing variants not represented in the PRG (“PRG-Mapped”). These are also reported as VCF.
Simulations
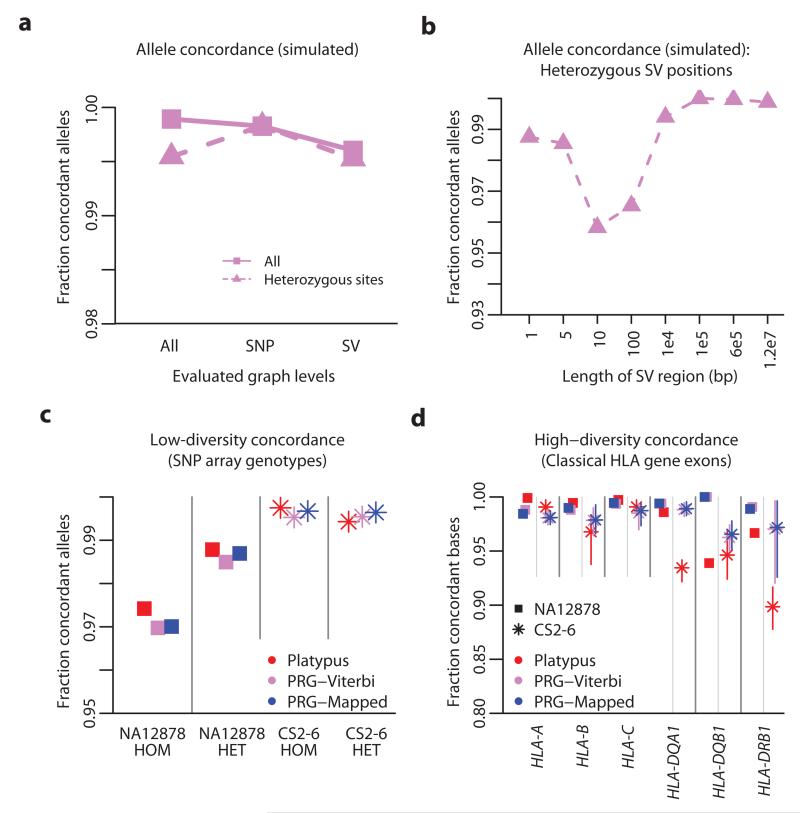
To verify that the method and implementation can work, prior to validation based on empirical data, we simulated high coverage HTS data (101bp paired-end reads from a 30x genome with an empirical error distribution) for 20 individuals. Each simulated diploid genome consists of two random paths through the PRG for the extended MHC (xMHC). The simulated genomes carry a mixture of recombination events between the original eight MHC haplotypes, SNPs and structural variants of varying size (insertions and deletions from 1 – 125,000bp). From the simulated data, we infer, for each sample, the pair of paths through the PRG using the HMM and measure allele concordance with the simulated paths at each level within the PRG (Supplementary Table 2). Across all levels (broadly corresponding to positions in the sequence), 99.89% of alleles are correctly recovered. Accuracy at heterozygous SNP positions is similar (99.83%) and drops slightly for INDEL positions (ranging from 95.8% to 100%, Figs. 3a, 3b).
Figure 3. Simulation study and empirical validation.
a. Allele concordance between simulated data (20 simulated diploid individuals; 101bp reads at 30x diploid coverage with empirical error distribution) and Viterbi path through the PRG stratified by simulated variant type (SNP or structural variant; SV) and genotype. b. Allele concordance in simulations at sites heterozygous for structural variants of different lengths. c. Allele concordance between SNP array genotypes and chromotypes from each method for NA12878 (squares; Illumina Omni 2.5M array) and the CS2-6 samples (stars; Illumina 1M array), stratified by whether the array specifies the genotype as homozygous or heterozygous. Results are shown for the mapping-based approach (Platypus, red), the Viterbi-path through the PRG (PRG-Viterbi, pink) and after mapping to the reference chromotype (PRG-Mapped, blue). d. Allele concordance between classical HLA genotypes at HLA-A, HLA-B, HLA-C, HLA-DQA1, HLA-DQB1 and HLA-DRB1 (measured at a per-base level) and chromotypes from each method for NA12878 and the CS2-6 samples (range of accuracy across CS2-6 displayed as vertical bars). Classical HLA genotypes were inferred from sequence-based HLA typing (see Methods).
Experiment 1: Comparison to SNP array data
To assess the ability of the PRG approach to genotype variation at sites of high uniqueness within the genome, we measured allele concordance at SNP positions within the xMHC region independently interrogated through array genotyping and HTS: one sample (NA12878) at 60x coverage with 100bp paired end-reads / Illumina Omni 2.5M array data and five clinical samples (CS2-6) at 30x coverage with 90bp paired-end reads / Illumina 1M array data (Methods).
The accuracy of all approaches is high (Fig. 3a); ≥97.38% allele concordance with the Illumina Omni 2.5M array (NA12878) and ≥99.53% allele concordance with the Illumina 1M array (CS2-6). Comparing the array genotype concordance of Platypus-generated genotypes and PRG-generated genotypes (PRG-Viterbi and PRG-Mapped), we find that both approaches yield comparable accuracies (97.75% vs 97.45% for the 2.5M array and 99.57% versus 99.66% for the 1M array; Fig. 3c, Supplementary Table 3).
Of the 285 sites at which the array genotypes for NA12878 disagree with the Viterbi chromotype, in 55 cases this difference is driven by the Viterbi chromotype specifying a gap character suggesting the presence of an indel that could interfere with array genotyping. We manually inspected the alignment1 of NA12878 reads for these sites, and found clear evidence for the presence of a deletion in 33 of the 55 cases (visualizations of read mapping at all positions are provided as Supplementary Data). These findings suggest that a significant fraction of the discrepancy between array and PRG approaches results from array errors at polymorphic indels. The cause of the remaining discrepancies is not understood.
Because almost all variant sites reported in NA12878 are present within the PRG, we also assessed accuracy of variant detection and genotyping for sites by comparing calls to an independent call set on the same data generated through de novo assembly with Cortex23. At sites within the graph we find that all methods perform well (allele concordance for Platypus: 96.7%, PRG-Viterbi: 96.7%, PRG-Mapped: 97.2%). At sites not in the PRG, all methods show poorer performance, though the mapped step improves accuracy substantially (Platypus: 65.9%, PRG-Viterbi: 40.2%, PRG-Mapped: 55.1%).
Experiment 2: Comparison to classical HLA data
In regions of high sequence diversity, such as the classical HLA alleles, single-reference mapping and variant calling methods may perform poorly because of the density of mismatches to the reference. To assess the accuracy of different methods at the classical HLA loci, we compared the per-base diploid genotypes inferred by mapping and PRG approaches to those expected from the results of sequence-based typing of the highly polymorphic exons of Class I (HLA-A, -B and –C) and Class II (HLA-DQA1, -DQB1 and –DRB1) genes in NA12878 and CS2-6. We analyzed agreement with the reference sequence for the reported allele (in HLA nomenclature this means XX:XX:01 or XX:XX:01:01 at 6 or 8 digit resolution respectively, though we note that typing was not carried out at this resolution). This analysis is distinct from classical HLA typing, where the presence of a particular set of haplotypes is inferred.
For Class I loci (HLA-A, -B and –C), we find comparable and high (typically ≥ 99%) accuracy for all methods (no comparison has P < 0.01 by paired t-test; Fig. 3d, Supplementary Table 4). In contrast, for Class II loci, PRG methods are significantly more accurate at HLA-DQB1 (P = 0.001) and HLA-DRB1 (P = 0.002) than Platypus, with no difference between the PRG-Viterbi and PRG-Mapped methods. For example, at DRB1, we find 97.19% allele concordance with the PRG-Mapped genotypes versus 89.85% concordance with mapping-based genotypes in the CS2-6 samples). The main difference between Class I and Class II loci is the existence of polymorphic paralogues and pseudogenes, which are likely to confuse approaches that map to a single reference, but which are represented in the GRCh37 ALT haplotypes. The very modest gain in accuracy from the mapping step (<1%) likely reflects the very extensive characterization of genetic variation within classical HLA alleles.
Experiment 3: kmer recovery from high coverage samples
A key notion of the PRG is that it contains the majority of sequence likely to be found in any individual. In the absence of full and independent de novo assemblies, we can nevertheless assess chromotype accuracy by measuring the recovery of kmers from HTS data; i.e. the proportion of kmers implied by the inferred chromotype that are found in the sample’s sequence data. We apply this benchmark to NA12878 and the CS2-6 samples.
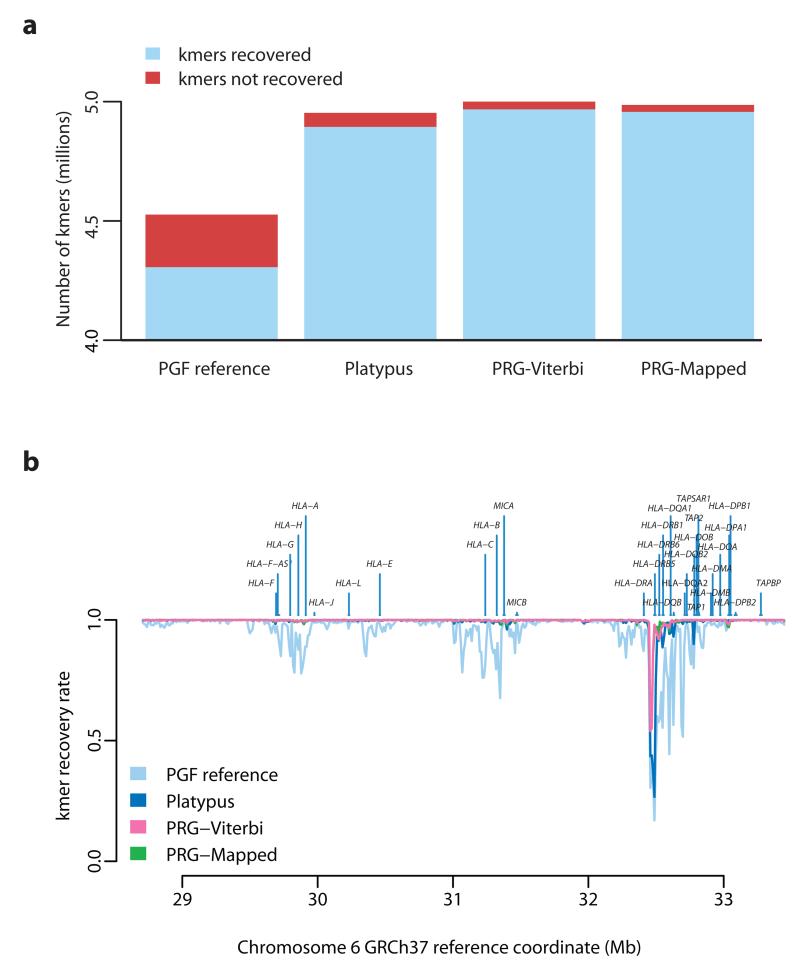
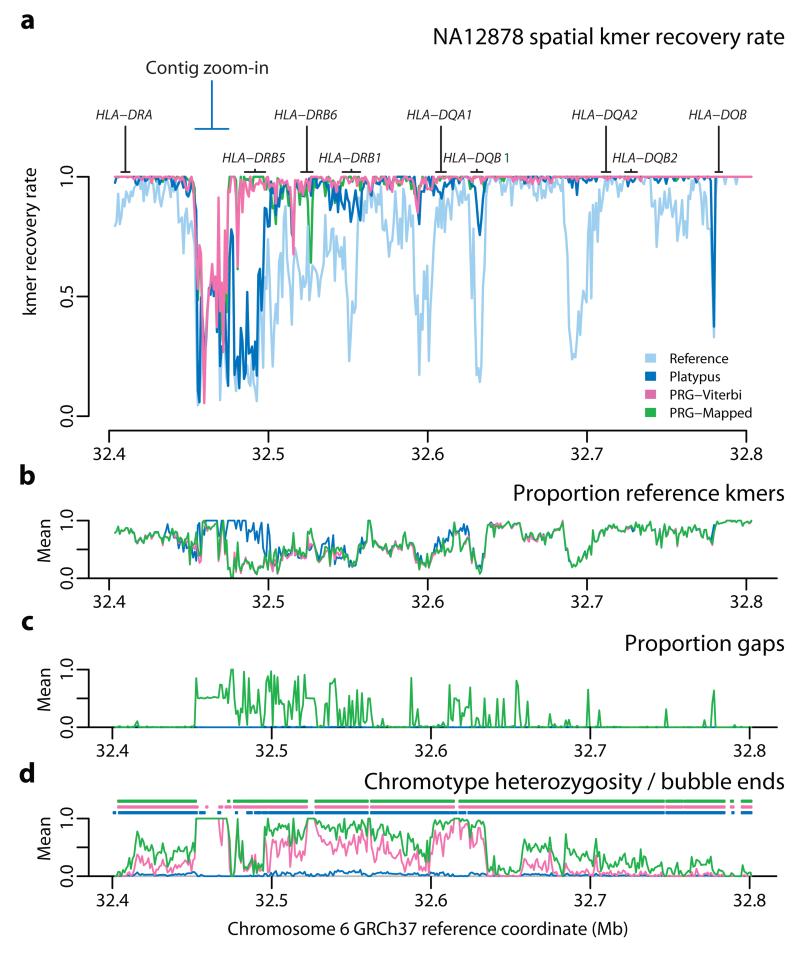
Across the 4.75 Mb xMHC region, the PGF reference contains 4.52M distinct kmers (k = 31) of which 4.8% are not recovered in the HTS data from NA12878 (Fig. 4a). The mapping approach (Platypus) predicts 4.94M distinct kmers, of which 1.2% are not recovered, while the two PRG approaches predict 4.98M (PRG-Viterbi) and 4.97M (PRG-Mapped) distinct kmers respectively and 0.63% and 0.57% are not recovered. Results are comparable though slightly lower for all methods in the CS2-6 samples (Supplementary Table 5). Consequently, the PRG approaches both predict greater sequence diversity than the mapping approach and achieve a higher rate of sequence recovery. Although the majority of the xMHC is accessible to all methods, there is substantial spatial heterogeneity in the rate of kmer recovery (Fig. 4b). Particularly, in the HLA class II region, the PRG approaches considerably outperform mapping (Fig. 5), consistent with knowledge of genomic complexity involving the HLA-DRB paralogues. We also note that in some regions, in particular around HLA-DRB5, all approaches perform poorly in terms of kmer recovery (Fig. 5), suggesting that current catalogues of sequence within the xMHC are substantially incomplete.
Figure 4. Recovery of chromotype kmers from high throughput sequencing data.
a. Number of recovered (blue) and non-recovered (red) kmers present in chromotypes inferred by the four methods (as for Fig. 3c with addition of single reference represented by the PGF MHC haplotype). A kmer is counted as recovered if it appears in HTS data from NA12878 (c. 60x coverage of 100bp paired-end reads represented by an un-cleaned Cortex graph; k = 31). Chromotypes within regions of clustered variants are disentangled using a greedy algorithm prior to evaluation, optimizing for the disentangled haplotypes to contain as many kmers recovered in the sample as possible (see Supplementary Note). b . Spatial pattern of kmer recovery along the extended MHC region for each of the four chromotypes showing the location of classical HLA loci. Recovery fraction averaged over 1 kb windows.
Figure 5. Spatial recovery of kmers within the HLA Class II region.
a. Blow-up of kmer recovery in Fig 4b in the MHC Class II region for the chromotypes inferred by the four approaches. b. Fraction of kmers predicted to be present along region that are also presented in the PGF reference haplotype (1 kb windows; PGF reference not shown). c. Fraction of positions in chromotype that correspond to gaps in the multiple sequence alignment used to construct the PRG (1 kb windows). Note that PRGComplete chromotype is effectively identical to the PRG-Viterbi path. d. Fraction of positions in inferred chromotypes that are heterozygous (lines; note this includes sites where one allele is a gap character) and the ending points of chromotype bubbles (points).
Within the classical HLA loci, all methods perform well for class I loci, recovering 98-100% of kmers compared to 80-95% from the PGF reference haplotype (Supplementary Fig. 2). At Class II loci, however, the advantage of the PRG methods (PRG-Viterbi and PRG-Mapped) is pronounced, with approximately 99% of all kmers recovered for HLA-DQA1, -DQB1 and -DRB1 (for both PRG methods), compared to 88-95% for Platypus (against a base-line of 37-85% for the PGF reference haplotype).
Experiment 4: Comparison to synthetic long-read Moleculo data
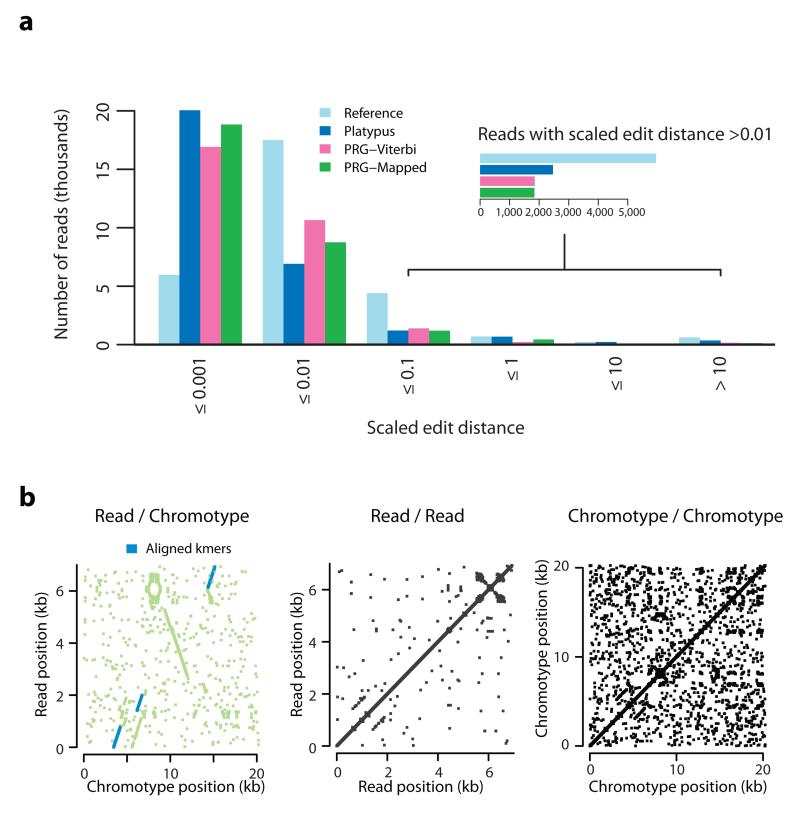
To assess accuracy over longer physical distances than kmers, we analyzed alignments of synthetic long-read Moleculo data (25x coverage) from NA12878 to chromotypes generated by each approach (Methods). We first identified 29,429 reads (median read length 3,165 bp; for convenience, we refer to the Moleculo sequences as reads, although they involve an assembly procedure) likely to have arisen from the xMHC region through the presence of diagnostic kmers (Online Methods). Read-to-chromotype alignment was performed with a Needleman-Wunsch-like algorithm that aligns to gapped graphs instead of sequence, implemented using dynamic programming (see Supplementary Note). We measure the scaled edit distance between reads and the chromotype (the number of non-identical characters in read to chromotype global alignment, including gap characters, divided by read length in kmers) as an indicator of chromotype accuracy.
We find that the mapping (Platypus) approach achieves the highest number of read alignments with zero mismatches (11,338 versus 10,071 for PRG-Mapped). However, both PRG approaches result in significantly fewer reads with many mismatches and/or gaps (Fig. 6a, Supplementary Table 6). For example, the total number of alignment columns indicating a deletion in the chromotype decreases from 1,017,231 (Platypus) to 586,852 (PRG-Mapped). Likewise, the number of reads with very bad alignments (more than 150,000 gaps in the aligned read or ≥33% of the aligned chromotype string consisting of novel gaps) decreases from 303 to 134. The modified chromotype (PRG-Mapped) has a modest benefit over the Viterbi chromotype (PRG-Viterbi), increasing the number of perfectly mapped reads from 8,359 to 10,071. Across the DRB5 region (identified from the kmer recovery analysis as being most poorly represented by the PRG) we find reads that suggest the presence of an inversion relative to known sequence (Fig. 6b).
Figure 6. Alignment of synthetic long-read data to chromotypes.
a. Histogram of scaled edit distance (the number of non-concordant columns in the alignment between read and chromotype, divided by the total number of bases in the read) between long-read data (Illumina NA12878 Moleculo xMHC-specific reads) to chromotypes inferred by four methods. Lower boundary for each interval omitted for clarity. Inset shows a blow-up for reads with scaled edit distance >0.01. b. Dot-plot between the sequence of a Moleculo contig and the sequence of the non-gap branch of the Viterbi chromotype for NA12878 over the region highlighted in Fig. 5a. There is a point (x, y) if and only if the 10-mer beginning at position x in the chromotype segment is identical to the 10-mer (or its reverse complement) beginning at position y in the read. Green indicates the region of the read which, according to the alignment, is matched to the target region (i.e. each green point represents a read kmer between the leftmost and the rightmost read kmers aligned to the target region). Blue indicates that the match between the kmer found at positions x in the chromotype and y in the read can be recovered from the alignment. Middle, right: Analogous dot-plots for the read and the chromotype against themselves, showing that there is no large-scale self-similarity along either sequence.
Discussion
Within a species (or even within an individual), the effects of mutation, recombination and selection can result in a great diversity of genomes, differing through events ranging from single nucleotide changes to major rearrangements and gains or losses of sequence. Our hypothesis was that using information about known diversity would aid in the reconstruction of individual genomes from HTS data, particularly within regions of high sequence and structural variation. To this end, we devised a graph structure for representing such reference variation, a method for using the structure to interrogate short read HTS data so as to infer the diploid sequence of an individual and a series of benchmarking tests to evaluate accuracy compared to a standard mapping pipeline. By applying the approach to variation within the MHC region, we identified regions where genome inference is improved, sometimes substantially. This work demonstrates the feasibility and potential of using known variation in genome inference from HTS data and represents an important intermediary between mapping to a single reference and full de novo assembly23,27,28. Our method has immediate application for researchers looking to understand the role of genetic variation within the MHC for disease risk and drug response, and also builds a framework for analyzing complex variation more generally, not least those regions with alternative assemblies in humans.
There are, however, many important choices concerning how to represent known variation, the set of variants to be included and how best to use such information in genome inference. These choices must be taken in the light of potential applications, ranging from microbial populations with highly mobile accessory genomes to common rearrangements in cancer. Below, we discuss the key considerations and how the approach described here could be extended or modified.
Choices about the structure and construction of a reference variation graph are intimately linked to its desired functions. Fundamentally, we see two functions of such a structure. First, it should provide a general and intuitive way of referring to variation, in a manner analogous to that of an rsID for SNPs and in a manner that is incremental over the current state (e.g. where an rsID or HGVS description retain a precise meaning). Second, it should be a generative model for new genomes, reflecting recombination between sequences in a manner that closely matches the true distribution of genomes. Our approach was to base the structure on a multiple sequence alignment of known material, allowing for recombination between sequences at aligned regions of identity. As such, the structure makes no attempt to model explicitly events such as duplication or rearrangement that lead to difficulties or ambiguities in alignment. For example, an inversion would be represented by a bubble in the same way as would a region of high divergence. Similarly, the homology within a copy-number-variable region would also not be recognized explicitly. To represent such events, and the more complex rearrangements and amplifications observed in bacteria and cancer, alternative structures would need to be developed, such as the A-Bruijn16 or cactus graphs17. However, whether such structures are well-suited to the problem of inference from HTS remains to be explored.
In constructing the PRG, we chose to include a wide catalogue of information including short variants, long haplotypes and lists of alleles at classical HLA loci. The comparison with the standard mapping approach suggests that over much of the xMHC, the use of such information under the current implementation leads to little or no gain in accuracy. The choice about what material to include in a graph is a balance between wanting to describe the space of genomic variation most fully and the practical issue of building and using a graph that represents many small and/or rare events, whose inclusion is not necessary and potentially damaging to inference (for example a duplication seen just once of an otherwise unique region). A pragmatic approach is to say that material should be included if, on average, it leads to better genome inference (here, for example, structural variation in the Class II region and Class II alleles). However, there are other possible advantages to including more sequence, for example in reducing the heterogeneity in how complex variants, or those in low complexity regions, are reported.
Perhaps the greatest limitation of the approach developed here is in terms of the inference algorithm. By summarizing HTS data as kmers, we lose longer range information within a read and between read pairs. In addition, while the HMM for inferring the underlying reference chromotype is efficient, the approach of mapping reads separately to each of the arbitrarily phased haplotypes is ad hoc. Ideally reads should be aligned directly to the graph structure, keeping track of the quality of mapping both within and across different levels in the graph. In principle, as demonstrated with the Moleculo data, graph mapping is feasible. However there is a major challenge in making the process comparable in efficiency to algorithms for to mapping to a linear reference. However, if direct mapping of reads to the graph can be achieved, the same HMM structure can be used for genome inference, though we note that the current structure is not well suited to analyzing polymorphic regions with extended identity where the ability to reconstruct the exact underlying sequence (as opposed to some summary, such as copy number) is limited. In theory, it would also be possible to use longer-range information about haplotype structure as a prior on paths through the PRG (such as is used in imputation29 and refinement of low-coverage sequencing data30).
Finally, we wished to know how unusual the Class II MHC region is within the human genome in being poorly served by the paradigm of mapping to a single reference. To assess this, we calculated a genome-wide kmer recovery map for the Platypus call set on NA12878 (provided as a track for the UCSC browser; see Methods). We find that 1% of the human genome has low kmer recovery (10kb regions with <90% predicted kmers recovered; Supplementary Fig. 3) and these regions affect multiple genes and gene families (Supplementary Table 7). Although some of these regions may reflect large homozygous deletions with respect to the reference, these results suggest that there is an important minority of the genome where identification and representation of alternative sequences would substantially improve genome inference.
Online Methods
Algorithms
A full description of the PRG algorithms can be found in the Supplementary Note, including (i) the algorithms used to build PRGs from a set of reference data, (ii) the algorithmic and statistical methods for inferring a best diploid path (chromotype) through the PRG, (iii) the algorithm to discover novel variation not presented in the PRG, (iv) the graph-mapping algorithm used for the contig analysis.
Data for PRG construction
We define the extended MHC (xMHC) as the genomic region spanned by the “PGF” xMHC haplotype (identical to the primary human reference in the region – in GRCh37 coordinates: chr6:28,702,185-33,451,429, GenBank ID for GRCh37 chromosome 6: CM000668.1). In addition to the PGF xMHC haplotype, we used seven xMHC haplotypes from the MHC haplotype project5 (GRCh37, ALT_REF_LOCI_1 – ALT_REF_LOCI_7). We created a multiple sequence alignment (MSA) for the eight haplotypes using the programs FSA25 and MAFFT for refinement24. We used the SNPs identified by the 1000 Genomes Project, Phase 1 release 3, to augment the MHC haplotypes.
We also included all available aligned genomic (i.e. .X_gen.txt files) HLA allele sequences from IMGT/HLA31 for the classical HLA alleles at the loci HLA-A, -B, -C, -DQA1, -DQB1, -DRB1 as additional scaffold haplotypes. These haplotypes cover all exons and introns of the genes. For many alleles the genetic sequences are not completely specified over all exons and introns; however, the PRG construction algorithm removes most of the wildcard characters found at the unspecified positions.
The edge probability distributions at each vertex in the PRG are improper; specifically, we assign probability 1 to each edge. This is motivated by the downstream parts of our pipeline, which rely on the Viterbi algorithm for inferring Maximum Likelihood personalized haplotypes. With the improper parameterization, each path through the model is equally likely under the Viterbi algorithm, irrespective of how many potential branching points (vertices where there is more than one possible edge to follow) it contains. We use kmer length k = 31 for creating the kmer-PRG.
In the process of examining the eight xMHC haplotypes, we discovered an inconsistency in the Ensembl database 32. On the SSTO haplotype, HLA-DRB1 and HLA-DRB4 were mapped to the same start coordinate, likely caused, according to Ensembl, by a mis-mapping of exonic sequence of the two transcripts ENST00000549627 and ENST00000548105 (HLA-DRB4 and HLA-DRB1 exon sequence is similar). The two transcripts will be deleted in release 72 or release 73 (pers. comm.).
Whole genome sequencing data
Subjects CS1 and CS2-6 were from four GSK sponsored clinical studies; EGF100151, EGF30008, EGF105485 and EGF106708. Germ-line DNA was extracted from peripheral blood samples collected from consented clinical trial subjects, previously determined to have evidence of a Class II HLA risk marker for drug induced liver injury33. DNA was fragmented and size selected to create 2 × 180 base pair (bp) libraries and 2 × 800 bp libraries. These libraries were sequenced on a HiSeq 2000 to generate 90bp paired end (PE) reads at the BGI (Shenzen, China). For the CS1 sample approximately 200 Gb, and for each of the 5 samples in CS2-6 approximately 100Gb, of sequence was generated. Access to anonymized patient-level data underlying this study can be made available to independent researchers, following review by an independent panel, and execution of a data sharing agreement. Applications will be considered if they aim to understand the variation in the MHC region in these individuals so as to determine if additional variants in this region may contribute to a specific adverse drug response. To submit a request or enquiry, please visit www.clinicalstudydatarequest.com.
For Fig. 1, CS1 data were initially aligned to GRCh37 (excluding the alternative loci) on the CLC Genomics Workbench (version 6.5.1) and coverage and intact and broken PE read numbers determined for ~180 kb surrounding HLA-DRB1. This process was repeated with the addition of the MANN alternative MHC haplotype (identifier ‘ALT_REF_LOCI_4′, Genbank ID GL000253.1). For all remaining analyses on CS2-6, reads were mapped to GRCh37 (excluding the alternative loci) using Stampy3 following BWA1 and variants were called using Platypus 0.1.819.
Read data for NA12878 from the Illumina Platinum genomes project (HiSeq 2000, ~60x coverage, 100bp paired-end reads) was obtained from the EBI. Reads were aligned to GRCh37 (excluding the alternative loci) using BWA 0.6.21 and variants were called with Platypus 0.1.819.
For Platypus 0.1.8-based variant calling in the MHC, we used the command line
python ${platypus_executable} callVariants --bamFiles=${bam_path} --
output=${output_VCF} --refFile=${HGref} --regions=6:28000000-
34000000 --logFileName=$logfile --nCPU=12 --mergeClusteredVariants=1
, with the variables substituted with their per-samples values.
For NA12878 mapping with BWA 0.6.2 (and for the re-mapping step), we used BWA-backtrack (aln / sampe) with parameter -q10 for aln (all other parameters standard values).
For Stampy alignment, we used (in addition to file input and output parameters) the following command line options:
--bwaoptions=-t 2 -q10 /tmp/hs37d5 --keepreforder -v0 -solexa
Simulations
Genomes were simulated from the PRG by independently sampling two paths with uniform choices at junctions. We concatenated the edge labels induced by each path, removed “gap” characters and used the strings thus generated as a sample’s two haplotypes from which to generate reads. The number of starting reads (read length 101bp) at each position is Poisson distributed with mean such that the average depth is 30x. Accuracy was assessed by comparing the true underlying genotype at each level of the PRG with the genotype inferred from the Viterbi path. Specifically, we used the scoring system shown in Supplementary Table 8 to measure the number of correct alleles:
Allele concordance is the sum over sites of the score obtained divided by the maximum possible score (i.e. 2x the number of sites analyzed). The same table was used to measure ‘accuracy’ in the empirical data analysis. In the ‘reads with error’ case, we used an empirical error model based on the PCR-free data from NA12878, which achieved and average per-base error rate c. 0.1%. Our simulations are limited in that we treat the simulated paths as a sample’s complete genome; i.e. we do not include additional variation.
Validation data
SNP arrays
Individual SNP array data were provided by GSK for samples CS1-6 (Illumina 1M array). We used publically-available Illumina Omni 2.5M SNP array data from the 1000 Genomes Project for NA12878.
HLA genotypes
Individual HLA genotypes (reported to 4-digit accuracy using ‘g’ nomenclature) are given In Supplementary Table 9.
Kmer recovery from short read data
See Supplementary Note for details of how kmer recovery was estimated.
Synthetic long-read Moleculo data
For the Moleculo-based validation, identified contigs likely to have originated from the xMHC region using the following strategy
We computed the set of all kmers (k = 31) occurring in the kmerified xMHC PRG. We call all kmers occurring in this set “xMHC kmers”.
We computed the set of all kmers (k = 31) occurring in the human reference genome, excluding the region covered by the xMHC PRG. We call all kmers in this set “reference kmers”. Note that some kmers are both xMHC kmers and reference kmers. We call kmers which are xMHC kmers but not reference kmers “xMHC-unique kmers”.
- We filtered Moleculo reads according to the following criteria:
- Fraction of xMHC kmers >= 0.8.
- ≥ 2 xMHC-unique kmers spanning a stretch of at least 50 bases (in between the two kmers). For each read, we select the maximum stretch MAXSTRETCH spanned by two such xMHC-unique kmers.
- Within MAXSTRETCH, fraction of xMHC-unique kmers >= 0.5.
- Within MAXSTRETCH, fraction of reference kmers <= 0.3
- If a read passed these tests, we truncated the read to MAXSTRETCH and aligned it to the PRG.
Runtime
Most algorithms are multithreaded (openMP); hence total effective runtime depends on local system configuration. We give example runtimes for generating the VCFs for NA12878 on a multi-core machine: VCF generation (PRG-Viterbi) takes 1.8h wall time (6.6h CPU time), while VCF generation (PRG-Mapped) takes c. 5h wall time (5h CPU time). Note that this does not include the actual whole-genome re-mapping process (2x), which is typically carried out on a cluster.
Genome-wide analysis of kmer recovery in NA12878
Genome-wide kmer recovery from the Platypus VCF was measured as for xMHC-specific kmer recovery, with the exception that we counted kmers that contained undefined characters (‘N’s) as recovered, whereas we counted them as absent for xMHC validation. We provide a wiggle plot with results from the genome-wide kmer recovery analysis (200bp bins) (see URLs) that can be used within the UCSC genome browser.
Supplementary Material
Acknowledgements
Funded by grants from GSK and 100956/Z/13/Z from the Wellcome Trust to GM, a Nuffield Department of Medicine Fellowship to ZI, and a Sir Henry Dale Fellowship jointly awarded by the Wellcome Trust and the Royal Society to ZI (102541/Z/13/Z). We thank Mike Eberle and colleagues at Illumina for early access to the Moleculo data.
Footnotes
URLs High coverage data on NA12878 from the Platinum Genomes Project: http://www.ebi.ac.uk/ena/data/view/ERP001775
Platinum Genomes Project: http://www.illumina.com/platinumgenomes
Synthetic Long-read Moleculo data on NA12878: http://ftp.1000genomes.ebi.ac.uk/vol1/ftp/phase3/integrated_sv_map/supporting/NA12878/moleculo
Genome Reference Consortium: http://www.ncbi.nlm.nih.gov/projects/genome/assembly/grc/
GRCh37.p13 accession: http://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.25
IMGT/HLA database at EBI: http://www.ebi.ac.uk/ipd/imgt/hla
Source code and additional data in the PRG and input alignments: https://github.com/AlexanderDilthey/MHC-PRG
Genome-wide recovery of kmers from NA12878 in wiggle format, suitable for display on the UCSC genome browser: http://oxfordhla.well.ox.ac.uk/VCF.bw
Competing financial interests All authors with GlaxoSmithKline (GSK) affiliations are employed by GSK and may own GSK stock. GSK does not sell or market any software or services related to genetic analysis or the generation of genetic data. GM is a founder and shareholder of Genomics Ltd. GM and AD are partners in Peptide Groove LLP.
Source code Source code is available from https://github.com/AlexanderDilthey/MHC-PRG. MHC-PRG is available under the GNU General Public License v3.
References
- 1.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lunter G, Goodson M. Stampy: A statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 2011;21:936–939. doi: 10.1101/gr.111120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zook JM, et al. Integrating human sequence data sets provides a resource of benchmark SNP and indel genotype calls. Nat Biotechnol. 2014;32:246–251. doi: 10.1038/nbt.2835. [DOI] [PubMed] [Google Scholar]
- 5.Horton R, et al. Variation analysis and gene annotation of eight MHC haplotypes: the MHC Haplotype Project. Immunogenetics. 2008;60:1–18. doi: 10.1007/s00251-007-0262-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang W, et al. Copy number variation leads to considerable diversity for B but not A haplotypes of the human KIR genes encoding NK cell receptors. Genome Res. 2012;22:1845–1854. doi: 10.1101/gr.137976.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trask BJ, et al. Large multi-chromosomal duplications encompass many members of the olfactory receptor gene family in the human genome. Hum Mol Genet. 1998;7:2007–2020. doi: 10.1093/hmg/7.13.2007. [DOI] [PubMed] [Google Scholar]
- 8.Steinberg KM, et al. Structural diversity and African origin of the 17q21.31 inversion polymorphism. Nat Genet. 2012;44:872–880. doi: 10.1038/ng.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boettger LM, Handsaker RE, Zody MC, McCarroll SA. Structural haplotypes and recent evolution of the human 17q21.31 region. Nat Genet. 2012;44:881–885. doi: 10.1038/ng.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stefansson H, et al. A common inversion under selection in Europeans. Nat Genet. 2005;37:129–137. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 11.Lupski JR, Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005;1:e49. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee C, Grasso C, Sharlow MF. Multiple sequence alignment using partial order graphs. Bioinformatics. 2002;18:452–464. doi: 10.1093/bioinformatics/18.3.452. [DOI] [PubMed] [Google Scholar]
- 16.Raphael B, Zhi D, Tang H, Pevzner P. A novel method for multiple alignment of sequences with repeated and shuffled elements. Genome Res. 2004;14:2336–2346. doi: 10.1101/gr.2657504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paten B, et al. Cactus graphs for genome comparisons. J Comput Biol. 2011;18:469–481. doi: 10.1089/cmb.2010.0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paten B, Novak A, Haussler D. Mapping to a reference genome structure. 2014 http://arxiv.org/abs/1404.5010.
- 19.Rimmer A, et al. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat Genet. 2014;46:912–918. doi: 10.1038/ng.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garrison EP, Marth G. Haplotype-based variant detection from short-read sequencing. 2012 http://arxiv.org/abs/1207.3907.
- 21.Huang L, Popic V, Batzoglou S. Short read alignment with populations of genomes. Bioinformatics. 2013;29:i361–370. doi: 10.1093/bioinformatics/btt215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneeberger K, et al. Simultaneous alignment of short reads against multiple genomes. Genome Biol. 2009;10:R98. doi: 10.1186/gb-2009-10-9-r98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iqbal Z, Caccamo M, Turner I, Flicek P, McVean G. De novo assembly and genotyping of variants using colored de Bruijn graphs. Nat Genet. 2012;44:226–232. doi: 10.1038/ng.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katoh K, Frith MC. Adding unaligned sequences into an existing alignment using MAFFT and LAST. Bioinformatics. 2012;28:3144–3146. doi: 10.1093/bioinformatics/bts578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bradley RK, et al. Fast statistical alignment. PLoS Comput Biol. 2009;5:e1000392. doi: 10.1371/journal.pcbi.1000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lefranc MP, et al. IMGT, the international ImMunoGeneTics information system. Nucleic acids research. 2009;37:D1006–1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H. Exploring single-sample SNP and INDEL calling with whole-genome de novo assembly. Bioinformatics. 2012;28:1838–1844. doi: 10.1093/bioinformatics/bts280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weisenfeld NI, et al. Comprehensive variation discovery in single human genomes. Nat Genet. 2014;46:1350–1355. doi: 10.1038/ng.3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Sidore C, Kang HM, Boehnke M, Abecasis GR. Low-coverage sequencing: implications for design of complex trait association studies. Genome Res. 2011;21:940–951. doi: 10.1101/gr.117259.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods only references
- 31.Holdsworth R, et al. The HLA dictionary 2008: a summary of HLA-A, -B, -C, -DRB1/3/4/5, and -DQB1 alleles and their association with serologically defined HLA-A, -B, -C, -DR, and -DQ antigens. Tissue Antigens. 2009;73:95–170. doi: 10.1111/j.1399-0039.2008.01183.x. [DOI] [PubMed] [Google Scholar]
- 32.Flicek P, et al. Ensembl 2013. Nucleic Acids Res. 2013;41:D48–55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spraggs CF, Parham LR, Hunt CM, Dollery CT. Lapatinib-induced liver injury characterized by class II HLA and Gilbert’s syndrome genotypes. Clin Pharmacol Ther. 2012;91:647–652. doi: 10.1038/clpt.2011.277. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.