Abstract
To assess the relative impact of inherited and de novo variants on autism risk, we generated a comprehensive set of exonic single nucleotide variants (SNVs) and copy number variants (CNVs) from 2,377 autism families. We find that private, inherited truncating SNVs in conserved genes are enriched in probands (odds ratio=1.14, p=0.0002) compared to unaffected siblings, an effect with significant maternal transmission bias to sons. We also observe a bias for inherited CNVs, specifically for small (<100 kbp), maternally inherited events (p=0.01) that are enriched in CHD8 target genes (p=7.4×10−3). Using a logistic regression model, we show that private truncating SNVs and rare, inherited CNVs are statistically independent autism risk factors, with odds ratios of 1.11 (p=0.0002) and 1.23 (p=0.01), respectively. This analysis identifies a second class of candidate genes (e.g., RIMS1, CUL7, and LZTR1) where transmitted mutations may create a sensitized background but are unlikely to be completely penetrant.
INTRODUCTION
Autism spectrum disorder (ASD) is a common neurodevelopmental disorder diagnosed in approximately 1/88 children 1 and manifests as deficits in social behavior and language development, as well as restricted or stereotyped interests. ASD is highly heritable with consensus estimates suggesting that ~50–60% of ASD etiologies are genetic in origin 2,3. In particular, de novo mutations have been implicated as an underlying genetic cause in autism, and these mutations have provided a rich source for understanding pathogenic genes and neurobiological mechanisms of ASD 4–10. However, de novo mutations are rare, and previous work suggests that they could account for the development of ASD in only 25–30% of cases 9, a fraction of the cases likely to be genetic. This suggests that other genetic factors contribute to ASD, including both rare and common inherited genetic variation 2,11.
Previous reports have put forward genetic models for ASD in which rare, inherited copy number variants (CNVs) or disruptive single nucleotide variants (SNVs) are disproportionally inherited by affected probands when compared to their unaffected siblings 11–16. Specifically, it has been posited that autism risk factors must exist that are essentially non-penetrant in females but that are transmitted preferentially to affected sons. While CNVs show some evidence of this 12,17, conclusive evidence from SNVs has been lacking 18. We sought to test this by reanalyzing exome sequence data from a family-based study design, where there are sequence data from a single autism proband, unaffected sibling, and both parents. Our goals were to assess and quantify this SNV transmission disequilibrium, identify potential candidate ASD risk genes, and integrate both inherited and de novo factors to create a unified ASD risk model for rare, disruptive SNV and CNV mutations.
RESULTS
SNV discovery and quality control
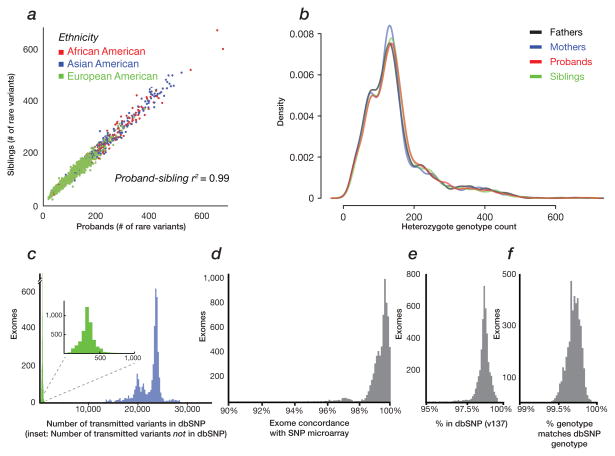
In order to generate a standard callset of inherited variants for analysis, we reprocessed 8,917 exomes sequenced at three different genome centers 4,5,7–9. The set includes 2,377 families from the Simons Simplex Collection (SSC)—of which 1,786 consisted of exome sequence data from both parents, an affected child, and unaffected sibling (referred to here as “quads”). Combined, we identified a total of 1,303,385 transmitted variants called by both GATK HaplotypeCaller and FreeBayes and passing our quality filters (Table 1, Online Methods). Of these, 31% of the variants were not observed in dbSNP (v137). As a quality control, we generated a principal component analysis (PCA) of the transmitted variants and compared to the self-identified ethnicity of the samples (Supplementary Figure 1). As expected, the number of rare variant alleles in probands and siblings were highly correlated (Figure 1a, r2 = 0.99) with no significant difference in heterozygosity being observed between proband and sibling (Figure 1b). Using the FreeBayes and GATK intersection set, we found a median of 23,055 transmitted variants per exome for probands and siblings (Figure 1c; 95% Confidence Interval [CI] 15,885–27,845). A median of 377 (95% CI 154–692) sites per family were novel and not observed in dbSNP (v137); conversely, a median of 98.6% of sites were in dbSNP and 99.7% of those were in agreement with respect to the alternate allele. The intersection set of variants had a median Ti/Tv ratio of 2.94 (95% CI 2.79–3.03) for all sites, 2.95 (95% CI 2.83–3.04) for dbSNP sites, and 1.94 (95% CI 1.05–2.75) for novel sites. In addition, we compared SNPs from exome calls with SNP calls from existing Illumina single nucleotide polymorphism (SNP) microarray data 19 (Sanders personal communication) and found the median genotype-level concordance to be 99.4% (for a median of 17,731 overlapping SNPs in 3,052 offspring in 1,796 families for which microarray data was available).
Table 1.
SNV and CNV discovery.
| Variants | Quads (n=1,786) | Trios (n=591) | All (n=2,377) |
|---|---|---|---|
| All | 1123040 | 614190 | 1737230 |
|
| |||
| SNVs | 1060422 | 581154 | 1641576 |
| Indels | 56008 | 31501 | 87509 |
| Private SNVs/Indels | 52279 | 12634 | 64913 |
|
| |||
| CNVs | 6610 | 1535 | 8145 |
| Deletions | 2289 | 492 | 2781 |
| Duplications | 4321 | 1043 | 5364 |
| <500 kbp | 6369 | 1480 | 7849 |
| >500 kbp | 241 | 55 | 296 |
Summary of SNVs, indels, and CNVs from exome sequence data from 2,377 (1,786 quads, 591 trios) families from the Simons Simplex Collection (SSC), including transmitted SNV and indel calls from the intersection of GATK HaplotypeCaller and FreeBayes and all CNVs with orthogonal validation. Note: All events in each category are given as the sum of quad and trio numbers resulting in ~1.6 million SNVs and indels (the unique independent sites is ~1.3 million).
Figure 1. SNV quality assessment.
SNVs were identified based on the intersection of FreeBayes and GATK variant callers. The panels display a) proband-sibling concordance for number of private SNVs (Pearson’s r2 > 0.99) and stratified by population (calculated by PCA using EIGENSTRAT on SNV markers); b) the number and distribution of private, heterozygous genotypes that do not differ significantly between mothers, fathers, probands or siblings (p-values > 0.3 for all comparisons); c) the number of transmitted SNVs per exome in dbSNP (blue) or novel (green); d) concordance between exome and SNP microarray calls 19 (Sanders unpublished); e) fraction of events per exome found in dbSNP; f) genotype concordance of SNVs found in dbSNP, per exome.
Although discovery of de novo events was not the primary goal of this study, our use of independent SNV callers allowed us to identify additional de novo mutations (Table 2). Our reanalysis pipeline predicted 1,544 de novo SNVs not previously reported (Supplementary Table 1). We selected a subset of 141 events for Sanger-based validation because they represented either new recurrences or likely gene-disruptive (LGD) events. Of these new sites, 55% (77) confirmed as de novo as well as an additional 132 events that had been called but not confirmed in previous studies (Supplementary Table 2). Post hoc analysis using three different classifiers (support vector machine (SVM), decision tree and random forest) suggested that the proband’s allele balance was the best individual predictor of de novo variant validation and that classification models could accurately predict which events would be most likely to validate (Supplementary Figure 2 and Online Methods). Extrapolating the proband’s allele balance across all untested candidate variants in probands (n = 771) suggests that there are 463 (60%) additional true de novo variants in probands (at an allele balance cutoff > 0.3); similarly, the predictions generated by the random forest model suggest 445 (58%) additional de novo variants in probands would validate.
Table 2.
Additional genes with recurrent de novo mutations.
| Gene | Krumm proband count | Iossifov proband count | De Rubeis proband count | Mutation Type | Number of valid mutations | Number of sibling mutations | p-value |
|---|---|---|---|---|---|---|---|
| ASH1L | 1 | 1 | 1 | 2 n, 1 fs | 3 | 0 | 5.70×10−6 |
| CCDC88C | 1 | 1 | 0 | 2 m | 2 | 0 | 0.07 |
| CDC42BPB | 1 | 1 | 1 | 1 n, 2 m | 3 | 0 | 8.45×10−4 |
| CGNL1 | 1 | 1 | 0 | 2 m | 2 | 0 | 0.04 |
| CUL7 | 1 | 1 | 0 | 2 m | 2 | 0 | 0.04 |
| DMXL2 | 1 | 1 | 0 | 2 m | 2 | 0 | 0.07 |
| FAM92B | 1 | 1 | 0 | 2 m | 2 | 0 | 7.96×10−3 |
| GIGYF2 | 1 | 1 | 1 | 1 n, 2 m | 3 | 0 | 3.40×10−4 |
| GRIK5 | 1 | 1 | 1 | 3 m | 3 | 0 | 0.01 |
| HECW2 | 1 | 1 | 0 | 2 m | 2 | 0 | 0.05 |
| P4HA2 | 1 | 1 | 1 | 3 m | 3 | 0 | 1.21×10−4 |
| PHRF1 | 1 | 1 | 1 | 3 m | 3 | 0 | 0.20 |
| PYHIN1* | 1 | 1 | 1 | 1 n, 2 m | 3 | 0 | 5.53×10−4 |
| RAB43 | 1 | 1 | 0 | 2 m | 2 | 0 | 1.10×10−3 |
| RBM27 | 1 | 1 | 0 | 2 m | 2 | 0 | 4.63×10−3 |
| SCN4A* | 1 | 1 | 0 | 2 m | 2 | 0 | 0.17 |
| TBC1D31 | 1 | 1 | 0 | 2 m | 2 | 0 | 7.29×10−3 |
| TET2 | 1 | 1 | 0 | 2 m | 2 | 0 | 0.02 |
| XIRP1* | 1 | 1 | 0 | 2 m | 2 | 0 | 0.07 |
| ZNF462 | 1 | 1 | 1 | 1 fs, 2 m | 3 | 0 | 4.03×10−3 |
| SSPO | 2 | 0 | 0 | 1 s, 1 m | 2 | 0 | 0.91 |
New validated de novo events (Krumm) are compared to previously discovered events (Iossifov 9 = SSC and De Rubeis 10 = Autism Sequencing Consortium [non-SSC probands]). The total number of events in probands (n = 2,377) is contrasted to the total number of de novo events in siblings (n = 1,786). All genes except those with an asterisk are brain expressed according to the GTEx Portal. P-value based on O’Roak et al. 2012 6; recurrence in genes with marginal- or non-significant p-values are potentially chance recurrences. (n = nonsense, fs = frameshift, m = missense, s = splice-site)
After validation, we identified 21 novel recurrently hit genes (Table 2). Notably, these validated mutations established recurrent de novo mutations for GIGYF2 and SSPO (a brain-secreted protein involved in axon growth) as well as added a new LGD mutation to GIGYF1 and ASH1L for a total of three LGD de novo mutations each.
SNV transmission disequilibrium
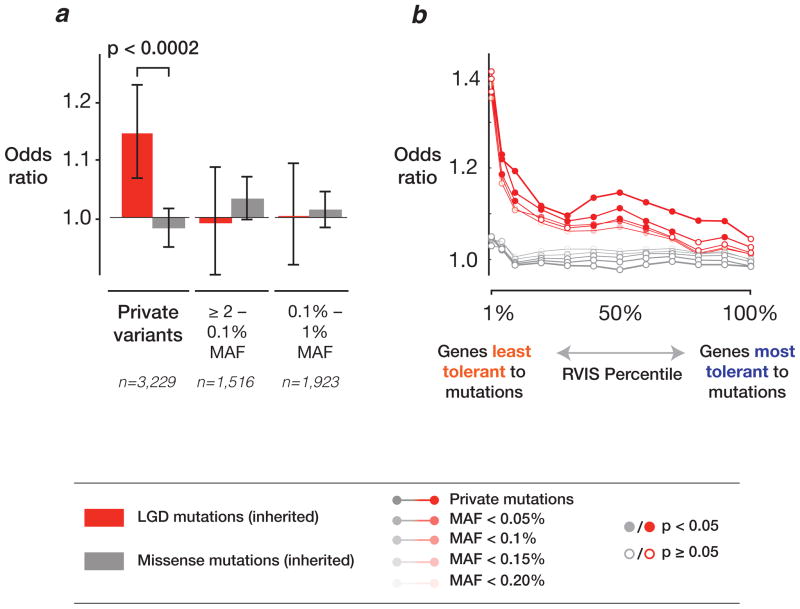
We tested for transmission disequilibrium between probands and siblings using Fisher’s exact and Mann-Whitney U tests and by logistic regression (where the dependent variable was the presence of a variant found in a proband or sibling). We considered only transmitted variants reported using both FreeBayes and GATK and defined private events as those unique to a single family. When considering all rare or private protein-altering mutations (LGD + missense) together, we observed no statistically significant difference in the overall burden between proband and sibling. Under the assumption that LGD mutations in genes intolerant to deleterious mutations would be more likely to be pathogenic, we repeated the analysis using residual variation intolerance score (RVIS) values 20,21. Restricting our analysis to private LGD mutations in genes in the lower 50% of RVIS values, we observed a significant enrichment in probands when compared to siblings (OR = 1.14, p = 0.0002, Fisher’s exact test) and at a family level (p < 0.0001, two-tailed paired t-test; Figure 2a).
Figure 2. Transmission disequilibrium of SNVs in ASD.
a) Private, inherited LGD (red bars) SNVs in genes not tolerant to functional variation were significantly enriched in probands. The analysis examines only SNVs in genes with an RVIS in the lower 50%. Non-private, rare variants or missense-inherited (gray bars) SNVs are not enriched in probands. Bar heights are Fisher’s exact test odds ratio and whiskers represent 95% confidence interval bounds. b) The RVIS is a critical determinant for enrichment in probands. Burden was highest (reaching OR = 1.4) for private, inherited LGD SNVs amongst genes with the lowest RVIS values (<1%). MAF = minor allele frequency.
This signal persists for all LGD mutations in genes (regardless of frequency) with RVIS values < 50% (OR = 1.06, p = 0.03, Fisher’s exact test; p = 0.02, two-tailed paired t-test). Furthermore, the RVIS was a significant predictor of proband or sibling inheritance in a logistic regression model built on all LGD mutations (p = 0.028, OR = 1.01 per RVIS percentage point). As suggested by this model, the burden of private LGD mutations in genes with progressively lower RVIS values continues to increase (Figure 2b). At the extreme, the burden between probands and siblings in genes with the lowest 1% of all RVIS values reaches an odds ratio of 1.4 (although this comparison is not statistically significant due to the small number of mutations present at this threshold in the current dataset). When we examined the fraction of probands and siblings that inherited LGD SNVs in highly conserved genes (RVIS < 10th percentile), we found that 50.6% of probands (903/1,786 quads) and 47.9% of siblings (855/1,786 quads) contained such events, respectively, a difference of 2.7%. Finally, we performed extensions of the rare variant-transmission disequilibrium test (RV-TDT) 22 at the individual gene level comparing transmission of rare variants to probands and siblings within the SSC families. Several promising candidate genes emerged (Supplementary Table 3) although none survived a multiple-testing correction (Online Methods).
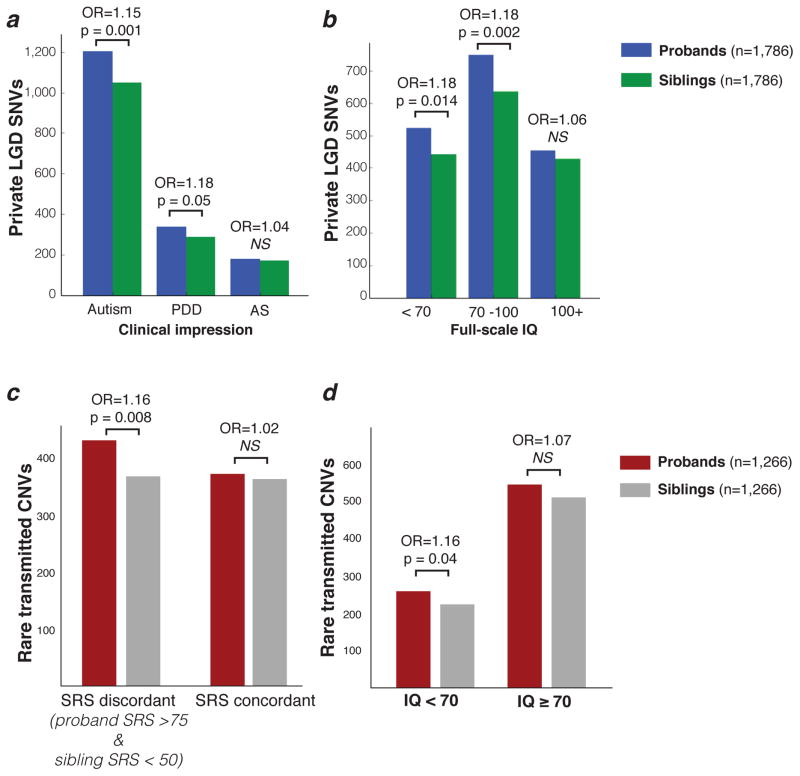
We considered the relationship between the set of private LGD mutations in RVIS-restricted genes and phenotypic features of the SSC families (Figure 3). First, we examined how inherited burden correlated with the overall clinical diagnosis. For the 1,575 probands with a diagnosis of “autism” or “pervasive developmental disorder” (PDD), the odds ratio was 1.15 and 1.18 (p = 0.001 and 0.05), respectively. In contrast, probands (n = 205) with a diagnosis of “Asperger’s” showed a lower odds ratio of 1.04 (p > 0.7; Figure 3a) for inherited gene-disruptive mutations. Consistent with this, we found that probands with full-scale IQ between 70–100 had an odds ratio of 1.18 (p = 0.002), whereas those with an IQ above 100 had a lower, non-significant odds ratio of 1.06 (Figure 3b). For probands in the SSC, IQ and clinical diagnosis are weakly correlated (Supplementary Figure 3; r2 = 0.18, p < 1×10−10), but we note that burden of private LGD mutations in RVIS-restricted genes in probands depends on both IQ and clinical diagnosis: for probands diagnosed with “autism” or “PDD” and a full-scale IQ above the median for the SSC probands at large (IQ = 84), the odds ratio was 1.1, while burden for “Asperger’s” probands of similar IQ was 1.03. Similarly, the odds ratio of this burden for probands with “autism” and IQ above 100 was 1.19, while that for “PDD” and “Asperger’s” at this threshold was less than 1 (Supplementary Table 4).
Figure 3. Transmitted mutations and their effect on phenotype.
a) Private, inherited LGD SNVs enriched in probands with autism and pervasive developmental disorder (PDD) diagnoses, but not Asperger’s syndrome (AS). b) Private, inherited LGD SNVs primarily enriched in probands with lower IQ than average (<100). c) We observe transmission disequilibrium of rare, inherited CNVs in SRS (Social Responsiveness Scale) discordant families (proband SRS score > 75, sibling < 50) but not in families where the SRS score is mild or more balanced between proband and sibling. d) Rare, inherited CNVs are enriched in probands (versus their siblings) with IQ < 70, but the effect is not significant in probands with IQ > 70. All tests and reported p-values are paired t-tests based on proband-sibling pairs. All analyses were restricted to genes with RVIS < 50.
Our previous work with CNVs suggested that simplex families could be distinguished into two groups depending on their overall Social Responsiveness Scale (SRS) T-scores 23. Probands and siblings with very different SRS scores (“discordant SRS sib-pairs”) should show stronger transmission disequilibrium when compared to unaffected siblings showing elevated ASD symptomatology (“concordant SRS sib-pairs”; see Online Methods). Using our previous threshold definitions (see 12), we observed a stronger proband-sibling differential of 3.7% for private LGD SNVs in conserved genes (RVIS < 10) for SRS discordant quads only (484 probands with events of 923 discordant quads, siblings 450/923), while SRS concordant quads had only a 1.6% differential (probands 419/863 and siblings 405/863).
CNV discovery and validation
Because exome and SNP microarray data provide the opportunity to accurately detect a subset of smaller CNVs within exonic regions of genes 12, we also revisited the burden of both inherited and de novo CNVs with respect to autism. We characterized CNVs from 1,266 quads with available SNP microarray data (validation shown in Supplementary Figure 4) and tested an additional 50 samples with CNVs of interest by array comparative genomic hybridization (CGH). We focused in particular on validating smaller CNV events that affected genes recurrently hit by de novo SNVs, such as DSCAM, CHD2, ARID1B and TNRC6B (Supplementary Table 5). We identified a total of 2,891 CNVs with an excess of autosomal proband events compared to siblings (854 vs. 743; OR = 1.25, p = 0.006, binomial two-sided test). The overall ratio of duplications to deletions was 1.6, consistent with previous results for a smaller SSC dataset 12. Restricting the analysis to de novo CNVs, we identified, as expected, a more significant 2.4-fold excess (p = 6.7×10−5, paired t-test) in probands (n = 79) when compared to siblings (n = 33) driven primarily by deletions (p = 4.2×10−5, paired t-test) and not duplications (p = 0.18, paired t-test) (Table 3). Overall, de novo CNVs were larger in probands than in siblings (p = 0.03, Wilcoxon) and carried genes with significantly lower total RVIS values (p = 0.02, Wilcoxon). Both FMRP and CHD8 targets were enriched in de novo CNVs (OR = 3.1, p = 6.6×10−4 and OR = 2.7, p = 1.7×10−3, Fisher’s exact test and p=1.4×10−4 and p=2.6×10−4, paired t-test, respectively) and this is likely due, in part, to the larger size of de novo events among probands.
Table 3.
CNV burden and transmission.
| Dataset | Inheritance | Odds Ratio | t-test | t-test mean of the differences | Number of CNV events |
|---|---|---|---|---|---|
| All | de novo | 1.90 [1.32,Inf] | 6.7×10−5 | 0.46 [0.28,Inf] | Pro (n=79) vs. Sib (n=33) |
| all inherited | 1.10 [0.95,Inf] | 0.03 | 0.08 [0.02,Inf] | Pro (n=775) vs. Sib (n=710) | |
| maternal inherited | 1.15 [1.00,Inf] | 0.01 | 0.11 [0.04,Inf] | Pro (n=411) vs. Sib (n=357) | |
| paternal inherited | 1.02 [0.89,Inf] | 0.59 | 0.02 [−0.05,Inf] | Pro (n=364) vs. Sib (n=353) | |
|
| |||||
| Deletions | de novo | 3.07 [1.79,Inf] | 4.2×10−5 | 0.68 [0.43,Inf] | Pro (n=49) vs. Sib (n=13) |
| all inherited | 1.11 [0.96,Inf] | 0.05 | 0.09 [0.01,Inf] | Pro (n=297) vs. Sib (n=262) | |
| maternal inherited | 1.08 [0.90,Inf] | 0.20 | 0.08 [−0.02,Inf] | Pro (n=156) vs. Sib (n=139) | |
| paternal inherited | 1.09 [0.90,Inf] | 0.14 | 0.09 [−0.01,Inf] | Pro (n=141) vs. Sib (n=123) | |
|
| |||||
| Duplications | de novo | 1.20 [0.74,Inf] | 0.18 | 0.21 [−0.05,Inf] | Pro (n=30) vs. Sib (n=20) |
| all inherited | 1.12 [0.98,Inf] | 0.20 | 0.05 [−0.02,Inf] | Pro (n=478) vs. Sib (n=448) | |
| maternal inherited | 1.16 [0.99,Inf] | 0.03 | 0.11 [0.03,Inf] | Pro (n=255) vs. Sib (n=218) | |
| paternal inherited | 1.00 [0.86,Inf] | 0.67 | −0.02 [−0.11,Inf] | Pro (n=223) vs. Sib (n=230) | |
|
| |||||
| Duplications <100 kbp | de novo | 0.60 [0.27,Inf] | 0.55 | −0.15 [−0.57,Inf] | Pro (n=9) vs. Sib (n=12) |
| all inherited | 1.12 [0.97,Inf] | 0.38 | 0.04 [−0.04,Inf] | Pro (n=315) vs. Sib (n=298) | |
| maternal inherited | 1.19 [0.99,Inf] | 0.01 | 0.15 [0.05,Inf] | Pro (n=177) vs. Sib (n=143) | |
| paternal inherited | 0.98 [0.81,Inf] | 0.22 | −0.08 [−0.19,Inf] | Pro (n=138) vs. Sib (n=155) | |
Test results (one-sided paired t-test) are shown for all CNV events, deletions, duplications, and small (<100 kbp) duplications. Square brackets indicate 95% confidence intervals.
The validated, inherited CNV dataset (frequency < 0.8%) consisted of a total of 1,485 events (n = 775 in probands, n = 710 in siblings) from 1,266 quads. We replicated the previously reported 12 transmission disequilibrium of CNVs to probands when compared to siblings (p = 0.03, paired t-test). This effect was driven almost exclusively by smaller (<100 kbp) maternally inherited events (p = 0.01, paired t-test). In contrast to de novo events, there was no difference in size of CNVs transmitted in probands versus those in siblings (p = 0.59, Wilcoxon). Similar to our observations of SNV mutations in conserved genes, we found that genes within proband CNV intervals had a borderline significantly lower average RVIS (p = 0.05, Wilcoxon).
In order to more fully understand the potential biology of these inherited CNV events, we tested if the CNVs were enriched in either FMRP 24 or CHD8 targets 25. Although no overall enrichment of FMRP (p = 0.22) or CHD8 (p = 0.19) targets was observed among inherited CNVs, when we restricted the analysis to maternally inherited duplications a significant enrichment was observed for CHD8 targets (OR = 1.5, p = 0.02, Fisher’s exact test, p = 3.9×10−3, paired t-test). In particular, this enrichment was strongest for small duplications (<100 kbp) (OR = 1.5, p = 0.05, Fisher’s exact test, p = 7.4×10−3, paired t-test). Since truncating mutations of CHD8 have been associated with a subtype of autism characterized by macrocephaly 26, we tested whether patients carrying CNVs that intersected CHD8 target genes showed any deviation in head circumference. We specifically stratified the patient population into two groups: those containing a maternally inherited CNV with a CHD8 target and those that have a maternally inherited CNV without a CHD8 target. We then tested whether or not there was an enrichment of macrocephalic or microcephalic patients in CNV carriers of CHD8 targets. Interestingly, we observed a modest enrichment for macrocephaly in maternally inherited autosomal deletions containing CHD8 targets (OR = 2.9, p = 0.03, Fisher’s exact test), including smaller deletions (<100 kbp, OR = 3.5, p = 0.04, Fisher’s exact test). The reciprocal was also observed with borderline significance for microcephaly in maternally inherited autosomal duplications containing CHD8 targets (OR = Inf, p = 0.04, Fisher’s exact test) (Supplementary Figure 5). As a control, we repeated the same analysis for inherited CNVs carrying FMRP targets, which we did not expect to have any relevance for head circumference and found no statistical enrichment for targets and head size.
SNV and CNV integration and gender bias
We jointly examined SNVs and CNVs at a gene level in order to identify potentially new ASD candidate genes (Supplementary Table 6). Based on our findings, we considered all de novo CNVs, private, inherited SNVs in genes with an RVIS lower than 50%, and rare, inherited CNVs where at least one gene had an RVIS lower than 50%; we then created a combined gene-level table identifying several candidate genes. In particular, the three highest ranked genes—RIMS1, CUL7 and CSMD1—each display brain-specific patterns or have identified neural functions (Supplementary Figure 6). The highest ranked gene, RIMS1, has two de novo LGD mutations and two private LGD-inherited mutations in probands. Additionally, there were six rare, inherited LGD mutations in probands (two are shared with siblings and one is found in a trio), and one mutation found in a sibling alone. CUL7 has two de novo and two LGD-inherited mutations in probands (none in siblings). Finally, CSMD1 has three de novo missense mutations in probands, four LGD SNVs (one is shared with a sibling and one is found in a trio), and five rare, inherited CNVs (where one is shared with a sibling and one is found in a trio).
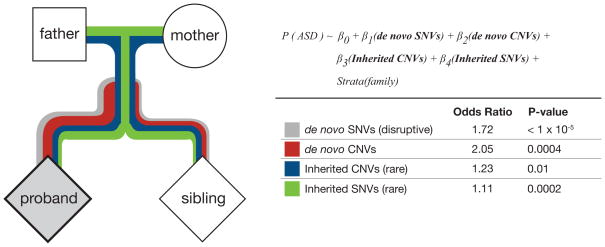
We quantified the risk for ASD by examining de novo and inherited CNVs and SNVs using a conditional logistic regression model (Figure 4; see Online Methods). In this model, the binary outcome of an ASD proband or unaffected sibling is predicted by four independent counts: 1) the number of de novo CNVs, 2) the number of LGD de novo SNVs, 3) the set of rare, inherited CNVs, and 4) the set of private LGD-inherited SNVs in genes in the lower 50% percentile of RVIS values (Supplementary Table 6). Additionally, we accounted for familial stratification effects by adding a family-level stratum to the model. Using data from the 1,786 quads, we found robust effects for de novo events (Supplementary Table 7): each de novo CNV increased the risk for ASD by 2.05-fold, while each de novo SNV increased risk by 1.72-fold (p = 0.0004 and p < 1 ×10−7, respectively). In addition, the results from this analysis reveal a significant role for inherited mutations in ASD risk: rare, inherited CNVs contribute an increased risk of 1.23 (p = 0.01), and private LGD SNVs have an odds ratio of 1.11 (p = 0.0002). These results suggest that each of the four types of mutations modeled additively contribute to the risk of ASD and that they do so in a statistically independent manner.
Figure 4. Combined risk model for SNVs and CNVs: inherited and de novo.
Integrative risk model for ASD, based on de novo and inherited events, and covering both SNVs and CNVs. The model used is a stratified logistic regression model, which utilizes proband-sibling pairs to estimate the odds ratio (i.e., risk of ASD) for each type of event (see also Supplementary Table 7).
Finally, by calculating the attributable fraction in the population (Supplementary Figure 7), we were able to identify the contribution of each variant type as follows: de novo LGD SNVs (6.62% [4.18%, 8.99%]), private, inherited LGD SNVs (8.54% [−24.23%, 32.66%]), de novo CNVs (2.92% [1.37%, 4.44%]), and rare, inherited CNVs (3.18% [−3.71%, 9.6%]). Whereas these values give high confidence for de novo events, the inherited events are less clear. By stratifying by inheritance and RVIS for private, inherited LGD SNV events, the results become much tighter and show a clear contribution for maternal events (7.15% [−0.25%, 14.01%]) and not for the paternal events (1.01% [−6.56%, 8.04%]). The same was found for maternal duplications (2.99% [−0.45%, 6.31%]) especially those under 100 kbp (2.65% [−0.16%, 5.38%]).
Specifically, we extended the work to investigate the role of maternally transmitted events to males and females. First, we assessed the attributable fractions in all quad families and subsequently in male proband and separately female proband quads. For the LGD SNVs, we were able to identify the maternally private, inherited LGD SNVs with RVIS < 50 as the category with the highest attributable fraction (estimated) in the population (8.32% [0.56%, 15.48%]) in the male proband families whereas the female proband families had a value of −2.33% [−29.06%, 18.87%] in this same category. No effect was observed for paternally inherited LGD events. This is in stark contrast to de novo LGD events, which contribute to 5.7% [−2.26%, 13.04%] of the attributable fraction in females (Supplementary Figure 7). While larger sample sizes will be required, these findings are consistent with the maternal bias observed for large and small CNVs and now extend the observation to maternal SNVs. To further examine this difference, we examined all four possible quad types based on gender: male proband / male sibling, male proband / female sibling, female proband / male sibling, and female proband / female sibling. These observations for maternal LGD SNVs held true regardless of sibling gender (Supplementary Figure 7, Supplementary Table 8) 27.
DISCUSSION
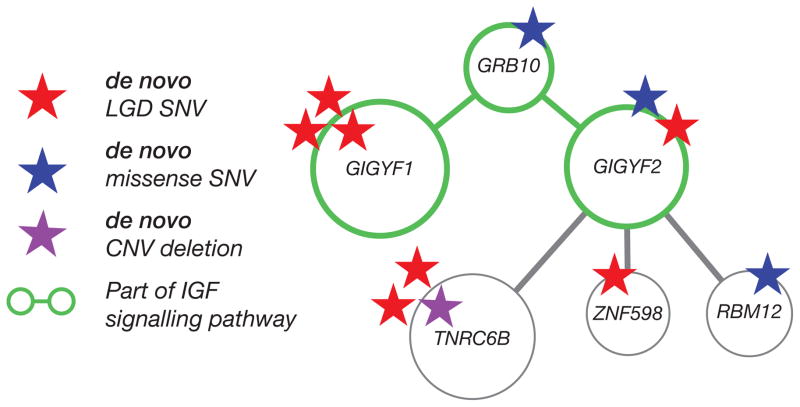
In this study, we have explored the effect of rare, inherited variation on the risk of autism. Our results provide some of the first genetic evidence that private, inherited SNVs that truncate proteins are enriched in autism probands. Remarkably, this effect is only observed for truncating SNVs that disrupt genes intolerant to functional variation and shows bias in transmission from mothers to their sons. The effect becomes more pronounced the more intolerant the gene is to mutation consistent with the notion that such genes are subject to strong selective pressure. While the effect is strongest for individuals with a diagnosis of autism, it is most significant for SRS-discordant quads and probands with an IQ between 70 and 100. Extending previous work 12–14 on the role of rare, inherited CNVs, we report that smaller maternally inherited duplications show the largest bias towards transmission to probands, and these duplications are enriched for gene targets of CHD8. The reciprocal shift in macrocephaly and microcephaly when comparing CHD8 target gene duplications and deletions, respectively, is intriguing but warrants further investigation. In addition, the application of two SNV callers identified 77 additional de novo SNVs previously missed 9. The recurrent hits highlight potentially new pathways such as the insulin-like growth factor protein-interaction network (Figure 5). This is interesting because variable levels of IGF1 are considered a biomarker of autism 28 and are of potential therapeutic relevance 29.
Figure 5. Networks and pathways.
A highly interconnected network was identified based on novel de novo mutations identified in this study (Note: one additional de novo missense mutation was recently identified in 10). Gene ontology annotation of the genes in this network suggests involvement of the insulin-like growth factor signaling pathway (GIGYF1, GIGYF2, GRB10; accession GO:0048009), which has been previously implicated in the development of ASD 29. Furthermore, GIGYF2 and ZNF598 form part of the m4EHP mRNA binding complex and have widespread translational repression roles, especially in the brain and lungs 57. Red stars: de novo LGD mutations (frameshift, stop-gained, splice-site); Blue stars: de novo missense mutations; Purple star: CNV deletion.
In some cases, inherited and de novo mutations of both SNVs and CNVs converge on the same gene (Supplementary Table 6). RIMS1 has been previously suggested as an ASD candidate as a result of recurrent de novo truncating mutations 4,32. In this analysis, we also find a nominally significant transmission disequilibrium of private, disruptive events of RIMS1 to probands (p = 0.013, TDT-Combined Multivariate and Collapsing (CMC) analytical 22) but not siblings (p = 0.841) (Supplementary Table 3). The gene displays brain-specific brain expression, and disruption of the gene in mice leads to increased postsynaptic density and impaired learning. CUL7 has two de novo and two LGD-inherited mutations in probands (none in siblings); functionally, it is an E3 ligase with high cerebellar brain expression and a selective role in neural dendrite patterning and growth 33. For the highly conserved gene CSMD1, there are three de novo missense mutations, one shared inherited LGD SNV, and four rare, inherited focal CNVs (one shared with siblings). Overall, there are eight events in ASD probands and two in siblings clustered at the exon-dense 3′ end of CSMD1, a region nearly devoid of exonic CNVs in the Database of Genomic Variants (DGV, Supplementary Figure 8). Functionally, CSMD1 exhibits strong and specific brain expression; this gene has been associated with schizophrenia 34 and damaging variants of the gene segregated in two ASD families with distantly related probands 35.
We also identified candidate genes for which no de novo events have yet been reported despite evidence of over-transmission of private LGD events to affected probands within the SSC families (Supplementary Tables 6 and 3). Using the RV-TDT on rare, inherited events, we identified candidates that have not yet reached locus-specific significance, including LZTR1 (commonly deleted in DiGeorge syndrome30) and CENPJ (gene with autosomal recessive mutations known to cause microcephaly and intellectual disability31). While these genes and genes like RIMS1 may represent important risk factors for ASD, the fact that gene-disruptive events are inherited from normal parents and/or occasionally transmitted to unaffected siblings argues that they are not necessary and sufficient to cause autism. This stands in contrast to other genes such as ADNP, CHD8 or DYRK1A where de novo LGD mutations have been observed almost exclusively in probands. In fact, genes enriched for de novo LGD mutations have significantly fewer inherited LGD mutations than expected from randomly sampled gene sets (empirical p < 1 × 10−4, see Online Methods and Supplementary Figure 9) suggesting that inherited and de novo mutation risk factors may often target different genes.
We hypothesize the second class of inherited-LGD genes simply predisposes an individual to ASD, requiring additional genetic or non-genetic factors to reach a disease state. Notably, the largest effect appears to be for maternal transmission to sons consistent with other recent findings17 and models of autism11. Such oligogenic models have been proposed previously for CNVs 36 as well as other forms of severe mutation associated other human diseases 37,38. The availability of CNVs and SNVs from exome sequence data is the first step to obtaining a more complete genetic picture at an individual level in the context of autism. In this light, it is interesting that our analysis uncovered a paternally inherited two-exon intragenic deletion of NRXN3 and a de novo missense mutation of NLGN2 in proband 13367.p1 (Supplementary Figure 10). Both of these genes have been identified as ASD risk factors, but crucially, they are also protein-protein interacting partners. The neuroligin-neurexin interaction has long been hypothesized to be a key underlying pathway in ASD pathology but, to our knowledge, this is the first identification of a case with mutations in both binding partners. As the genetic profile of the SSC becomes more complete through full genome sequencing, it is likely that examples of an oligogenic model for ASD will become more prevalent and informative to our understanding of the genetic etiology.
ONLINE METHODS
Datasets
We analyzed exome data from 2,377 ASD families (2,391 before quality control) from the Simons Simplex Collection or SSC 39, including 1,786 quads and 591 trios (total 8,917 exomes). These exomes were recently analyzed for de novo variants 4,5,8,9 but were reanalyzed here to increase sensitivity and to create a unified callset for private variants (Table 1). The raw sequence data for these exomes are available in the National Database for Autism Research (NDAR) at DOI: 10.15154/1151812, and the reanalyzed data, including the complete variant call format (VCF) files from the SNV callset, and bioinformatics pipelines for this study are available (see URLs). We used Illumina 1M, 1MDuo or Omni2.5 SNP microarray data for 1,266 complete quads for CNV validation 19 (Sanders personal communication). Relevant phenotype scores were extracted for both the SRS (parent-assessed T-scores 23) and the full-scale IQ (as in Krumm et al. 2013 12) from the SSC Simons Foundation Autism Research Initiative (SFARI) Base. Normalized head circumference scores were determined as previously described 40. Published databases of FMRP 24 and CHD8 target genes 25 were used to assess enrichment of targets within CNVs. The institutional review board (IRB) of the University of Washington approved this study (IRB # 46179).
Sequence data processing
Reads from all 8,917 exomes were realigned using BWA-MEM 41 (v0.7.5a, options -k 17) to the 1000 Genomes Phase 1 reference genome (hg19/GRCh37). We mapped all available libraries for samples, including single-ended and paired-end where appropriate. Mapped BAMs were processed according to GATK 42 best practices, including duplicate marking and mate fixing. We applied GATK (v. 2.7–4) Indel Realignment in a family-aware manner, ensuring that each member of a family was realigned at the same positions across the family. Base qualities were recalibrated using GATK. Next, we used QPLOT 43 and computed 24 read- and exome-level statistics (Supplementary Table 9) for QC assessment. Finally, to ensure we did not have any sample, family or data mix-ups, we used a custom-developed (Sanders, personal communication) tool to identify and match 287 polymorphic SNPs in each exome to an existing database of “SNP fingerprints” derived from Illumina SNP microarray data 19 and 96 SNP fingerprints collected by the Rutgers sample distribution center. We excluded 14 families for sample identity issues and concordance by center is shown in Supplementary Table 10.
SNV discovery
To identify SNVs, we batched families into groups of 16–20 families, or approximately 70 exomes, in order to ensure better sensitivity for events. We called SNVs and indels with both GATK HaplotypeCaller (v 2.7–4) and FreeBayes 44 (v0.99) to within 20 bp of the NimbleGen EZ-SeqCap v2.0 targets. Family-level VCF files from FreeBayes and GATK were merged into a union set. Merged VCF files were annotated using SnpEff 45 (v 3.4i), dbNSFP 46 (v2.1), CADD score 47, dbSNP (v137), tandem repeats and segmental duplications. Allele frequency was estimated by counting non-reference alleles across all parents (n = 4,754).
We called SNVs and indels with both GATK HaplotypeCaller (v 2.7–4) and FreeBayes (v0.99) to within 20 bp of the exon targets; calls were annotated using SnpEff and merged into union and intersection sets. Allele frequency was estimated by counting non-reference alleles across all parents (n = 4,754). For de novo events, we applied a minimum read-depth of six alternate alleles in offspring and a depth of >10 reference reads in parents and allowed for no more than two low-quality bases of the de novo variant. Because FreeBayes and GATK SNV-calling routines report only the number of high-quality reads supporting the alternate or reference allele, we queried the original BAM files at each site to include the count of low-quality bases in these filters. To exclude common artifacts, we only considered de novo sites that were private to a family. Inherited events were derived from the intersection set of both algorithms, with a minimum depth filter (DP > 20) and quality filter (QUAL > 50) for all events (Figure 1, Supplementary Figure 11). In addition, we applied a batch exclusion filter, which filtered variants found at high frequency exclusively in one batch (three or more times among 16–20 families). Using the FreeBayes and GATK intersection set, we found a median of 23,055 transmitted variants per exome for probands and siblings (Figure 1c; 95% CI 15,885–27,845) and a median of 26,920 transmitted variants per family (95% CI 23,394–31,401). A median of 377 (95% CI 154–692) sites per family were novel and not observed in dbSNP (v137); conversely, a median of 98.6% of sites were in dbSNP and 99.7% of those were in agreement with respect to the alternate allele. Overall, 81% of all transmitted variants were found by both FreeBayes and GATK, 12% by FreeBayes alone, and 7% by GATK. The intersection set of variants had a median Ti/Tv ratio of 2.94 (95% CI 2.79–3.03) for all sites, 2.95 (95% CI 2.83–3.04) for dbSNP sites, and 1.94 (95% CI 1.05–2.75) for novel sites. Of all inherited mutations in the intersection set, an average of 341 (95% CI 133–632) sites were novel and not observed in dbSNP (v137); 98.6% of sites were in dbSNP with a concordance rate of 99.7% (for all transmitted variants, 93.4% of variants were found in dbSNP and 99.5% were concordant). In addition, we compared SNPs from exome calls with SNP calls from existing Illumina SNP microarray data 19 (Sanders personal communication) and found the median genotype-level concordance to be 99.4% (for a median of 17,731 overlapping SNPs in 3,052 offspring in 1,796 families for which microarray data was available).
Modeling de novo SNV validation efficiency
We utilized the Sanger sequencing validation results from our 141 tested de novo SNV events to better understand which SNV calls would be the most likely to validate. In this post-hoc analysis, we constructed a feature matrix of 77 validated (truly de novo) and 63 “invalidated” (which turned out to be inherited, or otherwise not present) events, along with event- or site-level quality data emitted by GATK and/or FreeBayes (Supplementary Table 1). This quality information included data such as the QUAL (phred-scale quality score for the assertion made for the alternate allele), BaseQRankSum, MQ (mapping quality), MQ0 (number of reads with mapping quality equal to 0 covering the variant), MQRankSum (Z-score From Wilcoxon rank sum test of Alt vs. Ref read mapping qualities), as well as sample-specific data for allele depths, allele quality, GATK-specific fields such as the PL (phred likelihood score) score. As many of the invalidated events were found to present in one of the parents (i.e., they were inherited heterozygous SNVs), we also included the maximum (or minimum, when appropriate) values of both parents for PL and GQ (genotype quality). For values that were not outputted by both FreeBayes and GATK, we imputed values based on the mean of all values not missing.
Using this feature matrix, we investigated three types of classifiers present in the Python Scikit-Learn package 48: a support vector machine (SVM), a decision tree, and a random forest. We estimated all accuracy statistics by cross-validation (scores are reported average from cross validation; Supplementary Table 11). Using these data, the random forest had the best overall performance across most performance metrics, though the SVM had slightly better recall. For the decision tree and random forest methods, we were able to compute matrices corresponding to individual feature importance (Supplementary Table 12; Note: this is not possible using the SVM implementation). For both classifiers, we found that the most important feature was the proband’s allele balance and this was recapitulated when observing the AB values directly (Supplementary Figure 2).
CNV discovery
We used the CoNIFER 49 and XHMM 50 algorithms to discover copy number variation from exome data at a single-exon resolution. Identification with CoNIFER was done as described in Krumm et al. 2013 12. Briefly, we split reads into 36mers and aligned using mrsFAST 51 to the NimbleGen EZ-SeqCap v.2 targets and flanking sequence. Using CoNIFER, we processed all samples with the specific setting of --components-removed equal to 40). CNV calls were made using the CoNIFER tools package, which implements DNAcopy 52. In parallel, XHMM was applied using best-practice guidelines. GATK was used to calculate depth-of-coverage (from BWA-MEM alignments) for each individual and then all individuals were combined into one composite file. The XHMM-specific steps included hard filtering of samples and targets, PCA on the data, filtering based on PCA results, and discovery of CNVs. Post-discovery CNVs were genotyped by family and ultimately a score cutoff of 10 was used for determining inheritance in families based on SQ and NQ values 50.
Using the union of XHMM and CoNIFER callsets, we first genotyped all loci across family members to recover false negative calls and then identified transmitted and de novo CNVs. CNVs were clustered into copy number variable regions or CNVRs (as previously described in Krumm et al. 2013 12) and then annotated with family frequency across the entire cohort. In order to focus our analysis on those CNVs most likely relevant to ASD pathogenesis, we restricted our analysis to rare CNVs found at < 0.8% frequency (<10 events/1,266 families) mapping outside of repetitive genomic elements (Supplementary Figure 12).
Validation experiments
Our reanalysis pipeline identified 1,544 novel candidate de novo SNVs not detected by previous analyses of the same dataset (Supplementary Table 1). Using Sanger sequencing, we attempted validation of 141 (of the 1,544) previously unidentified de novo variants, including all novel LGD (stop-gain, frameshift and splice-site) events as well as recurrent missense mutations in autism candidate genes 55. We were able to validate 77 new sites (55%). These SNVs included various functional classes: 1 codon change plus codon deletion, 11 frameshift, 51 missense, 3 splice-site acceptor, 4 splice-site donor, 6 stop-gained, and 1 stop-lost. In addition, we also validated by Sanger sequencing another 132 previously called 9 but not confirmed events, resulting in a total of 209 validated de novo SNVs and indels (Supplementary Table 2). This new analysis identified 21 novel recurrently hit genes not identified in previous studies (Table 2). We did not attempt validation of any inherited SNVs but rather used the intersection of FreeBayes and GATK to get the highest quality variants events (all rare SNVs shown in Supplementary Dataset 2).
We used SNP microarray data (available for 1,266 quads) for validation of CNV events discovered using CoNIFER and XHMM. Probe-level copy number estimates were generated for each array sample using the Corrected Robust Linear Model with Maximum Likelihood Classification (CRLMM) software 53,54. A permutation-based method examined the mean copy number of all probes in each CNVR versus random sampling of the same number of probes from the genome (n = 10,000) in order to assess event confidence. Events with a permutation p-value < 0.01 and with a percentile rank < 30 or > 70 were considered validated deletions and duplications, respectively (full validated events for all quads and trios are shown in Supplementary Table 13). To further validate and genotype de novo events, we employed the CRLMM method to recall genotypes in trios and were able to recover events that were truly de novo as well as those inherited from a parent but missed in the exome analysis. Our final CNV dataset for all statistical testing consisted of validated events in the 1,266 quads for which SNP microarray data were available. We further tested an additional 50 de novo CNVs in individuals lacking SNP microarray data by array CGH 12 using a customized Agilent microarray. In this design, we targeted events and flanking genomic regions (up to 5 kbp or 3 exons) where probe density ranged from 150 bp to 5 kbp spacing, depending on the size of the event. Of these CNV events, 26 CNVs were validated, of which 21 were confirmed to be de novo while 5 CNVs were transmitted events (Supplementary Table 5).
Statistical analyses
We tested for transmission disequilibrium between probands and siblings in aggregate with a Fisher’s exact test (by comparing summed proband and sibling variant counts for LGD versus non-LGD events) and at the level of each proband-sibling pair using the Mann-Whitney U test (by comparing the variant counts in each proband-sibling pair). In addition, we utilized a logistic regression model in which the dependent variable was the presence of a variant in a proband (True or False), and independent variables were characteristics of the variant (such as its frequency or conservation score; see “SNV transmission disequilibrium”). Note that we applied a different conditional logistic regression to assess the risk by variant class to affected and unaffected individuals within the families). We utilized the RVIS 20 to identify genes that were not tolerant of functional or deleterious mutation in control populations (defined here as RVIS percentile < 50) and hypothesized that the score may have similar relevance to ASD genes (see also 21). We examined the RVIS profile of genes in a protein-protein interaction network of based on published de novo mutations in ASD 55 and found that these ASD-related genes have an average RVIS percentile of 26.3. This average was significantly lower than randomly picked sets of genes, suggesting that the RVIS percentile is a relevant predictor of ASD genes (p < 1×10−6, permutation testing, Supplementary Figure 13). In order to integrate both CNV and SNV data for specific genes, events were tabulated based on variation type (SNV/CNV) and inheritance class as presented throughout this manuscript. In particular, we counted all de novo CNVs and LGD or missense SNV events, private LGD-inherited SNVs in genes with an RVIS < 50%, and rare, inherited CNVs in which at least one gene had an RVIS < 50%. From these values, we calculated p-values for de novo SNVs 7 and inherited SNVs and CNVs (binomial test). Genes were ranked based on a Fisher’s combined p-value test (see Supplementary Table 6; family-based aggregation shown in Supplementary Table 14). We also applied the RV-TDT 22 using trio data (parents and either the affected proband or the unaffected sibling) to test for an association between rare, inherited LGD events in conserved genes (RVIS <50 and 20th percentile) and ASD.
Combined gene-level ranking
For each gene (Supplementary Table 6), we calculated the difference in counts between proband and sibling delta score in their number of de novo LGD and missense SNVs. The delta score was adjusted for gene size and gene-specific mutation rate as described previously 6. Due to the rarity of de novo CNVs and the difficulty in assigning gene-specific p-values to large CNVs, we did not include de novo CNVs in this calculation. For inherited SNVs, we also calculated the proband-sibling delta score based on private LGD SNVs and used a simple binomial test to rank genes. The de novo- and inherited-specific p-values were integrated using Fisher’s combined p-value test.
Conditional logistical regression
We estimated the contribution of genetic risk to ASD for both inherited and de novo CNVs and SNVs using an additive conditional logistic regression model, and adding a strata for families (or proband-sibling pairs). This model took the form:
Each term is composed of the total number of events in each individual. We included all de novo CNVs, all LGD de novo SNVs, the set of private LGD-inherited SNV mutations in genes with RVIS values < 50%, and the set of rare, inherited CNVs with a minimum RVIS of 50% or lower. We counted only autosomal events for all domains. The model was run with the survival.clogit function in the R language.
To test for nonlinear—or exponential—effects, we contrasted two simplified logistic regression models. In the first, we predicted proband (ASD) or sibling (unaffected) status based simply on the summed number of mutations defined above (and again including family-level stratums). The OR for each mutation (regardless of type) in this model was 1.17 (p < 1×10−8). In the second model, we added a term consisting of the total number of mutations squared. In this model, the simple sum was again significant (OR = 1.20, p = 0.002), but the squared sum term was not (OR = 1.00, p = 0.59).
Overlap between genes enriched for de novo and inherited events
We examined if genes enriched for de novo mutations were also enriched for the class of inherited, private LGD mutations. Using data from Supplementary Table 6, we ranked all genes by their enrichment for de novo mutations (via the ‘de.novo.SNV.p.value’ column). We took the top 100 genes in this sorted list and compared the summed gene counts for all inherited CNVs and SNVs in this group against 10,000 iterations of 100 randomly selected genes (without replacement) from the list. Observation of the resulting histogram and observed values suggests that genes enriched for de novo mutations do not overlap with genes enriched for inherited LGD mutations or disruptive rare CNVs (Supplementary Figure 9).
Population attributable risk
We assessed the contribution of different variant types to risk in the population. Included in the variant types were SNVs of the following classes: inheritance (de novo, private inherited), RVIS (no cutoff, 50, 20), and transmission (all, maternal, paternal). CNV classes tested included: inheritance (de novo, rare (<0.8%) inherited), type (deletion, duplication), and size (no cutoff, <100 kb). To assess the attributable fraction (estimated) in exposed and attributable fraction (estimated) in the population, we used the epi.2by2 function in epiR 56. We calculated population attributable risk using the method detailed in Taylor et al. 1977 27. For a given variant type, the attributable fraction in the exposed gives the fraction of cases with the variant type that have autism because of that variant type, the attributable fraction in the population is the number of cases with the variant type who have autism because of the variant type, and the population attributable risk is the proportion of autism relevant to the variant type 27. Complete results for all categories are listed in Supplementary Table 8.
Supplementary Material
Acknowledgments
We thank David Obenshain, Dan Hall, Brian Koser and Svetlana Novikova for providing support for usage of the Amazon Cloud and for assistance in the deposition of SNV and CNV callsets into the National Database for Autism Research (NDAR). We are grateful to the laboratories of Drs. Mike Wigler and Matt State for providing early access to exome sequencing data as well as access to SNP microarray data. We also thank Tonia Brown for assistance in editing this manuscript. Funding for this study was provided by the National Institutes for Mental Health (#R01MH101221 to E.E.E. and #R01MH100047 to R.B.) and by the Simons Foundation (SFARI #89368 to R.B. and SFARI #137578 to E.E.E.) E.E.E. is an investigator of the Howard Hughes Medical Institute. We are grateful to all of the families at the participating Simons Simplex Collection (SSC) sites, as well as the principal investigators (A. Beaudet, R. Bernier, J. Constantino, E. Cook, E. Fombonne, D. Geschwind, R. Goin-Kochel, E. Hanson, D. Grice, A. Klin, D. Ledbetter, C. Lord, C. Martin, D. Martin, R. Maxim, J. Miles, O. Ousley, K. Pelphrey, B. Peterson, J. Piggot, C. Saulnier, M. State, W. Stone, J. Sutcliffe, C. Walsh, Z. Warren, E. Wijsman). We appreciate obtaining access to phenotypic data on SFARI Base. Approved researchers can obtain the SSC population dataset described in this study by applying at https://base.sfari.org.
Footnotes
URLs
Deposited raw data (including BAM files, VCF files, additional data files, and software pipelines): https://ndar.nih.gov/study.html?id=353 or http://dx.doi.org/10.15154/1151812
ACCESSION CODES
NDAR Study 353
DOI 10.15154/1151812
COMPETING FINANCIAL INTERESTS
E.E.E. is on the scientific advisory board (SAB) of DNAnexus, Inc. and is a consultant for Kunming University of Science and Technology (KUST) as part of the 1000 China Talent Program.
AUTHOR CONTRIBUTIONS
NK, TNT, EEE designed experiments, wrote and edited the manuscript; NK performed sequence data reanalysis and created and analyzed the SNV callset; TNT created and analyzed the CNV callset, analyzed SNP microarray data, performed statistical analyses for SNV and CNV quality control, and examined epidemiological features of the full dataset; CB, LV, KM, KW and HAS performed validation experiments and sample handling. AR and BPC provided additional computational support; ZH and SZM performed the TDT tests and statistical analyses; RB provided phenotypes and additional SSC variables where needed.
References
- 1.Autism, I. MMWR Surveill Summ. 2012:1–9. [Google Scholar]
- 2.Gaugler T, et al. Most genetic risk for autism resides with common variation. 2014;46:881–5. doi: 10.1038/ng.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hallmayer J, et al. Genetic heritability and shared environmental factors among twin pairs with autism. Arch Gen Psychiatry. 2011;68:1095–102. doi: 10.1001/archgenpsychiatry.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iossifov I, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–99. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Roak BJ, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–9. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Roak BJ, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–22. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Roak BJ, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–50. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanders SJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–41. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iossifov I, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014 doi: 10.1038/nature13908. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Rubeis S, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014 doi: 10.1038/nature13772. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao X, et al. A unified genetic theory for sporadic and inherited autism. Proc Natl Acad Sci U S A. 2007;104:12831–6. doi: 10.1073/pnas.0705803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krumm N, et al. Transmission disequilibrium of small CNVs in simplex autism. Am J Hum Genet. 2013;93:595–606. doi: 10.1016/j.ajhg.2013.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poultney CS, et al. Identification of small exonic CNV from whole-exome sequence data and application to autism spectrum disorder. Am J Hum Genet. 2013;93:607–19. doi: 10.1016/j.ajhg.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pinto D, et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am J Hum Genet. 2014;94:677–94. doi: 10.1016/j.ajhg.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Rubeis S, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–15. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pinto D, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–72. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacquemont S, Coe BP, Hersch M, Duyzend M, Krumm N, Reymond A, Beckmann S, Bermann S, Rosenfeld JA, Eichler EE. A higher mutational burden in females supports a “female protective model” in neurodevelopmental disorders. Am J Hum Genet. doi: 10.1016/j.ajhg.2014.02.001. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ronemus M, Iossifov I, Levy D, Wigler M. The role of de novo mutations in the genetics of autism spectrum disorders. Nat Rev Genet. 2014;15:133–41. doi: 10.1038/nrg3585. [DOI] [PubMed] [Google Scholar]
- 19.Sanders SJ, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–85. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samocha KE, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46:944–50. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He Z, et al. Rare-variant extensions of the transmission disequilibrium test: application to autism exome sequence data. Am J Hum Genet. 2014;94:33–46. doi: 10.1016/j.ajhg.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Constantino JN, et al. Validation of a brief quantitative measure of autistic traits: comparison of the social responsiveness scale with the autism diagnostic interview-revised. J Autism Dev Disord. 2003;33:427–33. doi: 10.1023/a:1025014929212. [DOI] [PubMed] [Google Scholar]
- 24.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–61. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subtil-Rodriguez A, et al. The chromatin remodeller CHD8 is required for E2F-dependent transcription activation of S-phase genes. Nucleic Acids Res. 2014;42:2185–96. doi: 10.1093/nar/gkt1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bernier R, et al. Disruptive CHD8 mutations define a subtype of autism early in development. Cell. 2014;158:263–76. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taylor JW. Simple estimation of population attributable risk from case-control studies. American Journal of Epidemiology. 1977;106:260. doi: 10.1093/oxfordjournals.aje.a112461. [DOI] [PubMed] [Google Scholar]
- 28.Steinman G, Mankuta D. Insulin-like growth factor and the etiology of autism. Med Hypotheses. 2013;80:475–80. doi: 10.1016/j.mehy.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 29.Bozdagi O, Tavassoli T, Buxbaum JD. Insulin-like growth factor-1 rescues synaptic and motor deficits in a mouse model of autism and developmental delay. Mol Autism. 2013;4:9. doi: 10.1186/2040-2392-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurahashi H, et al. Isolation and characterization of a novel gene deleted in DiGeorge syndrome. Human Molecular Genetics. 1995;4:541–549. doi: 10.1093/hmg/4.4.541. [DOI] [PubMed] [Google Scholar]
- 31.Kaindl AM, et al. Many roads lead to primary autosomal recessive microcephaly. Prog Neurobiol. 2010;90:363–83. doi: 10.1016/j.pneurobio.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 32.Dong S, et al. De novo insertions and deletions of predominantly paternal origin are associated with autism spectrum disorder. Cell Rep. 2014;9:16–23. doi: 10.1016/j.celrep.2014.08.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Litterman N, et al. An OBSL1-Cul7Fbxw8 ubiquitin ligase signaling mechanism regulates Golgi morphology and dendrite patterning. PLoS Biol. 2011;9:e1001060. doi: 10.1371/journal.pbio.1001060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Havik B, et al. The complement control-related genes CSMD1 and CSMD2 associate to schizophrenia. Biol Psychiatry. 2011;70:35–42. doi: 10.1016/j.biopsych.2011.01.030. [DOI] [PubMed] [Google Scholar]
- 35.Cukier HN, et al. Exome sequencing of extended families with autism reveals genes shared across neurodevelopmental and neuropsychiatric disorders. Mol Autism. 2014;5:1. doi: 10.1186/2040-2392-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Girirajan S, et al. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet. 2010;42:203–9. doi: 10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bena F, et al. Molecular and clinical characterization of 25 individuals with exonic deletions of NRXN1 and comprehensive review of the literature. Am J Med Genet B Neuropsychiatr Genet. 2013;162b:388–403. doi: 10.1002/ajmg.b.32148. [DOI] [PubMed] [Google Scholar]
- 38.Lupski JR. Digenic inheritance and Mendelian disease. Nat Genet. 2012;44:1291–2. doi: 10.1038/ng.2479. [DOI] [PubMed] [Google Scholar]
- 39.Fischbach GD, Lord C. The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron. 2010;68:192–5. doi: 10.1016/j.neuron.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 40.Stessman HA, Bernier R, Eichler EE. A genotype-first approach to defining the subtypes of a complex disease. Cell. 2014;156:872–7. doi: 10.1016/j.cell.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li B, et al. QPLOT: a quality assessment tool for next generation sequencing data. Biomed Res Int. 2013;2013:865181. doi: 10.1155/2013/865181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garrison E, MG . Haplotype-based variant detection from short-read sequencing. 2012. arXiv preprint arXiv: 1207.3907 [q-bio.GN] [Google Scholar]
- 45.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu X, Jian X, Boerwinkle E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat. 2013;34:E2393–402. doi: 10.1002/humu.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pedregosa FaVG, Gramfort A, Michel V, Thirion BaGO, Blondel M, Prettenhofer P, Weiss RaDV, Vanderplas J, Passos A, Cournapeau DaBM, Perrot M, Duchesnay E. Scikit-learn: Machine Learning in {P}ython. Journal of Machine Learning Research. 2011;12:2825–2830. [Google Scholar]
- 49.Krumm N, et al. Copy number variation detection and genotyping from exome sequence data. Genome Res. 2012;22:1525–32. doi: 10.1101/gr.138115.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fromer M, et al. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am J Hum Genet. 2012;91:597–607. doi: 10.1016/j.ajhg.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hach F, et al. mrsFAST: a cache-oblivious algorithm for short-read mapping. Nat Methods. 2010;7:576–7. doi: 10.1038/nmeth0810-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–63. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 53.Ritchie ME, Carvalho BS, Hetrick KN, Tavare S, Irizarry RA. R/Bioconductor software for Illumina’s Infinium whole-genome genotyping BeadChips. Bioinformatics. 2009;25:2621–3. doi: 10.1093/bioinformatics/btp470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scharpf RB, Irizarry RA, Ritchie ME, Carvalho B, Ruczinski I. Using the R Package crlmm for Genotyping and Copy Number Estimation. J Stat Softw. 2011;40:1–32. [PMC free article] [PubMed] [Google Scholar]
- 55.Krumm N, O’Roak BJ, Shendure J, Eichler EE. A de novo convergence of autism genetics and molecular neuroscience. Trends Neurosci. 2014;37:95–105. doi: 10.1016/j.tins.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stevenson M. epiR: Tools for the Analysis of Epidemiological Data. 2015 [Google Scholar]
- 57.Morita M, et al. A novel 4EHP-GIGYF2 translational repressor complex is essential for mammalian development. Mol Cell Biol. 2012;32:3585–93. doi: 10.1128/MCB.00455-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.