Abstract
Background
The application of the Gal4/UAS system to enhancer and gene trapping screens in zebrafish has greatly increased the ability to label and manipulate cell populations in multiple tissues, including the central nervous system (CNS). However the ability to select existing lines for specific applications has been limited by the lack of detailed expression analysis.
Results
We describe a Gal4 enhancer trap screen in which we used advanced image analysis, including 3D confocal reconstructions and documentation of expression patterns at multiple developmental timepoints. In all, we have created and annotated 98 lines exhibiting a wide range of expression patterns, most of which include CNS expression. Expression was also observed in non-neural tissues such as muscle, skin epithelium, vasculature, and neural crest derivatives. All lines and data are publicly available from the Zebrafish International Research Center (ZIRC) from the Zebrafish Model Organism Database (ZFIN).
Conclusions
Our detailed documentation of expression patterns, combined with the public availability of images and fish lines, provides a valuable resource for researchers wishing to study CNS development and function in zebrafish. Our data also suggest that many existing enhancer trap lines may have previously uncharacterized expression in multiple tissues and cell types.
Keywords: CNS, Gal4, zebrafish, neural circuit, confocal
Introduction
The function of specific neuronal populations forms the basis of most animal behaviors, but in vertebrates many of these populations remain unstudied. Although new resources such as transgenic lines are becoming available (Satou et al., 2013), a detailed analysis of central nervous system (CNS) neuronal and glial identities, morphology, connectivity, and physiological properties has been hampered by a lack of reagents and tools suitable for such approaches. Similarly, developmental studies of CNS cells and nuclei have been limited as many enhancers and transgenic fish remain undefined or are not available.
Zebrafish represent an ideal vertebrate system in which to study CNS development and function. Physically small and transparent at larval stages, the entire zebrafish CNS can be imaged at high resolution in wholemount. A wide range of reporter lines can be used to characterize neuronal shape using fluorescent proteins (Mumm et al., 2006), synaptic connectivity using FP fusions directed to presynaptic or postsynaptic sites (Campbell et al., 2007), or neuronal activity using genetically-encoded calcium indicators such as cameleon (Higashijima et al., 2003) or inverse pericam (Li et al., 2005). One can test whether neurons function in particular behaviors by killing them through conversion of a prodrug to a cytotoxin using nitroreductase (Curado et al., 2007; Pisharath et al., 2007), or by silencing their synaptic output using tetanus toxin light chain (Asakawa et al., 2008). Recently, it has even become possible to activate neurons with light, by expressing the light-gated ionotropic glutamate receptor (Szobota et al., 2007) or the light-activated cation channel, channelrhodopsin-2 (Douglass et al., 2008).
Until recently, only a handful of neuronal populations were accessible to these types of manipulation, either because they could be identified by anatomical position (e.g. retinal cells), or were labeled by one of a few neuron-specific reporter transgenics. In recent years, the use of the Gal4-enhancer trap screens has gained traction in zebrafish. Gal4 is a yeast transcriptional activator that binds to an Upstream Activating Sequence (UAS), leading to transcription of associated genes (Brand and Perrimon, 1993). Used effectively to create neural specific enhancer trap lines in Drosophila (Otsuna and Ito, 2006), the Gal4/UAS system has been adapted to zebrafish (Scheer and Campos-Ortega, 1999; Koster and Fraser, 2001; Scheer et al., 2001; Halpern et al., 2008) employing previously identified gene enhancers, or in unbiased enhancer trap (ET) (Davison et al., 2007; Scott et al., 2007; Asakawa et al., 2008; Ogura et al., 2009) and gene trap (GT) (Asakawa et al., 2008; Asakawa et al., 2013; Balciuniene and Balciunas, 2013) screens. These screens identified several hundred lines of fish with defined expression patterns, many of which have been made available to the community at large.
As a resource, Gal4 enhancer trap lines still have several shortcomings. First, they represent a small slice of potential expression patterns. Particularly in the CNS, the vast majority of cell populations remain “un-trapped” such that there are no Gal4 or other transgenic lines available with which to manipulate them. Second, in previous screens, a number of lines have background (non-enhancer driven) expression in heart, muscle, and lens thought to be due to the minimal promoters used in the Gal4 enhancer trap constructs (Scott et al., 2007; Asakawa et al., 2008). This background could obscure neuronal morphology and projections in the retina and spinal cord. Third, and most importantly, the collections that do exist are often difficult to search for desired expression patterns as limited detail in images can omit or obscure signals in specific CNS cell types. It is thus often unclear whether existing enhancer trap lines are truly specific, as higher resolution analysis may uncover cell populations expressing reporter proteins at lower levels.
Using a newly engineered Gal4 enhancer trap construct, we have carried out a screen to address these shortcomings. To maximize the usefulness of our lines, we have greatly expanded the visual documentation such that researchers can more easily find enhancer trap lines with expression patterns of interest. We have produced compound images at several timepoints: 1 day post-fertilization (dpf), 2dpf, and 5–6dpf, to cover most of the development of the embryonic and larval zebrafish. These compound images are represented as Maximum Intensity Projections where appropriate, so that all structures can be identified in a single image regardless of focal plane. Additionally, we have produced confocal projections through the 5dpf larval brain and rendered these into 3D movies. Consisting of 98 lines, our screen effectively labels neuronal and glial cells in the CNS, as well as cells and tissues elsewhere in the developing zebrafish. This screen together with our high resolution and 3-dimensional data will allow researchers interested in neuronal development and function to identify and utilize truly appropriate lines, all of which are freely available to the community from ZFIN/ZIRC.
Results and Discussion
Generation of Gal4 constructs for use in enhancer trap screens
For our screen, we engineered new Tol2 based Gal4 enhancer trap constructs (Fig 1A, genbank accession #KC800697). The full length Tol2 was replaced with a truncated miniTol2 (Balciunas et al., 2006). To minimize nonspecific background expression, we used a short minimal promoter consisting of an 11bp fragment containing an adenovirus E1b TATA box, followed by a 30 bp fragment spanning the transcriptional start site of the carp beta-actin gene (Koster and Fraser, 2001). This promoter produces transient background expression in the notochord of some lines at 24 hpf stage (42 lines), which disappears by day 2 in most lines. Otherwise, our Gal4 lines showed no reproducible background expression patterns. We also used an optimized Gal4-VP16 (413–470) fusion for transcriptional activation (Ogura et al., 2009). Two additional Gal4 enhancer trap constructs were created in which Gal4 was fused to the FF activation sequence (genbank accession #KC800698 and #KC800699). This activation sequence uses two VP16 transcription activation modules and has previously been used without any reports of toxicity (Asakawa, 08). Ten pilot lines were established using these constructs (zc1080A, zc1081A, zc1082A, zc1083A, zc1084A, zc1087A, zc1088A, zc1090A, zc1091A, and zc1093A), but since we did not observe any appreciable advantages relative to our Gal4-VP16 construct in terms of expression level, we did not continue to use it in our screen. All other lines were created with the Gal4-VP16 (413–470) construct.
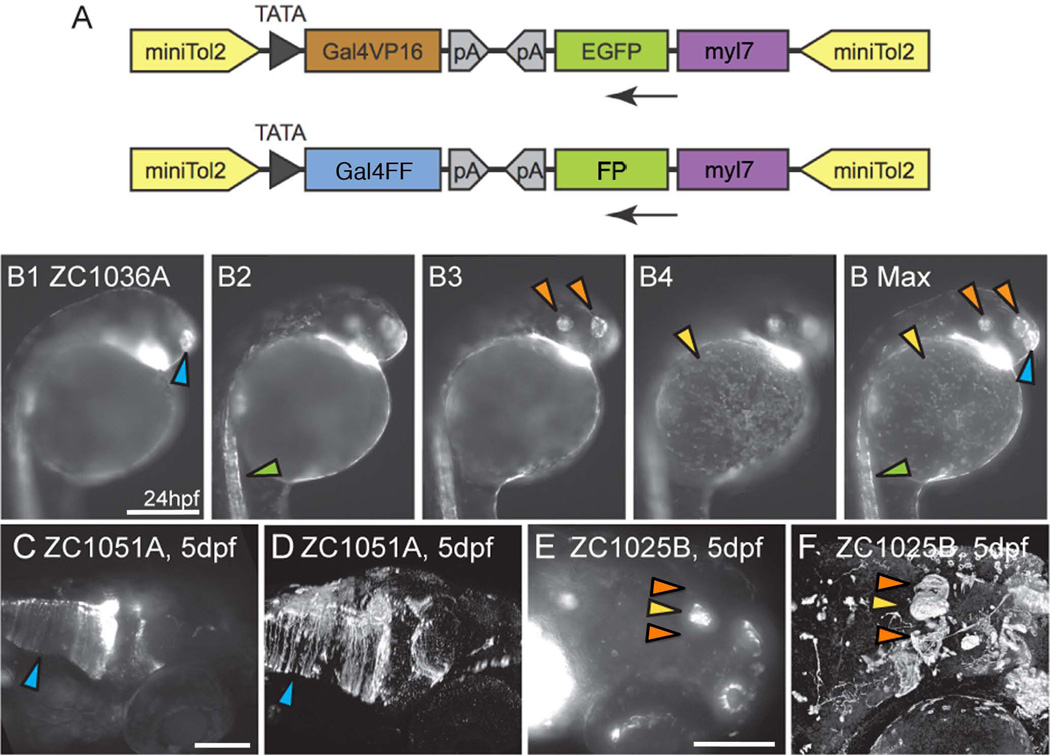
Figure 1. Gal4 enhancer trap constructs and imaging.
(A) Enhancer trap constructs used to generate all lines. (B1-B Max) To convey as much information as possible, compound microscopic images were taken at multiple depths (B1–B4) and used to generate Maximum Intensity Projections (B Max) in which signal from multiple focal depths is visible. Arrowheads mark the olfactory placode (blue), spinal neurons (green), otic vesicle and retina (orange) and epithelial cells (yellow). Images B1-B Max are lateral views. (C–F) Confocal images enhance fine details relative to compound microscopic images. Glial endfeet are clearly visible in the confocal image (D, Arrowhead) but are not clear in the compound scope image (C, Arrowhead). Habenulae are visible in confocal (F, Orange arrowheads) but not compound (E, Orange arrowheads) image. (E,F) Pineal gland marked by yellow arrowhead. Images C–F are dorso-lateral views. Scale bars: B1-B MAX 250um. C, D 250um. E, F 100um.
Finally, using the zebrafish Tol2kit (Kwan et al., 2007), we engineered a cardiac specific myl7 reporter cassette driving either eGFP or TagRFP (a monomeric ref fluorophore with 555nm excitation maximum, Evrogen). This marker allowed us to rapidly screen for embryos that contained a successful Tol2 integration. One must note, however, that the presence or absence of the myl7:FP marker does not indicate the number of integrations in F0 fish. Furthermore, the number of inherited Gal4 expression patterns, indicative of successful enhancer traps, is not equivalent to transgene insertions.
Isolation of 98 lines, nearly all with expression patterns in the CNS
A screen was performed by mating wild type (TU) fish and injecting F0 embryos at the 1-cell stage with 25pg of Gal4 enhancer trap construct DNA (Figure 1a) and 25–50pg transposase mRNA (Kwan et al., 2007). F0 fish were raised to maturity and mated to UAS reporter fish to produce F1 offspring. In most cases, the UAS-reporter fish Tg(UAS-E1b:NTR-mCherry) c264, (Davison et al., 2007) was used. Occasionally, either UAS-reporter fish Tg(UAS-E1b:Kaede)s1999t (Davison et al., 2007) or Tg(5×UAS:eGFP)zf82 (Asakawa et al., 2008) was used instead. F1 embryos exhibited one of the following patterns: 1) no expression, most likely indicating lack of germline mTol2 integration, 2) heart expression only, indicating vector integration but no enhancer trap, 3) heart expression and a UAS-reporter expression pattern, indicating a successful enhancer trap, or 4) heart expression and multiple UAS-reporter expression patterns indicating multiple successful enhancer traps. Importantly, all lines exhibiting tissue-specific UAS-reporter expression also had expression of the heart marker, indicating that it represents an effective tool to screen for transgene insertions.
Based on the interests of our laboratory, 86 F1 lines were established that expressed a UAS-driven reporter in unique patterns that included the CNS. As seen in Table 1, use of our Gal4 enhancer trap construct resulted in a very high rate of single enhancer traps as defined by expression of a single pattern (67%). Most of the remaining F1 lines had two independent expression patterns, with a small number exhibiting 3 or 4 patterns (Table 1). After outcrossing, we generated 120 individual Gal4 enhancer trap lines. A number of lines were lost as we were unable to maintain them or obtain sperm for cryopreservation. Thus, we were left with 98 unique lines, almost all of which show expression in CNS tissues under the control of trapped enhancers (Tables 2 and 3).
Table 1.
Rates of multiple expression patterns derived from enhancer trap screen using mTol2 enhancer trap construct. Number and frequency of single, double, triple, and quadruple patterns in F1 lines exhibiting reporter expression, and resulting number of unique enhancer trap lines after outcrossing to F3 generation.
| Expression patterns derived from Gal4 ET screen | ||
|---|---|---|
| Initial F1 | Outcrossed F3 | |
| Single pattern | 58 (67%) | 58 |
| double pattern | 23 (27%) | 46 |
| triple pattern | 4 (5%) | 12 |
| quadruple pattern | 1 (1%) | 4 |
| totals | 86 | 120 |
Table 2.
Tissue specific expression patterns derived from Gal4 enhancer trap screen.
| Expression by tissue | |||
|---|---|---|---|
| CNS | # lines | Other | # lines |
| amacrine cell | 3 | blood vessel | 2 |
| anterior commissure | 2 | epidermal cells | 30 |
| epiphysis | 8 | extraocular muscle | 10 |
| forebrain | 28 | fin | 4 |
| glia | 26 | hatching gland | 1 |
| habenula | 5 | lateral line | 8 |
| hindbrain | 39 | lens | 1 |
| hypothalamus | 13 | muscle | 35 |
| MHB | 7 | notochord | 42 |
| midbrain | 43 | pancreas | 2 |
| Muller glia | 22 | pronephros | 1 |
| olfactory placode/bulb | 31 | tail | 3 |
| otic vesicle | 12 | vasculature | 8 |
| pituitary | 1 | ||
| retina | 43 | ||
| RGC | 4 | ||
| Rohon-Beard cells | 3 | ||
| spinal neuron | 29 | ||
| tectum | 22 | ||
| trigeminal ganglion | 4 | ||
Table 3. Gal4 Enhancer Trap Lines.
Full list of enhancer trap lines deposited with ZIRC/ZFIN. Expression patterns are categorized by presence inside or outside the CNS
| Line | CNS | Other |
|---|---|---|
| 1000A | retina, MHB | notochord, epidermal cells |
| 1002A | olfactory placode/bulb | lens, muscle |
| 1003A | forebrain, midbrain, habenula retina, olfactory placode/bulb | notochord, epidermal cells extraocular muscle, fin |
| 1004A | Amacrine cells, hindbrain retina, midbrain | epidermal cells, notochord trigeminal ganglion, muscle |
| 1006A | hindbrain, retina, otic vesicle | muscle |
| 1007A | spinal neurons, hindbrain | muscle |
| 1008A | Amacrine cells, hypothalamus midbrain, otic vesicle, retina | epidermal cells muscle, lateral line |
| 1009A | midbrain, hindbrain olfactory placode/bulb | lateral line, muscle, epidermal cells blood vessels, pancreas, fin |
| 1010A | olfactory placode/bulb, pituitary | epidermal cells, notochord, hatching gland |
| 1011A | epidermal cells | |
| 1012A | midbrain, tectum, otic vesicle hindbrain, glia, Muller glia | epidermal cells notochord, blood vessel |
| 1013A | otic vesicle amacrine cell, retina | muscle, trigeminal ganglion epidermal cells |
| 1015A | Muller glia, retina, otic vesicle | muscle, epidermal cells |
| 1016A | retina, RGC, Muller glia Rohon-Beard cells, otic vesicle olfactory placode/bulb | fin |
| 1016C | olfactory placode/bulb, glia, midbrain spinal neuron, hypothalamus, tectum hindbrain, forebrain, Muller glia | notochord muscle vasculature |
| 1016D | Muller glia, glia | epidermal cells, notochord extraocular muscle |
| 1017A | spinal motor neuron, midbrain | epidermal cells |
| 1018A | retina, forebrain, hindbrain olfactory placode/bulb, glia | notochord, epidermal cells extraocular muscle |
| 1019A | forebrain, retina, RGC midbrain, tectum, hindbrain | lateral line |
| 1019B | spinal neuron, Muller glia | |
| 1020A | spinal neuron | muscle, trigeminal ganglion notochord |
| 1020B | forebrain, Muller glia, retina midbrain, hindbrain, glia | |
| 1021A | otic vesicle, hindbrain | epidermal cells, vasculature |
| 1022A | olfactory placode/bulb | notochord |
| 1022B | midbrain, hypothalamus anterior commissure, MHB olfactory placode/bulb | |
| 1023A | midbrain, forebrain, retina, Muller glia | epidermal cells, muscle |
| 1023B | midbrain, tectum, retina, forebrain, olfactory placode/bulb, hindbrain, MHB | muscle |
| 1023D | notochord, vasculature, epidermal cells, fin | |
| 1024A | midbrain, retina, bipolar cells | |
| 1025A | hypothalamus, retina, glia, hindbrain tectum, midbrain, forebrain | notochord |
| 1025B | anterior commissure, retina, habenula olfactory placode/bulb, epiphysis | epidermal cells muscle |
| 1025D | olfactory placode/bulb | epidermal cells, notochord |
| 1026A | epiphysis, midbrain, hypothalamus glia, olfactory placode/bulb, forebrain | |
| 1029E | glia, tectum, hypothalamus | |
| 1030A | Muller glia, spinal neuron olfactory placode/bulb, retina | notochord |
| 1031A | Muller glia, glia, spinal neurons tectum, midbrain, forebrain, hindbrain | |
| 1032A | glia, olfactory placode/bulb midbrain, forebrain, spinal neurons | muscle epidermal cells |
| 1033A | glia, midbrain, MHB, RGC, hindbrain | pronephros, lateral line |
| 1034A | forebrain, hindbrain, midbrain spinal neuron, habenula | notochord extraocular muscle |
| 1035A | olfactory placode/bulb, retina | vasculature, epidermal cells |
| 1036A | olfactory placode/bulb | epidermal cells, muscle, notochord |
| 1037A | extraocular muscle, muscle, notochord | |
| 1037B | spinal neuron, Rohon-Beard cells | trigeminal ganglion |
| 1037C | glia, midbrain, Muller glia, retina hindbrain, tectum, forebrain | muscle |
| 1038A | glia, olfactory placode/bulb retina, Muller glia | epidermal cells pancreas |
| 1039A | forebrain, midbrain, retina Muller glia, hindbrain, glia spinal neurons, hypothalamus | notochord |
| 1039B | hypothalamus, spinal neurons, retina tectum, midbrain, hindbrain, forebrain | |
| 1040A | spinal neuron | muscle, notochord |
| 1040B | spinal neurons, retina, Rohon-Beard cells | vasculature notochord |
| 1041A | olfactory placode/bulb | muscle, notochord |
| 1042A | forebrain, midbrain, retina, hindbrain hypothalamus, spinal neurons | |
| 1044A | glia, midbrain, hindbrain hypothalamus, spinal neurons | |
| 1046A | spinal neuron | vasculature |
| 1047A | olfactory placode/bulb, epiphysis, hindbrain, otic vesicle spinal neurons, forebrain | epidermal cells muscle notochord |
| 1048A | retina, Muller glia, olfactory placode/bulb | lateral line extraocular muscle |
| 1049A | otic vesicle | |
| 1050A | spinal neurons, midbrain, retina, glia tectum, olfactory placode/bulb | notochord |
| 1051A | midbrain, glia MHB, hindbrain | |
| 1052A | tectum, midbrain | |
| 1052B | forebrain, midbrain | muscle, notochord |
| 1053A | glia, Muller glia, midbrain, forebrain spinal neurons, hindbrain, otic vesicle | notochord |
| 1054A | midbrain, spinal neurons, tectum | |
| 1055A | forebrain, midbrain, tectum, hindbrain spinal neurons, retina, RGC | |
| 1055B | otic vesicle, Muller glia, retina, glia | extraocular muscle, epidermal cells |
| 1056A | epiphysis, hindbrain, spinal neurons | extraocular muscle, notochord |
| 1057A | retina, olfactory placode/bulb tectum, midbrain, spinal neuron | notochord |
| 1060A | olfactory placode/bulb, glia | lateral line, notochord, muscle |
| 1062A | lateral line, notochord | |
| 1062B | forebrain, midbrain, hindbrain spinal neuron, glia | |
| 1063A | forebrain, hypothalamus, hindbrain Muller glia, retina, epiphysis, midbrain | notochord muscle |
| 1063B | olfactory placode/bulb | |
| 1065A | tectum, midbrain, hindbrain olfactory placode/bulb, glia | muscle notochord |
| 1066A | olfactory placode/bulb, hypothalamus forebrain, midbrain, tectum | |
| 1067A | olfactory placode/bulb | notochord, tail, extraocular muscle |
| 1068A | dorsal retina, Muller glia, midbrain forebrain, otic vesicle, habenula | notochord |
| 1069A | retina, Muller glia | muscle, extraocular muscle |
| 1070A | midbrain, spinal neurons, tectum, glia | notochord |
| 1071A | midbrain, hindbrain | muscle |
| 1072A | vasculature, notochord, epidermal cells | |
| 1074B | retina, Muller glia | vasculature, epidermal cells |
| 1075A | glia, tectum, midbrain | notochord |
| 1076A | forebrain, retina, hindbrain | notochord, muscle, tail |
| 1077A | retina, hindbrain, MHB | notochord, muscle |
| 1078A | tectum | |
| 1080A | hindbrain, tectum, glia | |
| 1081A | retina, Muller glia | muscle |
| 1082A | hindbrain | muscle |
| 1083A | retina, Muller glia | |
| 1084A | retina, hindbrain | epidermal cells |
| 1085A | epiphysis | epidermal cells |
| 1086A | olfactory placode/bulb | muscle, epidermal cells |
| 1087A | glia | tail, muscle |
| 1088A | retina, epiphysis, hindbrain olfactory placode/bulb, hypothalamus | muscle |
| 1090A | hindbrain | |
| 1091A | notochord | |
| 1093A | notochord | |
| 1094A | forebrain, midbrain hindbrain, spinal neuron |
Imaging of expression patterns
Complex expression patterns from successful enhancer traps are often difficult to capture in a single image. Images obtained from a stereomicroscope lack resolution while images obtained from a compound microscope lack the necessary focal depth for a vertebrate organism. We attempted to address these problems by acquiring compound serial images at different focal depths, and then merging them into maximum intensity projections. As seen in Fig 1 (B1-Bmax), we were able to create a single image that captures all the expression elements from a wide range of focal planes. Thus, more data can be presented in fewer images, facilitating the search for desired lines and expression patterns.
While intensely labeled structures are clearly identifiable in both compound and confocal images (Fig 1C–F), compound images often fail to capture less intense expression typical of individual cells and axons, particularly in live embryos (Fig 1C–F, arrowheads). Thus, while the compound image in Fig 1C shows expression in hindbrain cells, the confocal images allow clear identification of glial expression as indicated by mCherry+ glial end feet (Fig 1D). Similarly, other structures only weakly labeled are often missing in the live-embryo compound image. In figure 1E and 1F, the epiphysis (yellow arrowhead) is clearly visible. The habenula, located laterally on both sides of the epiphysis (orange arrowheads) is only visible in the antibody stained confocal image (Fig 1F).
Lines exhibit a wide range of temporal and spatial expression patterns
As with prior Gal4 enhancer trap screens, we observed expression in a wide variety of tissues. Our focus being on generation of lines that are useful for analysis of CNS development and function, a majority of the lines that we isolated and maintained exhibit expression in this tissue (89/98), including all divisions of the brain and spinal cord. Sensory elements of the PNS (neuromasts, olfactory epithelium) were also trapped. Many of the lines also showed expression in non-neural tissues including the skin epithelium, vasculature, and musculature (Table 2). Additionally, we were surprised to find many lines with expression in at least a subset of radial glial cells. For instance, 11 lines showed expression in retinal Müller glial cells, 15 lines had expression in other CNS radial glia, and 11 lines showed expression in both cell populations (Table 2). Whether this represents a natural bias towards glial gene expression in the zebrafish genome, an inherent bias in our Gal4 construct, or a reflection of the expertise of our laboratory in identifying this cell type is unclear. Regardless, the presence of so many radial glial-targeted lines opens up exciting possibilities to study glia and glial-neuronal interactions in the developing, functioning, or regenerating vertebrate CNS.
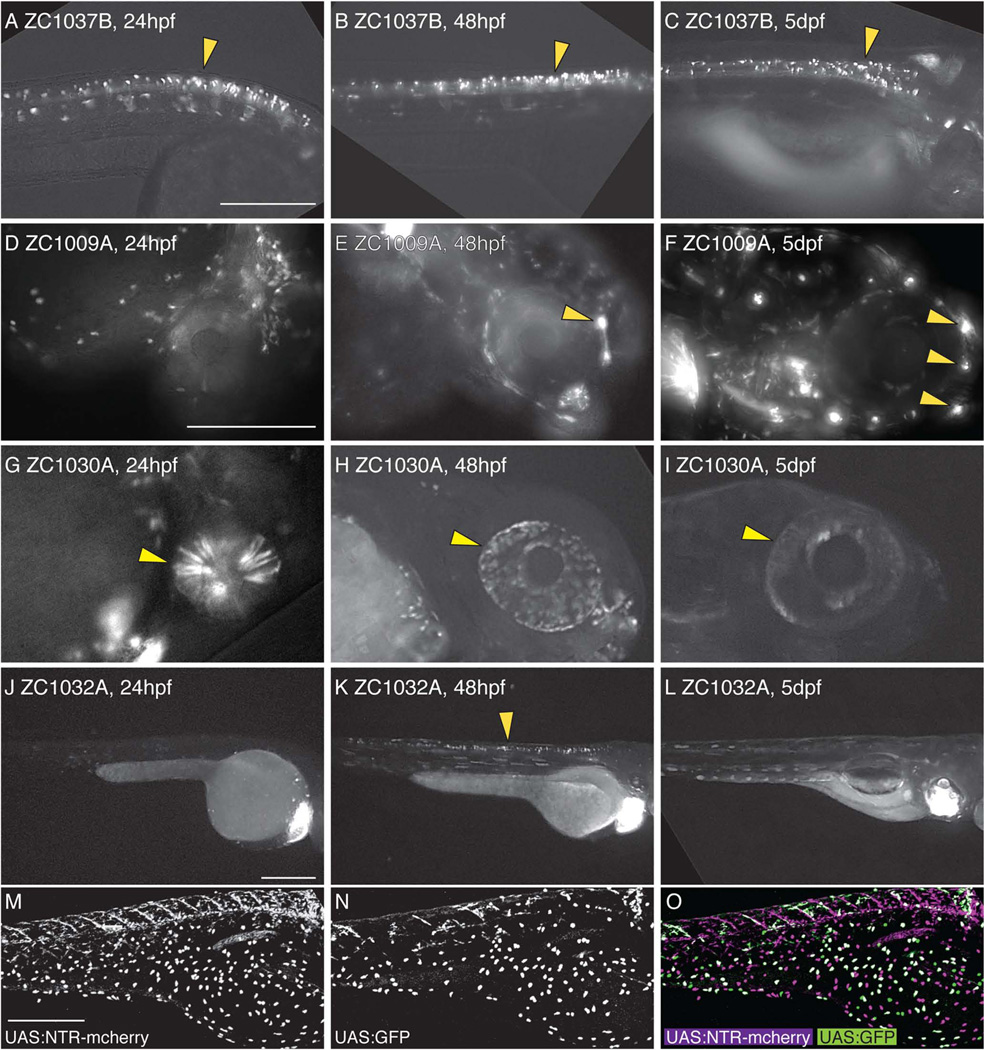
Because expression patterns are often spatially complex and can be transient in development, individual images at single developmental timepoints are not always informative. To make our screen more useful to the community, we chose to obtain images at three different developmental timepoints: 24hpf, 48hpf, and 5 or 6 dpf. We found many examples of expression patterns in which a tissue was labeled throughout development. For example line 1037B shows expression in spinal neurons at all three timepoints (Fig 2A–C). In contrast, other lines show expression that is absent at earlier timepoints but initiates or expands at later stages. zc1009A shows labeling of neuromasts starting at 48hpf and expanded expression by 5dpf when additional neuromasts have been deposited (Fig 2D–F). Additionally, some lines such as zc1030A show strong expression at 24 and 48hpf but greatly reduced expression by 5dpf (Fig 2G–I), while others such as zc1032A show transient expression only at a single timepoint (Figure 2J–L). By analyzing reporter expression at multiple timepoints, we were thus able to more accurately capture dynamic Gal4 expression profiles, something that is particularly useful for developmental studies.
Figure 2. Dynamic temporal expression patterns in enhancer trap screen.
Compound images taken at 24hpf (A,D,G,J), 48hpf (B,E,H,K), or 5dpf (C,F,I,L) show examples of neural expression that is maintained at all ages (A–C), is activated over time (D–F), is inactivated over time (G–I), or is only present at one timepoint (J–L). Arrowheads mark spinal neurons (A–C, K), lateral line neuromasts (E, F), and the eye (G–I). UAS reporters exhibit transgene-specific variegation, leading to partially overlapping but distinct expression patterns in a single animal (M–O). All images are lateral views. Scale bar: for A–C is 250um, for D–I is 250um, for J–L is 250um, and for M–O is 300um.
Even within an individual Gal4 enhancer trap line, we observed variable expression patterns. By crossing zc1016B to a line simultaneously expressing both Tg(5×UAS:eGFP) zf82 and Tg(UAS-E1b:NTR-mCherry) c264, we can show this variability in a single animal (Figure 2M–O). Both reporters were expressed in skin epithelial cells, but the majority of cells were labeled for only one reporter, clearly illustrating the variegation associated with UAS reporters (Davison et al., 2007; Scott, 2009; Akitake et al., 2011). Thus, in cases where the cells targeted by a specific enhancer trap need to be precisely defined, it is critical that either the Gal4 line be crossed to a number of different UAS reporters or the location of the insertion be mapped to better define the trapped enhancer. Importantly though, despite this background of variegation, we did not observe variability in the tissues or cell types expressing Gal4-driven reporters between individual embryos or multiple generations of a given line. We therefore believe that our Gal4 insertions are stable and reproducible.
3D confocal imaging shows detailed neural expression
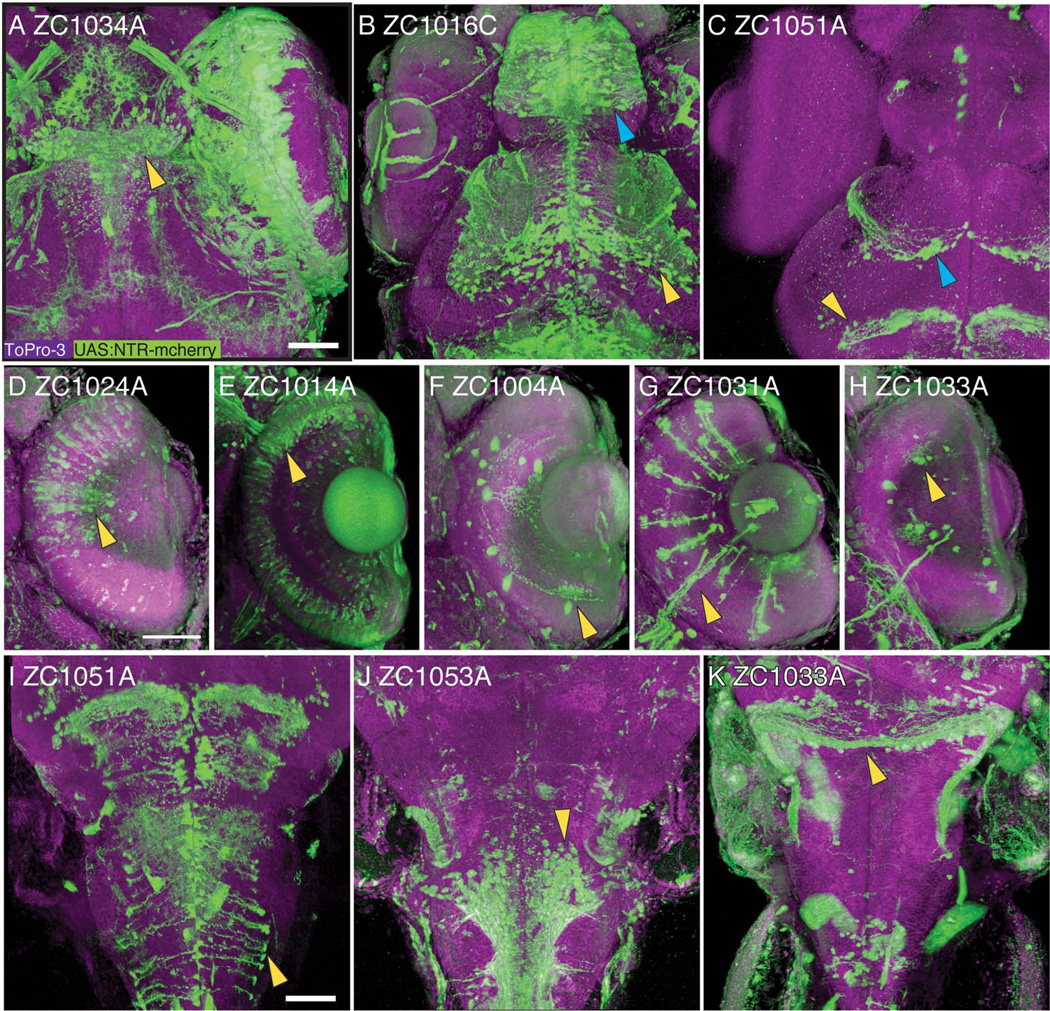
Although maximum intensity projections greatly increase the amount of information one can display in a single image, for complex three-dimensional structures such as the CNS these images are often still insufficient. Single-plane images do not convey three-dimensional information, nor do they allow the viewer to isolate and follow neuronal projections. To overcome this problem and make our screen more useful for study of the CNS, we have used confocal imaging together with the 3D rendering program FluoRender (Wan et al., 2009). 5dpf embryos were stained with antibodies to the respective UAS reporter (generally mCherry), and ToPro3 to label nuclei and provide a counterstain for better anatomical localization of the expression region. Rendered 3D reconstructions show highly detailed expression patterns in the CNS including the habenula, telencephalon, and ventral tectum (Fig 3A–C). In the retina, our screen has isolated lines with expression in a wide range of cell types including bipolar cells, photoreceptors, amacrine cells, Müller glia, and ganglion cells (Fig 3D–H). In the brainstem, examples of lines we have isolated show expression in radial glia, ventral motor neurons, and the rostral cerebellum (Fig 3I–K). In many cases, the morphology of substructures such as glial end feet, arborizations of individual cells, and axonal projections is clearly visible.
Figure 3.
Confocal 3-D reconstruction images of 5dpf embryos. Rostral CNS expression patterns are visible in the habenula (A, arrowhead), the telencephalon (B, blue arrowhead) and ventral tectum (B, yellow arrowhead), glial cells in the tectum (C, blue arrowhead) and midbrain-hindbrain boundary (C, yellow arrowhead). Expression is visible in major classes of retinal neurons including bipolar cells (D), photoreceptors (E), amacrine cells (F), Mueller glia (G), and ganglion cells (H). In the brainstem, expression is visible in radial glia (I), ventral neurons (J), and the rostral cerebellum (K). All images show UAS reporter (green), ToPro3 nuclear stain (magenta), and show dorsal view, with D–H showing a section through the retina. Scale bar 50um.
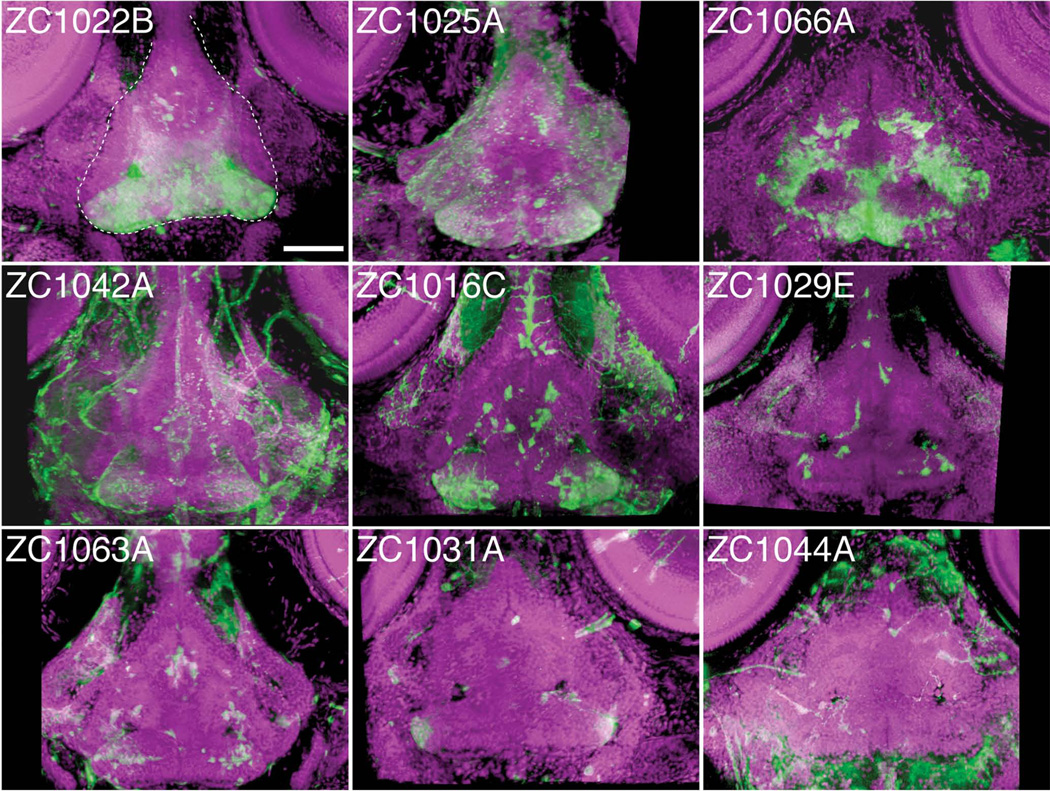
To fully take advantage of our data, rotatable 3D reconstructions were created and saved in MPEG file format. This allows us to convey with even greater clarity the types of cells labeled, their position in the CNS, and their axonal projections. For instance, in Figure 4, line zc1022B shows clear expression in the hypothalamus. When viewing an MPEG of the 3D reconstruction of confocal images for this line, it becomes clear that the expressing cells are located in the ventral portion of the hypothalamus, with axonal projections that extend between a more dorsal level of the hypothalamus and a small cluster of cells located in the telencephalon (Supplemental Movie 1 - MPEG of zc1022B). Line zc1016C shows expression clearly in neurons of the telencephalon and tectum, while expression in the hypothalamus is obscured by hindbrain expression in the dorsal view (Fig 3B). When viewed in MPEG format, the 3D rendering shows reporter-expressing cells are in the ventral tectum, and they project axons to the ventrolateral border of the neuropil layer (Supplemental Movie 2 - MPEG of zc1016C). Likewise, the labeled telencephalon neurons whose cell bodies lay along the dorsal midline extend axons towards the lateral border of the telencephalon. By rotating the 3D reconstruction, the hypothalamic expression in the ventral CNS is also clearly seen as two clusters of cells visible at the very bottom of the image (Supplemental Movie 2). Analysis of images in 3D thus greatly enhances the ability to determine exactly where in the CNS a given set of cells resides and projects.
Figure 4.
Individual enhancer traps drive various hypothalamic expression patterns. 3D reconstructed confocal images, dorsal view, of 5dpf whole mount embryos stained for UAS reporter (green) and counterstained with ToPro-3 for nuclei (magenta). Partial 3D reconstruction of these images has been limited to ventral confocal planes to highlight hypothalamic expression while removing more dorsal expressing cells. Dashed white line in panel zc1022B outlines the hypothalamus. Scale bar 50um.
Use of lines to define and manipulate CNS cells
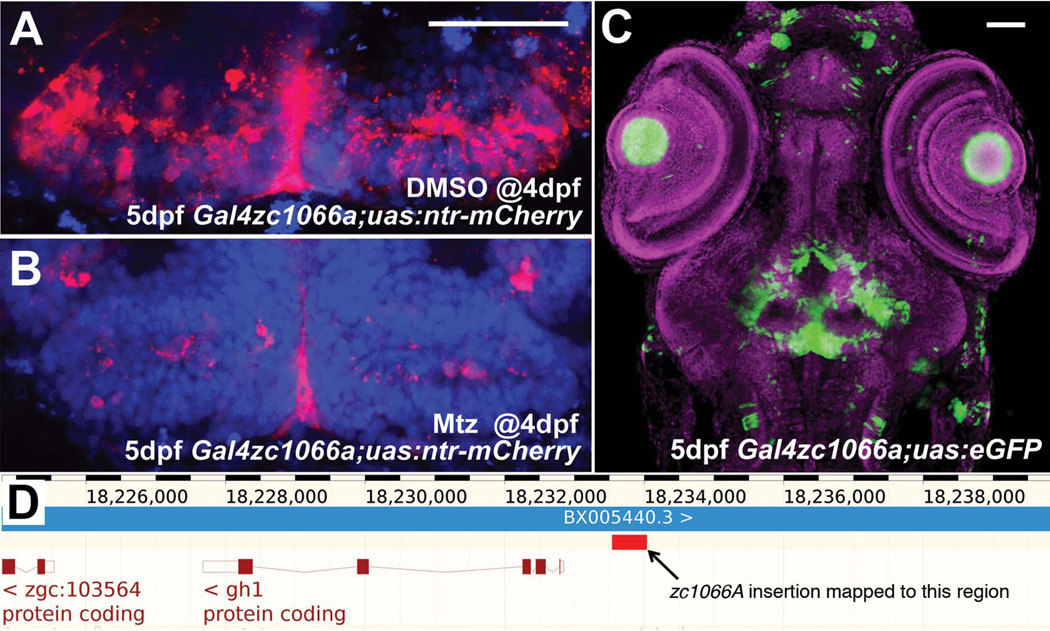
Within the CNS, many structures contain a complex assortment of cell types and functions. For instance, the hypothalamus contains neurons that respond to several different neurotransmitters, and both receive and send information to a wide range of targets. Investigating the development and circuitry of the hypothalamus requires genetic tools that are specific to functional and anatomical subsets of neurons. In this screen, we have identified zc1022B, zc1016C, and 7 other lines that express the UAS reporter in the hypothalamus under the control of trapped enhancers (Figure 4). Expression patterns range from just a few individual neurons to large regional subsets of the hypothalamus. The zc1066A allele shows expression in the posterior recess of the hypothalamus. We have found that this enhancer trap marks radial glia based on coexpression with BLBP and Glutamine synthetase (Wang et al., 2012). We have also used this allele to perform cell ablations, by crossing to fish expressing Tg(UAS-E1b:NTR-mCherry) c264, Following treatment with the pro-drug metrodinazole (Mtz), almost all labeled radial glia in the hypothalamus were ablated (Fig 5A,B), illustrating the efficacy of this line as a genetic driver. Because of the relatively limited expression pattern of the allele (Fig 5C), the enhancer trap can thus be used to selectively manipulate hypothalamic radial glia. Using an inverse-PCR strategy we mapped the insertion to only one position, approximately 800bp upstream of the growth hormone 1 (gh1) gene start site (Fig 5D, red box). Interestingly, the resulting enhancer trap does not recapitulate the gh1 expression pattern, which is limited to the pituitary. This serves as a cautionary note that, enhancer-trap lines may not necessarily reflect the expression of even closely neighboring genes.
Figure 5.
Use and characterization of a single enhancer trap line. (A) Line zc1066a drives expression of NTR-mCherry in hypothalamic radial glia. (B) Following incubation in metronidazole for 24 hours, most mCherry+ cells are ablated and only debris remains. (C) A confocal projection through the entire head of a zc1066a embryo at 5dpf shows the specificity of this enhancer trap expression pattern. (D) Using inverse-PCR, we have mapped the insertion to a genomic region immediately upstream of the gh1 mRNA. Scale bars 50um.
Expression in non-neural tissues
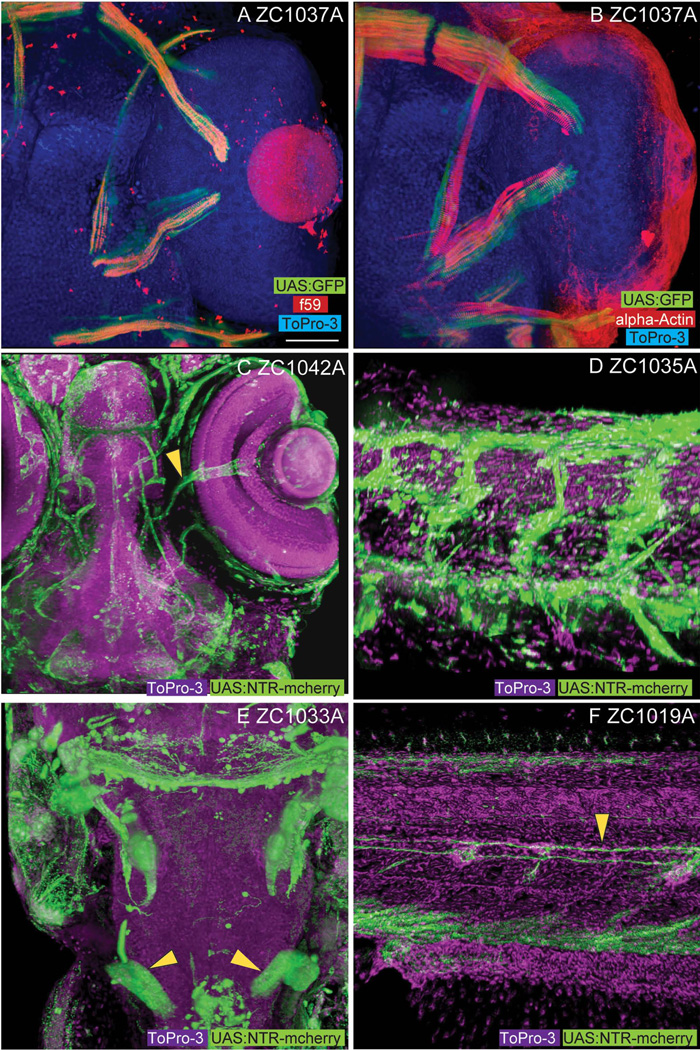
Although the focus of our work is centered on the CNS, the unbiased nature of enhancer trap screens is such that other expression patterns are also trapped. Among tissues in which we have observed expression are somatic muscle, including slow ocular muscles (zc1037A, Fig 6A, B), vasculature of the head and trunk (Fig 6C, D), developing pronephric tubules (Fig 6E), and glia of the lateral line (Fig 6F). For some, this non-neural expression will be useful. Recently, zc1044A was used for experimental analysis of defects in epidermal cell extrusion (Eisenhoffer et al., 2012).
Figure 6.
Non-CNS enhancer trap expression patterns. Among the non-CNS tissues represented in our screen, line zc1037A shows expression in slow (A, f59 positive) but not fast (B, alpha-actin) extraocular myofibers. Vascular expression is visible in both the head (C, arrowhead marks central retinal vessels, zc1042A) and body (D, zc1035A). Labeling of the developing pronephros (arrowheads) is visible in line zc1033A (E). Labeling of lateral line glial fibers (arrowhead) is visible in line zc1019A (F). Images A, B, C, and E show dorsal views. Images D and F show lateral views. Scale bar for images A–F are 50um.
Conclusions
Our enhancer trap screen has produced 98 lines with unique expression patterns, many of which encompass portions of the nervous system. Maximum intensity projections at multiple developmental stages and high resolution 3D image rendering were used to create a database of unparalleled detail, greatly enhancing the usefulness of this database to the community. However, with the ability to visualize enhancer trap expression at increased resolution comes the observation that many patterns are not specific to a single cell population even within a given tissue. This finding leads to the cautionary speculation that many previously characterized enhancer trap lines may also exhibit broad low-level expression in additional tissues and cells. Thus, care should be taken to confirm the specificity of expression, tools should be used to limit the function of Gal4-driven effector molecules where possible, and experimental results should be interpreted with this consideration.
All lines listed in Table 3 will be stored as sperm stocks at ZIRC, and the entire database of images available at ZFIN (http://zfin.org/action/figure/all-figureview/ZDB-PUB-130724-1). These lines, together with past screens and ongoing enhancer trap efforts will produce an increasingly large number of useful lines. As the number of documented Gal4 lines available to the scientific community increases, it will become increasingly possible to investigate previously intractable questions in vertebrates.
Experimental Procedures
Cloning of transgenic constructs
Gal4 enhancer trap constructs were created using multisite Gateway cloning to insert 5’, middle, and 3’ clones into mTol2 backbones. For ET(E1bTATA;Gal4VP16;mTol2CG3)(Genbank accession #KC800697), the 5’ clone encoded a TATA basal promoter, the middle clone encoded the Gal4VP16(413–470) fusion, and the 3’ clone encoded the polyadenylation sequence. Gateway LR recombination was performed to insert these clones into the pDestmTol2CG3 vector that contains a myl7 promoter/enhancer driving eGFP in reverse orientation (Kwan et al., 2007). ET(E1bTATA:Gal4FF;mTol2CG3) (Genbank accession #KC800698) and ET(E1bTATA:Gal4FF;mTol2CR3) (Genbank accession #KC800699) were created in a similar manner except that a Gal4FF fusion was used, and the pDestmTol2CR3 vector contains a TagRFP reporter driven by the myl7 promoter/enhancer, placed on the opposite strand.
Injections and establishment of lines
All fish were used and handled in accordance with University of Utah IACUC guidelines. Wild type TU strain fish were crossed to generate embryos. Injections were performed at the 1-cell stage by injecting 25pg of enhancer trap DNA with 50pg of Transposase mRNA. Injected F0 embryos were raised until maturity and then crossed to UAS-reporter lines. F1 embryos were scored for the presence or absence of both the heart specific myl7 reporter of transgenic integration as well as novel UAS-reporter expression patterns. F1 embryos positive for UAS-reporter expression were raised and outcrossed to the F3 generation to isolate individual expression patterns in cases where multiple productive enhancer trap integrations occurred in the same fish. Individual enhancer trap lines were given allele designations zc1000 – zc1094. At the end of each allele, an A, B, C, or D was appended to indicate that a specific line was the 1st, 2nd, 3rd, or 4th line obtained from a given F0 founder after outcrossing. Thus, both zc1062A and zc1062B were derived from the same F0 founder, but were isolated through outcrossing as separate insertions/enhancer traps. Outcrossed Gal4 lines were propagated with crosses to Tg(5×UAS:eGFP)zf82, (Asakawa et al., 2008)) and Tg(UAS-E1b:NTR-mCherry) c264 (Davison et al., 2007).
Imaging and analysis
Stable enhancer trap lines were analyzed first on an Olympus SZX16 fluorescent dissecting microscope to validate successful outcrossing/isolation of single enhancer traps. Live embryos were imaged at 4×–20× magnification on an Olympus BX51WI fluorescent compound microscope at 1dpf, 2dpf, and 5dpf to document expression patterns over a developmental timecourse. Embryos were embedded in FEP tubes (Cole-Parmer #EW-06406-60) and sealed with water to optically clear the tube. Confocal imaging was performed using an Olympus FV1000 confocal microscope with a 30× silicon oil objective (UPLSAPO30×S NA1.05). Antibody stained samples were embedded in 100% glycerol. 3D reconstructions were performed with FluoRender using 3-degree increments throughout a full 360-degree rotation. Serial images were imported into ImageJ and exported as non-compressed AVI movies. Movies were compressed by using TMPGEnc 4.0 Xpress (Pegasys).
Immunostaining
For confocal imaging, 5–6dpf embryos were fixed for 2 hours at room temperature (4%PFA), washed with PBS/Triton (0.5% Triton), permeabilized with 0.5U Dispase (Gibco #17105-041) for 45 minutes, then blocked with goat serum for 30 minutes. Next, embryos were incubated in primary antibodies: f59 (Developmental Studies Hybridoma Bank), 1:10; DsRed Polyclonal Antibody (#632496 - Clontech, 1:300; alpha-actin – Sigma #A2547), 1:100; in block for 3 days at 4C. Embryos were washed in PBS/Triton for 5 hours, then incubated in secondary antibodies and counterstain: Cy3-AffiniPure Goat Anti-Rabbit IgG (H+L) (111-165-003, Jackson) 1:300; Alexa Fluor® 488 Goat Anti-Mouse IgG (H+L) (A-1100, Life Technologies). 1:300; and TO-PRO®-3 Iodide (642/661) (T3605, Life Technologies). 1:300, for 3 days at 4C. Following staining, embryos were washed for 5 hours in PBS/Triton, cleared in 50:50 glycerol/PBS for 2 hours at 4C, and cleared in 80:20 glycerol/water overnight. At this point, embryos were stored at −20C until imaging.
Cell ablation
For cell ablations, 4dpf fish were placed in media containing 1mM Metronidazole (1M stock in DMSO; Sigma) for 24 hours. Control fish were placed in media containing an equivalent concentration of DMSO. After fixation, dissected whole brains were imaged for native mCherry expression using confocal microscopy.
Insertion mapping
Mapping by inverse PCR was carried out by phenol:chloroform extraction of genomic DNA from F3 generation embryos. Initial PCR used primers Tol2INV5A (5’-CTTGAGTATTAAGGAAGTAAAAGT-3’) and Tol2INV5B (5’-TTACTCAAGTAAGATTCTAGCC-3’) and was followed by a nested PCR with primers Tol2INV6A (5’-AAGCAAGAAAGAAAACTAGAG-3’) and Tol2INV6B (5’-TGAGTAAAATTTTCCCTAAGT-3’). PCR products were sequenced and BLASTN against Zebrafish Zv8 build was performed.
Supplementary Material
Supplemental Movie 1 – 3D reconstruction of confocal stack for line zc1022B. Reporter expression is visible in the ventrolateral hypothalamus. MPEG format. Movie starts with a dorsal view.
Supplemental Movie 2 – 3D reconstruction of confocal stack for line zc1016C. Reporter expression is visible in the telencephalon, ventral tectum, and hypothalamus. MPEG format. Movie starts with a dorsal view.
Acknowledgements
We thank Maurine Hobbs and Sharon Johnson of the University of Utah’s Centralized Zebrafish Animal Resource center. This work was funded by NIH RO1MH092256 and NIH R01GM098151.
References
- Akitake CM, Macurak M, Halpern ME, Goll MG. Transgenerational analysis of transcriptional silencing in zebrafish. Dev Biol. 2011;352:191–201. doi: 10.1016/j.ydbio.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa K, Abe G, Kawakami K. Cellular dissection of the spinal cord motor column by BAC transgenesis and gene trapping in zebrafish. Front Neural Circuits. 2013;7:100. doi: 10.3389/fncir.2013.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa K, Suster ML, Mizusawa K, Nagayoshi S, Kotani T, Urasaki A, Kishimoto Y, Hibi M, Kawakami K. Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc Natl Acad Sci U S A. 2008;105:1255–1260. doi: 10.1073/pnas.0704963105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balciunas D, Wangensteen KJ, Wilber A, Bell J, Geurts A, Sivasubbu S, Wang X, Hackett PB, Largaespada DA, McIvor RS, Ekker SC. Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLoS Genet. 2006;2:e169. doi: 10.1371/journal.pgen.0020169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balciuniene J, Balciunas D. Gene trapping using gal4 in zebrafish. J Vis Exp. 2013:e50113. doi: 10.3791/50113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Campbell DS, Stringham SA, Timm A, Xiao T, Law MY, Baier H, Nonet ML, Chien CB. Slit1a inhibits retinal ganglion cell arborization and synaptogenesis via Robo2-dependent and -independent pathways. Neuron. 2007;55:231–245. doi: 10.1016/j.neuron.2007.06.034. [DOI] [PubMed] [Google Scholar]
- Curado S, Anderson RM, Jungblut B, Mumm J, Schroeter E, Stainier DY. Conditional targeted cell ablation in zebrafish: a new tool for regeneration studies. Dev Dyn. 2007;236:1025–1035. doi: 10.1002/dvdy.21100. [DOI] [PubMed] [Google Scholar]
- Davison JM, Akitake CM, Goll MG, Rhee JM, Gosse N, Baier H, Halpern ME, Leach SD, Parsons MJ. Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev Biol. 2007;304:811–824. doi: 10.1016/j.ydbio.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglass AD, Kraves S, Deisseroth K, Schier AF, Engert F. Escape behavior elicited by single, channelrhodopsin-2-evoked spikes in zebrafish somatosensory neurons. Curr Biol. 2008;18:1133–1137. doi: 10.1016/j.cub.2008.06.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhoffer GT, Loftus PD, Yoshigi M, Otsuna H, Chien CB, Morcos PA, Rosenblatt J. Crowding induces live cell extrusion to maintain homeostatic cell numbers in epithelia. Nature. 2012;484:546–549. doi: 10.1038/nature10999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern ME, Rhee J, Goll MG, Akitake CM, Parsons M, Leach SD. Gal4/UAS transgenic tools and their application to zebrafish. Zebrafish. 2008;5:97–110. doi: 10.1089/zeb.2008.0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashijima S, Masino MA, Mandel G, Fetcho JR. Imaging neuronal activity during zebrafish behavior with a genetically encoded calcium indicator. J Neurophysiol. 2003;90:3986–3997. doi: 10.1152/jn.00576.2003. [DOI] [PubMed] [Google Scholar]
- Koster RW, Fraser SE. Tracing transgene expression in living zebrafish embryos. Dev Biol. 2001;233:329–346. doi: 10.1006/dbio.2001.0242. [DOI] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Li J, Mack JA, Souren M, Yaksi E, Higashijima S, Mione M, Fetcho JR, Friedrich RW. Early development of functional spatial maps in the zebrafish olfactory bulb. J Neurosci. 2005;25:5784–5795. doi: 10.1523/JNEUROSCI.0922-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumm JS, Williams PR, Godinho L, Koerber A, Pittman AJ, Roeser T, Chien CB, Baier H, Wong RO. In vivo imaging reveals dendritic targeting of laminated afferents by zebrafish retinal ganglion cells. Neuron. 2006;52:609–621. doi: 10.1016/j.neuron.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura E, Okuda Y, Kondoh H, Kamachi Y. Adaptation of GAL4 activators for GAL4 enhancer trapping in zebrafish. Dev Dyn. 2009;238:641–655. doi: 10.1002/dvdy.21863. [DOI] [PubMed] [Google Scholar]
- Otsuna H, Ito K. Systematic analysis of the visual projection neurons of Drosophila melanogaster. I. Lobula-specific pathways. J Comp Neurol. 2006;497:928–958. doi: 10.1002/cne.21015. [DOI] [PubMed] [Google Scholar]
- Pisharath H, Rhee JM, Swanson MA, Leach SD, Parsons MJ. Targeted ablation of beta cells in the embryonic zebrafish pancreas using E. coli nitroreductase. Mech Dev. 2007;124:218–229. doi: 10.1016/j.mod.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou C, Kimura Y, Hirata H, Suster ML, Kawakami K, Higashijima S. Transgenic tools to characterize neuronal properties of discrete populations of zebrafish neurons. Development. 2013;140:3927–3931. doi: 10.1242/dev.099531. [DOI] [PubMed] [Google Scholar]
- Scheer N, Campos-Ortega JA. Use of the Gal4-UAS technique for targeted gene expression in the zebrafish. Mech Dev. 1999;80:153–158. doi: 10.1016/s0925-4773(98)00209-3. [DOI] [PubMed] [Google Scholar]
- Scheer N, Groth A, Hans S, Campos-Ortega JA. An instructive function for Notch in promoting gliogenesis in the zebrafish retina. Development. 2001;128:1099–1107. doi: 10.1242/dev.128.7.1099. [DOI] [PubMed] [Google Scholar]
- Scott EK. The Gal4/UAS toolbox in zebrafish: new approaches for defining behavioral circuits. J Neurochem. 2009;110:441–456. doi: 10.1111/j.1471-4159.2009.06161.x. [DOI] [PubMed] [Google Scholar]
- Scott EK, Mason L, Arrenberg AB, Ziv L, Gosse NJ, Xiao T, Chi NC, Asakawa K, Kawakami K, Baier H. Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nat Methods. 2007;4:323–326. doi: 10.1038/nmeth1033. [DOI] [PubMed] [Google Scholar]
- Szobota S, Gorostiza P, Del Bene F, Wyart C, Fortin DL, Kolstad KD, Tulyathan O, Volgraf M, Numano R, Aaron HL, Scott EK, Kramer RH, Flannery J, Baier H, Trauner D, Isacoff EY. Remote control of neuronal activity with a light-gated glutamate receptor. Neuron. 2007;54:535–545. doi: 10.1016/j.neuron.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Wan Y, Otsuna H, Chien CB, Hansen C. An interactive visualization tool for multi-channel confocal microscopy data in neurobiology research. IEEE Trans Vis Comput Graph. 2009;15:1489–1496. doi: 10.1109/TVCG.2009.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kopinke D, Lin J, McPherson AD, Duncan RN, Otsuna H, Moro E, Hoshijima K, Grunwald DJ, Argenton F, Chien CB, Murtaugh LC, Dorsky RI. Wnt signaling regulates postembryonic hypothalamic progenitor differentiation. Dev Cell. 2012;23:624–636. doi: 10.1016/j.devcel.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Movie 1 – 3D reconstruction of confocal stack for line zc1022B. Reporter expression is visible in the ventrolateral hypothalamus. MPEG format. Movie starts with a dorsal view.
Supplemental Movie 2 – 3D reconstruction of confocal stack for line zc1016C. Reporter expression is visible in the telencephalon, ventral tectum, and hypothalamus. MPEG format. Movie starts with a dorsal view.