Summary
The membrane-spanning α-helices of single-pass receptors play crucial roles in stabilizing oligomeric structures and transducing biochemical signals across the membrane. Probing intermolecular transmembrane interactions in single-pass receptors presents unique challenges, reflected in a gross underrepresentation of their membrane-embedded domains in structural databases. Here we present two high-resolution structures of transmembrane assemblies from a eukaryotic single-pass protein crystallized in a lipidic membrane environment. Trimeric and tetrameric structures of the immunoreceptor signaling module DAP12, determined to 1.77 Å and 2.14 Å resolution, respectively, are organized by the same polar surfaces that govern intramembrane assembly with client receptors. We demonstrate that both trimeric and tetrameric products form in cells and that formation of products larger than dimers is competitive with receptor association in the ER. The polar transmembrane sequences therefore act as primary determinants of oligomerization specificity through interplay between charge-shielding and sequestration of polar surfaces within helix interfaces.
Introduction
The α-helical transmembrane (TM) domains of eukaryotic single-pass membrane proteins can engage in specific interactions that are critical to the structure and activity of receptors governing cell adhesion and signaling pathways. Notable examples include the control of dimer formation in the erythrocyte cell-surface protein glycophorin A (Lemmon et al., 1992; MacKenzie et al., 1997), stabilization of the low-affinity conformation of αβ integrins (Lau et al., 2009; Luo et al., 2004; Zhu et al., 2009) and transfer of conformational changes through the cell membrane in cytokine and growth hormone receptors (Arkhipov et al., 2013; Bocharov et al., 2008; Brooks et al., 2014; Lu et al., 2006). In a large group of modular activating immune receptors, ligand-binding subunits and signal-transducing subunits are assembled into hetero-oligomeric complexes via polar interactions among their TM domains (Call and Wucherpfennig, 2007). The lymphoid/myeloid receptor signaling module DAP12 (Lanier et al., 1998b) has no structured ectodomain and forms both homo- and hetero-oligomeric interfaces through its TM domain (Call et al., 2010; Feng et al., 2006; Feng et al., 2005; Lanier et al., 1998a) during assembly in the endoplasmic reticulum (ER). The homodimeric DAP12 interface was first directly observed in NMR studies of DAP12 TM peptides reconstituted in detergent micelles (Call et al., 2010), revealing how its composite surface accommodates a single receptor TM helix containing a central lysine residue aligned with aspartic acid/threonine motifs that form the receptor-assembly site on DAP12. This has become an important model system for studies of immunoreceptor assembly (Cheng and Im, 2012; Sharma and Juffer, 2013; Wei et al., 2014; Wei et al., 2013) because similar arrangements of polar residues are believed to form the core TM structures of more complex receptor systems such as the hexameric NKG2D-DAP10 receptor implicated in anti-tumor immune responses (Garrity et al., 2005; Raulet et al., 2013) and the octameric T cell antigen receptor (TCR) that occupies a central position in adaptive immunity (Call et al., 2002). Importantly, no detailed structures of these intact complexes have been experimentally determined, and the mechanisms of signal transmission through the cell membrane remain poorly understood for the entire class of multi-subunit activating immune receptors.
Assembly of DAP12-receptor TM complexes in the endoplasmic reticulum (ER) is thought to involve at least three steps: (1) co-translational translocation of all subunits into the ER, (2) formation of DAP12 homodimers and (3) assembly with a receptor TM domain. These steps likely occur cooperatively (Feng et al., 2005), but how the charge state of ionizable TM residues and their shielding from the apolar lipid bilayer interior influence the selective formation of homo- and hetero-oligomeric TM interfaces is unknown. To gain further insight into the structural features governing these interactions in a lipid bilayer environment, we crystallized DAP12-TM peptides in a lipidic cubic phase (LCP) medium. In the monoolein lipid bilayer, DAP12-TM crystallized in trimeric and tetrameric arrangements around a polar core composed of the aspartic acid/threonine motifs, which are further stabilized by coordinated cations obtained from the precipitant solutions. These higher-order oligomeric forms had not been identified in previous studies, yet our biochemical analysis reveals that together they represent a substantial fraction of the full-length DAP12 protein generated during synthesis in the ER, establishing a strong parallel between TM helix oligomerization in LCP conditions and in native cellular membranes. We further demonstrate that the formation of DAP12 homotrimers in particular is competitive with formation of the DAP12-receptor heterotrimer in the ER and may play a role as an intermediate assembly product. Thus the polar TM sequences in DAP12 not only direct its association with receptors within the membrane, but rather more broadly govern the distribution of homo- and hetero-oligomeric protein complexes whose interplay determines the final outcome of DAP12-receptor co-assembly in the ER.
Results
Human DAP12 comprises a 12-amino-acid extracellular region containing two cysteines with only one intervening residue, a 24-amino-acid TM domain (Figure 1A) and a 49-amino-acid cytoplasmic tail (sequence not shown). Receptor-associated DAP12 is recovered from the cell surface as a disulfide-linked homodimer (Lanier et al., 1998b), and both the homodimeric DAP12 interface and the heterotrimeric interface DAP12 forms with its associated receptors have been attributed to TM domain interactions. We produced a 33-amino-acid DAP12-TM peptide beginning from the second extracellular cysteine (Figure 1A) in its disulfide-linked dimeric form as previously described (Call et al., 2010; Sharma et al., 2013) and reconstituted this into monoolein by co-dissolution in hexafluoroisopropanol (HFIP) followed by removal of solvent under vacuum and mixing with water (Hofer et al., 2010) (see methods). Small-angle X-ray scattering (SAXS) analysis confirmed that this procedure generated a distribution of gyroid cubic, diamond cubic and fluid lamellar phases, sometimes coexisting, across a variety of precipitant conditions including those that produced crystals (Figure S1). Two crystals forms were identified in this screen: small oval discs were visible after 5–7 days in 10% cholesterol, 12.4 % (w/v) PEG 3350 and 0.149 M potassium thiocyanate; and star-like clusters of crystals were visible after 1–3 days in 0.1 M bis-tris propane chloride (pH 6.07), 19.7 % (w/v) PEG 3350 and 0.269 M calcium chloride.
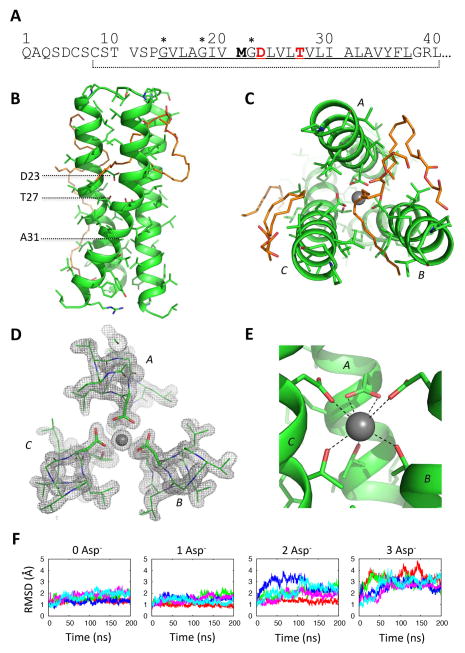
Figure 1. Crystal structure of a DAP12-TM trimer with coordinated potassium at 1.77 Å resolution.
(A) Sequence of human DAP12 from the amino-terminus of the mature polypeptide to the intracellular juxtamembrane region. The predicted TM domain is underlined; the 33-amino-acid peptide construct used for crystallization is indicated by the dotted line. The residues responsible for assembly with receptors are in red, and the bold methionine was changed to valine to avoid internal cleavage by cyanogen bromide. A potential glycine zipper (GxxxGxxxG) motif is marked by (*). (B) Side and (C) top views of the DAP12-TM trimer structure. Structured monoolein molecules are shown in orange stick representation. (D) Electron density depicted by a 2mFo-DFc map (sigma level 1.5) around the aspartic acids and threonines. (E) Coordination of a potassium ion by aspartic acid 23 and threonine 27. Distances from the centre of the potassium ion to oxygen atoms are: (aspartic acids, left to right) 2.7 Å, 2.8 Å, 2.8 Å, 3.1 Å and (threonines, left to right) 2.8 Å, 2.9 Å, 2.8 Å. (F) 200-ns fully atomistic molecular dynamics simulations of trimer structures in a POPC bilayer with 150 mM KCl in the bulk aqueous phase. The system was simulated with coordinated potassium and 0–3 ionized aspartic acids (Asp−). Each plot shows a time-series of the RMSD deviation from the starting structure for five independent trajectories (represented in different colors).
Structure of a DAP12-TM trimer
The first of these two crystal forms diffracted to 1.77 Å and contains three parallel α helices in the asymmetric unit, arranged in a right-handed trimeric coiled-coil (Figure 1; data collection and refinement statistics, Table S1). Four well-ordered monoolein molecules are also apparent in the electron density. The helices are tightly packed in the lower half of the structure, facilitated by the small side-chain at alanine 31 (Figure 1B). In the core of the trimeric interface, the aspartic acid and threonine side chains are oriented towards the center, where their polar groups are well shielded from the otherwise hydrophobic environment (Figure 1C–D). A residual peak in the density at this site was modeled as a potassium ion based on the best fit to the density and the presence of potassium thiocyanate in the precipitant. The cation is coordinated by seven side-chain oxygen atoms that are all within the range of 2.7 to 3.1 Å from its center (see Figure 1D–E). This network involves the aspartic acids and threonines from all three DAP12 chains in the trimer, but their contributions are asymmetrical (Figure 1E): the aspartic acid from chain A makes a bidentate contribution while those of chains B and C make monodentate contributions. Based on this observation, we postulated that aspartic acid A is deprotonated while those of chains B and C remain protonated, resulting in a net peptide-derived charge of −1e at this site.
To validate this model, we analyzed the stability of the trimeric structure in different charge states with coordinated potassium using fully atomistic molecular dynamics simulations in a palmitoyl-oleoyl phosphatidylcholine (POPC) bilayer (Figure 1F). Consistent with prediction, the structure with one ionized aspartic acid (1 Asp−1) was the most stable, experiencing only small fluctuations in RMSD over the 200 ns simulation. Structures containing two (2 Asp−1) or three (3 Asp−1) ionized aspartic acids were less stable, experiencing large increases in RMSD over the 200 ns simulation. Interestingly, the structure with no ionized aspartic acids (0 Asp−1) also remained stable despite the rapid loss of potassium into bulk solvent in three of five replica trajectories (Figure S2). This suggested that an intramembrane hydrogen-bonding network (Zhou et al., 2000) could provide a sufficiently stable polar core for this structure even in the absence of cation coordination (see below).
Structure of a DAP12-TM tetramer
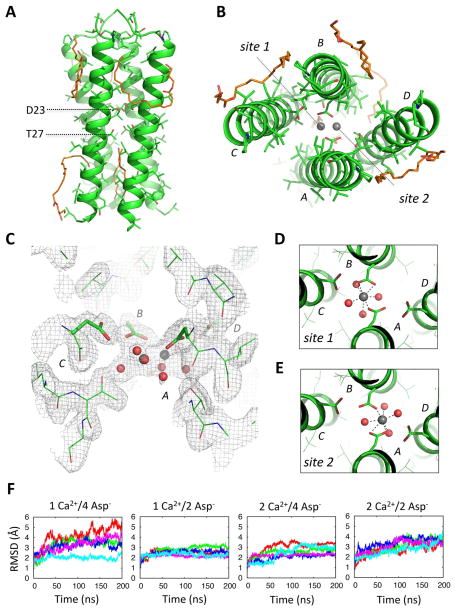
A second crystal form diffracted to 2.14 Å, containing four parallel α helices in the asymmetric unit (Table S1; Figure 2). As in the trimer, the aspartic acid/threonine motifs from all chains are sequestered in the interior of the complex (Figure 2A–B). Additional electron density in the polar core of this structure (Figure 2C) was interpreted as a single Ca2+ ion (from CaCl2 in the precipitant) present in two different configurations (Figure 2D–E), each with approximately 50% occupancy. MD simulations supported this interpretation (Figure 2F), showing that one Ca2+ ion coordinated through two ionized aspartic acids (1 Ca2+/2 Asp−1) provided the most stable structure in the lipid bilayer. In this arrangement, two of the aspartic acids (from chains A and B) are directly involved in cation coordination, while aspartic acids C and D and all four threonine residues provide further stabilization indirectly, through establishment of a water network recruiting additional coordinating ligands (Figure 2C–E). Consistent with the requirement for additional ligands, several water molecules were also recruited to this site in the MD simulations (not shown).
Figure 2. Crystal structure of a DAP12-TM tetramer with coordinated calcium at 2.14 Å resolution.
Side (A) and top (B) views of the tetrameric DAP12-TM crystal structure. Structured monoolein molecules are shown in orange stick representation. Two possible calcium coordination sites are indicated in (B). (C) Electron density depicted by a 2mFo-DFc map (sigma level 1.0) around the aspartic acid and threonines. (D–E) The extra density at this site was modelled as two overlapping Ca2+ coordination sites with pentagonal bipyramidal geometry, each with 50% occupancy, that included a network of additional water molecules. (F) 200-ns fully atomistic molecular dynamics simulations of the tetrameric structure with one or two Calcium ions (1Ca2+, 2Ca2+) and four or two ionized aspartic acid residues (4Asp−, 2Asp−) were run in an explicit bilayer composed of POPC molecules with 50 mM CaCl2 in the bulk solvent. Each plot shows the time-series of the RMSD deviation from the starting structure for five independent trajectories.
Molecular organization in LCP crystals
Because few precedents exist for crystallographic analysis of small helical peptides in lipid bilayers, we carefully examined the organization of DAP12 helices in the three-dimensional crystalline arrays. In both trimer and tetramer crystals, the lattices feature tightly packed DAP12 chains in parallel and antiparallel orientations with solvent contents of 27% and 35%, respectively. In each crystal form there is only one arrangement of DAP12 chains that shields the polar aspartic acids, and this guided our choice of asymmetric unit (Figure S3). This arrangement also satisfies the physiological criterion that oligomers must contain parallel DAP12 chains, consistent with enforcement of directionality during insertion into the ER membrane.
Analysis of the interfaces between individual DAP12 chains using PDBePISA (Krissinel and Henrick, 2007) (see Tables S2 and S3) indicates that the trimeric interface is dominated by coordination of the potassium ion, and the lattice is extended primarily by two contacts to one parallel and one anti-parallel neighboring asymmetric unit (red arrows, Figure S3A). Within the tetramer, the largest buried surface areas are between chains A and D, and between B and C within one asymmetric unit (see Figure 2B), and we interpret this arrangement as a dimer of dimers held together primarily by coordination of calcium. The only significant inter-asymmetric-unit interface is between anti-parallel chains C and D from neighboring asymmetric units (red arrows, Figure S3B). This stabilizes the crystal along the y-axis, but contacts along the other dimension in the plane of the lipid bilayer (x-axis of the crystal) are mediated entirely by structured lipids.
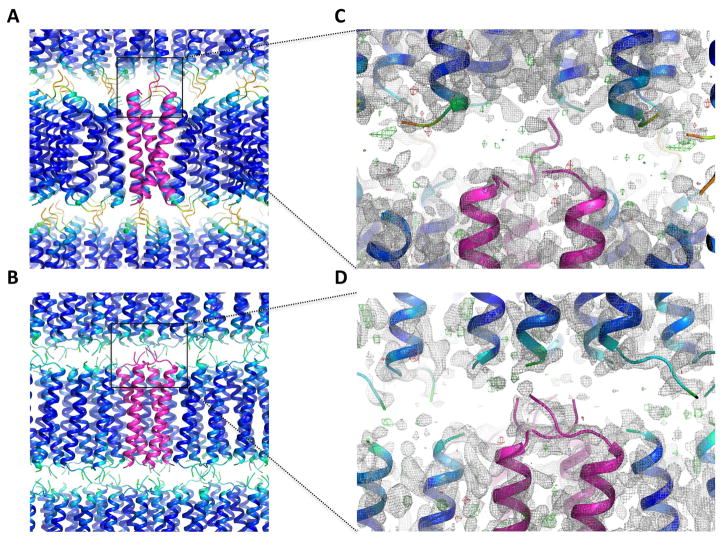
Density between layers is weak in both crystals (Figure 3), indicating high thermal motion in the N-terminus. While this is consistent with previously reported NMR structures (Call et al., 2010), it raises the question of what mediates alignment of the lamellae to enforce crystallographic symmetry. In the trimer structure, a network of hydrogen bonds involving the chain A terminal amine and three atoms in the layer above (see Figure S4) provides the only identifiable inter-lamellar contacts. This is the only chain with assignable density all the way to the N-terminus, and further density to unambiguously assign disulfide connectivity was not apparent. As the peptide used for crystallization was pure disulfide-linked dimer (verified by HPLC, mass spectrometry and SDS-PAGE; not shown), this leaves one chain in the asymmetric unit that must disulfide bond with another chain from a neighboring asymmetric unit. If the two unresolved N-terminal segments are in a fully extended conformation (covering approximately 24 Å distance), there are a total of 11 neighboring chains to which connections could be made (four in the same lamellar plane and seven in the plane above) and these may exist in a mixture of conformations. In the tetramer crystal, the density in the aqueous space is too weak to identify any inter-lamellar contacts and disulfide connectivity is not traceable.
Figure 3. Packing of trimeric and tetrameric DAP12-TM complexes in LCP crystals.
For the trimer (A) and tetramer (B), one asymmetric unit is shown in magenta, and symmetry mates are colored by B-factor. In (C) and (D) the zoom regions are shown as 2mFo-DFc maps (sigma level 1.0; gray) and mFo-DFc maps (sigma level 3.0, green; sigma level −3.0, red) of the poorly ordered space between layers of packed helices.
Distribution of DAP12 oligomers in cells and isolated ER
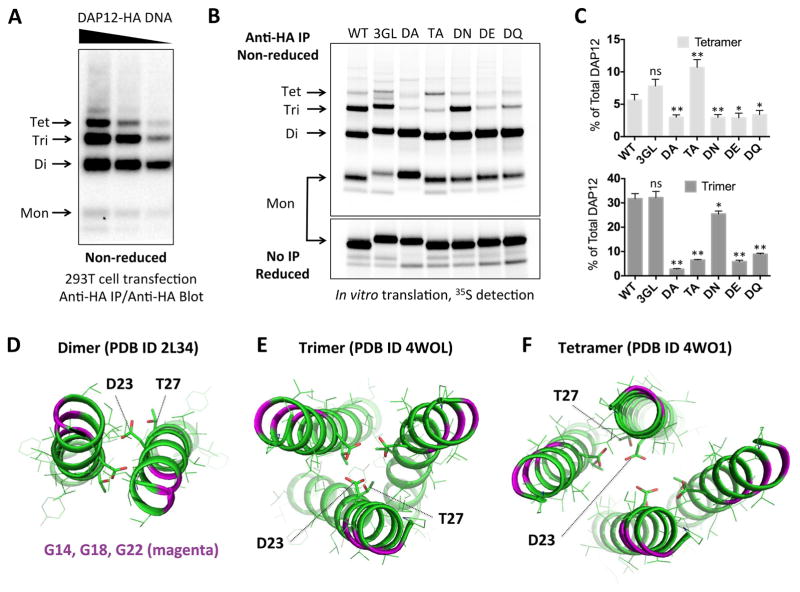
The full-length extracellular sequence of human DAP12 extends seven residues beyond the construct used for crystallization and contains an additional cysteine (see Figure 1A). Because previous biochemical studies used a mutant containing a serine substitution at the first cysteine position (Call et al., 2010; Feng et al., 2006; Feng et al., 2005), the distribution of products formed by the native sequence in total cell lysates had never been carefully examined. In 293T cells transfected with full-length human DAP12 containing both native cysteines (Figure 4A), we observed disulfide-linked dimers, trimers and tetramers. To evaluate the relationship between these products and the LCP crystal structures, we examined the effects of mutations on the distribution of DAP12 products formed during biosynthesis in isolated ER microsomes (Figure 4B–C). In both trimeric and tetrameric structures, the exterior surfaces of the DAP12-TM helices feature a triplet of glycines spaced with helical periodicity that form a potential “glycine zipper” motif (Kim et al., 2005) (marked with * in Figure 1A; colored magenta in Figure 4D–F). We reasoned that simultaneous substitution of all three glycines with bulky hydrophobic residues would be non-disruptive if the ER-derived oligomers were assembled around a polar core as in the crystal structures. Accordingly, neither triple-leucine (3GL, Figure 4B) nor triple-phenylalanine (not shown) replacements significantly altered the distribution of products (see quantitation in Figure 4C).
Figure 4. Correspondence of cellular DAP12 products with experimentally determined structures.
(A) HA-tagged human DAP12 was transiently transfected into 293T cells using serial dilutions of plasmid DNA. Cells were lysed 48 hours later in RIPA buffer and total DAP12 was immunoprecipitated using mouse anti-HA mAb. Recovered products were separated by non-reducing SDS-PAGE and immunoblotted using biotinylated rat anti-HA Fab with streptavidin-peroxidase. (B) HA-tagged human DAP12 was in vitro-translated in the presence of ER microsomes and 35S-methionine/cysteine and allowed to assemble for one hour. The ER membrane fraction was extracted in 0.5% digitonin and immunoprecipitated with anti-HA mAb. Recovered products were separated by non-reducing SDS-PAGE and visualized using a phosphorimager. Sequences were wild-type (WT), triple glycine-to-leucine (3GL), threonine-to-alanine (TA) and aspartic acid-to-alanine (DA), asparagine (DN), glutamic acid (DE) or glutamine (DQ). Half of the samples were run reduced with no IP to show equal total DAP12 signal (lower panel). (C) Quantitation of tetramer (top) and trimer (bottom) products expressed as % of total DAP12 signal in the non-reducing lanes for each mutant. Bar graphs show the mean ± standard deviation for three independent experiments. Statistical significance in unpaired t-test comparing each mutant to WT is indicated by ns (not significant), * (p<0.05) or ** (p< 0.01). (D–F) Structures of DAP12 complexes solved by solution NMR (dimer, panel D) and crystallography (trimer, panel E; tetramer, panel F). Glycines are shown in magenta ribbons, aspartic acid and threonines are shown in stick representation.
Conversely, substitutions of the polar residues had marked effects: while all mutants efficiently formed dimers, several exhibited defects in trimer and/or tetramer formation. Alanine substitution of the aspartic acid severely impaired the formation of both trimers and tetramers (10-fold and 2-fold reduction compared to WT, respectively; Figure 4B–C). Interestingly, alanine substitution of the TM threonine impaired trimer formation (5-fold decrease) but shifted the distribution towards tetramer (2-fold increase). This is consistent with the critical role of the threonine hydroxyl oxygens in cation coordination within the trimer structure (see Figure 1E) but not in the tetramer (see Figure 2C). While we cannot determine whether the multimers formed in the ER contain coordinated cations, a hydrogen-bonding network among non-ionized polar groups (Zhou et al., 2000) could provide sufficient stabilization in the absence of cation coordination, as indicated by our MD simulation results (Figure 1F; Figure S2) and by the partial tolerance of asparagine substitution (DN mutant) in the ER assembly assay (Figure 4B–C). Glutamine, asparagine, glutamic acid and aspartic acid have all been reported to favor trimer formation in model TM peptides (Gratkowski et al., 2001). However, we found that glutamic acid (DE) and glutamine (DQ) substitutions strongly disfavored DAP12 trimers and tetramers (Figure 4B–C), presumably due to a poor fit of the longer side-chains within the polar core.
DAP12 trimer formation is competitive with receptor assembly
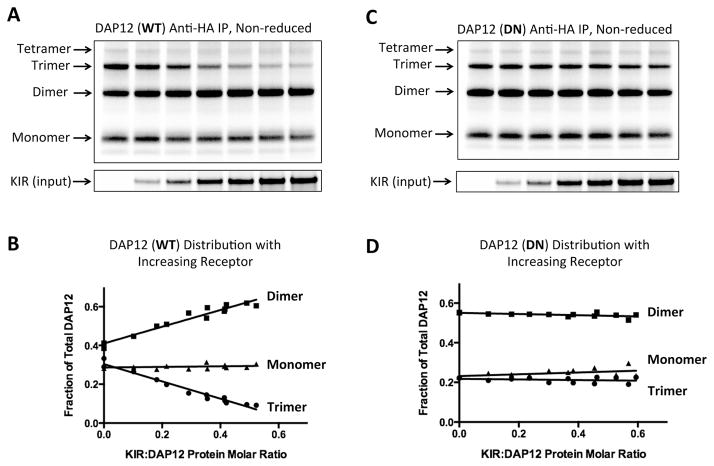
To evaluate the relationship between formation of higher-order oligomers and assembly with client receptors in the ER, we examined the distribution of DAP12 products when varying amounts of receptor were provided (Figure 5). Previous studies had demonstrated that the presence of a central TM lysine is the key feature of DAP12-associated receptors (Feng et al., 2005) and that assembly and surface expression proceed unimpeded even when the lysine is placed in a poly-valine or poly-leucine TM sequence (Feng et al., 2006). We therefore used the natural killer (NK) cell activating receptor KIR2DS2 with a poly-leucine TM domain (KIRpLeu) containing a single, centrally-located lysine residue to represent a “generic” receptor in our assembly assay. As shown in Figure 5A and B, provision of increasing amounts of this receptor resulted in recovery of less DAP12 trimer and a concomitant increase in the dimeric form. This effect is directly attributable to DAP12-receptor assembly because the distribution of the aspartic acid-to-asparagine mutant (DN), which forms trimers (Figure 4B, C) but cannot assemble with receptor (Feng et al., 2005), was unperturbed by the presence of KIR (Figure 5C, D). Thus the balance between DAP12 homo-oligomerization and assembly with receptor is governed both by the chemical nature of the polar side-chains in DAP12 and the relative availability of “preferred” complementary ligands.
Figure 5. DAP12 oligomer formation and receptor assembly in the ER.
DAP12-WT (A and B) or DN mutant (C and D) mRNA (150 ng per reaction) was distributed equally among seven assembly reactions containing increasing amounts of KIR-pLeu mRNA (0–900 ng). After a 2-hour assembly period, 25% of the ER fraction was run on a reducing SDS-PAGE gel to determine input receptor amounts (lower panel). The remaining ER fraction was boiled in 0.5% SDS to dissociate non-covalent DAP12-KIR complexes and diluted 20-fold in RIPA solution to allow antibody capture. DAP12 products were isolated by anti-HA IP and separated by non-reducing SDS-PAGE. (B and D) Monomeric, dimeric and trimeric DAP12 products were quantitated by densitometry and the proportion of each was plotted as a function of KIR:DAP12 protein molar ratio (measured from the input control gel and adjusted for number of methionine and cysteine labeling positions in each protein). The data for each plot were taken from two independent experiments, including the ones shown in (A) and (C), covering a range of KIR:DAP12 ratios.
Discussion
As increasingly sophisticated biochemical experiments have revealed the importance of TM domains in assembling oligomeric complexes and transducing signals through the membrane, understanding the structural features governing these functions has come to the fore as a critical problem in receptor biology. Very few structures of TM complexes from eukaryotic single-pass receptors are currently available (see for example (Bocharov et al., 2008; Call et al., 2006; Lau et al., 2009; MacKenzie et al., 1997)), and those that are available have been determined by solution NMR in detergent micelles, lipid-detergent mixtures or small isotropic bicelles (reviewed in (Call and Chou, 2010)). In the particular case of TM domains containing strongly polar groups, it is unclear how well these conditions represent a continuous lipid bilayer environment, where hydration and charge distribution can be quite different (Cross et al., 2013). Our new structures demonstrate that for DAP12, crystallization from a lipidic membrane environment favors different oligomeric forms (trimers and tetramers) than reconstitution in detergent micelles (dimers), and this difference correlates closely with the energetic favorability of charge-shielding at the site of the aspartic acid/threonine motifs that mediate assembly with receptors. The presence of intramembrane ion coordination sites in structures of proteins that do not function explicitly as ion channels or transporters is unusual, and the association of potassium and calcium with two different DAP12 oligomers demonstrates high sensitivity of the DAP12 TM sequence to the charge state of the available ligands. While it is difficult to experimentally determine whether coordinated ions are present in the ER-derived oligomers, it is tempting to speculate that the type of cationic ligands available plays a role in determining which structures are formed in cells, and we note that potassium is abundant at the cytosolic face of the ER membrane (>100 mM) while free calcium is found at near millimolar concentration on the luminal side. The capture of these cations by the monoolein bilayer-embedded DAP12-TM complexes suggests that a similar phenomenon could occur during incorporation into the ER membrane.
How do we reconcile the trimeric and tetrameric structures with the disulfide-linked dimer used for crystallization and the distribution of products observed in the ER? The appearance of a homotrimeric form was initially surprising, and while the disulfide connectivity is not traceable in the density maps, this arrangement clearly requires that trimers be formed from arrays of dimers within the crystal. The covalent trimeric structure formed by full-length DAP12 (with two extracellular cysteines) in the ER contains multiple inter-chain disulfide bonds, and different configurations of this unstructured region may interconvert in the presence of ER-resident redox chaperones. The tetramer is more easily conceptualized as a dimer of dimers, and this product may be significantly underrepresented in our biochemical assay because non-covalent tetramers would appear as smaller forms by SDS-PAGE. A similar phenomenon is observed in the proton-conducting influenza M2 protein, which contains two cysteines in its short luminal domain that are positioned identically to those found in DAP12. M2 forms a stable and constitutive tetramer, yet less than half of the native protein runs as a disulfide-linked tetramer by non-reducing SDS-PAGE (Holsinger and Lamb, 1991), indicating that the pattern of disulfide connectivity is flexible and a fully covalent tetrameric configuration is neither required for function nor necessarily favored.
The trimeric and tetrameric complexes crystallized from LCP readily formed in the ER when very low levels of partner receptors were available (Figure 4). The competition between formation of these products and assembly with receptor (Figure 5) indicates that these structures represent either alternative oligomerization pathways or intermediates in the receptor assembly process. While these two possibilities cannot be distinguished from the data presented here, a possible role as assembly intermediates may be related to the need to adequately shield strongly polar groups during exit from the Sec61 translocon and equilibration into the ER membrane (Heinrich et al., 2000; Hessa et al., 2007). These intermediate complexes could then sample other translocons for partner receptors that also cannot enter the lipid phase without charge-pairing their basic TM residues. Thus while stabilization of the DAP12 dimer has been proposed to underpin the apparent cooperativity of receptor complex assembly (Feng et al., 2005) (i.e., more disulfide-linked DAP12 dimer is recovered when receptor is present than when it is absent), this could also result from reassortment of larger intermediate complexes into the final dimeric form upon receptor association.
The propensity to form TM structures that fully sequester polar groups within helix interfaces also sheds light on an unexplained feature of the hetero-trimeric DAP12-receptor TM structure determined in detergent micelles by solution NMR (Call et al., 2010). In this structure, only one aspartic acid-threonine pair contacts the ε-amino group of the receptor TM lysine. The second polar motif faces out of the interface where it remains largely exposed to the aliphatic micelle interior (see Figure S5), an arrangement that is unlikely to be favorable in the membrane. Several groups have recently suggested, based on molecular dynamics simulations, that in a bilayer environment a more stable structure may be reached by rotation of the second DAP12 helix such that all polar groups are in the heterotrimer interface (Cheng and Im, 2012; Sharma and Juffer, 2013; Wei et al., 2014). We note that the conformational flexibility apparent in the suite of DAP12 structures now available is consistent with this model, as is the specific arrangement of the crystallized DAP12-TM trimer in which the third DAP12 helix with coordinated potassium resembles a receptor TM helix with positively charged lysine (Figure S5).
TM peptides have long been considered poor candidates for crystallization, and LCP methods offer the attractive prospect of examining natural α-helix associations at high resolution within a lipid bilayer using well-established crystallographic tools. However, prior to this study only two crystal structures of LCP-associated peptides had been published. The 15-residue bacterial toxin gramicidin A crystallized in an intertwined, double-stranded helix conformation (PDB ID 2XDC) (Hofer et al., 2010) that does not represent the α-helical structures formed by TM domains derived from integral membrane proteins. A 25-residue synthetic peptide designed to form a tetra-helical zinc transporter was also crystallized from monoolein LCP (PDB ID 4P6K) (Joh et al., 2014), but this peptide does not appear be in a trans-bilayer orientation and therefore does not represent how typical TM helices integrate into lipid membranes. Our new DAP12 structures now demonstrate that the rapidly developing LCP technique can be successfully applied to isolated α-helical TM domains derived from naturally self-associating eukaryotic single-pass receptors. These results raise the enticing prospect that LCP techniques could be applied to more complex hetero-oligomeric assemblies such as the hexameric NKG2D-DAP10 receptor (Garrity et al., 2005; Raulet et al., 2013) and the octameric T cell receptor (Call et al., 2002), or to systems like the human growth hormone receptor in which conversion from the resting to the activated state is thought to involve exchange between two different TM interfaces (Brooks et al., 2014).
Experimental Procedures
Production and purification of disulfide-linked human DAP12-TM peptide
The DAP12-TM peptide CSTVSPGVLAGIVVGDLVLTVLIALAVYFLGRL was produced as a trpLE fusion in E. coli, disulfide-linked through the N-terminal cysteine, cyanogen bromide digested and HPLC purified following the published procedure (Sharma et al., 2013) without modification. The disulfide-linked peptide was stored as a lyophilized product until used.
Crystallization and structure determination
Monoolein LCP with incorporated peptide was prepared following the detergent-free method of Caffrey and colleagues (Hofer et al., 2010). Protocol variations and screening conditions are given in Supplemental Information. The DAP12 trimer structure was solved in Phaser (McCoy et al., 2007) by molecular replacement using Glycophorin A (MacKenzie et al., 1997) (PDB1AFO, chain A, state 1) as the search model. Chain A from the DAP12 trimer structure was subsequently used as a molecular replacement model to solve the tetramer structure. Additional details on data collection and structure refinement can be found in Supplemental Information. The structures reported here have been deposited in the Protein Data Bank with PDB ID 4WOL (trimer) and 4WO1 (tetramer).
Molecular dynamics simulations
DAP12 trimer and tetramer structures were used to build the initial systems in a POPC bilayer by following the general procedure of bilayer system building and equilibration in Membrane Builder (Jo et al., 2007; Jo et al., 2008; Jo et al., 2009). The initial systems were composed of each DAP12 model with coordinated ions, 48 POPC lipids in each leaflet, bulk water, and the ions in the bulk aqueous phase (150 mM KCl and 50 mM CaCl2 for trimers and tetramers, respectively). After equilibration, a 200-ns simulation was performed for each system without any restraints under the constant temperature and pressure condition (NPT) at 303.15 K and 1 bar, respectively. Further details of system equilibration and simulation parameters are provided in Supplemental Information
Cellular and in vitro translation DAP12 analysis
Construct designs, cellular transfection conditions and in vitro translation reaction conditions can be found in Supplemental Information. DAP12-HA was isolated from cell/ER microsome lysates by immunoprecipitation with anti-HA agarose beads (clone HA-7, Sigma-Aldrich), separated by SDS-PAGE and transferred to PVDF for western blot or phosphor imaging analysis. Blots were probed with anti-HA-bio (3F10, Roche Life Science) followed by streptavidin horseradish peroxidase and bands were visualized using a chemiluminescent substrate (Western Lightning Plus ECL, Perkin Elmer).
Supplementary Material
Highlights.
TM peptides from immunoreceptor subunit DAP12 crystallized in lipid bilayers
Trimeric and tetrameric structures were solved to 1.77 Å and 2.14 Å resolution
Complex formation is driven by polar sequences that also direct receptor assembly
These newly identified products compete with receptor assembly in the ER
Acknowledgments
We thank David Aragao for helpful advice on LCP crystallization, Pooja Sharma for solution NMR analysis of DAP12-TM peptides, Emilia Wu for assistance with MD simulations, Peter Colman and Jacqui Gulbis for assistance with data processing and refinement and for much helpful discussion, and members of the Call laboratory for reading the manuscript. This work was supported by grants from the National Health and Medical Research Council (NHMRC) of Australia (GNT1011352 to MJC and MEC, GNT1059331 to PEC; Infrastructure Support [IRIISS] to WEHI), the Victorian Government (VESKI Innovation Fellowship VIF12 to MEC; Operational Infrastructure Support to WEHI), XSEDE MCB070009 (to WI), the National Science Foundation (NSF MCB-1157677 to WI) and the National Institutes of Health (NIH R01-GM092950 to WI). MEC is supported by a QEII Fellowship (DP110104369) from the Australian Research Council (ARC). MJC is supported by an ARC Future Fellowship (FT120100145). Part of this research was carried out at the MX2 and SAXS/WAXS beamlines of the Australian Synchrotron, and we thank the beamline scientists for their technical support. We also acknowledge the use of the CSIRO Collaborative Crystallisation Centre.
Footnotes
Author Contributions: MJC and MEC designed the study. KK produced and crystallized DAP12-TM peptides. MJC, MEC, KK, SAS and JN designed the crystallization and optimization strategies. LVH and CEC performed SAXS measurements. KK, MJC and PEC collected data and PEC assisted with initial data processing. MJC solved and refined the final structures. ML, MJC and MEC performed biochemical experiments. SP and WI performed MD simulations. MEC and MJC wrote the paper. All authors contributed to editing the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arkhipov A, Shan Y, Das R, Endres NF, Eastwood MP, Wemmer DE, Kuriyan J, Shaw DE. Architecture and membrane interactions of the EGF receptor. Cell. 2013;152:557–569. doi: 10.1016/j.cell.2012.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocharov EV, Mineev KS, Volynsky PE, Ermolyuk YS, Tkach EN, Sobol AG, Chupin VV, Kirpichnikov MP, Efremov RG, Arseniev AS. Spatial structure of the dimeric transmembrane domain of the growth factor receptor ErbB2 presumably corresponding to the receptor active state. The Journal of biological chemistry. 2008;283:6950–6956. doi: 10.1074/jbc.M709202200. [DOI] [PubMed] [Google Scholar]
- Brooks AJ, Dai W, O’Mara ML, Abankwa D, Chhabra Y, Pelekanos RA, Gardon O, Tunny KA, Blucher KM, Morton CJ, et al. Mechanism of activation of protein kinase JAK2 by the growth hormone receptor. Science. 2014;344:1249783. doi: 10.1126/science.1249783. [DOI] [PubMed] [Google Scholar]
- Call ME, Chou JJ. A view into the blind spot: solution NMR provides new insights into signal transduction across the lipid bilayer. Structure. 2010;18:1559–1569. doi: 10.1016/j.str.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Call ME, Pyrdol J, Wiedmann M, Wucherpfennig KW. The organizing principle in the formation of the T cell receptor-CD3 complex. Cell. 2002;111:967–979. doi: 10.1016/s0092-8674(02)01194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Call ME, Schnell JR, Xu C, Lutz RA, Chou JJ, Wucherpfennig KW. The structure of the zetazeta transmembrane dimer reveals features essential for its assembly with the T cell receptor. Cell. 2006;127:355–368. doi: 10.1016/j.cell.2006.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Call ME, Wucherpfennig KW. Common themes in the assembly and architecture of activating immune receptors. Nat Rev Immunol. 2007;7:841–850. doi: 10.1038/nri2186. [DOI] [PubMed] [Google Scholar]
- Call ME, Wucherpfennig KW, Chou JJ. The structural basis for intramembrane assembly of an activating immunoreceptor complex. Nat Immunol. 2010;11:1023–1029. doi: 10.1038/ni.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Im W. NMR observable-based structure refinement of DAP12-NKG2C activating immunoreceptor complex in explicit membranes. Biophysical journal. 2012;102:L27–29. doi: 10.1016/j.bpj.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross TA, Murray DT, Watts A. Helical membrane protein conformations and their environment. European biophysics journal : EBJ. 2013;42:731–755. doi: 10.1007/s00249-013-0925-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Call ME, Wucherpfennig KW. The assembly of diverse immune receptors is focused on a polar membrane-embedded interaction site. PLoS Biol. 2006;4:e142. doi: 10.1371/journal.pbio.0040142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Garrity D, Call ME, Moffett H, Wucherpfennig KW. Convergence on a distinctive assembly mechanism by unrelated families of activating immune receptors. Immunity. 2005;22:427–438. doi: 10.1016/j.immuni.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity D, Call ME, Feng J, Wucherpfennig KW. The activating NKG2D receptor assembles in the membrane with two signaling dimers into a hexameric structure. Proc Natl Acad Sci U S A. 2005;102:7641–7646. doi: 10.1073/pnas.0502439102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratkowski H, Lear JD, DeGrado WF. Polar side chains drive the association of model transmembrane peptides. Proc Natl Acad Sci U S A. 2001;98:880–885. doi: 10.1073/pnas.98.3.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich SU, Mothes W, Brunner J, Rapoport TA. The Sec61p complex mediates the integration of a membrane protein by allowing lipid partitioning of the transmembrane domain. Cell. 2000;102:233–244. doi: 10.1016/s0092-8674(00)00028-3. [DOI] [PubMed] [Google Scholar]
- Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, Lerch-Bader M, Nilsson I, White SH, von Heijne G. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature. 2007;450:1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- Hofer N, Aragao D, Caffrey M. Crystallizing transmembrane peptides in lipidic mesophases. Biophys J. 2010;99:L23–25. doi: 10.1016/j.bpj.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsinger LJ, Lamb RA. Influenza virus M2 integral membrane protein is a homotetramer stabilized by formation of disulfide bonds. Virology. 1991;183:32–43. doi: 10.1016/0042-6822(91)90115-r. [DOI] [PubMed] [Google Scholar]
- Jo S, Kim T, Im W. Automated Builder and Database of Protein/Membrane Complexes for Molecular Dynamics Simulations. PloS one. 2007;2:e880. doi: 10.1371/journal.pone.0000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: A web-based graphical user interface for CHARMM. Journal of Computational Chemistry. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- Jo S, Lim JB, Klauda JB, Im W. CHARMM-GUI Membrane Builder for Mixed Bilayers and Its Application to Yeast Membranes. Biophysical journal. 2009;97:50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joh NH, Wang T, Bhate MP, Acharya R, Wu Y, Grabe M, Hong M, Grigoryan G, DeGrado WF. De novo design of a transmembrane Zn(2)(+)-transporting four-helix bundle. Science. 2014;346:1520–1524. doi: 10.1126/science.1261172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Jeon TJ, Oberai A, Yang D, Schmidt JJ, Bowie JU. Transmembrane glycine zippers: physiological and pathological roles in membrane proteins. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:14278–14283. doi: 10.1073/pnas.0501234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. Journal of molecular biology. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Lanier LL, Corliss B, Wu J, Phillips JH. Association of DAP12 with activating CD94/NKG2C NK cell receptors. Immunity. 1998a;8:693–701. doi: 10.1016/s1074-7613(00)80574-9. [DOI] [PubMed] [Google Scholar]
- Lanier LL, Corliss BC, Wu J, Leong C, Phillips JH. Immunoreceptor DAP12 bearing a tyrosine-based activation motif is involved in activating NK cells. Nature. 1998b;391:703–707. doi: 10.1038/35642. [DOI] [PubMed] [Google Scholar]
- Lau TL, Kim C, Ginsberg MH, Ulmer TS. The structure of the integrin alphaIIbbeta3 transmembrane complex explains integrin transmembrane signalling. The EMBO journal. 2009;28:1351–1361. doi: 10.1038/emboj.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Flanagan JM, Hunt JF, Adair BD, Bormann BJ, Dempsey CE, Engelman DM. Glycophorin A dimerization is driven by specific interactions between transmembrane alpha-helices. J Biol Chem. 1992;267:7683–7689. [PubMed] [Google Scholar]
- Lu X, Gross AW, Lodish HF. Active conformation of the erythropoietin receptor: random and cysteine-scanning mutagenesis of the extracellular juxtamembrane and transmembrane domains. The Journal of biological chemistry. 2006;281:7002–7011. doi: 10.1074/jbc.M512638200. [DOI] [PubMed] [Google Scholar]
- Luo BH, Springer TA, Takagi J. A specific interface between integrin transmembrane helices and affinity for ligand. PLoS biology. 2004;2:e153. doi: 10.1371/journal.pbio.0020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie KR, Prestegard JH, Engelman DM. A transmembrane helix dimer: structure and implications. Science. 1997;276:131–133. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of applied crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raulet DH, Gasser S, Gowen BG, Deng W, Jung H. Regulation of ligands for the NKG2D activating receptor. Annual review of immunology. 2013;31:413–441. doi: 10.1146/annurev-immunol-032712-095951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Kaywan-Lutfi M, Krshnan L, Byrne EF, Call MJ, Call ME. Production of disulfide-stabilized transmembrane peptide complexes for structural studies. Journal of visualized experiments : JoVE. 2013:e50141. doi: 10.3791/50141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Juffer AH. An atomistic model for assembly of transmembrane domain of T cell receptor complex. Journal of the American Chemical Society. 2013;135:2188–2197. doi: 10.1021/ja308413e. [DOI] [PubMed] [Google Scholar]
- Wei P, Xu L, Li CD, Sun FD, Chen L, Tan T, Luo SZ. Molecular Dynamic Simulation of the Self-Assembly of DAP12-NKG2C Activating Immunoreceptor Complex. PloS one. 2014;9:e105560. doi: 10.1371/journal.pone.0105560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P, Zheng BK, Guo PR, Kawakami T, Luo SZ. The association of polar residues in the DAP12 homodimer: TOXCAT and molecular dynamics simulation studies. Biophysical journal. 2013;104:1435–1444. doi: 10.1016/j.bpj.2013.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou FX, Cocco MJ, Russ WP, Brunger AT, Engelman DM. Interhelical hydrogen bonding drives strong interactions in membrane proteins. Nature structural biology. 2000;7:154–160. doi: 10.1038/72430. [DOI] [PubMed] [Google Scholar]
- Zhu J, Luo BH, Barth P, Schonbrun J, Baker D, Springer TA. The structure of a receptor with two associating transmembrane domains on the cell surface: integrin alphaIIbbeta3. Molecular cell. 2009;34:234–249. doi: 10.1016/j.molcel.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.