Abstract
Cardiolipin (CL), the signature phospholipid of mitochondria, is involved in a plethora of cellular processes and is crucial for mitochondrial function and architecture. The de novo synthesis of CL in the mitochondria is followed by a unique remodeling process, in which CL undergoes cycles of deacylation and reacylation. Specific fatty acyl composition is acquired during this process, and remodeled CL contains predominantly unsaturated fatty acids. The importance of CL remodeling is underscored by the life-threatening genetic disorder Barth syndrome (BTHS), caused by mutations in tafazzin, which reacylates monolysocardiolipin (MLCL) generated from the deacylation of CL. Just as CL-deficient yeast mutants have been instrumental in elucidating functions of this lipid, the recently characterized CL-phospholipase mutant cld1Δ and the tafazzin mutant taz1Δ are powerful tools to understand the functions of CL remodeling. In this review, we discuss recent advances in understanding the role of CL in mitochondria with specific focus on the enigmatic functions of CL remodeling.
Keywords: Cardiolipin, Cardiolipin remodeling, Mitochondria, Bioenergetics, Tafazzin, Phospholipase, Barth syndrome
Introduction
In eukaryotes, cardiolipin (CL) is predominantly localized and exclusively synthesized de novo in the mitochondria (Hostetler et al. 1972; Joshi et al. 2009). As the signature phospholipid of mitochondria, CL is involved in numerous mitochondrial functions, including bioenergetics, apoptosis, mitochondrial dynamics, and mitochondrial structure (Joshi et al. 2009; Ren et al. 2014; Houtkooper & Vaz 2008; Schug & Gottlieb 2009; Schlame & Ren 2009). The diverse functions of CL in the mitochondria highlight the unique nature of this anionic phospholipid. CL contains four fatty acyl chains and two phosphatidyl moieties bridged by a glycerol (Pangborn 1947; Lecocq & Ballou 1964). The hydrophobicity of four acyl groups and negative charges of two phosphate groups confer a wide variety of interactions with mitochondrial proteins (Schlame & Ren 2009; Schlame et al. 2000; Claypool 2009; Klingenberg 2009; Gohil & Greenberg 2009). In addition, the dimeric structure confers a conical shape that favors a hexagonal HII phase in membranes (Cullis et al. 1986). Regulation of CL de novo synthesis, acyl remodeling, degradation, oxidation, and trafficking controls CL content, acyl composition, and membrane distribution. Recent reviews have described cellular functions (Joshi et al. 2009; Ren et al. 2014; Houtkooper & Vaz 2008) and physicochemical properties of CL (Schlame & Ren 2009; Lewis & McElhaney 2009). In this review, we focus on the mechanisms and functions of CL remodeling.
CL biosynthesis
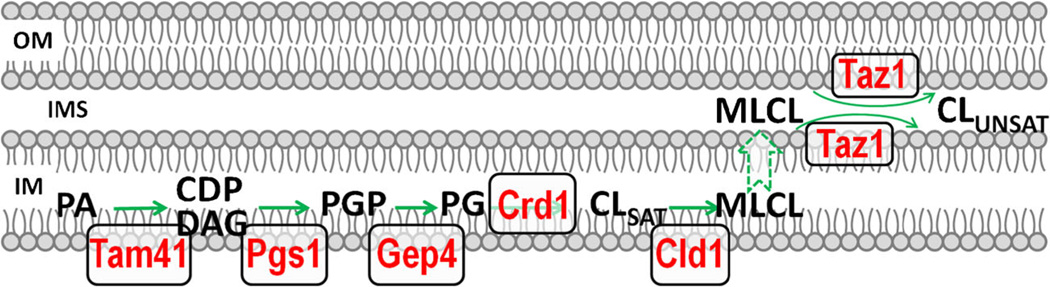
Most membrane lipids in the mitochondria are synthesized in the endoplasmic reticulum. In contrast, CL biosynthesis occurs exclusively in the mitochondrial inner membrane. The de novo synthesis of CL (Fig. 1) is a highly conserved pathway from yeast to mammals (Tian et al. 2012) and is well-characterized in the yeast Saccharomyces cerevisiae. The first reaction of CL biosynthesis is the conversion of mitochondrial phosphatidic acid (PA) to CDP-diacylglycerol (CDP-DAG) by the mitochondrial CDP-DAG synthase Tam41 (Tamura et al. 2013; Kutik et al. 2008). Pgs1 catalyzes the committed step of CL synthesis by transferring the phosphatidyl group of CDP-DAG to glycerol-3-phosphate, generating phosphatidylglycerolphosphate (PGP) (Chang et al. 1998a). The subsequent dephosphorylation of PGP to phosphatidylglycerol (PG) is catalyzed by the PGP phosphatase Gep4 in yeast (Osman et al. 2010; Kelly & Greenberg 1990) and PTPMT1 in mammals (Zhang et al. 2011; Xiao et al. 2011). CL synthase (Crd1) catalyzes the final reaction of the de novo synthesis of CL by adding a phosphatidyl group from CDP-DAG to PG (Tamai & Greenberg 1990; Jiang et al. 1997; Tuller et al. 1998; Chang et al. 1998b). Thus, CL contains two phosphatidyl groups that are linked by a glycerol molecule. The final reaction of CL synthesis in the prokaryote Escherichia coli is different from that in eukaryotes. In E. coli, PG incorporates the other phosphatidyl group from phosphatidylethanolamine (PE) (Tan et al. 2012) or another PG (Hirschberg & Kennedy 1972) to generate CL. The distinct mechanisms of action of CL synthase suggest that CL synthesis has evolutionarily differentiated paths (Tian et al. 2012).
Fig. 1.
CL de novo synthesis and remodeling in S. cerevisiae. The first reaction of CL de novo synthesis is the conversion of phosphatidic acid (PA) to CDP-diacylglycerol (CDP-DAG) by the mitochondrial CDP-DAG synthase Tam41. The committed step of CL synthesis is catalyzed by Pgs1, which converts CDP-DAG to phosphatidylglycerolphosphate (PGP). PGP is subsequently dephosphorylated to phosphatidylglycerol (PG) by the GEP4-encoded PGP phosphatase. CL synthase, encoded by CRD1, condenses PG and CDP-DAG to form CL. CL synthesized de novo has primarily saturated acyl chains (CLSAT). CLSAT is deacylated by the CL-specific phospholipase Cld1 to monolysocardiolipin (MLCL), which is reacylated by tafazzin (the TAZ1 gene product) to CL containing more unsaturated acyl chains (CLUNSAT). All the CL biosynthetic enzymes are localized in the mitochondrial inner membrane (IM), whereas tafazzin is localized in the outer face of the (IM) and the inner face of the outer membrane (OM). IMS: intermembrane space
CL remodeling enzymes
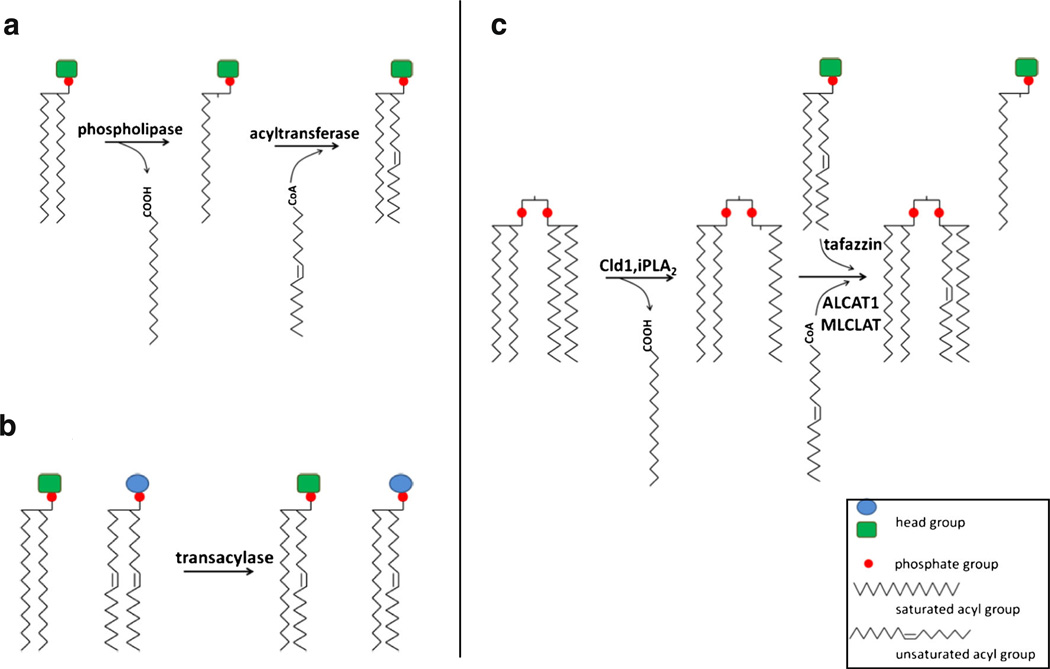
In yeast and metazoans, a few species of unsaturated acyl groups predominate in CL (Houtkooper et al. 2009a; Schlame et al. 2002), which is highly symmetric (Schlame et al. 2005). Because the enzymes for de novo CL synthesis do not exhibit acyl specificity (Ren et al. 2014; Houtkooper & Vaz 2008), remodeling in which acyl chains are removed and replaced with other acyl chains plays a key role in generating symmetric CL with characteristic fatty acid residues following de novo synthesis. The remodeling of phospholipids can occur by two mechanisms. In the two-step deacylation-reacylation Lands cycle (Fig. 2a), a phospholipid is deacylated by phospholipase to lysophospholipid. Subsequently, acyl groups from acyl-CoA or neighboring phospholipids are added to the lysophospholipid by acyltransferases or transacylases (Lands 1960). In an alternative mechanism of acyl remodeling, acyl chains are exchanged between adjacent phospholipids in a single transacylase-catalyzed step (Xu et al. 2003; Yamashita et al. 1997) (Fig. 2b). CL remodeling may occur by both mechanisms (Fig. 2c). In the two-step mechanism, CL is remodeled via the deacylation-reacylation cycle, in which CL is deacylated by phospholipases, and MLCL is reacylated to CL by either transacylase or acyltransferase activity. One-step CL remodeling is the reaction that remodels CL via only transacylation, in which deacylation of phospholipids is not necessary. The importance of CL remodeling is underscored by the life-threatening genetic disorder Barth syndrome (BTHS), which results from mutations in tafazzin, the transacylase that remodels CL (Bione et al. 1996; Barth et al. 1983).
Fig. 2.
phospholipid remodeling. The remodeling of phospholipids can occur by two mechanisms, (a) a two-step deacylation-reacylation Lands cycle, and (b) a single-step transacylation. CL remodeling (c) can occur by both mechanisms. Tafazzin-catalyzed transacylation can remodel CL in a single step or reacylate MLCL to CL in the two-step mechanism
Tafazzin
Tafazzin was identified in 1996 as the gene that is defective in BTHS (Bione et al. 1996; Barth et al. 1983). Homology to acyltransferase sequence motifs suggested that the enzyme was an acyltransferase (Neuwald 1997). Tafazzin is now well-characterized as a phospholipid-lysophospholipid transacylase (Xu et al. 2003) and can catalyze CL remodeling by both mechanisms (Schlame et al. 2002; Xu et al. 2003; Vreken et al. 2000). As a transacylase, tafazzin can catalyze CL acyl remodeling in a single step, in which acyl groups are exchanged between phospholipid and lysophospholipid. In the one-step transacylation of CL remodeling, CL phospholipases are not required for generating MLCL, as a transacylation reaction between a lyso-phospholipid and a CL can generate a phospholipid and MLCL. The subsequent transacylation between MLCL and an adjacent phospholipid generates a remodeled CL and a monolyso-phospholipid. Tafazzin can also participate in a deacylation-reacylation cycle to remodel CL (Fig. 2c). The involvement of tafazzin in this type of remodeling is underscored by the fact that tafazzin deficiency, which abrogates the reacylation step, leads to decreased total CL and increased MLCL, the intermediate generated from deacylation of CL (Vreken et al. 2000; Gu et al. 2004; Schlame et al. 2003; Valianpour et al. 2002; Xu et al. 2006; Acehan et al. 2011; Houtkooper et al. 2009b). The decreased CL/MLCL ratio illustrates that remodeling is initiated by deacylation, which consumes total CL and accumulates MLCL in the absence of tafazzin-mediated reacylation. Coincident with a decreased CL/MLCL ratio, unsaturated CL is also decreased in the absence of tafazzin. These changes in CL profiles are observed in all studied eukaryotes with tafazzin mutations, including yeast (Gu et al. 2004), fruit fly (Xu et al. 2006), mouse (Acehan et al. 2011; Soustek et al. 2011), and human (Vreken et al. 2000; Schlame et al. 2003; Valianpour et al. 2002; Houtkooper et al. 2009b).
Tafazzin-mediated CL remodeling shifts acyl composition towards unsaturation. Interestingly, initial findings showed that the purified recombinant tafazzin enzyme lacks acyl specificity and can react with nearly all phospholipids and lysophospholipids (Xu et al. 2009; Testet et al. 2005; Malhotra et al. 2009a). How, then, does tafazzin remodel CL with specific acyl groups in vivo? Recent studies indicate that the substrate specificity of tafazzin is affected by the membrane lipid phase (Schlame et al. 2012; Gawrisch 2012). In vitro, tafazzin-catalyzed CL transacylation occurs only in nonbilayer lipid micelles and is probably limited to mitochondrial microdomains (Schlame et al. 2012). This underlying mechanism suggests that CL remodeling may be spatially activated at specific mitochondrial membrane domains where tafazzin localizes to shape mitochondrial architecture.
Cld1, the CL-specific phospholipase
Although tafazzin alone can mediate CL remodeling in the presence of lyso-phospholipids, leading to acyl specificity via the transacylation reaction (Ren et al. 2014; Schlame et al. 2012), the dramatic decrease in the CL/MLCL ratio in the tafazzin mutant underscores the importance of CL remodeling via the deacylation-reacylation cycle. In the yeast S. cerevisiae, deletion of the cardiolipin-specific phospholipase Cld1, identified in 2009 (Beranek et al. 2009), reduces acyl unsaturation specifically in CL and also prevents MLCL accumulation in tafazzin mutants (Beranek et al. 2009; Baile et al. 2014). These findings indicate that Cld1 is the only CL-specific phospholipase in yeast that initiates CL remodeling by deacylation. Blocking Cld1-mediated deacylation of CL in tafazzin mutants prevents the decreased CL/MLCL ratio, while unremodeled CL remains primarily saturated. Therefore, the cld1Δ mutant is a powerful tool with which to distinguish between decreased CL/MLCL and decreased unsaturated (remodeled) CL as the cause of cellular defects in tafazzin-deficient cells. Deletion of CLD1 rescues respiratory growth defects (Ye et al. 2014; Baile et al. 2014) and decreased chronological lifespan (Ye et al. 2014) in taz1Δ, indicating that the decreased CL/MLCL ratio, but not decreased unsaturated CL, leads to the defects in tafazzin-deficient cells. These findings have implications for BTHS, because if a decreased CL/MLCL ratio is the cause of the pathology, attenuation of CL-specific phospholipase activity may be a potential strategy to treat BTHS patients.
Interestingly, CLD1 is the most highly regulated gene in the CL biosynthetic and remodeling pathways (Ye et al. 2014). Its transcription is repressed in logarithmically growing cells and upregulated about 30-fold in the stationary phase (Ye et al. 2014). This regulation is coordinated with cellular respiratory activity, as CLD1 transcriptional expression is not increased in the stationary phase in cells lacking mitochondrial DNA (Ye et al. 2014). In addition, respiratory conditions such as growth in non-fermentable carbon sources or glucose depletion also induce CLD1 expression (Ye et al. 2014; Baile et al. 2013). Controlling CLD1 expression in response to physiological conditions is important, as increased expression of CLD1 impairs mitochondrial integrity (Ye et al. 2014).
Cld1 has a higher affinity for saturated than unsaturated acyl groups; thus, Cld1-catalyzed CL remodeling contributes to a more unsaturated CL acyl composition (Beranek et al. 2009; Baile et al. 2014). Interestingly, the cld1Δ mutant contains more unsaturated CL in the stationary phase than the logarithmic phase (Ye et al. 2014), suggesting that additional mechanisms alter CL acyl composition in the absence of Cld1-mediated CL remodeling. A likely possibility is that CL is remodeled by tafazzin alone via the one-step transacylation, which is independent of Cld1-involved deacylation-reacylation cycles. Another possibility is that CL precursors PA or PG may be remodeled in the absence of Cld1, leading to changes in CL acyl profiles. Further studies are needed to distinguish among these possibilities.
Mammalian phospholipases that deacylate CL
CL-specific phospholipases in mammals have not been identified, although several mammalian phospholipases are reported to have CL-hydrolyzing activities. These CL phospholipases, including iPLA2β, iPLA2γ, cPLA2, and sPLA2 (Buckland et al. 1998; Hsu et al. 2013; Dennis et al. 2011), belong to a large phospholipase A2 superfamily. Among these enzymes, sPLA2 (the secreted phospholipase A2) displays the highest CL deacylase activity in vitro (Hsu et al. 2013).
iPLA2β (Group VIA PLA2; PNPLA9) and iPLA2γ (Group VIB; PNPLA8) are calcium-independent phospholipase A2s. They are both found in membrane fractions, including mitochondria (Dennis et al. 2011). iPLA2β is required for integrity of the mitochondrial membrane and influences the release of cytochrome c under oxidative stress (Gadd et al. 2006; Seleznev et al. 2006). Genetic suppression of this enzyme partially restores the CL/MLCL ratio in Drosophila tafazzin mutants (Malhotra et al. 2009b). Interestingly, male sterility caused by tafazzin deficiency in Drosophila is also rescued by inactivation of iPLA2β (Malhotra et al. 2009b).
iPLA2γ plays critical roles in mitochondrial lipid metabolism and mitochondrial function. Genetic ablation of iPLA2γ in mice leads to tissue-specific phenotypes. In the heart, it decreases CL content and increases CL containing arachidonic (20:4) and docosahexenoic acids (22:6) (Mancuso et al. 2007), accompanied by decreased mitochondrial bioenergetics and defective myocardial function (Mancuso et al. 2007). However, deletion of iPLA2γ in tafazzin knockdown mice increases immature CL species containing 16:0, 16:1, and 18:1 acyl chains and decreases MLCL species containing 18:2–18:2–18:2 and 18:2–18:2–18:1. Although deletion of iPLA2γ in tafazzin knockdown mice partially rescues the abnormal CL profile caused by tafazzin deficiency, it does not prevent a decrease in the predominant CL species (tetra-18:2) or increase in MLCL, suggesting that iPLA2γ is not the only phospholipase that participates in CL remodeling (Kiebish et al. 2013). This finding also suggests that iPLA2γ-catalyzed deacylation of CL is partially responsible for the decreased CL/MLCL ratio in tafazzin mutants. Interestingly, the decreased CL/MLCL ratio in tafazzin-deficient BTHS lymphoblast cells is almost restored to normal after treatment with bromoenol lactone (Malhotra et al. 2009b), an iPLA2 chemical inhibitor that can inhibit both iPLA2β and iPLA2γ. Therefore, it is probably safe to extrapolate that both iPLA2β and iPLA2γ participate in initiating CL remodeling by deacylation of CL and contribute to the decreased CL/MLCL ratio in tafazzin-deficient cells. Interestingly, in the hippocampus, deletion of iPLA2γ increases CL content and increases CL with shorter saturated acyl groups (Mancuso et al. 2009), coinciding with enlarged and degenerating mitochondria and cognitive dysfunction (Mancuso et al. 2009). What remains unclear are the specificities of these enzymes, how they are regulated, and their physiological functions. Furthermore, the CL phospholipase activities of the other PLA2s have not been studied.
Mammalian CL acyltransferases: ALCAT1 and MLCLAT1
In addition to tafazzin, two other acyltransferases can reacylate MLCL in mammalian cells, including acyl-CoA:lysocardiolipin acyltransferase (ALCAT1) and MLCL acyltransferase (MLCLAT1). ALCAT1 is an acyl-CoA-dependent acyltransferase that localizes to the mitochondria-associated membrane, which is a sub-compartment of the endoplasmic reticulum (Cao et al. 2004; Li et al. 2010). It preferentially transfers acyl groups from linoleoyl-CoA or oleoyl-CoA to polyglycerophospholipids that are metabolic intermediates for the synthesis of CL, including MLCL, dilysocardiolipin (DLCL), phosphatidylglycerol (PG), and bis (monoacylglycero) phosphate (Cao et al. 2004; Cao et al. 2009). Therefore, ALCAT1-involved remodeling can convert saturated CL into more unsaturated CL. ALCAT1 expression is increased in response to oxidative stress (Li et al. 2010). Interestingly, overexpression of ALCAT1 causes oxidative stress (Liu et al. 2012), mitochondrial fragmentation, and instability of mitochondrial DNA (Li et al. 2012). Mitochondrial DNA depletion was also observed in yeast cells overexpressing CLD1 (Ye et al. 2014). It is possible that both ALCAT1 and CLD1-regulated CL remodeling share a common function of modulating CL composition for mitochondrial activity.
MLCLAT1-catalyzed acylation of MLCL was first detected in rat heart mitochondria (Ma et al. 1999) and subsequently purified from pig liver mitochondria (Taylor & Hatch 2003). Purified recombinant MLCLAT1 has a higher affinity for linoleoyl-CoA than oleoyl-CoA or palmitoyl-CoA for the acylation of MLCL to CL. Interestingly, increased expression of MLCLAT1 in BTHS lymphoblast cells increases both incorporation of linoleate into CL and total CL levels (Taylor & Hatch 2009). Although ALCAT1 and MLCLAT1 can reacylate MLCL to CL, the accumulation of MLCL in the absence of tafazzin suggests that ALCAT1 and MLCLAT1-mediated acylation of MLCL is not functionally redundant with tafazzin.
Localization of CL remodeling enzymes
CL de novo synthesis occurs on the matrix side of the mitochondrial inner membrane where all the biosynthetic enzymes are located (Tamura et al. 2013; Osman et al. 2010; Tamai & Greenberg 1990; Dzugasova et al. 1998). The post-synthetic remodeling of CL is dependent on the localization of specific remodeling enzymes. Yeast tafazzin is an integral membrane protein of the mitochondria (Claypool et al. 2006), where it binds to the outer face of the inner membrane and the inner face of the outer membrane (Claypool et al. 2006). Submitochondrial fractionation analysis indicates that yeast tafazzin is associated with the mitochondrial inner and outer membrane as well as the contact sites of the mitochondrial inner and outer membrane (Claypool et al. 2006). Therefore, tafazzin can preferentially remodel CL facing the intermembrane space or in the contact sites. In contrast, Cld1 is located in the inner membrane facing the matrix side (Baile et al. 2013). How MLCL generated by Cld1 deacylation on the matrix side of the mitochondrial inner membrane is reacylated by tafazzin in the mitochondrial inner and outer membranes facing the intermembrane space is not understood. In mammals, CL remodeling enzymes tafazzin, iPLA2β, iPLA2γ, and MLCLAT1 are present in the mitochondria, and ALCAT1 is in the mitochondria-associated membrane (Li et al. 2010). The sub-mitochondrial localization of these enzymes has not been characterized. Understanding the precise localization of mammalian CL remodeling enzymes will help to clarify the mechanism of CL remodeling.
The function of CL remodeling
CL is important for mitochondrial architecture and organization, as it is required for cristae biogenesis (Xu et al. 2005; Acehan et al. 2009) and mitochondrial fusion and fission (Joshi et al. 2012; Xu et al. 2010). As a result of CL-protein interactions, CL promotes respiratory supercomplex formation and optimizes mitochondrial bioenergetics (Pfeiffer et al. 2003; Zhang et al. 2002; Claypool et al. 2008a; Mileykovskaya & Dowhan 2014; Paradies et al. 2014). In addition, CL functions in mitochondrial protein import (Jiang et al. 2000; Gebert et al. 2009), iron-sulfur biogenesis (Patil et al. 2013), and the TCA cycle (Patil et al. 2013; Raja & Greenberg 2014) and acts as a signal for apoptosis (Schug & Gottlieb 2009) and mitophagy (Chu et al. 2013). The diverse mitochondrial functions of CL highlight its important role as a structural component of the organelle as well as a signaling lipid. A major unanswered question is how CL is regulated during all these processes. CL remodeling may be a pivotal regulatory hub for CL functions. First, CL remodeling can occur in discrete mitochondrial domains where CL remodeling enzymes are localized. Specific CL functions may thus be controlled by regulation of the remodeling enzymes, which may be spatially restricted. Second, CL remodeling can alter CL unsaturation and CL content and generate MLCL and free fatty acids, any or all of which may be important for specific CL functions. Third, the physicochemical properties of CL are subject to change during remodeling, which is likely to affect the affinity of CL-protein interactions. These and possibly other mechanisms that regulate CL remodeling may affect CL-dependent functions. Currently, little is known about the function of CL remodeling, primarily because defective CL remodeling is coupled with deficiency in both CL acyl composition and CL/MLCL production. Thus, it is difficult to distinguish cellular defects caused by decreased CL content and CL remodeling. While the cld1Δ mutant may facilitate these studies in yeast, addressing the question awaits the identification of CL-specific phospholipase in mammalian cells.
Unremodeled and remodeled CL
As discussed above, deletion of the CL-specific yeast phospholipase CLD1 blocks CL remodeling and leads to decreased unsaturated acyl species in CL (Beranek et al. 2009; Baile et al. 2014; Ye et al. 2014). Nevertheless, respiratory growth (Baile et al. 2014; Ye et al. 2014), fermentative growth (Ye et al. 2014), and lifespan (Ye et al. 2014) are similar in cld1Δ and wild type cells, suggesting that unremodeled CL in cld1Δ and remodeled CL in wild type cells can similarly support these functions. Mitochondrial morphology and cristae size, respiratory supercomplex stability, mitochondrial respiration, and the mitochondrial membrane potential are also similar in cld1Δ and wild type cells (Baile et al. 2014). These findings indicate that CL remodeling is not essential in yeast, particularly with respect to bioenergetics. Although unsaturated CL is greatly diminished when CL is not remodeled (Beranek et al. 2009; Baile et al. 2014; Ye et al. 2014), we cannot conclude that Cld1-mediated alteration of acyl composition has no effect on cellular functions, as other genes and pathways may compensate for the loss of Cld1.
CL remodeling and bioenergetics
The role of CL in bioenergetics is well-studied, including mitochondrial respiration, electron transport, and oxidative phosphorylation. CL interacts with respiratory complexes (Lange et al. 2001; Palsdottir et al. 2003; Eble et al. 1990; Shinzawa-Itoh et al. 2007) and ADP/ATP carrier (Claypool et al. 2008a; Beyer & Klingenberg 1985), and the CL-protein interactions stabilize respiratory supercomplexes (Pfeiffer et al. 2003; Zhang et al. 2002; Claypool et al. 2008a). It is thus not surprising that mitochondrial respiration and energy production are correlated with CL biosynthesis (Claypool et al. 2008a; Jiang et al. 2000; Gohil et al. 2004), and that CL deficiency resulting from abnormal CL remodeling disrupts energy dynamics. For example, tafazzin mutations lead to decreased CL content and defective bioenergetics in yeast (Ma et al. 2004; Claypool et al. 2008b) and mammals (Xu et al. 2005; McKenzie et al. 2006). As discussed above, this defect in yeast taz1Δ is due to the decrease in CL/MLCL, not to the decrease in unsaturated CL (Baile et al. 2014; Ye et al. 2014).
The interplay between CL remodeling and bioenergetics is further underscored by the regulation of the yeast remodeling enzyme Cld1. CLD1 expression increases under respiratory conditions (Ye et al. 2014; Baile et al. 2013). It is also controlled by the Hap2/3/4/5 transcription factor complex that mediates activation of respiratory gene expression (Ye et al. 2014). Furthermore, dissipating the mitochondrial membrane potential in taz1Δ decreases the CL/MLCL ratio, suggesting that the activity of Cld1-catalyzed CL deacylation is associated with energy coupling (Baile et al. 2013). The respiratory control of Cld1 activity emphasizes the importance of Cld1-mediated remodeling in energy dynamics. Interestingly, CLD1 overexpression increases ATP content, and the energy supply is shifted from respiration to glycolysis (Ye et al. 2014). However, how alterations in CL properties resulting from Cld1-mediated remodeling influence energy dynamics remains unclear.
CL remodeling and mitochondrial architecture
Mitochondrial morphology is disrupted in CL remodeling-deficient mutants (Xu et al. 2005). BTHS lymphoblast cells that contain tafazzin mutations display clusters of fragmented mitochondria and dysmorphic cristae (Acehan et al. 2007). However, how tafazzin-catalyzed remodeling shapes mitochondrial architecture is unclear. While tafazzin-catalyzed transacylation is activated by specific physical properties of the membrane (Schlame et al. 2012), tafazzin-mediated remodeling is proposed to reshuffle acyl groups between CL and adjacent membrane lipids and create tightly packed membrane curvature (Schlame et al. 2012). In yeast, tafazzin is localized in the outer face of the inner membrane and the inner face of the outer membrane (Claypool et al. 2006). Therefore, tafazzin-mediated remodeling may participate in the biogenesis of curvature for both the mitochondrial inner and outer membranes. In addition, tafazzin is also localized to the contact sites (Claypool et al. 2006) where the mitochondrial inner and outer membranes are closely tethered together. The mitochondrial contact sites are a newly recognized feature of mitochondrial architecture and are organized by the large protein complex, MICOS. Although CL is enriched in the contact sites (de Kroon et al. 1997; Daum 1985; Zinser et al. 1991; Hovius et al. 1990), it is unclear if CL remodeling is important for their formation. Interestingly, when the integrity of the MICOS complex is disrupted, CL remodeling is also deficient (Harner et al. 2014). Specifically, AIM24 encodes a yeast inner membrane protein that is required for maintenance of MICOS subunit levels (Harner et al. 2014). Genetic manipulation of MICOS subunits in aim24Δ depletes mitochondrial cristae and decreases tafazzin protein, resulting in CL profiles similar to those of tafazzin-deficient mutants (Harner et al. 2014). This finding suggests that tafazzin may function at the contact sites. Although the mechanisms linking remodeling to mitochondrial architecture have not been investigated, tafazzin-catalyzed remodeling is probably important for many features of mitochondrial structure, including the mitochondrial inner and outer membrane, the contact sites, and the cristae.
CL remodeling, CL oxidation, and apoptosis
CL is susceptible to oxidative damage because it is predominantly localized in the mitochondrial inner membrane, where reactive oxygen species (ROS) are generated during respiration (Paradies et al. 2002; Tyurina et al. 2006). Fatty acyl groups of CL containing two or more double bonds are susceptible to oxygenation or CL peroxidation. Tetralinoleoyl-CL (18:2) is the most abundant CL in the heart and most other tissues, whereas brain CL is more complex and contains polyunsaturated fatty acids such as arachidonic (20:4) and docosahexaenoic acids (22:6) (Houtkooper et al. 2009a; Samhan-Arias et al. 2012). These CL species are subject to passive or selective oxidation. Passive oxidation of CL is a non-selective process in which CL is peroxidized, leading to decreased activity of complexes I, III, and IVas well as decreased supercomplex formation (Paradies et al. 2002; Paradies et al. 2001; Paradies et al. 2000; Genova et al. 2008). Selective CL peroxidation by ROS occurs during apoptosis, and CL is the only mitochondrial phospholipid that undergoes peroxidation during this process (Kagan et al. 2005). The release of cytochrome c is an apoptogenic step. CL is bound to cytochrome c by insertion of one acyl chain into a hydrophobic pocket of the protein (Bayir et al. 2006; Ott et al. 2007; Kalanxhi & Wallace 2007). Binding is fortified by electrostatic interactions between the phosphate groups of CL and lysine residues of cytochrome c (Sinibaldi et al. 2008). The tertiary structure of cytochrome c is altered by binding to CL, and the CL-bound cytochrome c complex manifests peroxidase activity (Kagan et al. 2005; Kagan et al. 2009), which is responsible for CL peroxidization by the superoxide dismutation product H2O2 (Kagan et al. 2005). Oxidation of CL decreases the CL-cytochrome c interaction, resulting in a decrease in membrane-bound cytochrome c (Iverson et al. 2004; Ostrander et al. 2001; Nomura et al. 2000; Ott et al. 2002). Thus, CL peroxidation is critical for the release of cytochrome c during apoptosis (Belikova et al. 2007; Paradies et al. 2009). Interestingly, double bonds in CL acyl chains increase the CL-cytochrome c binding affinity (Belikova et al. 2006). Thus, cytochrome c preferentially binds to polyunsaturated CL, which is highly susceptible to oxidation by ROS (Belikova et al. 2006). CL remodeling can both modulate the affinity of cytochrome c for CL and replace oxidized fatty acids with non-oxidized acyl groups, restoring the interaction. Therefore, CL remodeling may be important for controlling cytochrome c release during apoptosis.
CL remodeling and disease
BTHS is the most direct example of a human disorder resulting from perturbation of CL remodeling. BTHS is a severe X-linked genetic disorder caused by mutations in tafazzin, resulting in cardiomyopathy, skeletal myopathy, growth retardation, and neutropenia (Barth et al. 1999; Christodoulou et al. 1994; Clarke et al. 2013). Cardiac problems prevalent in BTHS patients include dilated, hypertrophic, and noncompaction cardiomyopathy and heart failure. Abnormal CL profiles resulting from tafazzin deficiency (Vreken et al. 2000; Schlame et al. 2003; Valianpour et al. 2002; Houtkooper et al. 2009b) affect energy metabolism and heart function, as CL is required for optimal activity of oxidative phosphorylation. As discussed above, defects in yeast tafazzin mutants are rescued by blocking CL deacylation. If these findings are conserved in mammalian cells, the identification and characterization of human CL phospholipases may help to develop therapeutic strategies for BTHS patients. BTHS is also characterized by a wide disparity of clinical presentations, ranging from severe incapacitating disease to nearly asymptomatic, even in patients carrying identical tafazzin mutations (Joshi et al. 2009; Ren et al. 2014; Clarke et al. 2013). The lack of genotype-phenotype correlation suggests that genetic modifiers may play a role in BTHS pathology. While the molecular basis underlying the pathology of BTHS is not understood, potential modifiers that inhibit CL deacylation may prevent the decreased CL/MLCL. Identifying such modifiers may shed light on other avenues for BTHS treatment.
The clinical presentations in BTHS are similar to those of an autosomal recessively inherited human disorder, dilated cardiomyopathy with ataxia (DCMA), which is caused by mutations in DNAJC19 (Davey et al. 2006; Ojala et al. 2012). DNAJC19 encodes a mitochondrial inner membrane chaperone protein that is thought to function in mitochondrial protein import (Davey et al. 2006). Interestingly, a recent study suggested that the DNAJC19 protein may function in CL remodeling by regulating tafazzin activity via the association with prohibitin, a ring-like scaffold protein located in the inner membrane of the mitochondria (Richter-Dennerlein et al. 2014). Defective CL remodeling has also been implicated in both Type I and II diabetes (Han et al. 2005; Watkins et al. 2002; He & Han 2014). At the very early stage of pathological development in the diabetic mouse model, cardiac CL undergoes abnormal remodeling, resulting in a decrease in total CL content, depletion of the major cardiac CL, tetralinoleoyl-CL (18:2), and an increase in CL species containing longer and polyunsaturated fatty acids (Han et al. 2007). Interestingly, these abnormal CL presentations are ameliorated by treatment with the antidiabetic drug rosiglitazone, which increases both total CL and tetralinoleoyl-CL but decreases polyunsaturated CL in diabetic mice (Watkins et al. 2002; Pan et al. 2006). In summary, CL remodeling plays a role in the pathology of human disease. Therefore, elucidating the functions of CL remodeling in metabolic and physiological process may provide novel strategies for treating human disorders.
Coda
CL was first isolated from beef heart in 1947 (Pangborn 1947). After more than six decades, a plethora of cellular and mitochondrial functions have been linked to CL through genetic, biochemical, and cellular studies. CL is now recognized as a crucial phospholipid for optimal cellular physiology and human health. Important as CL homeostasis is, CL remodeling may be a critical regulatory hub for controlling CL content, acyl composition, and distribution and, thus, for its versatile functions. The functions of CL remodeling are just beginning to be explored. While it is involved in bioenergetics, mitochondrial structure, and apoptosis, additional functions and detailed mechanisms of CL remodeling have yet to be elucidated.
Acknowledgments
The Greenberg laboratory acknowledges support from the Barth Syndrome Foundation, Barth Syndrome Foundation of Canada, Association Barth France, and the National Institutes of Health (HL117880).
Footnotes
Conflict of interest The authors declare that there are no conflicts of interest.
References
- Acehan D, Xu Y, Stokes DL, Schlame M. Comparison of lymphoblast mitochondria from normal subjects and patients with Barth syndrome using electron microscopic tomography. Lab Investig. 2007;87(1):40–48. doi: 10.1038/labinvest.3700480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acehan D, Khuchua Z, Houtkooper RH, Malhotra A, Kaufman J, Vaz FM, et al. Distinct effects of tafazzin deletion in differentiated and undifferentiated mitochondria. Mitochondrion. 2009;9(2):86–95. doi: 10.1016/j.mito.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acehan D, Vaz F, Houtkooper RH, James J, Moore V, Tokunaga C, et al. Cardiac and skeletal muscle defects in a mouse model of human Barth syndrome. J Biol Chem. 2011;286(2):899–908. doi: 10.1074/jbc.M110.171439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baile MG, Whited K, Claypool SM. Deacylation on the matrix side of the mitochondrial inner membrane regulates cardiolipin remodeling. Mol Biol Cell. 2013;24(12):2008–2020. doi: 10.1091/mbc.E13-03-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baile MG, Sathappa M, Lu YW, Pryce E, Whited K, McCaffery JM, et al. Unremodeled and remodeled cardiolipin are functionally indistinguishable in yeast. J Biol Chem. 2014;289(3):1768–1778. doi: 10.1074/jbc.M113.525733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth PG, Scholte HR, Berden JA, Van der Klei-Van Moorsel JM, Luyt-Houwen IE, Van’t Veer-Korthof ET, et al. An X-linked mitochondrial disease affecting cardiac muscle, skeletal muscle and neutrophil leucocytes. J Neurol Sci. 1983;62(1–3):327–355. doi: 10.1016/0022-510x(83)90209-5. [DOI] [PubMed] [Google Scholar]
- Barth PG, Wanders RJ, Vreken P, Janssen EA, Lam J, Baas F. X-linked cardioskeletal myopathy and neutropenia (Barth syndrome) (MIM 302060) J Inherit Metab Dis. 1999;22(4):555–567. doi: 10.1023/a:1005568609936. [DOI] [PubMed] [Google Scholar]
- Bayir H, Fadeel B, Palladino MJ, Witasp E, Kurnikov IV, Tyurina YY, et al. Apoptotic interactions of cytochrome c: redox flirting with anionic phospholipids within and outside of mitochondria. Biochim Biophys Acta. 2006;1757(5–6):648–659. doi: 10.1016/j.bbabio.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Belikova NA, Vladimirov YA, Osipov AN, Kapralov AA, Tyurin VA, Potapovich MV, et al. Peroxidase activity and structural transitions of cytochrome c bound to cardiolipin-containing membranes. Biochemistry. 2006;45(15):4998–5009. doi: 10.1021/bi0525573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belikova NA, Jiang J, Tyurina YY, Zhao Q, Epperly MW, Greenberger J, et al. Cardiolipin-specific peroxidase reactions of cytochrome C in mitochondria during irradiation-induced apoptosis. Int J Radiat Oncol Biol Phys. 2007;69(1):176–186. doi: 10.1016/j.ijrobp.2007.03.043. [DOI] [PubMed] [Google Scholar]
- Beranek A, Rechberger G, Knauer H, Wolinski H, Kohlwein SD, Leber R. Identification of a cardiolipin-specific phospholipase encoded by the gene CLD1 (YGR110W) in yeast. J Biol Chem. 2009;284(17):11572–11578. doi: 10.1074/jbc.M805511200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer K, Klingenberg M. ADP/ATP carrier protein from beef heart mitochondria has high amounts of tightly bound cardiolipin, as revealed by 31P nuclear magnetic resonance. Biochemistry. 1985;24(15):3821–3826. doi: 10.1021/bi00336a001. [DOI] [PubMed] [Google Scholar]
- Bione S, D’Adamo P, Maestrini E, Gedeon AK, Bolhuis PA, Toniolo D. A novel X-linked gene, G4.5. is responsible for Barth syndrome. Nat Genet. 1996;12(4):385–389. doi: 10.1038/ng0496-385. [DOI] [PubMed] [Google Scholar]
- Buckland AG, Kinkaid AR, Wilton DC. Cardiolipin hydrolysis by human phospholipases A2. The multiple enzymatic activities of human cytosolic phospholipase A2. Biochim Biophys Acta. 1998;1390(1):65–72. doi: 10.1016/s0005-2760(97)00170-7. [DOI] [PubMed] [Google Scholar]
- Cao J, Liu Y, Lockwood J, Burn P, Shi Y. A novel cardiolipin-remodeling pathway revealed by a gene encoding an endoplasmic reticulum-associated acyl-CoA:lysocardiolipin acyltransferase (ALCAT1) in mouse. J Biol Chem. 2004;279(30):31727–31734. doi: 10.1074/jbc.M402930200. [DOI] [PubMed] [Google Scholar]
- Cao J, Shen W, Chang Z, Shi Y. ALCAT1 is a polyglycerophospholipid acyltransferase potently regulated by adenine nucleotide and thyroid status. Am J Physiol Endocrinol Metab. 2009;296(4):E647–E653. doi: 10.1152/ajpendo.90761.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SC, Heacock PN, Clancey CJ, Dowhan W. The PEL1 gene (renamed PGS1) encodes the phosphatidylglycero-phosphate synthase of Saccharomyces cerevisiae. J Biol Chem. 1998a;273(16):9829–9836. doi: 10.1074/jbc.273.16.9829. [DOI] [PubMed] [Google Scholar]
- Chang SC, Heacock PN, Mileykovskaya E, Voelker DR, Dowhan W. Isolation and characterization of the gene (CLS1) encoding cardiolipin synthase in Saccharomyces cerevisiae. J Biol Chem. 1998b;273(24):14933–14941. doi: 10.1074/jbc.273.24.14933. [DOI] [PubMed] [Google Scholar]
- Christodoulou J, McInnes RR, Jay V, Wilson G, Becker LE, Lehotay DC, et al. Barth syndrome: clinical observations and genetic linkage studies. Am J Med Genet. 1994;50(3):255–264. doi: 10.1002/ajmg.1320500309. [DOI] [PubMed] [Google Scholar]
- Chu CT, Ji J, Dagda RK, Jiang JF, Tyurina YY, Kapralov AA, et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol. 2013;15(10):1197–1205. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke SL, Bowron A, Gonzalez IL, Groves SJ, Newbury-Ecob R, Clayton N, et al. Barth syndrome. Orphanet J Rare Dis. 2013;8:23. doi: 10.1186/1750-1172-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claypool SM. Cardiolipin, a critical determinant of mitochondrial carrier protein assembly and function. Biochim Biophys Acta. 2009;1788(10):2059–2068. doi: 10.1016/j.bbamem.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claypool SM, McCaffery JM, Koehler CM. Mitochondrial mislocalization and altered assembly of a cluster of Barth syndrome mutant tafazzins. J Cell Biol. 2006;174(3):379–390. doi: 10.1083/jcb.200605043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claypool SM, Oktay Y, Boontheung P, Loo JA, Koehler CM. Cardiolipin defines the interactome of the major ADP/ATP carrier protein of the mitochondrial inner membrane. J Cell Biol. 2008a;182(5):937–950. doi: 10.1083/jcb.200801152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claypool SM, Boontheung P, McCaffery JM, Loo JA, Koehler CM. The cardiolipin transacylase, tafazzin, associates with two distinct respiratory components providing insight into Barth syndrome. Mol Biol Cell. 2008b;19(12):5143–5155. doi: 10.1091/mbc.E08-09-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullis PR, Hope MJ, Tilcock CP. Lipid polymorphism and the roles of lipids in membranes. Chem Phys Lipids. 1986;40(2–4):127–144. doi: 10.1016/0009-3084(86)90067-8. [DOI] [PubMed] [Google Scholar]
- Daum G. Lipids of mitochondria. Biochim Biophys Acta. 1985;822(1):1–42. doi: 10.1016/0304-4157(85)90002-4. [DOI] [PubMed] [Google Scholar]
- Davey KM, Parboosingh JS, McLeod DR, Chan A, Casey R, Ferreira P, et al. Mutation of DNAJC19, a human homologue of yeast inner mitochondrial membrane co-chaperones, causes DCMA syndrome, a novel autosomal recessive Barth syndrome-like condition. J Med Genet. 2006;43(5):385–393. doi: 10.1136/jmg.2005.036657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kroon AI, Dolis D, Mayer A, Lill R, de Kruijff B. Phospholipid composition of highly purified mitochondrial outer membranes of rat liver and Neurospora crassa. Is cardiolipin present in the mitochondrial outer membrane? Biochim Biophys Acta. 1997;1325(1):108–116. doi: 10.1016/s0005-2736(96)00240-4. [DOI] [PubMed] [Google Scholar]
- Dennis EA, Cao J, Hsu YH, Magrioti V, Kokotos G. Phospholipase A2 enzymes: physical structure, biological function, disease implication, chemical inhibition, and therapeutic intervention. Chem Rev. 2011;111(10):6130–6185. doi: 10.1021/cr200085w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzugasova V, Obernauerova M, Horvathova K, Vachova M, Zakova M, Subik J. Phosphatidylglycerolphosphate synthase encoded by the PEL1/PGS1 gene in Saccharomyces cerevisiae is localized in mitochondria and its expression is regulated by phospholipid precursors. Curr Genet. 1998;34(4):297–302. doi: 10.1007/s002940050399. [DOI] [PubMed] [Google Scholar]
- Eble KS, Coleman WB, Hantgan RR, Cunningham CC. Tightly associated cardiolipin in the bovine heart mitochondrial ATP synthase as analyzed by 31P nuclear magnetic resonance spectroscopy. J Biol Chem. 1990;265(32):19434–19440. [PubMed] [Google Scholar]
- Gadd ME, Broekemeier KM, Crouser ED, Kumar J, Graff G, Pfeiffer DR. Mitochondrial iPLA2 activity modulates the release of cytochrome c from mitochondria and influences the permeability transition. J Biol Chem. 2006;281(11):6931–6939. doi: 10.1074/jbc.M510845200. [DOI] [PubMed] [Google Scholar]
- Gawrisch K. Tafazzin senses curvature. Nat Chem Biol. 2012;8(10):811–812. doi: 10.1038/nchembio.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebert N, Joshi AS, Kutik S, Becker T, McKenzie M, Guan XL, et al. Mitochondrial cardiolipin involved in outer-membrane protein biogenesis: implications for Barth syndrome. Curr Biol. 2009;19(24):2133–2139. doi: 10.1016/j.cub.2009.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genova ML, Baracca A, Biondi A, Casalena G, Faccioli M, Falasca AI, et al. Is supercomplex organization of the respiratory chain required for optimal electron transfer activity? Biochim Biophys Acta. 2008;1777(7–8):740–746. doi: 10.1016/j.bbabio.2008.04.007. [DOI] [PubMed] [Google Scholar]
- Gohil VM, Greenberg ML. Mitochondrial membrane biogenesis: phospholipids and proteins go hand in hand. J Cell Biol. 2009;184(4):469–472. doi: 10.1083/jcb.200901127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohil VM, Hayes P, Matsuyama S, Schagger H, Schlame M, Greenberg ML. Cardiolipin biosynthesis and mitochondrial respiratory chain function are interdependent. J Biol Chem. 2004;279(41):42612–42618. doi: 10.1074/jbc.M402545200. [DOI] [PubMed] [Google Scholar]
- Gu Z, Valianpour F, Chen S, Vaz FM, Hakkaart GA, Wanders RJ, et al. Aberrant cardiolipin metabolism in the yeast taz1 mutant: a model for Barth syndrome. Mol Microbiol. 2004;51(1):149–158. doi: 10.1046/j.1365-2958.2003.03802.x. [DOI] [PubMed] [Google Scholar]
- Han X, Yang J, Cheng H, Yang K, Abendschein DR, Gross RW. Shotgun lipidomics identifies cardiolipin depletion in diabetic myocardium linking altered substrate utilization with mitochondrial dysfunction. Biochemistry. 2005;44(50):16684–16694. doi: 10.1021/bi051908a. [DOI] [PubMed] [Google Scholar]
- Han X, Yang J, Yang K, Zhao Z, Abendschein DR, Gross RW. Alterations in myocardial cardiolipin content and composition occur at the very earliest stages of diabetes: a shotgun lipidomics study. Biochemistry. 2007;46(21):6417–6428. doi: 10.1021/bi7004015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harner ME, Unger AK, Izawa T, Walther DM, Ozbalci C, Geimer S, et al. Aim24 and MICOS modulate respiratory function, tafazzinrelated cardiolipin modification and mitochondrial architecture. Elife (Cambridge) 2014;3:e01684. doi: 10.7554/eLife.01684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Han X. Cardiolipin remodeling in diabetic heart. Chem Phys Lipids. 2014;179:75–81. doi: 10.1016/j.chemphyslip.2013.10.007. [DOI] [PubMed] [Google Scholar]
- Hirschberg CB, Kennedy EP. Mechanism of the enzymatic synthesis of cardiolipin in Escherichia coli. Proc Natl Acad Sci U S A. 1972;69(3):648–651. doi: 10.1073/pnas.69.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostetler KY, van den Bosch H, van Deenen LL. The mechanism of cardiolipin biosynthesis in liver mitochondria. Biochim Biophys Acta. 1972;260(3):507–513. doi: 10.1016/0005-2760(72)90065-3. [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Vaz FM. Cardiolipin, the heart of mitochondrial metabolism. Cell Mol Life Sci. 2008;65(16):2493–2506. doi: 10.1007/s00018-008-8030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Turkenburg M, Poll-The BT, Karall D, Perez-Cerda C, Morrone A, et al. The enigmatic role of tafazzin in cardiolipin metabolism. Biochim Biophys Acta. 2009a;1788(10):2003–2014. doi: 10.1016/j.bbamem.2009.07.009. [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Rodenburg RJ, Thiels C, van Lenthe H, Stet F, Poll-The BT, et al. Cardiolipin and monolysocardiolipin analysis in fibroblasts, lymphocytes, and tissues using high-performance liquid chromatography-mass spectrometry as a diagnostic test for Barth syndrome. Anal Biochem. 2009b;387(2):230–237. doi: 10.1016/j.ab.2009.01.032. [DOI] [PubMed] [Google Scholar]
- Hovius R, Lambrechts H, Nicolay K, de Kruijff B. Improved methods to isolate and subfractionate rat liver mitochondria. Lipid composition of the inner and outer membrane. Biochim Biophys Acta. 1990;1021(2):217–226. doi: 10.1016/0005-2736(90)90036-n. [DOI] [PubMed] [Google Scholar]
- Hsu YH, Dumlao DS, Cao J, Dennis EA. Assessing phospholipase A2 activity toward cardiolipin by mass spectrometry. PLoS ONE. 2013;8(3):e59267. doi: 10.1371/journal.pone.0059267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iverson SL, Enoksson M, Gogvadze V, Ott M, Orrenius S. Cardiolipin is not required for Bax-mediated cytochrome c release from yeast mitochondria. J Biol Chem. 2004;279(2):1100–1107. doi: 10.1074/jbc.M305020200. [DOI] [PubMed] [Google Scholar]
- Jiang F, Rizavi HS, Greenberg ML. Cardiolipin is not essential for the growth of Saccharomyces cerevisiae on fermentable or non-fermentable carbon sources. Mol Microbiol. 1997;26(3):481–491. doi: 10.1046/j.1365-2958.1997.5841950.x. [DOI] [PubMed] [Google Scholar]
- Jiang F, Ryan MT, Schlame M, Zhao M, Gu Z, Klingenberg M, et al. Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. J Biol Chem. 2000;275(29):22387–22394. doi: 10.1074/jbc.M909868199. [DOI] [PubMed] [Google Scholar]
- Joshi AS, Zhou J, Gohil VM, Chen S, Greenberg ML. Cellular functions of cardiolipin in yeast. Biochim Biophys Acta. 2009;1793(1):212–218. doi: 10.1016/j.bbamcr.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AS, Thompson MN, Fei N, Huttemann M, Greenberg ML. Cardiolipin and mitochondrial phosphatidylethanolamine have overlapping functions in mitochondrial fusion in Saccharomyces cerevisiae. J Biol Chem. 2012;287(21):17589–17597. doi: 10.1074/jbc.M111.330167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan VE, Tyurin VA, Jiang J, Tyurina YY, Ritov VB, Amoscato AA, et al. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat Chem Biol. 2005;1(4):223–232. doi: 10.1038/nchembio727. [DOI] [PubMed] [Google Scholar]
- Kagan VE, Bayir HA, Belikova NA, Kapralov O, Tyurina YY, Tyurin VA, et al. Cytochrome c/cardiolipin relations in mitochondria: a kiss of death. Free Radic Biol Med. 2009;46(11):1439–1453. doi: 10.1016/j.freeradbiomed.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalanxhi E, Wallace CJ. Cytochrome c impaled: investigation of the extended lipid anchorage of a soluble protein to mitochondrial membrane models. Biochem J. 2007;407(2):179–187. doi: 10.1042/BJ20070459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly BL, Greenberg ML. Characterization and regulation of phosphatidylglycerolphosphate phosphatase in Saccharomyces cerevisiae. Biochim Biophys Acta. 1990;1046(2):144–150. doi: 10.1016/0005-2760(90)90181-v. [DOI] [PubMed] [Google Scholar]
- Kiebish MA, Yang K, Liu X, Mancuso DJ, Guan S, Zhao Z, et al. Dysfunctional cardiac mitochondrial bioenergetic, lipidomic, and signaling in a murine model of Barth syndrome. J Lipid Res. 2013;54(5):1312–1325. doi: 10.1194/jlr.M034728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg M. Cardiolipin and mitochondrial carriers. Biochim Biophys Acta. 2009;1788(10):2048–2058. doi: 10.1016/j.bbamem.2009.06.007. [DOI] [PubMed] [Google Scholar]
- Kutik S, Rissler M, Guan XL, Guiard B, Shui G, Gebert N, et al. The translocator maintenance protein Tam41 is required for mitochondrial cardiolipin biosynthesis. J Cell Biol. 2008;183(7):1213–1221. doi: 10.1083/jcb.200806048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lands WE. Metabolism of glycerolipids. 2. The enzymatic acylation of lysolecithin. J Biol Chem. 1960;235:2233–2237. [PubMed] [Google Scholar]
- Lange C, Nett JH, Trumpower BL, Hunte C. Specific roles of protein-phospholipid interactions in the yeast cytochrome bc1 complex structure. EMBO J. 2001;20(23):6591–6600. doi: 10.1093/emboj/20.23.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecocq J, Ballou CE. On the structure of cardiolipin. Biochemistry. 1964;3:976–980. doi: 10.1021/bi00895a023. [DOI] [PubMed] [Google Scholar]
- Lewis RN, McElhaney RN. The physicochemical properties of cardiolipin bilayers and cardiolipin-containing lipid membranes. Biochim Biophys Acta. 2009;1788(10):2069–2079. doi: 10.1016/j.bbamem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- Li J, Romestaing C, Han X, Li Y, Hao X, Wu Y, et al. Cardiolipin remodeling by ALCAT1 links oxidative stress and mitochondrial dysfunction to obesity. Cell Metab. 2010;12(2):154–165. doi: 10.1016/j.cmet.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liu X, Wang H, Zhang W, Chan DC, Shi Y. Lysocardiolipin acyltransferase 1 (ALCAT1) controls mitochondrial DNA fidelity and biogenesis through modulation of MFN2 expression. Proc Natl Acad Sci U S A. 2012;109(18):6975–6980. doi: 10.1073/pnas.1120043109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ye B, Miller S, Yuan H, Zhang H, Tian L, et al. Ablation of ALCAT1 mitigates hypertrophic cardiomyopathy through effects on oxidative stress and mitophagy. Mol Cell Biol. 2012;32(21):4493–4504. doi: 10.1128/MCB.01092-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma BJ, Taylor WA, Dolinsky VW, Hatch GM. Acylation of monolysocardiolipin in rat heart. J Lipid Res. 1999;40(10):1837–1845. [PubMed] [Google Scholar]
- Ma L, Vaz FM, Gu Z, Wanders RJ, Greenberg ML. The human TAZ gene complements mitochondrial dysfunction in the yeast taz1Delta mutant. Implications for Barth syndrome. J Biol Chem. 2004;279(43):44394–44399. doi: 10.1074/jbc.M405479200. [DOI] [PubMed] [Google Scholar]
- Malhotra A, Xu Y, Ren M, Schlame M. Formation of molecular species of mitochondrial cardiolipin. 1. A novel transacylation mechanism to shuttle fatty acids between sn-1 and sn-2 positions of multiple phospholipid species. Biochim Biophys Acta. 2009a;1791(4):314–320. doi: 10.1016/j.bbalip.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra A, Edelman-Novemsky I, Xu Y, Plesken H, Ma J, Schlame M, et al. Role of calcium-independent phospholipase A2 in the pathogenesis of Barth syndrome. Proc Natl Acad Sci U S A. 2009b;106(7):2337–2341. doi: 10.1073/pnas.0811224106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso DJ, Sims HF, Han X, Jenkins CM, Guan SP, Yang K, et al. Genetic ablation of calcium-independent phospholipase A2gamma leads to alterations in mitochondrial lipid metabolism and function resulting in a deficient mitochondrial bioenergetic phenotype. J Biol Chem. 2007;282(48):34611–34622. doi: 10.1074/jbc.M707795200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso DJ, Kotzbauer P, Wozniak DF, Sims HF, Jenkins CM, Guan S, et al. Genetic ablation of calcium-independent phospholipase A2 {gamma} leads to alterations in hippocampal cardiolipin content and molecular species distribution, mitochondrial degeneration, autophagy, and cognitive dysfunction. J Biol Chem. 2009;284(51):35632–35644. doi: 10.1074/jbc.M109.055194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie M, Lazarou M, Thorburn DR, Ryan MT. Mitochondrial respiratory chain supercomplexes are destabilized in Barth Syndrome patients. J Mol Biol. 2006;361(3):462–469. doi: 10.1016/j.jmb.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Mileykovskaya E, Dowhan W. Cardiolipin-dependent formation of mitochondrial respiratory supercomplexes. Chem Phys Lipids. 2014;179:42–48. doi: 10.1016/j.chemphyslip.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF. Barth syndrome may be due to an acyltransferase deficiency. Curr Biol. 1997;7(8):R465–R466. doi: 10.1016/s0960-9822(06)00237-5. [DOI] [PubMed] [Google Scholar]
- Nomura K, Imai H, Koumura T, Kobayashi T, Nakagawa Y. Mitochondrial phospholipid hydroperoxide glutathione peroxidase inhibits the release of cytochrome c from mitochondria by suppressing the peroxidation of cardiolipin in hypoglycaemia-induced apoptosis. Biochem J. 2000;351(Pt 1):183–193. doi: 10.1042/0264-6021:3510183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala T, Polinati P, Manninen T, Hiippala A, Rajantie J, Karikoski R, et al. New mutation of mitochondrial DNAJC19 causing dilated and noncompaction cardiomyopathy, anemia, ataxia, and male genital anomalies. Pediatr Res. 2012;72(4):432–437. doi: 10.1038/pr.2012.92. [DOI] [PubMed] [Google Scholar]
- Osman C, Haag M, Wieland FT, Brugger B, Langer T. A mitochondrial phosphatase required for cardiolipin biosynthesis: the PGP phosphatase Gep4. EMBO J. 2010;29(12):1976–1987. doi: 10.1038/emboj.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrander DB, Sparagna GC, Amoscato AA, McMillin JB, Dowhan W. Decreased cardiolipin synthesis corresponds with cytochrome c release in palmitate-induced cardiomyocyte apoptosis. J Biol Chem. 2001;276(41):38061–38067. doi: 10.1074/jbc.M107067200. [DOI] [PubMed] [Google Scholar]
- Ott M, Robertson JD, Gogvadze V, Zhivotovsky B, Orrenius S. Cytochrome c release from mitochondria proceeds by a two-step process. Proc Natl Acad Sci U S A. 2002;99(3):1259–1263. doi: 10.1073/pnas.241655498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M, Zhivotovsky B, Orrenius S. Role of cardiolipin in cytochrome c release from mitochondria. Cell Death Differ. 2007;14(7):1243–1247. doi: 10.1038/sj.cdd.4402135. [DOI] [PubMed] [Google Scholar]
- Palsdottir H, Lojero CG, Trumpower BL, Hunte C. Structure of the yeast cytochrome bc1 complex with a hydroxyquinone anion Qo site inhibitor bound. J Biol Chem. 2003;278(33):31303–31311. doi: 10.1074/jbc.M302195200. [DOI] [PubMed] [Google Scholar]
- Pan HJ, Lin Y, Chen YE, Vance DE, Leiter EH. Adverse hepatic and cardiac responses to rosiglitazone in a new mouse model of type 2 diabetes: relation to dysregulated phosphatidylcholine metabolism. Vasc Pharmacol. 2006;45(1):65–71. doi: 10.1016/j.vph.2005.11.011. [DOI] [PubMed] [Google Scholar]
- Pangborn MC. The composition of cardiolipin. J Biol Chem. 1947;168(1):351–361. [PubMed] [Google Scholar]
- Paradies G, Petrosillo G, Pistolese M, Ruggiero FM. The effect of reactive oxygen species generated from the mitochondrial electron transport chain on the cytochrome c oxidase activity and on the cardiolipin content in bovine heart submitochondrial particles. FEBS Lett. 2000;466(2–3):323–326. doi: 10.1016/s0014-5793(00)01082-6. [DOI] [PubMed] [Google Scholar]
- Paradies G, Petrosillo G, Pistolese M, Ruggiero FM. Reactive oxygen species generated by the mitochondrial respiratory chain affect the complex III activity via cardiolipin peroxidation in beef-heart submitochondrial particles. Mitochondrion. 2001;1(2):151–159. doi: 10.1016/s1567-7249(01)00011-3. [DOI] [PubMed] [Google Scholar]
- Paradies G, Petrosillo G, Pistolese M, Ruggiero FM. Reactive oxygen species affect mitochondrial electron transport complex I activity through oxidative cardiolipin damage. Gene. 2002;286(1):135–141. doi: 10.1016/s0378-1119(01)00814-9. [DOI] [PubMed] [Google Scholar]
- Paradies G, Petrosillo G, Paradies V, Ruggiero FM. Role of cardiolipin peroxidation and Ca2+ in mitochondrial dysfunction and disease. Cell Calcium. 2009;45(6):643–650. doi: 10.1016/j.ceca.2009.03.012. [DOI] [PubMed] [Google Scholar]
- Paradies G, Paradies V, De Benedictis V, Ruggiero FM, Petrosillo G. Functional role of cardiolipin in mitochondrial bioenergetics. Biochim Biophys Acta. 2014;1837(4):408–417. doi: 10.1016/j.bbabio.2013.10.006. [DOI] [PubMed] [Google Scholar]
- Patil VA, Fox JL, Gohil VM, Winge DR, Greenberg ML. Loss of cardiolipin leads to perturbation of mitochondrial and cellular iron homeostasis. J Biol Chem. 2013;288(3):1696–1705. doi: 10.1074/jbc.M112.428938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer K, Gohil V, Stuart RA, Hunte C, Brandt U, Greenberg ML, et al. Cardiolipin stabilizes respiratory chain supercomplexes. J Biol Chem. 2003;278(52):52873–52880. doi: 10.1074/jbc.M308366200. [DOI] [PubMed] [Google Scholar]
- Raja V, Greenberg ML. The functions of cardiolipin in cellular metabolism-potential modifiers of the Barth syndrome phenotype. Chem Phys Lipids. 2014;179:49–56. doi: 10.1016/j.chemphyslip.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren M, Phoon CK, Schlame M. Metabolism and function of mitochondrial cardiolipin. Prog Lipid Res. 2014;55C:1–16. doi: 10.1016/j.plipres.2014.04.001. [DOI] [PubMed] [Google Scholar]
- Richter-Dennerlein R, Korwitz A, Haag M, Tatsuta T, Dargazanli S, Baker M, et al. DNAJC19, a Mitochondrial Cochaperone Associated with Cardiomyopathy, Forms a Complex with Prohibitins to Regulate Cardiolipin Remodeling. Cell Metab. 2014;20(1):158–171. doi: 10.1016/j.cmet.2014.04.016. [DOI] [PubMed] [Google Scholar]
- Samhan-Arias AK, Ji J, Demidova OM, Sparvero LJ, Feng W, Tyurin V, et al. Oxidized phospholipids as biomarkers of tissue and cell damage with a focus on cardiolipin. Biochim Biophys Acta. 2012;1818(10):2413–2423. doi: 10.1016/j.bbamem.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlame M, Ren M. The role of cardiolipin in the structural organization of mitochondrial membranes. Biochim Biophys Acta. 2009;1788(10):2080–2083. doi: 10.1016/j.bbamem.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlame M, Rua D, Greenberg ML. The biosynthesis and functional role of cardiolipin. Prog Lipid Res. 2000;39(3):257–288. doi: 10.1016/s0163-7827(00)00005-9. [DOI] [PubMed] [Google Scholar]
- Schlame M, Towbin JA, Heerdt PM, Jehle R, DiMauro S, Blanck TJ. Deficiency of tetralinoleoyl-cardiolipin in Barth syndrome. Ann Neurol. 2002;51(5):634–637. doi: 10.1002/ana.10176. [DOI] [PubMed] [Google Scholar]
- Schlame M, Kelley RI, Feigenbaum A, Towbin JA, Heerdt PM, Schieble T, et al. Phospholipid abnormalities in children with Barth syndrome. J Am Coll Cardiol. 2003;42(11):1994–1999. doi: 10.1016/j.jacc.2003.06.015. [DOI] [PubMed] [Google Scholar]
- Schlame M, Ren M, Xu Y, Greenberg ML, Haller I. Molecular symmetry in mitochondrial cardiolipins. Chem Phys Lipids. 2005;138(1–2):38–49. doi: 10.1016/j.chemphyslip.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Schlame M, Acehan D, Berno B, Xu Y, Valvo S, Ren M, et al. The physical state of lipid substrates provides transacylation specificity for tafazzin. Nat Chem Biol. 2012;8(10):862–869. doi: 10.1038/nchembio.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug ZT, Gottlieb E. Cardiolipin acts as a mitochondrial signalling platform to launch apoptosis. Biochim Biophys Acta. 2009;1788(10):2022–2031. doi: 10.1016/j.bbamem.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Seleznev K, Zhao C, Zhang XH, Song K, Ma ZA. Calcium-independent phospholipase A2 localizes in and protects mitochondria during apoptotic induction by staurosporine. J Biol Chem. 2006;281(31):22275–22288. doi: 10.1074/jbc.M604330200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzawa-Itoh K, Aoyama H, Muramoto K, Terada H, Kurauchi T, Tadehara Y, et al. Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J. 2007;26(6):1713–1725. doi: 10.1038/sj.emboj.7601618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinibaldi F, Fiorucci L, Patriarca A, Lauceri R, Ferri T, Coletta M, et al. Insights into cytochrome c-cardiolipin interaction. Role played by ionic strength. Biochemistry. 2008;47(26):6928–6935. doi: 10.1021/bi800048v. [DOI] [PubMed] [Google Scholar]
- Soustek MS, Falk DJ, Mah CS, Toth MJ, Schlame M, Lewin AS, et al. Characterization of a transgenic short hairpin RNA-induced murine model of Tafazzin deficiency. Hum Gene Ther. 2011;22(7):865–871. doi: 10.1089/hum.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamai KT, Greenberg ML. Biochemical characterization and regulation of cardiolipin synthase in Saccharomyces cerevisiae. Biochim Biophys Acta. 1990;1046(2):214–222. doi: 10.1016/0005-2760(90)90192-z. [DOI] [PubMed] [Google Scholar]
- Tamura Y, Harada Y, Nishikawa S, Yamano K, Kamiya M, Shiota T, et al. Tam41 is a CDP-diacylglycerol synthase required for cardiolipin biosynthesis in mitochondria. Cell Metab. 2013;17(5):709–718. doi: 10.1016/j.cmet.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan BK, Bogdanov M, Zhao J, Dowhan W, Raetz CR, Guan Z. Discovery of a cardiolipin synthase utilizing phosphatidylethanolamine and phosphatidylglycerol as substrates. Proc Natl Acad Sci U S A. 2012;109(41):16504–16509. doi: 10.1073/pnas.1212797109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor WA, Hatch GM. Purification and characterization of monolysocardiolipin acyltransferase from pig liver mitochondria. J Biol Chem. 2003;278(15):12716–12721. doi: 10.1074/jbc.M210329200. [DOI] [PubMed] [Google Scholar]
- Taylor WA, Hatch GM. Identification of the human mitochondrial linoleoyl-coenzyme A monolysocardiolipin acyltransferase (MLCL AT-1) J Biol Chem. 2009;284(44):30360–30371. doi: 10.1074/jbc.M109.048322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testet E, Laroche-Traineau J, Noubhani A, Coulon D, Bunoust O, Camougrand N, et al. Ypr140wp, ‘the yeast tafazzin’, displays a mitochondrial lysophosphatidylcholine (lyso-PC) acyltransferase activity related to triacylglycerol and mitochondrial lipid synthesis. Biochem J. 2005;387(Pt 3):617–626. doi: 10.1042/BJ20041491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian HF, Feng JM, Wen JF. The evolution of cardiolipin biosynthesis and maturation pathways and its implications for the evolution of eukaryotes. BMC Evol Biol. 2012;12:32. doi: 10.1186/1471-2148-12-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller G, Hrastnik C, Achleitner G, Schiefthaler U, Klein F, Daum G. YDL142c encodes cardiolipin synthase (Cls1p) and is non-essential for aerobic growth of Saccharomyces cerevisiae. FEBS Lett. 1998;421(1):15–18. doi: 10.1016/s0014-5793(97)01525-1. [DOI] [PubMed] [Google Scholar]
- Tyurina YY, Kini V, Tyurin VA, Vlasova II, Jiang J, Kapralov AA, et al. Mechanisms of cardiolipin oxidation by cytochrome c: relevance to pro- and antiapoptotic functions of etoposide. Mol Pharmacol. 2006;70(2):706–717. doi: 10.1124/mol.106.022731. [DOI] [PubMed] [Google Scholar]
- Valianpour F, Wanders RJ, Barth PG, Overmars H, van Gennip AH. Quantitative and compositional study of cardiolipin in platelets by electrospray ionization mass spectrometry: application for the identification of Barth syndrome patients. Clin Chem. 2002;48(9):1390–1397. [PubMed] [Google Scholar]
- Vreken P, Valianpour F, Nijtmans LG, Grivell LA, Plecko B, Wanders RJ, et al. Defective remodeling of cardiolipin and phosphatidylglycerol in Barth syndrome. Biochem Biophys Res Commun. 2000;279(2):378–382. doi: 10.1006/bbrc.2000.3952. [DOI] [PubMed] [Google Scholar]
- Watkins SM, Reifsnyder PR, Pan HJ, German JB, Leiter EH. Lipid metabolome-wide effects of the PPARgamma agonist rosiglitazone. J Lipid Res. 2002;43(11):1809–1817. doi: 10.1194/jlr.m200169-jlr200. [DOI] [PubMed] [Google Scholar]
- Xiao J, Engel JL, Zhang J, Chen MJ, Manning G, Dixon JE. Structural and functional analysis of PTPMT1, a phosphatase required for cardiolipin synthesis. Proc Natl Acad Sci U S A. 2011;108(29):11860–11865. doi: 10.1073/pnas.1109290108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Kelley RI, Blanck TJ, Schlame M. Remodeling of cardiolipin by phospholipid transacylation. J Biol Chem. 2003;278(51):51380–51385. doi: 10.1074/jbc.M307382200. [DOI] [PubMed] [Google Scholar]
- Xu Y, Sutachan JJ, Plesken H, Kelley RI, Schlame M. Characterization of lymphoblast mitochondria from patients with Barth syndrome. Lab Investig. 2005;85(6):823–830. doi: 10.1038/labinvest.3700274. [DOI] [PubMed] [Google Scholar]
- Xu Y, Condell M, Plesken H, Edelman-Novemsky I, Ma J, Ren M, et al. A Drosophila model of Barth syndrome. Proc Natl Acad Sci U S A. 2006;103(31):11584–11588. doi: 10.1073/pnas.0603242103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhang S, Malhotra A, Edelman-Novemsky I, Ma J, Kruppa A, et al. Characterization of tafazzin splice variants from humans and fruit flies. J Biol Chem. 2009;284(42):29230–29239. doi: 10.1074/jbc.M109.016642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu FY, McBride H, Acehan D, Vaz FM, Houtkooper RH, Lee RM, et al. The dynamics of cardiolipin synthesis post-mitochondrial fusion. Biochim Biophys Acta. 2010;1798(8):1577–1585. doi: 10.1016/j.bbamem.2010.04.007. [DOI] [PubMed] [Google Scholar]
- Yamashita A, Sugiura T, Waku K. Acyltransferases and transacylases involved in fatty acid remodeling of phospholipids and metabolism of bioactive lipids in mammalian cells. J Biochem. 1997;122(1):1–16. doi: 10.1093/oxfordjournals.jbchem.a021715. [DOI] [PubMed] [Google Scholar]
- Ye C, Lou W, Li Y, Chatzispyrou IA, Huttemann M, Lee I, et al. Deletion of the cardiolipin-specific phospholipase Cld1 rescues growth and life span defects in the tafazzin mutant: implications for Barth syndrome. J Biol Chem. 2014;289(6):3114–3125. doi: 10.1074/jbc.M113.529487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Mileykovskaya E, Dowhan W. Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J Biol Chem. 2002;277(46):43553–43556. doi: 10.1074/jbc.C200551200. [DOI] [PubMed] [Google Scholar]
- Zhang J, Guan Z, Murphy AN, Wiley SE, Perkins GA, Worby CA, et al. Mitochondrial phosphatase PTPMT1 is essential for cardiolipin biosynthesis. Cell Metab. 2011;13(6):690–700. doi: 10.1016/j.cmet.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinser E, Sperka-Gottlieb CD, Fasch EV, Kohlwein SD, Paltauf F, Daum G. Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J Bacteriol. 1991;173(6):2026–2034. doi: 10.1128/jb.173.6.2026-2034.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]