Abstract
Background
Rice mutant, spl5 (spotted leaf 5), has spontaneous hypersensitive-like lesions on its leaves and shows enhanced resistance to pathogens, indicating that SPL5 plays a role in programmed cell death (PCD) and disease resistance. To understand the molecular mechanism of SPL5 gene, we investigated the transcriptome profiles of the spl5 mutant leaves with few lesions (FL) and leaves with many lesions (ML) compared to the wild-type (WT) leaves respectively by microarray.
Results
The data from microarray revealed that 243 and 896 candidate genes (Fold change ≥ 3.0) were up- or down-regulated in the spl5-FL and spl5-ML, respectively, and a large number of these genes involved in biotic defense responses or reactive oxygen species (ROS) metabolism. Interestingly, according to our microarray and real-time PCR assays, the expressions of a transcription factor OsWRKY14 and genes responsible for the biosynthesis of serotonin, anthranilate synthase (AS), indole-3-glycerolphosphate synthase (IGPS), tryptophan synthase (TS) and tryptophan decarboxylase (TDC) were significantly up-regulated in the spl5 mutant. It has been reported previously that TS and TDC expressions are regulated by OsWRKY14 in rice, which raises the possibility that OsWRKY14 regulates serotonin production through the up-regulation of TS and TDC. Our HPLC analysis further confirmed that serotonin levels were higher in the leaves of spl5 mutant than that in WT.
Conclusions
Since the serotonin plays a critical role in inducing disease-resistance, the increased serotonin level may contribute, at least partly, to the disease resistance in spl5. The SPL5 gene may act as a negative regulatory factor activating the serotonin metabolic pathway, and these results might provide a new insight into the spl5-induced defense response mechanisms in plants.
Electronic supplementary material
The online version of this article (doi:10.1186/s12284-015-0052-7) contains supplementary material, which is available to authorized users.
Keywords: spl5, Lesion mimic, Disease resistance, Microarray, Serotonin, Rice
Background
In response to pathogen attack, plants have evolved an elaborate defense system with a complex signaling network. One of the most efficient resistance responses in plants is the hypersensitive response (HR), which is characterized by the rapid induction of local cell death around the infection site (Morel and Dangl 1997). Previous research into the molecular mechanisms behind HR has led to the discovery of mutants that display HR-like cell death in plant species such as Arabidopsis (Lorrain et al. 2003), maize (Johal et al. 1995), barley (Wolter et al. 1993), rice (Takahashi et al. 1999; Yin et al. 2000; Mizobuchi et al. 2002), wheat (Nair and Tomar 2001) and groundnut (Badigannavar et al. 2002). These mutants are referred to as lesion mimic mutants (lmms) because they spontaneously induce cell death and exhibit HR-like lesions in the absence of pathogen attack (Moeder and Yoshioka 2008). Many lmms spontaneously activate immune responses, such as reactive oxygen species (ROS) bursts, callose deposition and the induction of pathogenesis-related (PR) genes (Staskawicz et al. 1995). Therefore, the lmms can be used to investigate the molecular mechanisms behind HR and disease resistance in plants.
At least 43 lmms have been isolated in rice (Wu et al. 2008) and most show enhanced resistance to blast and/or bacterial blight pathogens (Jung et al. 2005; Mori et al. 2007; Qiao et al. 2010). Genetic analysis has indicated that the rice lmms phenotypes are mostly controlled by a single recessive or dominant gene (Huang et al. 2010) and many lmm genes have been cloned and characterized, such as spl7 (Yamanouchi et al. 2002), spl11 (Zeng et al. 2004), NPR1 (Chern et al. 2005), lsd1 (Wang et al. 2005), Spl18 (Mori et al. 2007), ttm1 (Takahashi et al. 2007), acdr1 (Kim et al. 2009), spl28 (Qiao et al. 2010), sl (Fujiwara et al. 2010), edr1 (Shen et al. 2011), rlin1 (Sun et al. 2011), lms (Jerwin et al. 2012) and spl5 (Chen et al. 2012). However, the proteins encoded by these LMM genes have different functions. For example, SPL7 is a heat stress transcription factor (Yamanouchi et al. 2002); SPL11 is an E3 ubiquitin ligase (Zeng et al. 2004); SPL28 is a clathrin-associated adaptor protein complex 1 medium subunit 1 (AP1M1), which is important in the post-Golgi trafficking pathway (Qiao et al. 2010). These findings indicate that numerous proteins, with distinct functions in multiple signaling pathways, are involved in the regulation of HR cell death and disease resistance.
The rice lesion mimic, spotted leaf 5 (spl5), created by γ-ray radiation, has spontaneous HR-like lesions on its leaves (Iwata et al. 1978) and shows enhanced resistance to rice blast and bacterial blight pathogens (Yin et al. 2000; Mizobuchi et al. 2002). Previously, we cloned the SPL5 gene using a map-based cloning strategy and showed that this gene encoded a novel protein that was homologous with human splicing factor 3b subunit 3 (SF3b3) (Chen et al. 2012). SF3b3 is a subunit of the SF3b multi-subunit complex, which is required, together with SF3a, when binding U2 snRNA to the branch site of pre-mRNA (Brosi et al. 1993; Das et al. 1999). Therefore, it is likely that SPL5 post-transcriptionally regulates cell death and resistance responses. According to our proteomic assay, many proteins involved in pre-mRNA splicing, amino acid metabolism, photosynthesis, glycolysis, ROS metabolism and defense responses were significantly up or down-regulated in the spl5 mutant (Chen et al. 2013). However, the molecular mechanisms controlling SPL5 and its signaling pathway have not been fully investigated.
Serotonin (5-hydroxytryptamine) is a well-known neurotransmitter in mammals and is widely distributed in plants (Pelagio-Flores et al. 2011). Recently, serotonin has been reported to activate intracellular defense mechanisms during immune responses by the rice lesion mimic mutant sl (Fujiwara et al. 2010). The sl mutant did not produce serotonin in its leaves and showed increased susceptibility to fungal infection, and treating the sl mutant with serotonin suppressed fungal growth; SL gene encodes a cytochrome P450 monooxygenase that has tryptamine 5-hydroxylase enzyme activity and catalyzes the conversion of tryptamine to serotonin (Fujiwara et al. 2010). These results indicated that activation of serotonin production is involved in the establishment of effective disease defenses in rice.
In this study, we compared the expression profiles of the spl5 mutant and the wild type by microarray analysis and found that many candidate genes were involved in defense response regulation. In particular, genes that encoded enzymes for serotonin biosynthesis were significantly up-regulated in the spl5 mutant. As a result, we also detected the over-accumulation of serotonin and its precursor, tryptophan, in the spl5 mutant. Previously, it had been reported that tryptophan and serotonin play direct roles in plant defense response regulation (Elaine et al. 1995). Therefore, we suggest that SPL5 may negatively regulate the biosynthesis of tryptophan and serotonin, which, in turn, affects defense responses in rice.
Results
Transcriptome profiles in the spl5 mutant
To investigate the effect of spl5 mutation on the genes expression in rice, we analyzed the transcriptome profiles in spl5-FL (few lesions) and spl5-ML (many lesions) and WT leaves. The results revealed that 243 (176 up-regulated; 67 down-regulated) and 896 (445 up-regulated; 451 down-regulated) genes were differentially expressed in the spl5-FL and spl5-ML compared to the WT, respectively (FC ≥ 3.0; Table 1; Additional file 1: Table S1; Additional file 2: Table S2). According to GO annotation, these genes could be classified into 20 different functional categories (Table 1). It was clear that the known-functional categories with the large number of differentially expressed genes were involved in defense response and oxidation-reduction process both in the spl5-FL and spl5-ML. Interestingly, among the differentially expressed genes, 208 genes were found to be both in spl5-FL and spl5-ML (Additional file 1: Table S1; Additional file 2: Table S2), and these 208 common genes may play important roles in SPL5 signaling and function. Genes that were probably associated with the spl5 phenotype are listed in Table 2.
Table 1.
Functional classification of differentially expressed genes identified by the microarray analysis
| Function type | spl5 -FL | spl5 -ML | a Common | |||
|---|---|---|---|---|---|---|
| Up | Down | Up | Down | Up | Down | |
| Defense response | 16 | 4 | 33 | 12 | 15 | 3 |
| Oxidation-reduction process | 13 | 6 | 33 | 35 | 12 | 5 |
| Response to stress | 5 | 1 | 14 | 5 | 4 | 1 |
| Cellular amino acid metabolic process | 6 | 2 | 15 | 6 | 6 | 1 |
| Cellular cell wall/membrance organization | 4 | 0 | 4 | 5 | 4 | 0 |
| Hormone-mediated signaling pathway | 4 | 2 | 8 | 10 | 4 | 0 |
| Fatty acid biosynthetic process | 7 | 3 | 12 | 10 | 6 | 1 |
| Development | 3 | 0 | 7 | 8 | 3 | 0 |
| Carbohydrate metabolic process | 4 | 1 | 16 | 18 | 4 | 1 |
| Transport | 6 | 3 | 17 | 21 | 6 | 2 |
| ATP biosynthetic process | 5 | 1 | 8 | 6 | 4 | 0 |
| Photosynthesis | 1 | 1 | 5 | 23 | 1 | 1 |
| RNA/DNA | 9 | 3 | 25 | 22 | 9 | 1 |
| Apoptosis | 1 | 0 | 2 | 0 | 1 | 0 |
| Transcription factor | 5 | 0 | 16 | 10 | 5 | 0 |
| Kinase | 11 | 5 | 32 | 14 | 10 | 3 |
| Signal transduction | 5 | 0 | 14 | 4 | 4 | 0 |
| Protein modification process | 6 | 0 | 15 | 15 | 6 | 0 |
| Metal ion | 2 | 2 | 3 | 7 | 2 | 2 |
| Others | 15 | 4 | 54 | 78 | 12 | 3 |
| Unknow | 48 | 29 | 112 | 142 | 43 | 23 |
| Total | 176 | 67 | 445 | 451 | 161 | 47 |
aDifferentially expressed genes which were both in spl5-FL and spl5-ML.
Table 2.
Differentially expressed genes that are likely to be associated with the spl5 phenotype according to the microarray analysis
| Function type | a Accession | b Annotation | c spl5 -FL/WT | d spl5 -ML/WT |
|---|---|---|---|---|
| Defense response | Os07g0251200 | Harpin-induced 1 domain containing protein | 22.04 | 25.34 |
| Os10g0464000 | Hypersensitive-induced response protein | 17.80 | 44.48 | |
| Os11g0514500 | Leucine-rich repeat-containing extracellular glycoprotein precursor | 11.75 | 13.44 | |
| Os01g0944900 | Beta-1,3-glucanase precursor | 8.75 | 15.88 | |
| Os01g0687400 | Chitinase | 8.50 | 11.16 | |
| Os12g0127200 | Harpin-induced 1 domain containing protein | 8.23 | 9.83 | |
| Os04g0349700 | Leucine-rich repeat, typical subtype containing protein | 7.80 | 4.08 | |
| Os03g0207400 | Protein phosphatase 2C-like | 5.67 | 6.58 | |
| Os06g0136000 | Hypersensitive-induced reaction protein 4 | 3.38 | 6.28 | |
| ROS metabolism | Os07g0665200 | Superoxide dismutase [Cu-Zn] 2 | 7.33 | 7.76 |
| Os04g0434800 | Aseorbate peroxidase 7 | 4.67 | 7.71 | |
| Os01g0962700 | Peroxidase 12 precursor | 4.58 | 4.21 | |
| Transcription factor | Os01g0730700 | WRKY transcription factor 14 | 11.77 | 18.74 |
| Os03g0335200 | WRKY transcription factor 17 | 5.66 | 7.45 | |
| Os03g0321700 | WRKY transcription factor 55 | 5.67 | 10.10 | |
| Amino acid metabolism | Os09g0255400 | Indole-3-glycerol phosphate synthase | 13.76 | 8.79 |
| Os08g0140500 | Tryptophan decarboxylase | 10.19 | 9.25 | |
| Os07g0182100 | Tryptophan synthase alpha chain | 5.22 | 4.49 | |
| Os03g0718000 | Anthranilate synthase beta chain | 3.64 | 5.10 |
aGenBank Accession (http://www.ncbi.nlm.nih.gov/); bFunction annotation; Fold change of gene expression between spl5-FLc or spl5-MLd to WT using the average normalized intensity of microarray.
Defense response
The expressions of many genes involved in the defense response, such as Chitinase and β-1, 3-glucanases, were induced in spl5 mutant (Table 2). Chitinase and β-1, 3-glucanasesare are the important hydrolytic enzymes in plants and show in vitro antifungal activity (Sela-Buurlage et al. 1993; Hwang et al. 2007). In addition, genes encoding harpin/hypersensitive-induced response protein, leucine-rich repeat (LRR) protein and protein phosphatase 2Cs (PP2Cs) were also induced in spl5 mutant (Table 2). These genes have been shown to play critical roles in the regulation of plant disease resistance (Choi et al. 2011; Andi et al. 2001; Hu et al. 2009).
ROS metabolism
ROS (O2− and H2O2) are toxic metabolic products that can effectively kill infected cells and activate the defense response in plants, but the over-accumulated ROS must be scavenged in time to avoid damage to other cells (Lee et al. 1999). Three genes encoding different ROS scavengers, Superoxide dismutase (SOD) [Cu-Zn] 2, Peroxidase (POD) 12 and Aseorbate peroxidase (APX) 7 were up-regulated in the spl5 mutant (Table 2). SOD is the first enzyme in the detoxification process which converts very harmful O2− into less reactive H2O2, then POD eliminate H2O2. APX is considered the most important H2O2 scavengers, using ascorbate as the reducing agent (Kim et al. 2012).
Transcription factor
The WRKY transcription factor gene family have been identified in a range of biological processes, and many WRKY genes are transcriptionally regulated under conditions of biotic and/or abiotic stress (Berri et al. 2009). Three WRKY genes, OsWRKY14, OsWRKY17 and OsWRKY55, were also induced in the spl5 mutant (Table 2). OsWRKY14 was a transcription factor which was also induced by environmental stresses and some plant hormones, such as: JA, ABA and ET (Yang 2007). OsWRKY17 can be induced under a number of adverse stresses, such as drought, cold damage and high temperature (Wang et al. 2012). OsWRKY55 was strongly induced by the rice blast fungus and may be a common component in the signal transduction pathway of defense response (Zhang et al. 2008). These transcription factors may follow in the signal transduction of SPL5 for regulation of some candidate genes expressions in rice.
Amino acid metabolism
Four genes involved in the biosynthesis of tryptophan and serotonin were up-regulated in the spl5 mutant (Table 2). They are anthranilate synthase (AS), indole-3-glycerolphosphate synthase (IGPS), tryptophan synthase (TS) and tryptophan decarboxylase (TDC). It is known that AS catalyzes chorismate to anthranilate, and IGPS catalyzes anthranilate to indole-3-glycerol phosphate (IGP); then TS catalyzes IGP to tryptophan, which forms serotonin catalyzed by TDC. Recent research showed that OsWRKY14 was a transcription factor for TS and TDC gene in the regulation of the serotonin biosynthetic pathway in rice (Kang et al. 2011). This gene was also up-regulated in spl5 mutant (Table 2).
Serotonin biosynthesis was enhanced in the spl5 mutant
It is likely that the biosynthesis pathway of serotonin was enhanced in the spl5 mutant. To verify the microarray results and to improve our hypothesis, we analyzed the expression profiles of genes OsWRKY14, AS, IGPS, TS and TDC by real-time PCR (Figure 1), and detected the level of tryptophan and serotonin by High-performance liquid chromatography (HPLC) (Figure 2), in the leaves of WT, spl5-NL (No lesion), spl5-FL and spl5-ML, respectively.
Figure 1.
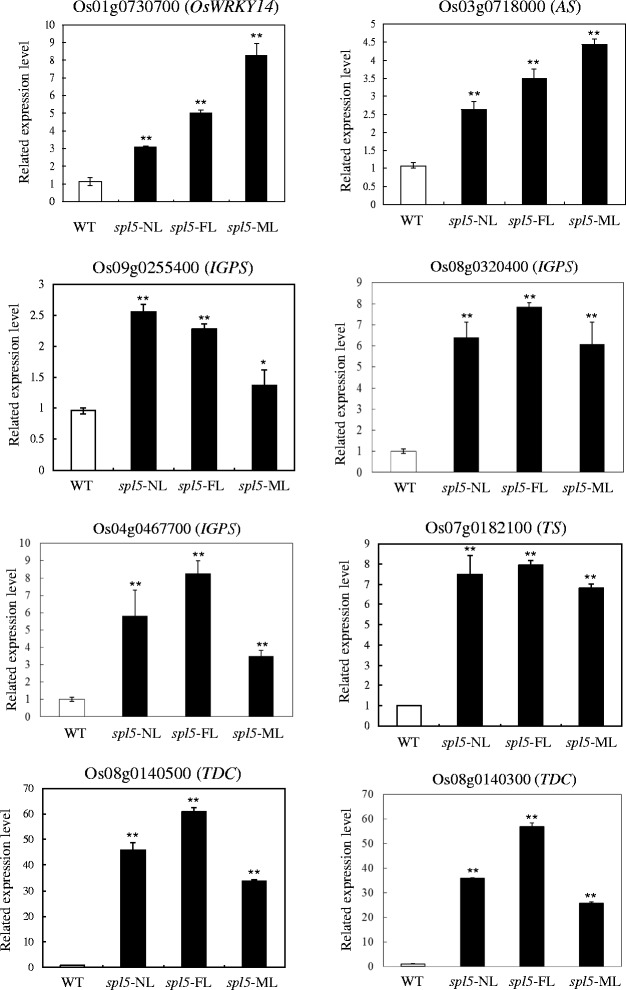
Expressions of OsWRKY14, AS, IGPS, TS and TDC by real-time PCR analysis. The OsWRKY14, AS (anthranilate synthase), IGPS (indole-3-glycerolphosphate synthase), TS (tryptophan synthase) and TDC (tryptophan decarboxylase) genes expressions are shown for the WT leaves and the spl5 leaves with different degrees of lesion development: NL (no lesions), FL (few lesions) and ML (many lesions). The accession number of gene is from the NCBI database (http://www.ncbi.nlm.nih.gov/). The significance of expression compared to WT with the P value less than 0.05 and 0.01 are marked by * and **, respectively.
Figure 2.
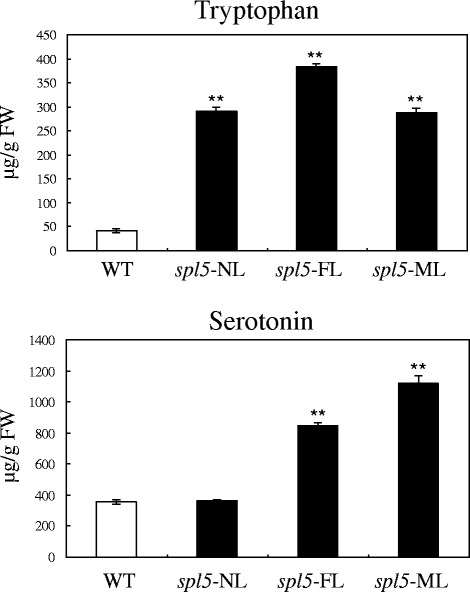
In vivo tryptophan and serotonin levels by HPLC analysis. The tryptophan and serotonin concentrations are shown for the WT leaves and the spl5-NL, spl5-FL, and spl5-ML leaves, respectively. The significance of expression compared to WT with the P value less than 0.05 and 0.01 are marked by * and **, respectively.
OsWRKY14, AS, IGPS, TS, and TDC expressions by real-time PCR
According to the results of real-time PCR (Figure 1), these five genes of OsWRKY14 (Os01g0730700), AS (Os03g0718000), IGPS (Os09g0255400), TS (Os07g0182100) and TDC (Os08g0140500) were all significantly induced in the spl5-FL and -ML compared to the WT, and this result is consistent with the microarray data. Even in the spl5-NL leaves with no lesions, the expressions of the five genes were higher than that in WT. However, except for OsWRKY14, the protein of AS, IGPS, TS or TDC is not encoded by a single gene in the rice genome. According to the NCBI database (http://www.ncbi.nlm.nih.gov/), we found that there are additional 3 AS, 2 IGPS, 8 TS and 1 TDC genes in rice genome (Additional file 3: Table S3). The expression of these 14 genes were also analyzed in the rice WT and spl5 mutant by real-time PCR. As shown in Figure 1, all additional IGPS (Os08g0320400, Os04g0467700) and TDC (Os08g0140300) genes were induced in spl5 mutant (Figure 1), but only 1 out 3 AS and 4 out of 8 TS genes were significantly induced in the spl5 mutant, compared to the WT respectively (Additional file 4: Figure S1).
Serotonin and tryptophan level analysis by HPLC
The serotonin and its precursor tryptophan levels were also determined in spl5-NL, spl5-FL, spl5-ML and WT by HPLC. Figure 2 shows that the tryptophan levels significantly increased in spl5 leaves, including spl5-NL where there were no lesions. Except in spl5-NL, serotonin levels also significantly increased in spl5-FL and -ML compared to the WT, and it is likely that the catalytic reaction from tryptophan to serotonin was not actively increased in spl5-NL. This result was consistent with the induced or enhanced expressions of AS, IGPS and TS genes in the tryptophan and serotonin biosynthetic pathway, and the lesion mimic phenotype of spl5 mutant was positive correlated with the content of serotonin.
Discussion and conclusions
Lesion mimic mutants, which display HR-like cell death and enhance disease resistance, are useful genetic tools for study on the molecular mechanisms of HR and disease resistance in plants. Though many lesion mimic genes have been cloned in rice (Yamanouchi et al. 2002; Zeng et al. 2004; Kim et al. 2009; Qiao et al. 2010; Shen et al. 2011; Jerwin et al. 2012), the signal pathways of most these genes have not been reported up till now. Here, in order to reveal the signal pathway of SPL5 gene in regulation of disease resistance, we analyzed the transcriptional profiling of spl5 mutant and WT using the microarray. Totally, 243 and 896 up- or down-regulated genes (FC ≥ 3.0) were identified from spl5-FL and spl5-ML, respectively (Additional file 1: Table S1; Additional file 2: Table S2). Among them, there were many genes involved in defense response (Chitinase, β-1, 3-glucanases), ROS metabolism (SOD, POD and APX) or transcription regulation in stress response (OsWRKY14, OsWRKY17, OsWRKY55). We speculated that these may involve in the SPL5 mediated resistance in rice.
We also have clearly demonstrated that the serotonin biosynthetic pathway was up-regulated in the spl5 mutant. Firstly, the expression of genes involved in this pathway: AS, IGPS, TS and TDC were significantly induced in spl5 (Figure 1) and secondly, the tryptophan and serotonin concentration increased in the spl5 leaves (Figure 2). Interestingly, another rice lesion-mimic mutant sl was from the mutation of SL gene, which encodes a cytochrome P450 monooxygenase and catalyzes biosynthesis of serotonin (Fujiwara et al. 2010). Therefore, the expression of SL gene was examined in spl5 mutant, and results showed this gene also significantly induced in the spl5 mutant compared to the WT (Figure 3), suggesting that SL may contribute to the increased accumulation of serotonin in spl5 mutant plants. It has been reported that the tryptophan pathway plays a direct role in regulating plant defense responses, plant-insect interactions and plant development (Elaine et al. 1995). Serotonin is one of the most important secondary metabolites from tryptophan, and has been implicated in several important physiological and developmental functions, such as senescence, flowering and seed germination (Kang et al. 2009; Murch et al. 2001; Ishihara et al. 2008). Recent research showed that in infected rice leaves, serotonin can serve as a substrate for peroxidase in the presence of hydrogen peroxide, forming a complex mixture of oligomerics that function as a physical barrier against the spread of pathogen infections (Ishihara et al. 2011). Treating the sl mutant with serotonin it can effectively suppress the growth of fungal, and activate the expression of some resistance genes, such as probenazol 1 (PBZ1), phenylalanine ammonia-lyase 1 (PAL1), chitinase 1 (Cht1) and chitinase 3 (Cht3) (Fujiwara et al. 2010). In our previous research, we have also proved that the OsChib2a was increased in spl5 mutant (Chen et al. 2013). In order to further confirm whether the defense responses were activated in spl5 mutant, the expression of OsPR1a, a marker gene of systemic acquired resistance (Durrant and Dong, 2004), was tested by our Real-time PCR (Figure 3). The result showed that the OsPR1a was also induced in spl5 mutant. So, it is likely that the serotonin may play a key role in the defense responses of spl5 mutant. In addition, the microarray data analysis showed that OsWRKY14, a key transcription factor for serotonin biosynthesis, was also induced in the spl5 mutant. Based on our experimental results, we have proposed a model for the serotonin biosynthetic signaling pathway in rice that is mediated by the SPL5 gene (Figure 4). In this model, the SPL5 gene may act as a negative regulatory factor activating the serotonin metabolic pathway, which was mediated by OsWRKY14. The accumulation of serotonin may lead to pathogen resistance in rice. However, we could not confirm if the SPL5 gene mutation affects other biological pathways that trigger similar phenotypes. We have shown that the SPL5 gene encodes a SF3b3 protein that presumably has a role in the pre-mRNA splicing process (Chen et al. 2012). So far, there have been no reports about SF3b3 being involved in rice defense responses. The spl5 mutant is a useful tool that can be used to study the mechanisms behind SF3b3 defense regulation in plants and to apply in molecular breeding for crop disease resistance.
Figure 3.
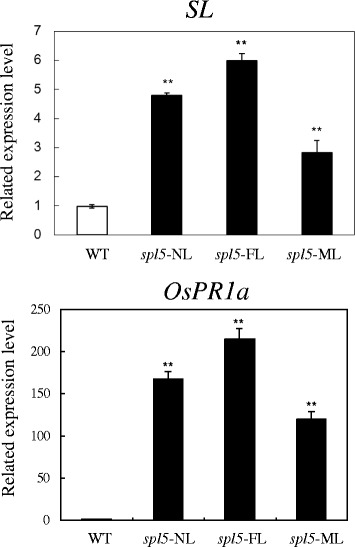
SL and OsPR1a expressions by real-time PCR analysis. The SL and OsPR1a gene expressions are shown for the spl5 leaf parts with different lesion-mimic numbers and for WT. The significance of expression compared to WT with the P value less than 0.05 and 0.01 are marked by * and **, respectively.
Figure 4.
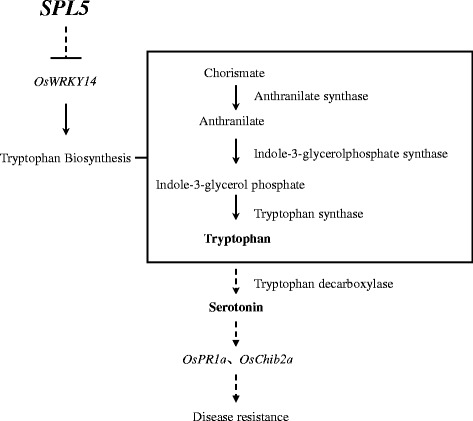
A hypothetical model for the SPL5 signaling pathway in rice. The SPL5 may act as a negative regulatory factor activating the serotonin metabolic pathway, which is mediated by OsWRKY14. The accumulation of serotonin may lead to pathogen resistance in rice.
Methods
Plant materials
Rice seeds from the spl5 mutant and its wild-type (WT) control, Zhefu802 (a near-isogenic line of spl5), were germinated in an incubator at 28°C, transferred to the nutrient solution used by Yoshida (Yoshida et al. 1976), and then cultivated in a growth chamber at 28/24°C (12 h day/12 h night). The nutrient solution was maintained at pH 5.6 and refreshed every 5 d. At 60 days old, the fully developed leaves were collected from each spl5 mutant and WT plant. Further, the leaf blades of spl5 mutant were split into three groups, based on the degree of lesion formation: no lesion (NL), leaf area without any lesions; few lesions (FL), leaf area with 10–20% lesions and many lesions (ML), leaf area with 70–80% lesions as the method described by Chen (Chen et al. 2013). All the leaf tissues were immediately frozen in liquid nitrogen and stored at −80°C.
RNA extraction
Total RNAs of leaves were extracted using TRIzol Reagent (Life technologies, USA) following the manufacturer’s instructions, and further purified by RNeasy mini kit (QIAGEN, Germany) and RNase-Free DNase Set (QIAGEN).
Microarray assay
RNAs Samples were analyzed using Affymetrix (USA) rice 44 k gene chips with two biological replicates by the Shanghai Biotechnology Co. Ltd. China. The data of this microarray was deposited in GEO database (http://www.ncbi.nlm.nih.gov/geo/), and the accession number is GSE61952. The signal intensity emitted by each probe on the microarray was scanned using a GeneChip® Scanner 3000 and analyzed with Command Console Software 3.1 using the default settings. The raw data for all the arrays were normalized by the MAS 5.0 algorithm using Gene Spring Software 11.0. Genes with a fold change (FC) of ≥ 3.0 between the spl5 mutant and the WT were identified as the differential expression genes, but those which have poor microarray signals with the Flag value of A (absent) or with the normalized intensity ≤ 500 were manually eliminated. Gene function prediction was carried out using the NCBI database (http://www.ncbi.nlm.nih.gov/gene). The Gene Ontology database (http://www.geneontology.org/) was used for gene functional classification.
Real-time PCR
The leaf total RNAs were isolated by TRIzol Reagent and treated with RNase-free DNase I (Promega, USA) to eliminate any contamination by genomic DNA. The first-strand synthesis of the cDNAs was carried out by M-MLV Reverse Transcriptase (Promega, USA) according to the manufacturer’s instructions. Real-time PCR was performed by a Step One™ Real-Time PCR System (Applied Biosystems, USA) using the Fast SYBR Green Master Mix reagent (Applied Biosystems). The thermal cycle used was as follows: 95°C for 20 s; 40 cycles of 95°C for 3 s and 60°C for 30 s. A rice housekeeping gene Actin (GenBank accession: X16280) was used for the standardization control, and the primer pair was 5′-TGGCATCTCTCAGCACATTCC-3′ and 5′-TGCACAATG GATGGGTCAGA-3′. The gene-specific primers for the candidate genes used in the real-time PCR analysis are listed in Table 3 and Additional file 3: Table S3. Each sample was independently tested by three times. Finally, the real-time PCR data was analyzed using the delta-delta Ct method (Livak and Schmittgen 2001).
Table 3.
Gene-specific primers used for the real-time PCR
| Gene | a Accession | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|---|
| AS | Os03g0718000 | GGCTCCTCCCCAAGATCCAGTCC | TGCGTTTTCACCTTCCACCACAG |
| IGPS | Os09g0255400 | CGCCGCCTCTTCCTCTCTC | GGACTTGCCGCTCTCCCAC |
| TS | Os07g0182100 | AGCTGTGGCTGTTGGGTTCGGTAT | GCTTCTTCAATCCTTCTTCGGGTG |
| TDC | Os08g0140500 | TCAAGAACCACGCCAGCGACTC | GTAGGTGCGCATGACCATCCAG |
| OsWRKY14 | Os01g0730700 | AGCACAACCACTCCGCCAC | CCTCCTCCCATCTCCAGCC |
| OsPR1a | Os07g0129200 | TATGCTATGCTACGTGTTTATGC | CACTAAGCAAATACGGCTGACA |
aGenBank Accession (http://www.ncbi.nlm.nih.gov/).
HPLC analysis
For each sample, 100 mg leaf tissue was ground with liquid nitrogen into a powder and soaked in 2 ml 100% methanol. The homogenates were centrifuged at 10,000 × g for 10 min and the supernatant was filtered through a syringe with a 0.2 μm cellulose acetate membrane filter (Pall, USA). Then the filtrate was evaporated to dryness under vacuum and dissolved in 500 μl 50% methanol. The final sample was analyzed by reversed-phase HPLC (Waters, USA) so that the tryptophan and serotonin contents could be quantified. The samples were separated on an XTerra RP C18 column (250 × 4.6 mm, 5 μm, Waters) with an isocratic elution of 50% methanol in water containing 0.3% trifluoroacetic acid at a flow rate of 0.4 ml/min. A UV wavelength of 280 nm was used for detection. The standard samples for tryptophan and serotonin were made by Sigma (USA).
Acknowledgements
We thank Dr. Jianzhong Liu and Dr. Kewei Zhang for critical reading of this manuscript. This work was supported by the National Key Programs for Transgenic Crops (2014ZX08009003-001); the National Natural Science Foundation of China (31171519, 31371593); the Natural Science Foundation of Zhejiang Province (LZ14C130001, LY13C130008, LY14C130004).
Abbreviations
- spl5
Spotted leaf 5
- HR
Hypersensitive response
- lmms
Lesion mimic mutants
- ROS
Reactive oxygen species
- PR
Pathogenesis-related
- WT
Wild type
- NL
No lesion
- FL
Few lesions
- ML
Many lesions
- AS
Anthranilate synthase
- IGPS
Indole-3-glycerolphosphate synthase
- TS
Tryptophan synthase
- TDC
Tryptophan decarboxylase
- HPLC
High-performance liquid chromatography
- SOD
Superoxide dismutase
- POD
Peroxidase
- APX
Ascorbate peroxidase
- FC
Fold change
Additional files
Differentially expressed genes between spl5-FL with WT (FC ≥ 3.0).
Differentially expressed genes between spl5-ML with WT (FC ≥ 3.0).
Gene-specific primers of additional AS, IGPS, TS and TDC genes used for real-time PCR.
Gene expressions of additional AS and TS by real-time PCR.
Footnotes
Bin Jin and Xinru Zhou contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JB (Jin B.) and ZX carried out the microarray analysis and drafted the manuscript. JB (Jiang B.) carried out the Real-time PCR, and participated in preparation of data. GZ participated in the design of the study and performed the statistical analysis. ZP carried out the assay of HPLC. QQ provided the plant materials of this study. CX and MB conceived the study, and participated in its design and coordination and revised the manuscript. All authors read and approved the final manuscript.
Contributor Information
Bin Jin, Email: 659195358@qq.com.
Xinru Zhou, Email: yorisep9098@163.com.
Baolin Jiang, Email: 1051493958@qq.com.
Zhimin Gu, Email: zhmgu@zjnu.cn.
Pinghua Zhang, Email: zph@zjnu.cn.
Qian Qian, Email: qianqian188@hotmail.com.
Xifeng Chen, Email: xfchen@zjnu.cn.
Bojun Ma, Email: mbj@zjnu.cn.
References
- Andi S, Taguchi F, Toyoda K, Shiraishi T, Ichinose Y. Effect of methyl jasmonate on harpin-induced hypersensitive cell death, generation of hydrogen peroxide and expression of PAL mRNA in tobacco suspension cultured BY-2 cells. Plant Cell Physiol. 2001;42:446–449. doi: 10.1093/pcp/pce056. [DOI] [PubMed] [Google Scholar]
- Badigannavar AM, Kale DM, Eapen S, Murty GSS. Inheritance of disease lesion mimic leaf trait in groundnut. J Hered. 2002;93:50–52. doi: 10.1093/jhered/93.1.50. [DOI] [PubMed] [Google Scholar]
- Berri S, Abbruscato P, Faivre-Rampant O, Brasileiro AC, Fumasoni I, Satoh K, Kikuchi S, Mizzi L, Morandini P, Pè ME, Piffanelli P. Characterization of WRKY co-regulatory networks in rice and Arabidopsis. BMC Plant Biol. 2009;9:120. doi: 10.1186/1471-2229-9-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosi R, Groning K, Behrens SE, Luhrmann R, Kramer A. Interaction of mammalian splicing factor SF3a with U2 snRNP and relation of its 60-kD subunit to yeast PRP9. Science. 1993;262:102–105. doi: 10.1126/science.8211112. [DOI] [PubMed] [Google Scholar]
- Chen X, Hao L, Pan J, Zheng X, Jiang G, Jin Y, Gu Z, Qian Q, Zhai W, Ma B. SPL5, a cell death and defense-related gene, encodes a putative splicing factor 3b subunit 3 (SF3b3) in rice. Mol Breeding. 2012;30:939–949. doi: 10.1007/s11032-011-9677-4. [DOI] [Google Scholar]
- Chen X, Fu S, Zhang P, Gu Z, Liu J, Qian Q, Ma B. Proteomic analysis of a disease-resistance-enhanced lesion mimic mutant spotted leaf 5 in rice. Rice. 2013;6:1. doi: 10.1186/1939-8433-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chern M, Fitzgerald HA, Canlas PE, Navarre DA, Ronald PC. Overexpression of a rice NPR1 homolog leads to constitutive activation of defense response and hypersensitivity to light. Mol Plant Microbe Interact. 2005;18:511–520. doi: 10.1094/MPMI-18-0511. [DOI] [PubMed] [Google Scholar]
- Choi HW, Kim YJ, Hwang BK. The hypersensitive induced reaction and leucine-rich repeat proteins regulate plant cell death associated with disease and plant immunity. Am Phytopathol Soc. 2011;24:68–78. doi: 10.1094/MPMI-02-10-0030. [DOI] [PubMed] [Google Scholar]
- Das BK, Xia L, Palandjian L, Gozani O, Chyung Y, Reed R. Characterization of a protein complex containing spliceosomal proteins SAPs 49, 130, 145, and 155. Mol Cell Biol. 1999;19:6796–6802. doi: 10.1128/mcb.19.10.6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant WE, Dong X. Systemic acquired resistance. Annu Rev Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- Elaine R, Radwanski L, Robert L. Tryptophan biosynthesis and metabolism: biochemical and molecular genetics. Plant Cell. 1995;7:921–934. doi: 10.1105/tpc.7.7.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Maisonneuve S, Isshiki M, Mizutani M, Chen L, Wong HL, Kawasaki T, Shimamoto K. Sekiguchi lesiongene encodes a cytochrome P450 monooxygenase that catalyzes conversion of tryptamine to serotonin in rice. J Biol Chem. 2010;285:11308–11313. doi: 10.1074/jbc.M109.091371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu XB, Zhang HJ, Li GJ, Yang YX, Zheng Z, Song FM. Ectopic expression of a rice protein phosphatase 2C gene OsBIPP2C2 in tobacco improves disease resistance. Plant Cell. 2009;28:985–995. doi: 10.1007/s00299-009-0701-7. [DOI] [PubMed] [Google Scholar]
- Huang QN, Yang Y, Shi YF, Chen J, Wu JL. Recent advances in research on spotted-leaf mutants of rice (Oryza sativa) Rice. 2010;24:108–115. [Google Scholar]
- Hwang DH, Kim ST, Kim SG, Kang KY. Comprehensive analysis of the expression of twenty-seven β-1, 3-glucanase genes in rice (Oryza sativa L.) Mol Cells. 2007;23:207–214. [PubMed] [Google Scholar]
- Ishihara A, Hashimoto Y, Tanaka C, Dubouzet JG, Nakao T, Matsuda F, Nishioka T, Miyagawa H, Wakasa K. The tryptophan pathway is involved in the defense responses of rice against pathogenic infection via serotonin production. Plant J. 2008;54:481–495. doi: 10.1111/j.1365-313X.2008.03441.x. [DOI] [PubMed] [Google Scholar]
- Ishihara A, Nakao T, Mashimo Y, Murai M, Ichimaru N, Tanaka C, Nakajima H, Wakasa K, Miyagawa H. Probing the role of tryptophan-derived secondary metabolism in defense responses against Bipolaris oryzae infection in rice leaves by a suicide substrate of tryptophan decarboxylase. Phytochemistry. 2011;72:7–13. doi: 10.1016/j.phytochem.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Iwata N, Omura T, Satoh H. Linkage studies in rice (Oryza sativa L.) on some mutants for physiological leaf spots. J Fac Agr Kyushu Univ. 1978;22:243–251. [Google Scholar]
- Jerwin R, Muluneh T, Akira A, Kentaro Y, Shunichi K, Hiroki T, Kakoto Y, Hiroyuki K, Hiromasa S, Rym F, Shailendra S, Jesusa U, Masahiro Y, Ryohei T. Mutation in OsLMS, a gene encoding a protein with two double-stranded RNA binding motifs, causes lesion mimic phenotype and early senescence in rice (Oryza sativa L.) Genes Genet. 2012;87:169–179. doi: 10.1266/ggs.87.169. [DOI] [PubMed] [Google Scholar]
- Johal GS, Hulbert S, Briggs SP. Disease lesion mimics of maize: a model for cell death in plants. Bio Essays. 1995;17:685–692. [Google Scholar]
- Jung YH, Lee JH, Agrawal GK, Rakwal R, Kim JA, Shim JK, Lee SK, Jeon JS, Koh HJ, Lee YH, Iwahashi H, Jwa NS. The rice (oryza sativa) blast lesion mimic mutant, blm, may confer resistance to blast pathogens by triggering multiple defense-associated signaling pathways. Plant Physiol Biochem. 2005;43:397–406. doi: 10.1016/j.plaphy.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Kang K, Kim YS, Park S, Back K. Senescence-induced serotonin biosynthesis and its role in delaying senescence in rice leaves. Plant Physiol. 2009;150:1380–1393. doi: 10.1104/pp.109.138552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K, Park S, Natsagdorj U, Kim YS, Back K. Methanol is an endogenous elicitor molecule for the synthesis of tryptophan and tryptophan-derived secondary metabolites upon senescence of detached rice leaves. Plant J. 2011;66:247–257. doi: 10.1111/j.1365-313X.2011.04486.x. [DOI] [PubMed] [Google Scholar]
- Kim JA, Cho K, Singh R, Jung YH, Jeong SH, Kim SH, Lee JE, Cho YS, Agrawal GK, Rakwal R, Tamogami S, Kersten B, Jeon JS, An G, Jwa NS. Rice OsACDR1 (Oryza sativa accelerated cell death and resistance 1 is a potential positive regulator of fungal disease resistance. Mol Cells. 2009;28:431–439. doi: 10.1007/s10059-009-0161-5. [DOI] [PubMed] [Google Scholar]
- Kim SH, Song M, Lee KJ, Hwang SG, Jang CS, Kim JB, Kim SH, Ha BK, Kang SY, Kim DS. Genome-wide transcriptome profiling of ROS scavenging and signal transduction pathways in rice (Oryza sativa L.) in response to different types of ionizing radiation. Mol Biol Rep. 2012;39:11231–11248. doi: 10.1007/s11033-012-2034-9. [DOI] [PubMed] [Google Scholar]
- Lee SC, Kang BG, Oh SE. Induction of ascorbate peroxidase isoenzymes by ethylene and hydrogen peroxide during growth of cultured soybean cells. Mol Cells. 1999;9:166–171. [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lorrain S, Vailleau F, Balague C, Roby D. Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants. Trends Plant Sci. 2003;8:263–271. doi: 10.1016/S1360-1385(03)00108-0. [DOI] [PubMed] [Google Scholar]
- Mizobuchi R, Hirabayashi H, Kaji R, Nishizawa Y, Yoshimura A, Satoh H, Ogawa T, Okamoto M. Isolation and characterization of rice lesion mimic mutants with enhanced resistance to rice blast and bacterial blight. Plant Sci. 2002;163:345–353. doi: 10.1016/S0168-9452(02)00134-6. [DOI] [Google Scholar]
- Moeder W, Yoshioka K. Lesion mimic mutants: A classical, yet still fundamental approach to study programmed cell death. Plant Signal Behav. 2008;3(10):764–767. doi: 10.4161/psb.3.10.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel JB, Dangl JL. The hypersensitive response and the induction of cell death in plants. Cell Death Differ. 1997;4:671–683. doi: 10.1038/sj.cdd.4400309. [DOI] [PubMed] [Google Scholar]
- Mori M, Tomita C, Sugimoto K, Hasegawa M, Hayashi N, Dubouzet JG, Ochiai H, Sekimoto H, Hirochika H, Kikuchi S. Isolation and molecular characterization of a Spotted leaf 18 mutant by modified activation-tagging in rice. Plant Mol Biol. 2007;63:847–860. doi: 10.1007/s11103-006-9130-y. [DOI] [PubMed] [Google Scholar]
- Murch SJ, Campbell SSB, Saxena P. The role of serotonin and melatonin in plant morphogenesis: regulation of auxin-induced root organogenesis in in vitro-cultured explants of St. John’s wort (Hypericum perforatum L.) Cell Dev Biol Plant. 2001;37:786–793. doi: 10.1007/s11627-001-0130-y. [DOI] [Google Scholar]
- Nair SK, Tomar SMS. Genetical and anatomical analyses of a leaf flecking mutant in wheat. Euphytica. 2001;121:53–58. doi: 10.1023/A:1012061016783. [DOI] [Google Scholar]
- Pelagio-Flores R, Ortız-Castro R, Mendez-Bravo A, Macıas-Rodrıguez L, Lopez-Bucio J. Serotonin, a tryptophan-derived signal conserved in plants and animals, regulates root system architecture probably acting as a natural auxin inhibitor in Arabidopsis thaliana. Plant Cell Physiol. 2011;52(3):490–508. doi: 10.1093/pcp/pcr006. [DOI] [PubMed] [Google Scholar]
- Qiao Y, Jiang W, Lee J, Park B, Choi MS, Piao R, Woo MO, Roh JH, Han L, Paek NC, Seo HS, Koh HJ. SPL28 encodes a clathrin-associated adaptor protein complex 1, medium subunit micro 1 (AP1M1) and is responsible for spotted leaf and early senescence in rice (oryza sativa) New Phytol. 2010;185:258–274. doi: 10.1111/j.1469-8137.2009.03047.x. [DOI] [PubMed] [Google Scholar]
- Sela-Buurlage MB, Ponstein AS, Bres-Vloemans SA, Melchers LS, van den Elzen PJM, Comelissen BJC. Only specific tobacco (nicotiana tabacum) chitinases and [beta]-1,3-glucanase exhibit antifungal activity. Plant Physiol. 1993;101:857–863. doi: 10.1104/pp.101.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Liu H, Yuan B, Li X, Xu C, Wang S. OsEDR1 negatively regulates rice bacterial resistance via activation of ethylene biosynthesis. Plant Cell Environ. 2011;34:179–191. doi: 10.1111/j.1365-3040.2010.02219.x. [DOI] [PubMed] [Google Scholar]
- Staskawicz BJ, Ausubel FM, Baker BJ, Ellis JG, Jones JD. Molecular genetics of plant disease resistance. Science. 1995;268:661–667. doi: 10.1126/science.7732374. [DOI] [PubMed] [Google Scholar]
- Sun C, Liu L, Tang J, Lin A, Zhang F, Fang J, Zhang G, Chu C. RLIN1, encoding a putative coproporphyrinogen III oxidase, is involved in lesion initiation in rice. J Genet Genomics. 2011;38:29–37. doi: 10.1016/j.jcg.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Takahashi A, Kawasaki T, Henmi K, Shii K, Kodama O, Satoh H, Shimamoto K. Lesion mimic mutants of rice with alterations in early signaling events of defense. Plant J. 1999;17:535–5453. doi: 10.1046/j.1365-313X.1999.00405.x. [DOI] [PubMed] [Google Scholar]
- Takahashi A, Agrawal GK, Yamazaki M, Onosato K, Miyao A, Kawasaki T, Shimamoto K, Hirochika H. Rice Pti1a negatively regulates RAR1-dependent defense responses. Plant Cell. 2007;19:2940–2951. doi: 10.1105/tpc.106.047142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Pei Z, Tian Y, He C. OsLSD1, a rice zinc finger protein, regulates programmed cell death and callus differentiation. Mol Plant Microbe Interact. 2005;18:375–384. doi: 10.1094/MPMI-18-0375. [DOI] [PubMed] [Google Scholar]
- Wang XL, Tang X, Liu ZY. Construction of OsWRKY17 specific expression vector in rice. J Age Sci Tech-Iran. 2012;13:79–81. [Google Scholar]
- Wolter M, Hollricher K, Salaminip F, Schulze-Lefert P. The mlo resistance alleles to powdery mildew infection in barley trigger a developmentally controlled defense mimic phenotype. Mol Gen Genet. 1993;239:122–128. doi: 10.1007/BF00281610. [DOI] [PubMed] [Google Scholar]
- Wu C, Bordeos A, Madamba MR, Baraoidan M, Ramos M, Wang GL, Leach JE, Leung H. Rice lesion mimic mutants with enhanced resistance to diseases. Mol Genet Genomics. 2008;279:605–619. doi: 10.1007/s00438-008-0337-2. [DOI] [PubMed] [Google Scholar]
- Yamanouchi U, Yano M, Lin H, Ashikari M, Yamada K. A rice spotted leaf gene, Spl7, encodes a heat stress transcription factor protein. Proc Natl Acad Sci U S A. 2002;99:7530–7535. doi: 10.1073/pnas.112209199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WY (2007) Separation of rice OsWRKY14 and preliminary analysis of gene function. Dissertation, China Agricultural University
- Yin Z, Chen J, Zeng L, Goh M, Leung H, Khush GS, Wang GL. Characterizing rice lesion mimic mutants and identifying a mutant with broad-spectrum resistance to rice blast and bacterial blight. Mol Plant Microbe Interact. 2000;13:869–876. doi: 10.1094/MPMI.2000.13.8.869. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Forno DA, Cook JH, Gomes KA. Routine procedure for growing rice plants in culture solution. Los Banos, Philippines: IRRI Laboratory manual for physiological studies of rice; 1976. [Google Scholar]
- Zeng L, Qu S, Bordeos A, Yang C, Baraoidan M, Yan H, Xie Q, Nahm BH, Leung H, Wang G. Spotted leaf 11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell. 2004;16:2795–2808. doi: 10.1105/tpc.104.025171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Peng YL, Guo ZJ. Constitutive expression of pathogen-inducible OsWRKY31 enhances disease resistance and affects root growth and auxin response in transgenic rice plants. Cell Res. 2008;18:508–521. doi: 10.1038/cr.2007.104. [DOI] [PubMed] [Google Scholar]


