Abstract
Aerobic glycolysis involves increased glycolysis and decreased oxidative catabolism of glucose even in the presence of an ample oxygen supply. Aerobic glycolysis, a common metabolic pattern in cancer cells, was recently discovered in both the healthy and diseased human brain, but its functional significance is not understood. This metabolic pattern in the brain is surprising because it results in decreased efficiency of adenosine triphosphate (ATP) production in a tissue with high energetic demands. We report that highly aggressive honey bees (Apis mellifera) show a brain transcriptomic and metabolic state consistent with aerobic glycolysis, i.e. increased glycolysis in combination with decreased oxidative phosphorylation. Furthermore, exposure to alarm pheromone, which provokes aggression, causes a metabolic shift to aerobic glycolysis in the bee brain. We hypothesize that this metabolic state, which is associated with altered neurotransmitter levels, increased glycolytically derived ATP and a reduced cellular redox state, may lead to increased neuronal excitability and oxidative stress in the brain. Our analysis provides evidence for a robust, distinct and persistent brain metabolic response to aggression-inducing social cues. This finding for the first time associates aerobic glycolysis with naturally occurring behavioral plasticity, which has important implications for understanding both healthy and diseased brain function.
Keywords: Aerobic glycolysis, aggression, brain metabolism, metabolomics, neurogenomics, transcriptomics
Aerobic glycolysis (AG) is characterized by elevated glycolysis relative to oxidative phosphorylation despite adequate oxygen availability to completely metabolize glucose to carbon dioxide and water (Vaishnavi et al. 2010). This metabolic shift leads to a relative decrease in the efficiency of adenosine triphosphate (ATP) production as the cell becomes more reliant on glycolysis-derived ATP (Pfeiffer et al. 2001). Aerobic glycolysis, also known as the Warburg effect, is observed in cancer cells and is predicted to provide precursors for cellular biomass production (Lunt & Vander Heiden 2011; Warburg 1956). Another common site of AG is the healthy adult human brain; brain activity is characterized not only by increased blood flow and glucose uptake in particular brain regions, but also by a shift in metabolic flux toward AG (Fox et al. 1988; Madsen et al. 1995; Phelps & Barrio 2010). Previous studies estimate that 10–12% of glucose taken up by cells in the brain is metabolized by AG (Boyle et al. 1994; Raichle et al. 1970). Given the high energetic demands for neural function, it is surprising to see this less efficient metabolic state (Goyal et al. 2014). Aerobic glycolysis varies regionally throughout the vertebrate brain, though the causes of this variation are unknown (Vaishnavi et al. 2010). Furthermore, there is a correlation between AG and beta amyloid plaque deposition (a symptom of Alzheimer’s Disease), and aberrant AG has been linked to Huntington’s Disease (Powers et al. 2007; Vlassenko et al. 2010). Thus AG is critical for brain health, but its function is unresolved (Goyal et al. 2014). The study of brain AG would benefit from new experimental systems that are amenable to manipulation.
Here we evaluate evidence that natural variation in aggressive behavior is associated with a shift toward AG in the brain of the honey bee (Apis mellifera). A relationship between AG and a distinct, experimentally inducible and evolutionarily conserved behavioral phenotype like aggression in an insect model organism could provide a tractable system to study the neural signaling and cognitive outcomes of AG in the brain.
Previous studies in the honey bee associated heightened aggression with a distinct brain metabolic phenotype, specifically decreased expression and activity of genes and enzymes involved in oxidative phosphorylation (OXPhos; Alaux et al. 2009). A combination of pharmacological treatments in bees and genetic manipulations in fruit flies demonstrated a causal relationship between brain OXPhos inhibition and increased aggression (Li-Byarlay et al. 2014). In honey bees, topical treatments with insecticides that target OXPhos complex I and V enzymes resulted in a 50% increase in aggression in a small group behavioral assay, an effect that scales to substantial changes in aggressive response at the colony level. Furthermore, in Drosophila melanogaster, a modest degree of RNAi knockdown of a single OXPhos NADH dehydrogenase mRNA in neurons, but not in glia, resulted in an increase in male territorial aggression (Li-Byarlay et al. 2014). Taken together, these results provide strong evidence of a causal relationship between decreased OXPhos levels and increased aggression. Because whole-body metabolic rate increases for aggressively aroused bees (Harrison & Hall 1993), a negative relationship between aggression and OXPhos activity in the brain is surprising, and suggests that the brain has specific metabolic requirements when individuals are in a state of heightened aggression.
Prior work shows a consistent relationship between OXPhos and aggression, but no study has assessed the relationship between aggression and other pathways associated with energy metabolism. In the current study we reanalyzed microarray data and took new metabolomics measurements to evaluate the possibility that decreased OXPhos in the aggressive brain may be more accurately described as a shift toward AG. Furthermore, we used metabolomics data to hypothesize functional outcomes of a shift toward AG in the brain.
Materials and methods
Transcriptomics
Gene expression analysis
Statistical analyses to identify differentially expressed genes were performed separately for each experiment as described in (Chandrasekaran et al. 2011). Briefly, effects were evaluated with an F-test statistic and the P-values were adjusted for multiple hypothesis testing by using a FDR criterion. The resulting lists of differentially expressed genes below a given cutoff (FDR < 0.1) were used for further enrichment analysis. Through enrichment analysis, we measured over-representation of genes in Glycolysis and OxPhos pathway among the genes that were up or downregulated. All enrichment analyses were performed with the hypergeometric test (hygecdf function in MATLAB). Given an overlap (L) between a pathway of interest and a list of upregulated or downregulated genes, we calculated the enrichment P-value by summing over hypergeometric probabilities for all values of overlap greater than L. Pathways annotations for honey bee genes were obtained from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa et al. 2008).
Metabolomics
Honey bee sample collection and preparation
Bees (n = 60; female workers) from two different colonies were collected serially in the afternoon. Undisturbed middle-aged bees were collected at the colony entrance (Control), and then isopentyl acetate (IPA), the active compound in the honey bee alarm pheromone that induces aggressive behavior, was presented at the colony entrance. Aroused bees were collected and flash-frozen (5 min post-stimulus), or caged and flash-frozen (60 min post-stimulus; following Alaux et al. 2009). Bee heads were lyophilized at −80°C and brains were dissected under 100% ethanol chilled on dry ice. Aqueous metabolites were extracted in a solvent composed of methanol, chloroform and water. Non-aqueous portions of the extraction were then analyzed for protein content using the BCA assay.
Assay
Metabolomics measurements were done using a LC-QQQ-MS platform in the MRM mode targeting 156 metabolites in 25 important metabolic pathways in both positive MS and negative MS modes. Among the 156 measured metabolites we were able to detect 122 metabolites consistently across all the bees. The mean variance of metabolites was 7% across samples. We used standards to validate the compound identities and to optimize the platform.
Analysis
Metabolomics profiles for each sample were normalized to protein concentration to account for variability in the input tissue levels. The data were then quantile normalized using MATLAB quantilenorm function, and standardized to have zero mean and unit variance. Data normality for statistical tests was assessed through quantile–quantile plots and histograms using MATLAB. Differential metabolites were inferred using a two-sample t-test with unequal variance.
Clustering
Unsupervised hierarchical clustering was performed using MATLAB. The data were standardized along the rows to have zero mean and unit variance prior to clustering to enable easier interpretation across metabolites. Pearson’s Correlation was used to compute the distances between metabolomics profiles to build the clustergram. We chose all metabolites that were differentially expressed between any of the conditions (P-value < 0.1). The numbers of metabolites in each case were 10 (T5 vs. control), 30 (T60 vs. control) and 23 (T5 vs. T60). There were 40 total unique metabolites among these and they were used as input data to build the clustergram.
Results
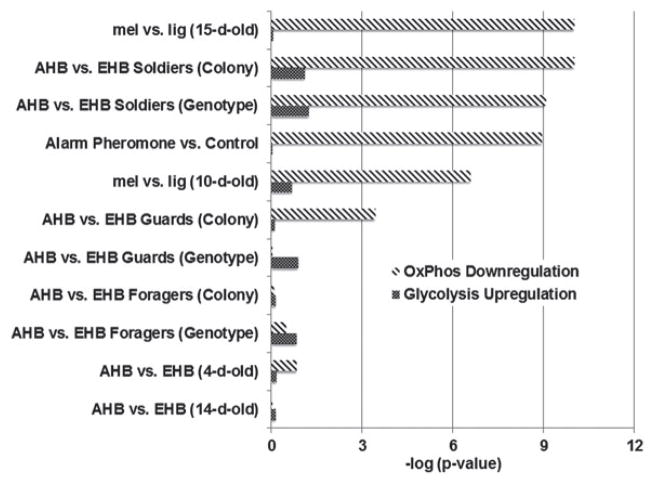
To explore the brain metabolic correlates of aggression, we analyzed transcriptomic data from 11 previously published microarray experiments (277 individual brain transcriptomes; Table 1). Each of these experiments measured whole-brain transcriptomic differences associated with genetic or socially induced variation in aggressive behavior (Chandrasekaran et al. 2011), but not all were analyzed for metabolic patterns by Alaux et al. (2009). In this expanded set of aggression studies, we found that OXPhos gene expression was consistently significantly lower in high-aggression behavioral states (P < 10−9; Table 1). Specifically, transcripts encoding protein constituents of four of the five enzyme complexes of the OXPhos pathway were significantly downregulated, including NADH dehydrogenase (complex I), succinate dehydrogenase (complex II), cytochrome c (complex IV) and ATP synthase (complex V) (S. Table 1). This pattern was robust across contexts of genetic, developmental and socially induced differences in aggression: highly aggressive vs. more docile subspecies; older, more aggressive adult bees vs. younger adults; and bees exposed to alarm pheromone, an aggression-inducing social cue, vs. unexposed control.
Table 1.
Summary of the bee microarray experiments used to explore the relationship between aggression and brain AG
| Study | High-aggression state | Low-aggression state | Source of variation | No. of samples | OXPhos downregulation, differential genes/total genes | Glycolysis upregulation, differential genes/total genes | Ref |
|---|---|---|---|---|---|---|---|
| mel vs. lig (15-day-old) | 15 day old mel | 15 day old lig | Genetic | 40 | 57/79 | 2/30 | 2 |
| AHB vs. EHB Soldiers (Colony) | AHB and EHB soldier bees kept in an AHB colony | AHB and EHB soldiers kept in an EHB colony | Social | 19 | 32/79 | 6/30 | 1 |
| AHB vs. EHB Soldiers (Genotype) | AHB soldiers kept in EHB and AHB colonies | EHB soldiers kept in EHB and AHB colonies | Genetic | 19 | 28/79 | 5/30 | 1 |
| Alarm Pheromone vs. Control | Bees exposed to alarm pheromone | Unexposed bees | Social | 40 | 24/79 | 0/30 | 1 |
| mel vs. lig (10-day-old) | 10 day old mel | 10 day old lig | Genetic | 40 | 6/79 | 6/30 | 2 |
| AHB vs. EHB Guards (Colony) | AHB and EHB guard bees kept in an AHB colony | AHB and EHB guards kept in an EHB colony | Social | 20 | 20/79 | 1/30 | 1 |
| AHB vs. EHB Guards (Genotype) | AHB guards kept in EHB and AHB colonies | EHB guards kept in EHB and AHB colonies | Genetic | 20 | 2/79 | 4/30 | 1 |
| AHB vs. EHB Foragers (Colony) | AHB and EHB foragers kept in an AHB colony | AHB and EHB foragers kept in an EHB colony | Social | 20 | 3/79 | 3/30 | 1 |
| AHB vs. EHB Foragers (Genotype) | AHB foragers kept in EHB and AHB colonies | EHB foragers kept in EHB and AHB colonies | Genetic | 20 | 2/79 | 2/30 | 1 |
| AHB vs. EHB (4-day-old) | 4 day old AHB | 4 day old EHB | Genetic | 20 | 17/79 | 4/30 | 2 |
| AHB vs. EHB (14-day-old) | 14 day old AHB | 14 day old EHB | Genetic | 19 | 4/79 | 5/30 | 2 |
Each study, described in Chandrasekaran et al. (2011), compared brain transcriptomic profiles in high- vs. low-aggression states, based on differences in genotype, age or social environment. ‘Soldiers’ and ‘Guards’ are bees that specialize in colony defense. ‘Foragers’ are food collectors. ‘AHB’ stands for Africanized honey bee, a particularly aggressive hybrid of the subspecies Apis mellifera ligustica (a European sub-species) and scutellata (an African sub-species). ‘EHB’ stands for European honey bee (a mixture of EHB sub-species, but primarily Apis mellifera ligustica). AHB is more aggressive than EHB. The ‘lig’ vs. ‘mel’ comparison compares two sub-species of EHB, Apis mellifera ligustica (‘lig’) and Apis mellifera mellifera (‘mel’). ‘mel’ is more aggressive than ‘lig’. The number of transcripts that were up- or downregulated in the pathway (FDR < 0.1), along with the total transcripts in the pathway are also provided for both glycolysis and OxPhos. References: 1 – Alaux et al. (2009); 2 – Chandrasekaran et al. (2011).
In contrast, glycolysis gene expression, especially for key rate-limiting reactions, was higher in the brains of more aggressive bees, and this pattern was significant for the highest aggression state (P = 0.05; Table 1; S. Table 1). Thus, contrary to the implication of the OXPhos data alone, aggressive bee brain metabolism is not globally decreased, but is shifted toward AG, i.e. glucose metabolism by glycolysis in excess of OXPhos (Vaishnavi et al. 2010). The states of highest aggression (‘soldier’ bees) showed the strongest AG pattern, with strong OXPhos downregulation and an increase in glycolysis. Moreover, no datasets showed a decrease in glycolysis, suggesting glucose metabolism in excess of OxPhos was a general pattern (Fig. 1). Overall we observed this pattern across aggressive states caused by both genetic and social factors, implying that AG is a general feature of the aggressive brain.
Figure 1. Differential expression of glycolysis and OXPhos genes in the aggressive brain.
Each category, described in Table 1, compares genetic and socially induced differences in aggression; all comparisons involve a more aggressive vs. a less aggressive state. AHB, Africanized honey bee (hybrids of Apis mellifera ligustica and scutellata); EHB, European honey bee (primarily Apis mellifera ligustica); lig, Apis mellifera ligustica; mel, Apis mellifera mellifera. ‘Colony’ label indicates effects of colony genotype on gene expression, independent of individual genotype (cross-fostering design). P-values based on hypergeometric test of enrichment are shown (Materials and methods). The P-value for mel v. lig. (15-day-old) was floored to 10−10 for visual clarity.
To further support the link between aggression and AG, we performed a brain metabolomic analysis on newly collected samples. To evaluate the time course of brain metabolic changes, we collected 20 aggressive bees 5 min (T5) and 60 min (T60) after exposure to alarm pheromone, and compared these groups to control bees unexposed to pheromone. We selected the 5 min sampling point to capture rapid changes in metabolic flux that may occur prior to transcriptional changes, which typically peak around 30 min (Cummings et al. 2008, Ellis & Carney 2011). We selected the 60 min sampling time based on previous empirical data, which gave ample evidence of a robust shift in OXPhos (manifesting both in terms of mRNA abundance and mitochondrial enzyme activity) at this time point (Alaux et al. 2009).
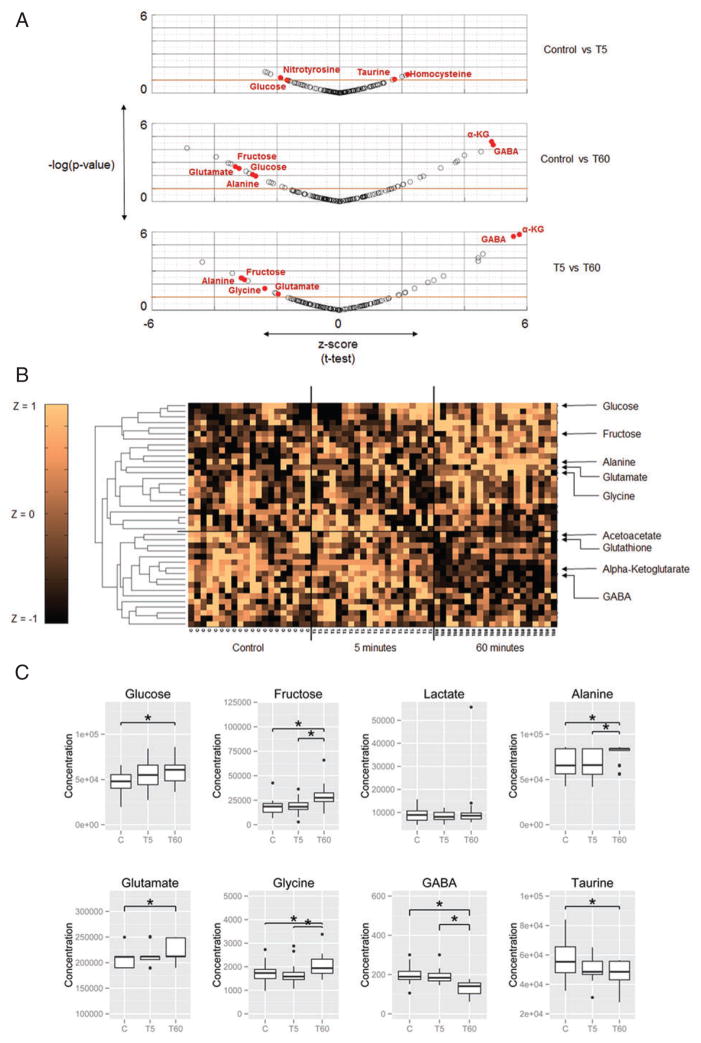
We measured a targeted set of 156 metabolites for each brain. Though the strongest differences in metabolite concentrations occurred comparing the T60 bees to control, 21 of the 25 metabolites showing significant differences between T60 and Control showed the same direction of change comparing T5 to Control and T60 to T5 (S. Table 2 and 3). This suggests that the brain metabolic response to an aggression-inducing cue is initiated within 5 min, increases in severity, and is maintained for at least 60 min (Fig. 2a,b).
Figure 2. Metabolomics data.
(a) Scatter plot of z-scores and P-values of all measured metabolites. Scatter plot shows differentially expressed metabolites and outliers between Control individuals and individuals 5 or 60 min after the start of the aggressive response. Outliers and differentially expressed metabolites (P-value < 0.05) mentioned in the text are highlighted. Metabolites above the red line (P-value < 0.1) are plotted in the clustergram in (b). Abbreviations: Alpha-ketoglutarate –αKG, Gamma-Amino Butryic Acid – GABA. (b) Unsupervised hierarchical clustering of metabolite levels in the aggressive brain. Clustergram shows differentially expressed (P < 0.1; t-test) metabolite levels (standardized to z-scores with zero mean and unit variance to enable comparison across metabolites) across the 60 samples from Control individuals and individuals 5 or 60 min after the start of the aggressive response. The horizontal line separates upregulated from downregulated metabolites at 60 min. (c) Levels of key metabolites connected to AG. Glucose and fructose are energy substrates, and lactate and alanine are end products of AG. The bottom row shows levels of excitatory and inhibitory neurotransmitters connected to AG. Asterisks indicate significant differences (P < 0.05). Refer to Figure S3 and Table S2 for full list of metabolite labels and P-values.
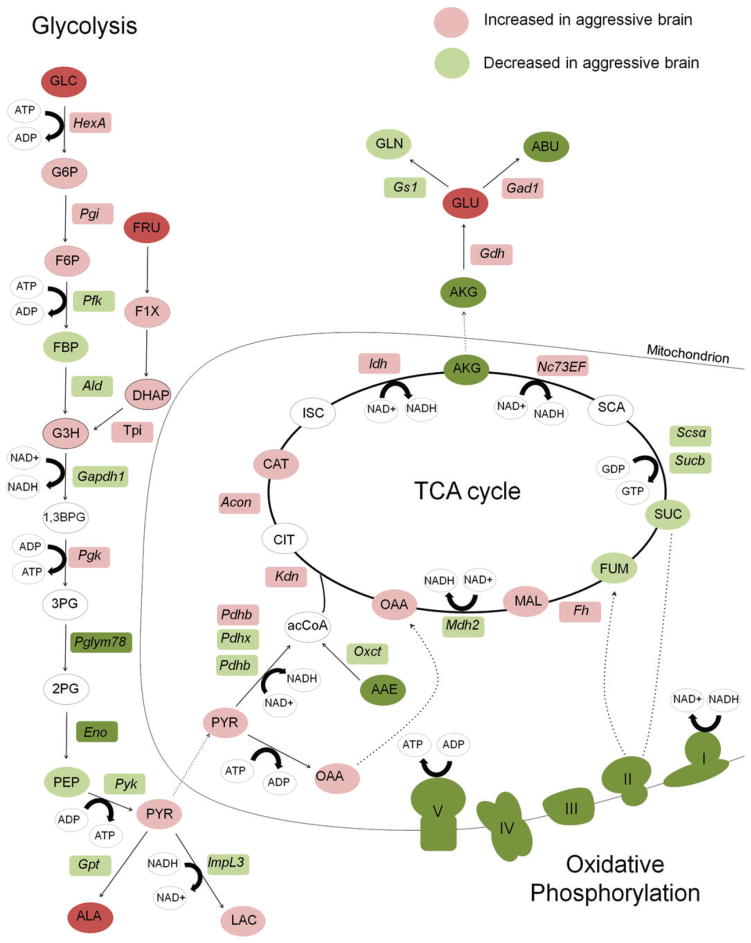
Indicators of AG in cancer include increased concentrations of lactate, glucose and alanine (Agathocleous et al. 2012; Lunt & Vander Heiden 2011; Vander Heiden et al. 2009). We observed a significant increase in two major glycolytic precursors, glucose and fructose, in the aggressive brain (P < 0.01; t(37.6) = 2.78). Moreover, alanine, an end product of glycolysis, was significantly higher in aggressive brains (P < 0.01; t(33.7) = 2.68; Fig. 2c). The direct precursor to alanine, pyruvate, does not accumulate in neurons and glia of bees (Wegener 1988). Therefore alanine levels depend on de novo pyruvate production from glycolysis, and thus alanine is an accepted indicator of glycolytic flux in insects (Ragland et al. 2010). High levels of alanine suggest that increased glycolytic flux occurs without a concordant increase in tri carboxylic acid (TCA) cycle and OXPhos activity (Lunt & Vander Heiden 2011), consistent with AG. Lactate is another common indicator of AG, and in our analyses, lactate increased only slightly. This finding however is consistent with previous studies showing very little lactate production at rest or following increased activity in the brain and flight muscles of honey bees and other invertebrates (Tsacopoulos et al. 1994; Wegener 1988). Finally, we observed a slight, non-significant increase in 4 of 6 other measured glycolytic intermediates – glucose-6-phosphate, fructose-6-phosphate, glyceraldehyde-3-phosphate and dihydroxyacetone Phosphate, as well as a slight increase in pyruvate (Fig. 3; Table S2 and Fig. S1, Supporting Information). These data, coupled with the transcriptomic analysis, suggest that increased aggression is associated with increased glycolysis and decreased OXPhos, i.e. a shift to AG in the brain.
Figure 3. Relative levels of central metabolism metabolites and genes in the aggressive brain.
Metabolites are listed in circles and genes are in rectangles. Metabolites in white were not measured. Dark green and dark red colors represent changes in metabolites and mRNA that were statistically significant (a decrease or increase, respectively) according to gene expression and metabolite data from honey bees treated with alarm pheromone vs. control. Light green and light red colors represent a non-significant decrease or increase, respectively. Protein complexes in the oxidative phosphorylation pathway, which are composed of variable numbers of protein subunits, are drawn as cartoons that represent the physical shapes of the complexes. Complexes are colored green to represent the general down regulation of the transcripts that encode protein constituents. Circles with black and dotted borders were compounds that were indistinguishable from one another in the metabolomics analysis. GLC, glucose; PYR, pyruvate; OAA, oxaloacetate; CIT, citrate; AKG, alpha-ketoglutarate; SUC, succinate; FUM, fumarate; MAL, malate; SCA, succinyl-CoA; GLU, glutamate; GLN, glutamine; LAC, lactate; ALA, alanine; G6P, glucose 6 phosphate; F6P, fructose 6 phosphate; FBP, fructose 1,6 bis phosphate; GADP, glyceraldehyde 3 phosphate; DHAP, dihydroxyacetone phosphate; 1,3BPG, 1,3 bisphosphoglycerate; 3PG, 3 phosphoglycerate; 2PG, 2 phosphoglycerate; PEP, phosphoenolpyruvate.
The metabolomics results identified additional molecular features that distinguish the aggressive brain and may be connected to AG. We detected significant decreases in the predominantly inhibitory neurotransmitters GABA and taurine (P < 0.05; t(34) = −4.8; Fig. 2c), and increases in glutamate and glycine (P < 0.05; t(33.6) = 3.2; Fig. 2c), two excitatory neurotransmitters. Alpha-ketoglutarate, a TCA cycle intermediate and glutamate precursor, also significantly decreased (Fig. 3). Furthermore, we detected evidence of oxidative stress in the aggressive brain, i.e. a significant increase in the marker nitrotyrosine (P < 0.01; t(34) = 3.56) and decreases in the antioxidants taurine and glutathione (Macone et al. 2011) (P < 0.09; t(23.8) = −1.74; Fig. S2).
Discussion
Our data suggest a shift toward brain AG in high aggression states. Other features of the aggressive brain include changes in neurotransmitter levels and indicators of oxidative stress. Here we discuss possible functions of AG in the aggressive brain that are consistent with our transcriptomic and metabolomics data.
Our data support the previously described negative relationship between aggression and brain OXPhos activity (Alaux et al. 2009; Li-Byarlay et al. 2014). We found that this relationship was most robust in the highest aggression comparisons, but the general pattern was consistent across all eleven contexts for aggression in our study. Based on OXPhos data alone, a previous study hypothesized that energy metabolism may globally decrease in association with heightened aggression (Alaux et al. 2009). However, our analyses suggest that energy metabolism in the aggressive brain is not decreased overall, but rather shifted toward AG, with increased glycolysis in addition to decreased OXPhos activity. Glycolysis gene expression was increased in the most aggressive states, and never decreased in concert with OXPhos.
The functional implications of a shift toward AG in the aggressive brain are unknown. Our data support one possibility, which is that AG alters concentrations of metabolites that serve as precursors for neurotransmitter synthesis; we detected significant decreases in the predominantly inhibitory neurotransmitters GABA and taurine (El Hassani et al. 2008; Jia et al. 2008; Papandreou et al. 2006), and significant increases in the predominantly excitatory neurotransmitters glutamate and glycine (Lopez-Cocuera et al. 2001; Signorotti et al. 2014). Moreover, alpha-ketoglutarate, a TCA cycle intermediate and glutamate precursor, also significantly decreased, suggesting this metabolite may be diverted away from the TCA cycle and OXPhos toward glutamate production, altering neurotransmitter levels in the aggressive brain. Decreased inhibitory neurotransmission, and specifically decreased GABA:glutamate ratios, have been implicated in aggression (Wnuk et al. 2013), and our findings provide a possible mechanism to explain how the decreased ratio occurs.
Our data suggest that AG may also be linked to an oxidative stress response, as we detected evidence of oxidative stress in the aggressive brain. This does not conflict with the previous hypothesis, as AG could enable the concurrent production of antioxidants and neurotransmitters, and a combination of different cellular demands may drive AG in the bee brain. Oxidative stress could indicate that the aggressive brain is oxygen deprived (i.e. hypoxic), which is known to induce glycolysis (Lunt & Vander Heiden 2011) and thus could drive our observed metabolic patterns. However, the prolonged time-scale of the aggression-induced metabolic state, persisting for at least 60 min, suggests oxygen deprivation is probably not the primary driver. Similarly, AG could provide a short burst of ATP (Pfeiffer et al. 2001) for enhanced synaptic activity during the relatively brief defensive event, but this does not explain the persistence of AG beyond the initial defensive response. Finally, the long time-scale of the metabolic response differentiates it from the immediate behavioral response to alarm pheromone, which may activate a constrained set of target neurons mediating the immediate aggressive response.
Previous studies suggest other possible functions of AG in the brain, which could also be important in the aggressive context. AG could lead to NADH accumulation (Lunt & Vander Heiden 2011), reducing cellular redox state and increasing neural excitability by reducing voltage-gated membrane potassium channel conductance (Wang et al. 2012; Yang et al. 2014). Increased excitability, e.g. in glutamatergic circuits, could enhance aggressive behavior as these circuits are known to potentiate response to stressors (Herman et al. 2004). Aerobic glycolysis could also facilitate increased synaptic activity; membrane-associated glycolytic enzymes provide ATP for ion channels and glutamate transporters (Sattler & Tymianski 2001). Finally, although cell proliferation is not a relevant goal in the bee brain because there is no known adult neurogenesis (Fahrbach et al. 1995), AG might support synapse formation and growth, which, similar to cancer, have strong biosynthetic demands (Goyal et al. 2014). Our data cannot rule out these hypotheses, which may not be mutually exclusive. Testing these different hypotheses, which involves detailed electrophysiological, behavioral and metabolomics analysis, is beyond the scope of the present study. Furthermore, our analyses are at the whole-brain level, and do not account for the possibility that the metabolic response we have observed is limited to particular brain regions or circuits.
The metabolic shift in the aggressive brain is itself likely mediated by neurotransmitter signaling. Similar to the predicted role of lactate in memory and neural plasticity, ala-nine may serve as a signaling molecule in the bee brain (Barros 2013; Tsacopoulos et al. 1994; Yang et al. 2014) or as a long-range paracrine signal of metabolic state (Bergersen & Gjedde 2012). Brain metabolic changes may be induced by octopamine, the insect ‘fight or flight’ neurohormone that is commonly associated with changes in aggression and mediates shifts in muscle energy metabolism. Our results could provide a neural mechanism to link known relationships between octopamine signaling and experience-dependent modulation of aggressive behaviors (Stevenson et al. 2005). Finally, transcription factors likely coordinate the neurogenomic changes we observe. The transcription factor dorsal, the insect ortholog of NF-Kappa B, was implicated in the general control of socially regulated behaviors, including foraging and aggression, in honeybees (Chandrasekaran et al. 2011). dorsal has been shown to regulate the balance between glycolysis and mitochondrial respiration in mammalian cells (Mauro et al. 2011). Thus tuning the metabolic state of the brain could be a general mechanism used to modulate socially responsive behaviors.
Aberrant brain metabolic flux is associated with neurological, behavioral and mood disorders. Aerobic glycolysis occurs in regions of beta-amyloid deposition in Alzheimer’s cases (Vlassenko et al. 2010), an AG-like brain metabolic state has been observed in Schizophrenia (Prabakaran et al. 2004), and Huntington’s Disease has been associated with deficits in AG (Powers et al. 2007). Decreased GABA levels are also associated with impulsivity, an aggression-related behavioral disorder (Boy et al. 2011). Thus the connection between honey bee aggression and AG provides an interesting context to address the neural and cognitive consequences of human brain AG.
Supplementary Material
Acknowledgments
This research was supported by the National Science Foundation grant IOS-1256705 (G.E.R. and N.D.P.), and additional funding through the National Institutes of Health (NIH) Center for Systems Biology/2P50GM076547 and 1U01AG046139-01 (N.D.P.), an NIH Pioneer Award DP1 OD006416 (G.E.R.), and an International Predoctoral Fellowship from the Howard Hughes Medical Institute (HHMI) to S.C. The authors declare that they have no competing interests.
Footnotes
S.C., C.C.R., N.D.P. and G.E.R. designed research. S.C. and C.C.R. performed research, analyzed the data and wrote the manuscript. D.D., H.G. and D.R. performed the metabolomics analysis. All authors read and approved the final manuscript.
Additional supporting information may be found in the online version of this article at the publisher’s web-site:
Figure S1: (a) Levels of key metabolites connected to AG. glucose, fructose and sorbitol are energy substrates and lactate and alanine are end products of AG. Acetoacetate is an alternate carbon source that is metabolized at elevated levels under starvation conditions. (b) Levels of amino acids in the Control, T5 and T60 samples. Asterisks indicate significant differences (P < 0.05).
Figure S2: Levels of oxidative stress markers (3-Nitrotyrosine) and antioxidants (taurine and glutathione) in the brain. Note that taurine is also a neurotransmitter. Asterisks indicate significant differences (P < 0.05).
Figure S3: (Fig. 2 with all the metabolite labels): Unsupervised hierarchical clustering of metabolite levels. Clustergram shows metabolite levels (represented as z-scores) across the 60 samples from Control, T5 and T60 for metabolites that were differentially expressed (P < 0.1; t-test) between any two states (C, T5, T60).
Table S1: Genes upregulated (a) or downregulated (b) in Glycolysis, TCA cycle and Oxidative phosphorylation in the highest aggression state comparisons (FDR < 0.1). Important rate-limiting enzymes in Glycolysis are highlighted in gray boxes. Transcripts encoding protein constituents of four of the five enzyme complexes of the OXPhos pathway were significantly downregulated, including NADH dehydrogenase (complex I), succinate dehydrogenase (complex II), cytochrome c (complex IV) and ATP synthase (complex V).
Table S2: Normalized honey bee brain metabolomics data, p-values and t-scores for all metabolites measured in this study.
Table S3: (a) The list of 25 metabolites differentially expressed between T60 and control (P < 0.05) and their direction of change in T5 compared to control (21/25 have the same direction). (b) The corresponding list of 10 metabolites differentially expressed between T5 and control (P < 0.1) and their direction of change in T60 (9/10 have the same direction). (c) The list of 19 metabolites differentially expressed between T5 and T60 (P < 0.05) and their direction of change in T60 vs. Control (13/19 have the same direction).
References
- Agathocleous M, Love NK, Randlett O, Harris JJ, Liu J, Murray AJ, Harris WA. Metabolic differentiation in the embryonic retina. Nat Cell Biol. 2012;14:859–864. doi: 10.1038/ncb2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaux C, Sinha S, Hasadsri L, Hunt GJ, Guzman-Novoa E, DeGrandi-Hoffman G, Uribe-Rubio JL, Southey BR, Rodriguez-Zas S, Robinson GE. Honey bee aggression supports a link between gene regulation and behavioral evolution. Proc Natl Acad Sci USA. 2009;106:15400–15405. doi: 10.1073/pnas.0907043106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros LF. Metabolic signaling by lactate in the brain. Trends Neurosci. 2013;36:396–404. doi: 10.1016/j.tins.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Bergersen LH, Gjedde A. Is lactate a volume transmitter of metabolic states of the brain? Front Neuroenergetics. 2012;4:5. doi: 10.3389/fnene.2012.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boy F, Evans CJ, Edden RA, Lawrence AD, Singh KD, Husain M, Sumner P. Dorsolateral prefrontal gamma-aminobutyric acid in men predicts individual differences in rash impulsivity. Biol Psychiatry. 2011;70:866–872. doi: 10.1016/j.biopsych.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle PJ, Scott JC, Krentz AJ, Nagy RJ, Comstock E, Hoffman C. Diminished brain glucose metabolism is a significant determinant for falling rates of systemic glucose utilization during sleep in normal humans. J Clin Invest. 1994;93:529. doi: 10.1172/JCI117003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran S, Ament SA, Eddy JA, Rodriguez-Zas SL, Schatz BR, Price ND, Robinson GE. Behavior-specific changes in transcriptional modules lead to distinct and predictable neurogenomic states. Proc Natl Acad Sci USA. 2011;108:18020–18025. doi: 10.1073/pnas.1114093108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings ME, Larkins-Ford J, Reilly CR, Wong RY, Ramsey M, Hofmann HA. Sexual and social stimuli elicit rapid and contrasting genomic responses. Proc R Soc B Biol Sci. 2008;275:393–402. doi: 10.1098/rspb.2007.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hassani AK, Giurfa M, Gauthier M, Armengaud C. Inhibitory neurotransmission and olfactory memory in honeybees. Neurobiol Learn Mem. 2008;90:589–595. doi: 10.1016/j.nlm.2008.07.018. [DOI] [PubMed] [Google Scholar]
- Ellis LL, Carney GE. Socially-responsive gene expression in male Drosophila melanogaster is influenced by the sex of the interacting partner. Genetics. 2011;187:157–169. doi: 10.1534/genetics.110.122754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrbach SE, Strande JL, Robinson GE. Neurogenesis is absent in the brains of adult honey bees and does not explain behavioral neuroplasticity. Neurosci Lett. 1995;197:145–148. doi: 10.1016/0304-3940(95)11913-h. [DOI] [PubMed] [Google Scholar]
- Fox PT, Raichle ME, Mintun MA, Dence C. Nonoxidative glucose consumption during focal physiologic neural activity. Science. 1988;241:462–464. doi: 10.1126/science.3260686. [DOI] [PubMed] [Google Scholar]
- Goyal Manu S, Hawrylycz M, Miller Jeremy A, Snyder Abraham Z, Raichle Marcus E. Aerobic glycolysis in the human brain is associated with development and neotenous gene expression. Cell Metab. 2014;19:49–57. doi: 10.1016/j.cmet.2013.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison JF, Hall HG. African-European honeybee hybrids have low nonintermediate metabolic capacities. Nature. 1993;363:258–260. [Google Scholar]
- Herman JP, Mueller NK, Figueiredo H. Role of GABA and glutamate circuitry in hypothalamo-pituitary-adrenocortical stress integration. Ann N Y Acad Sci. 2004;1018:35–45. doi: 10.1196/annals.1296.004. [DOI] [PubMed] [Google Scholar]
- Jia F, Yue M, Chandra D, Keramidas A, Goldstein PA, Homanics GE, Harrison NL. Taurine is a potent activator of extrasynaptic GABA(A) receptors in the thalamus. J Neurosci. 2008;28:106–115. doi: 10.1523/JNEUROSCI.3996-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Byarlay HR, Rittschof CC, Massey JH, Pittendrigh BR, Robinson GE. Socially responsive effects of brain oxidative metabolism on aggression. Proc Natl Acad Sci USA. 2014;111:12533–12537. doi: 10.1073/pnas.1412306111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Cocuera B, Geerlings A, Aragon C. Glycine neurotransmitter transporters: an update. Mol Membr Biol. 2001;18:13–20. [PubMed] [Google Scholar]
- Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- Macone A, Fontana M, Barba M, Botta B, Nardini M, Ghirga F, Calcaterra A, Pecci L, Matarese RM. Antioxidant properties of aminoethylcysteine ketimine decarboxylated dimer: a review. Int J Mol Sci. 2011;12:3072–3084. doi: 10.3390/ijms12053072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen PL, Hasselbalch SG, Hagemann LP, Olsen KS, Bulow J, Holm S, Wildschiodtz G, Paulson OB, Lassen NA. Persistent resetting of the cerebral oxygen/glucose uptake ratio by brain activation: evidence obtained with the Kety-Schmidt technique. J Cereb Blood Flow Metab. 1995;15:485–491. doi: 10.1038/jcbfm.1995.60. [DOI] [PubMed] [Google Scholar]
- Mauro C, Leow SC, Anso E, Rocha S, Thotakura AK, Tornatore L, Moretti M, De Smaele E, Beg AA, Tergaonkar V, Chandel NS, Franzoso G. NF-kappaB controls energy homeostasis and metabolic adaptation by upregulating mitochondrial respiration. Nat Cell Biol. 2011;13:1272–1279. doi: 10.1038/ncb2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papandreou I, Cairns RA, Fontana L, Lim AL, Denko NC. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Pfeiffer T, Schuster S, Bonhoeffer S. Cooperation and competition in the evolution of ATP-producing pathways. Science. 2001;292:504–507. doi: 10.1126/science.1058079. [DOI] [PubMed] [Google Scholar]
- Phelps ME, Barrio JR. Correlation of brain amyloid with “aerobic glycolysis”: a question of assumptions? Proc Natl Acad Sci USA. 2010;107:17459–17460. doi: 10.1073/pnas.1012684107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers WJ, Videen TO, Markham J, McGee-Minnich L, Antenor-Dorsey JV, Hershey T, Perlmutter JS. Selective defect of in vivo glycolysis in early Huntington’s disease striatum. Proc Natl Acad Sci USA. 2007;104:2945–2949. doi: 10.1073/pnas.0609833104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabakaran S, Swatton JE, Ryan MM, Huffaker SJ, Huang JT, Griffin JL, Wayland M, Freeman T, Dudbridge F, Lilley KS, Karp NA, Hester S, Tkachev D, Mimmack ML, Yolken RH, Webster MJ, Torrey EF, Bahn S. Mitochondrial dysfunction in schizophrenia: evidence for compromised brain metabolism and oxidative stress. Mol Psychiatry. 2004;9:684–697. 643. doi: 10.1038/sj.mp.4001511. [DOI] [PubMed] [Google Scholar]
- Ragland GJ, Denlinger DL, Hahn DA. Mechanisms of suspended animation are revealed by transcript profiling of diapause in the flesh fly. Proc Natl Acad Sci USA. 2010;107:14909–14914. doi: 10.1073/pnas.1007075107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME, Posner JB, Plum F. Cerebral blood flow during. Arch Neurol. 1970;23:394. doi: 10.1001/archneur.1970.00480290014002. [DOI] [PubMed] [Google Scholar]
- Sattler R, Tymianski M. Molecular mechanisms of glutamate receptor-mediated excitotoxic neuronal cell death. Mol Neurobiol. 2001;24:107–129. doi: 10.1385/MN:24:1-3:107. [DOI] [PubMed] [Google Scholar]
- Signorotti L, Jaisson P, d’Ettorre P. Larval memory affects adult nest-mate recognition in the ant Aphaenogaster senilis. Proc R Soc B Biol Sci. 2014;281:20132579. doi: 10.1098/rspb.2013.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson PA, Dyakonova V, Rillich J, Schildberger K. Octopamine and experience-dependent modulation of aggression in crickets. J Neurosci. 2005;25:1431–1441. doi: 10.1523/JNEUROSCI.4258-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsacopoulos M, Veuthey AL, Saravelos SG, Perrottet P, Tsoupras G. Glial cells transform glucose to alanine, which fuels the neurons in the honeybee retina. J Neurosci. 1994;14:1339–1351. doi: 10.1523/JNEUROSCI.14-03-01339.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnavi SN, Vlassenko AG, Rundle MM, Snyder AZ, Mintun MA, Raichle ME. Regional aerobic glycolysis in the human brain. Proc Natl Acad Sci. 2010;107:17757–17762. doi: 10.1073/pnas.1010459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Sci Signall. 2009;324:1029. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlassenko AG, Vaishnavi SN, Couture L, Sacco D, Shannon BJ, Mach RH, Morris JC, Raichle ME, Mintun MA. Spatial correlation between brain aerobic glycolysis and amyloid-beta (Abeta ) deposition. Proc Natl Acad Sci USA. 2010;107:17763–17767. doi: 10.1073/pnas.1010461107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TA, Yu YV, Govindaiah G, Ye X, Artinian L, Coleman TP, Sweedler JV, Cox CL, Gillette MU. Circadian rhythm of redox state regulates excitability in suprachiasmatic nucleus neurons. Science. 2012;337:839–842. doi: 10.1126/science.1222826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Wegener G. Oxygen Sensing in Tissues. Springer; 1988. Oxygen availability, energy metabolism, and metabolic rate in invertebrates and vertebrates; pp. 13–35. [Google Scholar]
- Wnuk A, Kostowski W, Korczynska J, Szczuka A, Symonowicz B, Bienkowski P, Mierzejewski P, Godzinska EJ. Brain GABA and glutamate levels in workers of two ant species (Hymenoptera: Formicidae): interspecific differences and effects of queen presence/absence. Insect Sci. 2013;21:647–658. doi: 10.1111/1744-7917.12076. [DOI] [PubMed] [Google Scholar]
- Yang J, Ruchti E, Petit JM, Jourdain P, Grenningloh G, Allaman I, Magistretti PJ. Lactate promotes plasticity gene expression by potentiating NMDA signaling in neurons. Proc Natl Acad Sci USA. 2014;111:12228–12233. doi: 10.1073/pnas.1322912111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.