Abstract
Studies are emerging in support of the cancer stem cells (CSCs) theory which considers that a tiny subset of cancer cells is exclusively responsible for the initiation and malignant behavior of a cancer. This cell population, also termed CSCs, possesses the capacity both to self-renew, producing progeny that have the identical tumorigenic potential, and to differentiate into the bulk of cancer cells, helping serve the formation of the tumor entities, which, altogether, build the hierarchically organized structure of a cancer. In this review, we try to articulate the complicated signaling pathways regulating the retention of the characteristics of pancreatic CSCs, and in the wake of which, we seek to offer insights into the CSCs-relevant targeted therapeutics which are, in the meantime, confronted with bigger challenges than ever.
Keywords: Pancreatic cancer, cancer stem cells, signaling pathway, microRNA, CSC-targeted therapeutic
Introduction
Pancreatic duct adenocarcinoma (PDAC), currently the fourth most frequent cause of cancer-related deaths, is responsible for an estimated number of 227,000 deaths each year. Surgical resection is still the most effective therapeutic method and takes the hope of the curing this malignancy. However, at the time of diagnosis, less than 20% of the patients with PDAC are clinically amenable to surgical resection. Furthermore, most of patients inevitably develop distant metastases and local recurrence even after radical resection, thus, the overall survival rate of PDAC is dismally below 5% [1]. This could be explained in part by its high resistance to chemotherapy, early metastasis and inclination to recurrence. Whereas, little has the treatment strategies been advanced and no significant improvement in overcoming the chemo-resistant nature of pancreatic tumors has been achieved in the recent decades, with gemcitabine still being the first-line chemotherapy drug. Thus, radical therapeutic strategies are needed for PDAC to improve its prognosis. Presently, evidences show CSCs have been investigated in emerging studies as potentially valid candidates for therapeutic targets.
Cancer stem cells, as a specific cell type uniquely and identifiably existing in cancers, were first identified in acute myelogenous leukemia [2,3], following by more validations in some solid cancers including pancreatic cancer. More recently, evidence has emerged that suggests a crucial role that cancer stem cells may play not only in the initiation, but in the malignant nature of various human tumors [4-7].
Pancreatic cancer stem cells were first explored by Li and colleagues [8]. They successfully isolated a subset of cancer cells displaying a high expression of CD24, CD44 and ESA (epithelia-specific antigen) and bearing a highly carcinogenic potential from a successfully established xenograft model in which immunodeficient mice were implanted with primary human pancreatic cancer cells. Afterwards, more studies have sprung up shedding light on the mechanisms controlling the survival, proliferation and invasion of this subpopulation of pancreatic cancer cells, which will, without a doubt, illuminate our way to develop novel therapeutics that may target the CSCs in pancreatic cancer, as opposed to mainly targeting the more differentiated cancer cells when treated with chemodrugs or/and radiation. This offers a more promising prosperity for completely curing human pancreatic adenocarcinoma.
Surface markers for pancreatic CSCs
Cancer stem cells help lay the foundation and further complete the constitution of the whole architecture of cancer entity. An effective way, thus, to single out this minute subpopulation for better understanding is urgently needed. Previous studies have offered several methods and basic concepts concerning CSCs markers. Dick et al, in 1997, first reported the identification of CSCs in myeloid leukemia by using cell surface markers , and Al-Hajj et al initially found it also appropriate to use such markers as CD44, CD24, and epithelial-specific antigen (ESA) to isolate CSCs from solid organ epithelial cancer in breast [9]. They reported that the CD44+CD24-/low ESA+ cancer cells are CSC candidates with a high tumorinitiating potential and cancer formation ability in immune-compromised mice. Fluorescence-activated cell sorting (FACS) and human tumor xenograft models in immune-deficient mice are methods playing a crucial role in both studies above and were later also recognized as valid to identify CSCs in pancreatic cancer. In seeking to isolate CSCs from pancreatic cancer cells, Lee and colleagues [9] demonstrated that the CD44+CD24+ESA+ cells, which account for 0.2%-0.8% of all human pancreatic cancer cells showed a 100-fold greater potential of tumorigenesis (100 triple positive cells were enough to form new tumors in 6 of all 12 experimental NOD/SCID mice) than the CD44-CD24-ESA- cancer cells.
CD133 has been well characterized in the past a few years in cancers of brain [10-12], colon [13], lung [14], breast [15], and prostate [16]. Hermann et al [17] demonstrated that the CD133+ cells (cytokeratin negative) isolated by means of flow cytometry from patient-derived pancreatic cancer tissues displayed high and permanent carcinogenic potential. In this study, 500 CD133+ cells singled out and purified by using magnetic bead sorting and flow cytometry were capable of initiating new tumors in athymic mice (secondary tumor) from which another 500 CD133+ cells were implanted into secondary mice. In addition, CD133+ tumor cells obtained from the secondary xenografts were subsequently transplanted into third-generation mice. The serial passaging of CD133+ pancreatic cancer cells built up new tumors where cytokeratin positive differentiated progeny cells and cytokeratin negative CD133+ cells were similarly detectable. Of interestsX, their study also denoted the presence of a uniquely specific subpopulation of CSCs, termed migrating CSCs, in the invasive front of human pancreatic cancer samples. These cells are not only morphologically but also molecularly definable by the expression of CXCR4 receptor which, together with its specific ligand SDF-1, mediates the metastasis of pancreatic cancer. However, a few findings in brain tumors exemplified a common problem concerning the cancer stem cell model. CD133+ tumor cells were previously verified in brain tumors [18,19] as potent in distinguishing tumor initiating from non-tumor initiating cells, but were questioned by discoveries indicating that tumor initiating capability was also conferred on CD133- cancer cells in glioblastoma [18,20,21]. It remains interesting to investigate whether these findings can be testified in pancreatic cancer.
Recently, a study by Bailey and colleagues [22] found that the DCLK1 (a member of the microtubule regulator family) positive subpopulations of cells collected from the KCPdx1, KPCPdx1, and KCiMist1 mouse models of pancreatic intraepithelial neoplasia (PanIN) bore a high potential to generate tumors and resistance to chemotherapy. Thus DCLKT1 could be used as a cancer stem cell marker. These stem cell-like cells were sorted out using such methods of confocal and electron microscopy, lineage tracing, and fluorescence-activated cell sorting. Their study, in some way, illuminated our way to detect pancreatic cancer at PanIN stage and to prevent the progression of PanIN, and offered new targets for treatments. Additionally, there are studies showing other CSC markers such as the nestin [23,24], aldehyde dehydrogenase (ALDH-1) [23-26], ABCG2 and c-Met [24] pancreatic adenocarcinoma [25].
However, it still remains to answer whether CD44+CD24+ESA+ and CD133+ pancreatic cancer cells represent distinct cancer stem cell populations or whether even increased or decreased CSC enrichment can be obtained by selecting all 4 (or even more markers, ALDH-1, etc.) markers. In each of these studies, CSCs were identified by a set of specific criteria in order to demonstrate their role in tumor initiation and progression. Whereas, interestingly, studies showed that the CD44+CD24+ESA+ subpopulation obviously overlapped with the CD133+ population. Cancer stem cells were reported to be characterized by alterations in the frequency and distribution of cells with stem-cell like and more differentiated cell features among different cancer subtypes and patients and tumor progression stages [27]. In other words, there may be different markers in the different stages of CSCs’ differentiation into tumor cells. So, CSCs may have their own heterogeneity, which, if proved true, will definitely apply more pressure on researchers endeavoring to develop therapeutic agents targeting CSC markers.
Self-renewal pathways in pancreatic cancer stem cells
Analysis of the complicated signaling pathway networks from normal stem cells serves as well-established models, paving the way for the elucidation of CSC signaling systems. Signaling pathways, including Notch, Wnt, PTEN, SHH (Sonic hedgehog), BMP-4, and BMI-1, have already been verified in cancers from solid organs [7,28-31]. Phosphoinositide-3 kinase (PI3K)/Akt signaling has been implicated in the regulation of diverse cellular functions, including cell proliferation, growth, survival, migration, metabolism, angiogenesis, and tumorigenesis [32,33].
Hedgehog
Hedgehog signaling, a crucial pathway for embryonic development, the dysregulation of which has been implicated in several forms of cancer, may also be of great significance in human pancreatic carcinoma [34].
Three Hedgehog (Hh) ligand proteins exist (Sonic, Indian, and Desert Hh), which are recognized and bound by Patched (Ptch1 and 2, cell surface receptors). Here is the way Hedgehog signaling is activated (Figure 1): Binding of Hh to Ptch halts the repression of its trans-membrane protein Smoothened (Smo), which ultimately initiates nuclear translocation and activation of the Gli family (transcription factors, Gli1,2,3). Lee and colleagues [7], together with other studies, also found that the expression of SHH transcripts in CD44+CD24+ESA+ pancreatic cancer cells was 46-fold more significantly increased compared that in CD44-CD24-ESA- pancreatic cancer cells or normal pancreatic epithelial cells, which means in other words that pancreatic cancer stem cells bear a higher SHH activity.
Figure 1.
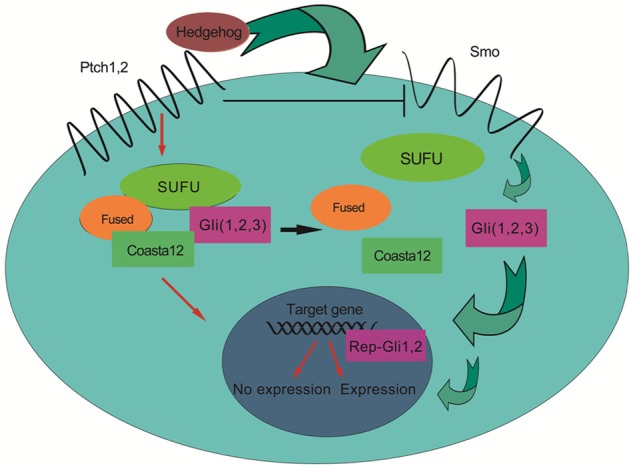
Hedgehog signaling. When engaged with Hedgehog protein, the Ptch receptor loses its repression of the Smo and then disassembles the complex composed of Fused, SUFU and Glis. The detailed mechanism remains to be elucidated. The separated Glis travel into the nucleus, initiating the expression of target genes.
In seeking to explain the indispensability of Hedgehog signaling for pancreatic cancer stem cell renewal, Sarah and colleagues investigated the role of the sonic hedgehog, a secreted hedgehog ligand abnormally expressed, in the initiation of pancreatic cancer and its cancer precursor lesions: pancreatic intraepithelial neoplasia (PanIN) [34]. They reported the over-expression of Ptch1 and Smo protein in transgenic mice models in which Shh misexpression was driven by the pancreatic-specific Pdx-1 promoter. And they further verified the function of misregulation of Hh signaling in human pancreatic cancer by using cyclopamine, a Smo antagonist, in cell lines collected either from primary tumors (the Panc series Panc 01.28 to 10.05) or from metastases in liver (CFPAC1), lymph node (Hs 766T) and spleen (SW 1990), (all of which were tested to express at least two components of the Hh signaling pathway system) all of which are induced to apoptosis compared with no effect in the control.
The Wnt pathway
Wnt proteins are capable of activating at least three pathways, the canonical (Wnt–β-catenin), the planar cell polarity (PCP; also known as non-canonical) and the Wnt-Ca2+ pathways. And they were explained elsewhere [35]. Here, we discuss the initiation of the canonical pathway which was testified to be aberrantly activated in pancreatic cancer [36]. In the absence of Wnt ligands, the downstream intracellular protein β-catenins are phosphorylated at sites of Ser and Thr residues of amino terminus by CKI and GSK3, kinases attaching to the destruction complex that consists of APC (the tumour suppressors adenomatous polyposis coli) and axin, and that functions as a disintegrator of β-catenin. While as soon as the Wnt receptor complex (composed of a seven-transmembrane receptor of the Frizzled family and Lrp5/6, a member of the LDL receptor family) are occupied by Wnt ligands, the interaction of axin and APC or/and the axin-binding molecule Dishevelled will repress the phosphorylation process of the β-catenin, as a result of which, β-catenins are accumulated which then will translocate into the nucleus and combine with the amino terminus of DBPC (Tcf/Lef family) resulting in the aberrant activation of target genes and ultimately the generation of cancers (Figure 2 [37]).
Figure 2.
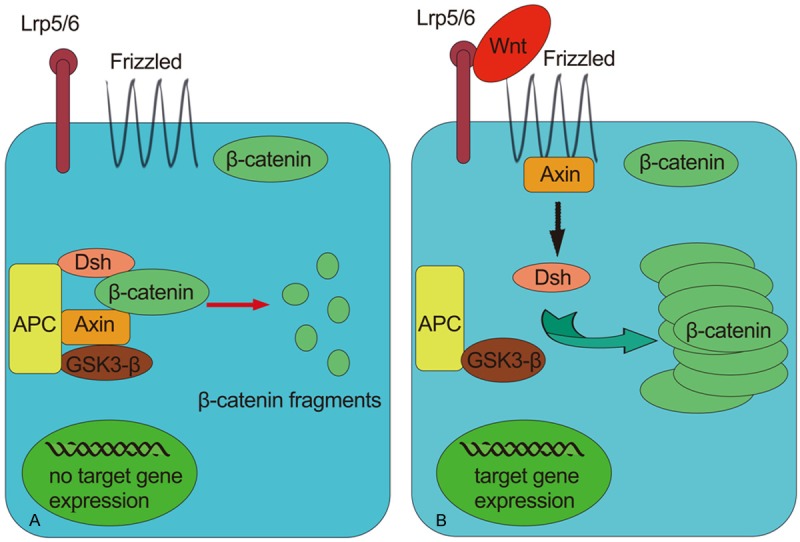
The activation of canonical (Wnt–β-catenin) pathway. A. In the absence of Wnt β-catenin is conventional phosphorylated by kinases CKI and GSK3 which are components of the destruction complex also consisting of APC and Axin. The β-catenin is subsequently degraded into pieces with no expression of Wnt target genes. B. The binding of Wnt to its receptors Lrp5/6 and Frizzled disintegrates the destruction complex. The interaction of APC and Axin or Dsh through an unknown mechanism puts a halt to the phosphorylation of β-catenins which are therefore accumulated in the cytoplasm and are then translocated into the nucleus and bind to Tcf/Lef, causing the expression of target genes.
An important role of Wnt signaling in maintaining the tumorigenesis, resistance to therapy of pancreatic cancer has also been verified in numerous studies [36,38,39]. And many have devoted themselves to the development of new targets in Wnt signaling for novel therapeutics. The inhibition effect of the secreted Frizzled-related proteins (SFRPs), for instance, were substantiated recently in a study [40]. WIFs (Wnt inhibitory factors) inhibiting Wnt ligands were also proved as effective to attenuate the proliferation of several types of cancers [41].
The Notch pathway
Also intensively investigated is the Notch pathway whose role of inducing the EMT and initiate several solid cancers, including pancreatic cancer has been verified, and potential to be therapeutic targets in CSCs has gained support and applause from many researchers.
Notch signaling is highly conserved in human beings that functions as a regulator of intercellular communication and a determinant of cell fate through the cell-cell interaction. There are four membrane Notch receptor proteins (Notch1–4) and 5 canonical transmembrane ligands (Delta family, DII-1, Dll-3, Dll-4, Jagged1 and Jagged2). The receptor–ligand interaction induces proteolytic cleavages of Notch receptors, which finally translocate the mature notch receptor engaged with membrane-associated ligands from the cell membrane into the nucleus, where the receptor complexes with the transcriptional repressor CSL (RBP-Jκ) and co-activators, and then initiate the transcription of such target genes as Hey and Hes families of transcriptional repressors (Figure 3 [42]).
Figure 3.
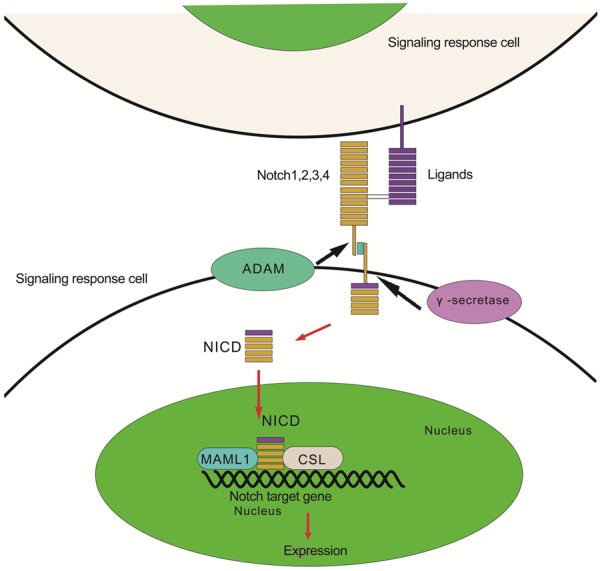
Schematic of Notch signaling. The activation of the Notch signaling is initiated by the bind of Notch receptors to their ligands sent out from the neighboring cell. The Notch-ligand interaction then trigger the enzymolysis of ADAM and γ-secretase to Notch receptors which afterward go through three times of cleaves that make NICD separated from the Notch complex. Finally, the NICD is translocated into the nucleus where it binds to CSL (transcription repressor) and co-activator (MAML1) and initiate the expression of target genes such Hey and Hes family.
Researchers in the recent a few years summarized the involvement of Notch signaling in myeloid malignancies as a tumor suppressor or an oncogene, indicating the tissue type dependent feature of the Notch signaling [43-45]. Whereas, few studies are currently available to explain the exact mechanisms concerning the activation of different pathways and the complicated interaction among them, based on which, novel and effective targeting strategies may be developed and bring about a hopeful promise for patients.
However, one of the major challenges in eliminating CSCs by targeting one or all of the above self-renewal pathways is the involvement of distinct signaling pathways in the crosstalk between the complicated networking pathways. A study has found that PI3K/AKT (phosphoinositide 3-kinase inhibitor/protein kinase B) pathway was key to regulate the Hh signaling pathway by blocking the degradation of the main components of Hh signaling, and the blockade of PI3K/AKT was effective to sensitize the breast cancer to tamoxifen treatment [46]. Future studies should focus more on the clarification of different signaling pathways and the complex relationships between them, thus to construct the basis for combination therapies in cancers.
Pancreatic CSCs and tumor metastasis
Hermann and colleagues [17] has identified two distinct subsets of CSCs in pancreatic cancer: the quiescent and migrating CSCs, the latter has been proven to be in existence by the obviously detectable expression of CXCR4, a chemokine receptor for the ligand SDF-1 (stromal derived factor-1), a well characterized mediator of cell migration. Hermann et al reported that CXCR4 was coexpressed in CD133+ pancreatic CSCs and that CD133+ CXCR4- and CD133+CXCR4+ cells were both capable of forming primary tumors, but only CD133+ cells with the expression of CXCR4+ were endowed the ability to metastasize. Notably, what has caught so much attention in their study was that the CXCR4 inhibition prevented metastasis in established tumor models [17]. These findings may be therapeutically hopeful when scheming drug agents to inhibit metastasis of CSCs.
Whereas, for CSCs in a static biological state to acquire the metastatic and chemotherapy-resistant phenotype, an EMT (epithelial-to-mesenchymal transition) process, in which epithelial cells lose their owned traits such as cell polarity and adhesion between cells and assume a mesenchymal cell phenotype [47] bearing properties including migration and invasion abilities that are crucial for cancer genesis and progression, is of the essence. The transition of epithelial to mesenchymal was thought to be regulated by many factors and proteins such as TGF-beta [48-51], Id protein [47], Smad4 [52], mi-RNA [53], ZEB-1 [53] and mTORC [54] et al.
Numerous genes (Bmi1, BMP4, BST2, BTG1, FOLR1, FoxQ1, PRKAR1A, Sox4, TACSTD2, and Wnt3a), in a research paper by Bao et al [55] were reported to be differentially expressed in CSCs (marked by CD44+/CD133+/EpCAM+) which were isolated from human pancreatic cancer cells (MiaPaCa-2 and L3.6pl cells) by using FACS method. One of these genes is FoxQ1, a member of the forkhead transcription factor family known to be critically involved in the regulation of gene expression during different biological processes such as early development, metabolism, and immune function, the inhibition of which was found to attenuate the invasiveness, renewal ability, and growth potential of CSCs. This study suggested that FoxQ1 was crucial in the regulation of CSC phenotypes and functions and that the over-expression of FoxQ1 was mediated by TGF-β which might lead to the EMT process.
Epithelial-mesenchymal transition (EMT) is a phenotypic changing process in which epithelial cells lose cell-cell and cell-extracellular matrix contacts and then migrate to distant sites. Once these mesenchymal cells reach a site, they can undergo a mesenchymal-epithelial transition (MET) process. The EMT process functions necessarily in the embryonic development by differentiating into different tissues, facilitating the morphological formation of various organs and regenerating tissues after injury. The induction of EMT can be triggered by many factors ranging from cytokines including TGF-β, and types of transcription factors such as Twist [56-58], Snail [59-61], Slug [62,63] and KLF8 [64-66], to various signaling pathways encompassing the ones we have discussed above, and certain exogenous drug agents will be discussed below.
Unfortunately, there is an increasingly accepted consensus suggesting that the inappropriate activation of the EMT processes contributes in part to the increased invasive and metastatic potential of various tumors [67-74]. There are also evidences exist associating the EMT process with the origin of CSCs and considering EMT as a precondition for cancer metastasis. Based on this, many researchers are striving to develop EMT as a target for cancer treatment [49,51,69,70]. resveratrol on human lung cancer, Wang H et al [75] found that resveratrol (trans-3,4,5 trihydroxy stilbene, found in several plants) was effective to increase the expression of epithelial phenotype marker E-cadherin and to downregulate the mesenchymal marker Fibronectin and Vimentin, which led them to conclude that the resveratrol could be used as an inhibitor of TGF-β1-induced EMT, in that it triggered the process of MET reversal in in virto conditions with A549 lung cancer cells whose invasiveness and metastasis were largely reduced [75]. Salinomycin was identified in breast cancer model by Gupta and colleagues to show selective toxicity for epithelial CSCs with a much more efficacy than paclitaxel, a common chemotherapeutic agent used in breast cancers [76], and was later found therapeutically promising in pancreatic cancer [77,78].
Marko and colleagues [79] showed that Id expression rendered breast cancer cells a stem-like phenotype with the epithelial properties retained. And Id1’s role in inducing MET in lung colonization was only taken in cells who had already taken the EMT switch. Additionally, they revealed that the knockdown of Id1 in migrating cells initiated the inhibition of MET and the reduction of lung colonization.
MicroRNAs (miRNAs), small non-coding RNAs whose critical roles have been confirmed in development, have also emerged as significant regulators of CSCs in types of malignancies and have been substantiated as inspiring potential targets for cancer therapy [72,80-85]. Downregulated expression of micro-RNA494 (miR494) mediated by the loss of SMAD4, as demonstrated in a recent study, was correlated to the enhanced level of FoxM1 and the increased activity of beta-catenin signaling in pancreatic cancer cells [86]. Li and colleagues further verified that the transgenic expression of MIR494 in PDAC cells, the leveled-up expression of FOXM1 or the inhibited nuclear translocation of beta-catenin similarly reduced cell proliferation, migration, and invasion and increased their sensitivity to gemcitabine, which finally brought them the conclusion that reduced expression of miR494 correlated with pancreatic cancer metastasis and the shortened survival period of patients and that the miR494 might serve as a prognostic marker for patients with pancreatic carcinoma [86]. It has also been observed that miR200 miRNAs and miR205 are obviously downregulated in cancer cell experiencing EMT and in metastatic breast cancer samples [87,88]. Here we dare propose that miRNAs could offer clues for bettered targeting strategies aiming for CSCs in pancreatic adenocarcinoma.
Pancreatic CSCs and resistance to therapy
One of the major reasons leading to an extremely dismal prognosis for patients with pancreatic cancer is the resistance of CSCs to current chemo-radiation therapy, and the mechanism involved still remains to be elucidated.
Todaro and colleagues [89] demonstrated that CD133+ colon CSCs were rendered resistant to oxiplatin and fluorouracil, which was mediated by the expression of IL-4 in the CD133+ colon CSCs. IL-4 receptor antagonist or anti-IL-4 neutralizing antibody were found to attenuate the chemotherapy tolerance and to bring about more promising outcomes for other types of human cancers. And interestingly, some other studies suggest that aberrantly activated developmental pathways may contribute to CSCs’ resistance to chemotherapy and radiation. For example, Wnt/-catenin signaling has been reported to result in increased tolerance of DNA damage, rendering CSCs resistant to radiation [90,91]. And Notch pathway has been implicated in the failure of cancers in breast, pancreas to response with high efficacy to chemo- and radiation therapy [30,92,93]. In addition, TGF-beta pathway was found to play a role in the metastasis of breast cancer by David and colleagues [50]. However, as observed in their study, TGF-beta has a complicated role in cancer progression: TGF-beta was able to either act as a cancer suppressor or promoter depending on cancer type and stage in process of cancer progression. According to Wang et al [94] in a recent published study, ATDC (Ataxia-Telangiectasia Group D Associated gene) was over-expressed in human pancreatic cancer and was able to contribute to the radio-resistant phenotype of pancreatic cancer. In a word, CSCs are the core of chemotherapy and radiotherapy resistance, which is the root of recurrence; it thus should be effectively targeted to eliminate cancer.
CSCs and circulating cancer cells
Many have accepted the assertion that circulating tumor cells in peripheral blood are, much alike to those residing in cancer tissues, heterogeneous and capable of initiating new cancers which are also hierarchically organized. But the number of the circulating cancer cells is very small even in metastatic cancers and it’s nearly undetectable in normal humans. In the interest of verifying the potential predictive value of these rare circulating cancer cells, also termed circulating tumor cells or isolated tumor cells (CTCs or ITCs), in prejudging the prognosis of intractable metastatic malignancies, investigators of generations have been striving to accelerate the technological and theoretical progress of people’s knowledge to CTCs, the detection and enumeration of which were assumed as novel indicators monitoring the response to anti-cancer therapeutics of various types of cancers [95-97]. Main methods that are currently available for the detection of CTCs in the blood of patients are CellSearch technology and AdnaTest BreastCancer Select/Detect. Both are dependent on EpCAM-positive cancer cells captured in peripheral blood [98]. As we discussed in this review, the phenotypic shift EMT endows a CSC with the migrating phenotype and then to initiate metastasis, which also implies whether or not CSCs could become migrating or circulating CSCs is depended on the specific expression pattern of chemokine receptors, adhesion molecules on these cells and certain stimuli from the CSC niche. And it still remains unclear whether the EpCAM-negative CTCs also contain populations of migrating CSCs that evade the detection by the CellSearch and that can also initiate metastasis. Current studies can only identify CTCs from a EpCAM level with an quite low efficiency. Xenograft assays, therefore, was suggested as a hopeful method for characterizing CTCs [99]. However, it was also noted that this is highly challenging to realize because of the extremely small number of CTCs and the metastasis initiating CSCs that can be obtained directly from patients with metastatic cancer.
CSCs-targeted treatments in pancreatic cancer
The competence of CSCs to evade the traditional chemotherapy agents often results in the recurrence of cancer. Thus CSCs-specified therapeutic tactics are urgently needed for potential complete eradication of such cancers as human pancreatic cancer. Therapeutics targeting CSC-related signaling pathways and EMT process have already been investigated in emerging studies [28,100-103]. Numerous in-depth studies still remain desperately necessary to determine which targets are best to kill CSCs in pancreatic cancer without assassinating the normal stem cells and to be developed as efficient drugs for future clinical usage. Stem cell surface markers such as CD44, CD133, ESA, CD24 et al were identified to have a high level of expression in human pancreatic carcinoma CSCs compared to the rest of the cancer cells. We assume that CSCs cell surface molecules-targeted treatments might provide an effective avoidance of harming the normal stem cells and leaving out CSC subpopulations. CD44, for example, was used as a therapeutic target in the treatment aiming for fully eliminating human hematological malignancies [104]. We speculate that there may be markers that are exclusively expressed in CSCs which are uniquely specific and highly distinguishable, and such markers are perfect candidates for targeted strategies and are what we ultimately expect to find. But as we get to know more about the pathophysiological traits of CSCs, we find ourselves confronted with a more challenging task. Pancreatic cancer stem cells may have many genetic or epigenetic types. So, if a treatment project is effective to one type of pancreatic cancer stem cells, it may be ineffective to another. Furthermore, the CSCs plasticity is an obstacle for targeted therapy, such as interconversion between cancer stem and non-stem phenotypes, in this case, existing CSCs are killed but newly produced CSC populations through activators from the tumor microenvironment or EMT are regenerating cancer tissues. We propose that novel design of CSCs-targeting agents should take into account the exact controlling mechanisms of the formation and maintenance of the equilibrium between cancer stem cells and non-stem cells.
Conclusions
As evidenced in emerging studies, a subpopulation of cells that locate within a hierarchically organized structure of a cancer and that are functionally termed as CSCs is exclusively responsible for the initiation, recurrence, metastasis and resistance to current therapies that fail to effectually target CSCs. This failure causes a much shortened median survival period for patients, which is true with pancreatic cancer. Present data has given a clue for developing therapeutics targeting CSCs by taking advantage of their controlling signaling pathways and specific expression of certain surface markers such as CD44, CD133 in human pancreatic cancer. Besides, mountains of genes and molecules such as miRNA200 and miRNA34 family members have been verified to be interwound with the regulatory networks of CSCs in pancreatic carcinoma and investigated as potential targets for helping eradicating CSCs.
Therefore, a bettered understanding of the mechanisms involved in malignant behaviors of CSCs brings with a great promise for future exploitation of new curative approaches to life-threatening pancreatic adenocarcinoma, although it is without a doubt that there are challenges and difficulties still remaining to be overcome. Now that basic concepts and characteristics, identification and isolation of pancreatic cancer stem cells have been extensively and thoroughly investigated, new methods will be springing up to serve this end with the efforts of the whole world. All fundamental researches funded by various organizations should be designed to offer new and better approaches to eradicate human pancreatic cancer. We expect there will be a breakthrough in therapeutic agents targeting not only the existing CSCs but also the non-CSCs.
Acknowledgements
Thanks are due to Zhou Yuan for assistance with the draft writing and valuable discussion. And both authors read and approved the final manuscript.
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Bonnet D, Dick J. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 3.Lapidot T, Sirard C, Vormoor J. A cell initiating human acute myeloid leukemia after transplantation into SCID mice. Nature. 1994:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 4.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, Bruns CJ, Heeschen C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Van den Broeck A, Vankelecom H, Van Delm W, Gremeaux L, Wouters J, Allemeersch J, Govaere O, Roskams T, Topal B. Human pancreatic cancer contains a side population expressing cancer stem cell-associated and prognostic genes. PLoS One. 2013;8:e73968. doi: 10.1371/journal.pone.0073968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kopp JL, Sander M. New insights into the cell lineage of pancreatic ductal adenocarcinoma: evidence for tumor stem cells in premalignant lesions? Gastroenterology. 2014;146:24–26. doi: 10.1053/j.gastro.2013.11.023. [DOI] [PubMed] [Google Scholar]
- 7.Lee CJ, Dosch J, Simeone DM. Pancreatic Cancer Stem Cells. J. Clin. Oncol. 2008;26:2806–2812. doi: 10.1200/JCO.2008.16.6702. [DOI] [PubMed] [Google Scholar]
- 8.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 9.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3938. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan X, Ma L, Yi D. A CD133-related gene expression signature identifies an aggressive glioblastoma subtype with excessive mutations. Proc Natl Acad Sci U S A. 2011;108:1591–1596. doi: 10.1073/pnas.1018696108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beier D, Hau P, Proescholdt M. CD133(+) and CD133(-) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67:4010–4015. doi: 10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- 12.Boman BM, Wicha MS. Cancer stem cells: a step toward the cure. J. Clin. Oncol. 2008;26:2795–2799. doi: 10.1200/JCO.2008.17.7436. [DOI] [PubMed] [Google Scholar]
- 13.Shmelkov SV, Butler JM, Hooper AT. CD133 expression is not restricted to stem cells, and both CD133+ and CD133- metastatic colon cancer cells initiate tumors. J Clin Invest. 2008;118:2111–2120. doi: 10.1172/JCI34401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bertolini G, Roz L, Perego P, Tortoreto M, Fontanella E. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc Natl Acad Sci U S A. 2009;106:16281–16286. doi: 10.1073/pnas.0905653106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meyer MJ, Fleming JM, Lin AF, Hussnain SA, Ginsburg E, Vonderhaar BK. CD44posCD49fhiCD133/2hi defines xenograft-initiating cells in estrogen receptor-negative breast cancer. Cancer Res. 2010;70:4624–4633. doi: 10.1158/0008-5472.CAN-09-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective Identification of Tumorigenic Prostate Cancer Stem Cells. Cancer Res. 2005;65:10946–10951. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- 17.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, Bruns CJ, Heeschen C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 18.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 19.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 20.Beier D, Hau P, Proescholdt M, Lohmeier A, Wischhusen J, Oefner PJ, Aigner L, Brawanski A, Bogdahn U, Beier CP. CD133+ and CD133− Glioblastoma-Derived Cancer Stem Cells Show Differential Growth Characteristics and Molecular Profiles. Cancer Res. 2007;67:4010–4015. doi: 10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Sakariassen PØ, Tsinkalovsky O, Immervoll H, Bøe SO, Svendsen A, Prestegarden L, Røsland G, Thorsen F, Stuhr L, Molven A, Bjerkvig R, Enger PØ. CD133 negative glioma cells form tumors in nude rats and give rise to CD133 positive cells. Int J Cancer. 2008;122:761–768. doi: 10.1002/ijc.23130. [DOI] [PubMed] [Google Scholar]
- 22.Bailey JM, Alsina J, Rasheed ZA, McAllister FM, Fu YY, Plentz R, Zhang H, Pasricha PJ, Bardeesy N, Matsui W. DCLK1 Marks a Morphologically Distinct Subpopulation of Cells With Stem Cell Properties in Preinvasive Pancreatic Cancer. Gastroenterology. 2014;146:245–256. doi: 10.1053/j.gastro.2013.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su HT, Weng CC, Hsiao PJ, Chen LH, Kuo TL, Chen YW, Kuo KK, Cheng KH. Stem cell marker nestin is critical for TGF-beta1-mediated tumor progression in pancreatic cancer. Mol Cancer Res. 2013;11:768–779. doi: 10.1158/1541-7786.MCR-12-0511. [DOI] [PubMed] [Google Scholar]
- 24.Matsuda Y, Kure S, Ishiwata T. Nestin and other putative cancer stem cell markers in pancreatic cancer. Med Mol Morphol. 2012;45:59–65. doi: 10.1007/s00795-012-0571-x. [DOI] [PubMed] [Google Scholar]
- 25.Duong HQ, Hwang JS, Kim HJ, Kang HJ, Seong YS, Bae I. Aldehyde dehydrogenase 1A1 confers intrinsic and acquired resistance to gemcitabine in human pancreatic adenocarcinoma MIA PaCa-2 cells. Int J Oncol. 2012;41:855–861. doi: 10.3892/ijo.2012.1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van den Hoogen C, van der Horst G, Cheung H, Buijs JT, Lippitt JM, Guzman-Ramirez N, Hamdy FC, Eaton CL, Thalmann GN, Cecchini MG, Pelger RC, vander Pluijm G. High aldehyde dehydrogenase activity identifies tumor-initiating and metastasis-initiating cells in human prostate cancer. Cancer Res. 2010;70:5163–5173. doi: 10.1158/0008-5472.CAN-09-3806. [DOI] [PubMed] [Google Scholar]
- 27.Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501:328–337. doi: 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Onishi H, Katano M. Hedgehog signaling pathway as a new therapeutic target in pancreatic cancer. World J Gastroenterol. 2014;20:2335–2342. doi: 10.3748/wjg.v20.i9.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buchler P, Gazdhar A, Schubert M, Giese N, Reber HA, Hines OJ, Giese T, Ceyhan GO, Muller M, Buchler MW, Friess H. The Notch signaling pathway is related to neurovascular progression of pancreatic cancer. Ann Surg. 2005;242:791–800. doi: 10.1097/01.sla.0000189115.94847.f1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abel EV, Kim EJ, Wu J, Hynes M, Bednar F, Proctor E, Wang L, Dziubinski ML, Simeone DM. The Notch pathway is important in maintaining the cancer stem cell population in pancreatic cancer. PLoS One. 2014;9:e91983. doi: 10.1371/journal.pone.0091983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee JY, Song SY, Park JY. Notch pathway activation is associated with pancreatic cancer treatment failure. Pancreatology. 2014;14:48–53. doi: 10.1016/j.pan.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 32.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 33.Manning B, Cantley L. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thayer SP, di Magliano MP, Heiser PW, Nielsen CM, Roberts DJ, Lauwers GY, Qi YP, Gysin S, Fernández-del Castillo C, Yajnik V, Antoniu B, McMahon M, Warshaw AL, Hebrok M. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425:851–856. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bovolenta P, Rodriguez J, Esteve P. Frizzled/RYK mediated signalling in axon guidance. Development. 2006;133:4399–4408. doi: 10.1242/dev.02592. [DOI] [PubMed] [Google Scholar]
- 36.Pasca di Magliano M, Biankin AV, Heiser PW, Cano DA, Gutierrez PJ, Deramaudt T, Segara D, Dawson AC, Kench JG, Henshall SM, Sutherland RL, Dlugosz A, Rustgi AK, Hebrok M. Common activation of canonical Wnt signaling in pancreatic adenocarcinoma. PLoS One. 2007;2:e1155. doi: 10.1371/journal.pone.0001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 38.Lai SL, Chien AJ, Moon RT. Wnt/Fz signaling and the cytoskeleton: potential roles in tumorigenesis. Cell Res. 2009;19:532–545. doi: 10.1038/cr.2009.41. [DOI] [PubMed] [Google Scholar]
- 39.Jiang H, Li Q, He C, Li F, Sheng H, Shen X, Zhang X, Zhu S, Chen H, Chen X, Yang C, Gao H. Activation of the Wnt pathway through Wnt2 promotes metastasis in pancreatic cancer. Am J Cancer Res. 2014;4:537–544. [PMC free article] [PubMed] [Google Scholar]
- 40.Bovolenta P, Esteve P, Ruiz JM, Cisneros E, Lopez-Rios J. Beyond Wnt inhibition: new functions of secreted Frizzled-related proteins in development and disease. J Cell Sci. 2008;121:737–746. doi: 10.1242/jcs.026096. [DOI] [PubMed] [Google Scholar]
- 41.Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13:11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 42.Espinoza I, Pochampally R, Xing F, Watabe K, Miele L. Notch signaling: targeting cancer stem cells and epithelial-to-mesenchymal transition. Onco Targets Ther. 2013;6:1249–1259. doi: 10.2147/OTT.S36162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lobry C, Oh P, Mansour MR, Look AT, Aifantis I. Notch signaling: switching an oncogene to a tumor suppressor. Blood. 2014;123:2451–2459. doi: 10.1182/blood-2013-08-355818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Radtke F, Raj K. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat Rev Cancer. 2003;3:756–767. doi: 10.1038/nrc1186. [DOI] [PubMed] [Google Scholar]
- 45.Lobry C, Oh P, Aifantis I. Oncogenic and tumor suppressor functions of Notch in cancer: it’s NOTCH what you think. J Exp Med. 2011;208:1931–1935. doi: 10.1084/jem.20111855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramaswamy B, Lu Y, Teng KY, Nuovo G, Li X, Shapiro CL, Majumder S. Hedgehog signaling is a novel therapeutic target in tamoxifen-resistant breast cancer aberrantly activated by PI3K/AKT pathway. Cancer Res. 2012;72:5048–5059. doi: 10.1158/0008-5472.CAN-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lasorella A, Benezra R, Iavarone A. The ID proteins: master regulators of cancer stem cells and tumour aggressiveness. Nat Rev Cancer. 2014;14:77–91. doi: 10.1038/nrc3638. [DOI] [PubMed] [Google Scholar]
- 48.Oft M, Peli J, Rudaz C, Schwarz H, Beug H, Reichmann E. TGF-beta1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes Dev. 1996;10:2462–2477. doi: 10.1101/gad.10.19.2462. [DOI] [PubMed] [Google Scholar]
- 49.Xu Z, Shen MX, Ma DZ, Wang LY, Zha XL. TGF-beta1-promoted epithelial-to-mesenchymal transformation and cell adhesion contribute to TGF-beta1-enhanced cell migration in SMMC-7721 cells. Cell Res. 2003;13:343–350. doi: 10.1038/sj.cr.7290179. [DOI] [PubMed] [Google Scholar]
- 50.Padua D, Massague J. Roles of TGF [beta] in metastasis. Cell Res. 2009;19:89–102. doi: 10.1038/cr.2008.316. [DOI] [PubMed] [Google Scholar]
- 51.Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19:156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen YW, Hsiao PJ, Weng CC, Kuo KK, Kuo TL, Wu DC, Hung WC, Cheng KH. SMAD4 Loss triggers the phenotypic changes of pancreatic ductal adenocarcinoma cells. BMC Cancer. 2014;14:181. doi: 10.1186/1471-2407-14-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wellner U, Brabletz T, Keck T. ZEB1 in Pancreatic Cancer. Cancers (Basel) 2010;2:1617–1628. doi: 10.3390/cancers2031617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gulhati P, Bowen KA, Liu J, Stevens PD, Rychahou PG, Chen M, Lee EY, Weiss HL, O’Connor KL, Gao T. mTORC1 and mTORC2 regulate EMT, motility, and metastasis of colorectal cancer via RhoA and Rac1 signaling pathways. Cancer Res. 2011;71:3246–3256. doi: 10.1158/0008-5472.CAN-10-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bao B, Azmi A, Aboukameel A, Ahmad A, Bolling-Fischer A, Sethi S, Ali S, Li Y, Kong D, Banerjee S. Pancreatic cancer stem-like cells display aggressive behavior mediated via activation of FoxQ1. J Biol Chem. 2014;289:14520–33. doi: 10.1074/jbc.M113.532887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang MH, Chen CL, Chau GY, Chiou SH, Su CW, Chou TY, Peng WL, Wu JC. Comprehensive analysis of the independent effect of twist and snail in promoting metastasis of hepatocellular carcinoma. Hepatology. 2009;50:1464–1474. doi: 10.1002/hep.23221. [DOI] [PubMed] [Google Scholar]
- 57.Ishikawa D, Shimada M, Utsunomiya T, Morine Y, Imura S, Ikemoto T, Arakawa Y, Kanamoto M, Iwahashi S, Saito Y, Yamada S, Miyake H. Effect of Twist and Bmi-1 on intraductal papillary mucinous neoplasm of the pancreas. J Gastroenterol Hepatol. 2014;29:2032–2037. doi: 10.1111/jgh.12652. [DOI] [PubMed] [Google Scholar]
- 58.Shi J, Wang Y, Zeng L, Wu Y, Deng J, Zhang Q, Lin Y, Li J, Kang T, Tao M, Rusinova E, Zhang G, Wang C, Zhu H, Yao J, Zeng YX, Evers BM, Zhou MM, Zhou BP. Disrupting the interaction of BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell. 2014;25:210–225. doi: 10.1016/j.ccr.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun M, Guo X, Qian X, Wang H, Yang C, Brinkman KL, Serrano-Gonzalez M, Jope RS, Zhou B, Engler DA, Zhan M, Wong ST, Fu L, Xu B. Activation of the ATM-Snail pathway promotes breast cancer metastasis. J Mol Cell Biol. 2012;4:304–315. doi: 10.1093/jmcb/mjs048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hahn S, Jackstadt R, Siemens H, Hunten S, Hermeking H. SNAIL and miR-34a feed-forward regulation of ZNF281/ZBP99 promotes epithelial-mesenchymal transition. EMBO J. 2013;32:3079–3095. doi: 10.1038/emboj.2013.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng H, Shen M, Zha YL, Li W, Wei Y, Blanco MA, Ren G, Zhou T, Storz P, Wang HY, Kang Y. PKD1 phosphorylation-dependent degradation of SNAIL by SCF-FBXO11 regulates epithelial-mesenchymal transition and metastasis. Cancer Cell. 2014;26:358–373. doi: 10.1016/j.ccr.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carpenter RL, Paw I, Dewhirst MW, Lo HW. Akt phosphorylates and activates HSF-1 independent of heat shock, leading to Slug overexpression and epithelial-mesenchymal transition (EMT) of HER2-overexpressing breast cancer cells. Oncogene. 2015;34:546–557. doi: 10.1038/onc.2013.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li A, Morton JP, Ma Y, Karim SA, Zhou Y, Faller WJ, Woodham EF, Morris HT, Stevenson RP, Juin A, Jamieson NB, MacKay CJ, Carter CR, Leung HY, Yamashiro S, Blyth K, Sansom OJ, Machesky LM. Fascin is regulated by slug, promotes progression of pancreatic cancer in mice, and is associated with patient outcomes. Gastroenterology. 2014;146:1386–1396. e1381–1317. doi: 10.1053/j.gastro.2014.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang X, Lu H, Urvalek AM, Li T, Yu L, Lamar J, DiPersio CM, Feustel PJ, Zhao J. KLF8 promotes human breast cancer cell invasion and metastasis by transcriptional activation of MMP9. Oncogene. 2011;30:1901–1911. doi: 10.1038/onc.2010.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang H, Liu L, Wang Y, Zhao G, Xie R, Liu C, Xiao X, Wu K, Nie Y, Zhang H, Fan D. KLF8 involves in TGF-beta-induced EMT and promotes invasion and migration in gastric cancer cells. J Cancer Res Clin Oncol. 2013;139:1033–1042. doi: 10.1007/s00432-012-1363-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin F, Shen Z, Tang LN, Zheng SE, Sun YJ, Min DL, Yao Y. KLF8 knockdown suppresses proliferation and invasion in human osteosarcoma cells. Mol Med Rep. 2014;9:1613–1617. doi: 10.3892/mmr.2014.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rosanò L, Spinella F, Di Castro V, Nicotra MR, Dedhar S, de Herreros AG, Natali PG, Bagnato A. Endothelin-1 Promotes Epithelial-to-Mesenchymal Transition in Human Ovarian Cancer Cells. Cancer Res. 2005;65:11649–11657. doi: 10.1158/0008-5472.CAN-05-2123. [DOI] [PubMed] [Google Scholar]
- 68.Kitagawa K, Murata A, Matsuura N, Tohya K, Takaichi S, Monden M, Inoue M. Epithelial-mesenchymal transformation of a newly established cell line from ovarian adenosarcoma by transforming growth factor-beta1. Int J Cancer. 1996;66:91–97. doi: 10.1002/(SICI)1097-0215(19960328)66:1<91::AID-IJC16>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 69.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 70.Petersen OW, Nielsen HL, Gudjonsson T, Villadsen R, Rank F, Niebuhr E, Bissell MJ, Ronnov-Jessen L. Epithelial to mesenchymal transition in human breast cancer can provide a nonmalignant stroma. Am J Pathol. 2003;162:391–402. doi: 10.1016/S0002-9440(10)63834-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yokoyama K, Kamata N, Fujimoto R, Tsutsumi S, Tomonari M, Taki M, Hosokawa H, Nagayama M. Increased invasion and matrix metalloproteinase-2 expression by Snail-induced mesenchymal transition in squamous cell carcinomas. Int J Oncol. 2003;22:891–898. [PubMed] [Google Scholar]
- 72.Rosano L, Spinella F, Di Castro V, Nicotra MR, Dedhar S, de Herreros AG, Natali PG, Bagnato A. Endothelin-1 promotes epithelial-to-mesenchymal transition in human ovarian cancer cells. Cancer Res. 2005;65:11649–11657. doi: 10.1158/0008-5472.CAN-05-2123. [DOI] [PubMed] [Google Scholar]
- 73.Xu J, Wang R, Xie ZH, Odero-Marah V, Pathak S, Multani A, Chung LWK, Zhau HE. Prostate cancer metastasis: Role of the host microenvironment in promoting epithelial to mesenchymal transition and increased bone and adrenal gland metastasis. Prostate. 2006;66:1664–1673. doi: 10.1002/pros.20488. [DOI] [PubMed] [Google Scholar]
- 74.Yang AD, Camp ER, Fan F, Shen L, Gray MJ, Liu W, Somcio R, Bauer TW, Wu Y, Hicklin DJ, Ellis LM. Vascular endothelial growth factor receptor-1 activation mediates epithelial to mesenchymal transition in human pancreatic carcinoma cells. Cancer Res. 2006;66:46–51. doi: 10.1158/0008-5472.CAN-05-3086. [DOI] [PubMed] [Google Scholar]
- 75.Wang H, Zhang H, Tang L, Chen H, Wu C, Zhao M, Yang Y, Chen X, Liu G. Resveratrol inhibits TGF-β1-induced epithelial-to-mesenchymal transition and suppresses lung cancer invasion and metastasis. Toxicology. 2013;303:139–146. doi: 10.1016/j.tox.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 76.Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, Lander ES. Identification of Selective Inhibitors of Cancer Stem Cells by High-Throughput Screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang GN, Liang Y, Zhou LJ, Chen SP, Chen G, Zhang TP, Kang T, Zhao YP. Combination of salinomycin and gemcitabine eliminates pancreatic cancer cells. Cancer Lett. 2011;313:137–144. doi: 10.1016/j.canlet.2011.05.030. [DOI] [PubMed] [Google Scholar]
- 78.He L, Wang F, Dai WQ, Wu D, Lin CL, Wu SM, Cheng P, Zhang Y, Shen M, Wang CF, Lu J, Zhou YQ, Xu XF, Xu L, Guo CY. Mechanism of action of salinomycin on growth and migration in pancreatic cancer cell lines. Pancreatology. 2013;13:72–78. doi: 10.1016/j.pan.2012.11.314. [DOI] [PubMed] [Google Scholar]
- 79.Marko S, Svetlana P, Yvette C, Edi B, David P, Larry N, Joan M, Robert B. TGF-β-Id1 Signaling Opposes Twist1 and Promotes Metastatic Colonization via a Mesenchymal-to-Epithelial Transition. Cell Reports. 2013;5:1228–1242. doi: 10.1016/j.celrep.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu S, Patel SH, Ginestier C, Ibarra I, Martin-Trevino R, Bai S, McDermott SP, Shang L, Ke J, Ou SJ, Heath A, Zhang KJ, Korkaya H, Clouthier SG, Charafe-Jauffret E, Birnbaum D, Hannon GJ, Wicha MS. MicroRNA93 regulates proliferation and differentiation of normal and malignant breast stem cells. PLoS Genet. 2012;8:e1002751. doi: 10.1371/journal.pgen.1002751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ji Q, Hao X, Zhang M, Tang W, Yang M, Li L, Xiang D, Desano JT, Bommer GT, Fan D, Fearon ER, Lawrence TS, Xu L. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS One. 2009;4:e6816. doi: 10.1371/journal.pone.0006816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shi Y, Liu C, Liu X, Tang DG, Wang J. The microRNA miR-34a inhibits non-small cell lung cancer (NSCLC) growth and the CD44hi stem-like NSCLC cells. PLoS One. 2014;9:e90022. doi: 10.1371/journal.pone.0090022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 84.Lynam-Lennon N, Maher SG, Reynolds JV. The roles of microRNA in cancer and apoptosis. Biol Rev Camb Philos Soc. 2009;84:55–71. doi: 10.1111/j.1469-185X.2008.00061.x. [DOI] [PubMed] [Google Scholar]
- 85.Kent OA, Mendell JT. A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene. 2006;25:6188–6196. doi: 10.1038/sj.onc.1209913. [DOI] [PubMed] [Google Scholar]
- 86.Li L, Li Z, Kong X, Xie D, Jia Z, Jiang W, Cui J, Du Y, Wei D, Huang S, Xie K. Down-regulation of microRNA-494 via loss of SMAD4 increases FOXM1 and beta-catenin signaling in pancreatic ductal adenocarcinoma cells. Gastroenterology. 2014;147:485–497. e418. doi: 10.1053/j.gastro.2014.04.048. [DOI] [PubMed] [Google Scholar]
- 87.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 88.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Todaro M, Alea MP, Di Stefano AB. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell. 2007;1:389–402. doi: 10.1016/j.stem.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 90.Ayyanan A, Civenni G, Ciarloni L, Morel C, Mueller N, Lefort K, Mandinova A, Raffoul W, Fiche M, Dotto GP, Brisken C. Increased Wnt signaling triggers oncogenic conversion of human breast epithelial cells by a Notch-dependent mechanism. Proc Natl Acad Sci U S A. 2006;103:3799–3804. doi: 10.1073/pnas.0600065103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen MS, Woodward WA, Behbod F, Peddibhotla S, Alfaro MP, Buchholz TA, Rosen JM. Wnt/β-catenin mediates radiation resistance of Sca1+ progenitors in an immortalized mammary gland cell line. J Cell Sci. 2007;120:468–477. doi: 10.1242/jcs.03348. [DOI] [PubMed] [Google Scholar]
- 92.Wang Z, Banerjee S, Ahmad A, Li Y, Azmi AS, Gunn JR, Kong D, Bao B, Ali S, Gao J, Mohammad RM, Miele L, Korc M, Sarkar FH. Activated K-ras and INK4a/Arf deficiency cooperate during the development of pancreatic cancer by activation of Notch and NF-kappaB signaling pathways. PLoS One. 2011;6:e20537. doi: 10.1371/journal.pone.0020537. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 93.Pukazhendhi G, Gluck S. Circulating tumor cells in breast cancer. J Carcinog. 2014;13:8. doi: 10.4103/1477-3163.135578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang L, Yang H, Palmbos PL, Ney G, Detzler TA, Coleman D, Leflein J, Davis M, Zhang M, Tang W, Hicks JK, Helchowski CM, Prasad J, Lawrence TS, Xu L, Yu X, Canman CE, Ljungman M, Simeone DM. ATDC/TRIM29 Phosphorylation by ATM/MAPKAP Kinase 2 Mediates Radioresistance in Pancreatic Cancer Cells. Cancer Res. 2014;74:1778–1788. doi: 10.1158/0008-5472.CAN-13-2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LWMM, Hayes DF. Circulating Tumor Cells, Disease Progression, and Survival in Metastatic Breast Cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 96.Cristofanilli M, Hayes DF, Budd GT, Ellis MJ, Stopeck A, Reuben JM, Doyle GV, Matera J, Allard WJ, Miller MC, Fritsche HA, Hortobagyi GN, Terstappen LWMM. Circulating Tumor Cells: A Novel Prognostic Factor for Newly Diagnosed Metastatic Breast Cancer. J. Clin. Oncol. 2005;23:1420–1430. doi: 10.1200/JCO.2005.08.140. [DOI] [PubMed] [Google Scholar]
- 97.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Andreopoulou E, Yang LY, Rangel KM, Reuben JM, Hsu L, Krishnamurthy S, Valero V, Fritsche HA, Cristofanilli M. Comparison of assay methods for detection of circulating tumor cells in metastatic breast cancer: AdnaGen AdnaTest BreastCancer Select/Detect™ versus Veridex CellSearch™ system. Int J Cancer. 2012;130:1590–1597. doi: 10.1002/ijc.26111. [DOI] [PubMed] [Google Scholar]
- 99.Baccelli I, Schneeweiss A, Riethdorf S, Stenzinger A, Schillert A, Vogel V, Klein C, Saini M, Bauerle T, Wallwiener M, Holland-Letz T, Hofner T, Sprick M, Scharpff M, Marme F, Sinn HP, Pantel K, Weichert W, Trumpp A. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol. 2013;31:539–544. doi: 10.1038/nbt.2576. [DOI] [PubMed] [Google Scholar]
- 100.Boman BM, Wicha MS. Cancer stem cells: a step toward the cure. J. Clin. Oncol. 2008;26:2795–2799. doi: 10.1200/JCO.2008.17.7436. [DOI] [PubMed] [Google Scholar]
- 101.Wei P, Niu M, Pan S, Zhou Y, Shuai C, Wang J, Peng S, Li G. Cancer stem-like cell: a novel target for nasopharyngeal carcinoma therapy. Stem Cell Res Ther. 2014;5:44. doi: 10.1186/scrt433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Onishi H, Katano M. Hedgehog signaling pathway as a therapeutic target in various types of cancer. Cancer Sci. 2011;102:1756–1760. doi: 10.1111/j.1349-7006.2011.02010.x. [DOI] [PubMed] [Google Scholar]
- 103.Zhu Z, Khan MA, Weiler M, Blaes J, Jestaedt L, Geibert M, Zou P, Gronych J, Bernhardt O, Korshunov A, Bugner V, Lichter P, Radlwimmer B, Heiland S, Bendszus M, Wick W, Liu HK. Targeting self-renewal in high-grade brain tumors leads to loss of brain tumor stem cells and prolonged survival. Cell Stem Cell. 2014;15:185–198. doi: 10.1016/j.stem.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 104.Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med. 2006;12:1167–1174. doi: 10.1038/nm1483. [DOI] [PubMed] [Google Scholar]


