Abstract
Gastric cancer (GC) remains a serious threat to many people, representing the second leading cause of cancer-related death worldwide. The lack of early diagnostic biomarkers, effective prognostic indicators and therapeutic targets all account for the poor prognosis of GC. Therefore, the identification of novel molecular biomarkers for early diagnosis, therapeutic response, and prognosis are urgently needed. High-throughput sequencing has identified a large number of transcribed long non-coding RNAs (lncRNAs) throughout the human genome. Accumulating evidence demonstrates that these lncRNAs play multiple roles in regulating gene expression at the transcriptional, post-transcriptional, and epigenetic levels. Aberrant expression of lncRNAs occurs in various pathological processes, including GC. Many dysregulated lncRNAs in GC have been significantly associated with a larger tumor size, higher degree of tumor invasion, lymph node and distant metastasis, and poorer survival outcome. In this review, we will provide an overview of the pathogenesis of GC, the characteristics and regulatory functions of lncRNAs, and the versatile mechanisms of lncRNAs in GC development, as well as evaluate the translational potential of lncRNAs as novel diagnostic and prognostic biomarkers and therapeutic targets in GC.
Keywords: Gastric cancer, lncRNA, diagnosis, prognosis, therapy, biomarker
Introduction
Gastric cancer (GC) is one of the most common malignancies and represents the second leading cause of cancer-associated mortality worldwide [1,2]. Many factors are implicated in the carcinogenesis of GC, including genetic factors, Helicobacter pylori infection, unhealthy diet (for example, high intake of salt and nitrates), and smoking [3-6]. Most GC cases are diagnosed at an advanced stage due to a lack of typical early symptoms. Presently, surgical resection and chemoradiotherapy are the main treatment approaches for GC [7], but relapse, distant metastasis and chemo-resistance frequently occur, and the overall 5-year survival for GC is only approximately 25% [8]. Despite great efforts to understand the biological properties of cancer, there have been minimal improvements in the clinical outcome of GC. The lack of diagnostic biomarkers, prognostic indicators and effective therapeutic targets account for the poor outcome of GC. Thus, the underlying mechanisms of GC must be interrogated so that novel promising diagnostic and prognostic molecular biomarkers can be identified and developed to improve the quality of life and survival of GC patients.
Comprehensive investigations over the past few decades have mainly concentrated on the function of critical protein-coding genes and genome alterations in the pathogenesis of GC [9,10]. Recent progress in sequencing technology and genome-wide analysis has revealed that protein-coding genes only account for less than 2% of the total transcriptome, whereas most genes are transcribed to non-protein-coding RNAs (ncRNAs) [11]. Small ncRNAs have lengths shorter than 200 nucleotides (nt) and include microRNAs (miRNAs), small-interfering RNAs (siRNAs) and Piwi-interacting RNAs (piRNAs), and their functions and regulatory mechanisms have been extensively studied, especially the miRNAs [12]. Through complete or incomplete base-pairing complementary to the 3’-untranslated regions (3’-UTRs) of mRNAs, miRNAs mediate targeted mRNA degradation or translational inhibition [13,14]. As the regulators of gene expression, miRNAs play important roles in cancer, and various dysregulated miRNAs are associated with GC [15,16]. In addition to these small ncRNAs, long ncRNAs (lncRNAs) with lengths of more than 200 nt are abundant in the human genome and have attracted increasing scientific interest. Despite being initially regarded as “transcriptional noise”, accumulating evidence has found that lncRNAs can manipulate local or global gene expression via transcriptional, post-transcriptional and epigenetic regulation [17]. The lncRNAs that have been characterized are implicated in diverse physiological and pathological processes, such as X-chromosome inactivation, stem cell pluripotency, development, immune response, cell differentiation, apoptosis, and cancer metastasis and invasion. lncRNAs can interact with proteins, DNA and RNA transcripts to control alternative splicing, chromosome remodeling, nuclear import and mRNA decay, and lncRNAs participate in almost every aspect of gene expression programs [18]. Aberrant lncRNA expression has been discovered in many types of cancers, including GC [19,20]; these dysregulated lncRNAs can function as oncogenes or tumor suppressors to alter cellular pathways [21]. Some lncRNAs are expressed in disease- or tissue-specific patterns, which make lncRNAs attractive as diagnostic, prognostic and therapeutic biomarkers in cancers.
In this review, we summarize the pathogenesis of GC and the functional activity of lncRNAs, especially the underlying mechanisms of lncRNAs in GC development. The aim is to evaluate the significant potential of lncRNAs as novel biomarkers for early diagnosis or prognosis or as therapeutic targets in GC.
Pathogenesis of GC
Environmental risk factors of GC development
GC remains an ongoing serious threat to the public health. There are an estimated 989,000 new cases and approximately 738,000 patients dying from GC throughout the world annually [2,22]. Although GC is the fourth most common malignancy, it is the second most frequent cancer-related cause of death worldwide [2,22]. GC carcinogenesis is a complex and multistep process with high molecular heterogeneity. Environmental factors and genetic alterations within the host play important roles in the etiology of GC development.
Stomach infections by the gram-negative, microaerophilic bacterium H. pylori is a well-established risk factor for GC [23]. The World Health Organization (WHO) defined H. pylori as a class I carcinogen in 1994, and approximately 50% of the population are infected [24]. H. pylori infection can induce chronic inflammation, reactive oxygen species (ROS) accumulation and DNA damage in the gastric mucosa, which induces the normal gastric epithelium to develop atrophic gastritis, intestinal metaplasia, and dysplasia, with eventual progression to carcinoma [25]. Furthermore, H. pylori infection has been found to enhance aberrant promoter methylation modification in the gastric mucosa and gastric epithelial cells, leading to the silencing of certain tumor suppressor genes and the promotion of gastric carcinogenesis [26-28]. Clinical studies suggest that H. pylori eradication can effectively reduce precancerous lesions and GC [29]. In addition to H. pylori infection, Epstein-Barr virus (EBV) infection has also been found to cause GC development [30].
Unhealthy dietary habits and lifestyles are also important factors that increase the risk of GC development. Epidemiological data suggest that a diet rich in salt, N-nitroso compounds and fat is a risk factor for GC development [31]. Salt and N-nitroso ingestion can cause mechanical damage to the gastric mucosa, induce gastritis and promote persistent H. pylori infection [32]. Fresh fruits and vegetables are full of carotenoids, vitamin C, folate, phytochemicals, and fiber, which can modestly reduce GC susceptibility [33]. Cigarette smoking and alcohol intake are established risk factors for GC [34]. Alcohol can stimulate the gastric mucosa, and tobacco may induce precursor gastric lesions and increase H. pylori infection. Drinking green tea may prevent carcinogenesis, as green tea is the most abundant source of epigallocatechin gallate (EGCG), which protects gastric epithelial cells from H. pylori-induced cytotoxicity [35].
Molecular mechanisms of GC based on mRNA and protein alterations
Most GC patients suffer from malignant gastric epithelial lesions. However, GC has highly heterogeneous properties that can be classified into various subgroups based on histological, anatomical, epidemiological and molecular characteristics. Currently, according to gene expression profiling with consensus hierarchical clustering selections, three major subtypes of GC have been identified: proliferative, metabolic and mesenchymal, each of which exhibits characteristic variations in molecular and genetic properties and responses to chemotherapy [36]. Proliferative GC presents high levels of genomic instability, including tumor suppressor p53 (TP53) mutations and DNA methylation. Metabolic patients appear to respond better to 5-fluorouracil treatment. The mesenchymal subtype may have cancer stem-like cells that are sensitive to inhibitors targeting the PI3K/AKT/mTOR signaling pathway [36]. These molecular classifications for GC provide the rationale for more effective personalized therapy.
With advances in high-resolution sequencing technology, a wide range of somatic alterations have been studied in GC [10]. There are frequent gene mutations in TP53, PIK3CA and ARID1A. Mutations in cell adhesion genes, such as FAT4 and the chromatin remodeling genes ARID1A, MLL3 and MLL, are a common occurrence in GC [10]. Among the numerous gene mutations implicated, TP53 alterations have been extensively studied. TP53 plays important roles in cell fate determination and has been described as “the guardian of the genome”. Inactivation mutations and loss of heterozygosity (LOH) of TP53 occur at high frequency and seem to be early events in GC [37]. A comprehensive genomic analysis of 233 GC samples revealed that receptor tyrosine kinase (RTK)/RAS alterations collectively occurred in up to 37% of GC patients, mainly comprising alterations in FGFR2, KRAS, EGFR, HER2 and MET [38]. HER2 is a well-characterized oncogene with tyrosine kinase activity that belongs to the EGFR family. The dimerization of HER2 can induce the autophosphorylation of tyrosine residues within the cytoplasmic domain, which can activate many cellular signaling cascades to trigger proliferation and carcinogenesis. Amplification or overexpression of HER2 is found in approximately 10-30% of gastric/gastroesophageal cancers and could serve as a prognostic and predictive marker. Trastuzumab monoclonal antibody directed at HER-2, which has been shown to decrease shedding of the HER-2 extracellular domain and reduce its dimerization, has now become a standard first-line treatment option for advanced HER-2-positive GC [39].
Our previous studies found several examples of aberrant expression of mRNAs and proteins in GC, which play vital roles in GC development. For example, VEGF-C and CNTN1 levels were significantly correlated with tumor size [40]; overexpressed JMJD2A was positively associated with tumor stage, nodal status and poor prognosis; and PCBP2 upregulation was linked to shorter survival time in GC [41,42]. JMJD2A can catalyze the demethylation of histone H3 lysines 9 and 36, and PCBP2 is an RNA-binding protein. Our findings suggested that both JMJD2A and PCBP2 could reduce the expression of pro-apoptotic miR-34a to promote cell proliferation and suppress apoptosis in GC cells [41,42]. Recently, the upregulated oncogenic protein AEG-1 was shown to associate with proinflammatory signaling in GC. Bacterial lipopolysaccharide (LPS) could induce AEG-1 expression, enhancing nuclear translocation of the NF-κB p65 subunit and, in turn, decreasing the TLR4 negative regulator SOCS. Eventually, AEG-1 promotes uncontrolled inflammation in the GC microenvironment and aggravates malignant progression [43]. AEG-1 depletion has been found to inhibit invasion and the epithelial-mesenchymal transition (EMT) program in cervical cancer [44] and to suppress cell migration in hepatocellular carcinoma [45]. In GC, AEG-1 also exerts critical roles and is a potential prognostic biomarker and therapeutic target [43]. Additionally, several single-nucleotide polymorphisms (SNP) have been associated with GC risk in genome-wide association studies (GWASs). For example, SNPs at 8q24.3 located in PSCA and SNPs at 1q22 within MUC1 are significantly associated with GC susceptibility according to three independent GWASs: two studies that examined a Chinese population [46,47] and one that examined Japanese and Korean populations [48].
Many diagnostic, prognostic and therapeutic biomarkers have been developed. For example, serum-based carcinoembryonic antigens (CEA) and tissue-based HER2 have been suggested as potential GC markers and used in clinical practice. However, low sensitivity and specificity constrain their application. So far, there are no good markers for early GC detection and prognosis. This situation indicates that our current knowledge of the complexities of GC remains limited, and novel pathogenic mechanisms should be explored to identify novel biomarkers for better clinical applications in GC.
The versatile long non-coding RNAs
Over the past decade, gene-tiling array and RNA deep sequencing (RNA-seq) studies have revealed that the human genome is pervasively transcribed and produces thousands of transcripts that lack obvious coding capacities [11]. These non-coding transcripts were initially regarded as genomic noise, but emerging evidence has demonstrated that the proverbial “dark matter” actually has important effects on the regulation of gene expression [12]. These findings have greatly challenged the conventional dogma that only proteins can perform these cellular functions and that RNA serves only as a template between DNA sequences and proteins. In the past, intensive efforts have concentrated on the functions of proteins in the pathogenesis of cancer. Recently, non-coding RNAs have attracted more attention, not only shorter transcripts (< 200 nt, which include siRNAs, piRNAs, and miRNAs and have been well documented) but also long non-coding RNAs (lncRNAs) that possess more complex regulatory mechanisms and take part in multiple biological processes, including cancer [21].
Biological characteristics of lncRNAs
LncRNAs are commonly defined as transcripts with lengths ranging from 200 nt to 100 kb that have little or no protein coding capacity. LncRNAs share many characteristics with mRNAs, are transcribed by one of three DNA-dependent RNA polymerases (Pol I, II, or III), have 5’ capping and polyadenylation, and undergo alternative splicing. The tertiary structures play important roles in lncRNA functions [17]. LncRNAs can be classified into diverse subgroups based on various criteria, which may help to elucidate their regulatory mechanisms. According to their functional relevance, lncRNAs can be divided into “housekeeping” and “regulatory” lncRNAs. The “housekeeping” lncRNAs are constitutively expressed, such as the tRNA and rRNA involved in protein biosynthesis. Regulatory lncRNAs may display dysregulated expression under certain physiological and pathological conditions. Based on their orientation and locations relative to neighboring genes, lncRNAs can be further classified as sense, antisense, divergent, convergent, intronic and intergenic [11]. The five broad categories of lncRNAs are described as follows: (1) “sense” and (2) “antisense” describe lncRNA transcripts that overlap with one or more exons of another transcript on the same or opposite strands, respectively; (3) “bidirectional” indicates that lncRNA expression and the neighboring coding transcript on the opposite strand is initiated in close genomic proximity; (4) “intronic” indicates a lncRNA derived from the intron of a second transcript; and (5) “intergenic” describes a lncRNA that lies within the genomic intervals between two genes as an independent unit. Additionally, some lncRNAs can modulate the expression of neighboring genes within the same chromosome in a cis manner or control distant gene expression located on the same or different chromosomes in a trans manner by affecting RNA polymerase complex recruitment or chromatin remodeling. Another classification system is based on the role of lncRNA in cancer development and comprises oncogenic lncRNAs and tumor suppressor lncRNAs.
LncRNA subcellular localization has been studied using RNA fluorescence in situ hybridization (FISH). Identifying the subcellular localization of lncRNAs provides some insight into their functions and potential interacting partners. Similar to mRNA, lncRNAs have been shown to localize across a wide range of subcellular organelles. A subset of lncRNAs are selectively localized in the nucleus [49-51], some have been visualized specifically in the cytoplasm [52], and some seem to appear in both the nuclear and the cytoplasmic compartments [53].
The evolutionary conservation of lncRNAs has been compared across species. Some lncRNAs exhibit a high level of nucleotide sequence identity within vertebrates, as exemplified by MALAT1 and its murine ortholog hepcarcin [54]. Moreover, certain lncRNAs are derived from ultra-conserved genomic regions (UCR) and are fully conserved among orthologous regions in human, rat, and mouse genomes [55]. A subset of lncRNAs show tissue-specific expression patterns in equivalent genome loci across species despite the absence of a conserved nucleotide sequence [56].
All of the abovementioned features imply the functional importance of lncRNAs. Several lncRNAs have been annotated; however, only a few of them have well-characterized functions. Multiple lncRNA databases have been constructed to understand lncRNAs more systematically. Here we summarized them in Table 1.
Table 1.
The databases for functional annotation of lncRNAs
| Database name | Description | URL |
|---|---|---|
| LncRNA2Target | A database for differentially expressed genes after lncRNA knockdown or overexpression | http://www.lncrna2target.org |
| LNCipedia | A database for annotated human lncRNA transcript sequences and structures | http://www.lncipedia.org |
| lncRNAdb | Expanding the reference database for functional long noncoding RNAs | http://lncrnadb.org |
| LncRNAWiki | Harnessing community knowledge in collaborative curation of human long non-coding RNAs | http://lncrna.big.ac.cn |
| LncRNADisease | A database for long-non-coding RNA-associated diseases | http://cmbi.bjmu.edu.cn/lncrnadisease |
| lncRNASNP | A database of SNPs in lncRNAs and their potential functions in human and mouse | http://bioinfo.life.hust.edu.cn/lncRNASNP/ |
| DIANA-LncBase | Experimentally verified and computationally predicted microRNA targets on long non-coding RNAs | www.microrna.gr/LncBase |
| NONCODE | Exploring the world of long non-coding RNA genes | http://www.bioinfo.org/noncode/ |
| ChIPBase | A database for decoding the transcriptional regulation of long non-coding RNA and microRNA genes from ChIP-Seq data | http://deepbase.sysu.edu.cn/chipbase/ |
| starBase v2.0 | Decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data | http://starbase.sysu.edu.cn/ |
| Rfam | The universally acclaimed database of RNA families, as well as several databases on long non-coding RNA, microRNA and their targets | http://rfam.sanger.ac.uk |
| Linc2GO | Functional annotation of human lincRNA based on the ceRNA hypothesis | http://www.bioinfo.tsinghua.edu.cn/~liuke/Linc2GO/index.html |
| circBase | Database of circular RNAs | http://www.circbase.org |
Mechanisms of lncRNA-mediated regulation of gene expression
LncRNAs play important roles in regulating gene expression via various mechanisms and at multiple levels. LncRNAs have the capacity to interact with proteins, DNAs or RNAs to act versatile functions. Moreover, certain lncRNAs can also function as the precursors for small RNAs to produce mature miRNAs.
LncRNA interacts with proteins
LncRNA can serve as a scaffold to recruit protein complexes and influence gene expression. Recent studies have revealed that a lncRNA known as HOTAIR can bind polycomb repressive complex 2 (PRC2) with its 5’ domain and interact with the LSD1/CoREST/REST complex with its 3’ end. PRC2 is composed of methylase EZH2, SUZ12 and EED, which are responsible for histone H3 lysine 27 trimethylation (H3K27me3); whereas the LSD1 complex serves as the demethylase that mediates histone H3 lysine 4 demethylation (H3K4me2). HOTAIR mediates two distinct complex assemblies, enabling PRC2 and LSD1 to target specific gene sites to induce H3K27me3 and H3K4me2 modification and ultimately repress gene transcription across 40 kb of the HOX D locus, despite the fact that HOTAIR itself is transcribed from the HOX C region [57,58]. The aberrant gene expression induced by HOTAIR increases the invasiveness and metastasis of cancer [57,59,60]. Overexpression of HOTAIR has been observed in various cancer types, such as breast, hepatocellular, gastric, colorectal and pancreatic cancers, and affects patient survival and prognosis [59].
LncRNA can also regulate nuclear trafficking. Nuclear factor of activated T cells (NFAT), a sensitive transcriptional factor response to the alteration of calcium signals, is essential for immune response mediated by the T cell receptor [61]. When intracellular calcium levels increase, the calcium-mediated phosphatase calcineurin dephosphorylates NFAT complex subunits in the cytoplasm, which promotes NFAT translocation into the nucleus to become transcriptionally active. The lncRNA NRON was identified in a complex with nuclear import factors that specifically modulated the nuclear trafficking of NFAT. NRON inhibited the nuclear accumulation of NFAT, which prevented the active transcription of NFAT, although the precise mechanisms are unclear [62]. The lncRNA 5’aHIF-1α is distributed at the perinuclear area of kidney cancer cells and co-localizes with the nuclear pore complex protein Nup62. Antitumor inhibitor camptothecin (CPT) treatment can elevate the level of 5’aHIF-1α and is associated with a decrease in HIF-1α mRNA levels. Localization within the perinuclear area and interaction with the nuclear pore complex suggests that 5’aHIF-1α may be involved in the export of novel mRNAs into the cytoplasm [63].
LncRNAs are involved in alternative splicing via interactions with splicing factors. The lncRNA MALAT1, also known as NEAT2, is upregulated in many solid cancers and is correlated with cancer metastasis and recurrence [64]. MALAT1 is highly conserved in mammals and is localized to nuclear speckles. MALAT1 interacts with splicing factors, including the serine/arginine (SR) splicing proteins, and decreases the cellular levels of active SR splicing proteins, which repress the association of SR splicing factors with pre-mRNA [65]. A recent study found that MALAT1 could promote cell cycle progression by enhancing expression of the oncogenic transcription factor B-MYB. MALAT-1 mainly induced proliferation by attenuating the affinity of splicing factors to B-MYB pre-mRNA, leading to an aberrant alternative splicing process [66].
LncRNA interacts with DNA, RNA or miRNA
Natural antisense lncRNA can form an RNA duplex with its sense mRNA, which may alter mRNA stability and translation efficiency. The overlapping region of the RNA duplex may protect the mRNA from endo- or exonucleolytic degradation [67]. Moreover, antisense lncRNAs may mask the miRNA-targeting site on sense mRNA and prevent the mRNA degradation by miRNA. The BACE1 natural antisense transcript BACE1-AS increases the stability of BACE1 mRNA through the mechanisms described above [68]. This RNA duplex formation has the capacity to affect epigenetic silencing. The INK4b-ARF-INK4a locus in the human genome encodes p15INK4b, p14ARF and p16INK4a, which are three tumor suppressors known to inhibit malignant cell proliferation and promote senescence and apoptosis. The natural antisense lncRNA ANRIL can form heterochromatin with INK4b-ARF-INK4a transcripts to recruit the polycomb repressor complexes PRC1 and PRC2 and establish repressive epigenetic marks on the chromatin, thereby silencing these tumor suppressors in prostate cancer [69,70].
LncRNA can mediate ribosomal DNA (rDNA) silencing by binding and methylating the rDNA promoter region. Mammalian genomes have several clusters of tandem rDNA, most of which are silenced by heterochromatic histone modifications and CpG methylation within the rDNA promoter. RNA polymerase I produces lncRNA transcripts from rDNA promoters (termed pRNA, promoter-associated RNA). As a subset of lncRNA, pRNA is complementary to the rDNA promoter and interacts with transcription factor TTF-I to form a DNA:RNA triplex that is preferentially recognized by DNMT3b to induce CpG methylation of the rDNA promoter and silence rDNA [71].
Many studies have revealed that some lncRNAs can act as miRNA sponges by competitively interacting with miRNAs to reduce miRNA availability to their target mRNAs. The tumor suppressor gene PTEN has a pseudogene lncRNA PTENP1. PTENP1 can act as a “decoy” to sequester the miRNAs that target PTEN, thus protecting PTEN from silencing by miRNAs and exerting a growth-suppression function; however, the PTENP1 locus is selectively lost in human cancer [72].
LncRNA serves as a miRNA precursor
Several unannotated lncRNAs have the potential to produce natural precursors for miRNA-like small RNAs. For example, the imprinted lncRNA H19 has been discovered as a precursor for miR-675 [73]. MiR-675 is derived from the first exon of H19 and is involved in controlling the expression of developmental genes. Additionally, H19-derived miR-675 has an impact on tumorigenesis and tumor progression by targeting tumor suppressor RB in colorectal cancer [74] and by silencing tumor suppressor RUNX1 in GC [75]. The lncRNA PVT1 locus located on 8q24 is amplified in multiple cancers. The PVT1 locus has been found to encode several miRNAs, such as miR-1204, and these miRNAs are important for T lymphomagenesis [76].
LncRNAs are associated with cancers
Cancer is a multistep process in which normal cells progressively evolve to a neoplastic state by acquiring particular capacities to disturb cellular homeostasis. Weinberg et al. proposed the following hallmarks of the malignant transformation process of cancer: (1) sustaining proliferative signaling; (2) evading growth suppressors; (3) resisting cell death; (4) enabling replicative immortality; (5) inducing angiogenesis; and (6) activating invasion and metastasis [77]. Many lncRNAs have been found to be dysregulated in cancers, and lncRNAs play important roles in each of the hallmarks of cancer [21]. Yang et al. reported that two lncRNAs, PRNCR1 and PCGEM1, which are highly overexpressed in aggressive prostate cancers could strongly enhance androgen receptor (AR)-induced transcriptional activation programs to promote cell proliferation. Their study involved an intricate series of events leading to the formation of a complex among PRNCR1, PCGEM1 and AR. PRNCR1 first bound to the C-terminally acetylated AR on the enhancers and recruited the DOT1L enzyme to methylate the N-terminus of AR, which was required for the association of PCGEM1 with AR. Subsequently, PCGEM1 interacted with the Pygo2 protein, which recognized and bound the methylated histone H3 marker H3K4me3 at the gene promoter. These successive complex associations eventually facilitated the formation of a “loop” between the enhancer and promoter sequences, which resulted in transcriptionally activated AR-targeted genes [78,79]. The regulatory patterns of lncRNA make cancer pathogenesis more intricate and complicated, but the multi-functional and tissue-specific properties of lncRNAs provide new avenues for the development of novel diagnostic, prognostic and therapeutic biomarkers for cancers.
LncRNAs play important roles in GC
There has been increasing interest in the role of lncRNAs in GC pathogenesis. Recently, several research teams have performed lncRNA microarray profiling or RNA-seq analysis and identified many dysregulated lncRNAs in GC [20,80-82]. These aberrantly expressed lncRNAs play critical roles in gastric carcinogenesis and aggressive progression. The upregulated lncRNAs may function as oncogenes to expedite the acquisition of the malignant cancer hallmarks, whereas the downregulated lncRNAs may possess tumor suppressor features in GC.
The oncogenic functions of lncRNAs in GC
A recent study found that the lncRNA GAPLINC (gastric adenocarcinoma predictive long intergenic noncoding RNA) was highly expressed in GC specimens according to in situ hybridization (ISH) analysis [83]. The aberrant expression of GAPLINC strongly correlated with alterations in CD44, and CD44 is a well-known cancer stem cell marker that drives cancer proliferation, migration and angiogenesis. The cell migration and proliferation functions of GAPLINC could be attenuated by CD44 repression. The in vitro and in vivo data demonstrated that GAPLINC acted as a molecular decoy for miR211-3p to protect CD44 from degradation by miR211-3p [83]. In fact, there is intricate crosstalk among lncRNAs, miRNAs and mRNAs to control gene expression in GC. LncRNA can serve as a competing endogenous RNA (ceRNA) to antagonize the repressive role of miRNAs on their target mRNAs. According to lncRNA microarray data, the bioinformatics algorithm miRcode, and the miRNA targets database TarBase, the first lncRNA-miRNA-mRNA networks have been constructed for GC [84].
HOTAIR is one of the most extensively studied oncogenic lncRNAs and is upregulated in various cancers, including GC. HOTAIR overexpression has been correlated with tumor stage, venous invasion, lymph node metastasis, peritoneal dissemination, and poor overall survival rate in GC [60,85-87]. Ectopic expression of HOTAIR promoted proliferation, the EMT program, and migration and invasion of GC cells, while HOTAIR knockdown effectively inhibited these malignant phenotypes, reduced the expression of MMP1, MMP3 and Snail proteins, and suppressed tumor growth and peritoneal metastasis in the xenograft mouse model [85,87]. The well-studied mechanism for HOTAIR is mediated through interaction with the PRC2 and LSD1/CoREST/REST complexes, thus leading to gene silencing in trans via H3K27 methylation and H3K4 demethylation. Recent investigation has found that HOTAIR can impose a further level of post-transcriptional regulation by acting as a competing endogenous RNA [60]. MiR-331-3p can directly bind HOTAIR, and HOTAIR functions as the endogenous decoy to disrupt the repression of HER2 by miR-331-3p. The increased HER2 expression may account for the more aggressive properties and poor survival associated with GC [60]. A case-control study in a northern Chinese population indicated that the T allele of rs12826786 might increase gastric cardia adenocarcinoma (GCA) risk, and this SNP has a genotype-specific influence on HOTAIR expression [88].
The oncogenic lncRNA ANRIL has also been found to be overexpressed in GC tissues, and higher ANRIL expression has been significantly correlated with an aggressive TNM stage and larger tumor size and has served as an independent predictor of poor overall survival. ANRIL exerts critical roles in cell proliferation both in vitro and in vivo [89]. The transcriptional activator E2F1 can induce ANRIL expression and promote ANRIL-mediated rapid cell growth. ANRIL can recruit and bind to the PRC2 complex to epigenetically repress miR-99a/miR-449a in trans, which activates the miR-99a/miR-449a target (the mTOR and CDK6/E2F1 pathway), thereby partially accounting for aberrant cell proliferation. Moreover, the formation of a positive feedback loop between ANRIL and E2F1 enables cell proliferation to be maintained continuously [89].
Overexpression of the imprinted lncRNA H19 gene is associated with GC development and poor prognosis. MiR-675 derived from H19 is a pivotal mediator in H19-induced GC development by silencing certain tumor suppressors. RUNX1 is a direct target of miR-675, and H19/miR-675-mediated RUNX1 depletion triggers cell proliferation and inhibits apoptosis in GC [75]. Independent of the miR-675 product, H19 has been found to interact with the tumor suppressor p53, abolishing p53 activity and thereby suppressing the expression of p53 targets, such as Bax, and leading to GC cell proliferation [90]. In addition, a coexpression network has revealed that ISM1 is the binding protein of H19, and its expression positively correlates with that of H19. CALN1 has been identified as another target of H19-derived miR-675. By directly upregulating ISM1 and indirectly repressing CALN1 expression by miR-675, H19 can promote cell proliferation, anti-apoptosis, migration, invasion and metastasis in GC. GC patients in the high H19 expression subgroup had shorter survival times [91]. H19 expression can be activated by the oncogene c-Myc in GC [90].
Multidrug resistance (MDR) is responsible for chemotherapy failure during GC treatment. The lncRNA MRUL (MDR-related and upregulated lncRNA) has a significant effect on MDR. MRUL is located 400 kb downstream of ABCB1, which is an ATP-dependent efflux pump that eliminates toxic intracellular metabolic products. MRUL has been found to be significantly upregulated in adriamycin- or vincristine-resistant SGC7901 cells [92]. MRUL could positively impact ABCB1 expression in an orientation- and position-independent manner. MRUL depletion in multidrug-resistant cells enhanced the accumulation of adriamycin or vincristine, decreased adriamycin release, and promoted apoptosis in MDR GC cell lines. The high levels of MRUL in GC tissues were negatively correlated with the growth inhibition rates of GC specimens treated with chemotherapy drugs in vitro and predicted a poor prognosis [92].
The lncRNA GHET1 (gastric carcinoma high expressed transcript-1) is enhanced in GC tissues and is correlated with larger tumor size, increased tumor invasion and a poor survival rate. Gain-of-function and loss-of-function analyses have revealed that GHET1 can drive gastric carcinoma cell proliferation. A mechanistic investigation demonstrated that GHET1 physically interacted with the IGF2BP1 protein and facilitated the association of c-Myc mRNA with IGF2BP1, thus increasing the mRNA stability and expression of the oncogene c-Myc to promote proliferation [93].
The lncRNA CCAT1 has a higher expression in GC tissues than in normal counterparts, and ectopic expression of CCAT1 promotes cell proliferation and migration [94]. Computational screen and chromatin immunoprecipitation (ChIP) assays have confirmed that c-Myc binds directly to the E-box element within the promoter region of CCAT1, thereby increasing transcriptional activity of the CCAT1 promoter and CCAT1 levels. CCAT1 and c-Myc expression showed a strong correlation in GC [94].
MALAT1 was highly expressed in GC cell lines and induced SF2/ASF proteins to localize to the nucleolus. SF2/ASF is an important member of the serine/arginine-rich protein (SR) family and is involved in alternative splicing. MALAT1 depletion can arrest the cell cycle in the G0/G1 phase, inhibit proliferation, and impair the nuclear distribution and expression of SF2/ASF [95]. SF2/ASF-silencing can repress the cell cycle and proliferation, but SF2/ASF overexpression fails to rescue the effects induced by MALAT1 knockdown. These findings indicate that MALAT1 may modulate GC cell proliferation partly by regulating SF2/ASF expression and distribution [95].
The lncRNA HULC (highly upregulated in liver cancer) was originally identified in hepatocellular carcinoma (HCC) and acts as an oncogene to promote HCC development and progression [96]. Recently, HULC was shown to be significantly elevated in GC tissues and positively associated with lymph node metastasis, distant metastasis and advanced TNM stages. HULC overexpression enhances proliferation, invasion, and EMT and suppresses apoptosis in GC cells. Additionally, ectopic HULC expression could induce autophagy, whereas HULC silencing or treatment with an autophagic inhibitor increased apoptosis in SGC7901 GC cells. These results suggest that HULC might function as a molecular switch between autophagy and apoptosis during GC pathogenesis [97].
The tumor suppressor roles of lncRNAs in GC
The lncRNA FENDRR (FOXF1 adjacent non-coding developmental regulatory RNA) is an essential regulator of heart and body development in the mouse [98]. Similar to HOTAIR, FENDRR can bind to PRC2 to control chromatin structure and gene activity [99]. In GC tissues and cell lines, FENDRR expression has been shown to be lower, which is associated with tumor invasion, advanced tumor stage, lymphatic metastasis, and poor prognosis. Histone deacetylation is involved in the downregulation of FENDRR in GC cells [100]. The lncRNA FENDRR has no obvious effect on cell proliferation but plays important roles in cell invasion and migration. Forced FENDRR overexpression is able to effectively reduce the number of metastatic nodules in xenograft mouse models. Further investigation revealed that the reduction of fibronectin1 and MMP2/MMP9 expression accounted for the FENDRR-induced inhibition of GC cell metastasis [100].
The lncRNA GAS5 (growth arrest-specific transcript 5) has been identified as a tumor-suppressor lncRNA in bladder, pancreatic and breast cancer, as GAS5 is downregulated and exhibits the capacity for growth arrest in these cancers [101-103]. Sun et al. found that GAS5 exhibited decreased expression in GC tissues, and the reduced expression of GAS5 was significantly associated with larger tumor size, advanced pathological stage, and poorer survival and overall survival. Increasing GAS5 expression was able to inhibit GC cell proliferation and induce apoptosis partly via the regulation of E2F1, P21, and cleaved caspase3 expression, although the mechanism is unclear [104]. Our ongoing investigations also demonstrated that GAS5 is downregulated in GC specimens, and GAS5 depletion triggers GC cell cycle arrest at the G1 phase. The results from the RNA-pull down and RNA immunoprecipitation (RIP) assays revealed that GAS5 interacted with the transcriptional activator YBX1 and regulated YBX1 protein abundance without affecting its mRNA level. The depletion of GAS5 and subsequent reduction in YBX1 protein decreased the expression of P21 transactivated by YBX1, eventually abolishing P21-induced cell cycle arrest in GC cells. Our findings suggest that the lncRNA GAS5/YBX1/p21 pathway plays critical roles in GC cell proliferation (Liu YC and Zhao J. et al. accepted).
The imprinted gene MEG3 (maternally expressed gene3) acts as a tumor suppressor lncRNA. MEG3 levels are markedly decreased in GC tissues and are correlated with an advanced TNM stage, increased invasion depth, larger tumor size and poor prognosis. DNA methylation is involved in MEG3 expression [105]. MEG3 overexpression can inhibit cell proliferation, drive apoptosis, and increase p53 expression in GC cells [105].
The lncRNA LincBM742401 is markedly downregulated in GC according to RNA-seq and public microarray data analyses. Reduced levels of BM742401 have been closely associated with poor survival in GC patients [106]. Forced expression of BM742401 in GC cells can block cell metastasis and decrease extracellular MMP9 secretion. Moreover, mice injected with BM742401-overexpressing cells had a significant reduction in tumor size and fewer lung metastatic foci [106].
The lncRNA ncRuPAR can increase protease-activated receptor-1 (PAR-1) levels during embryonic growth [107]. A recent study of GC tissues demonstrated that ncRuPAR was significantly downregulated, and the level of ncRuPAR was positively correlated with the tumor invasion depth, lymph node metastasis, distant metastasis, tumor size, and TNM stage. Moreover, decreased ncRuPAR expression was inversely associated with the mRNA level and immunohistochemical signal intensity of PAR-1 and VEGF in GC tissues [107], but the underlying mechanisms remain to be elucidated.
There is accumulating evidence that lncRNAs participate in GC development. A small fraction of lncRNAs have well-characterized mechanisms (Figure 1), but many dysregulated lncRNAs are not well understood and are correlated with clinical significance in GC. The GC-related lncRNAs have been summarized in Table 2.
Figure 1.
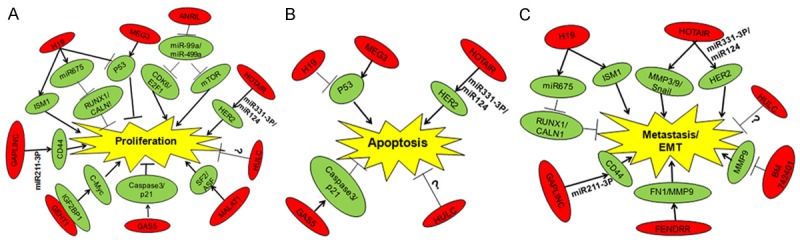
The functions and regulatory mechanisms of dysregulated lncRNAs in GC. The red ellipse diagrams indicate the dysregulated lncRNAs in GC, the green ellipse diagrams are their downstream proteins, and the yellow schematics show the aberrant biological processes regulated by lncRNAs in GC.
Table 2.
The potential clinical applications of dysregulated lncRNAs in GC
| Oncogenic LncRNAs | Sample type and size | Methods | Clinical significance | Potential Application | Ref |
|
| |||||
| H19↑ | 74 paired GC tissues | qRT-PCR | poor prognosis | diagnostic, prognostic biomarker and therapeutic target | [91] |
| 80 paired GC tissues | advanced TNM, poor prognosis | [117] | |||
| 24 paired GC tissues | - | [75] | |||
| 22 paired GC tissues | - | [90] | |||
| 43 pre-operative, 20 post-operative plasma, and 33 healthy plasma | Plasma H19 levels higher in GC patients and reduced in postoperative sample | [108] | |||
| 77 paired GC tissues | - | [20] | |||
| HOTAIR↑ | 78 paired GC tissues | qRT-PCR | tumor size, advanced TNM, metastasis, and shorter survival | diagnostic, prognostic biomarker and therapeutic target | [60] |
| 68 paired GC tissues | venous invasion, lymph node metastases and lower survival | [87] | |||
| 31 paired GC tissues | TNM stage and lymph node metastasis | [115] | |||
| 50 paired GC tissues | Lymphovascular invasion and lymph node metastasis | [86] | |||
| 150 paired GC tissues | as independent prognostic and risk factor for peritoneal dissemination | [85] | |||
| GAPLINC↑ | 90 GC tissues | ISH | poor survival | prognosis biomarker | [83] |
| MRUL↑ | 40 paired GC tissues, and SGC7901 cell line resistant VCR or ADR | qRT-PCR | in vitro growth inhibition rates of GC specimens treated with chemotherapy drugs and a poor prognosis | chemotherapy prediction and prognosis biomarker | [92] |
| ANRIL↑ | 120 paired GC tissues | qRT-PCR | TNM stage, tumor size and poor survival | prognostic biomarker and therapeutic target | [89] |
| GHET↑ | 42 paired GC tissues | qRT-PCR | tumor size, tumor invasion and poor survival | prognostic biomarker and therapeutic target | [93] |
| CCAT↑ | 20 paired GC tissues | qRT-PCR | - | therapeutic target | [94] |
| HULC↑ | 58 paired GC tissues | qRT-PCR | lymph node metastasis, distant metastasis and advanced TNM stages | diagnosis biomarker and therapeutic target | [97] |
| MALAT1↑ | 150 paired GC tissues | qRT-PCR | correlated with peritoneal metastasis in GC patients | diagnosis and prognosis biomarker, and therapeutic target | [85] |
| Linc00152↑ | 79 GC plasma, and paired pre- and postoperative plasma | qRT-PCR | Linc00152 levels higher in GC patients plasmas | diagnosis as blood-based biomarker | [110] |
| 71 paired GC tissues, 17 gastric juice of GC and 16 normal mucosa or minimal gastritis | correlated with invasion, diagnostic potential | [109] | |||
| ABHD11-AS1↑ | 75 paired GC tissues | qRT-PCR | differentiation, Lauren histological classification | diagnosis biomarker | [121] |
| AC130710 (GACAT3)↑ | 78 paired GC tissues | qRT-PCR | tumor size, TNM and distal metastasis | prognosis biomarker | [122] |
| SUMO1P3↑ | 96 paired GC tissues | qRT-PCR | tumor size, differentiation, lymphatic metastasis, and invasion | diagnosis biomarker | [123] |
| PVT-1↑ | 31 paired GC tissues | qRT-PCR | lymph node invasion and paclitaxel-resistant in SGC7901 | prognosis biomarker | [81] |
|
| |||||
| Tumor suppressor LncRNAs | Sample type and size | Methods | Clinical significance | Potential Application | Ref |
|
| |||||
| FENDRR↓ | 158 paired GC tissues | qRT-PCR | tumor invasion, TNM, lymphatic metastasis and poor prognosis | prognosis biomarker | [100] |
| GAS5↓ | 89 paired GC tissues | qRT-PCR | tumor size, advanced TNM and prognosis | prognosis biomarker and therapeutic target | [104] |
| 55 paired GC tissues | |||||
| ncRuPAR↓ | 138 paired GC tissues | qRT-PCR | tumor invasion depth, lymph node metastasis, distant metastasis, tumor size, and TNM stage | diagnosis biomarker | [107] |
| uc001lsz↓ | 77 paired GC tissues | qRT-PCR | TNM stage | early diagnosis marker | [20] |
| MEG3↓ | 72 paired GC tissues | qRT-PCR | TNM stages, depth of invasion, tumor size and poor prognosis | prognostic biomarker | [105] |
| BM742401↓ | 113 paired GC tissues | qRT-PCR | poor survival | prognostic biomarker and therapeutic target | [106] |
| AC096655.1-002 (GACAT1)↓ | 78 paired GC tissues | qRT-PCR | lymph node metastasis, distant metastasis, TNM, and differentiation | diagnosis biomarker | [124] |
| AA174084↓ | 134 paired GC tissues, 127 gastric mucosal tissues, 335 plasma, and 130 gastric juice | qRT-PCR | Borrmann type, perineural invasion and lymphatic metastasis, dropped in post-operation | early diagnosis biomarker | [111] |
| FER1L4↓ | 61 paired GC tissues, 80 healthy control plasma and 83 paired pre- and postoperative plasma | qRT-PCR | tumor size, histologic grade, general classification, depth of invasion, lymphatic metastasis, distant metastasis , TNM stage , vessel or nerve invasion | prognosis biomarker | [125] |
| HMlincRNA717 (GACAT2)↓ | 107 paired GC tissues, 37 healthy gastric mucosa, 34 gastritis mucosa, and 28 gastric precancerous lesions | qRT-PCR | cancer distal metastasis, venous invasion and nervous invasion- | early diagnosis biomarker | [126] |
| AC138128.1↓ | 94 paired GC tissues | qRT-PCR | - | diagnosis biomarker | [127] |
|
| |||||
| LncRNAs SNP | Sample type and size | Methods | Clinical significance | Potential Application | Ref |
|
| |||||
| HOTAIR | 515 GCA and 654 healthy control blood | PCR-RFLP, qRT-PCR | T allele of rs12826786 was risk for GCA and associated with advanced TNM | Screening and prognosis biomarker | [88] |
| CASC8 | 940 GC tissues | SNaP shot | GG genotype of rs10505477 survived for a longer time | prognosis biomarker | [113] |
upregulated;
downregulated;
GC: gastric cancer; GCA: gastric cardia adenocarcinoma; PCR-RFLP: polymerase chain reaction-restriction fragment length polymorphism; qRT-PCR: quantitative real-time reverse transcription PCR; ISH: in situ hybridization.
The diagnosis and prognosis potential of lncRNAs in GCs
Early detection and diagnosis and effective prognostic indicators are necessary for improving the survival of GC patients. However, conventional tumor markers, such as CEA and CA 19-9, have limited sensitivity and specificity in routine screening for GC. Therefore, novel biomarkers are urgently needed for GC. As lncRNAs play versatile roles in the regulation of gene expression via transcription, post-transcription and especially epigenetic modulations, aberrant lncRNA expression may therefore occur during carcinogenesis and disease progression, and dysregulated lncRNA levels may be better indicators of the intrinsic properties of cancer. Furthermore, lncRNAs have tissue-specific expression patterns and can be detected in body fluids. These advantages make lncRNAs promising biomarkers for diagnosis, prognosis and therapy in various cancers, including GC.
The diagnostic potential of lncRNAs in GC
The ideal biomarker should be easily and non-invasively accessible. Test samples from body fluids, such as blood and gastric juice, are good choices. A previous study evaluated levels of the lncRNAs H19, HOTAIR and MALAT1 in the plasma of GC patients, and H19 was markedly stable and could be successfully amplified with qRT-PCR, suggesting the potential of evaluating circulating lncRNAs as biomarkers. The authors found that the H19 plasma levels were significantly higher in 43 GC patients than in 34 healthy controls [108]. Moreover, H19 levels were markedly lower in 16 one-month post-operative patients than in the pre-operative plasma from those same 16 patients, indicating that the circulating lncRNAs might be released from primary GCs [108]. These findings suggest that circulating lncRNAs, such as H19, could become new complementary tumor biomarkers for GC.
The lncRNA linc00152 is another GC-related lncRNA that is significantly upregulated in GC tissues compared with adjacent normal tissues [109]. The linc00152 plasma levels in 79 GC patients were significantly higher than those in 81 healthy controls. The receiver operating characteristic (ROC) curve assessing the diagnostic value of plasma linc00152 showed that the area under the ROC curve reached 0.657 (p < 0.001), with a sensitivity of 48.1% and a specificity of 85.2%. It is interesting to note that the sensitivity of linc00152 was better than that of the traditional markers CEA and CA19-9 [110]. This study also indicated that the stable existence of lncRNAs in the blood might result from exosome protection, as there were also no differences in the linc00152 levels between the plasma and exosomes isolated from the same plasma [110]. These results suggest that plasma linc00152 has great potential for GC diagnosis. Recently, levels of the lncRNA AA174084 were assessed in 134 paired GC tissues, 127 gastric mucosal tissues, 335 plasma samples, and 130 gastric juice samples at various stages of GC [111]. It was found that AA174084 was significantly reduced in 95 of 134 GC tissues (71%) compared with paired normal samples, and the reduced expression of AA174084 was negatively associated with Bormann type GC and perineural invasion [111]. The plasma AA174084 levels decreased significantly on day 15 after surgery compared with preoperative samples and were correlated with invasion and lymphatic metastasis [111]. Furthermore, AA174084 could be detected in gastric juice by qRT-PCR, and gastric juice from GC patients had significantly higher AA174084 levels than those from the patients with normal mucosa or with minimal gastritis, gastric ulcers, and atrophic gastritis. The area under the ROC curve was up to 0.848 (p < .001), with a sensitivity of 46% and a specificity of 93%, which were higher than those obtained using the tissue level of AA174084 as the biomarker. These data demonstrated that the AA174084 gastric juice level may be a potential screening biomarker for the early diagnosis of GC [111].
Recently, Guo et al. performed a case-control study in northern China to evaluate the association between haplotype-tagging SNPs (htSNPs) of HOTAIR and the susceptibility to gastric cardia adenocarcinoma (GCA). PCR-RFLP was used to detect the htSNP genotype for HOTAIR (rs12826786 C > T, rs4759314 A > G, and rs10783618 C > T) in 515 GCA patients (blood from before surgery) and 654 healthy controls. They found that the T allele of rs12826786 increased the susceptibility to GCA and was associated with smoking and the TNM stage. qRT-PCR results revealed that the rs12826786 SNP had a genotype-specific effect on HOTAIR expression. This study indicated that the rs12826786 SNP of HOTAIR could be a useful candidate biomarker for high-risk GCA populations [88].
There are other dysregulated lncRNAs in GC tissues that have the potential to be candidate diagnostic biomarkers, including GACAT1, ncRuPAR, HULC, and FER1L4, among others (see Table 2). Whether the altered expression of these lncRNAs can be validated in body fluids remains to be elucidated.
The prognostic potential of lncRNAs in GC
Great advances have been made in lncRNA-based prognosis biomarker research. For instance, the prostate-specific lncRNA PCA3 has become the first FDA-approved lncRNA-based cancer biomarker to predict the prognosis of prostate cancer [112]. In GC, there are many dysregulated lncRNAs that are closely associated with a poor prognosis, and these lncRNAs could become desirable candidates for monitoring high-risk populations and predicting GC outcomes. An effective prognosis prediction may help GC patients gain access to more appropriate treatments.
The high cancer risk gene desert region 8q24 has a genetic variant SNP rs10505477 located within the intron of the lncRNA CASC8 that may affect the folding structures of CASC8. Ma et al. found that GC patients with rs10505477 GG survived for a longer time compared with those carrying the GA and AA genotypes in 940 surgical GC specimens [113]. This risk effect was more significant among patients with a tumor size ≤ 5 cm, diffuse-type GC, lymph node metastasis, no distant metastasis and TNM stage III and IV. These findings suggest that SNP rs10505477 may be a potential prognostic biomarker for GC [113].
MALAT1 and HOTAIR were expressed in 150 GC patients at higher levels than in corresponding adjacent normal mucosa and were markedly correlated with peritoneal metastasis in GC patients [85]. Additionally, HOTAIR overexpression was not only an independent prognostic indicator but also a risk factor for peritoneal dissemination [85]. Lee et al. reported that higher levels of HOTAIR were associated with lymphovascular invasion, lymph node metastasis, advanced TNM stage, and inferior disease-free survival in 50 GC samples [86]. Increased HOTAIR expression has been further confirmed to be positively correlated with an aggressive clinical significance in GC patients in many independent studies [60,87,114-116]. H19 is another extensively studied lncRNA with the potential to be a prognostic biomarker. Increased levels of H19 have been significantly correlated with lymph nodes metastasis, advanced TNM stage and poorer survival, and have been regarded as an independent predictor of overall survival in GC patients [91,117].
High levels of MRUL in GC tissues were negatively associated with rates of growth inhibition in GC specimens treated with the chemotherapy drugs adriamycin or vincristine in vitro [92]. Furthermore, upregulated MRUL indicated a poor prognosis for GC patients. Thus, MRUL is a potential target for reversing multidrug-resistance in GC [92].
Variations in the lncRNAs GAPLINC, ANRIL, GHET1, HULC GAS5, MEG3, BM742401, and ncRuPAR have also been found to be significantly correlated with an aggressive TNM stage, increased invasion depth, lymph node metastasis, distant metastasis, and poorer survival rate (Table 2).
In addition, certain lncRNAs that are specifically overexpressed in cancer tissues, such as H19, are promising therapeutic targets for decreasing the “off-target” effects of gene therapy for cancers. Plasmids containing diphtheria toxin subunit A under the H19 promoter have been studied for curing colon, pancreatic, bladder and ovarian cancers via intratumoral injection [118], as the high H19 levels in cancer cells could induce subunit A expression to kill cancer cells while simultaneously protecting normal cells from this destruction. H19 is upregulated in GC, and the effect of the H19 vector on GC therapy should be explored.
Despite some progress, the clinical translation of lncRNAs in GC remains in its infancy. There are still many puzzles that must be solved via elaborate investigations. For example, cancer stem cells played important roles in cancer progression in our previous studies [119,120], and it remains to be determined whether lncRNAs are involved in the maintenance of cancer stem cells in GC. In addition, it is unclear whether the circulating lncRNA alterations in the body fluids of GC patients account for GC development and if they are secreted from the GC cells or from other cell types. The stability of lncRNAs as biomarkers remains largely unknown but should be addressed to determine the general utility of lncRNAs in clinical practice.
Conclusions
LncRNAs can regulate gene expression at the transcriptional, post-transcriptional, and epigenetic architectures and play important roles in carcinogenesis and aggressive progression. Many dysregulated lncRNAs in GC have been significantly associated with increased tumor size, invasion and metastasis, and poor survival outcomes. Certain dysregulated lncRNAs are promising candidate molecular biomarkers in GC. Future studies are needed to identify additional cancer-specific lncRNAs, validate the utility of lncRNA-based biomarkers for diagnosis and prognosis, and address the roles of lncRNAs in cancer biology, as well determine whether lncRNAs have the potential to serve as improved therapeutic targets.
Acknowledgements
This study was supported by the Natural Science Foundation of China (81201521), the China Postdoctoral Science Foundation Funded Project (2013M541467), and the Foundation from Science and Technology Commission of Shanghai (13XD1401200).
Disclosure of conflict of interest
None.
References
- 1.Ferro A, Peleteiro B, Malvezzi M, Bosetti C, Bertuccio P, Levi F, Negri E, La Vecchia C, Lunet N. Worldwide trends in gastric cancer mortality (1980-2011), with predictions to 2015, and incidence by subtype. Eur J Cancer. 2014;50:1330–1344. doi: 10.1016/j.ejca.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Saeki N, Ono H, Sakamoto H, Yoshida T. Genetic factors related to gastric cancer susceptibility identified using a genome-wide association study. Cancer Sci. 2013;104:1–8. doi: 10.1111/cas.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shin CM, Kim N, Lee HS, Lee DH, Kim JS, Jung HC, Song IS. Intrafamilial aggregation of gastric cancer: a comprehensive approach including environmental factors, Helicobacter pylori virulence, and genetic susceptibility. Eur J Gastroenterol Hepatol. 2011;23:411–417. doi: 10.1097/MEG.0b013e328343b7f5. [DOI] [PubMed] [Google Scholar]
- 5.Nan HM, Song YJ, Yun HY, Park JS, Kim H. Effects of dietary intake and genetic factors on hypermethylation of the hMLH1 gene promoter in gastric cancer. World J Gastroenterol. 2005;11:3834–3841. doi: 10.3748/wjg.v11.i25.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamajima N, Naito M, Kondo T, Goto Y. Genetic factors involved in the development of Helicobacter pylori-related gastric cancer. Cancer Sci. 2006;97:1129–1138. doi: 10.1111/j.1349-7006.2006.00290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Macdonald JS, Smalley SR, Benedetti J, Hundahl SA, Estes NC, Stemmermann GN, Haller DG, Ajani JA, Gunderson LL, Jessup JM, Martenson JA. Chemoradiotherapy after surgery compared with surgery alone for adenocarcinoma of the stomach or gastroesophageal junction. N Engl J Med. 2001;345:725–730. doi: 10.1056/NEJMoa010187. [DOI] [PubMed] [Google Scholar]
- 8.De Angelis R, Sant M, Coleman MP, Francisci S, Baili P, Pierannunzio D, Trama A, Visser O, Brenner H, Ardanaz E, Bielska-Lasota M, Engholm G, Nennecke A, Siesling S, Berrino F, Capocaccia R. Cancer survival in Europe 1999-2007 by country and age: results of EUROCARE--5-a population-based study. Lancet Oncol. 2014;15:23–34. doi: 10.1016/S1470-2045(13)70546-1. [DOI] [PubMed] [Google Scholar]
- 9.Nagarajan N, Bertrand D, Hillmer AM, Zang ZJ, Yao F, Jacques PE, Teo AS, Cutcutache I, Zhang Z, Lee WH, Sia YY, Gao S, Ariyaratne PN, Ho A, Woo XY, Veeravali L, Ong CK, Deng N, Desai KV, Khor CC, Hibberd ML, Shahab A, Rao J, Wu M, Teh M, Zhu F, Chin SY, Pang B, So JB, Bourque G, Soong R, Sung WK, Tean Teh B, Rozen S, Ruan X, Yeoh KG, Tan PB, Ruan Y. Whole-genome reconstruction and mutational signatures in gastric cancer. Genome Biol. 2012;13:R115. doi: 10.1186/gb-2012-13-12-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zang ZJ, Cutcutache I, Poon SL, Zhang SL, McPherson JR, Tao J, Rajasegaran V, Heng HL, Deng N, Gan A, Lim KH, Ong CK, Huang D, Chin SY, Tan IB, Ng CC, Yu W, Wu Y, Lee M, Wu J, Poh D, Wan WK, Rha SY, So J, Salto-Tellez M, Yeoh KG, Wong WK, Zhu YJ, Futreal PA, Pang B, Ruan Y, Hillmer AM, Bertrand D, Nagarajan N, Rozen S, Teh BT, Tan P. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44:570–574. doi: 10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- 11.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H, Giresi PG, Goldy J, Hawrylycz M, Haydock A, Humbert R, James KD, Johnson BE, Johnson EM, Frum TT, Rosenzweig ER, Karnani N, Lee K, Lefebvre GC, Navas PA, Neri F, Parker SC, Sabo PJ, Sandstrom R, Shafer A, Vetrie D, Weaver M, Wilcox S, Yu M, Collins FS, Dekker J, Lieb JD, Tullius TD, Crawford GE, Sunyaev S, Noble WS, Dunham I, Denoeud F, Reymond A, Kapranov P, Rozowsky J, Zheng D, Castelo R, Frankish A, Harrow J, Ghosh S, Sandelin A, Hofacker IL, Baertsch R, Keefe D, Dike S, Cheng J, Hirsch HA, Sekinger EA, Lagarde J, Abril JF, Shahab A, Flamm C, Fried C, Hackermuller J, Hertel J, Lindemeyer M, Missal K, Tanzer A, Washietl S, Korbel J, Emanuelsson O, Pedersen JS, Holroyd N, Taylor R, Swarbreck D, Matthews N, Dickson MC, Thomas DJ, Weirauch MT, Gilbert J, Drenkow J, Bell I, Zhao X, Srinivasan KG, Sung WK, Ooi HS, Chiu KP, Foissac S, Alioto T, Brent M, Pachter L, Tress ML, Valencia A, Choo SW, Choo CY, Ucla C, Manzano C, Wyss C, Cheung E, Clark TG, Brown JB, Ganesh M, Patel S, Tammana H, Chrast J, Henrichsen CN, Kai C, Kawai J, Nagalakshmi U, Wu J, Lian Z, Lian J, Newburger P, Zhang X, Bickel P, Mattick JS, Carninci P, Hayashizaki Y, Weissman S, Hubbard T, Myers RM, Rogers J, Stadler PF, Lowe TM, Wei CL, Ruan Y, Struhl K, Gerstein M, Antonarakis SE, Fu Y, Green ED, Karaoz U, Siepel A, Taylor J, Liefer LA, Wetterstrand KA, Good PJ, Feingold EA, Guyer MS, Cooper GM, Asimenos G, Dewey CN, Hou M, Nikolaev S, Montoya-Burgos JI, Loytynoja A, Whelan S, Pardi F, Massingham T, Huang H, Zhang NR, Holmes I, Mullikin JC, Ureta-Vidal A, Paten B, Seringhaus M, Church D, Rosenbloom K, Kent WJ, Stone EA, Batzoglou S, Goldman N, Hardison RC, Haussler D, Miller W, Sidow A, Trinklein ND, Zhang ZD, Barrera L, Stuart R, King DC, Ameur A, Enroth S, Bieda MC, Kim J, Bhinge AA, Jiang N, Liu J, Yao F, Vega VB, Lee CW, Ng P, Yang A, Moqtaderi Z, Zhu Z, Xu X, Squazzo S, Oberley MJ, Inman D, Singer MA, Richmond TA, Munn KJ, Rada-Iglesias A, Wallerman O, Komorowski J, Fowler JC, Couttet P, Bruce AW, Dovey OM, Ellis PD, Langford CF, Nix DA, Euskirchen G, Hartman S, Urban AE, Kraus P, Van Calcar S, Heintzman N, Kim TH, Wang K, Qu C, Hon G, Luna R, Glass CK, Rosenfeld MG, Aldred SF, Cooper SJ, Halees A, Lin JM, Shulha HP, Xu M, Haidar JN, Yu Y, Iyer VR, Green RD, Wadelius C, Farnham PJ, Ren B, Harte RA, Hinrichs AS, Trumbower H, Clawson H, Hillman-Jackson J, Zweig AS, Smith K, Thakkapallayil A, Barber G, Kuhn RM, Karolchik D, Armengol L, Bird CP, de Bakker PI, Kern AD, Lopez-Bigas N, Martin JD, Stranger BE, Woodroffe A, Davydov E, Dimas A, Eyras E, Hallgrimsdottir IB, Huppert J, Zody MC, Abecasis GR, Estivill X, Bouffard GG, Guan X, Hansen NF, Idol JR, Maduro VV, Maskeri B, McDowell JC, Park M, Thomas PJ, Young AC, Blakesley RW, Muzny DM, Sodergren E, Wheeler DA, Worley KC, Jiang H, Weinstock GM, Gibbs RA, Graves T, Fulton R, Mardis ER, Wilson RK, Clamp M, Cuff J, Gnerre S, Jaffe DB, Chang JL, Lindblad-Toh K, Lander ES, Koriabine M, Nefedov M, Osoegawa K, Yoshinaga Y, Zhu B, de Jong PJ. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 13.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 14.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 15.Tchernitsa O, Kasajima A, Schafer R, Kuban RJ, Ungethum U, Gyorffy B, Neumann U, Simon E, Weichert W, Ebert MP, Rocken C. Systematic evaluation of the miRNA-ome and its downstream effects on mRNA expression identifies gastric cancer progression. J Pathol. 2010;222:310–319. doi: 10.1002/path.2759. [DOI] [PubMed] [Google Scholar]
- 16.Xu X, Yang X, Xing C, Zhang S, Cao J. miRNA: The nemesis of gastric cancer (Review) Oncol Lett. 2013;6:631–641. doi: 10.3892/ol.2013.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 18.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du Z, Fei T, Verhaak RG, Su Z, Zhang Y, Brown M, Chen Y, Liu XS. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol. 2013;20:908–913. doi: 10.1038/nsmb.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song H, Sun W, Ye G, Ding X, Liu Z, Zhang S, Xia T, Xiao B, Xi Y, Guo J. Long non-coding RNA expression profile in human gastric cancer and its clinical significances. J Transl Med. 2013;11:225. doi: 10.1186/1479-5876-11-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 23.Houghton J, Wang TC. Helicobacter pylori and gastric cancer: a new paradigm for inflammation-associated epithelial cancers. Gastroenterology. 2005;128:1567–1578. doi: 10.1053/j.gastro.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 24.Schistosomes, liver flukes and Helicobacter pylori. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Lyon, 7-14 June 1994. IARC Monogr Eval Carcinog Risks Hum. 1994;61:1–241. [PMC free article] [PubMed] [Google Scholar]
- 25.Augusto AC, Miguel F, Mendonca S, Pedrazzoli J Jr, Gurgueira SA. Oxidative stress expression status associated to Helicobacter pylori virulence in gastric diseases. Clin Biochem. 2007;40:615–622. doi: 10.1016/j.clinbiochem.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 26.Nakajima T, Yamashita S, Maekita T, Niwa T, Nakazawa K, Ushijima T. The presence of a methylation fingerprint of Helicobacter pylori infection in human gastric mucosae. Int J Cancer. 2009;124:905–910. doi: 10.1002/ijc.24018. [DOI] [PubMed] [Google Scholar]
- 27.Niwa T, Tsukamoto T, Toyoda T, Mori A, Tanaka H, Maekita T, Ichinose M, Tatematsu M, Ushijima T. Inflammatory processes triggered by Helicobacter pylori infection cause aberrant DNA methylation in gastric epithelial cells. Cancer Res. 2010;70:1430–1440. doi: 10.1158/0008-5472.CAN-09-2755. [DOI] [PubMed] [Google Scholar]
- 28.Shin CM, Kim N, Jung Y, Park JH, Kang GH, Kim JS, Jung HC, Song IS. Role of Helicobacter pylori infection in aberrant DNA methylation along multistep gastric carcinogenesis. Cancer Sci. 2010;101:1337–1346. doi: 10.1111/j.1349-7006.2010.01535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Correa P, Piazuelo MB, Camargo MC. The future of gastric cancer prevention. Gastric Cancer. 2004;7:9–16. doi: 10.1007/s10120-003-0265-0. [DOI] [PubMed] [Google Scholar]
- 30.Iizasa H, Nanbo A, Nishikawa J, Jinushi M, Yoshiyama H. Epstein-Barr Virus (EBV)-associated gastric carcinoma. Viruses. 2012;4:3420–3439. doi: 10.3390/v4123420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsugane S, Sasazuki S. Diet and the risk of gastric cancer: review of epidemiological evidence. Gastric Cancer. 2007;10:75–83. doi: 10.1007/s10120-007-0420-0. [DOI] [PubMed] [Google Scholar]
- 32.Wang XQ, Terry PD, Yan H. Review of salt consumption and stomach cancer risk: epidemiological and biological evidence. World J Gastroenterol. 2009;15:2204–2213. doi: 10.3748/wjg.15.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gonzalez CA. Vegetable, fruit and cereal consumption and gastric cancer risk. IARC Sci Publ. 2002;156:79–83. [PubMed] [Google Scholar]
- 34.de Martel C, Forman D, Plummer M. Gastric cancer: epidemiology and risk factors. Gastroenterol Clin North Am. 2013;42:219–240. doi: 10.1016/j.gtc.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 35.Hou IC, Amarnani S, Chong MT, Bishayee A. Green tea and the risk of gastric cancer: epidemiological evidence. World J Gastroenterol. 2013;19:3713–3722. doi: 10.3748/wjg.v19.i24.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lei Z, Tan IB, Das K, Deng N, Zouridis H, Pattison S, Chua C, Feng Z, Guan YK, Ooi CH, Ivanova T, Zhang S, Lee M, Wu J, Ngo A, Manesh S, Tan E, Teh BT, So JB, Goh LK, Boussioutas A, Lim TK, Flotow H, Tan P, Rozen SG. Identification of molecular subtypes of gastric cancer with different responses to PI3-kinase inhibitors and 5-fluorouracil. Gastroenterology. 2013;145:554–565. doi: 10.1053/j.gastro.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 37.Fenoglio-Preiser CM, Wang J, Stemmermann GN, Noffsinger A. TP53 and gastric carcinoma: a review. Hum Mutat. 2003;21:258–270. doi: 10.1002/humu.10180. [DOI] [PubMed] [Google Scholar]
- 38.Deng N, Goh LK, Wang H, Das K, Tao J, Tan IB, Zhang S, Lee M, Wu J, Lim KH, Lei Z, Goh G, Lim QY, Tan AL, Sin Poh DY, Riahi S, Bell S, Shi MM, Linnartz R, Zhu F, Yeoh KG, Toh HC, Yong WP, Cheong HC, Rha SY, Boussioutas A, Grabsch H, Rozen S, Tan P. A comprehensive survey of genomic alterations in gastric cancer reveals systematic patterns of molecular exclusivity and co-occurrence among distinct therapeutic targets. Gut. 2012;61:673–684. doi: 10.1136/gutjnl-2011-301839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okines AF, Cunningham D. Trastuzumab: a novel standard option for patients with HER-2-positive advanced gastric or gastro-oesophageal junction cancer. Therap Adv Gastroenterol. 2012;5:301–318. doi: 10.1177/1756283X12450246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu YC, Zhao J, Hu CE, Gan J, Zhang WH, Huang GJ. Comprehensive analysis of vascular endothelial growth factor-C related factors in stomach cancer. Asian Pac J Cancer Prev. 2014;15:1925–1929. doi: 10.7314/apjcp.2014.15.5.1925. [DOI] [PubMed] [Google Scholar]
- 41.Hu CE, Liu YC, Zhang HD, Huang GJ. JMJD2A predicts prognosis and regulates cell growth in human gastric cancer. Biochem Biophys Res Commun. 2014;449:1–7. doi: 10.1016/j.bbrc.2014.04.126. [DOI] [PubMed] [Google Scholar]
- 42.Hu CE, Liu YC, Zhang HD, Huang GJ. The RNA-binding protein PCBP2 facilitates gastric carcinoma growth by targeting miR-34a. Biochem Biophys Res Commun. 2014;448:437–442. doi: 10.1016/j.bbrc.2014.04.124. [DOI] [PubMed] [Google Scholar]
- 43.Li G, Wang Z, Ye J, Zhang X, Wu H, Peng J, Song W, Chen C, Cai S, He Y, Xu J. Uncontrolled inflammation induced by AEG-1 promotes gastric cancer and poor prognosis. Cancer Res. 2014;74:5541–5552. doi: 10.1158/0008-5472.CAN-14-0968. [DOI] [PubMed] [Google Scholar]
- 44.Liu X, Wang D, Liu H, Feng Y, Zhu T, Zhang L, Zhu B, Zhang Y. Knockdown of astrocyte elevated gene-1 (AEG-1) in cervical cancer cells decreases their invasiveness, epithelial to mesenchymal transition, and chemoresistance. Cell Cycle. 2014;13:1702–1707. doi: 10.4161/cc.28607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao J, Wang W, Huang Y, Wu J, Chen M, Cui P, Zhang W, Zhang Y. HBx Elevates Oncoprotein AEG-1 Expression to Promote Cell Migration by Downregulating miR-375 and miR-136 in Malignant Hepatocytes. DNA Cell Biol. 2014;33:715–22. doi: 10.1089/dna.2014.2376. [DOI] [PubMed] [Google Scholar]
- 46.Shi Y, Hu Z, Wu C, Dai J, Li H, Dong J, Wang M, Miao X, Zhou Y, Lu F, Zhang H, Hu L, Jiang Y, Li Z, Chu M, Ma H, Chen J, Jin G, Tan W, Wu T, Zhang Z, Lin D, Shen H. A genome-wide association study identifies new susceptibility loci for non-cardia gastric cancer at 3q13.31 and 5p13.1. Nat Genet. 2011;43:1215–1218. doi: 10.1038/ng.978. [DOI] [PubMed] [Google Scholar]
- 47.Abnet CC, Freedman ND, Hu N, Wang Z, Yu K, Shu XO, Yuan JM, Zheng W, Dawsey SM, Dong LM, Lee MP, Ding T, Qiao YL, Gao YT, Koh WP, Xiang YB, Tang ZZ, Fan JH, Wang C, Wheeler W, Gail MH, Yeager M, Yuenger J, Hutchinson A, Jacobs KB, Giffen CA, Burdett L, Fraumeni JF Jr, Tucker MA, Chow WH, Goldstein AM, Chanock SJ, Taylor PR. A shared susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma and esophageal squamous cell carcinoma. Nat Genet. 2010;42:764–767. doi: 10.1038/ng.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sakamoto H, Yoshimura K, Saeki N, Katai H, Shimoda T, Matsuno Y, Saito D, Sugimura H, Tanioka F, Kato S, Matsukura N, Matsuda N, Nakamura T, Hyodo I, Nishina T, Yasui W, Hirose H, Hayashi M, Toshiro E, Ohnami S, Sekine A, Sato Y, Totsuka H, Ando M, Takemura R, Takahashi Y, Ohdaira M, Aoki K, Honmyo I, Chiku S, Aoyagi K, Sasaki H, Yanagihara K, Yoon KA, Kook MC, Lee YS, Park SR, Kim CG, Choi IJ, Yoshida T, Nakamura Y, Hirohashi S. Genetic variation in PSCA is associated with susceptibility to diffuse-type gastric cancer. Nat Genet. 2008;40:730–740. doi: 10.1038/ng.152. [DOI] [PubMed] [Google Scholar]
- 49.Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Redrup L, Branco MR, Perdeaux ER, Krueger C, Lewis A, Santos F, Nagano T, Cobb BS, Fraser P, Reik W. The long noncoding RNA Kcnq1ot1 organises a lineage-specific nuclear domain for epigenetic gene silencing. Development. 2009;136:525–530. doi: 10.1242/dev.031328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, Sementchenko V, Piccolboni A, Bekiranov S, Bailey DK, Ganesh M, Ghosh S, Bell I, Gerhard DS, Gingeras TR. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 53.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 54.Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- 55.Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 56.Khaitovich P, Kelso J, Franz H, Visagie J, Giger T, Joerchel S, Petzold E, Green RE, Lachmann M, Paabo S. Functionality of intergenic transcription: an evolutionary comparison. PLoS Genet. 2006;2:e171. doi: 10.1371/journal.pgen.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cai B, Song XQ, Cai JP, Zhang S. HOTAIR: a cancer-related long non-coding RNA. Neoplasma. 2014;61:379–391. doi: 10.4149/neo_2014_075. [DOI] [PubMed] [Google Scholar]
- 60.Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, Kong R, Xia R, Lu KH, Li JH, De W, Wang KM, Wang ZX. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003;17:2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- 62.Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 63.Bertozzi D, Iurlaro R, Sordet O, Marinello J, Zaffaroni N, Capranico G. Characterization of novel antisense HIF-1alpha transcripts in human cancers. Cell Cycle. 2011;10:3189–3197. doi: 10.4161/cc.10.18.17183. [DOI] [PubMed] [Google Scholar]
- 64.Gutschner T, Hammerle M, Diederichs S. MALAT1 - a paradigm for long noncoding RNA function in cancer. J Mol Med (Berl) 2013;91:791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- 65.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG, Prasanth KV. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A, Prasanth KV. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013;9:e1003368. doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G 3rd, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30:1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schmitz KM, Mayer C, Postepska A, Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010;24:2264–2269. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13:313–316. doi: 10.1261/rna.351707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung JJ, Kwok TT. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31:350–358. doi: 10.1093/carcin/bgp181. [DOI] [PubMed] [Google Scholar]
- 75.Zhuang M, Gao W, Xu J, Wang P, Shu Y. The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochem Biophys Res Commun. 2014;448:315–322. doi: 10.1016/j.bbrc.2013.12.126. [DOI] [PubMed] [Google Scholar]
- 76.Beck-Engeser GB, Lum AM, Huppi K, Caplen NJ, Wang BB, Wabl M. Pvt1-encoded microRNAs in oncogenesis. Retrovirology. 2008;5:4. doi: 10.1186/1742-4690-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 78.Yang L, Lin C, Jin C, Yang JC, Tanasa B, Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, Evans CP, Rosenfeld MG. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500:598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schmitt AM, Chang HY. Gene regulation: Long RNAs wire up cancer growth. Nature. 2013;500:536–537. doi: 10.1038/nature12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cao WJ, Wu HL, He BS, Zhang YS, Zhang ZY. Analysis of long non-coding RNA expression profiles in gastric cancer. World J Gastroenterol. 2013;19:3658–3664. doi: 10.3748/wjg.v19.i23.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ding J, Li D, Gong M, Wang J, Huang X, Wu T, Wang C. Expression and clinical significance of the long non-coding RNA PVT1 in human gastric cancer. Onco Targets Ther. 2014;7:1625–1630. doi: 10.2147/OTT.S68854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lin XC, Zhu Y, Chen WB, Lin LW, Chen DH, Huang JR, Pan K, Lin Y, Wu BT, Dai Y, Tu ZG. Integrated analysis of long non-coding RNAs and mRNA expression profiles reveals the potential role of lncRNAs in gastric cancer pathogenesis. Int J Oncol. 2014;45:619–628. doi: 10.3892/ijo.2014.2431. [DOI] [PubMed] [Google Scholar]
- 83.Hu Y, Wang J, Qian J, Kong X, Tang J, Wang Y, Chen H, Hong J, Zou W, Chen Y, Xu J, Fang JY. Long Noncoding RNA GAPLINC Regulates CD44-Dependent Cell Invasiveness and Associates with Poor Prognosis of Gastric Cancer. Cancer Res. 2014;74:6890–6902. doi: 10.1158/0008-5472.CAN-14-0686. [DOI] [PubMed] [Google Scholar]
- 84.Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi Y, Guo J. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep. 2014;4:6088. doi: 10.1038/srep06088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Okugawa Y, Toiyama Y, Hur K, Toden S, Saigusa S, Tanaka K, Inoue Y, Mohri Y, Kusunoki M, Boland CR, Goel A. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis. 2014;35:2731–2739. doi: 10.1093/carcin/bgu200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee NK, Lee JH, Park CH, Yu D, Lee YC, Cheong JH, Noh SH, Lee SK. Long non-coding RNA HOTAIR promotes carcinogenesis and invasion of gastric adenocarcinoma. Biochem Biophys Res Commun. 2014;451:171–178. doi: 10.1016/j.bbrc.2014.07.067. [DOI] [PubMed] [Google Scholar]
- 87.Endo H, Shiroki T, Nakagawa T, Yokoyama M, Tamai K, Yamanami H, Fujiya T, Sato I, Yamaguchi K, Tanaka N, Iijima K, Shimosegawa T, Sugamura K, Satoh K. Enhanced expression of long non-coding RNA HOTAIR is associated with the development of gastric cancer. PLoS One. 2013;8:e77070. doi: 10.1371/journal.pone.0077070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guo W, Dong Z, Bai Y, Guo Y, Shen S, Kuang G, Xu J. Associations between polymorphisms of HOTAIR and risk of gastric cardia adenocarcinoma in a population of north China. Tumour Biol. 2014 doi: 10.1007/s13277-014-2912-y. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 89.Zhang EB, Kong R, Yin DD, You LH, Sun M, Han L, Xu TP, Xia R, Yang JS, De W, Chen J. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget. 2014;5:2276–2292. doi: 10.18632/oncotarget.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J, Fang G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279:3159–3165. doi: 10.1111/j.1742-4658.2012.08694.x. [DOI] [PubMed] [Google Scholar]
- 91.Li H, Yu B, Li J, Su L, Yan M, Zhu Z, Liu B. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5:2318–2329. doi: 10.18632/oncotarget.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang Y, Zhang D, Wu K, Zhao Q, Nie Y, Fan D. Long non-coding RNA MRUL promotes ABCB1 expression in multidrug-resistant gastric cancer cell sublines. Mol Cell Biol. 2014;34:3182–93. doi: 10.1128/MCB.01580-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang F, Xue X, Zheng L, Bi J, Zhou Y, Zhi K, Gu Y, Fang G. Long non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stability. FEBS J. 2014;281:802–813. doi: 10.1111/febs.12625. [DOI] [PubMed] [Google Scholar]
- 94.Yang F, Xue X, Bi J, Zheng L, Zhi K, Gu Y, Fang G. Long noncoding RNA CCAT1, which could be activated by c-Myc, promotes the progression of gastric carcinoma. J Cancer Res Clin Oncol. 2013;139:437–445. doi: 10.1007/s00432-012-1324-x. [DOI] [PubMed] [Google Scholar]
- 95.Wang J, Su L, Chen X, Li P, Cai Q, Yu B, Liu B, Wu W, Zhu Z. MALAT1 promotes cell proliferation in gastric cancer by recruiting SF2/ASF. Biomed Pharmacother. 2014;68:557–64. doi: 10.1016/j.biopha.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 96.Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder R, Trauner M, Zatloukal K. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 97.Zhao Y, Guo Q, Chen J, Hu J, Wang S, Sun Y. Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: a clinical and in vitro investigation. Oncol Rep. 2014;31:358–364. doi: 10.3892/or.2013.2850. [DOI] [PubMed] [Google Scholar]
- 98.Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A, Macura K, Blass G, Kellis M, Werber M, Herrmann BG. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 100.Xu TP, Huang MD, Xia R, Liu XX, Sun M, Yin L, Chen WM, Han L, Zhang EB, Kong R, De W, Shu YQ. Decreased expression of the long non-coding RNA FENDRR is associated with poor prognosis in gastric cancer and FENDRR regulates gastric cancer cell metastasis by affecting fibronectin1 expression. J Hematol Oncol. 2014;7:63. doi: 10.1186/s13045-014-0063-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lu X, Fang Y, Wang Z, Xie J, Zhan Q, Deng X, Chen H, Jin J, Peng C, Li H, Shen B. Downregulation of gas5 increases pancreatic cancer cell proliferation by regulating CDK6. Cell Tissue Res. 2013;354:891–896. doi: 10.1007/s00441-013-1711-x. [DOI] [PubMed] [Google Scholar]
- 102.Liu Z, Wang W, Jiang J, Bao E, Xu D, Zeng Y, Tao L, Qiu J. Downregulation of GAS5 promotes bladder cancer cell proliferation, partly by regulating CDK6. PLoS One. 2013;8:e73991. doi: 10.1371/journal.pone.0073991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 104.Sun M, Jin FY, Xia R, Kong R, Li JH, Xu TP, Liu YW, Zhang EB, Liu XH, De W. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer. 2014;14:319. doi: 10.1186/1471-2407-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sun M, Xia R, Jin F, Xu T, Liu Z, De W, Liu X. Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumour Biol. 2014;35:1065–1073. doi: 10.1007/s13277-013-1142-z. [DOI] [PubMed] [Google Scholar]
- 106.Park SM, Park SJ, Kim HJ, Kwon OH, Kang TW, Sohn HA, Kim SK, Moo Noh S, Song KS, Jang SJ, Sung Kim Y, Kim SY. A known expressed sequence tag, BM742401, is a potent lincRNA inhibiting cancer metastasis. Exp Mol Med. 2013;45:e31. doi: 10.1038/emm.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu L, Yan B, Yang Z, Zhang X, Gu Q, Yue X. ncRuPAR inhibits gastric cancer progression by down-regulating protease-activated receptor-1. Tumour Biol. 2014;35:7821–7829. doi: 10.1007/s13277-014-2042-6. [DOI] [PubMed] [Google Scholar]
- 108.Arita T, Ichikawa D, Konishi H, Komatsu S, Shiozaki A, Shoda K, Kawaguchi T, Hirajima S, Nagata H, Kubota T, Fujiwara H, Okamoto K, Otsuji E. Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res. 2013;33:3185–3193. [PubMed] [Google Scholar]
- 109.Pang Q, Ge J, Shao Y, Sun W, Song H, Xia T, Xiao B, Guo J. Increased expression of long intergenic non-coding RNA LINC00152 in gastric cancer and its clinical significance. Tumour Biol. 2014;35:5441–5447. doi: 10.1007/s13277-014-1709-3. [DOI] [PubMed] [Google Scholar]
- 110.Li Q, Shao Y, Zhang X, Zheng T, Miao M, Qin L, Wang B, Ye G, Xiao B, Guo J. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour Biol. 2015;36:2007–12. doi: 10.1007/s13277-014-2807-y. [DOI] [PubMed] [Google Scholar]
- 111.Shao Y, Ye M, Jiang X, Sun W, Ding X, Liu Z, Ye G, Zhang X, Xiao B, Guo J. Gastric juice long noncoding RNA used as a tumor marker for screening gastric cancer. Cancer. 2014;120:3320–3328. doi: 10.1002/cncr.28882. [DOI] [PubMed] [Google Scholar]
- 112.Vlaeminck-Guillem V, Ruffion A, Andre J, Devonec M, Paparel P. Urinary prostate cancer 3 test: toward the age of reason? Urology. 2010;75:447–453. doi: 10.1016/j.urology.2009.03.046. [DOI] [PubMed] [Google Scholar]
- 113.Ma G, Gu D, Lv C, Chu H, Xu Z, Tong N, Wang M, Tang C, Xu Y, Zhang Z, Wang B, Chen J. Genetic variant in 8q24 is associated with prognosis for gastric cancer in a Chinese population. J Gastroenterol Hepatol. 2014;30:689–95. doi: 10.1111/jgh.12801. [DOI] [PubMed] [Google Scholar]
- 114.Emadi-Andani E, Nikpour P, Emadi-Baygi M, Bidmeshkipour A. Association of HOTAIR expression in gastric carcinoma with invasion and distant metastasis. Adv Biomed Res. 2014;3:135. doi: 10.4103/2277-9175.133278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hajjari M, Behmanesh M, Sadeghizadeh M, Zeinoddini M. Up-regulation of HOTAIR long non-coding RNA in human gastric adenocarcinoma tissues. Med Oncol. 2013;30:670. doi: 10.1007/s12032-013-0670-0. [DOI] [PubMed] [Google Scholar]
- 116.Xu ZY, Yu QM, Du YA, Yang LT, Dong RZ, Huang L, Yu PF, Cheng XD. Knockdown of long non-coding RNA HOTAIR suppresses tumor invasion and reverses epithelial-mesenchymal transition in gastric cancer. Int J Biol Sci. 2013;9:587–597. doi: 10.7150/ijbs.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang EB, Han L, Yin DD, Kong R, De W, Chen J. c-Myc-induced, long, noncoding H19 affects cell proliferation and predicts a poor prognosis in patients with gastric cancer. Med Oncol. 2014;31:914. doi: 10.1007/s12032-014-0914-7. [DOI] [PubMed] [Google Scholar]
- 118.Smaldone MC, Davies BJ. BC-819, a plasmid comprising the H19 gene regulatory sequences and diphtheria toxin A, for the potential targeted therapy of cancers. Curr Opin Mol Ther. 2010;12:607–616. [PubMed] [Google Scholar]
- 119.Zhou J, Zhang Y. Cancer stem cells: Models, mechanisms and implications for improved treatment. Cell Cycle. 2008;7:1360–1370. doi: 10.4161/cc.7.10.5953. [DOI] [PubMed] [Google Scholar]
- 120.Zhou J, Wulfkuhle J, Zhang H, Gu P, Yang Y, Deng J, Margolick JB, Liotta LA, Petricoin E 3rd, Zhang Y. Activation of the PTEN/mTOR/STAT3 pathway in breast cancer stem-like cells is required for viability and maintenance. Proc Natl Acad Sci U S A. 2007;104:16158–16163. doi: 10.1073/pnas.0702596104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lin X, Yang M, Xia T, Guo J. Increased expression of long noncoding RNA ABHD11-AS1 in gastric cancer and its clinical significance. Med Oncol. 2014;31:42. doi: 10.1007/s12032-014-0042-4. [DOI] [PubMed] [Google Scholar]
- 122.Xu C, Shao Y, Xia T, Yang Y, Dai J, Luo L, Zhang X, Sun W, Song H, Xiao B, Guo J. lncRNA-AC130710 targeting by miR-129-5p is upregulated in gastric cancer and associates with poor prognosis. Tumour Biol. 2014;35:9701–6. doi: 10.1007/s13277-014-2274-5. [DOI] [PubMed] [Google Scholar]
- 123.Mei D, Song H, Wang K, Lou Y, Sun W, Liu Z, Ding X, Guo J. Up-regulation of SUMO1 pseudogene 3 (SUMO1P3) in gastric cancer and its clinical association. Med Oncol. 2013;30:709. doi: 10.1007/s12032-013-0709-2. [DOI] [PubMed] [Google Scholar]
- 124.Xiao B, Guo J. Long noncoding RNA AC096655.1-002 has been officially named as gastric cancer-associated transcript 1, GACAT1. Tumour Biol. 2013;34:3271. doi: 10.1007/s13277-013-0916-7. [DOI] [PubMed] [Google Scholar]
- 125.Liu Z, Shao Y, Tan L, Shi H, Chen S, Guo J. Clinical significance of the low expression of FER1L4 in gastric cancer patients. Tumour Biol. 2014;35:9613–9617. doi: 10.1007/s13277-014-2259-4. [DOI] [PubMed] [Google Scholar]
- 126.Shao Y, Chen H, Jiang X, Chen S, Li P, Ye M, Li Q, Sun W, Guo J. Low expression of lncRNA-HMlincRNA717 in human gastric cancer and its clinical significances. Tumour Biol. 2014;35:9591–9595. doi: 10.1007/s13277-014-2243-z. [DOI] [PubMed] [Google Scholar]
- 127.Chen X, Sun J, Song Y, Gao P, Zhao J, Huang X, Liu B, Xu H, Wang Z. The novel long noncoding RNA AC138128.1 may be a predictive biomarker in gastric cancer. Med Oncol. 2014;31:262. doi: 10.1007/s12032-014-0262-7. [DOI] [PubMed] [Google Scholar]


