Abstract
Hyaluronan-binding protein 1 (HABP1) is a protein with high affinity for HA, and has been reported to be upregulated in cancer cells. In this study, we show that silencing HABP1 inhibits proliferation, and suppresses the migration and invasion ability of breast cancer cell lines. In addition, silencing HABP1 remarkably slows down tumor growth in mice. We examined the correlation between HABP1 expression and clinicopathological parameters using immunohistochemistry in patients with breast cancer. The results indicate that HABP1 is overexpressed in cancer tissue, and its high levels are related to lymph node metastasis (P = 0.032) and tumor stage (P = 0.041). Moreover, high HABP1 expression is correlated with poor overall survival in breast cancer patients (P = 0.018), and is a signifi cant independent prognostic indicator. Our fi ndings suggest that HABP1 regulates proliferation and migration of breast cancer cells. HABP1 may be a useful independent predictor of outcomes in patients with breast cancer.
Keywords: HABP1, breast cancer, metastasis
Introduction
Breast cancer is the most common malignancy and the second leading cancer-related cause of death in women [1]. Several prognostic markers, such as tumor size, lymph node metastasis and stage, have been useful for clinical therapy, but additional biomarkers are needed to improve the prognosis for patients. A number of genes and proteins have been identified to be abnormally expressed in tumors, and many molecules are known to be involved in tumor progression and migration. Therefore, it is necessary to explore these genes for their potential as effective biomarkers and the detailed molecular mechanisms in breast cancer.
Hyaluronan-binding protein 1 (HABP1/gC1qR/p32) was previously characterized as a protein with high affinity for HA[2], and as the receptor for globular complement subcomponent 1q [3]. It was originally isolated based on its co-purification with pre-mRNA splicing factor SF2 [4]. HABP1 has been reported to be upregulated in human cancers [5-8], and several studies suggest that abnormal expression of HABP1 plays an important role on tumor progression and migration. Extensive studies have shown that high levels of HABP1 promote tumorigenesis via HA synthesis and survival pathway activation [9]. HABP1 can regulate metabolism to maintain oxidative phosphorylation of cancer cell [10]. Moreover, siRNA targeting of HABP1 inhibits cell proliferation, and the treated cells become more sensitive to chemotherapeutic drugs [11]. It has also been reported that HABP1 enhances migration through EGF-induced cancer cell chemotaxis [12] and regulates lamellipodia formation [13], implicating its role in cell metastasis.
We previously demonstrated that a high level of HABP1mRNA positively correlates with metastasis and poor prognosis in patients with breast cancer [6]. However, HABP1 protein levels were unclear, and the role of HABP1 on breast cancer cell migration remains to be explored. The purpose of this study was to examine the effects of HABP1 on breast tumor progression and metastasis.
Materials and methods
Reagents
Antibodies specific for HABP1 and β-actin were purchased from Abcam (Cambridge, MA, USA). Secondary antibodies, HRP-conjugated Goat anti-Mouse IgG, and Goat anti-Rabbit IgG were obtained from Cell Signaling Technology (Beverly, MA, USA). Cell-culture media and matrigel were purchased from BD Biosciences (San Diego, CA, USA).
Cell culture
Human breast cancer cell lines BT549 and MDA-MB-231were purchased from Shanghai Cell bank, Type Culture Collection Committee, Chinese Academy of Sciences. BT549 cells and MDA-MB-231 cells were maintained in RPMI 1640 medium. All media were supplemented with 10% fetal bovine serum (FBS), penicillin (100 units/ml), and streptomycin (0.1 mg/ml). All cells were incubated at 37°C in a humidified 5% CO2 atmosphere.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted by using Trizol reagent (Invitrogen, Carlsbad, CA, USA), and reverse transcription was performed with a High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA). The mRNA expression was examined by real-time PCR using FastStart Universal SYBR Green Master (Roche, Mannheim, Germany) with gene-specific primers and the ABI 7500 Fast Real-time PCR Detection System (Applied Biosystems, Foster City, CA, USA). The thermal cycling conditions were 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 35 s. All reactions were performed in triplicate. The results of HABP1 mRNA expression were calculated using the 2-ΔΔCt method with minor revision [14]. GAPDH was applied as the internal reference. The sequences of the primers used were as follows: HABP1-F: 5’-GGACTGAAAGCTAACTTCCCTGAT-3’; HABP1-R: 5’-CCCAATTTCGTGGTTGAAGTTATA-3’; GAPDH-F: 5’-TGCACCACCAACTGCTTAGC-3’; GAPDH-R: 5’-TCTTCTGGGTGGCAGTGATG-3’.
Western blot
Cell lysates were prepared in RIPA buffer with protease inhibitors (Complete Mini Protease Inhibitor; Roche). The lysates were clarified by centrifugation at 12,000 g for 10 minutes at 4°C. Protein concentration was measured with the BCA assay (Thermo Scientific). Equivalent amounts of protein were subjected to SDS-PAGE analysis. Immunoblotting was performed using standard methods. Quantification of the protein was conducted by the FluorChem M (ProteinSimple, Santa Clara, CA, USA) according to the instructions of the manufacturer. Each sample was normalized to β-actin.
HABP1 silencing by lentiviral vector-mediated short hairpin RNA (shRNA)
ShRNA was obtained from Invitrogen (Shanghai, China). Oligonucleotides were annealed and inserted into digested pENTR/U6-GFP Cloning Vector plasmid. Production of the lentiviral particles was carried out according to the manufacturer’s protocol. Cells were infected with lentivirus particles containing the shRNA, and stable transfectants were isolated by way of selection with 3 μg/ml blasticidin. The pENTR/U6-GFP-scramble shRNA plasmid was packaged as a negative control.
Cell growth assay
Cells were seeded at a density of 2000 cells per well in 96-well plates, and incubated with MTS according to the manufacturer’s instructions (Promega, Beijing, China). Cell growth was monitored daily.
Migration assay and invasion assay
To measure cell migration and invasion, 24-well transwell plates (8-μm pore size, Corning Costar, Shanghai, China) were used. For transwell migration analysis, cells suspended in medium with serum or growth factors were plated in the top chamber underlined with a non-coated membrane. For invasion analysis, chamber inserts were coated with 200 mg/ml Matrigel and dried overnight under sterile conditions, after which cells in medium with serum or growth factors were plated in the top chamber. For both assays, medium containing serum was added in the lower chamber. After incubation at 37°C for 24 h, the top chambers were wiped with cotton wool to remove non-migratory or non-invasive cells. Invading cells on the underside of membrane were fixed in 100% methanol for 10 min, air-dried, stained in 0.1% crystal violet, and counted under microscopy.
Animal experiments
The animals used in this study were 6-week-old athymic female nude BALB/c-/- mice, which were provided by Shanghai Institute of Material Medical, Chinese Academy of Science. Twenty mice were divided into four groups, with five mice in each group. The breast cancer cells were harvested by trypsinization, and 2 × 106 cells with 100 μl of PBS were injected directly into the mammary fat pad. The primary tumor out growth was monitored weekly by taking measurements of the tumor length (L) and width (W). The animals were killed4 weeks after tumor inoculation. The tumor volumes were calculated by using the following formula: volume = 0.5 × width2 × length. All animal experiments were carried out with approval from the Medical Experimental Animal Care Commission, Harbin Medical University.
Immunohistochemistry (IHC)
This study used archival material from the Department of Pathology at the Third Affiliated Hospital of Harbin Medical University, including tissues from 233 consecutive patients with histologically confirmed breast cancer and 50 benign tissue samples, all from 2006. Neither chemotherapy nor radiotherapy was performed on any patient prior to surgery. All of the tissue specimens used in this study were obtained with patients’ written informed consent, and the Ethics Committee of Harbin Medical University granted approval for this measure as well as the research protocol.
Each section was incubated in a 1:200 dilution of anti-HABP1 antibody. The IHC reaction was quantified by multiplying staining intensity by the percentage of positive tumor cells. In cytoplasm, staining intensity was graded as 0 (no staining), 1 (weak staining), 2 (moderate staining), and 3 (strong staining). The percentage (0-100 %) of the extent of reactivity was scored as follows: 0 (no positive tumor cells), 1 (≤ 10%), 2 (10-50%), and 3 (≥ 50%) [15]. Each case was scored independently and in a blinded manner by two investigators. Scores ≤ 4 were regarded as negative expression, and the remainder were classified as positive expression.
Statistical analysis
Statistical analysis was assessed using statistical software (SPSS 17.0 for Windows; SPSS, Inc., Chicago, IL). Student T or Mann-Whitney U tests were used to determine the statistical significance of differences between experimental groups. Chi-square tests were used to assess associations between HABP1 expression and patients’ clinicopathological features. The Kaplan-Meier survival curves and log rank tests were used for survival curves. Prognostic factors were evaluated by univariate and multivariate analyses using Cox proportional hazard regression models. The level of significance was set at P < 0.05.
Results
Knockdown of HABP1 inhibits proliferation
BT549 and MDA-MB-231 cells were triple-negative breast cancer cell line with highly aggressive. To determine the function of HABP1 in breast cancer cells, we infected BT549 and MDA-MB-231 cells with a lentivirus-mediated system, to generate 549-KD cells and 231-KD cells, respectively. Cells infected with negative controls expressing scrambled shRNA were named 549-NC cells and 231-NC cells. Then, we carried out qRT-PCR and Western blot analyses to confirm knockdown of HABP1 (Figure 1A, 1B). To investigate the potential involvement of HABP1 in cancer cell proliferation, the MTS assay was performed after shRNA treatment. The MTS assay showed that there was a significant inhibitory effect on cell proliferation in 549-KD and 231-KD cells compared to cells treated with negative control shRNA (P < 0.01, Figure 1C, 1D).
Figure 1.
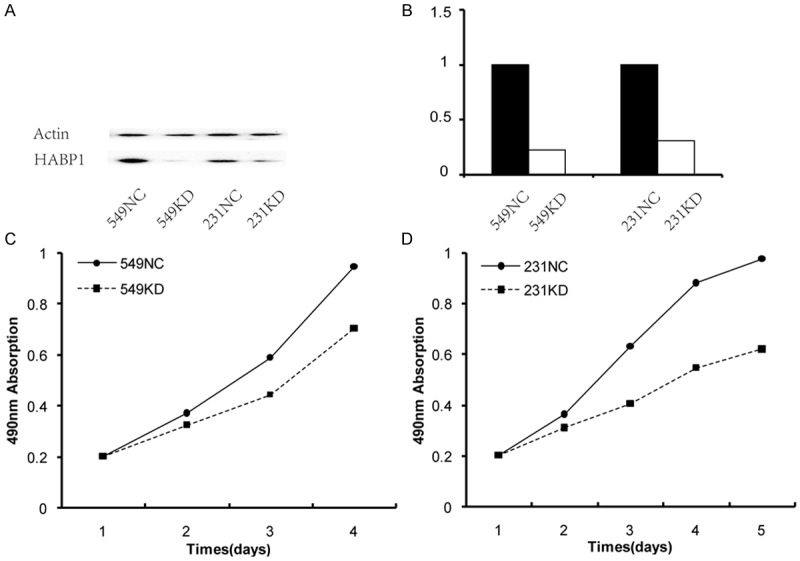
HABP1 promotes proliferation of breast cancer cells. A and B. The silencing efficiency of HABP1 shRNA examined by western blotting and quantility realtime PCR, respectively. C and D. MTS assay showed that HABP1 knocking down inhibited cell proliferation in BT549 and MDA-MB-231 cells, respectively.
HABP1 knockdown inhibits migration and invasion of breast cancer cells
Next, to determine the possible role of HABP1 in the breast cancer cells, the transwell assay was performed in BT549 and MDA-MB-231 cells. In a transwell assay, the 549-KD cells and 231-KD cells showed reduced migration and invasion ability compared with 549-NC cells and 231-NC cells (P < 0.05, Figure 2), suggesting that silencing HABP1 might inhibit cell migration and invasion in vitro.
Figure 2.

HABP1 promotes migration and invasion of breast cancer cells. Cells were plated on noncoated or gelatinous protein (Matrigel)-coated filters. (A and E) 549NC cells were significantly increase migrated or invade cells compared with 549KD cells (B and F). (C and G) 231NC cells were significantly increase migrated or invade cells compared with 231KD cells (D and H).
Knockdown of HABP1 inhibits tumor formation
To confirm the negative effect of HABP1 on tumor formation in vivo, we monitored a total of 20 BALB/c athymic mice, which were inoculated with either 549-KD, 549-NC, 231-KD or 231-NC cells for 4 weeks. We observed that depletion of HABP1 inhabited tumor growth in mice, and that the size of sh-HABP1 cell-derived tumors was smaller than the control cells (P < 0.01, Figure 3A, 3B). Overall, these results demonstrate that knockdown of HABP1 inhibits the growth of breast cancer cells.
Figure 3.
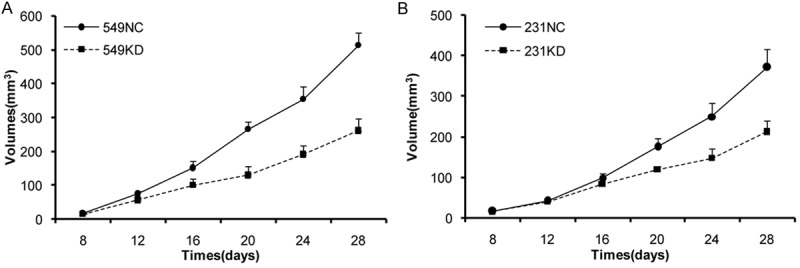
HABP1 knockdown abolishes the growth of established tumors. BT549 and MDA-MB-231 cells were inoculated subcutaneously into the mammary fat pad. The graph presents the growth of the tumors in the mice.
HABP1 expression is correlated with clinicopathological parameters in breast cancer patients
Positive HABP1 staining in breast cancer tissues appeared as brown particles which were mainly cytoplasmic. HABP1was overexpressed in cancer tissue compared with normal tissue (P = 0.001, Figure 4A). As shown in Table 1, 137 (58.8%) samples were HABP1 positive in our cohort of patients, which was similar to results from our previous study [6]. Statistical analysis showed that HABP1 expression was strongly associated with lymph node metastasis (P = 0.032) and tumor stage (P = 0.041). Expression of HABP1 was not associated with age, tumor size, estrogen receptor (ER), progesterone receptor (PR) or human epidermal growth factor receptor-2 (Her-2).
Figure 4.
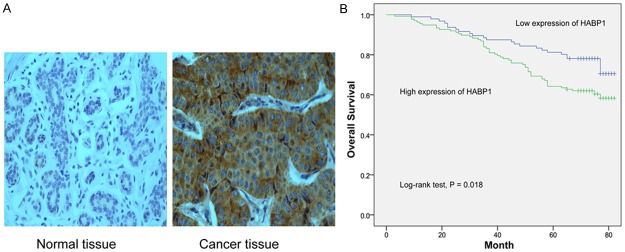
HABP1 high expression correlates with poor survival. A. Representative images of HABP1 immunohistochemistry staining. B. Kaplan-Meier survival analyses of breast cancer patients.
Table 1.
Correlation between clinical pathological factors and HABP1 expression
| Variables | No (n = 233) | HABP1 expression | P | |
|---|---|---|---|---|
|
| ||||
| Negative | Positive | |||
| Age | ||||
| < 50 y | 133 | 60 (45.1) | 73 (54.9) | 0.162 |
| ≥ 50 y | 100 | 36 (36) | 64 (64) | |
| Tumor | ||||
| ≤ 2 cm | 68 | 28 (41.2) | 40 (58.8) | 0.996 |
| >2 cm | 165 | 68 (41.2) | 97 (58.8) | |
| LNM | ||||
| Negative | 65 | 34 (52.3) | 31 (47.7) | 0.032 |
| Positive | 168 | 62 (36.9) | 106 (63.1) | |
| Stage | ||||
| I | 26 | 15 (57.7) | 11 (42.3) | 0.041 |
| II | 94 | 43 (45.7) | 51 (54.3) | |
| III | 113 | 38 (33.6) | 75 (66.4) | |
| ER | ||||
| Negative | 135 | 60 (44.4) | 75 (55.6) | 0.238 |
| Positive | 98 | 36 (36.7) | 62 (63.3) | |
| PR | ||||
| Negative | 98 | 44 (44.9) | 54 (55.1) | 0.329 |
| Positive | 135 | 52 (38.5) | 83 (61.5) | |
| HER-2 | ||||
| Negative | 179 | 71 (39.7) | 108 (60.3) | 0.386 |
| Positive | 54 | 25 (46.3) | 29 (53.7) | |
| P53 | ||||
| Negative | 45 | 18 (40) | 27 (60) | 0.855 |
| Positive | 188 | 78 (41.5) | 110 (58.5) | |
| Ki67 | ||||
| < 10% | 68 | 31 (45.6) | 37 (54.4) | 0.382 |
| ≥ 10% | 165 | 65 (39.4) | 100 (60.6) | |
| Molecular classification | ||||
| Luminal-A | 45 | 21 (46.7) | 24 (53.3) | 0.210 |
| Luminal-B | 116 | 40 (34.5) | 76 (65.5) | |
| Her-2(+) | 53 | 25 (47.2) | 28 (52.8) | |
| Triple negative | 19 | 10 (52.6) | 9 (47.4) | |
Chi-square test is used. P-value < 0.05 are considered statistically significant.
HABP1 expression is correlated with breast cancer patient survival
Kaplan-Meier analysis shown that HABP1 expression was significantly related to OS (log-rank test, P = 0.018; Figure 4B). HABP1 expression, along with tumor size, lymph node metastasis and Her-2, were found to be significantly associated with survival of patients (P = 0.02, 0.002, 0.006 and 0.008, respectively; Table 2). Next, we performed a Cox regression multivariate analysis with parameters including tumor size, lymph node metastasis, Her-2 and HABP1. The Cox proportional hazards model indicated that HABP1 was an independent prognostic factor for survival (hazard ratio [HR]: 1.709; 95% confidence interval [CI], 1.052-2.775; P = 0.03).
Table 2.
Prognostic factors in cox proportional hazards model
| Variables | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| HR | 95% CI | P | HR | 95% CI | P | |
| Age | ||||||
| < 50 y/≥ 50 y | 1.193 | 0.765-1.861 | 0.437 | |||
| Tumor size | ||||||
| ≤ 2 cm/> 2 cm | 2.603 | 1.407-4.816 | 0.002 | 2.281 | 1.224-4.251 | 0.009 |
| LNM | ||||||
| Positive/Negative | 2.354 | 1.272-4.356 | 0.006 | 1.95 | 1.047-3.633 | 0.035 |
| ER | ||||||
| Positive/Negative | 0.896 | 0.659-1.612 | 0.896 | |||
| PR | ||||||
| Positive/Negative | 0.711 | 0.456-1.109 | 0.132 | |||
| HER-2 | ||||||
| Positive/Negative | 1.909 | 1.186-3.074 | 0.008 | 1.772 | 1.097-2.863 | 0.019 |
| P53 | ||||||
| Positive/Negative | 0.773 | 0.514-1.162 | 0.216 | |||
| HABP1 | ||||||
| High/Low | 1.173 | 1.095-2.870 | 0.020 | 1.709 | 1.052-2.775 | 0.030 |
Cox proportional hazards regression analysis is used. P-value < 0.05 are considered statistically significant. AJCC: American Joint Committee on Cancer, CI: confidence interval.
Discussion
HABP1, located at 17p13.3, is an important gene that interacts specifically with HA , and has been implicated in cell adhesion and tumor invasion [16], sperm maturity and motility [17], and cellular signaling [18-20]. HABP1 has been detected in various cellular compartments, including the nucleus [21], mitochondria [22,23], Golgi [24] as well as secreted into the extracellular [25] in different cell types. Due to positive HABP1 staining mainly in the cytoplasmic, only a few sections showed nuclear and cell membrane staining in breast cancer tissues. Therefore, we directly compared the scores of cytoplasmic staining and clinical parameters in breast cancer patients. In the present study, we found that HABP1 was upregulated in breast cancer tissues, and its level was correlated with lymph node metastasis and stage. In addition, Kaplan Meier analysis revealed that HABP1 expression was significantly associated with poorer prognosis in breast cancer patients, univariate and multivariate analyses indicated that HABP1 expression was an independent prognostic factor.
Metastasis is the leading cause of death among cancer patients, but the mechanism of metastasis is complex and unclear. HABP1 may interact with αvβ3, resulting in MT1-MMP overexpression and MMP-2 activation by NFκB signaling in melanoma cell [26]. Rozanovet al. [27] suggest that MT1-MMP can directly bind to HABP1 in breast cancer cells, which may be involved in extracellular matrix degradation, thus inducing cancer cell invasion. In addition, HABP1 has been identified to be an essential component of lamellipodia in lung cancer cells [13], it may regulates the activity of PRKCZ and induces actin polymerization through GSK3β activation [12], resulting in increased cell mobility, also indicating that it may be a participant in tumor cell metastasis. Zhang et al. [12] demonstrated that high expression of HABP1 was associated with distant metastasis in patients with breast cancer, and silenced HABP1 cancer cells had a decreased number of lung surface metastatic foci in mice model. Our results suggest that shRNA against HABP1 influenced the migration and invasion abilities of breast cancer cells, consistent with those previous findings. However, the exact mechanism of HABP1 in breast cancer cell migration and invasion needs more study.
In this study, knockdown of HABP1 could inhibit cancer cell proliferation in vivo. High levels of HABP1 were observed to have an increased “HA pool”, and showed enhanced cell proliferation by regulating cyclin D1 promoter activity in HepG2 cells [9]. Fogal et al. [10] found that HABP1 knockdown cells reduce oxidative phosphorylation enzyme levels and activity, resulting decrease the cellular levels of energy and metabolism intermediates necessary for cancer cell growth. Orthotropic xenograft tumor models in mice further supported that HABP1 could involve in proliferation. It has been reported that HABP1 is an endogenous inhibitor of mitochondrial-dependent apoptosis, and that siRNA knockdown of HABP1 enhances staurosporine and doxorubicin-induced cell death, and reduces cellular proliferation in the MDA-MB-231 cell line [11]. However, silencing HABP1 has no influence on apoptosis of breast cancer cells in this study (data were not shown). This discrepancy may be explained by different environmental contexts, high level of HABP1 may tolerance for medicine stimulation.
In summary, the present findings provide evidence that HABP1 expression is higher in breast cancer tissues compared with normal breast tissues, and its higher levels correlate with lymph node metastasis and poor prognosis. Furthermore, HABP1 promote cell proliferation and metastasis, thus may be a novel and potential prognostic marker in patients with breast cancer.
Acknowledgements
The project was supported by Health and Family Planning Commission of the Heilongjiang Province (No. 645).
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Gupta S, Deb TB, Datta K. Evidence for the existence of hyaluronectin-binding proteins in the plasma membranes. FEBS Lett. 1993;336:511–515. doi: 10.1016/0014-5793(93)80866-s. [DOI] [PubMed] [Google Scholar]
- 3.Ghebrehiwet B, Lu PD, Zhang W, Lim BL, Eggleton P, Leigh LE, Reid KB, Peerschke EI. Identification of functional domains on gC1Q-R, a cell surface protein that binds to the globular “heads” of C1Q, using monoclonal antibodies and synthetic peptides. Hybridoma. 1996;15:333–342. doi: 10.1089/hyb.1996.15.333. [DOI] [PubMed] [Google Scholar]
- 4.Deb TB, Datta K. Molecular cloning of human fibroblast hyaluronic acid-binding protein confirms its identity with P-32, a protein co-purified with splicing factor SF2. Hyaluronic acid-binding protein as P-32 protein, co-purified with splicing factor SF2. J Biol Chem. 1996;271:2206–2212. doi: 10.1074/jbc.271.4.2206. [DOI] [PubMed] [Google Scholar]
- 5.Fogal V, Zhang L, Krajewski S, Ruoslahti E. Mitochondrial/cell-surface protein p32/gC1qR as a molecular target in tumor cells and tumor stroma. Cancer Res. 2008;68:7210–7218. doi: 10.1158/0008-5472.CAN-07-6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen YB, Jiang CT, Zhang GQ, Wang JS, Pang D. Increased expression of hyaluronic acid binding protein 1 is correlated with poor prognosis in patients with breast cancer. J Surg Oncol. 2009;100:382–386. doi: 10.1002/jso.21329. [DOI] [PubMed] [Google Scholar]
- 7.Amamoto R, Yagi M, Song Y, Oda Y, Tsuneyoshi M, Naito S, Yokomizo A, Kuroiwa K, Tokunaga S, Kato S, Hiura H, Samori T, Kang D, Uchiumi T. Mitochondrial p32/C1QBP is highly expressed in prostate cancer and is associated with shorter prostate-specific antigen relapse time after radical prostatectomy. Cancer Sci. 2011;102:639–647. doi: 10.1111/j.1349-7006.2010.01828.x. [DOI] [PubMed] [Google Scholar]
- 8.Rubinstein DB, Stortchevoi A, Boosalis M, Ashfaq R, Ghebrehiwet B, Peerschke EI, Calvo F, Guillaume T. Receptor for the globular heads of C1q (gC1q-R, p33, hyaluronan-binding protein) is preferentially expressed by adenocarcinoma cells. Int J Cancer. 2004;110:741–750. doi: 10.1002/ijc.20105. [DOI] [PubMed] [Google Scholar]
- 9.Kaul R, Saha P, Saradhi M, Prasad RL, Chatterjee S, Ghosh I, Tyagi RK, Datta K. Overexpression of hyaluronan-binding protein 1 (HABP1/p32/gC1qR) in HepG2 cells leads to increased hyaluronan synthesis and cell proliferation by up-regulation of cyclin D1 in AKT-dependent pathway. J Biol Chem. 2012;287:19750–19764. doi: 10.1074/jbc.M111.266270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fogal V, Richardson AD, Karmali PP, Scheffler IE, Smith JW, Ruoslahti E. Mitochondrial p32 protein is a critical regulator of tumor metabolism via maintenance of oxidative phosphorylation. Mol Cell Biol. 2010;30:1303–1318. doi: 10.1128/MCB.01101-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGee AM, Douglas DL, Liang Y, Hyder SM, Baines CP. The mitochondrial protein C1qbp promotes cell proliferation, migration and resistance to cell death. Cell Cycle. 2011;10:4119–4127. doi: 10.4161/cc.10.23.18287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, Zhang F, Guo L, Wang Y, Zhang P, Wang R, Zhang N, Chen R. Interactome analysis reveals that C1QBP (complement component 1, q subcomponent binding protein) is associated with cancer cell chemotaxis and metastasis. Mol Cell Proteomics. 2013;12:3199–3209. doi: 10.1074/mcp.M113.029413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim KB, Yi JS, Nguyen N, Lee JH, Kwon YC, Ahn BY, Cho H, Kim YK, Yoo HJ, Lee JS, Ko YG. Cell-surface receptor for complement component C1q (gC1qR) is a key regulator for lamellipodia formation and cancer metastasis. J Biol Chem. 2011;286:23093–23101. doi: 10.1074/jbc.M111.233304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Remmele W, Stegner HE. [Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue] . Pathologe. 1987;8:138–140. [PubMed] [Google Scholar]
- 16.Gupta S, Datta K. Possible role of hyaluronectin on cell adhesion in rat histiocytoma. Exp Cell Res. 1991;195:386–394. doi: 10.1016/0014-4827(91)90388-b. [DOI] [PubMed] [Google Scholar]
- 17.Ranganathan S, Ganguly AK, Datta K. Evidence for presence of hyaluronan binding protein on spermatozoa and its possible involvement in sperm function. Mol Reprod Dev. 1994;38:69–76. doi: 10.1002/mrd.1080380112. [DOI] [PubMed] [Google Scholar]
- 18.Das S, Deb TB, Kumar R, Datta K. Multifunctional activities of human fibroblast 34-kDa hyaluronic acid-binding protein. Gene. 1997;190:223–225. doi: 10.1016/s0378-1119(97)00035-8. [DOI] [PubMed] [Google Scholar]
- 19.Itahana K, Zhang Y. Mitochondrial p32 is a critical mediator of ARF-induced apoptosis. Cancer Cell. 2008;13:542–553. doi: 10.1016/j.ccr.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reef S, Shifman O, Oren M, Kimchi A. The autophagic inducer smARF interacts with and is stabilized by the mitochondrial p32 protein. Oncogene. 2007;26:6677–6683. doi: 10.1038/sj.onc.1210485. [DOI] [PubMed] [Google Scholar]
- 21.Brokstad KA, Kalland KH, Russell WC, Matthews DA. Mitochondrial protein p32 can accumulate in the nucleus. Biochem Biophys Res Commun. 2001;281:1161–1169. doi: 10.1006/bbrc.2001.4473. [DOI] [PubMed] [Google Scholar]
- 22.Muta T, Kang D, Kitajima S, Fujiwara T, Hamasaki N. p32 protein, a splicing factor 2-associated protein, is localized in mitochondrial matrix and is functionally important in maintaining oxidative phosphorylation. J Biol Chem. 1997;272:24363–24370. doi: 10.1074/jbc.272.39.24363. [DOI] [PubMed] [Google Scholar]
- 23.McGee AM, Baines CP. Complement 1q-binding protein inhibits the mitochondrial permeability transition pore and protects against oxidative stress-induced death. Biochem J. 2011;433:119–125. doi: 10.1042/BJ20101431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sengupta A, Banerjee B, Tyagi RK, Datta K. Golgi localization and dynamics of hyaluronan binding protein 1 (HABP1/p32/C1QBP) during the cell cycle. Cell Res. 2005;15:183–186. doi: 10.1038/sj.cr.7290284. [DOI] [PubMed] [Google Scholar]
- 25.Lim BL, Reid KB, Ghebrehiwet B, Peerschke EI, Leigh LA, Preissner KT. The binding protein for globular heads of complement C1q, gC1qR. Functional expression and characterization as a novel vitronectin binding factor. J Biol Chem. 1996;271:26739–26744. doi: 10.1074/jbc.271.43.26739. [DOI] [PubMed] [Google Scholar]
- 26.Prakash M, Kale S, Ghosh I, Kundu GC, Datta K. Hyaluronan-binding protein 1 (HABP1/p32/gC1qR) induces melanoma cell migration and tumor growth by NF-kappa B dependent MMP-2 activation through integrin alpha(v)beta(3) interaction. Cell Signal. 2011;23:1563–1577. doi: 10.1016/j.cellsig.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 27.Rozanov DV, Ghebrehiwet B, Ratnikov B, Monosov EZ, Deryugina EI, Strongin AY. The cytoplasmic tail peptide sequence of membrane type-1 matrix metalloproteinase (MT1-MMP) directly binds to gC1qR, a compartment-specific chaperone-like regulatory protein. FEBS Lett. 2002;527:51–57. doi: 10.1016/s0014-5793(02)03153-8. [DOI] [PubMed] [Google Scholar]


