Abstract
Nicotinamide adenine dinucleotide (NAD) is a crucial molecule of energy production and signal transduction processes that have been linked to ovarian cancer development. Notably, emerging evidence has led to considerable interest in the role of DNA methyltransferase 1 (DNMT1) in the initiation and progression of ovarian cancer. However, dynamic crosstalk between NAD and DNMT1 is poorly understood. Here, we show that DNMT1 levels are upregulated, along with increased NAD levels in non-BRCA1-mutated ovarian cancer cells. In contrast, DNMT1 levels are not affected by increasing NAD levels in BRCA1-mutated ovarian cancer cells. Mechanistically, BRCA1 inactivity-mediated loss of H3K9ac enrichment around the core promoter inhibits DNMT1 transcription. Consistent with this, BRCA1 levels correlate with DNMT1 levels (R = 0.534, R < 0.001) in human ovarian cancer specimens. Therefore, these results highlight a novel regulatory effect of NAD on DNMT1, and further correlate the physiological properties of NAD metabolism with DNMT1-mediated biological processes. All of this may improve our understanding of the basic molecular mechanism underlying NAD- and DNMT1-related ovarian cancer progression.
Keywords: NAD, DNMT1, BRCA1, ovarian cancer
Introduction
Ovarian cancer is the most prevalent malignancy, and is a leading cause of mortality in women worldwide [1]. To date, although the exact cause of ovarian cancer remains largely unknown, BRCA mutations are the main known hereditary factors [2]. BRCA1 is a tumor suppressor gene involved in multiple cellular processes [3]. Our previous study also indicated that there are wide ranges of transcriptional regulation, epigenetic patterns and metabolic differences between BRCA1 dysfunction and the basal phenotype [4-10]. Nicotinamide adenine dinucleotide (NAD) is a crucial molecule of energy production and signal transduction processes that have been linked to ovarian cancer development [9-12]. Notably, recent evidence suggests that NAD is an important transcription regulatory factor for BRCA1 [10].
DNA methyltransferase 1 (DNMT1) is a key enzyme involved in maintaining DNA methylation, gene regulation, and chromatin stability [13]. Emerging evidence has suggested a possible role of DNMT1 in ovarian cancer progression. For example, (i) DNMT1 is frequently overexpressed in ovarian cancer samples [14], and associated with clinicopathological and survival data in sporadic ovarian cancer patients [15]; (ii) DNMT1 variants may be risk factors of ovarian cancer [16]; (iii) DNMT1 has the potential to be an adjuvant therapeutic approach to overcome ovarian cancer chemoresistance [17,18]. Notably, our recent study showed that BRCA1-mutated breast cancer displayed a hypermethylated E2F transcription factor 1 (E2F1) motif , which is a key regulatory mechanism for DNMT1 transcription [7], and the observation is consistent with previous reports that DNMT1 may be a transcriptional target of BRCA1 [19]. However, to date, little is known about the direct link(s) between NAD and DNMT1. For this reason, the present study addresses the effects of NAD on DNMT1 through regulation of BRCA1 in ovarian cancer, and provides novel insight into the mechanisms involved in the regulation of DNMT1 expression.
Materials and methods
Ethical statements
The research was carried out in accordance with the Helsinki Declaration of 1975 and approved by the Institutional Review Board at China Medical University.
Patients and tissue collection
Serous ovarian cancer patients were enrolled between 2010 and 2012, and all patients gave informed consent. Fresh tumor samples, ascites, and blood samples were obtained at the time of primary surgery before any chemotherapy or radiotherapy (15 pairs of BRCA1-mutated or not, and 11 pairs with hypermethylated BRCA1 promoter or not). Hematoxylin and eosin staining of the samples for histopathologic diagnosis and grading were performed by three staff pathologists using World Health Organization criteria. All patients were screened for BRCA1 mutations by multiplex polymerase chain reaction (PCR). Their characteristics were shown in our previous studies [20].
Cell culture, lentiviral infection and cell proliferation assay
Primary ovarian cancer cells were obtained from the ascites of patients undergoing surgery for ovarian cancer and cultured in RPMI 1640 with 10% fetal bovine serum (Invitrogen, CA, USA) as previously reported [5,6,9,10,20]. The shRNA lentiviral particles of nicotinamide phosphoribosyltransferase (NAMPT) (sc-45843-V), histone acetyltransferase (GCN5) (sc-37946-V), p300/CBP-associated factor (PCAF) (sc-36198-V) were purchased from Santa Cruz Biotechnology (CA, USA). For overexpression of NAMPT, the open reading frame of NAMPT (NM_005746) were respectively cloned into the lentiviral vector GV287 (Ubi-MCS-3FLAG-SV40-EGFP; GeneChem Co., Ltd, Shanghai, China). Transfections were performed using polybrene and enhanced infection solution (GeneChem Co., Ltd) according to the manufacturer’s recommended protocol. The efficiency of NAMPT, GCN5 or PCAF knockdown, and NAMPT overexpression, and the procedures were as previously reported [8,10]. After 48 hours of infection, cell proliferation was determined using the Cell-LightTM EdU Apollo®643 In Vitro Imaging Kit (Ribobio, Guangzhou, China) following the instructions provided by the manufacturer.
Real-time quantitative PCR
Total RNA was extracted using Trizol reagents (Invitrogen) according to the manufacturer’s protocol. DNA contamination was removed by adding DNase I (Invitrogen) according to the manufacturer’s protocol. Total RNA was then reverse-transcribed from 2 μg of RNA using the PrimeScript RT Master Mix kit (TaKaRa, Dalian, China) and amplified by SYBR Premix Ex TaqTM II (TaKaRa) in a Roche LightCycler 2.0 instrument (Roche Diagnostics, Mannheim, Germany). The specific primer sequences for BRCA1, NAMPT and DNMT1 were shown in our previous studies [7,10]. GAPDH mRNA was amplified as an internal control for normalization of each sample. All samples were analyzed in triplicate using the 2-ΔΔCT method.
Bisulfite sequencing for BRCA1 promoter
All the tissues were used for bisulfite sequencing from the non-BRCA1-mutated cases. Genomic DNA extracted from ovarian cancer tissue with a TIANamp Genomic DNA kit (Tiangen Biotech, Beijing, China) was subjected to bisulfite conversion using the EZ DNA Methylation-Direct kit (Zymo Research, Orange, USA) following the manufacturer’s instructions; the conversion efficiency was estimated to be at least 99.6%. It was then amplified by nested PCR. After gel purification, cloning and transformation into E. coli Competent Cells JM109 (TaKaRa), ten positive clones of each sample were sequenced to ascertain the methylation patterns of each CpG locus. The primers were used for BRCA1 gene promoter (Accession number: NG_005905) were shown in our previous studies [6,9]. The conditions were as follows: 95°C for 2 min, 40 cycles of 30 s at 95°C, 30 s at 56°C and 45 s at 72°C, then 72°C for 7 min.
NAD incubation
Primary ovarian cancer cells were incubated with 0, 10, 50, 250 or 1250 μM NAD (Sigma, CA, USA) for 3 h at 37°C.
NAD levels assay
For NAD assay, 20 μl packed ovarian cancer cells were homogenized in 400 μl BioVsion NAD/NADH Extraction Buffer (BioVsion, CA, USA). The homogenate was ultrafiltered using BioVsion 10-kD cut-off filters (14000 g, 30 min, 4°C). Assays were performed using the NAD/NADH Quantification Kits according to the manufacturer’s instructions (BioVsion).
Chromatin immunoprecipitation assay (ChIP)
ChIP assays were performed using the EpiQuikTM Tissue Chromatin Immunoprecipitation kit (Epigentek Group Inc., Brooklyn, NY, USA), as previously reported [5,7,8]. Briefly, the small pieces of ovarian cancer tissues (1-2 mm3) were crosslinked with 1% formaldehyde. Cross-linking was terminated using 1.25 M glycine. The tissues and cells were added to homogenizing buffer, triturated, disaggregated, and centrifuged at 1000 g for 5 min at 4°C. After the removal of supernatants, the protease inhibitors were added and the disaggregated tissue pellet was resuspended. Chromatin was sheared by sonication. Immunoprecipitation was performed at room temperature for 90 min. The specific antibodies for ChIP were shown in our previous study [7]. Crosslinking was reversed at 65°C for 90 min. Eventually, genomic DNA was eluted for PCR analysis. The specific primer sequences for ChIP were shown in our previous study [7]. The thermocycle was 94°C for 2 min, then 30 cycles of 45 s at 94°C, 45 s at 56°C and 45 s at 72°C.
Statistical analysis
Regression analysis was used to examine the possible relationship between BRCA1 and DNMT1 levels in human ovarian cancer samples. The data are presented as means ± standard deviations (SD). Statistical differences in the data were evaluated by Student’s t-test or one-way analysis of variance as appropriate, and were considered significant at P < 0.05.
Results
NAD may regulate DNMT1 levels through BRCA1 in primary ovarian cancer cells
To confirm the role of NAD-mediated BRCA1 in the regulation of DNMT1 expression, the effects of incubation with different concentrations of NAD were evaluated in primary ovarian cancer cells with identified BRCA1 mutations or no BRCA1 mutations. BRCA1 expression was upregulated (Figure 1B), along with increased levels of intracellular NAD (Figure 1A) in non-BRCA1-mutated and BRCA1-mutated ovarian cancer cells. Notably, DNMT1 levels were also upregulated (Figure 1C), along with increased BRCA1 levels in non-BRCA1-mutated ovarian cancer cells (Figure 1B). In contrast, DNMT1 levels were not affected (Figure 1C) by increasing levels of BRCA1 (Figure 1B) in BRCA1-mutated ovarian cancer cells. It is well known that NAMPT is a rate-limiting enzyme in regenerating NAD in mammals. Similarly, knockdown or overexpression of NAMPT could effectively reduce or increase NAD levels (Figure 1D) and BRCA1 levels (Figure 1E) in primary non-BRCA1-mutated and BRCA1-mutated ovarian cancer cells. However, DNMT1 levels were downregulated or upregulated (Figure 1F), along with decreased or increased BRCA1 levels (Figure 1E) in non-BRCA1-mutated ovarian cancer cells, but DNMT1 levels were not affected (Figure 1F) by decreasing or increasing BRCA1 levels (Figure 1E) in BRCA1-mutated ovarian cancer cells. Therefore, these results suggest that BRCA1 may have an important role in NAD-related DNMT1 transcription.
Figure 1.
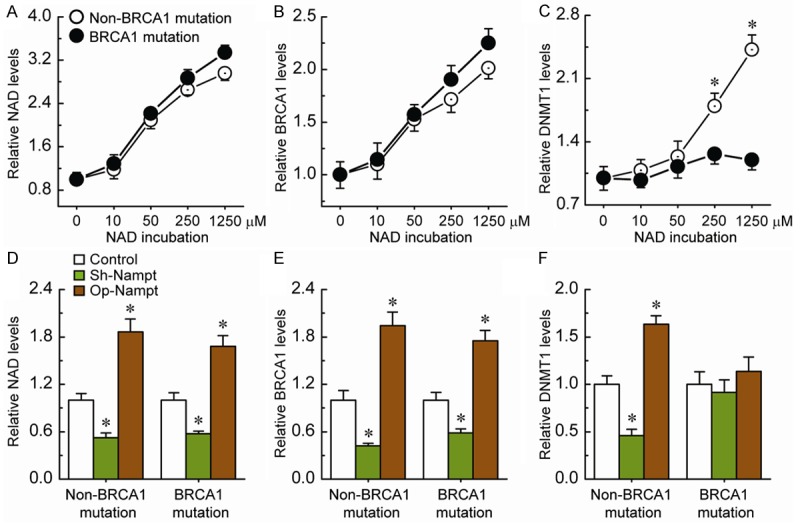
Effects of intracellular NAD-mediated BRCA1 on DNMT1 levels. A-C. Relative NAD, BRCA1 or DNMT1 levels after incubation with different concentrations of NAD in primary non-BRCA1-mutated and BRCA1-mutated ovarian cancer cells. Bar graphs show mean ± SD. *P < 0.05 vs. control. D-F. Relative NAD, BRCA1 or DNMT1 levels after knockdown or overexpression of NAMPT in primary non-BRCA1-mutated and BRCA1-mutated ovarian cancer cells. Bar graphs show mean ± SD. Each group, n = 15. Sh, shRNAs; Op, overexpression. *P < 0.05 vs. control.
Loss of H3K9ac enrichment around the DNMT1 core promoter region in BRCA1-mutated ovarian cancer
Our previous study suggested that specific histone modification patterns play an important part in regulating DNMT1 transcription [7]. To obtain further understanding of the regulatory potential of the histone modification in controlling DNMT1 transcription, we examined the active histone markers H3K9ac, H3K18ac, H3K27ac, H3K4me1, H3K4me2, H3K4me3, H3K36me3, and H3K79me, and the repressive histone markers H3K9me, H3K9me2, H3K9me3, H3K27me, H3K27me2, and H3K27me3 in the core promoter of DNMT1. We also focused on the enrichment of the transcription factor E2F1, due to the fact that the E2F1-binding site was found in this region. Chromatin immunoprecipitation analysis indicated that the levels of H3K9ac around the DNMT1 core promoter region were only significantly decreased in BRCA1-mutated ovarian cancer, compared with non-BRCA1-mutated ovarian cancer (Figure 2A and 2B). Chromatin-modifying enzymes (GCN5 and PCAF) that facilitate the creation of H3K9ac were also analyzed. Although there was no significant change in the expression of PCAF, the expression levels of GCN5 were reduced (Figure 2C). These results, together with Figure 1, suggest that BRCA1-related DNMT1 transcription may be associated with changes in decreased H3K9ac enrichment in ovarian cancer cells.
Figure 2.
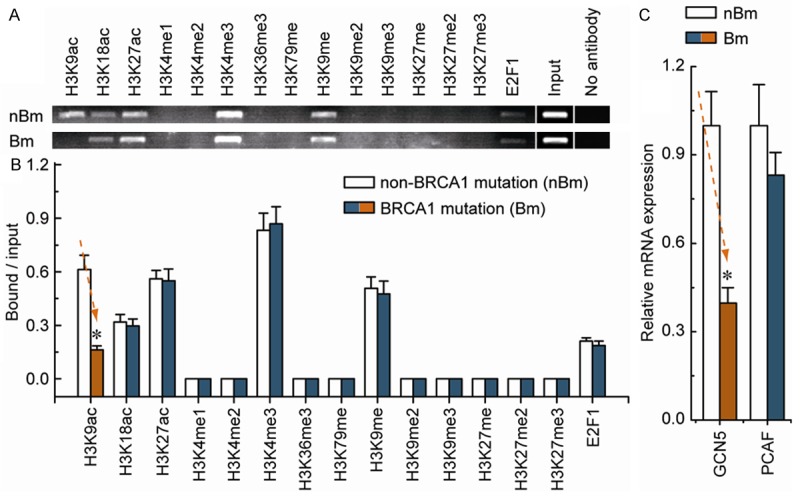
Comparative analysis of histone modification and E2F1 enrichment around the DNMT1 core promoter between non-BRCA1-mutated and BRCA1-mutated ovarian cancer tissues. A. Chromatin immunoprecipitation was performed using antibodies to H3K9ac, H3K18ac, H3K27ac, H3K4me1, H3K4me2, H3K4me3, H3K36me3, H3K79me, H3K9me, H3K9me2, H3K9me3, H3K27me, H3K27me2, H3K27me3 and E2F1. PCR was performed for regions within the core promoter of DNMT1. A negative control without antibodies is included for comparison. B. Representative results of primary non-BRCA1-mutated and BRCA1-mutated ovarian cancer tissues are shown. Bar graphs show mean ± SD. C. Expression levels of the chromatin-modifying enzymes GCN5 and PCAF in non-BRCA1-mutated and BRCA1-mutated ovarian cancer tissues. Bar graphs show mean ± SD. Each group, n = 15. *P < 0.05 vs. control.
H3K9ac present around the DNMT1 core promoter region are responsible for transcriptional regulation of DNMT1 in ovarian cancer
We observed that the knockdown of GCN5 and PCAF (Figure 3A and 3B) had no detectable effect on cell morphology and proliferation (Figure 3C and 3D). Combined knockdown of GCN5 and PCAF specifically induced a decrease in H3K9ac enrichment around the DNMT1 core promoter region (Figure 3E). Notably, after the deletion of H3K9ac, the DNMT1 levels were significantly down-regulated (Figure 3F).
Figure 3.
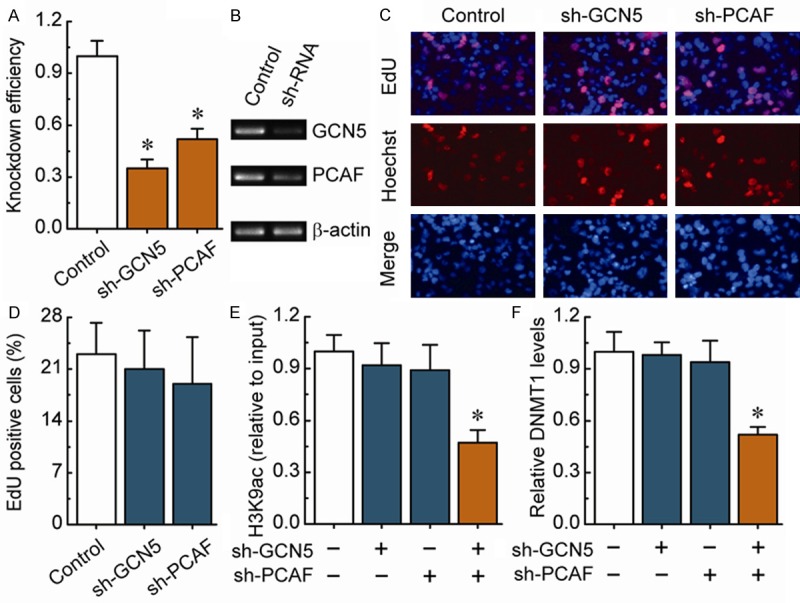
H3K9ac-mediated transcriptional regulation of DNMT1 in non-BRCA1-mutated ovarian cancer cells. A. RT-PCR showing GCN5 and PCAF levels before and after knockdown by shRNAs, and normalized to -actin expression (n = 3). B. Representative RT-PCR results are shown. C. EdU labeling showing the proliferation of GCN5- and PCAF-silenced in primary non-BRCA1-mutated ovarian cancer cells. Blue, Hoechst 33342 labeling of cell nuclei; red, EdU labeling of nuclei of proliferative cells. D. The EdU incorporation rate was expressed as the ratio of EdU-positive cells to total Hoechst 33342-positive cells. E. Analysis of histone modification H3K9ac enrichment around the core promoter region of DNMT1 after the deletion of GCN5 or/and PCAF in primary non-BRCA1-mutated ovarian cancer cells. F. DNMT1 levels after the deletion of H3K9ac around the core promoter region of DNMT1 in primary non-BRCA1-mutated ovarian cancer cells. Bar graphs show mean ± SD. Each group, n = 15. *P < 0.05 vs. control.
Hypermethylated BRCA1 promoter-mediated decreased BRCA1 levels are correlated with DNMT1 expression
In mammals, promoter methylation is an epigenetic modification involved in regulating gene expression [5,7,8]. Consistent with this idea, we showed that ovarian cancer with a hypermethylated BRCA1 promoter displayed decreased expression of BRCA1 in comparison with unmethylated BRCA1 promoter (Figure 4A-D). Based on these considerations, the low levels of BRCA1 appeared to be mediated by promoter hypermethylation, making this an appropriate model to investigate the physiological relationship between BRCA1 and DNMT1. Notably, the H3K9ac levels (Figure 4E) and DNMT1 levels (Figure 4F) were decreased markedly, along with hypermethylated promoter-mediated BRCA1 deficiency in ovarian cancer (Figure 4D). In addition, we analyzed the correlation between BRCA1 and DNMT1 levels in 22 ovarian cancer samples (Figure 4G). It is interesting to note a significantly positive correlation between levels of BRCA1 and DNMT1 (R = 0.534, P < 0.001; Figure 4G). These results further indicate that BRCA1 may be responsible for the regulation of DNMT1 in ovarian cancer cells.
Figure 4.
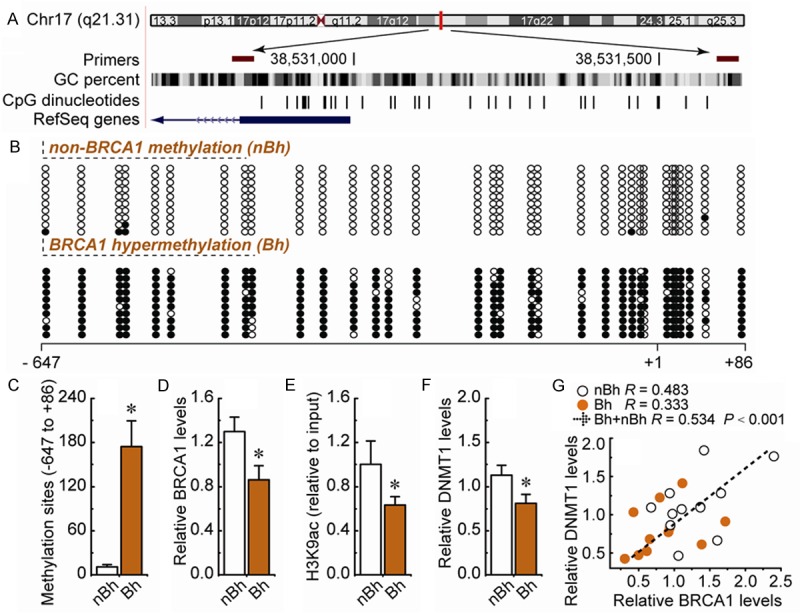
DNMT1 expression patterns in ovarian cancer tissues with hypermethylated promoter-mediated BRCA1 inactivation. A. Location of CpG sites in the core promoter region of BRCA1. Genomic coordinates are shown, along with the primer-amplified fragments, GC percentage, location of individual CpG dinucleotides (dashes) and BRCA1 RefSeq gene (exon 1 shown as a blue box and intron shown as an arrowed line). The arrow indicates the direction of transcription. B. Representative figures of unmethylated and hypermethylated BRCA1 promoter. The circles correspond to the CpG sites denoted by black dashes in (A). Closed circles, methylation; open circles, unmethylated. Ten individual clones were sequenced for each sample. C. Summary of the methylation levels of BRCA1 core promoter. D-F. Relative BRCA1, H3K9ac and DNMT1 levels were measured in ovarian cancer tissues with identified unmethylated and hypermethylated BRCA1 promoter. Bar graphs show mean ± SD. *P < 0.05 vs. control. G. Correlation between BRCA1 and DNMT1 levels in human ovarian cancer tissues with identified unmethylated and hypermethylated BRCA1 promoter. Each group, n = 11.
Discussion
In this study, we report for the first time that NAD may regulate DNMT1 levels through BRCA1 in ovarian cancer cells; the molecular mechanism may involve BRCA1-mediated alteration of histone modification H3K9ac enrichment around the core promoter of DNMT1. However, as shown in Figure 1, intracellular NAD levels may be responsible for the induction of BRCA1-related DNMT1 transcription in non-BRCA1-mutated ovarian cancer cells, but we observed that DNMT1 levels were not affected by NAD incubation, and knockdown or overexpression of NAMPT in BRCA1-mutated ovarian cancer cells. It may be due to the fact that BRCA1 itself, has no function along with BRCA1-mutation.
NAD, as well as its precursors, derivatives, and metabolic enzymes, has recently drawn much attention as regards a variety of biological functions including transcriptional regulation, DNA repair, cell cycle progression, metabolism, lifespan, and cell death [21]. Typically, (i) sirtuin 1 (SIRT1) is an NAD-dependent protein deacetylase [9]; (ii) poly(ADP-ribose) polymerase-1 (PARP1) as a sensor of DNA damage catalyzes the transfer of ADP-ribose units from NAD to target proteins [4]; (iii) NAMPT is the rate-limiting enzyme for NAD biosynthesis [10], which together comprise a system of regulatory networks, termed the ‘NAD World’, that maintains the balance of cellular metabolism and has essential roles in tumor development and therapy. To date, there have been few reports about the link(s) between NAD and DNMT1 in ovarian cancer cells. It is, however, interesting to note that the evidence accumulated suggests a possible link between NAD World and DNMT1. For example, (i) SIRT1 physically interacts with DNMT1, and SIRT1-mediated deacetylation of DNMT1 is responsible for the multiple effects of DNMT1-related gene silencing [22]; (ii) PARP1 can enhance DNMT1 expression by maintaining the unmethylated state of the DNMT1 promoter [23], and PARP1 can directly influence DNA methylation patterns through controlling the transcription and activity of DNMT1 [24]; (iii) NAD serves as the common substrate independent of its redox-carrier function by SIRT1 and PARP1, and NAMPT-related NAD synthesis is a key regulator of SIRT1 and PARP1 activity [4,9]. Therefore, the emerging picture from these studies prompted us to evaluate a possible link between NAD World and DNMT1. It seems beneficial for the understanding of DNMT1-related biological processes from NAD World aspects. For instance, both SIRT1 and PARP1 are consumers of NAD, SIRT1, and PARP1-related regulatory function of DNMT1, and may partly involve the direct effect of NAD on DNMT1.
Overall, these observations further correlate the physiological properties of NAD metabolism with DNMT1-mediated biological processes. All of this may improve our understanding of the basic molecular mechanism underlying NAD- and DNMT1-related ovarian cancer progression.
Acknowledgements
This work was supported by the 973 Program of China (No. 2013CB945201), Natural Science Foundation of China (No. 31171259, No. 31271364 and No. 81402130), and Doctoral Startup Foundation of Liaoning Province (No. 20141045).
Disclosure of conflict of interest
None.
References
- 1.Lech A, Daneva T, Pashova S, Gagov H, Crayton R, Kukwa W, Czarnecka AM, Szczylik C. Ovarian cancer as a genetic disease. Front Biosci(Landmark Ed) 2013;18:543–563. doi: 10.2741/4119. [DOI] [PubMed] [Google Scholar]
- 2.Pruthi S, Gostout BS, Lindor NM. Identification and Management of Women With BRCA Mutations or Hereditary Predisposition for Breast and Ovarian Cancer. Mayo Clin Proc. 2010;85:1111–1120. doi: 10.4065/mcp.2010.0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dacheux E, Vincent A, Nazaret N, Combet C, Wierinckx A, Mazoyer S, Diaz JJ, Lachuer J, Venezia ND. BRCA1-Dependent Translational Regulation in Breast Cancer Cells. PLoS One. 2013;8:e67313. doi: 10.1371/journal.pone.0067313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li D, Bi FF, Chen NN, Cao JM, Sun WP, Zhou YM, Li CY, Yang Q. A novel crosstalk between BRCA1 and poly (ADP-ribose) polymerase 1 in breast cancer. Cell Cycle. 2014;13:3442–3449. doi: 10.4161/15384101.2014.956507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li D, Bi FF, Cao JM, Cao C, Li CY, Liu B, Yang Q. Poly (ADP-ribose) polymerase 1 transcriptional regulation: a novel crosstalk between histone modification H3K9ac and ETS1 motif hypomethylation in BRCA1-mutated ovarian cancer. Oncotarget. 2014;5:291–297. doi: 10.18632/oncotarget.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li D, Bi FF, Cao JM, Cao C, Li CY, Yang Q. Effect of BRCA1 on epidermal growth factor receptor in ovarian cancer. J Exp Clin Cancer Res. 2013;32:102. doi: 10.1186/1756-9966-32-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li D, Bi FF, Cao JM, Cao C, Liu B, Yang Q. Regulation of DNA methyltransferase 1 transcription in BRCA1-mutated breast cancer: a novel crosstalk between E2F1 motif hypermethylation and loss of histone H3 lysine 9 acetylation. Mol Cancer. 2014;13:26. doi: 10.1186/1476-4598-13-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li D, Bi FF, Chen NN, Cao JM, Sun WP, Zhou YM, Cao C, Li CY, Yang Q. Epigenetic repression of phosphatidylethanolamine, N-methyltransferase (PEMT) in BRCA1-mutated breast cancer. Oncotarget. 2014;5:1315–1325. doi: 10.18632/oncotarget.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li D, Bi FF, Chen NN, Cao JM, Sun WP, Zhou YM, Li CY, Yang Q. A novel crosstalk between BRCA1 and sirtuin 1 in ovarian cancer. Sci Rep. 2014;4:6666. doi: 10.1038/srep06666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li D, Chen NN, Cao JM, Sun WP, Zhou YM, Li CY, Wang XX. BRCA1 as a nicotinamide adenine dinucleotide (NAD)-dependent metabolic switch in ovarian cancer. Cell Cycle. 2014;13:2564–2571. doi: 10.4161/15384101.2015.942208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan LZ, Ahn DG, Sharif T, Clements D, Gujar SA, Lee PW. The NAD+ synthesizing enzyme nicotinamide mononucleotide adenylyltransferase 2 (NMNAT-2) is a p53 downstream target. Cell Cycle. 2014;13:1041–1048. doi: 10.4161/cc.28128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schouten LJ, van Dijk BA, Lumey LH, Goldbohm RA, van den Brandt PA. Energy restriction during childhood and early adulthood and ovarian cancer risk. PLoS One. 2011;6:e27960. doi: 10.1371/journal.pone.0027960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein CJ, Botuyan MV, Wu Y, Ward CJ, Nicholson GA, Hammans S, Hojo K, Yamanishi H, Karpf AR, Wallace DC, Simon M, Lander C, Boardman LA, Cunningham JM, Smith GE, Litchy WJ, Boes B, Atkinson EJ, Middha S, B Dyck PJ, Parisi JE, Mer G, Smith DI, Dyck PJ. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat Genet. 2011;43:595–600. doi: 10.1038/ng.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu Y, Yang P, Shao Q, Liu X, Xia S, Zhang M, Xu H, Shao Q. Investigation of the expression patterns and correlation of DNA methyltransferases and class I histone deacetylases in ovarian cancer tissues. Oncol Lett. 2013;5:452–458. doi: 10.3892/ol.2012.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bai X, Song Z, Fu Y, Yu Z, Zhao L, Zhao H, Yao W, Huang D, Mi X, Wang E, Zheng Z, Wei M. Clinicopathological significance and prognostic value of DNA methyltransferase 1, 3a, and 3b expressions in sporadic epithelial ovarian cancer. PLoS One. 2012;7:e40024. doi: 10.1371/journal.pone.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mostowska A, Sajdak S, Pawlik P, Lianeri M, Jagodzinski PP. DNMT1, DNMT3A and DNMT3B gene variants in relation to ovarian cancer risk in the Polish population. Mol Biol Rep. 2013;40:4893–4899. doi: 10.1007/s11033-013-2589-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cacan E, Ali MW, Boyd NH, Hooks SB, Greer SF. Inhibition of HDAC1 and DNMT1 modulate RGS10 expression and decrease ovarian cancer chemoresistance. PLoS One. 2014;9:e87455. doi: 10.1371/journal.pone.0087455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiang Y, Ma N, Wang D, Zhang Y, Zhou J, Wu G, Zhao R, Huang H, Wang X, Qiao Y, Li F, Han D, Wang L, Zhang G, Gao X. MiR-152 and miR-185 co-contribute to ovarian cancer cells cisplatin sensitivity by targeting DNMT1 directly: a novel epigenetic therapy independent of decitabine. Oncogene. 2014;33:378–386. doi: 10.1038/onc.2012.575. [DOI] [PubMed] [Google Scholar]
- 19.Shukla V, Coumoul X, Lahusen T, Wang RH, Xu X, Vassilopoulos A, Xiao C, Lee MH, Man YG, Ouchi M, Ouchi T, Deng CX. BRCA1 affects global DNA methylation through regulation of DNMT1. Cell Res. 2010;20:1201–1215. doi: 10.1038/cr.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bi FF, Li D, Cao C, Li CY, Yang Q. Regulation of angiotensin II type 1 receptor expression in ovarian cancer: a potential role for BRCA1. J Ovarian Res. 2013;6:89. doi: 10.1186/1757-2215-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiarugi A, Dolle C, Felici R, Ziegler M. The NAD metabolome--a key determinant of cancer cell biology. Nat Rev Cancer. 2012;12:741–752. doi: 10.1038/nrc3340. [DOI] [PubMed] [Google Scholar]
- 22.Peng L, Yuan Z, Ling H, Fukasawa K, Robertson K, Olashaw N, Koomen J, Chen J, Lane WS, Seto E. SIRT1 deacetylates the DNA methyltransferase 1 (DNMT1) protein and alters its activities. Mol Cell Biol. 2011;31:4720–4734. doi: 10.1128/MCB.06147-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zampieri M, Passananti C, Calabrese R, Perilli M, Corbi N, De Cave F, Guastafierro T, Bacalini MG, Reale A, Amicosante G, Calabrese L, Zlatanova J, Caiafa P. Parp1 localizes within the Dnmt1 promoter and protects its unmethylated state by its enzymatic activity. PLoS One. 2009;4:e4717. doi: 10.1371/journal.pone.0004717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ciccarone F, Klinger FG, Catizone A, Calabrese R, Zampieri M, Bacalini MG, De Felici M, Caiafa P. Poly(ADP-ribosyl)ation acts in the DNA demethylation of mouse primordial germ cells also with DNA damage-independent roles. PLoS One. 2012;7:e46927. doi: 10.1371/journal.pone.0046927. [DOI] [PMC free article] [PubMed] [Google Scholar]


