Abstract
Alterations in mitochondrial DNA (mtDNA) copy number have been widely identified in many types of human cancers and are considered a common cancer hallmark. However, the prognostic value of altered mtDNA content in gliomas remains largely unknown. The aim of this study was to investigate mtDNA copy number in a cohort of gliomas (n = 124) and non-neoplastic brain tissues (control subjects; n = 27) and to explore the association between variable mtDNA content and clinical outcomes in glioma patients. Using real-time quantitative PCR assay, we demonstrated that glioma patients had an increased mtDNA content as compared with control subjects. In addition, our data showed that increased mtDNA copy number was significantly negatively associated with tumor grade, recurrence and cancer-related death, whereas there was a significantly positively relationship between increased mtDNA content and seizures. More importantly, increased mtDNA content were closely relevant to longer survival in glioma patients. Taken together, our data provide the strong evidences that high copy number of mtDNA may be a useful good prognostic factor in glioma patients.
Keywords: Glioma, mitochondrial DNA (mtDNA), copy number, real-time quantitative PCR, clinical outcomes
Introduction
Glioma is the most frequent primary brain tumors in adults. The main types of glioma include astrocytoma, oligodendroglioma, mixed glioma, and ependymoma, which account for 70% of adult malignant primary brain tumors [1]. Despite important advances in the understanding and management of gliomas, these tumors are typically associated with a dismal prognosis and poor quality of life [2-4]. Thus, there is a critical need to develop reliable biomarkers that can predict clinical outcomes and establish new therapeutic and preventive strategies for this disease.
Although genetic, cellular and animal modeling studies have identified a number of molecules implicated in the initiation and progression of malignant gliomas over the previous 20 years, most molecules are concerned with the roles of nuclear DNA (nDNA) alterations [5-7]. However, relatively less attention has focused on mitochondrial DNA (mtDNA) alterations. Mitochondria are organelles found in all nucleated cells. The major role of mitochondria is to generate cellular adenosine triphosphate (ATP) through oxidative phosphorylation [8]. Human mtDNA is a 16,569 base-pair, double-stranded, closed-circular DNA molecule, which encodes 13 polypeptides, 2 rRNAs, and one set of 22 tRNAs required for protein synthesis in mitochondria [9]. The displacement loop (D-loop) is a noncoding region essential for the replication and transcription of mtDNA. Thus, mutations in the D-loop may cause a reduction in mtDNA copy number or altered mtDNA gene expression [10,11]. In general, each human cell contains several hundred to 1,000 mitochondria, and each mitochondrion has 2 to 10 copies of mtDNA. The mitochondrial genome is more vulnerable to oxidative damage and undergoes a higher rate of mutation compared with nDNA [12,13]. Increasing evidence has demonstrated an association between increased mtDNA content in peripheral blood and an increased risk of non-Hodgkin lymphoma, lung cancer, pancreatic cancer, breast cancer, and colorectal cancer [13-16]. In contrast, an increased risk of renal cancer has been associated with decreased mtDNA content [17]. There are no consistent results regarding the mtDNA copy number in gliomas. Several studies have reported amplification in the mtDNA copy number in gliomas as compared with normal brain controls [18-20], whereas another study identified a deletion of the mtDNA copy number [21]. To date, the association between mtDNA content and clinical outcomes in glioma patients remains largely unknown, and the prognostic implications are unclear.
In the present study, we investigated mtDNA copy number in a cohort of gliomas and non-neoplastic brain tissues using a real-time quantitative PCR approach. Moreover, we also explored the effect of mtDNA content on clinical outcomes of glioma patients.
Material and methods
Patients and tissue samples
With the approval of our institutional review board and human ethics committee, 124 paraffin-embedded glioma tissues were randomly obtained at the First Affiliated Hospital of Xi’an Jiaotong University School of Medicine. Twenty-seven tissues from patients with non-neoplastic brain tissues were used as control subjects. None of these patients had received chemotherapy or radiotherapy prior to surgery. All samples were histologically examined by a senior pathologist in the Department of Pathology of the Hospital based on the World Health Organization (WHO) criteria. The clinicopathological data were collected retrospectively and were summarized in Table 1.
Table 1.
Clinicopathological characteristics of the glioma patients
| Characteristics | No. of patients (%) |
|---|---|
| Gender | |
| Male | 68 (54.8) |
| Female | 56 (45.2) |
| Age, years | |
| Mean | 45.1 |
| SD | 16.5 |
| WHO grade | |
| I/II | 79 (63.7) |
| III/IV | 45 (36.3) |
| Pathological diagnosis | 16.5 |
| DA | 96 (77.4) |
| OL | 6 (4.8) |
| AOA | 1 (0.8) |
| GBM | 12 (9.7) |
| EP | 9 (7) |
| Recurrence | |
| No | 30 (24.2) |
| Yes | 94 (75.8) |
| Radiotherapy | |
| No | 48 (38.7) |
| Yes | 76 (61.3) |
| Chemotherapy | |
| No | 74 (59.7) |
| Yes | 50 (40.3) |
| Seizures | |
| No | 68 (54.8) |
| Yes | 56 (45.2) |
| KPS | |
| ≤ 80 | 76 (61.3) |
| > 80 | 48 (38.7) |
| Smoking history | |
| No | 89 (71.8) |
| Yes | 35 (28.2) |
| Survival status | |
| Dead | 45 (41.1) |
| Alive | 73 (58.9) |
Abbreviations: Diffuseastrocytoma (DA); Oligodendroglioma (OL); Anaplastic oligoastrocytoma (AOA); Glioblastoma (GBM); Ependymoma (EP); Karnofsky performance status (KPS).
DNA preparation
All tissues sections were reviewed by board-certified pathologists to ensure that ≥ 50% of the cells used for DNA purification were neoplastic. DNA was extracted from paraffin-embedded tissues as previously described [15]. Briefly, the tissues were first treated with xylene for 12 h at room temperature to remove the paraffin, and were then subjected to digestion with 1% sodium dodecylsulfate (SDS) and proteinase K at 48°C for 48 to 72 h with the addition of several spiking aliquots of concentrated proteinase K to facilitate digestion. Genomic DNA was subsequently isolated from the digested tissues followed by a standard phenol-chloroform extraction and ethanol precipitation protocol. The samples were then stored at -80°C until use.
mtDNA copy number analysis
The relative mtDNA copy number was measured in a cohot of gliomas and control subjects using real-time quantitative PCR. The specific primers and TaqMan probes were designed using Primer Express 3.0 (Applied Biosystems, Foster City, CA, USA) to amplify the MT-ND1 gene and the internal reference gene β-actin. The TaqMan probes were labeled with 5’-FAM (6-carboxyfluorescein, fluorescent reporter) and 3’-TAMRA (6-carboxy-tetramethylrhodamine, fluorescent quencher). The primer and probe sequences for MT-ND1 and β-actin genes are presented in Table 2. Using a PCR protocol previously described [22], the PCR amplification was conducted in a buffer that contained 16.6 mM ammonium sulfate, 67 mM Tris base, 2.5 mM MgCl2, 10 mM 2-mercaptoethanol, 0.1% DMSO, 0.2 mM each of dATP, dCTP, dGTP and dTTP, 600 nM each of forward and reverse primers, 200 nM TaqMan probe, 0.6 unit Platinum Taq polymerase and 2% Rox reference dye. Each sample was run in triplicate, and β-actin was run in parallel to standardize the input DNA. The standard curves were established using serial dilutions of normal brain tissue DNA with a quantity range of 6.25 to 100 ng per 2 μL. The relative mtDNA copy number for each sample was calculated as previously described [17].
Table 2.
The primer and TaqMan probe sequences used in this study
| Genes | Forward primer sequence (5’→3’) | Probe sequence (5’→3’) | Reverse primer sequence (5’→3’) |
|---|---|---|---|
| MT-ND1 | CCCCTAAAACCCGCCACATC | 6FAM-ACCCTCTACATCACCGCCCCGACC-TAMRA | GTAGAAGAGCGATGGTGAGAGC |
| β-actin | TCACCCACACTGTGCCCATCTACGA | 6FAM-ATGCCCTCCCCCATGCCATCC-TAMRA | TCGGTGAGGATCTTCATGAGGTA |
Statistical analysis
The Mann-Whitney U test was used to compare the mtDNA copy number between the gliomas and control subjects. The association between the mtDNA copy number and the clinicopathological characteristics was univariately assessed using the SPSS statistical package (version 16.0, Chicago, IL, USA). Multivariate models were subsequently developed that adjusted for the most important covariates, including gender, age, WHO grade, recurrence and Karnofsky performance status (KPS). The survival length was determined from the day of the primary tumor surgery to the day of death or last clinical follow-up. The Kaplan-Meier method was used for survival analysis grouping with the copy number variations of mtDNA. The differences between the curves were analyzed using the log-rank test. A multivariate Cox regression analysis was used to evaluate the effects of mtDNA copy number on the independent survival of gender, age, WHO grade and radiotherapy. All statistical analyses were performed using the SPSS statistical package (version 16.0, Chicago, IL). P values < 0.05 were considered significant.
Results
Relative mtDNA copy number in gliomas
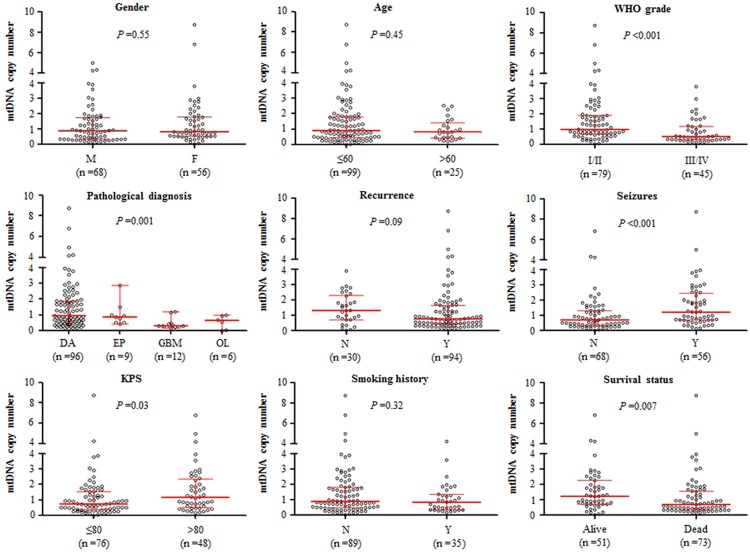
Real-time quantitative PCR assay was performed to analyze mtDNA copy number in 124 gliomas and 27 non-neoplastic brain tissues (control subjects). As shown in Figure 1, mtDNA copy number was higher in the glioma patients as compared with the control subjects with 0.86 copies (interquartile range = 0.45-1.76 copies) versus 0.28 copies (interquartile range = 0.14-0.71 copies), respectively, P < 0.001). This finding suggests the glioma patients had an amplication of mtDNA content as compared with the control subjects, which was consistent with previous studies [18,19]. We next evaluated whether mtDNA content differed by the selected clinicopathological characteristics. As shown in Figure 2, mtDNA copy number was significantly negatively associated with tumor grade (P < 0.001) and pathological diagnosis (P = 0.001). The patients with GBM, which is typically associated with poor outcomes, had a lower mtDNA content as compared to other glioma subtypes. Moreover, mtDNA copy number was significant higher in the patients with seizures than in the patients without seizures (P < 0.001). Also shown in Figure 2, mtDNA copy number was significantly negatively correlated with KPS (P = 0.03) and cancer-related death (P = 0.007). Notably, although no statistical significance was noted, recurrent cases appeared to have a lower mtDNA content as compared with non-recurrent cases (P = 0.09). We did not find significant associations of mtDNA copy number with gender, age and smoking history (Figure 2). Collectively, these observations suggest that high mtDNA content may predict good prognosis of glioma patients.
Figure 1.
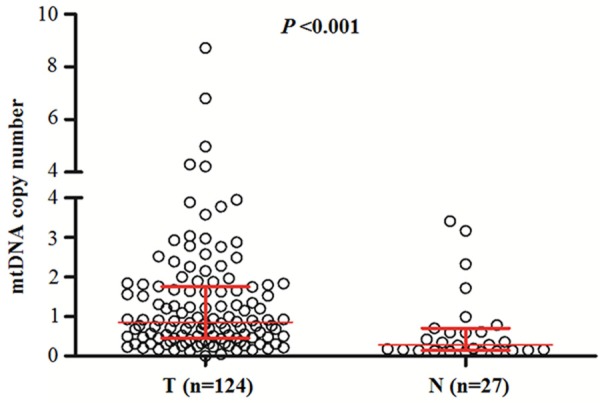
Copy number of mtDNA corresponding to each individual case of gliomas and non-neoplastic brain tissues (control subjects). A real-time quantitative PCR assay was performed to analyze mtDNA copy number in a cohort of gliomas and control subjects. The horizontal lines represent median ± interquartile range. T, tumor tissues; N, control subjects.
Figure 2.
Association of mtDNA copy number with clinicopathological characteristics in gliomas. Copy number of mtDNA was analyzed using real-time quantitative PCR approach. The circle represents mtDNA copy number of each glioma case. The horizontal lines represent median ± interquartile range. The sample medians were compared using the Mann-Whitney U test. M, male; F, female; DA, diffuseastrocytoma; EP, ependymoma; GBM, glioblastoma; OL, oligodendroglioma; N, No; Y, Yes; KPS, karnofsky performance status.
Associations between variable mtDNA contents and clinicopathological characteristics in glioma patients
Given frequently altered mtDNA content in gliomas, the associations between the mtDNA copy number and the clinicopathological characteristics were investigated in a cohort of clinically well-characterized gliomas. We chosed median mtDNA copy number (0.86 copies) in glioma patients as cutoff point. The glioma patients were subsequently categorized into two groups using cutoff point, including individuals with low (≤ 0.86 copies) (termed “low mtDNA content” hereafter) and high (> 0.86 copies) (termed “high mtDNA content” hereafter) category of mtDNA content. In univariate analyses, mtDNA content was increased in the patients with low tumor grades (I and II) as compared with the patients with high grades (III and IV) (OR = 0.34, 95% CI = 0.16-0.73; P = 0.006) (Table 3). Also shown in Table 3, increase mtDNA content was associated with a significantly decreased risk of tumor recurrence (OR = 0.40, 95% CI = 0.17-0.96; P = 0.04) and cancer-related death (OR = 0.36, 95% CI = 0.17-0.76; P = 0.007). Moreover, we found that there was a significantly positive association between mtDNA content and seizures (OR = 2.21, 95% CI = 1.07-4.54; P = 0.03) (Table 3). To assess the independent associations between mtDNA content and gender, age, tumor grade, recurrence, KPS and survival, we conducted a multivariable logistic regression. As shown in Table 4, similar to the univariate analysis, the mtDNA content remained negatively associated with tumor grade after the adjustment (OR = 0.42, 95% CI = 0.18-0.98; P = 0.04).
Table 3.
Copy number of mtDNA in gliomas: univariate associations with clinicopathological characteristics
| Characteristics | Copy number | P |
|---|---|---|
|
| ||
| OR* (95% CI) | ||
| Male vs. Female | 0.87 (0.55-1.38) | 0.55 |
| Age1 | 0.83 (0.67-1.04) | 0.11 |
| WHO grade2 | 0.34 (0.16-0.73) | 0.006 |
| Pathological diagnosis3 | 0.71 (0.47-1.07) | 0.10 |
| Recurrence | 0.40 (0.17-0.96) | 0.04 |
| Smoking history | 0.92 (0.42-2.02) | 0.84 |
| Seizures | 2.21 (1.07-4.54) | 0.03 |
| KPS4 | 1.73 (0.83-3.59) | 0.14 |
| Survival status5 | 0.36 (0.17-0.76) | 0.007 |
OR: odds ratio with 95% confidence interval (CI);
Age (per 10 years);
Tumor grade (I/II; III/IV);
Pathological diagnosis (DA; OL; AOA; GBM; EP);
KPS (≤ 80; > 80);
Survival status (Alive vs. Dead).
Table 4.
Copy number of mtDNA in gliomas: multivariable models that assessed gender, age, tumor grade, KPS, recurrence, KPS and survival status
| Characteristics | Copy number | P |
|---|---|---|
|
| ||
| OR* (95% CI) | ||
| Male vs. Female | 0.70 (0.32-1.54) | 0.38 |
| Age1 | 0.88 (0.69-1.11) | 0.27 |
| WHO grade2 | 0.42 (0.18-0.98) | 0.04 |
| Recurrence | 0.79 (0.24-2.66) | 0.71 |
| KPS3 | 1.65 (0.75-3.60) | 0.21 |
| Survival | 0.62 (0.21-1.79) | 0.37 |
OR: odds ratio with 95% confidence interval (CI);
Age (per 10 years);
WHO grade (I/II; III/IV);
KPS (≤ 80; > 80).
High mtDNA content predicts good survival in gliomas
The Kaplan-Meier estimator of the survivorship function was used to evaluate the impact of the mtDNA copy number on the survival of glioma patients. The survival times of glioma patients with variable mtDNA contents were compared using a log-rank test. As shown in Figure 3 (upper panel), the patients with high mtDNA content had significantly longer median survival times as compared with the patients with low mtDNA content (34.5 months vs. 13.0 months; P < 0.001). Similarly, univariate Cox regression analyses also showed that there was significant association of high mtDNA content with good survival in glioma patients (HR = 0.38, 95% CI = 0.23-0.62; P < 0.001) (Table 5). Moreover, Cox multivariate repression demonstrated that high mtDNA content (HR = 0.55, 95% CI = 0.33-0.93; P = 0.03) is a predictor of good survival for glioma patients as an independently variable with respect to gender, age, WHO grade and radiotherapy (Table 5).
Figure 3.
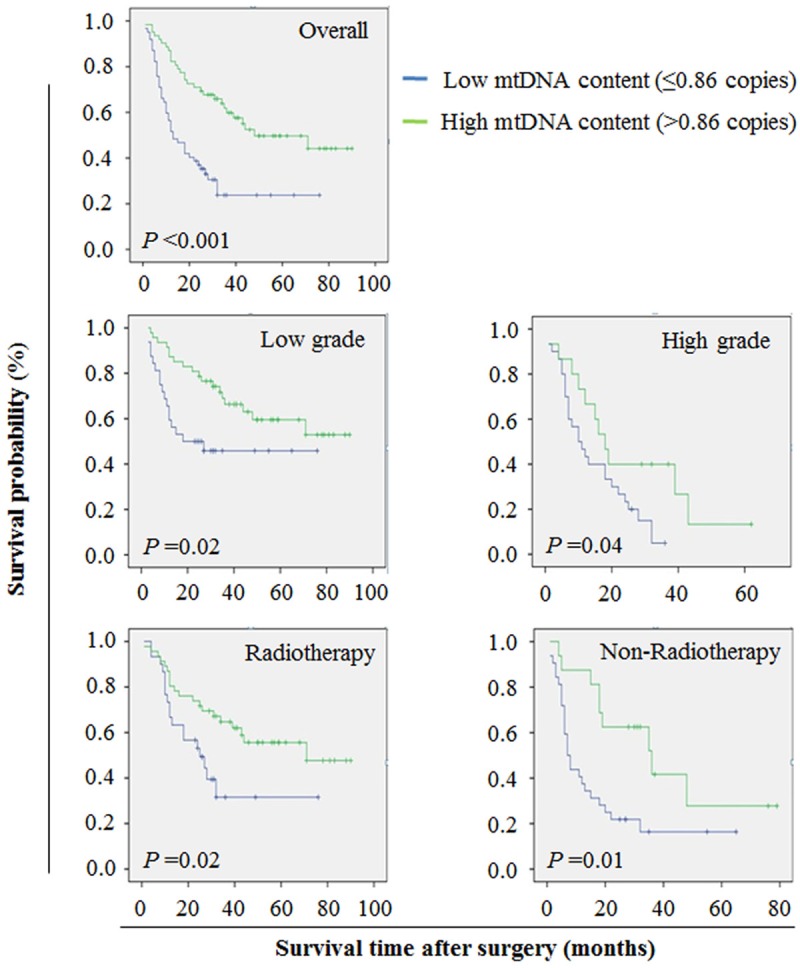
The effect of mtDNA content on the survival of glioma patients. The median mtDNA copy number (0.86 copies) of all glioma cases was set as cutoff point. Kaplan-Meier survival analysis was performed according to variable mtDNA content in a large cohort of gliomas. The results indicated that high mtDNA content was strongly associated with good survival in glioma patients (upper panel). When the data were further stratified based on tumor grade and the presence or absence of radiotherapy, increased mtDNA content still remained associated with good survival in gliomas (middle and lower panels).
Table 5.
Prognostic value of clinicopathological factors and copy number of mtDNA in univariate and multivariate Cox regression analyses (n = 124)
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
|
|
||||
| Variables | Hazard Ratio (95% CI) | P | Hazard Ratio (95% CI) | P |
| Copy number of mtDNA1 | 0.38 (0.23-0.62) | < 0.001 | 0.55 (0.33-0.93) | 0.03 |
| Gender | 0.81 (0.51-1.30) | 0.38 | 0.83 (0.55-1.34) | 0.45 |
| Age2 | 1.30 (1.11-1.51) | 0.001 | 1.22 (1.05-1.42) | 0.008 |
| WHO grade3 | 3.20 (1.99-5.15) | < 0.001 | 2.55 (1.54-4.24) | < 0.001 |
| Radiotherapy | 0.46 (0.29-0.73) | 0.001 | 0.54 (0.34-0.86) | 0.01 |
Copy number of mtDNA (≤ 0.86 vs. > 0.86);
Age (per 10 years);
Tumor grade (I/I; III/IV).
Given that tumor grade and radiotherapy were independent risk and protective factors for glioma patients, respectively, the data were stratified based on tumor grade and the presence or absence of radiotherapy. As shown in Figure 3 (middle panels), high mtDNA content was significantly associated with good survival whatever the patients who had low-grade or high-grade tumors. Similarly, there was significant association between high mtDNA content and longer survival times in the patients who received or did not receive the radiotherapy (Figure 3, lower panels). These findings further demonstrate a strong link between high mtDNA content and good prognosis in gliomas.
Discussion
Cancer arises from the accumulation of DNA and cytoplasmic abnormalities. Although many of the current findings are aligned with further understanding the functional details of the nuclear genome, the mitochondrion and its modest complement of DNA and protein is emerging as a crucial component of the biological networking of nuclear pathways [9]. There has been considerable evidences that have implicated mitochondria in many important physiological processes, including metabolism, signaling, apoptosis, cell cycle, differentiation and energy production [23]. Mitochondrial DNA has been demonstrated to be altered in cancers, which suggests mitochondria play vital roles in the tumorigenic phenotype [9]. Although mtDNA had been identified as structurally abnormal, no studies have reported the prognostic values of mtDNA copy number variations in gliomas.
In the present study, we investigated relative mtDNA copy number in a cohort of gliomas and non-neoplastic brain tissues (control subjects) using real-time quantitative PCR approach. Our data showed that the mtDNA content was increased in the glioma patients as compared with the control subjects. In line with these findings, previous studies have demonstrated mtDNA amplification in glioma patients [18,19], which implies an increased mtDNA content may be involved in glioma tumorigenesis. It is well known that the mtDNA in the oncocyte has been demonstrated to be functionally abnormal, and the aberrant proliferation is most likely because of a compensatory effect triggered by a shortage of energy [24,25]. Moreover, mtDNA amplification has suggested the possibility of mtDNA escape in cancer. In yeast, mtDNA escape occurs as a manifestation of communication between the nuclear and mitochondrial genomes [26,27]. It has been hypothesized that high mtDNA content may be required to transfer mitochondrial sequences from mitochondrial sources to the nucleus in cancer, which is similar to observations in yeast [18,28]. When evaluated by the interphase fluorescence in situ hybridization (FISH) technique using fluorescent probes of the entire mitochondrial genome, at least 69% of the nuclei exhibited mitochondrial sequence hybridization in the specimens investigated compared with two normal brain samples in which < 8% of the nuclei exhibited hybridization with the mtDNA fluorescent probes [19]. In addition, mtDNA sequences have been demonstrated to interrupt intronic regions of MADH2, a suppressor gene in colorectal cancer [29], as supported by two previous studies that promiscuous mtDNA can be frequently found in both intron and exon regions of the nuclear genome [30,31]. Taken together, it is reasonable to hypothesize that increased mtDNA content may represent an early ontological aspect of the pathway to malignant behavior [32-34].
Notably, we found the associations of mtDNA content with some of clinicopathological features in the present study, such as WHO grade, pathological diagnosis, recurrence, seizures, KPS and survival status. To further explore the relationships between the mtDNA content and the clinicalpathological characteristics and prognosis of the glioma patients, we categorized the gliomas into two groups based on one cutoff point (the median mtDNA content in the glioma patients), including low mtDNA content (≤ median) and high mtDNA content (> median). We determined that increased mtDNA copy number was negatively associated with WHO grade, recurrence and survial status. In addition, the multivariate analysis showed that mtDNA content exhibited a significantly negative association with WHO grade, implicating that it may represent an early molecular event and may play a key role in the initiation of glioma. These observations suggest that variable mtDNA content may contribute to clinical outcomes in glioma patients.
Given that mtDNA amplication plays a critical role in glioma tumorigenesis, we further explored the association between the mtDNA content and the survival of glioma patients. We surprisingly found that mtDNA copy number was significantly negative with survival times of glioma patients. The patients with high mtDNA content had significantly longer survival times than the patients with low mtDNA content. Even when the data were further stratified according to tumor grade and the presence or absence of radiotherapy, increased mtDNA content remained significantly associated with a longer survival in gliomas. This is supported by a previous study that high mtDNA content was associated with longer survial times in 10 GBM cases [20]. Collectively, these findings suggest variable mtDNA content should be a strong prognostic signature.
In summary, we investigated the relative mtDNA content in a large cohort of gliomas and demonstrated that mitochondria play a significant role in the tumorigenic phenotype. The mtDNA content was strongly associated with WHO grade, recurrence, seizures and cancer-related death. Importantly, increased mtDNA content is an independent prognostic factor in gliomas, which may serve as a highly valuable biomarker in the evaluation of clinical outcomes in glioma patients.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81171969, 81272933 and 81372217), and the Fundamental Research Funds for the Central Universities.
Disclosure of conflict of interest
The authors declare that they have no competing interests.
References
- 1.Ricard D, Idbaih A, Ducray F, Lahutte M, Hoang-Xuan K, Delattre JY. Primary brain tumours in adults. Lancet. 2012;379:1984–96. doi: 10.1016/S0140-6736(11)61346-9. [DOI] [PubMed] [Google Scholar]
- 2.Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310:1842–50. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- 3.Krex D, Klink B, Hartmann C, von Deimling A, Pietsch T, Simon M, Sabel M, Steinbach JP, Heese O, Reifenberger G, Weller M, Schackert G. Long-term survival with glioblastoma multiforme. Brain. 2007;130:2596–606. doi: 10.1093/brain/awm204. [DOI] [PubMed] [Google Scholar]
- 4.Lim SK, Llaguno SR, McKay RM, Parada LF. Glioblastoma multiforme: a perspective on recent findings in human cancer and mouse models. BMB Rep. 2011;44:158–64. doi: 10.5483/BMBRep.2011.44.3.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanson M, Marie Y, Paris S, Idbaih A, Laffaire J, Ducray F, El Hallani S, Boisselier B, Mokhtari K, Hoang-Xuan K, Delattre JY. Isocitrate dehydrogenase 1 codon 132 mutation is an important prognostic biomarker in gliomas. J. Clin. Oncol. 2009;27:4150–4. doi: 10.1200/JCO.2009.21.9832. [DOI] [PubMed] [Google Scholar]
- 6.Cohen AL, Holmen SL, Colman H. IDH1 and IDH2 mutations in gliomas. Curr Neurol Neurosci Rep. 2013;13:345. doi: 10.1007/s11910-013-0345-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bettegowda C, Agrawal N, Jiao Y, Sausen M, Wood LD, Hruban RH, Rodriguez FJ, Cahill DP, McLendon R, Riggins G, Velculescu VE, Oba-Shinjo SM, Marie SK, Vogelstein B, Bigner D, Yan H, Papadopoulos N, Kinzler KW. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science. 2011;333:1453–5. doi: 10.1126/science.1210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hatefi Y. The mitochondrial electron transport and oxidative phosphorylation system. Annu Rev Biochem. 1985;54:1015–69. doi: 10.1146/annurev.bi.54.070185.005055. [DOI] [PubMed] [Google Scholar]
- 9.Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12:685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shadel GS. Expression and maintenance of mitochondrial DNA: new insights into human disease pathology. Am J Pathol. 2008;172:1445–56. doi: 10.2353/ajpath.2008.071163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chinnery PF, Hudson G. Mitochondrial genetics. Br Med Bull. 2013;106:135–59. doi: 10.1093/bmb/ldt017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Copeland WC, Wachsman JT, Johnson FM, Penta JS. Mitochondrial DNA alterations in cancer. Cancer Invest. 2002;20:557–69. doi: 10.1081/cnv-120002155. [DOI] [PubMed] [Google Scholar]
- 13.Lan Q, Lim U, Liu CS, Weinstein SJ, Chanock S, Bonner MR, Virtamo J, Albanes D, Rothman N. A prospective study of mitochondrial DNA copy number and risk of non-Hodgkin lymphoma. Blood. 2008;112:4247–9. doi: 10.1182/blood-2008-05-157974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen J, Platek M, Mahasneh A, Ambrosone CB, Zhao H. Mitochondrial copy number and risk of breast cancer: a pilot study. Mitochondrion. 2010;10:62–8. doi: 10.1016/j.mito.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hosgood HD 3rd, Liu CS, Rothman N, Weinstein SJ, Bonner MR, Shen M, Lim U, Virtamo J, Cheng WL, Albanes D, Lan Q. Mitochondrial DNA copy number and lung cancer risk in a prospective cohort study. Carcinogenesis. 2010;31:847–9. doi: 10.1093/carcin/bgq045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lynch SM, Weinstein SJ, Virtamo J, Lan Q, Liu CS, Cheng WL, Rothman N, Albanes D, Stolzenberg-Solomon RZ. Mitochondrial DNA copy number and pancreatic cancer in the alpha-tocopherol beta-carotene cancer prevention study. Cancer Prev Res (Phila) 2011;4:1912–9. doi: 10.1158/1940-6207.CAPR-11-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xing J, Chen M, Wood CG, Lin J, Spitz MR, Ma J, Amos CI, Shields PG, Benowitz NL, Gu J, de Andrade M, Swan GE, Wu X. Mitochondrial DNA content: its genetic heritability and association with renal cell carcinoma. J Natl Cancer Inst. 2008;100:1104–12. doi: 10.1093/jnci/djn213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang BC. Evidence for association of mitochondrial DNA sequence amplification and nuclear localization in human low-grade gliomas. Mutat Res. 1996;354:27–33. doi: 10.1016/0027-5107(96)00004-8. [DOI] [PubMed] [Google Scholar]
- 19.Liang BC, Hays L. Mitochondrial DNA copy number changes in human gliomas. Cancer Lett. 1996;105:167–73. doi: 10.1016/0304-3835(96)04276-0. [DOI] [PubMed] [Google Scholar]
- 20.Marucci G, Maresca A, Caporali L, Farnedi A, Betts CM, Morandi L, de Biase D, Cerasoli S, Foschini MP, Bonora E, Vidone M, Romeo G, Perli E, Giordano C, d’Amati G, Gasparre G, Baruzzi A, Carelli V, Eusebi V. Oncocytic glioblastoma: a glioblastoma showing oncocytic changes and increased mitochondrial DNA copy number. Hum Pathol. 2013;44:1867–76. doi: 10.1016/j.humpath.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 21.Correia RL, Oba-Shinjo SM, Uno M, Huang N, Marie SK. Mitochondrial DNA depletion and its correlation with TFAM, TFB1M, TFB2M and POLG in human diffusely infiltrating astrocytomas. Mitochondrion. 2011;11:48–53. doi: 10.1016/j.mito.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 22.Hou P, Liu D, Shan Y, Hu S, Studeman K, Condouris S, Wang Y, Trink A, El-Naggar AK, Tallini G, Vasko V, Xing M. Genetic alterations and their relationship in the phosphatidylinositol 3-kinase/Akt pathway in thyroid cancer. Clin Cancer Res. 2007;13:1161–70. doi: 10.1158/1078-0432.CCR-06-1125. [DOI] [PubMed] [Google Scholar]
- 23.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gasparre G, Romeo G, Rugolo M, Porcelli AM. Learning from oncocytic tumors: Why choose inefficient mitochondria? Biochim Biophys Acta. 2011;1807:633–42. doi: 10.1016/j.bbabio.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 25.Iommarini L, Calvaruso MA, Kurelac I, Gasparre G, Porcelli AM. Complex I impairment in mitochondrial diseases and cancer: parallel roads leading to different outcomes. Int J Biochem Cell Biol. 2013;45:47–63. doi: 10.1016/j.biocel.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 26.Campbell CL, Thorsness PE. Escape of mitochondrial DNA to the nucleus in yme1 yeast is mediated by vacuolar-dependent turnover of abnormal mitochondrial compartments. J Cell Sci. 1998;111:2455–64. doi: 10.1242/jcs.111.16.2455. [DOI] [PubMed] [Google Scholar]
- 27.Thorsness PE, Weber ER. Escape and migration of nucleic acids between chloroplasts, mitochondria, and the nucleus. Int Rev Cytol. 1996;165:207–34. doi: 10.1016/s0074-7696(08)62223-8. [DOI] [PubMed] [Google Scholar]
- 28.Netter P, Robineau S. The differential overamplification of short sequences in the mitochondrial DNA of rho- petites in Saccharomyces cerevisiae stimulates recombination. Gene. 1989;83:25–38. doi: 10.1016/0378-1119(89)90400-9. [DOI] [PubMed] [Google Scholar]
- 29.Eppert K, Scherer SW, Ozcelik H, Pirone R, Hoodless P, Kim H, Tsui LC, Bapat B, Gallinger S, Andrulis IL, Thomsen GH, Wrana JL, Attisano L. MADR2 maps to 18q21 and encodes a TGFbeta-regulated MAD-related protein that is functionally mutated in colorectal carcinoma. Cell. 1996;86:543–52. doi: 10.1016/s0092-8674(00)80128-2. [DOI] [PubMed] [Google Scholar]
- 30.Borensztajn K, Chafa O, Alhenc-Gelas M, Salha S, Reghis A, Fischer AM, Tapon-Bretaudiere J. Characterization of two novel splice site mutations in human factor VII gene causing severe plasma factor VII deficiency and bleeding diathesis. Br J Haematol. 2002;117:168–71. doi: 10.1046/j.1365-2141.2002.03397.x. [DOI] [PubMed] [Google Scholar]
- 31.Turner C, Killoran C, Thomas NS, Rosenberg M, Chuzhanova NA, Johnston J, Kemel Y, Cooper DN, Biesecker LG. Human genetic disease caused by de novo mitochondrial-nuclear DNA transfer. Hum Genet. 2003;112:303–9. doi: 10.1007/s00439-002-0892-2. [DOI] [PubMed] [Google Scholar]
- 32.Jakupciak JP, Wang W, Markowitz ME, Ally D, Coble M, Srivastava S, Maitra A, Barker PE, Sidransky D, O’Connell CD. Mitochondrial DNA as a cancer biomarker. J Mol Diagn. 2005;7:258–67. doi: 10.1016/S1525-1578(10)60553-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ivanova R, Lepage V, Loste MN, Schachter F, Wijnen E, Busson M, Cayuela JM, Sigaux F, Charron D. Mitochondrial DNA sequence variation in human leukemic cells. Int J Cancer. 1998;76:495–8. doi: 10.1002/(sici)1097-0215(19980518)76:4<495::aid-ijc9>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 34.Shay JW, Werbin H. New evidence for the insertion of mitochondrial DNA into the human genome: significance for cancer and aging. Mutat Res. 1992;275:227–35. doi: 10.1016/0921-8734(92)90026-l. [DOI] [PubMed] [Google Scholar]