Highlight
Ubiquitin E3 ligase PUB17 functions in the nucleus to regulate transcriptional responses positively in PAMP-triggered immunity and programmed cell death following perception of specific elicitors at the plant cell surface.
Key words: Disease resistance, effector-triggered immunity, hypersensitive response, oomycete, plant defence, ubiquitination.
Abstract
Ubiquitination regulates many processes in plants, including immunity. The E3 ubiquitin ligase PUB17 is a positive regulator of programmed cell death (PCD) triggered by resistance proteins CF4/9 in tomato. Its role in immunity to the potato late blight pathogen, Phytophthora infestans, was investigated here. Silencing StPUB17 in potato by RNAi and NbPUB17 in Nicotiana benthamiana by virus-induced gene silencing (VIGS) each enhanced P. infestans leaf colonization. PAMP-triggered immunity (PTI) transcriptional responses activated by flg22, and CF4/Avr4-mediated PCD were attenuated by silencing PUB17. However, silencing PUB17 did not compromise PCD triggered by P. infestans PAMP INF1, or co-expression of R3a/AVR3a, demonstrating that not all PTI- and PCD-associated responses require PUB17. PUB17 localizes to the plant nucleus and especially in the nucleolus. Transient over-expression of a dominant-negative StPUB17V314I,V316I mutant, which retained nucleolar localization, suppressed CF4-mediated cell death and enhanced P. infestans colonization. Exclusion of the StPUB17V314I,V316I mutant from the nucleus abolished its dominant-negative activity, demonstrating that StPUB17 functions in the nucleus. PUB17 is a positive regulator of immunity to late blight that acts in the nucleus to promote specific PTI and PCD pathways.
Introduction
Plants are constantly exposed to pathogenic microorganisms such as bacteria, viruses, fungi, and oomycetes. To detect these microbes and to defend against infection, plants have an inducible innate immune system that minimizes the impact of pathogens on plant growth and development (Jones and Dangl, 2006). The first inducible layer of the plant immune system involves detection of conserved, secreted or surface-exposed molecules, called pathogen/microbe-associated molecular patterns (P/MAMPs), leading to the production of reactive oxygen species, activation of mitogen-activated protein kinases (MAPK), deposition of callose in the cell wall, and synthesis of pathogenesis-related (PR) proteins. This is collectively termed pattern-triggered immunity (PTI) (Jones and Dangl, 2006; Dodds and Rathjen, 2010; Thomma et al., 2011). Pathogens secrete effectors that suppress or otherwise manipulate PTI (Block and Alfano, 2011). In turn, effectors can be specifically recognized, directly or indirectly, by nucleotide-binding, leucine-rich repeat (NB-LRR) resistance (R) proteins that trigger a further layer of defence, effector-triggered immunity (ETI) (Jones and Dangl, 2006; Boller and He, 2009). ETI is an accelerated and amplified PTI response, resulting in disease resistance and usually accompanied by a rapid, localized programmed cell death (PCD) known as the hypersensitive response (HR) (Jones and Dangl, 2006).
The disease, late blight, caused by the oomycete pathogen Phytophthora infestans, is a significant threat to potato production worldwide (Birch et al., 2012). P. infestans secretes and delivers pathogenicity proteins into the plant cell cytosol, including potentially hundreds of effectors containing the conserved amino acid motif RXLR (Whisson et al., 2007; Birch et al., 2008; Haas et al., 2009; Cooke et al., 2012). Several RXLR effectors have been shown to be avirulence (AVR) proteins recognized by host cytoplasmic NB-LRR R proteins (Hein et al., 2009; Vleeshouwers et al., 2011). Recent activity has focused on the roles of these RXLR effectors in modifying host processes to the benefit of the pathogen. Amongst them, the effector AVR3a, which is recognized by potato R3a (Armstrong et al., 2005), is able to suppress PCD triggered by the P. infestans PAMP INF1 (Bos et al., 2006), and a range of other pathogen elicitors (Gilroy et al., 2011), by modifying the activity of the host ubiquitin E3 ligase CMPG1 (Bos et al., 2010). This establishes ubiquitination as a process targeted by P. infestans to promote the colonization of its hosts (Birch et al., 2009).
During pathogen infection, plant defence responses include remodelling the proteome by protein synthesis (transcription and translation), and post-translational modifications (PTMs) (Stone and Callis, 2007; Kwon et al., 2006). Ubiquitination is a PTM that plays a crucial role during the regulation of plant immune signalling (Lu et al., 2011; Hofmann, 2012; Sadanandom et al., 2012; Stegmann et al., 2012; Duplan and Rivas, 2014). It is initiated by activation of the ubiquitin-activating enzyme (E1) in an ATP-dependent manner, which is then transferred to a ubiquitin-conjugating enzyme (E2). E2 intermediates interact with E3 ligases to catalyse ubiquitin transfer to specific target proteins and/or to auto-ubiquitinate the E3 itself (Yee and Goring, 2009). In this pathway, E3 ligases play a central role in selecting target proteins for ubiquitination and have been shown to be involved in all steps of plant immunity (Dreher and Callis, 2007; Shirsekar et al., 2010; Trujillo and Shirasu, 2010). E3 ubiquitin ligases are classified into four main subfamilies depending on their subunit composition and mechanism of action: HECT, RING, U-box, and cullin-RING ligases (CRLs) (Vierstra, 2009).
A number of PUB (Plant U-Box) E3 ligases are documented as positive or negative regulators of plant immunity (Marino et al., 2012). The E3 ligase SPL11 negatively regulates PAMP-triggered signalling and PTI (Zeng et al., 2004; Li et al., 2012; Liu et al., 2012). In addition, PUB12 and PUB13 are recruited to the FLS2 receptor complex in a BAK1-dependent manner to target FLS2 for degradation (Lu et al., 2011; O’Neill, 2011). Moreover, PUB22 negatively regulates PTI by targeting a subunit of the exocyst complex, Exo70B2, for proteasomal degradation (Trujillo et al., 2008; Stegmann et al., 2012).
In contrast to the negative regulators above, the U-box E3 ubiquitin ligases CMPG1/AtPUB20 (González-Lamothe et al., 2006) and Arabidopsis AtPUB17 (Yang et al., 2006), and its functional orthologue in tobacco, NtACRE276 (hereafter referred to as NtPUB17), were demonstrated to act as positive regulators of plant disease resistance. They are required for the HR on perception of the C. fulvum Avr9 and AVR4 peptides by the Cf9 and Cf4 receptor-like proteins, respectively, in tomato (Rowland et al., 2005; González-Lamothe et al., 2006; Yang et al., 2006). NtPUB17 and AtPUB17 are functional orthologues as transient expression of AtPUB17 in NtPUB17-silenced lines of Cf-9 tobacco plants can restore the AVR9-triggered HR. Moreover, AtPUB17 is also required for HR mediated by the NB-LRR resistance proteins RPM1 and RPS4 (Yang et al., 2006). Potato PUB17 (StPUB17) was first identified by suppression subtractive hybridization analysis of transcriptional changes after P. infestans inoculation and it was found to be rapidly up-regulated during infection (Tian et al., 2003). Preliminary work showed that StPUB17-RNAi potato plants exhibited enhanced susceptibility to P. infestans (Ni et al., 2010), suggesting it potentially plays a role in basal immunity to P. infestans. Currently, it is unknown: (i) what aspects of plant immunity of potential relevance to the host-P. infestans interaction are positively regulated by PUB17; or (ii) where within the plant cell this E3 ligase acts to promote immune responses.
A deeper investigation is conducted here into the role of StPUB17 in immunity against P. infestans and the role of PUB17 from solanaceous plants in acting as a positive regulator of PCD. Given that StPUB17 is potentially involved in basal immunity to P. infestans (Ni et al., 2010), a further hypothesis to be tested was that it contributes to other aspects of pattern-triggered immunity. To that end, it is shown that stable silencing by RNAi of StPUB17 in potato leads to enhanced P. infestans colonization, confirming a negative impact of StPUB17 on late blight disease development. A relative of potato in the Solanaceae, Nicotiana benthamiana, acts as a host for P. infestans and thus an alternative system for studying P. infestans-host interactions (Bos et al., 2010; Saunders et al., 2012; McLellan et al., 2013; King et al., 2014). N. benthamiana is a model plant for virus-induced gene silencing, cell biology, and transient immune-associated assays, allowing more detailed functional studies to be performed than are feasible in potato. Silencing NbPUB17 in N. benthamiana using virus-induced gene silencing (VIGS) also led to enhanced P. infestans colonization and attenuated early PTI-associated transcriptional responses, indicating that it contributes to more than just cell death pathways. Whereas VIGS of NbPUB17, or RNAi silencing of NtPUB17 in tobacco, each compromised CF4/Avr4 cell death, there was no attenuation of cell death mediated by the P. infestans PAMP INF1, showing that not all cell-surface perception events leading to PCD require PUB17. In addition, silencing PUB17 in N. benthamiana, tobacco or potato had no impact on HR triggered by potato R3a-mediated recognition of AVR3a from P. infestans. Thus not all R gene–mediated PCD requires PUB17. These results implicate PUB17 involvement in the nucleus as a positive regulator of specific defence pathways activated by P. infestans infection.
Materials and methods
Plant materials and microbe strains
In vitro potato plantlets were propagated in sterile culture boxes containing MS medium supplemented with 4% sucrose and 0.7% agar and raised in a climate room under controlled conditions (16/8h light/dark cycle at 20 °). Three-week-old plantlets were transplanted and grown in individual pots in a greenhouse at 20–26 ° with humidity above 80%. Nicotiana benthamiana, Cf9, and ACRE276-RNAi tobacco lines were grown as described in Bos et al. (2010) and Yang et al. (2006). All Agrobacterium tumefaciens cultures were grown at 28 ℃ at 200rpm for 24–48h and spun at 4000 g. The pellet was re-suspended in sterile 10mM MES and 10mM MgCl2 buffer with 200 µM acetosyringone. The following bacterial optical densities at 600nm (OD600) were used for each assay: 0.1–0.01 for confocal imaging, 0.5 for Western Blot analyses and HR assays, and 0.1 for P. infestans virulence assays.
Isolation and sequence analysis of StPUB17 and NbPUB17 genes
A full-length StPUB17 gene was cloned with gene-specific primers from potato cDNA [synthesized from the RNA of E-potato 3 (E3) leaves inoculated with P. infestans for 36 h]. Primer sequences are shown in Supplementary Table S1 at JXB online. The single PCR products were gel purified, then digested with EcoRI and NotI, ligated into the EcoRI–NotI-digested pMD18-T vector (Takara, Dalian, China), and transformed into E. coli strain DH5a for sequencing, resulting in pMD18-T-StPUB17. Full-length NbPUB17 was cloned from N. benthamiana cDNA with gene-specific primers and then ligated into the pMD18-T vector, resulting in pMD18-T-NbPUB17. Sequence analysis was performed by BLAST (http://blast.ncbi.nlm.nih.gov/). Information on all genes, constructs, and their purpose is included in Supplementary Table S2 at JXB online.
Mutagenesis
The StPUB17V314I,V316I mutant was generated according to the manufacturer’s protocol QuickChange® Site-Directed Mutagenesis Kit (Stratagene) using pDonr201-StPUB17 as a template. The primer sequences used for mutation are shown in Supplementary Table S1 at JXB online. Two conserved amino acids of StPUB17, valines at positions 314 and 316, were substituted with isoleucine, resulting in the mutant StPUB17V314I,V316I. The mutant StPUB17 was recombined, using LR clonase, into pB7WGF2 or NES-pB7WGF2 for in planta assays.
Cloning of protein fusions
A full-length StPUB17 gene was cloned from a pMD18-T-StPUB17 plasmid with gene-specific primers modified to contain the Gateway® (Invitrogen) attB recombination sites. Primer sequences are shown in the Supplementary Table S1 at JXB online. PCR products were purified and recombined into pDONR201 (Invitrogen) to generate entry clones via BP reactions using Gateway® technology (Invitrogen). N terminal GFP fusions of StPUB17, StPUB17V314I,V316I, and NtPUB17 were made by recombining the entry clones with pB7WGF2 using using LR clonase® (Invitrogen). The NES-GFP-StPUB17V314I,V316I was made by recombining the entry clones with NES-pB7WGF2 (the NES signal sequence was inserted with annealed oligonucleotides into the unique SpeI site at the beginning of the GFP in the pB7WGF2 vector). They were then transformed into the Agrobacterium tumefaciens strain AGL1 virG for in planta assays.
Recombinant protein purification
The fragment of StPUB17 was cloned into the pET28a vector (N terminal HIS-tag), then transformed into the E. coli strain BL21. The culture was induced by 0.2mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 37 ℃ for about 3h according to the method described by Yang et al. (2006). Cells were harvested by centrifugation at 10 000rpm for 20min, and then re-suspended in 8ml of Lysis/Equilibration Buffer (50mM sodium phosphate, 300mM sodium chloride, 5mM imidazole, pH 8 with 0.1% Triton X-100). Then it was sonicated and centrifuged at 10 000rpm for 20–30min to remove genomic DNA, insoluble proteins, and cell debris. The supernatants were incubated and purified according to the protocol of PureProteome Nickel Magnetic Beads (Millipore Corporation). The fused HIS-StPUB17 proteins were eluted with Elution Buffer (50mM sodium phosphate, 300mM sodium, chloride, 300mM imidazole, pH 8) and used for the ubiquitination assays.
E3 ubiquitin ligase activity assay
The in vitro ubiquitination assays were performed according to the Auto-ubiquitinylation Kit (Instruction Manual BML-UW0970; Enzo Life Sciences). Each reaction (50 μl final volume) contained 2.5 μl 20× E1, 2.5 μl 20× E 2, 5 μl 10× Ub E3 ligase buffer, 5 μl 10× ubiquitin, 1 μl 50mM DTT, 2.5 μl Mg-ATP, and 2.5 μl of eluted/bead-bound HIS: StPUB17 proteins or 2.5 μl 20× E3 control (6 μM). Hdm2 RING domain (KW0200) is provided as a positive control of ubiquitin E3 ligase. The reactions were incubated at 37 ℃ for 1h. Quench assays were performed by addition of 50 μl 2× SDS-PAGE gel loading buffer (0.25M TRIS-Cl, pH 6.8, 4% SDS, 10% glycerol, 2% mercaptoethanol, 0.01% bromophenol blue), heated to 95 ℃ for 5min, and analysed by SDS-PAGE electrophoresis. Following blotting, hybridization was performed using ubiquitin antibody (the kit supplied) and Goat Anti-Rabbit IgG-peroxidase antibody (HRP-linked) (Sigma, A0545).
Western analysis
Leaf discs expressing GFP-StPUB17, GFP-StPUB17V314I,V316I, NES-GFP-StPUB17V314I,V316I, and GFP-NtPUB17 were harvested at 2 dpi, ground in liquid nitrogen, suspended in 200 μl 2× SDS-PAGE loading buffer, loaded onto a 4–12% TRIS NuPAGE Novex gel, and electrophoresed with 1× MOPS SDS running buffer for 1h at 80V (Invitrogen). All further steps for Western analyses are as described previously by McLellan et al. (2013).
Confocal microscopy
A. tumefaciens containing GFP-StPUB17 was pressure infiltrated into leaves of 4-week-old wild-type N. benthamiana plants and the transgenic N. benthamiana line CB157 (with an mRFP fusion to histone H2B), separately. A. tumefaciens containing GFP-StPUB17V314I,V316I, NES-GFP-StPUB17V314I,V316I, and NtPUB17 were pressure infiltrated into leaves of 4-week-old normal N. benthamiana plants. Cells expressing fluorescent protein fusions were observed using a Leica TCS-SP2 AOBS confocal microscope no more than 2 d post-infiltration, and use a low OD600 (start with 0.01) as described in McLellan et al. (2013).
TRV-based VIGS in N. benthamiana
Virus-induced gene silencing (VIGS) constructs were made by cloning 348bp and 222bp PCR fragments from NbPUB17 into pBinary Tobacco Rattle Virus (TRV) vectors (Ratcliff et al., 2001). Primer sequences are shown in Supplementary Table S1 at JXB online. A TRV construct expressing GFP, described previously, was used as a control (Gilroy et al., 2007). A. tumefaciens strains containing a mixture of RNA1 and each NbPUB17 VIGS construct at OD600=0.5 were pressure infiltrated into the two largest leaves of 4-leaf-stage N. benthamiana plants. Systemic leaves were detached, analysed by qRT-PCR, and used for P. infestans colonization and HR assays 3 weeks later.
Agrobacterium-mediated transient expression
Agrobacterium strains (expressing INF1, R3a/AVR3a, or Cf4/Avr4, Cf4/Avr4/GFP-pB7WGF2, Cf4/Avr4/GFP-StPUB17V314I,V316I Cf4/Avr4/NES-GFP- StPUB17V314I,V316I) were infiltrated into leaves of N. benthamiana wild-type or VIGS plants. Transient expression by agroinfiltration was performed as described previously (Bos et al., 2006; Gilroy et al., 2011). The number of positive HRs (i.e. more than 50% of the inoculated region produces clear cell death) were counted as described previously (Gilroy et al., 2011) and expressed as the mean percentage of total inoculations per plant. The error bars represent ±standard errors (SE) of combined data from at least three biological replicates. One-way ANOVA was performed to determine statistically significant differences.
Bacterial pathogen strains and pathology tests
Pathogen assays were performed as in Vleeshouwers et al. (1999). P. infestans Ljx18 (race 3.4.7.10.11) and HB09-14-2 race (race 1.2.3.4.5.6.7.8.9.10.11, collected from Hubei Province, China) were used for potato infection after cultured on Rye Agar at 19 ℃ for 2 weeks.
For N. benthamiana plants and leaves, P. infestans strain 88069-tdT was inoculated at a concentration of 4×104 sporangia ml–1, as previously described by McLellan et al. (2013). Sporangia counts with a haemocytometer were performed on 10 dpi leaves from VIGS plants which had been washed with in ddH2O to release sporangia and were expressed as sporangia ml–1. All the data were analysed by ANOVA.
Quantitative RT-PCRs
Total RNA from the leaves of potato transgenic lines, N. benthamiana VIGS plants, and plants that were treated with flg22 for 3h were extracted with TRIzol reagent according to the manufacturer’s recommendations. The cDNA was synthesized and the qRT-PCR conditions are as described previously (McLellan et al. 2013). Primers for Real-time PCR are in Supplementary Table S1 at JXB online. Gene expression levels were calculated by a comparative Ct method as described by Cikos et al. (2007).
Trypan blue staining
At least three leaves of each line of infected leaves at 5 dpi were collected and stained with trypan blue (0.25mg ml–1) solution in a heated water bath, boiling for 1min. Leaves were kept in the staining solution overnight and then de-stained with alcoholic lactophenol solution (95% ethanol:lactophenol=2:1 v/v; lactophenol,phenol:glycerol:lactic acid:water=1:1:1:1 by vol.). Leaves were cleared with ethanol (50%) and stored in glycerol (50%). The representative phenotypes were photographed.
Results
StPUB17 functions as an E3 ubiquitin ligase
A transcript (GenBank: EF091878) shown previously to accumulate during P. infestans colonization of potato by 2 d post-inoculation (dpi) (Ni et al., 2010) encodes a 724 amino acid protein of predicted size 79kDa which is a reciprocal best blast hit (RBBH) of AtPUB17 in Arabidopsis and ACRE276 (NtPUB17) in tobacco. Alignment of the corresponding StPUB17 protein with AtPUB17 and NtPUB17 reveals a shared U-box domain (amino acids 300–363 in StPUB17), and three ARM repeat domains (amino acids 430–470, 515–555, 556–594 in StPUB17) in the C-terminal halves of the proteins (see Supplementary Fig. S1 at JXB online). To test whether StPUB17 possesses E3 ubiquitin ligase activity, full-length StPUB17 was expressed in E. coli (BL21) as a HIS-tagged fusion protein and purified by affinity chromatography (see Supplementary Fig. S2A at JXB online). In the presence of E1, E2, ubiquitin, and ATP, ubiquitination activity was observed in immunoblots probed with monoclonal antibodies of ubiquitin (see Supplementary Fig. SB at JXB online). No ubiquitination was detected in the absence of E1, E2, ubiquitin, or StPUB17, indicating that the latter possesses E3 ligase activity (see Supplementary Fig. S2B at JXB online), as demonstrated for AtPUB17 and SlPUB17 previously (Yang et al., 2006).
P. infestans colonization is enhanced by RNAi of StPUB17 in potato and by virus-induced gene silencing of NbPUB17 in Nicotiana benthamiana
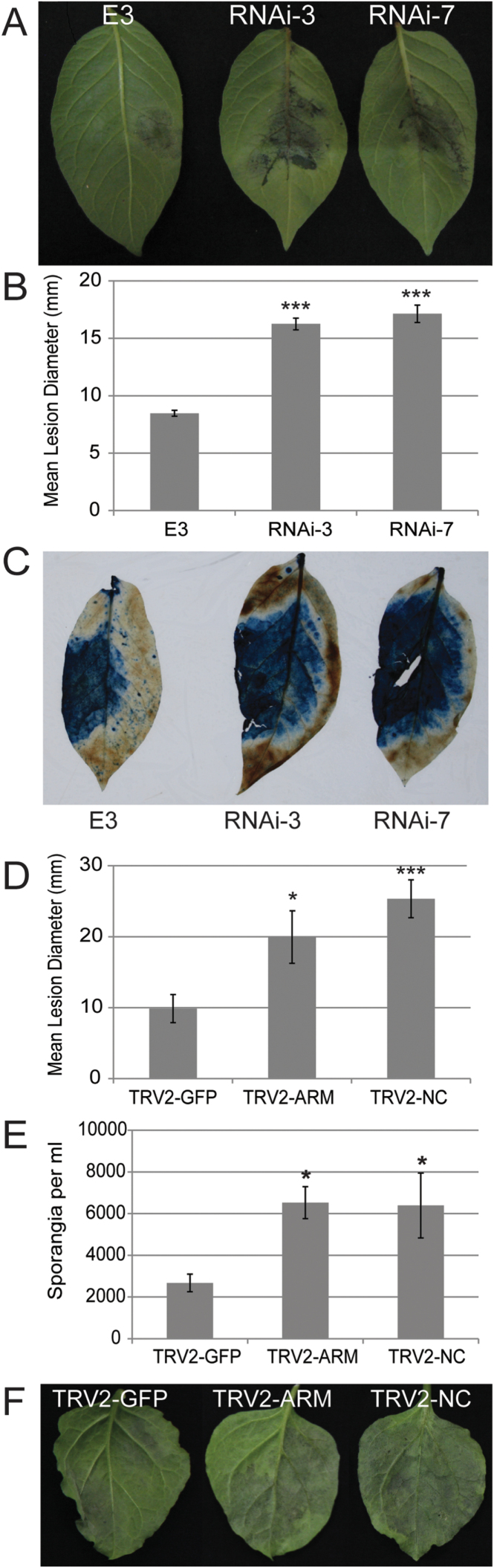
Previous preliminary results had indicated that RNAi of StPUB17, using a portion spanning the U-box-encoding region (see Supplementary Fig. S3A at JXB online) in susceptible cultivar E-potato-3 (E3) resulted in enhanced P. infestans colonization (Ni et al., 2010). As all RNAi lines generated previously had shown similar enhancement of P. infestans colonization (Ni et al., 2010), just two of these RNAi lines were selected (RNAi-3 and RNAi-7) for further analysis here. Each revealed an approximately 60% reduction in StPUB17 transcript accumulation (see Supplementary Fig. S3B at JXB online). In each line, P. infestans colonization, measured as lesion diameter, was significantly (P <0.001, one way ANOVA) more extensive (Fig. 1A, B), compared with the untransformed cultivar E3. This was strikingly apparent using trypan blue (Fig. 1C), which stains P. infestans mycelium to reveal the extent of pathogen colonization (Bos et al., 2010).
Fig. 1.
P. infestans colonization is enhanced by RNAi of StPUB17 in potato and by virus-induced gene silencing of NbPUB17 in N. benthamiana. (A) Representative images of leaves of the Control (E3) and StPUB17 RNAi potato lines at 4 d post infection (dpi) with P. infestans. (B) Graph showing mean lesion diameter at 4 dpi with P. infestans on the Control (E3), and the StPUB17 RNAi-3 and RNAi-7 potato lines. (C) Trypan blue staining of representative leaves of the Control (E3) and the StPUB17 RNAi-3 and RNAi-7 potato lines at 4 dpi. (D) Graph showing mean lesion diameter of P. infestans inoculations at 7 dpi on N. benthamiana plants silenced for NbPUB17 compared with the TRV-GFP control. (E) Graph shows the number of sporangia recovered ml–1 at 10 dpi from P. infestans-infected leaves silenced for NbPUB17 compared with the TRV-GFP control. (F) Representative leaves silenced for NbPUB17 and TRV-GFP at 10 dpi with P. infestans. Statistical analysis was carried out using ANOVA with pairwise comparisons performed with a Holm–Sidak test (Holm, 1979). Asterisks denote the P value as follows: *P ≤0.05, ***P ≤0.001; error bars show standard error. All experiments are the combination of at least three biological replicates, each using six or seven plants, and inoculations of at least four leaves from each plant.
To investigate the impact of PUB17 on P. infestans–host interactions further, StPUB17 was aligned with the corresponding RBBH in the N. benthamiana genome, designated NbPUB17, and two portions were selected, based on them sharing no significant homology with other sequences in potato or N. benthamiana, to avoid the potential for off-target silencing. Each portion was cloned into the Tobacco Rattle Virus (TRV) vector for virus-induced gene silencing.
Constructs TRV::ARM, containing a portion spanning the ARM repeat-encoding region, and TRV::NC, containing a ‘non-conserved’ portion upstream of the U-box (see Supplementary Fig. S4A at JXB online), together with TRV::GFP as a control, were inoculated into 3-week-old plants and subsequently challenged with P. infestans 21 d later. A combination of three experimental replicates, each involving inoculations in quadruple on three leaves on each of six VIGS plants revealed that silencing of NbPUB17 with either TRV::ARM or TRV::NC resulted in significantly (ANOVA, P <0.01) increased P. infestans colonization compared with TRV::GFP control plants, as measured by lesion diameter, and by number of sporangia recovered from lesions (Fig. 1D–F). QRT-PCR revealed a decrease of NbPUB17 by 60–80% with either TRV::ARM or TRV::NC constructs, compared with TRV::GFP, in each of the three replicate experiments (see Supplementary Fig. S4B–D at JXB online). Taken together, VIGS in N. benthamiana and RNAi in potato demonstrate that NbPUB17 and StPUB17, respectively, contribute to reducing P. infestans infection.
Silencing of NbPUB17 in N.benthamiana attenuates early up-regulation of PTI marker genes
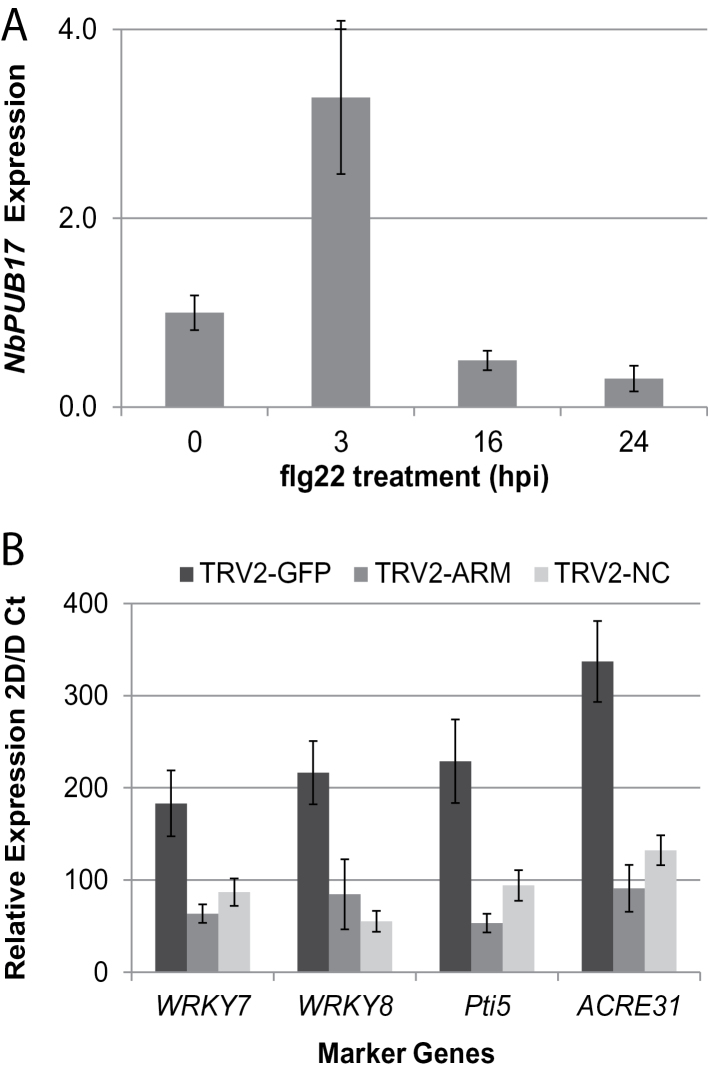
It has been reported previously that AtPUB17 is rapidly up-regulated following treatment with the PAMP flg22 (Navarro et al., 2004). It was therefore investigated whether this was also the case for NbPUB17 in N. benthamiana. Following flg22 infiltration into leaves as described previously (McLellan et al., 2013) a discrete peak of NbPUB17 transcript accumulation was noted specifically at 3h post-inoculation (Fig. 2A). This prompted an investigation into whether PUB17 contributes to the flg22-mediated PTI response, using the early-induced genes NbWRKY7, NbWRKY8, NbPti5, and NbACRE31 (McLellan et al., 2013). Following flg22 treatment of VIGS plants, remarkably, given that PUB17 function has only previously been associated with ETI (Yang et al., 2006), a significantly reduced accumulation of NbWRKY7, NbWRKY8, NbPti5, and NbACRE31 transcripts was observed in TRV::ARM or TRV::NC plants, compared with the TRV::GFP control (Fig. 2B). This indicates that PUB17 contributes to the activation of PTI as well as ETI.
Fig. 2.
Virus-induced gene silencing of NbPUB17 in N. benthamiana attenuates flg22-mediated transcriptional responses. (A) NbPUB17 transcript accumulation at 3h post-inoculation (hpi) of flg22. (B) Flg22-mediated transcript accumulation for NbWRKY7, NbWRKY8, NbPti5, and NbACRE31 at 3 hpi in TRV2-GFP, TRV2-ARM, and TRV2-NC expressing plants, relative to untreated TRV2-GFP.
Silencing of NbPUB17 in N.benthamiana, or NtPUB17 (NtACRE276) in tobacco, compromises Cf4/Avr4 cell death, but not that triggered by INF1 or by co-expression of R3a and AVR3aKI
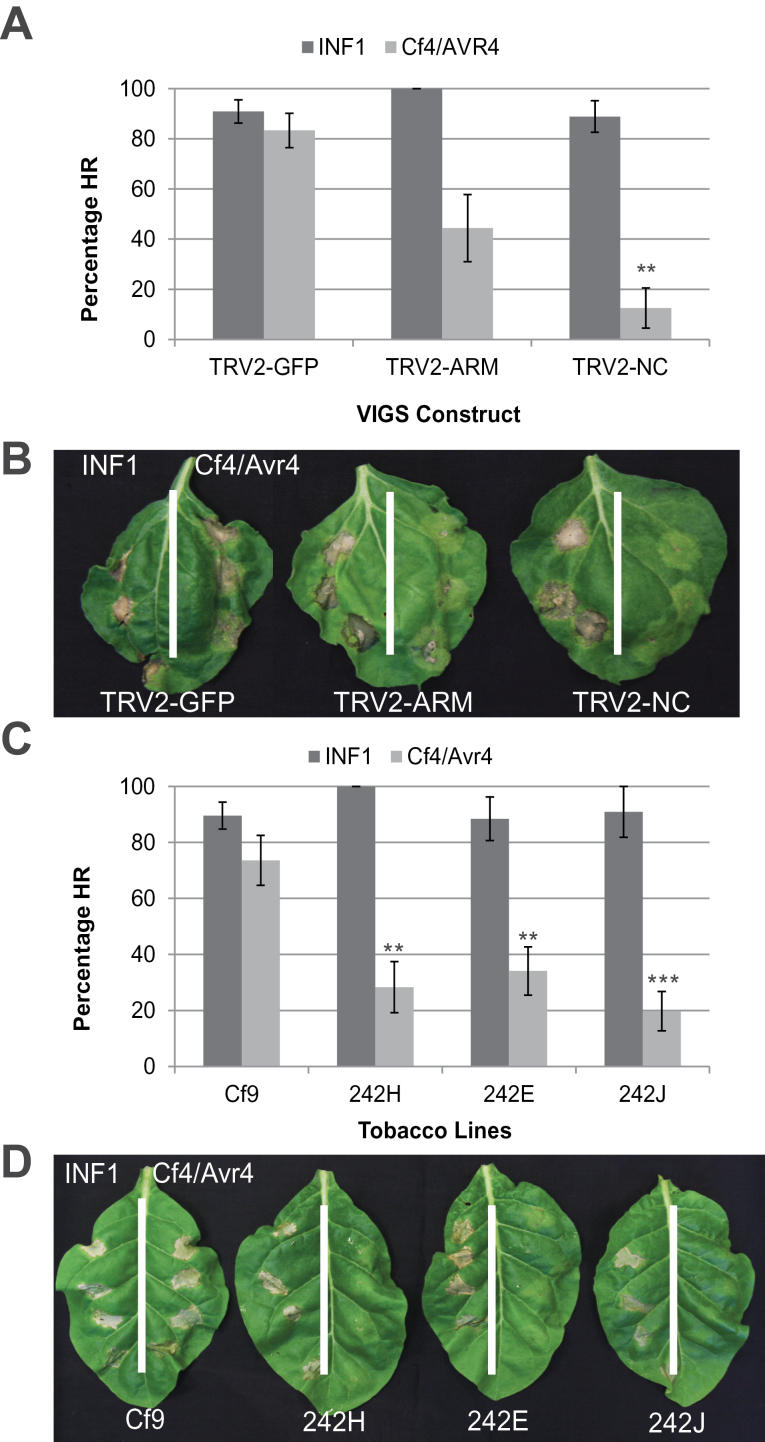
Previous work showed that PUB17 from either tomato or tobacco is a positive regulator of programmed cell death (PCD) triggered by a recognition of the apoplastic effectors Avr4 and AVR9 from C. fulvum by the corresponding receptor-like resistance proteins CF4 and CF9 (Yang et al., 2006). This analysis was extended to investigate whether PUB17 is also required for cell death triggered by perception of a PAMP from P. infestans, the elicitin INF1. In addition to VIGS of NbPUB17 in N. benthamiana, the previously published (Yang et al., 2006) tobacco NtPUB17 RNAi lines were also included. As expected, VIGS of NbPUB17 (Fig. 3A, B), or RNAi of NtPUB17 (Fig. 3C, D), significantly compromised CF4/AVR4 PCD. However, silencing of NbPUB17 or NtPUB17 failed to attenuate INF1 cell death in either plant, indicating that not all cell death events following perception of an elicitor at the cell surface require PUB17 (Fig. 3).
Fig. 3.
Silencing of PUB17 in N. benthamiana or tobacco compromises Cf4/Avr4 cell death but not INF1 cell death. (A) Graph shows the percentage HR elicited by INF1 or Cf4/Avr4 on N. benthamiana plants silenced for NbPUB17 using the TRV-ARM and NC constructs, respectively. Statistical analysis was carried out using ANOVA with pairwise comparisons performed with a Holm–Sidak test; **P ≤0.01 (n=11), error bars show standard error. (B) Representative leaves silenced for NbPUB17 and TRV-GFP showing INF1 HR on the left side of the leaf and Cf4/Avr4 HR on the right. (C) Graph shows the percentage HR elicited by INF1 or Cf4/Avr4 on N. tabacum plants silenced for NtPUB17 (lines 242H, E, and J) are shown beside the non-silenced control Cf9. Statistical analysis was carried out using ANOVA with pairwise comparisons performed with a Holm–Sidak test; **P ≤0.01, ***P ≤0.001 (n=22), error bars show standard error. (D) Representative leaves silenced for NtPUB17 and the control showing INF1 HR on the left side of the leaf and Cf4/Avr4 HR on the right.
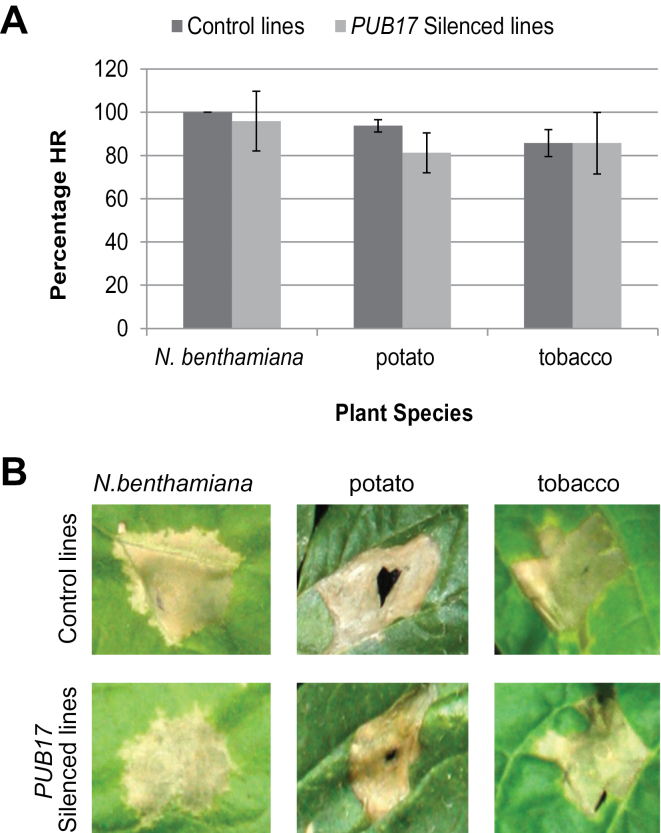
To investigate the potential roles of PUB17 orthologues as positive regulators of PCD further, the impact of silencing NbPUB17, NtPUB17 or StPUB17 on the HR triggered by the intracellular recognition of AVR3aKI from P. infestans by potato R3a was studied. This was shown previously to be independent of the E3 ligase, CMPG1, which is another positive regulator of CF4-AVR4-mediated PCD (Bos et al., 2010; Gilroy et al., 2011). It was observed that VIGS of NbPUB17 in N. benthamiana, or RNAi of NtPUB17 in tobacco or StPUB17 in potato, each failed to attenuate R3a-mediated HR (Fig. 4).
Fig. 4.
The HR mediated by R3a is not affected by silencing of PUB17. (A) Graph shows percentage HR elicited by Agro-infiltration of R3a/Avr3a constructs on N. benthamiana, potato, and tobacco plants with the control lines (respectively, TRV2-GFP, potato E3, and tobacco Cf9) compared with PUB17 silenced lines (respectively, TRV2-NC in N. benthamiana, RNAi-3 in potato, and 242J in tobacco). Statistical analysis carried out by ANOVA indicates no significant differences, error bars show standard error. (B) Representative images showing close up of the HR on the control and PUB17 silenced lines used for each plant species. All experiments are the combination of at least three biological replicates, each using six or seven plants, and inoculations of at least four leaves from each plant.
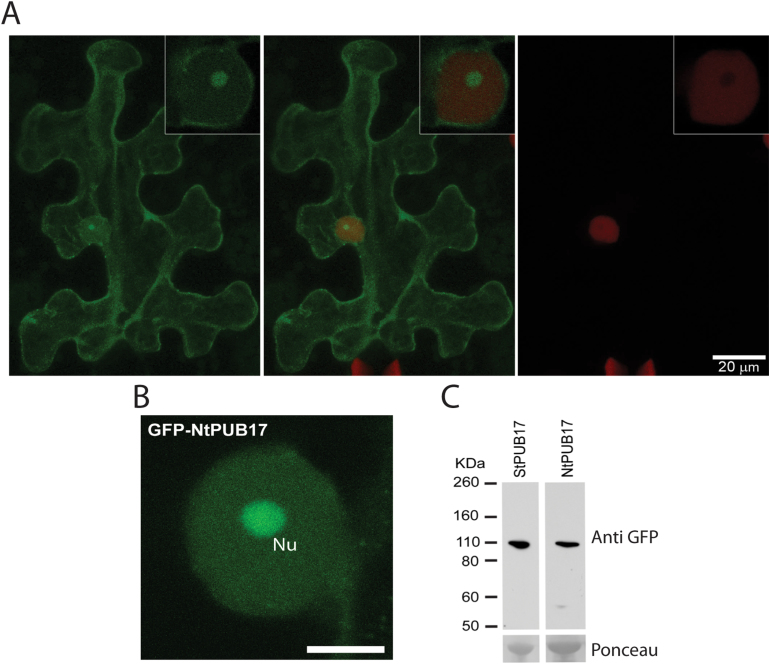
GFP-StPUB17 and GFP-NtPUB17 strongly accumulate in the plant nucleolus
StPUB17 was cloned in-frame with an N-terminal GFP fusion and transiently expressed in N. benthamiana to study sub-cellular localization. Strikingly, when expressed in transgenic N. benthamiana CB157 (expressing mRFP-H2B, which labels the nucleoplasm), it was noted that GFP-StPUB17 protein was observed in the nucleus and especially in the nucleolus (Fig. 5A), with background cytoplasmic fluorescence (see Supplementary Fig. S5 at JXB online). GFP-fusion to NtPUB17 demonstrated that it showed similar localizations (Fig. 5B; see Supplementary Fig. S5 at JXB online). Immunoblots demonstrated that each GFP-fusion protein was intact (Fig. 5C), indicating that the fluorescence accurately reflects the localization of PUB17 in each case.
Fig. 5.
GFP-StPUB17 and GFP-NtPUB17 strongly accumulate in the nucleolus in planta. (A) Representative CB157 N. benthamiana leaves expressing mRFP-H2B examined by confocal microscopy 48h after infiltration of 35S-GFP-StPUB17, indicating green (left), red (right), and merged (centre) channels, with close-up nuclear confocal images inset. (B) Representative close-up nuclear confocal image showing the sub-nuclear localization of GFP-NtPUB17. Nu indicates the nucleolus and the scale bar is 10 μm. (C) Western blots probed with a GFP antibody showing stable protein fusions of potato and tobacco GFP-PUB17 of the expected size. The lower panels show Ponceau staining of the membrane as a loading control.
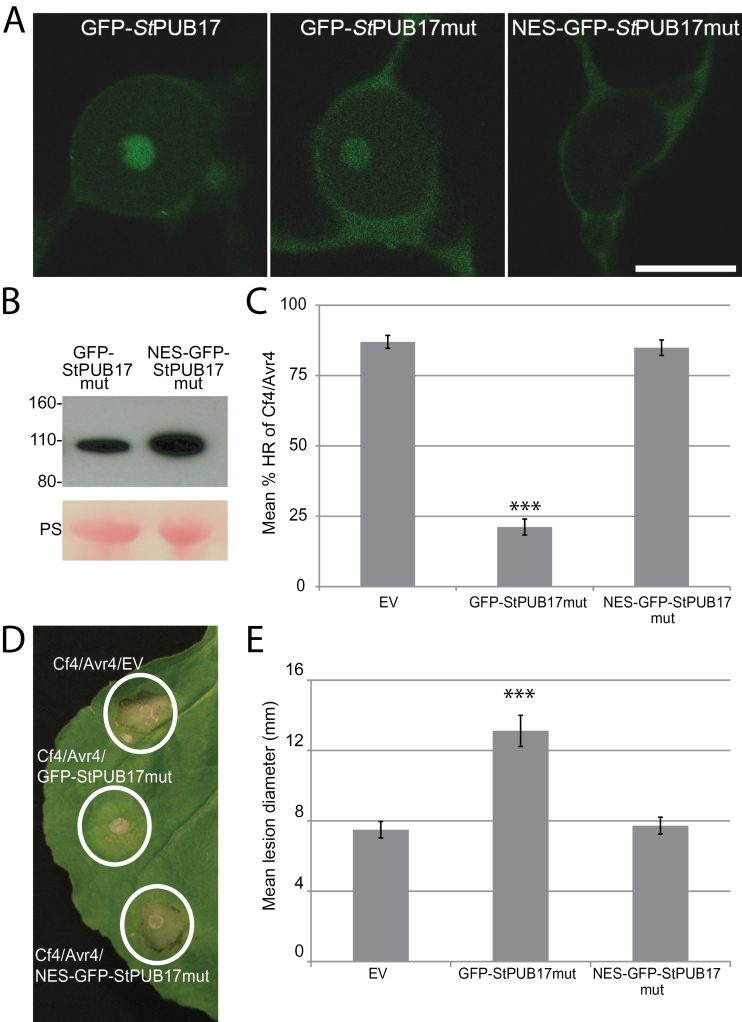
Dominant-negative activity of an StPUB17V314I,V316I mutant is prevented by nuclear exclusion
Previously, mutation of AtPUB17 to prevent U-box activity resulted in a mutant form that acted as a dominant-negative to suppress Cf9-mediated cell death (Yang et al., 2006). Therefore, equivalent valine residues in StPUB17 were mutated to isoleucines (V314I, V316I) to create StPUB17V314I,V316I (referred to as StPUB17mut). Similar to WT GFP-StPUB17, GFP-StPUB17mut localized to the nucleus and predominantly to the nucleolus, with a cytoplasmic background (Fig. 6A), and was stable in planta (Fig. 6B). Transient co-expression of GFP-StPUB17mut with Cf4 and Avr4 resulted in a significant suppression of cell death (Fig. 6C, D), indicating that our mutant form acts as a dominant-negative, and that this activity is not inhibited by the presence of the N-terminal GFP fusion. Moreover, expression of GFP-StPUB17mut supported significantly enhanced P. infestans colonization of N. benthamiana (Fig. 6E). In order to assess whether such dominant-negative activity requires nuclear localization, a nuclear exclusion signal (NES) was fused to the N-terminus of the fused GFP in this construct, creating NES-GFP-StPUB17mut. NES-GFP-StPUB17mut was excluded from the nucleus (Fig. 6A) and was again stable in planta (Fig. 6B). However, in contrast to the GFP-StPUB17mut, the NES-GFP-StPUB17mut failed to suppress CF4/Avr4 cell death (Fig. 6C, D) or enhance P. infestans colonization of N. benthamiana (Fig. 6E).
Fig. 6.
Dominant-negative activity of an StPUB17V314I,V316I mutant (StPUB17mut) is prevented by nuclear exclusion. (A) Representative nuclear images of GFP-STPUB17, GFP-StPUB17mut, and NES-GFP-StPUB17mut. Size marker is 10 μm. (B) Western blots probed with a GFP antibody showing stable protein fusions of GFP-StPUB17mut and NES-GFP-StPUB17mut of the expected size. The lower panels show Ponceau staining (PS) of the membrane as a loading control. (C) Mean % HR of Cf4/Avr4 co-expressed with empty vector (EV), GFP-StPUB17mut, and NES-GFP-StPUB17mut. (D) Images of HR of Cf4/Avr4 co-expressed with empty vector (EV), GFP-StPUB17mut, and NES-GFP-StPUB17mut. (E) Mean lesion diameter of P. infestans colonization following expression of empty vector (EV), GFP-StPUB17mut, and NES-GFP-StPUB17mut. Statistical analysis was carried out using ANOVA with pairwise comparisons performed with a Holm–Sidak test; ***P ≤0.001 [n=92 for (C) and n=106 for (E)]; error bars show standard error.
Discussion
Previously, the Arabidopsis AtPUB17 protein, and its functional orthologues in tomato and tobacco, SlPUB17 (SlACRE276) and NtPUB17 (NtACRE276), respectively, were shown to be functional ubiquitin E3 ligases that act as positive regulators of the HR triggered by R/AVR recognition events: RPM1/AvrB and RPS4/AvrRPS4 in Arabidopsis and Cf4/AVR4 in tomato and tobacco (Yang et al., 2006). In addition, using RNAi, it was shown that StPUB17 in potato is required for basal immunity to P. infestans (Ni et al., 2010). It was confirmed that silencing of PUB17 in potato enhances late blight infection and it is shown that this is also the case in the closely-related solanaceous host N. benthamiana, allowing detailed functional studies to be performed in this plant. The following new observations about PUB17 function have been made: (i) whereas PUB17 is required for CF4/AVR4 cell death, as previously shown (Yang et al., 2006), it is not required for cell death triggered by the P. infestans PAMP INF1, indicating that PUB17 is not involved in all cell death events triggered following pathogen perception by cell surface receptors; (ii) silencing PUB17 orthologues in potato, N. benthamiana or tobacco failed to attenuate R3a/AVR3aKI HR, indicating that PUB17 is not involved in all R/AVR-mediated cell death events; (iii) remarkably, however, it is demonstrated that PUB17 promotes the activation of PTI triggered by the PAMP flg22; and (iv) critically, using a dominant-negative mutant of StPUB17, it is shown that its activity requires nuclear localization. Each of these observations is discussed below.
It has been demonstrated that GFP fusions of StPUB17 and NtPUB17 show a similar nucleo-cytoplasmic location and strongly accumulate in the nucleolus, suggestive of a shared nucleolar function. Interestingly, the E3 ligase CMPG1, which is a target of the P. infestans effector AVR3a (Bos et al., 2010; Gilroy et al., 2011), also showed nucleolar accumulation when stabilized by the effector, even though AVR3a itself does not localize in the nucleolus. It is therefore likely that both positive regulators of cell death, PUB17 and CMPG1, have nuclear/nucleolar functions, although this has yet to be shown for CMPG1. Nevertheless, whereas CMPG1 is required for both INF1- and CF4-mediated cell death (Gilroy et al., 2011), PUB17 is not required for INF1 cell death, indicating that they may have distinct substrates for ubiquitination, and thus different regulatory roles.
By mutation of the key valine residue required for E3 ligase activity in the U-box, Yang et al. (2006) generated a dominant-negative version of AtPUB17 that suppresses Cf-mediated cell death. Therefore, this strategy was utilized to investigate the sub-cellular localization of PUB17 activity. An equivalent dominant- negative StPUB17 form was generated, showing that it also suppressed Cf4-mediated cell death. Importantly, the mutant retained predominantly nuclear/nucleolar localization. By excluding this mutant from the nucleus the dominant-negative activity was lost, indicating that at least some critical StPUB17 substrates for ubiquitination are potentially also located in the nucleus. By contrast, a number of PUB E3 ligases that negatively regulate immunity have been shown to act at the plasma membrane to target and ubiquitinate receptors such as FLS2 (PUB12 and PUB13), or to target components of vesicle trafficking (PUB22) (Lu et al., 2011; Stegmann et al., 2012). Future work will involve identifying the nuclear substrates of PUB17 to determine their fate following ubiquitination accurately.
Previously, AtPUB17 was shown to be required for HR mediated by NB-LRR resistance proteins RPM1 and RPS4, suggesting that it is a positive regulator of ETI (Yang et al., 2006). However, silencing of PUB17 in potato, tobacco, and N. benthamiana had no impact on R3a-AVR3aKI mediated HR, suggesting that it is not required for this ETI response, and that this is conserved within the Solanaceae. CMPG1 was also not required for the HR mediated by cytoplasmic NB-LRR proteins such as R3a, R2, and Rx, and seems to be associated with positively regulating cell death triggered by cell-surface recognition events (Gilroy et al., 2011). It is interesting that RPM1 is activated at, and signals from, the plasma membrane (Gao et al., 2011); hence it will be important to explore further any role of PUB17 in signal transduction from the cell surface. It is possible that, although PUB17 substrates probably reside in the nucleus, the protein itself could be activated from outside the nucleus.
StPUB17 was shown previously to be induced during a compatible potato–P. infestans interaction as early as 24 hpi (Ni et al., 2010). It is shown here that silencing StPUB17 in potato by RNAi, and VIGS of NbPUB17 in N. benthamiana, each significantly enhance colonization by P. infestans. Moreover, expression of the dominant-negative GFP-StPUB17mut also enhanced colonization. These results indicate that PUB17 probably contributes to basal immunity to late blight disease during the biotrophic stage of interaction. Silencing results with PUB17 in potato and N. benthamiana, in contrast to VIGS results with CMPG1 which resulted in a reduction in sporulating P. infestans lesions, further emphasize the different roles that these two E3 ubiquitin ligases play during late blight disease. The CMPG1 VIGS result was explained by the coincident reduction in AVR3a transcript accumulation and the increase in INF1 expression during the switch from biotrophy to necrotrophy. This implies that the pathogen promotes CMPG1 activity during the necrotrophic phase to trigger host cell death for the benefit of completing its infection cycle (Bos et al., 2010).
How does PUB17 contribute to immunity to late blight? Whilst PUB17 in the Solanaceae plants studied here is required for CF4-mediated HR, it is not involved in the HR triggered by perception of the P. infestans PAMP, INF1. P. infestans does not possess an effector related to the C. fulvum AVR4 and, to date. no CF-like proteins have been shown to provide resistance to late blight. However, it has recently been shown that the RXLR effector RD2 from P. infestans enhances colonization of N. benthamiana by targeting the kinase domain of the host MAP3Kɛ protein, which is required for CF4-mediated cell death, but not for INF1-mediated PCD. Moreover, VIGS of MAP3Kɛ led to significantly enhanced P. infestans colonization (King et al., 2014). This indicates that the signal transduction pathway associated with CF4-mediated immunity is likely also to be triggered by P. infestans perception, supporting the involvement of PUB17 in basal defence to late blight. Future studies, prompted by observations here and in King et al. (2014), will focus on identifying the host receptor(s) and the corresponding pathogen molecules that trigger the CF4/9-associated signal transduction pathway in the P. infestans–host interaction, and in identifying the substrates of PUB17 to understand better its regulatory role in immunity.
In addition to a potential role, during late blight infection, in promoting a response pathway associated with Cf-mediated cell death, it is shown that PUB17 also acts to promote PTI activated by the PAMP flg22; silencing of PUB17 significantly attenuated induction of flg22-triggered early marker genes. Whilst P. infestans does not possess the PAMP flg22, the associated, generic FLS2 signal transduction pathway has recently been shown to be relevant to late blight as it is inhibited, at different stages, by a range of P. infestans RXLR effectors (Zheng et al., 2014). Thus, the contribution of PUB17 to basal immunity to late blight is likely to include its roles in promoting both Cf- and FLS2-associated signalling pathways. Interestingly, the observation that PUB17 does not contribute to INF1-mediated cell death highlights the differences in signal transduction between cell-surface perception events and, indeed, between different PAMPs. It will be important to identify both the host receptors and P. infestans elicitors responsible for activating these pathways.
In conclusion, it has been shown that, whereas PUB17 has previously been shown positively to regulate cell death triggered from cell-surface receptors and from intracellular NB-LRR resistance proteins (Yang et al., 2006), it is not involved in all cell death pathways, as exemplified by INF1-mediated and R3a/AVR3a-mediated cell death. However, it has also been shown that it does play a role in early activation of PTI gene expression. Finally, it has been demonstrated that PUB17 acts as a positive regulator of immunity in the host nucleus.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Alignment of potato, tobacco, and Arabidopsis PUB17 amino acid sequences.
Supplementary Fig. S2. StPUB17 functions as an E3 ubiquitin ligase.
Supplementary Fig. S3. Expression analysis of StPUB17 RNAi lines.
Supplementary Fig. S4. Expression analysis of NbPUB17 levels in three replicates of virus-induced gene silencing.
Supplementary Fig. S5. Confocal images of potato and tobacco GFP-PUB17 fusion proteins.
Supplementary Table S1. Primers used in this study.
Supplementary Table S2. Genes, constructs, vectors and their use during this study
Acknowledgements
This work was supported by the National High Technology Research and Development Program of China of China (2013AA102603), the National Natural Science Foundation of China (31171603, 31471550), and the Research Fund for the Doctoral Program of Higher Education of China (20110146110019). We are grateful for funding from the Scottish Government Rural and Environmental Science and Analytical Services Department, and from BBSRC grant BB/G015244/1.
References
- Armstrong MR, Whisson SC, Pritchard L, et al. 2005. An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognised in the host cytoplasm. Proceedings of the National Academy of Sciences, USA 102, 7766–7771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch PRJ, Boevink PC, Gilroy EM, Hein I, Pritchard L, Whisson SC. 2008. Oomycete RXLR effectors: delivery, functional redundancy and durable disease resistance. Current Opinion in Plant Biology 11, 373–379. [DOI] [PubMed] [Google Scholar]
- Birch PRJ, Armstrong MR, Bos JI, et al. 2009. Towards understanding the virulence functions of RXLR effectors of the oomycete plant pathogen Phytophthora infestans . Journal of Experimental Botany 60, 1133–1140. [DOI] [PubMed] [Google Scholar]
- Birch PRJ, Bryan G, Fenton B, Gilroy EM, Hein I, Jones J, Taylor M, Torrance L, Toth IK. 2012. Crops that feed the world. Potato: are the trends of increased global production sustainable? Food Security 4, 477–508. [Google Scholar]
- Block A, Alfano JR. 2011. Plant targets for Pseudomonas syringae type III effectors: virulence targets or guarded decoys? Current Opinion in Microbiology 14, 39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, He SY. 2009. Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324, 742–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JIB, Armstrong MR, Gilroy EM, et al. 2010. Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proceedings of the National Academy of Sciences, USA 107, 9909–9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JIB, Kanneganti TD, Young C, Cakir C, Huitema E, Win J, Armstrong MR, Birch PRJ, Kamoun S. 2006. The C-terminal half of Phytophthora infestans RXLR effector AVR3a is sufficient to trigger R3a-mediated hypersensitivity and suppress INF1-induced cell death in Nicotiana benthamiana . The Plant Journal 48, 165–176. [DOI] [PubMed] [Google Scholar]
- Cikos S, Bukovska A, Koppel J. 2007. Relative quantification of mRNA: comparison of methods currently used for real-time PCR data analysis. BMC Molecular Biology 8, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke D, Cano L, Raffaele S, et al. 2012. Genome analyses of an aggressive and invasive lineage of the Irish Potato Famine pathogen. PLoS Pathogens 8, e1002940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP. 2010. Plant immunity: towards an integrated view of plant–pathogen interactions. Nature Reviews Genetics 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Dreher K, Callis J. 2007. Ubiquitin, hormones, and biotic stress in plants. Annals of Botany 99, 787–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplan V, Rivas S. 2014. E3 ubiquitin-ligases and their target proteins during the regulation of plant innate immunity. Frontiers in Plant Science 5, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Chung EH, Eitas TK, Dangl JL. 2011. Plant intracellular innate immune receptor resistance to Pseudomonas syringae pv. maculicola 1 (RPM1) is activated at, and functions on, the plasma membrane. Proceedings of the National Academy of Sciences, USA 108, 7619–7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy EM, Hein I, van der Hoorn R, et al. 2007. Involvement of cathepsin B in the plant disease resistance hypersensitive response. The Plant Journal 52, 1–13. [DOI] [PubMed] [Google Scholar]
- Gilroy EM, Taylor RM, Hein I, Boevink P, Sadanandom A, Birch PRJ. 2011. CMPG1-dependent cell death follows perception of diverse pathogen elicitors at the host plasma membrane and is suppressed by Phytophthora infestans RXLR effector AVR3a. New Phytologist 190, 653–666. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Lamothe R, Tsitsigiannis DI, Ludwig AA, Panicot M, Shirasu K, Jones JDG. 2006. The U-box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. The Plant Cell 18, 1067–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B, Kamoun S, Zody MC, et al. 2009. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans . Nature 461, 393–398. [DOI] [PubMed] [Google Scholar]
- Hein I, Gilroy EM, Armstrong MR, Birch PRJ. 2009. The zig-zag-zig in oomycete–plant interactions. Molecular Plant Pathology 10, 547–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann NR. 2012. Ubiquitination and exocytosis in plant immunity. The Plant Cell 24, 4312–4315. [Google Scholar]
- Holm S. 1979. A simple sequentially rejective multiple test procedure. Scandinavian Journal of Statistics 6, 65–70. [Google Scholar]
- Jones JD, Dangl JL. 2006. The plant immune system. Nature 444, 323–329. [DOI] [PubMed] [Google Scholar]
- King SR, McLellan H, Boevink PC, Armstrong MR, Bukharova T, Sukarta O, Win J, Kamoun S, Birch PR, Banfield MJ. 2014. Phytophthora infestans RXLR effector PexRD2 interacts with host MAPKKKε to suppress plant immune signaling. The Plant Cell 26, 1345–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SJ, Choi EY, Choi YJ, Ahn JH, Park OK. 2006. Proteomics studies of post-translational modifications in plants. Journal of Experimental Botany 57, 1547–1551. [DOI] [PubMed] [Google Scholar]
- Li W, Ahn IP, Ning YS, et al. 2012. The U-box/ARM E3 ligase PUB13 regulates cell death, defense, and flowering time in Arabidopsis . Plant Physiology 159, 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JL, Li W, Ning YS, Shirsekar G, Cai YH, Wang XL, Dai LY, Wang ZL, Liu WD, Wang GL. 2012. The U-box E3 ligase SPL11/PUB13 is a convergence point of defense and flowering signaling in plants. Plant Physiology 160, 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu DP, Lin WW, Gao XQ, Wu SJ, Cheng C, Avila J, Heese A, Devarenne TP, He P, Shan LB. 2011. Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332, 1439–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino D, Peeters N, Rivas S. 2012. Ubiquitination during plant immune signaling. Plant Physiology 160, 15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan H, Boevink PC, Armstrong MR, Pritchard L, Gomez S, Morales J, Whisson SC, Beynon JL, Birch PR. 2013. An RxLR effector from Phytophthora infestans prevents re-localisation of two plant NAC transcription factors from the endoplasmic reticulum to the nucleus. PLoS Pathogens 9, e1003670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Zipfel C, Rowland O, Keller I, Robatzek S, Boller T, Jones JD. 2004. The transcriptional innate immune response to flg22. Interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiology 135, 1113–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni XM, Tian ZD, Liu J, Song BT, Li JC, Shi XL, Xie CH. 2010. StPUB17, a novel potato UND/PUB/ARM repeat type gene, is associated with late blight resistance and NaCl stress. Plant Science 178, 158–169. [Google Scholar]
- O’Neill LA. 2011. Plant science. Innate immunity in plants goes to the PUB. Science 332, 1386–1387. [DOI] [PubMed] [Google Scholar]
- Ratcliff F, Martin‐Hernandez AM, Baulcombe DC. 2001. Technical advance: tobacco rattle virus as a vector for analysis of gene function by silencing. The Plant Journal 25, 237–245. [DOI] [PubMed] [Google Scholar]
- Rowland O, Ludwig AA, Merrick CJ, et al. 2005. Functional analysis of Avr9/Cf-9 rapidly elicited genes identifies a protein kinase, ACIK1, that is essential for full Cf-9-dependent disease resistance in tomato. The Plant Cell 17, 295–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadanandom A, Bailey M, Ewan R, Lee J, Nelis S. 2012. The ubiquitin–proteasome system: central modifier of plant signalling. New Phytologist 196, 13–28. [DOI] [PubMed] [Google Scholar]
- Saunders D, Breen S, Schornack S, et al. 2012. Host protein BSL1 associates with Phytophthora infestans RXLR effector PiAVR2 and the immune receptor R2 to mediate disease resistance. The Plant Cell 24, 3420–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirsekar G, Dai LY, Hu YJ, Wang XJ, Zeng LR, Wang GL. 2010. Role of ubiquitination in plant innate immunity and pathogen virulence. Journal of Plant Biology 53, 10–18. [Google Scholar]
- Stegmann M, Anderson RG, Ichimura K, Pecenkova T, Reuter P, Zarsky V, McDowell JM, Shirasu K, Trujillo M. 2012. The ubiquitin ligase PUB22 targets a subunit of the exocyst complex required for PAMP-triggered responses in Arabidopsis . The Plant Cell 24, 4703–4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Callis J. 2007. Ubiquitin ligases mediate growth and development by promoting protein death. Current Opinion in Plant Biology 10, 624–632. [DOI] [PubMed] [Google Scholar]
- Thomma BPHJ, Nurnberger T, Joosten MHAJ. 2011. Of PAMPs and effectors: the blurred PTI-ETI dichotomy. The Plant Cell 23, 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian ZD, Liu J, Xie CH. 2003. Isolation of resistance related-genes to Phytophthora infestans with suppression subtractive hybridization in the R-gene-free potato. Yi Chuan Xue Bao 30, 597–605. [PubMed] [Google Scholar]
- Trujillo M, Ichimura K, Casais C, Shirasu K. 2008. Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis . Current Biology 18, 1396–1401. [DOI] [PubMed] [Google Scholar]
- Trujillo M, Shirasu K. 2010. Ubiquitination in plant immunity. Current Opinion in Plant Biology 13, 402–408. [DOI] [PubMed] [Google Scholar]
- Vierstra RD. 2009. The ubiquitin–26S proteasome system at the nexus of plant biology. Nature Reviews Molecular Cell Biology 10, 385–397. [DOI] [PubMed] [Google Scholar]
- Vleeshouwers VG, van Dooijeweert W, Keizer LP, Sijpkes L, Govers F, Colon LT. 1999. A laboratory assay for Phytophthora infestans resistance in various Solanum species reflects the field situation. European Journal of Plant Pathology 105, 241–250. [Google Scholar]
- Vleeshouwers VG, Raffaele S, Vossen JH, et al. 2011. Understanding and exploiting late blight resistance in the age of effectors. Annual Review of Phytopathology 49, 507–531. [DOI] [PubMed] [Google Scholar]
- Whisson SC, Boevink PC, Moleleki L, et al. 2007. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450, 115–118. [DOI] [PubMed] [Google Scholar]
- Yang CW, Gonzalez-Lamothe R, Ewan RA, Rowland O, Yoshioka H, Shenton M, Ye H, O’Donnell E, Jones JDG, Sadanandom A. 2006. The E3 ubiquitin ligase activity of Arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. The Plant Cell 18, 1084–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee D, Goring DR. 2009. The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. Journal of Experimental Botany 60, 1109–1121. [DOI] [PubMed] [Google Scholar]
- Zeng LR, Qu S, Bordeos A, Yang C, Baraoidan M, Yan H, Xie Q, Nahm BH, Leung H, Wang GL. 2004. Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. The Plant Cell 16, 2795–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, McLellan H, Fraiture M, et al. 2014. Functionally redundant RXLR effectors from Phytophthora infestans act at different steps to suppress early flg22-triggered immunity. PLoS Pathogens 10, e1004057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.