Highlight
The ambiguous ripening nature of fig fruits contradicts the climacteric definition. One ethylene-responsive factor gene is potentially the ethylene-synthesis regulator responsible for the non-climacteric auto-inhibition of ethylene production in fig.
Key words: 1-Methylcyclopropene (1-MCP), ethylene, Ficus carica, fruit ripening, MADS-box, preharvest treatment.
Abstract
The traditional definition of climacteric and non-climacteric fruits has been put into question. A significant example of this paradox is the climacteric fig fruit. Surprisingly, ripening-related ethylene production increases following pre- or postharvest 1-methylcyclopropene (1-MCP) application in an unexpected auto-inhibitory manner. In this study, ethylene production and the expression of potential ripening-regulator, ethylene-synthesis, and signal-transduction genes are characterized in figs ripening on the tree and following preharvest 1-MCP application. Fig ripening-related gene expression was similar to that in tomato and apple during ripening on the tree, but only in the fig inflorescence–drupelet section. Because the pattern in the receptacle is different for most of the genes, the fig drupelets developed inside the syconium are proposed to function as parthenocarpic true fruit, regulating ripening processes for the whole accessory fruit. Transcription of a potential ripening regulator, FcMADS8, increased during ripening on the tree and was inhibited following 1-MCP treatment. Expression patterns of the ethylene-synthesis genes FcACS2, FcACS4, and FcACO3 could be related to the auto-inhibition reaction of ethylene production in 1-MCP-treated fruit. Along with FcMADS8 suppression, gene expression analysis revealed upregulation of FcEBF1, and downregulation of FcEIL3 and several FcERFs by 1-MCP treatment. This corresponded with the high storability of the treated fruit. One FcERF was overexpressed in the 1-MCP-treated fruit, and did not share the increasing pattern of most FcERFs in the tree-ripened fig. This demonstrates the potential of this downstream ethylene-signal-transduction component as an ethylene-synthesis regulator, responsible for the non-climacteric auto-inhibition of ethylene production in fig.
Introduction
In the last few years, fruit-ripening research has challenged the classical definitions of climacteric and non-climacteric fleshy fruits (Paul et al., 2012). A unique example of this controversy is the fig, Ficus carica L. The ripening process in fig fruit is categorized as climacteric, showing a rise in respiration rate and ethylene production at the onset of the ripening phase (Marei and Crane, 1971). Surprisingly, ripening-related ethylene production increases following pre- or postharvest 1-methylcyclopropene (1-MCP) application in an unexpected auto-inhibitory manner. Moreover, postharvest 1-MCP treatment does not affect the ripening parameters of the treated fruit (unlike other climacteric fruits), while application to fruit on the tree improves fruit-storage abilities, inhibiting deterioration with minor effects on fruit growth and ripening (Sozzi et al., 2005; Owino et al., 2006; Freiman et al., 2012). In addition to the auto-inhibitory reaction of ethylene production in the climacteric-classified fig, other unique characteristics of this fruit differentiate it from the well-studied Solanum lycopersicum (tomato) climacteric model. Not only does the fig develop to its final size during ripening, the ripening process in the main summer crop is rapid, taking less than 3 days. Fruit picked before optimal maturity never reach the desirable parameters of size, colour, flavour, or texture, while fruits harvested too late tend to perish due to over-ripening and high susceptibility to pathogens (Flaishman et al., 2008). By contrast, tomato, Malus domestica (apple), and Musa spp. (banana) fruits, for example, reach their final size at the mature green stage, and only then are the ripening processes initiated. Tomato fruit can be picked at the mature green stage and still develop to the red ripe stage within 10 days, whereas ripening of non-harvested tomato can take over 20 days (Yokotani et al., 2009; Van de Poel et al., 2013).
As with all Ficus species, the fig bears a unique closed inflorescence structure—the syconium—that is not present in any other fruit in the human diet. The multiple fig fruit is composed of small individual drupelets which develop from the ovaries enclosed in the succulent receptacle to form a single accessory fruit (Storey, 1977). Development of the fig’s female fruit is characterized by a double sigmoid growth curve comprising three phases (Marei and Crane, 1971). Phase I is characterized by a rapid growth in size; during phase II, the fruit remains nearly the same size, colour, and firmness. Phase III is the ripening phase and includes fruit growth, colour change, softening, and alteration of the pulp texture to an edible state. The parthenocarpic fruit of the purple female fig cultivar ‘Brown Turkey’ shows onset of ethylene production when the green hue of the peel starts to fade to yellow, at the transition from phase II to III. In attached ‘Brown Turkey’ figs, ethylene peaks during ripening at the commercially ripe fruit stage (50% purple peel), and declines toward the fully ripened fruit stage (100% purple peel). All ripening parameters—size, firmness, and inner texture—present differences between on-tree 1-MCP-treated and untreated fruit (Freiman et al., 2012). Fig ethylene-synthesis genes—three ACC-synthase (ACS) genes and a single ACC-oxidase (ACO) gene—were isolated by Owino et al. (2006), and their expression patterns were studied postharvest and following several treatments. The expression patterns of FcACS1, FcACS3, and FcACO1 were inhibited in figs following postharvest 1-MCP treatment, indicating positive regulation by ethylene (when ethylene is not sensed by the tissue, its synthesis genes are downregulated), whereas FcACS2 expression was induced by 1-MCP, indicating negative regulation (when ethylene is not sensed by the tissue, its synthesis genes are upregulated). Recently, a transcriptome analysis of caprifig (a hermaphroditic fruit that functions as male) and fig female fruit in late phase II was published. The study identified several unigenes encoding proteins involved in ethylene synthesis and signal transduction, assembled from combined data from female ‘Houraishi’ and hermaphroditic ecotypes (Ikegami et al., 2013). A transcriptome analysis of fig (‘Brown Turkey’) during phase III development was published by Freiman et al., 2014. Unlike the work on ‘Houraishi’ and caprifigs, which focused on the differences between the caprifig and female fig in phase II, the transcriptome analysis of ‘Brown Turkey’ targeted ripening processes in phase III and the transition toward this phase. In the latter work, a few ethylene-synthesis genes were identified and MADS-box genes were isolated. The genes were analysed for their expression in the receptacle and FcMADS genes were classified into different MADS-box family clades.
The ripening cascade in the climacteric fruit model, the tomato, can be divided into three levels: transcription factors controlling the transition to ripening phase, ethylene biosynthesis and perception networks regulating ripening, and the coordinated metabolic processes of ripening (Seymour et al., 2013). Compared to ethylene synthesis and regulation, which involves ACSs and ACOs, signal transduction of ethylene is much more complex. Ethylene binds to its receptors, releasing the activation of constitutive triple response1 (CTR1), which, in the absence of ethylene, phosphorylates ethylene-insensitive2 (EIN2). In the presence of ethylene, EIN2 is not phosphorylated and its C terminus is cleaved and moves to the nucleus to induce ethylene-insensitive 3-binding F-box (EBF)1/2 degradation activity. EBF1/2 targets EIN3 and EIN3-like (EIL) for protein degradation, and release of EIN2’s C terminus by ethylene thus stabilizes EIN3/EILs, which in turn bind to ethylene-responsive factor (ERF) promoters. This is the last step in the ethylene-signal-transduction pathway: ERFs bind to downstream genes involved in metabolic ripening processes, as well as in feedback regulation of ethylene synthesis (Merchante et al., 2013).
In the present work, ethylene production and the expression of potential ripening-regulator, ethylene-synthesis, and signal-transduction genes (Supplementary Table S1) are characterized in on-tree-ripening fig fruit. In addition, the effect of preharvest 1-MCP treatment on gene expression is examined. The association between ethylene profile and specific ethylene-synthesis genes is analysed in the context of ethylene-synthesis systems. Expression of ethylene-signal-transduction elements is compared to that in other climacteric fruits with respect to feedback regulation of ethylene synthesis. The study of both ethylene-synthesis and signal-transduction pathways, in on-tree-ripening fruit and in preharvest 1-MCP-treated fruit, indicates the specific genes incorporated in fig ripening and in the auto-inhibitory response to 1-MCP application. In addition, the study pinpoints genes that are assumed to be responsible for the positive effect of preharvest 1-MCP treatment on fig-ripening parameters after storage.
Materials and methods
Plant material
For the on-tree trial, fruit of the female fig (Ficus carica) cultivar ‘Brown Turkey’ were collected from a commercial orchard located near Be’er Tuvia in the southern coastal plain of Israel, 55 m above sea level. Fruits used for this study were from the summer crop, August 2013, with day temperatures of 26–32°C and night temperatures of 22–25°C. Five developmental stages were sampled for ethylene-production measurements and gene-expression quantification: green fruit – pre-ripening stage, end of phase II; yellow fruit – ripening-onset stage, transition from phase II to III; 10% purple peel – ripening-initiation stage, phase III; 50% purple peel – commercially mature stage, phase III; 100% purple peel – fully ripe stage, end of phase III (Fig. 1A).
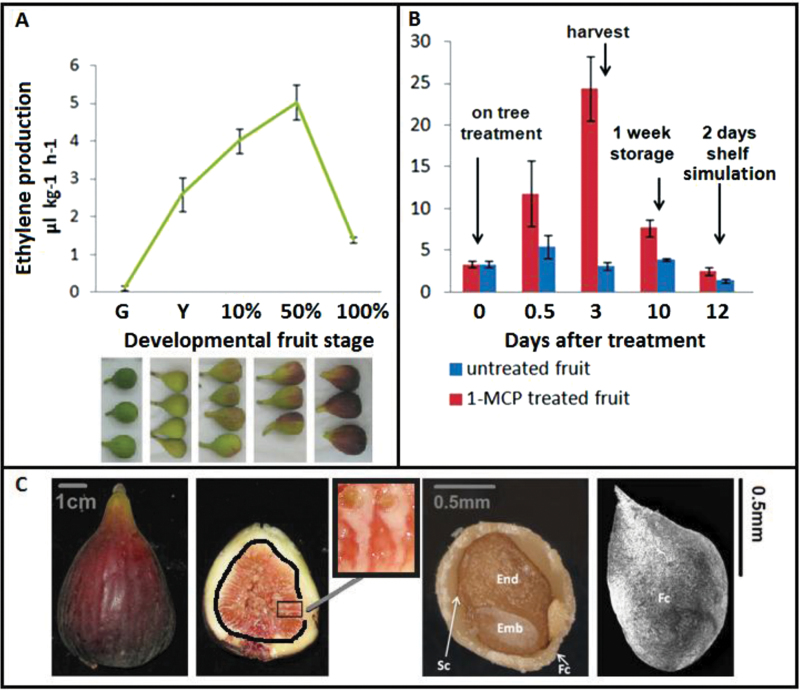
Fig. 1.
Studied fig fruit-ripening systems. (A) Ethylene production in fig fruit during on-tree ripening. Developmental fruit stages: G – green fruit, pre-ripening stage at the end of phase II; Y – yellow fruit, ripening-onset stage at the transition from phase II to phase III; 10% – 10% purple peel fruit, ripening-initiation stage; 50% – 50% purple peel fruit, commercially ripe stage; 100% – 100% purple peel fruit, fully ripe stage. Average of seven fruits per treatment/control group ± SE. (B) Ethylene production in preharvest 1-MCP-treated fruit (at ripening-onset fruit stage) followed by 1 week of cold storage and 2 days of shelf simulation Days after treatment: 0 – ripening-onset fruit before treatment; 0.5 – ripening-onset fruit after 1-MCP treatment on tree; 3 – commercially ripe fruit developed on tree; 10 – harvested commercially ripe fruit after 1 week of storage; 12 – harvested commercially ripe and stored fruit after 2 days of shelf simulation. Average of seven fruits per treatment/control group ± SE. (C) Fully ripe parthenocarpic ‘Brown Turkey’ fig syconium. Left to right – an external view of the syconium; an internal view of a longitudinal cross section of the syconium (the receptacle and inflorescence sections are separated by a thick line); a close view of the female flowers enclosed in the syconium; a stereoscope image of a longitudinal cross section of a single drupelet; a SEM image of the external of a single drupelet.
For the preharvest 1-MCP treatment and storage trial, figs were subjected to preharvest treatment with 1-MCP in a commercial orchard located near Karmei Yosef in the Judea plain of Israel, 137 m above sea level. Fruits used for this study were from the autumn crop, November 2011, with day temperatures of 20–25°C and night temperatures of 10–15°C. Samples subjected to ethylene-production measurements and gene-expression quantification were as follows: ripening-onset fruit before treatment; fruit after overnight 1-MCP treatment/untreated tagged control; commercially mature fruit harvested 3 days after treatment/control; treated/control fruit subjected to storage; treated/control fruit subjected to shelf simulation (Fig. 1B).
Parthenocarpic drupelet examination
For the demonstration of parthenocarpic drupelets, a longitudinal cross section of a fully ripe fig drupelet was examined under a stereoscope (Zeiss Stemi 2000-C, Zeiss, Oberkochen, Germany; Fig. 1C). An external view of the drupelet was obtained by scanning electron microscopy (SEM; Fig. 1C) as follows: parthenocarpic drupelets were fixed in Formalin-acetic acid-alcohol solution (100% acetic acid, 40% formalin, 95% ethanol at 1:2:10, v/v); dehydrated in a series of 25, 50, 75, 90 and 100% ethanol; dried in liquid CO2 (Bio-Rad 750 critical-point dryer, Hemel Hempstead, UK); then placed on SEM discs, coated with a 10-nm gold layer, and studied by SEM (JEOL, Tokyo, Japan) at an accelerating potential of 15kV (Kamenetsky, 1994).
Chemicals
Required concentrations of 1-MCP were obtained from Ethyl-Block powder (Floralife, Inc., Walterboro, SC, USA), with an active ingredient content of 0.14%. The preparation was a gift from Riesel Chackvet Ltd. (Petah Tikva, Israel). The release of 1-MCP from the preparation was tested by gas chromatography as described below and found comparable to that from the commercial product Smart-Fresh (AgroFresh, Spring House, PA, USA).
Preharvest 1-MCP treatment
Fig fruit were treated in the orchard with the 1-MCP gas as described by Freiman et al. (2012).
Ethylene-production measurements
Seven fruits per stage or treatment were sampled for ethylene-production analysis, performed as described by Freiman et al. (2012).
Storage trial
For the storage trial, 1-MCP was applied to the fruit after onset of chlorophyll loss, determined as a change in fruit colour from dull to light green. Fig fruit of approximately the same size (300 fruit, about 4cm in diameter) were marked on trees. Of these, 150 figs were treated with 1-MCP as described above and the other 150 were tagged as untreated controls. After the treatment, the fruit were left on the tree for an additional 3 days and then harvested and stored at the commercially mature stage, according to previously determined cultivar-specific visual and manual criteria (Rodov et al., 2002), namely purple coloration on 20–70% of their surface and slight elastic yielding to mild finger pressure. For analysis, figs with coloration on 50% of their surface were arranged in cartons with plastic-cavity insert trays, each fruit in a separate cavity. The cartons, each containing 10–20 commercially mature fruit, were stored for 7 days at 1–2°C and 90–95% relative humidity, and then for 2 days at 20°C and 85% relative humidity (shelf simulation).
RNA extraction, cDNA synthesis, and transcript isolation
For quantitative PCR analysis, total RNA was extracted according to Jaakola et al., 2001. RNA concentration was determined in a NanoDrop ND-1000 spectrophotometer (Wilmington, DE, USA), and its integrity was checked by running 1 µL in a 1% (w/v) agarose gel stained with bromophenol blue. Total RNA was digested with RQ-DNase (Promega, Madison, WI, USA). Complementary DNA was synthesized, using Oligo-dT primers, with the VERSO cDNA kit (Thermo Scientific, Waltham, MA, USA). The reaction was performed in a T-Gradient PCR system (Biometra, Goettingen, Germany). Newly isolated transcripts of FcACS1L (KP892658), FcACS4 (KP892659), FcACO2 (KP892660) and FcACO3 (KP892661) were sequenced with the use of specific primers flanking the coding sequence according to the published transcriptome (Supplementary Table S2; Freiman et al., 2014).
High-throughput real-time quantitative PCR
High-throughput real-time quantitative PCR was performed on a BioMark 96.96 Dynamic Array (Fluidigm Corp., San Francisco, CA, USA) with TaqMan Gene Expression Assays (Applied Biosystems, Carlsbad, CA, USA) at the Weizmann Institute of Science (Rehovot, Israel). Three biological replicates were used for each treatment and two technical replicates were analysed for each biological replicate. Primers (Supplementary Table S3) were designed with Primer3 software, and synthesized by Metabion (Steinkirchen, Germany) and Hylabs (Rehovot, Israel). Expression levels of the target genes were normalized to the control gene actin.
Results
Ethylene production of natural on-tree-ripening fig and stored fruit treated preharvest with 1-MCP
To investigate the function of ripening-regulator genes in fig fruit, two systems were examined. The first was natural on-tree fig ripening, with sampling of five fruit stages as presented in Fig. 1A. The second system examined was preharvest 1-MCP-treated fruit stored under commercial conditions (1–2°C) and subjected to shelf simulation (20°C) (Fig. 1B). As expected, in the natural on-tree-ripening fig, a rise in ethylene production was detected when fruit colour changed from green (0.11 µL kg-1 h-1) to yellow (2.6 µL kg-1 h-1). Ethylene production continued to increase toward the 50% purple stage (5 µL kg-1 h-1) and decreased at the 100% purple stage (1.4 µL kg-1 h-1). Following overnight preharvest 1-MCP treatment, ethylene production in treated yellow fruit (0.5 days after treatment) was 2.5 times higher than that in untreated fruit (Fig. 1B). This auto-inhibitory pattern following preharvest 1-MCP treatment has been previously documented, along with improved storability of the treated fruit (Freiman et al., 2012). In the present study, further ethylene measurements were taken: after fruit harvest, storage, and shelf simulation (Fig. 1B). High levels of ethylene were evident in the treated fruit 3 days after treatment (harvested commercially mature stage)—eight times higher than that in the untreated fruit (Fig. 1B). After 1 week of storage (10 days after treatment), ethylene levels of the treated fruit were twice as high as those of the untreated fruit and even after 2 days of shelf simulation (12 days after treatment), a difference in ethylene production was observed (1.8 times higher in treated versus untreated fruit, Fig. 1B).
Identification of MADS-box and ethylene-related genes
To determine the molecular components responsible for the unique ethylene characteristics in fig and the improved storability of preharvest 1-MCP-treated fruit, a matrix of ripening-related fig genes was established. Fifty-seven fig genes homologous to MADS-box, ethylene-synthesis and ethylene-signal-transduction genes were subjected to gene-expression analysis (Supplementary Table S1). Gene families included MADS-box, ACS, ETO1-like (EOL), ACO, ethylene receptors (ETR, EIN4, and ERS1), CTR/enhanced disease resistance 1 (EDR1; EDR1 and CTR1 are similar and share some functions; Frye et al., 2001), EIN2, EBF1, EIL, and the ERF family. Eight of the FcMADS-box genes have been isolated previously as potential regulators of ripening and ethylene production. Six of them were further examined here for their expression levels: FcMADS1 from the AGL6 subfamily, FcMADS2 and FcMADS3 from the SQUAMOSA subfamily, FcMADS4 from the STMADS11 subfamily, and FcMADS6 and FcMADS8 from the SEP subfamily (Freiman et al., 2014).
To explore ethylene synthesis, the expressions of four FcACS family members were analysed: FcACS1L, FcACS2, FcACS3, and FcACS4. Partial transcript sequences of FcACS1, FcACS2, and FcACS3 were previously published by Owino et al. (2006). A longer transcript of FcACS1 (FcACS1L, Supplementary Fig. S1) showed 99% identity to the published sequence, with a single nucleotide addition resulting in a frame shift to complete the full-length transcript. FcACS4 is newly presented here. Phylogenetic analysis revealed that FcACS1L and FcACS2 are type 1 ACSs, whereas FcACS3 and FcACS4 are type 2 (Supplementary Fig. S2). Homologues of ETO1 and EOL, which target type 2 ACSs for degradation via the proteasome (McClellan and Chang, 2008), were identified in the developing fig transcriptome: FcEOL1 and FcEOL2. The last group of ethylene-synthesis genes isolated was from the ACO family: FcACOL, FcACO2, and FcACO3. The isolated FcACOL was 96% identical to the FcACO1 sequence previously published by Owino et al. (2006) and 100% identical to the full-length FcACOL transcript sequence submitted to the National Center for Biotechnology Information (NCBI) with no additional information (mRNA GenBank accession no. AB307720.1 2007, Supplementary Fig. S3), while FcACO2 and FcACO3 are newly presented here.
To study ethylene-signal-transduction elements in fig fruit, genes from this pathway were identified in the developing fig transcriptome. Upstream components, acting as ethylene-response inhibitors, were identified, including four receptors (FcETR1, FcETR2, FcEIN4, and FcERS1) and four CTR1 genes (FcCTR1, FcCTR2, FcCTR3, and FcEDR1). The positive signal-transduction regulator FcEIN2 was identified as a single gene, as found in Arabidopsis and tomato (Gapper et al., 2013). Downstream of EIN2, the ethylene-response inhibitor FcEBF1 was also identified. Three possible targets of FcEBF1, namely EIN3/EIL, were detected: FcEIL1, FcEIL2, and FcEIL3. These components are targeted for degradation by EBF1 and are positive regulators of the ethylene response via activation of a large group of ERFs. Twenty-seven FcERFs were traced in the developing fig transcriptome, and their transcript quantification completes the set of ethylene-signal-transduction genes in fig fruit.
Expression of MADS-box and ethylene-synthesis-related genes in the fig inflorescence and receptacle during natural on-tree ripening
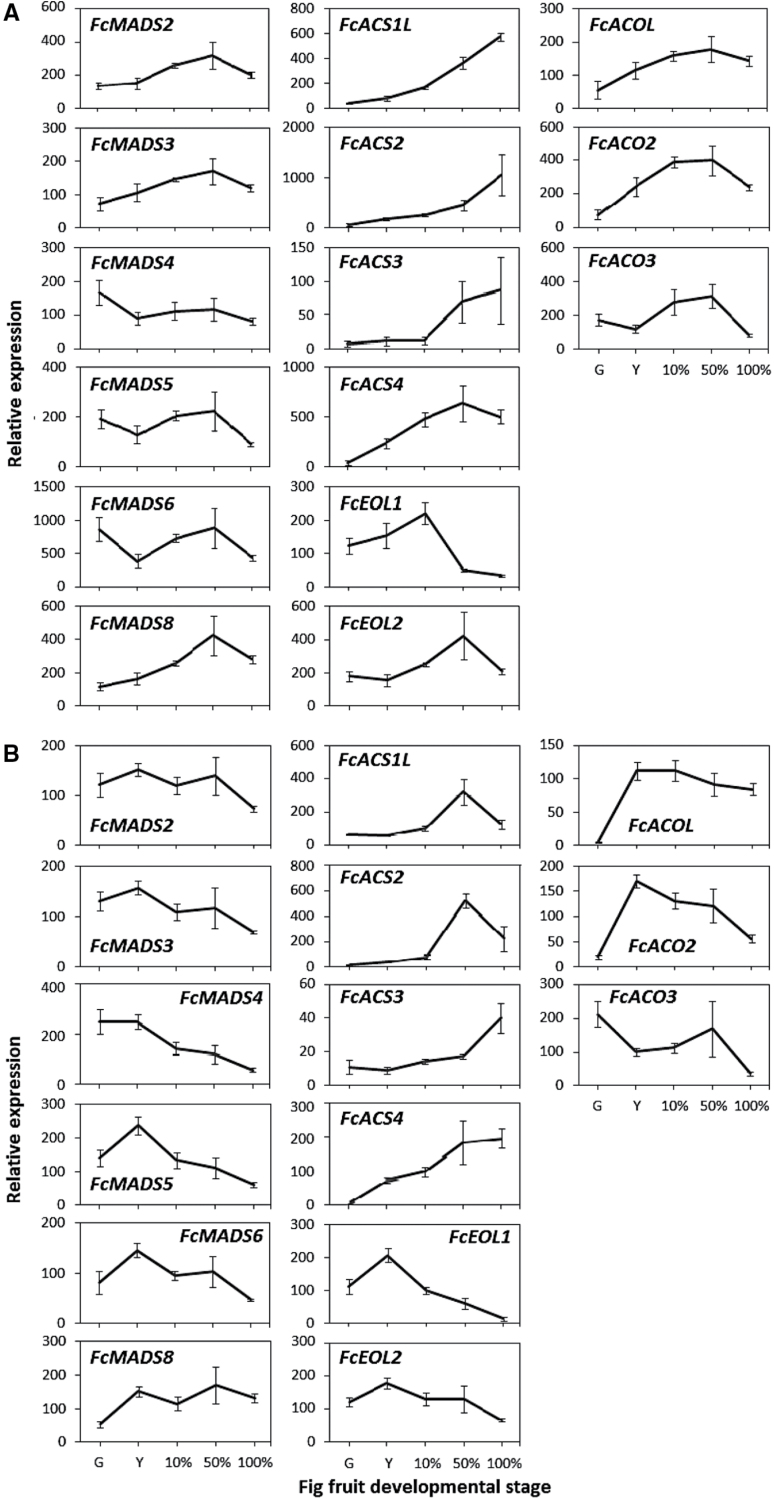
To find potential ripening-regulator genes in fig fruit, the expression of FcMADS and ethylene-synthesis-related genes was examined during natural on-tree ripening (Fig. 1A). For gene-expression analysis in natural ripening fig, the fruit inflorescence and receptacle were separated. Inflorescence tissue included non-pollinated drupelets, while the receptacle tissue included the coloured outer peel of the fruit (Fig. 1C). The exterior and interior of a single parthenocarpic drupelet can be seen in Fig. 1C. As shown in Fig. 2A, in the fig inflorescence, three MADS-box genes—FcMADS2, FcMADS3, and FcMADS8—were upregulated from the yellow stage toward the 50% purple stage, followed by a minor decrease in expression at the 100% purple stage. FcMADS8 showed a 3.5-fold change in expression from the green stage to the 50% purple stage, while both FcMADS2 and FcMADS8 exhibited higher expression levels than FcMADS3. Three genes (FcMADS4, FcMADS5, and FcMADS6) were downregulated from the green stage to the yellow stage, while FcMADS5 and FcMADS6 presented minor increases at the 10% purple stage and a decrease at the 100% purple stage. Constituting the first exclusive ethylene-synthesis step, all FcACS transcripts in the inflorescence showed low expression levels at the green stage, which were enhanced at some point during fig ripening. FcACS4 gene expression showed the first prominent rise at the yellow stage (fold change of 16.4), and continued to rise at the 10% stage. Both FcACS1L and FcACS2 exhibited their major changes at the latter stages of the ripening process (fold changes of 3.4 and 4, respectively). Expression levels of FcACS3 were lower than those of the other ACS genes; nevertheless, FcACS3 transcription was upregulated 5.7-fold at the 50% purple stage. Post-translational regulators of type 2 ACS proteins—FcEOL1 and FcEOL2—showed transcription peaks at the 10% and 50% purple stages, respectively. Like the FcACSs, FcACOs presented increasing transcription patterns as the fig ripened. Expression of both FcACOL and FcACO2 was enhanced from the green stage to the 50% purple stage (3.2- and 5.3-fold, respectively), while minor decreases were observed at the 100% purple stage. FcACO3 transcription displayed a different pattern, with a minor decrease at the yellow stage followed by upregulation at the 10% purple stage (fold change of 2.5). The high transcript level of FcACO3 was still observed at the 50% purple stage but decreased 3.7-fold at the 100% purple stage.
Fig. 2.
Expression patterns of FcMADS-box and ethylene-synthesis-related genes (ACSs, EOLs, and ACOs) in on-tree-ripening fig fruit. (A) Gene expression in the inflorescence. (B) Gene expression in the receptacle. Developmental fruit stages are as described in Fig. 1A. Average ± SE of three fruits per stage, each in two technical replicates.
In the fig receptacle (Fig. 2B), three MADS-box genes—FcMADS2, FcMADS3, and FcMADS4—were downregulated from the yellow stage toward the 100% purple stage. FcMADS5 and FcMADS6 showed similar patterns from the yellow stage on, but the decreases in transcript levels followed increases from the green to yellow stage for both genes. The expression of FcMADS8 remained high after rising at the yellow stage (fold change of 2.8). Though the rate of FcMADS8 upregulation in the receptacle resembled that in the inflorescence, transcript levels in the inflorescence were higher (Fig. 2A). Transcription of FcACS genes in the receptacle shared a common pattern with the inflorescence, i.e. low expression levels at the green stage and enhancement at some point during fig ripening. Here, too, FcACS4 expression showed the first prominent rise at the yellow stage (fold change of 77), but continued to rise toward the 100% purple stage with 215 times higher levels than at the green stage. Both FcACS1L and FcACS2 exhibited enhanced expression at the 50% purple stage (fold changes of 3.3 and 6.3, respectively) followed by a decrease at the 100% purple stage (fold changes of 2.5 and 2.3, respectively). Expression levels of FcACS3 rose at the 100% purple stage, albeit to lower levels than the other ACSs. In general, transcription levels of ACSs in the receptacle were lower than in the inflorescence. The post-translational regulators of type 2 ACS proteins—FcEOL1 and FcEOL2—both showed peak transcription at the 10% purple stage. Two FcACOs—FcACOL and FcACO2—presented low expression at the green stage and enhanced transcription levels at the yellow stage (fold changes of 22 and 9, respectively). FcACOL expression remained high, while that of FcACO2 decreased toward the 100% purple stage. FcACO3 expression displayed a different pattern, with downregulation at the yellow stage and again at the 100% purple stage.
Expression of ethylene-signal-transduction genes in the fig inflorescence and receptacle during natural on-tree ripening
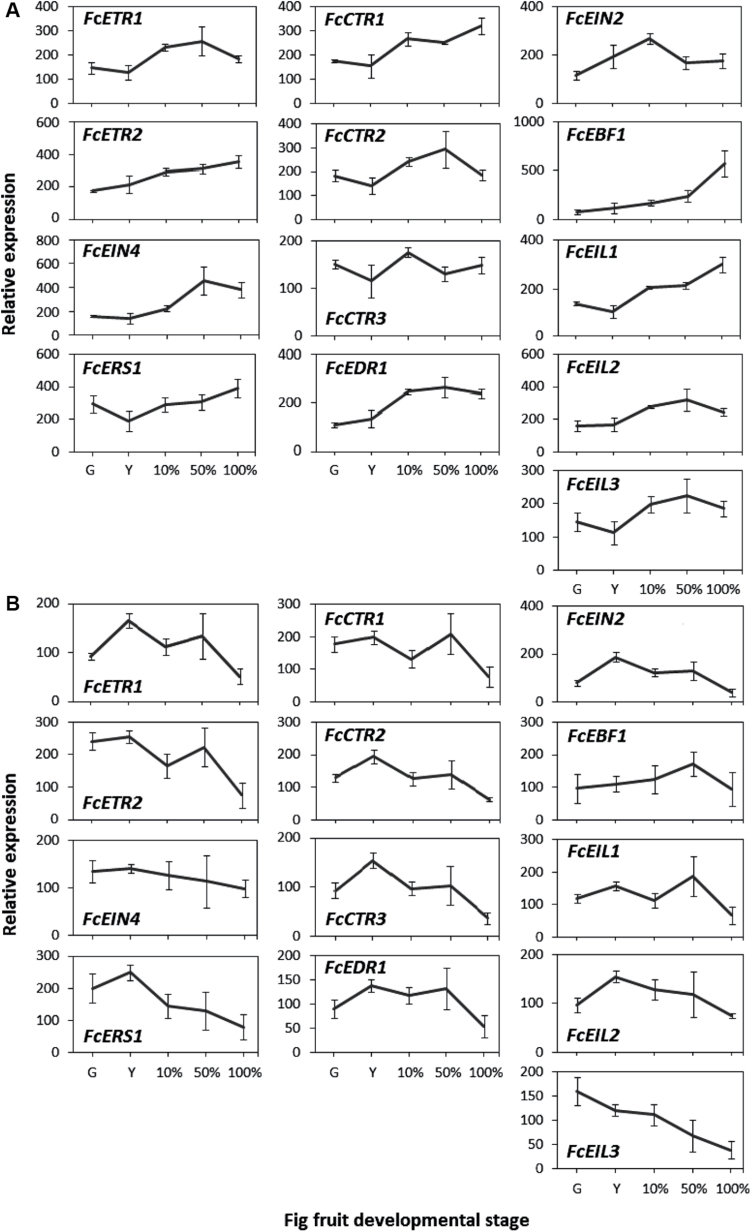
To determine the function of the ethylene-signal-transduction pathway during fig ripening, expression of its genes was examined (Fig. 3). In the inflorescence (Fig. 3A), ethylene receptors showed moderate increases in expression, with FcEIN4 presenting the largest change (3.3-fold) from the yellow stage to the 50% purple stage. The CTRs—FcCTR1, FcCTR2, and FcEDR1—also showed increased expression in the inflorescence during fig ripening. FcCTR1 exhibited the highest change (6-fold) from the yellow stage to the 100% purple stage, and FcCTR3 expression showed minor changes. FcEIN2 was upregulated from the green stage to the 10% stage (2.3-fold) and moderately downregulated at the 50% stage (1.6-fold). Transcription of FcEBF1 gradually increased with fig ripening, with the transcript level at the 100% purple stage being 7.5 times higher than at the green stage. FcEIL genes were also upregulated in the inflorescence during ripening, with FcEIL3 exhibiting the highest change (2.7-fold) from the yellow stage to the 50% purple stage.
Fig. 3.
Expression patterns of ethylene-signal-transduction-related genes in on-tree-ripening fig fruit. Ethylene-receptor genes (FcETR1, FcETR2, FcEIN4, and FcERS1), CTR1-like genes (FcEDR1 and FcCTR1–3), FcEIN2, FcEBF1, and EIN3/EIL (FcEIL1–3). (A) Gene expression in the inflorescence. (B) Gene expression in the receptacle. Developmental fruit stages are as described in Fig. 1A.
In the receptacle, the expression patterns of all ethylene-signal-transduction genes differed from their inflorescence profiles (Fig. 3B). With the exception of FcETR1, there was no detectable increase in expression of the ethylene receptors during ripening. A minor (1.7-fold) rise in FcETR1 expression in the receptacle was restricted to the yellow stage. Later on, the FcETR1 transcript level decreased: at the 100% purple stage, it was even lower than its starting point—the green stage. Both FcETR2 and FcERS1 shared this decrease from the yellow stage to the 100% purple stage (fold changes of 3.4 and 3.1, respectively). No apparent change was observed in FcEIN4 expression in the receptacle. The CTR genes—FcCTR2, FcCTR3, and FcEDR1—also showed gradual decreases in expression from the yellow stage to the 100% purple stage (fold changes of 3, 4, and 2.5, respectively). FcCTR1 showed a 2.7-fold decline in transcription from the 50% purple stage to the 100% purple stage. FcEIN2 was upregulated 2.3-fold from the green stage to the yellow stage and then gradually downregulated toward the 100% purple stage (4.7-fold). Transcription levels of FcEBF1 presented minor changes in the receptacle as the fig ripened, while FcEILs exhibited different transcription patterns. FcEIL1 expression was downregulated 2.7-fold at the 100% purple stage, FcEIL2 showed a minor expression peak at the yellow stage, and FcEIL3 transcription was 4 times lower at the 100% purple versus green stage.
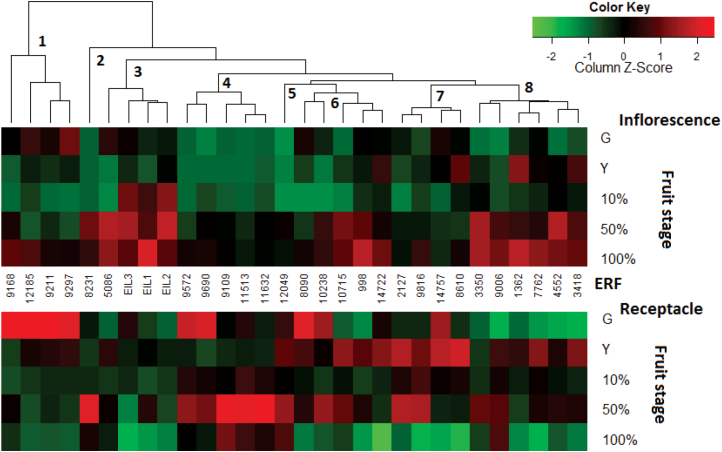
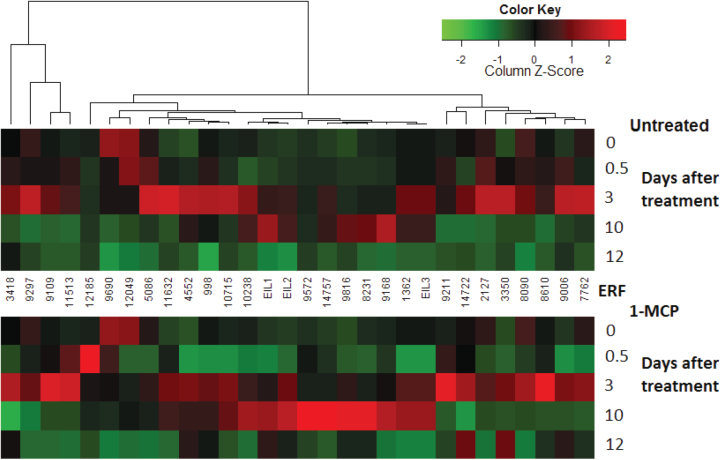
Comprising the last step in ethylene-signal transduction, the transcription patterns of the 27 examined FcERFs differed between the inflorescence and the receptacle (Fig. 4). The FcERFs and FcEILs were clustered according to their expression patterns. Cluster 1 consisted of FcERFs with higher expression levels in the receptacle than in the inflorescence at the green stage, but higher levels in the inflorescence at the 100% purple stage. Cluster 2 only contained FcERF8231, which showed similar patterns in both sampled tissues. Cluster 3 (which included the three FcEILs) and cluster 8 showed higher transcription levels in the inflorescence at later ripening stages, whereas cluster 4 exhibited the opposite trend, with higher expression levels in the receptacle at the later ripening stages. Cluster 5 only contained FcERF12049, which showed higher transcription levels in the receptacle during all ripening stages. Cluster 6 contained genes with elevated transcript levels toward ripening completion in the inflorescence compared to the receptacle, in which levels were high at the yellow and 50% purple stages. Minor changes in transcription in the inflorescence were evident for the genes in cluster 7, while in the receptacle, elevated transcription was evident at the yellow stage and at the 50% purple stage in the case of FcERF2127 and FcERF9816. To associate FcEILs and their targets, FcERFs, FcEILs were also clustered with FcERFs. As mentioned, all three EILs were designated to cluster 3.
Fig. 4.
Expression patterns of ethylene-responsive factor genes (ERFs) in on-tree-ripening fig fruit. Numbers indicate clusters. Developmental fruit stages are as described in Fig. 1A.
Gene expression of MADS-box and ethylene-synthesis-related genes following preharvest 1-MCP treatment, storage, and shelf simulation
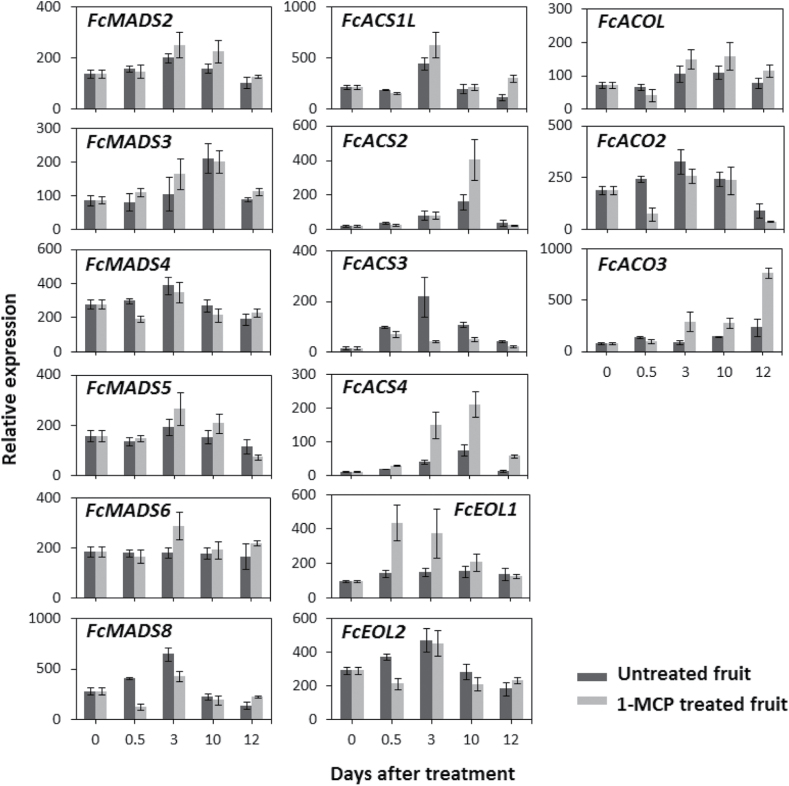
To determine ethylene sensitivity of ripening-regulation pathways in fig and their relationship with the improved storability of preharvest 1-MCP-treated fruit, the expression of FcMADS and ethylene-synthesis-related genes was examined. Gene-expression analysis in preharvest 1-MCP-treated stored fruit was performed on whole fruit; sampling was performed as presented in Fig. 1B. As shown in Fig. 5, out of the six MADS-box genes examined, only FcMADS3 was upregulated after storage in untreated as well as treated fruit. Expression of FcMADS2, FcMADS4, FcMADS5, and FcMADS8 declined after storage and even further after shelf simulation in untreated fruit, whereas the FcMAD6 expression level did not change. The effect of 1-MCP treatment was observed in the transcription patterns of FcMADS4, FcMADS6, and FcMADS8. While FcMADS4 and FcMADS8 expression was downregulated 0.5 days after treatment relative to untreated fruit, FcMADS6 expression was upregulated on day of harvest (3 days after treatment) following preharvest 1-MCP treatment. The decreased levels of FcMADS8 transcript in treated fruit were still evident 3 days later, on day of harvest. With respect to the ethylene-synthesis pathway after storage, two FcACSs—FcACS1L and FcACS3—exhibited decreased transcript levels after storage and 2 days later, after shelf simulation. On the other hand, the expression levels of FcACS2 and FcACS4 peaked after storage. As for 1-MCP treatment effects on FcACS transcripts, expression of FcACS2 and FcACS4 was upregulated and that of FcACS3 downregulated in treated versus untreated fruit 0.5 days after treatment. The upregulation of FcACS2 transcription was restricted to the post-storage sample (10 days after treatment), while FcACS4 upregulation was observed from day of harvest onwards. FcACS3 expression did not change following 1-MCP treatment, in contrast to the evident expression peak on day of harvest in untreated fruit. Post-translational regulators of type 2 ACS proteins—FcEOL1 and FcEOL2—showed different patterns following 1-MCP treatment (0.5 days after treatment) and in stored untreated fruit (10 days after treatment). FcEOL1 expression increased very little in untreated fruit during the experiment. Following 1-MCP treatment, a sharp upregulation in FcEOL1 transcription was observed and was still evident after storage. By comparison, FcEOL2 transcription levels peaked on day of harvest in untreated fruit, and 1-MCP treatment temporarily downregulated this gene’s transcription (0.5 days after treatment). FcACOL expression underwent a moderate change in the untreated fruit following storage, whereas FcACO2 expression decreased after storage. Compared to FcACOL transcription, which was not affected by 1-MCP treatment, FcACO2 transcription was temporarily downregulated by the treatment, but its levels then increased, reaching those in the untreated fruit. The moderate increase observed in FcACO3 expression in the untreated fruit after storage was greatly enhanced by 1-MCP application.
Fig. 5.
Expression patterns of FcMADS-box genes and ethylene-synthesis-related genes (ACSs, EOLs, and ACOs) in preharvest 1-MCP-treated and untreated fruit (at ripening-onset fruit stage) followed by 1 week in cold storage and 2 days of shelf simulation. Days after treatment are as described in Fig. 1B. Average ± SE of three fruits per treatment/control, each in two technical replicates.
Gene expression of ethylene-signal-transduction genes following preharvest 1-MCP treatment, storage, and shelf simulation
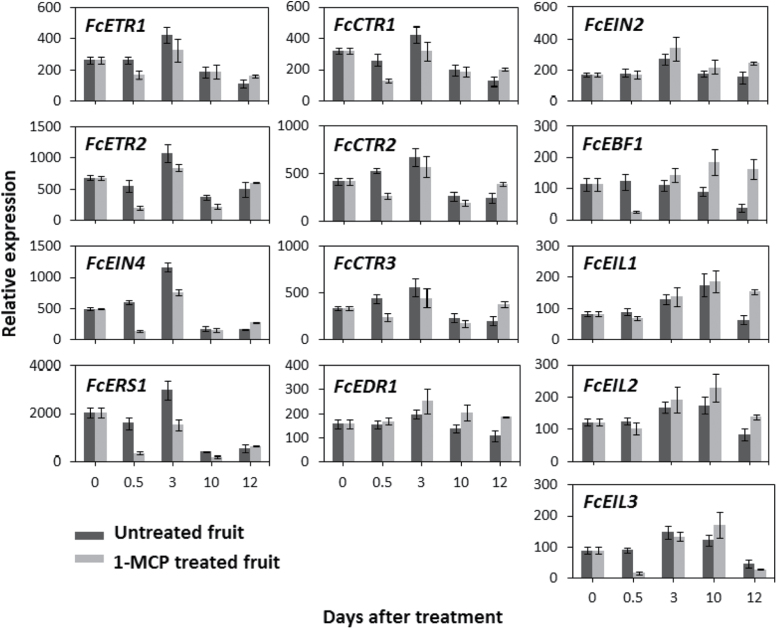
To investigate the ethylene sensitivity of the ethylene-signal-transduction pathway in fig and its relationship with the improved storability of preharvest 1-MCP-treated fruit, expression of ethylene-signal-transduction genes was examined. Following storage (10 days after treatment) of untreated fruit (Fig. 1B), transcription levels of ethylene-receptor genes and FcCTRs were reduced (Fig. 6). These upstream signal-transduction components, except for FcEDR1, were downregulated following preharvest 1-MCP treatment (0.5 day after treatment). The downregulation effect of 1-MCP on FcEIN4 and FcERS1 expression was still evident on day of harvest (3 days after treatment), whereas FcCTR1, FcCTR2, and FcCTR3 presented moderately higher levels in the treated versus untreated fruit after shelf simulation (12 days after treatment). As for FcEDR1 expression, higher levels were found in 1-MCP-treated fruit after storage and shelf simulation relative to untreated fruit. The transcription pattern of FcEIN2 was not significantly affected by 1-MCP treatment and generally presented a peak on day of harvest in both treated and untreated fruit; after storage, its expression level in treated fruit was moderately higher than in untreated fruit (1.5-fold change). Expression of FcEBF1 showed a unique reaction to 1-MCP treatment: at 0.5 days after treatment, its expression level in the treated fruit was 5 times lower than in the untreated fruit, but after storage its expression level in the treated fruit was twice that in the untreated fruit. The upregulation of FcEBF1 in treated fruit was even more pronounced after shelf simulation, when transcription in the treated fruit was 4.5 times higher than in the untreated fruit. FcEIN2, FcEIL1, and FcEIL2 presented similar trends in treated and untreated fruit, with minor upregulation in treated fruit, compared to untreated, after shelf simulation. FcEIL3, by contrast, showed transient downregulation of transcription in treated fruit 0.5 days after treatment.
Fig. 6.
Expression patterns of ethylene-signal-transduction-related genes in preharvest 1-MCP-treated and untreated fruit (at ripening-onset fruit stage) followed by 1 week in cold storage and 2 days of shelf simulation. Ethylene-receptor genes (FcETR1, FcETR2, FcEIN4, and FcERS1), CTR1-like genes (FcEDR1 and FcCTR1–3), FcEIN2, FcEBF1, and EIN3/EIL genes (FcEIL1–3). Days after treatment are as described in Fig. 1B. Experimental design is as described in Fig. 5.
Given that FcERFs are downstream regulators of fruit ripening, which respond to ethylene by definition, their reaction to preharvest 1-MCP treatment was surprisingly mild (Fig. 7). Transcription of most of the FcERFs was temporarily downregulated by the treatment in fruit after on-tree overnight exposure (0.5 days after treatment) compared to untreated fruit. Transcription in the treated fruit was restored on day of harvest at the 50% purple stage. One exception was FcERF12185, whose transcription increased sharply in treated fruit (0.5 days after treatment) compared to untreated fruit. Its higher transcript level in treated fruit continued to day of harvest, but was downregulated to the level in untreated fruit after 1 week of storage. To associate FcERF expression patterns and FcEIL transcription levels following preharvest 1-MCP application, FcEILs were clustered with FcERFs. Unlike the naturally on-tree-ripening fig, in which all FcEILs clustered together, 1-MCP treatment led to branching of the FcEILs into different clusters. FcEIL3 was located in a different cluster than FcEIL1 and FcEIL2, although the difference in expression patterns was moderate, because the difference between the clusters was moderate, as already noted.
Fig. 7.
Expression patterns of ethylene-responsive factor genes (ERFs) in preharvest 1-MCP-treated and untreated fruit (at ripening-onset fruit stage) followed by 1 week in cold storage and 2 days of shelf simulation. Days after treatment are as described in Fig. 1B. Experimental design is as detailed in Fig. 5.
Discussion
The ripening process in fig fruit is categorized as climacteric, showing a rise in respiration rate and ethylene production at the onset of the ripening phase. Surprisingly, ripening-related ethylene production increases following pre- or postharvest 1-MCP application in an unexpected auto-inhibitory manner (Sozzi et al., 2005; Owino et al., 2006; Freiman et al., 2012). This phenomenon supports the more recent notion that classification of fruits based on ethylene production is not very clear-cut (Paul et al., 2012). Unlike most climacteric fruits, such as apple, Pyrus communis (pear), and banana, fig harvested before ripening onset will not ripen postharvest; therefore, commercial crops are harvested at the cultivar’s specific stage to reach ripe but not overripe stage (Flaishman et al., 2008). In the current study, ethylene-related genes involved in the natural process of ripening in attached fruit were investigated. The analysis was expanded to preharvest 1-MCP-treated fruit at the ripening-onset stage and commercially mature fruit picked and stored following this treatment. The expression of fig MADS-box and ethylene-related genes was examined to understand their functions in fig fruit and to further explore ethylene- and ripening-regulation mechanisms in this unique fruit.
Members of the MADS-box gene family have been found to regulate ripening in several climacteric species: RIN and TAGL1 in tomato, PLENA in Prunus persica (peach), MADS1–5 in banana and MADS8, and MADS9 in apple (Vrebalov et al., 2002, 2009; Itkin et al., 2009; Tadiello et al., 2009; Elitzur et al., 2010; Ireland et al., 2013). MADS-box protein activity is not restricted to ethylene-pathway regulation; direct targets of RIN, FUL1, and FUL2 in tomato include downstream metabolic genes, such as those from the carotenoid-synthesis pathway, as well as several transcription factors (Fujisawa et al., 2014). In addition, genes from the MADS-box family are involved in non-climacteric fruit ripening, such as MADS9 in Fragaria ananassa (strawberry) and CaMADS-RIN in Capsicum annuum (pepper) (Seymour et al., 2011; Dong et al., 2014). In light of the central role of MADS-box genes in fruit ripening, eight MADS-box transcripts were previously identified and partially isolated from on-tree-developing fig fruit (Freiman et al., 2014). In the present study, the transcripts of six of those genes were quantified to establish their function in fig fruit-ripening regulation. Among the examined FcMADS-box genes, the deduced amino acid sequences of FcMADS6 and FcMADS8 showed the highest homology to SlRIN among FcMADS genes (Freiman et al., 2014). One of them, FcMADS8, presented increasing expression levels as ripening progressed in both the inflorescence and receptacle, resembling the pattern of SlRIN transcription in ripening tomato (Fig. 2; (Vrebalov et al., 2002; Martel et al., 2011; Yan et al., 2013)). Moreover, among the FcMADS genes with increasing transcription during fig ripening, inhibition of FcMADS8 transcription by 1-MCP continued until 3 days after treatment (Fig. 5), indicating ethylene-induced behaviour similar to SlRIN (Yan et al., 2013). Considering the high resemblance of the predicted FcMADS8 protein to SlRIN (75% similarity, Supplementary Fig. S4), and the parallel expression patterns of these genes, including the prolonged ethylene sensitivity of FcMADS8 transcription, this gene may well be the homologue of SlRIN, regulating the ripening process in fig fruit. As such, its downregulation following 1-MCP treatment could contribute to the improved storability of preharvest 1-MCP-treated fruit.
In climacteric fruits and senescing flowers, ethylene production is classified as system 2 autocatalytic ethylene synthesis (Yang and Hoffman, 1984). In the climacteric tomato model, ethylene-synthesis genes are associated with system 2 according to their upregulation during fruit ripening and downregulation in response to 1-MCP treatment (Yokotani et al., 2009; Van de Poel et al., 2012). This is also the case with ethylene-synthesis genes in apple (Yang et al., 2013). Surprisingly, in the climacteric fig, ripening-related ethylene production increased following pre- or postharvest 1-MCP application in an unexpected auto-inhibitory reaction. Fig ethylene-synthesis genes, three ACS genes, and one ACO gene were partially isolated by Owino et al. (2006), and their expression patterns were studied postharvest and following several treatments. All ethylene-synthesis genes in the study exhibited elevated transcript levels after harvest, while FcACS2 expression was induced by postharvest 1-MCP treatment. To expand our knowledge of the function of ethylene-synthesis genes during fig ripening, expression patterns of four ACS genes and three ACO genes were examined (FcACS4, FcACO2, and FcACO3 are newly presented here). All ethylene-synthesis genes seemed to be active during system 2 ethylene synthesis, increasing in the inflorescence during on-tree ripening (Fig. 2A). With the exception of FcACO3, expression of all of the genes increased in the receptacle as well, albeit to lower levels (Fig. 2B). Ethylene synthesis in fig, which is enhanced by 1-MCP treatment, can be related to FcACS2, FcACS4, and FcACO3, all of which were upregulated following treatment (Fig. 5). The unique combination of climacteric ripening alongside auto-inhibitory ethylene production has also been documented in postharvest ripening banana fruit (Inaba et al., 2007). The origin of the 1-MCP-enhanced ethylene levels was found to be the banana pulp; in the peel, ethylene production was inhibited by 1-MCP treatment. The expression pattern of MaACS1 was found to correspond to the difference between the tissues’ ethylene reactions to 1-MCP. In addition, MaACS1 was downregulated in the pulp once ethylene synthesis in the control fruit had started, as expected with this gene’s auto-inhibitory regulation. It is tempting to assume that, in fig, ethylene synthesis is differentially affected by 1-MCP in the inflorescence and receptacle. That said, ethylene-synthesis genes in the two tissues only showed distinct magnitudes of otherwise similar expression patterns in on-tree-ripening figs (Fig. 3). Moreover, examination of ethylene-synthesis genes in separate tissues following preharvest 1-MCP application showed that upregulation of ethylene-synthesis genes is not restricted to either tissue (data not shown). In light of these findings, the duality of the ethylene characteristics in fig is suspected to be a consequence of an overcomplicated feedback mechanism that is not specific to certain genes, or tissues. Ethylene synthesis is also influenced by ETO1 and EOL proteins that regulate type 2 ACSs post-translation (McClellan and Chang, 2008). Here, FcEOL1 and FcEOL2 were identified in addition to ACS and ACO genes. According to their expression patterns, these genes may serve as ethylene-synthesis regulators in ripening fruit but not in relation to the ethylene auto-inhibitory mechanism (Figs. 3 and 6). In fact, tomato ETO1 expression is restricted to the fully ripe fruit stage, whereas in fig both genes are active during the ripening period (Fig. 2), presumably regulating the type 2 ACSs FcACS3 and FcACS4. To the best of our knowledge, this is the first example of EOL genes that are active as fruit ripening progresses.
Compared to ethylene synthesis, ethylene-signal transduction is much more complex. Expression patterns of tomato ethylene receptors during fruit ripening have shown that three out of seven receptors have increased transcription levels in parallel to increasing levels of ethylene (Kevany et al., 2007). Because ethylene receptors function as ethylene-response inhibitors (Kevany et al., 2007), the elevation in receptor expression during ripening presents a feedback inhibitory mechanism of the hormone by signal-transduction components. As such, silencing of two of these receptors with a fruit-specific promoter causes an early-ripening phenotype (Kevany et al., 2007, 2008). Like ethylene receptors, CTRs function as ethylene-response inhibitors and their transcription induction during tomato ripening implies a role in the feedback inhibitory mechanism of that process (Merchante et al., 2013). The protein CTR phosphorylates EIN2 to inactivate it, while EIN2 is a positive regulator of ethylene response (Ji and Guo, 2013). In tomato, expression of EIN2, the only gene of its kind, increases at the onset of ripening (Gapper et al., 2013). In Arabidopsis, EIN2 is a target for protein turnover via the proteosome (Qiao et al., 2009) though, to date, no homologues for its targeting proteins have been found in tomato. Another downstream positive ethylene-response regulator is EIN3; as such, antisense suppression of tomato EIL1, EIL2, and EIL3 reduced ethylene sensitivity (Tieman et al., 2001). Positively regulating the ethylene response, EIL genes are upregulated in ripening tomato (Yokotani et al., 2003). Like EIN2, EIN3 is targeted for protein turnover; thus, repression of SlEBF1/SlEBF2—the EILs’ targeting proteins—resulted in a constitutive ethylene response and early ripening (Yang et al., 2010). The last step in the ethylene-signal-transduction pathway is ERF activation by EIN3/EILs. The ERF family is large, comprising both repressors and activators, with a degree of functional redundancy among its members (Klee and Giovannoni, 2011; Licausi et al., 2013). On the one hand, SlERF1 positively regulates fruit ripening and softening in tomato fruit; on the other hand, SlERF6 has been characterized as a negative regulator of ethylene and carotenoid biosynthesis, while SlERF.B3 has contrasting effects on tomato-ripening processes (Li et al., 2007; Lee et al., 2012; Liu et al., 2014). These studies demonstrate the balancing role of the ERF family, contributing to the complexity of ripening regulation in climacteric fruit. The ERF family will be discussed separately further on.
This study represents a first attempt to follow the expression patterns of a large group of genes involved in the fig’s ethylene-signal-transduction cascade. In general, fig ethylene-signal-transduction genes presented expression patterns similar to those in tomato and apple during ripening (Klee and Giovannoni, 2011; Yang et al., 2013). However, this resemblance was restricted to the inflorescence (Fig. 3A). In the receptacle, minor changes compared to those in the inflorescence resulted in opposing patterns in most genes (Fig. 3B). The correspondence between ethylene-signal-transduction genes in the fig inflorescence and tomato suggests a similar feedback mechanism for the hormone by its receptors and FcCTR components. Ethylene-signal transduction in fig is probably subjected to negative post-translational regulation by FcEBF1, as presented here. FcEBF1 expression levels rose in the fig inflorescence at the ripe stage, presumably to negatively control FcEIL proteins (Fig. 3A). Interestingly, this gene was downregulated following preharvest 1-MCP treatment, although it was upregulated again after storage (Fig. 6). FcEBF1 is assumed to be one of the key genes responsible for the improved storability conferred by preharvest 1-MCP application, along with FcEIL3, which is the only FcEIL gene that was downregulated following preharvest 1-MCP treatment (Fig. 6). Regarding the effect of preharvest 1-MCP treatment on ethylene production, the mentioned upstream members of the signal-transduction pathway (receptors, CTRs, EIN2, EBF1, and EILs) give few clues to the fig’s specific regulatory network. Downregulation of ethylene-receptor and CTR transcription is evident in 1-MCP-treated tomato and apple, both attached and detached, with both fruit showing the classical climacteric autocatalytic ethylene reaction, unlike the ambiguous reaction of fig fruit to 1-MCP (Varanasi et al., 2013; Yan et al., 2013; Yang et al., 2013). The auto-inhibitory nature of ethylene in ripening fig may therefore be a consequence of the downstream regulators of ethylene-signal transduction—the ERFs.
As noted above, the ERF family is highly complex, contributing to the fine regulation of climacteric fruit ripening. In the FcERFs analysis during on-tree-ripening, cluster 1 consists of FcERFs that regulate processes in the receptacle towards ripening onset, but not during the ripening period. Cluster 2 only contains FcERF8231, regulating processes towards the commercially ripe stage in both receptacle and inflorescence. Clusters 3 and 8 represent FcERFs controlling activities in the inflorescence after the initiation of fig colour change, whereas cluster 4 exhibited this trend in the receptacle. Clusters 5 and 7 are more active in the receptacle than in the inflorescence, while cluster 6 ERFs regulate processes towards ripening completion in the inflorescence, but in the receptacle this cluster is active at the yellow and 50% purple stages. The fact that all three EILs were designated to a single cluster may indicate the existence of additional EIL genes in the ripening fig or regulation that includes more than one component in a complex cascade. Only one FcERF, namely FcERF12185, was upregulated in response to preharvest 1-MCP treatment in yellow-stage figs, while its transcripts remained at higher levels than in the untreated fruit on harvest day, 3 days after treatment (Fig. 7). This ERF may well be responsible for the burst in ethylene synthesis in treated fruit following treatment, targeting ethylene-synthesis genes as well as activating other positive regulators of ethylene synthesis, resulting in higher ethylene levels after storage and shelf simulation. In the fruit that ripened naturally on the tree, FcERF12185 transcription only increased at the fully ripe stage, meaning that it is probably not responsible for the climacteric ethylene rise or metabolic ripening processes at early ripening stages. It may be associated with the regulation of system 1, rather than system 2 ethylene synthesis. None of FcERF12185 homologs found have specific functions related to fruit development, ethylene regulation, or otherwise (through BLASTX against the nr collection in NCBI, data not shown). Since ERF protein structures are diverse and their action depends on the promoter sequences of their target genes, further investigation of this unique FcERF, its targets, and its function should unravel its role in the non-climacteric behaviour of the ‘climacteric’ fig fruit. Regarding the temporary downregulation of most FcERFs following preharvest 1-MCP treatment (Fig. 7), one might ask whether this minor difference from the untreated fruit is responsible for the high quality of treated stored fruit. Though some FcERFs reach high expression levels after storage (Fig. 7), it is proposed that the ripening-retarded fig subjected to storage may be less vulnerable to storage damage than the untreated fruit, in which tissues are in a more advanced stage of ripening. As such, untreated fruit have a shorter shelf-life and their storability is lower.
The different structures of fleshy fruits are defined according to the flower organ’s fate in the developing fruit (Esau, 1977). The tomato fruit is an example of a true fruit, developed from the ovary with no accessory tissue. The fig, on the other hand, is an accessory multiple fruit composed of individual drupelets developed from the ovaries in a closed inflorescence, the syconium (Fig. 1C). The surrounding receptacle, which constitutes a large portion of the edible fruit flesh, is the visible part of the enclosed inflorescence/fruit as the syconium develops (Condit, 1947). A few studies have shown that the molecular events regulating ripening processes exhibit different profiles in the different tissues of accessory fruits. In apple, MADS-box and ethylene-synthesis genes present different transcription levels between the core and the cortex, and the same phenomenon has been documented for MADS-box gene expression in pear (Ireland et al., 2013; Ubi et al., 2013). Diverse gene-expression patterns in banana peel and pulp were mentioned above and even in ripening tomato fruit, tissue-specific trends of ethylene-synthesis activity have been recently found (Van de Poel et al., 2014). In this study, expression trends of most of the examined genes were dissimilar in the fig inflorescence and receptacle. In the inflorescence, as mentioned, FcMADS-box genes as well as ethylene-synthesis and signal-transduction genes showed patterns characteristic of climacteric fruits. In the receptacle, this was only true for FcMADS8 and the ethylene-synthesis genes (with the exception of FcACO3). That said, the fig drupelets developed inside the syconium are proposed to function as parthenocarpic true fruit, regulating ripening processes for the whole accessory fruit. As such, the inflorescence may produce higher ethylene levels than the receptacle. Given that ethylene can diffuse freely between cells, it is assumed that the activity of FcMADS8 and ethylene-synthesis genes in the receptacle is a reaction to the ethylene produced in the inflorescence at ripening onset.
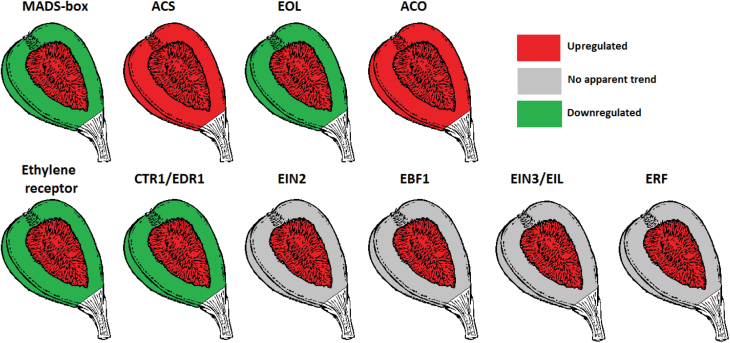
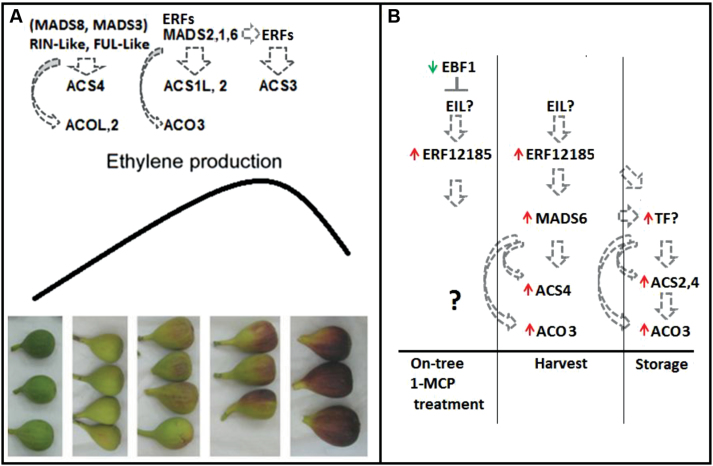
To conclude, the expression patterns of ethylene-synthesis and signal-transduction genes in fig were similar to those in tomato and apple during ripening, specifically in the fig inflorescence–drupelet section, as summarized in Fig. 8. FcMADS8 shares several features with the well-studied SlRIN; as such it is a potential key regulator of ripening onset and events. The auto-inhibition reaction of ethylene production may be related to the direct functions of FcACS2, FcACS4, and FcACO3, as detailed in Fig. 9. Genes of the ethylene-signal-transduction cascade in the fig inflorescence present expression patterns similar to those in tomato and apple during ripening, suggesting a similar feedback mechanism of the hormone by its negative regulators of signal transduction. FcMADS8, FcEBF1, and FcEIL3 are proposed to be the key genes responsible for the improved storability of preharvest 1-MCP-treated fruit. Several FcERFs were also shown to be suppressed following 1-MCP treatment. In addition to the association of ethylene-synthesis genes to the ethylene profile during ripening and following 1-MCP application, a possible regulator of the feedback reaction is proposed, namely FcERF12185 (Fig. 9B). This downstream component of ethylene-signal transduction could play a role in regulating ethylene-synthesis system 1 in reaction to 1-MCP, causing the non-climacteric behaviour of fig ethylene production.
Fig. 8.
Schematic representation of the different gene families’ activities in fig fruit during on-tree ripening. The general expression pattern was concluded from the majority of genes in a family that were up- or downregulated during the ripening process. No apparent trend was defined when expression patterns showed combined trends in one gene family or a changing pattern with ripening.
Fig. 9.
Proposed model of ethylene-regulation and synthesis genes in fig fruit. (A) Proposed ethylene-regulator and ethylene-synthesis gene activity during on-tree ripening. (B) Proposed ethylene-regulation and ethylene-synthesis gene activity following on-tree 1-MCP treatment of ripening-onset fruit, in commercially mature fruit harvested 3 days after treatment, and in stored fruit. Red arrow, upregulated genes in treated fruit compared to untreated fruit; green arrow, downregulated genes in treated fruit compared to untreated fruit.
Supplementary material
Table S1. Fig genes homologous to MADS-box, ethylene-synthesis, and ethylene-signal-transduction genes subjected to gene-expression analysis.
Table S2. Primers used for transcript isolation and sequencing.
Table S3. Primers used for high-throughput real-time quantitative PCR.
Fig. S1. Newly isolated FcACS1L transcript alignment with the published sequence of FcACS1.
Fig. S2. Phylogenetic analysis of FcACS predicted proteins.
Fig. S3. Isolated FcACOL transcript (published previously under NCBI accession number AB307720.1 2007) aligned with the published sequence of FcACO1 (Owino et al., 2006).
Fig. S4. Alignment of deduced FcMADS8 amino acid sequence and SlRIN protein.
Acknowledgements
Funding was provided by the Ministry of Agriculture, Bet Dagan, Israel.
References
- Condit IJ. 1947. The fig . Waltham, MA: Chronica Botanica Co. [Google Scholar]
- Dong T, Chen G, Tian S, Xie Q, Yin W, Zhang Y, Hu Z. 2014. A non-climacteric fruit gene CaMADS-RIN regulates fruit ripening and ethylene biosynthesis in climacteric fruit. PLoS One 9, e95559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elitzur T, Vrebalov J, Giovannoni JJ, Goldschmidt EE, Friedman H. 2010. The regulation of MADS-box gene expression during ripening of banana and their regulatory interaction with ethylene. Journal of Experimental Botany 61, 1523–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau K. 1977. Anatomy of seed plants . New York: John Wiley & Sons. [Google Scholar]
- Flaishman MA, Rodov V, Stover E. 2008. The fig: botany, horticulture, and breeding. Horticultural Reviews 34, 113–196. [Google Scholar]
- Freiman ZE, Doron-Faigenboim A, Dasmohapatra R, Yablovitz Z, Flaishman MA. 2014. High-throughput sequencing analysis of common fig (Ficus carica L.) transcriptome during fruit ripening. Tree Genetics & Genomes 10, 923–935. [Google Scholar]
- Freiman ZE, Rodov V, Yablovitz Z, Horev B, Flaishman MA. 2012. Preharvest application of 1-methylcyclopropene inhibits ripening and improves keeping quality of ‘Brown Turkey’ figs (Ficus carica L.). Scientia Horticulturae 138, 266–272. [Google Scholar]
- Frye CA, Tang D, Innes RW. 2001. Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proceedings of the National Academy of Sciences U S A 98, 373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa M, Shima Y, Nakagawa H, Kitagawa M, Kimbara J, Nakano T, Kasumi T, Ito Y. 2014. Transcriptional regulation of fruit ripening by tomato FRUITFULL homologs and associated MADS box proteins. The Plant Cell 26, 89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapper NE, McQuinn RP, Giovannoni JJ. 2013. Molecular and genetic regulation of fruit ripening. Plant Molecular Biology 82, 575–591. [DOI] [PubMed] [Google Scholar]
- Ikegami H, Habu T, Mori K, Nogata H, Hirata C, Hirashima K, Tashiro K, Kuhara S. 2013. De novo sequencing and comparative analysis of expressed sequence tags from gynodioecious fig (Ficus carica L.) fruits: caprifig and common fig. Tree Genetics & Genomes 9, 1075–1088. [Google Scholar]
- Inaba A, Liu X, Yokotani N, Yamane M, Lu W-J, Nakano R, Kubo Y. 2007. Differential feedback regulation of ethylene biosynthesis in pulp and peel tissues of banana fruit. Journal of Experimental Botany 58, 1047–1057. [DOI] [PubMed] [Google Scholar]
- Ireland HS, Yao J-L, Tomes S, Sutherland PW, Nieuwenhuizen N, Gunaseelan K, Winz RA, David KM, Schaffer RJ. 2013. Apple SEPALLATA1/2-like genes control fruit flesh development and ripening. The Plant Journal 73, 1044–1056. [DOI] [PubMed] [Google Scholar]
- Itkin M, Seybold H, Breitel D, Rogachev I, Meir S, Aharoni A. 2009. TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. The Plant Journal 60, 1081–1095. [DOI] [PubMed] [Google Scholar]
- Jaakola L, Pirttila AM, Halonen M, Hohtola A. 2001. Isolation of high quality RNA from bilberry (Vaccinium myrtillus L.) fruit. Molecular Biotechnology 19, 201–203. [DOI] [PubMed] [Google Scholar]
- Ji Y, Guo H. 2013. From endoplasmic reticulum (ER) to nucleus: EIN2 bridges the gap in ethylene signaling. Molecular Plant 6, 11–14. [DOI] [PubMed] [Google Scholar]
- Kamenetsky R. 1994. Life cycle, flower initiation, and propagation of the desert geophyte Allium rothii. International Journal of Plant Sciences 155, 597–605. [Google Scholar]
- Kevany BM, Taylor MG, Klee HJ. 2008. Fruit‐specific suppression of the ethylene receptor LeETR4 results in early‐ripening tomato fruit. Plant Biotechnology Journal 6, 295–300. [DOI] [PubMed] [Google Scholar]
- Kevany BM, Tieman DM, Taylor MG, Cin VD, Klee HJ. 2007. Ethylene receptor degradation controls the timing of ripening in tomato fruit. The Plant Journal 51, 458–467. [DOI] [PubMed] [Google Scholar]
- Klee HJ, Giovannoni JJ. 2011. Genetics and control of tomato fruit ripening and quality attributes. Annual review of genetics 45, 41–59. [DOI] [PubMed] [Google Scholar]
- Lee JM, Joung JG, McQuinn R, Chung MY, Fei Z, Tieman D, Klee H, Giovannoni J. 2012. Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. The Plant Journal 70, 191–204. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhu B, Xu W, Zhu H, Chen A, Xie Y, Shao Y, Luo Y. 2007. LeERF1 positively modulated ethylene triple response on etiolated seedling, plant development and fruit ripening and softening in tomato. Plant Cell Reports 26, 1999–2008. [DOI] [PubMed] [Google Scholar]
- Licausi F, Ohme-Takagi M, Perata P. 2013. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytologist 199, 639–649. [DOI] [PubMed] [Google Scholar]
- Liu M, Diretto G, Pirrello J, Roustan JP, Li Z, Giuliano G, Regad F, Bouzayen M. 2014. The chimeric repressor version of an Ethylene Response Factor (ERF) family member, Sl‐ERF.B3, shows contrasting effects on tomato fruit ripening. New Phytologist 203, 206–218. [DOI] [PubMed] [Google Scholar]
- Marei N, Crane JC. 1971. Growth and respiratory response of fig (Ficus carica L. cv. ‘Mission’) fruits to ethylene. Plant Physiology 48, 249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel C, Vrebalov J, Tafelmeyer P, Giovannoni JJ. 2011. The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiology 157, 1568–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan CA, Chang C. 2008. The role of protein turnover in ethylene biosynthesis and response. Plant Science 175, 24–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchante C, Alonso JM, Stepanova AN. 2013. Ethylene signaling: simple ligand, complex regulation. Current Opinion in Plant Biology 16, 554–560. [DOI] [PubMed] [Google Scholar]
- Owino WO, Manabe Y, Mathooko FM, Kubo Y, Inaba A. 2006. Regulatory mechanisms of ethylene biosynthesis in response to various stimuli during maturation and ripening in fig fruit (Ficus carica L.). Plant Physiology and Biochemistry 44, 335–342. [DOI] [PubMed] [Google Scholar]
- Paul V, Pandey R, Srivastava G. 2012. The fading distinctions between classical patterns of ripening in climacteric and non-climacteric fruit and the ubiquity of ethylene—an overview. Journal of Food Science and Technology 49, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Chang KN, Yazaki J, Ecker JR. 2009. Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes & Development 23, 512–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodov V, Percelan J, Horev B, Vinokur Y, Ben-Yehoshua S, Yablowich Z, Flaishman MA. 2002. Development of dark figs for export: I. Optimal picking criteria for the ‘Brazilian’ variety. Alon-Hanotea 56, 372–376 (in Hebrew). [Google Scholar]
- Seymour GB, Chapman NH, Chew BL, Rose JKC. 2013. Regulation of ripening and opportunities for control in tomato and other fruits. Plant Biotechnology Journal 11, 269–278. [DOI] [PubMed] [Google Scholar]
- Seymour GB, Ryder CD, Cevik V, Hammond JP, Popovich A, King GJ, Vrebalov J, Giovannoni JJ, Manning K. 2011. A SEPALLATA gene is involved in the development and ripening of strawberry (Fragaria x ananassa Duch.) fruit, a non-climacteric tissue. Journal of Experimental Botany 62, 1179–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzi GO, Abrajan-Villasenor MA, Trinchero GD, Fraschina AA. 2005. Postharvest response of ‘Brown Turkey’ figs (Ficus carica L.) to the inhibition of ethylene perception. Journal of the Science of Food and Agriculture 85, 2503–2508. [Google Scholar]
- Storey WB. 1977. The fig: its biology, history, culture, and utilization . Riverside, CA: Jurupa Mountains Cultural Center. [Google Scholar]
- Tadiello A, Pavanello A, Zanin D, Caporali E, Colombo L, Rotino GL, Trainotti L, Casadoro G. 2009. A PLENA-like gene of peach is involved in carpel formation and subsequent transformation into a fleshy fruit. Journal of Experimental Botany 60, 651–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman DM, Ciardi JA, Taylor MG, Klee HJ. 2001. Members of the tomato LeEIL (EIN3‐like) gene family are functionally redundant and regulate ethylene responses throughout plant development. The Plant Journal 26, 47–58. [DOI] [PubMed] [Google Scholar]
- Ubi BE, Saito T, Bai S, Nishitani C, Ban Y, Ikeda K, Ito A, Moriguchi T. 2013. Characterization of 10 MADS-box genes from Pyrus pyrifolia and their differential expression during fruit development and ripening. Gene 528, 183–194. [DOI] [PubMed] [Google Scholar]
- Van de Poel B, Bulens I, Markoula A, et al. 2012. Targeted systems biology profiling of tomato fruit reveals coordination of the Yang cycle and a distinct regulation of ethylene biosynthesis during postclimacteric ripening. Plant Physiology 160, 1498–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Poel B, Bulens I, Oppermann Y, Hertog ML, Nicolai BM, Sauter M, Geeraerd AH. 2013. S-adenosyl-l-methionine usage during climacteric ripening of tomato in relation to ethylene and polyamine biosynthesis and transmethylation capacity. Physiologia Plantarum 148, 176–188. [DOI] [PubMed] [Google Scholar]
- Van de Poel B, Vandenzavel N, Smet C, et al. 2014. Tissue specific analysis reveals a differential organization and regulation of both ethylene biosynthesis and E8 during climacteric ripening of tomato. BMC Plant Biology 14, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varanasi V, Shin S, Johnson F, Mattheis JP, Zhu Y. 2013. Differential suppression of ethylene biosynthesis and receptor genes in ‘Golden Delicious’ apple by preharvest and postharvest 1-MCP treatments. Journal of Plant Growth Regulation 32, 585–595. [Google Scholar]
- Vrebalov J, Pan IL, Arroyo AJM, et al. 2009. Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. The Plant Cell 21, 3041–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J. 2002. A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296, 343–346. [DOI] [PubMed] [Google Scholar]
- Yan R, Yokotani N, Yamaoka T, Ushijima K, Nakano R, Yano K, Aoki K, Kubo Y. 2013. Characterization of ripening-associated genes using a tomato DNA macroarray, 1-methylcyclopropene, and ripening-impaired mutants. Postharvest Biology and Technology 86, 159–170. [Google Scholar]
- Yang SF, Hoffman NE. 1984. Ethylene biosynthesis and its regulation in higher plants. Annual Review of Plant Physiology 35, 155–189. [Google Scholar]
- Yang X, Song J, Campbell-Palmer L, Fillmore S, Zhang Z. 2013. Effect of ethylene and 1-MCP on expression of genes involved in ethylene biosynthesis and perception during ripening of apple fruit. Postharvest Biology and Technology 78, 55–66. [Google Scholar]
- Yang Y, Wu Y, Pirrello J, Regad F, Bouzayen M, Deng W, Li Z. 2010. Silencing Sl-EBF1 and Sl-EBF2 expression causes constitutive ethylene response phenotype, accelerated plant senescence, and fruit ripening in tomato. Journal of Experimental Botany 61, 697–708. [DOI] [PubMed] [Google Scholar]
- Yokotani N, Nakano R, Imanishi S, Nagata M, Inaba A, Kubo Y. 2009. Ripening-associated ethylene biosynthesis in tomato fruit is autocatalytically and developmentally regulated. Journal of Experimental Botany 60, 3433–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokotani N, Tamura S, Nakano R, Inaba A, Kubo Y. 2003. Characterization of a novel tomato EIN3‐like gene (LeEIL4). Journal of Experimental Botany 54, 2775–2776. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.