Abstract
Background
Acinetobacter baumannii is a major health problem. The most common infection caused by A. baumannii is hospital acquired pneumonia, and the associated mortality rate is approximately 50 %. Neither in vivo nor ex vivo expression profiling has been performed at the proteomic or transcriptomic level for pneumonia caused by A. baumannii. In this study, we characterized the proteome of A. baumannii under conditions that simulate those found in the airways, to gain some insight into how A. baumannii adapts to the host and to improve knowledge about the pathogenesis and virulence of this bacterium. A clinical strain of A. baumannii was grown under different conditions: in the presence of bronchoalveolar lavage fluid from infected rats, of RAW 264.7 cells to simulate conditions in the respiratory tract and in control conditions. We used iTRAQ labelling and LC-MALDI-TOF/TOF to investigate how A. baumannii responds on exposure to macrophages/BALF.
Results
179 proteins showed differential expression. In both models, proteins involved in the following processes were over-expressed: (i) pathogenesis and virulence (OmpA, YjjK); (ii) cell wall/membrane/envelope biogenesis (MurC); (iii) energy production and conversion (acetyl-CoA hydrolase); and (iv) translation (50S ribosomal protein L9). Proteins involved in the following were under-expressed: (i) lipid metabolism (short-chain dehydrogenase); (ii) amino acid metabolism and transport (aspartate aminotransferase); (iii) unknown function (DNA-binding protein); and (iv) inorganic ion transport and metabolism (hydroperoxidase).
Conclusions
We observed alterations in cell wall synthesis and identified 2 upregulated virulence-associated proteins with >15 peptides/protein in both ex vivo models (OmpA and YjjK), suggesting that these proteins are fundamental for pathogenesis and virulence in the airways. This study is the first comprehensive overview of the ex vivo proteome of A. baumannii and is an important step towards identification of diagnostic biomarkers, novel drug targets and potential vaccine candidates in the fight against pneumonia caused by A. baumannii.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-015-1608-z) contains supplementary material, which is available to authorized users.
Keywords: Proteome, Acinetobacter baumannii, Host-pathogen interaction, Ex vivo, Virulence
Background
The World Health Organization has recently identified antimicrobial resistance as one of the three most important problems facing human health. The most common and serious multidrug-resistant pathogens have been encompassed within the acronym “ESKAPE”, which stands for Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa and Enterobacter spp. [1]. Acinetobacter baumannii is an important opportunistic nosocomial pathogen that is often associated with epidemic outbreaks of infection. This organism is frequently pandrug-resistant and is capable of causing substantial morbidity and mortality in patients with severe underlying disease, both in the hospital and in the community [2]. Acinetobacter baumannii is an uncommon but important cause of community-acquired pneumonia, which appears to be a unique clinical entity occurring predominantly in tropical climates. This community-acquired pneumonia appears to be characterized by a fulminant course, with acute onset of dyspnea, cough and fever followed by rapid progression to respiratory failure and shock. The mortality rate is high (40–64 %) [3, 4]. Nosocomial pneumonia is the most important infection caused by A. baumannii and is particularly associated with the application of mechanical ventilatory procedures [5]. The crude mortality rate associated with nosocomial Acinetobacter infections has been reported to range from 20 to 45 % [6, 7]. A prospective study of 240 A. baumannii infections showed that > 90 % of infections were nosocomially acquired and that only 4 % were community acquired; moreover, respiratory track infections were the most common (39.3 %) [8]. Acinetobacter baumannii has a propensity to cause outbreaks, probably because of its ability to survive desiccation and its multidrug resistance, amongst other reasons [9]. Although A. baumannii is only rarely isolated from soil, vegetables, animals, humans and inanimate surfaces that are often in contact with humans [10], the natural habitats of A. baumannii remain to be established [11]. In the community, A. baumannii is a rare colonizer of human skin in temperate climates, although skin carriage is more common in tropical environments [12, 13]. In Australia, wet-season throat carriage of A. baumannii was found in 10 % of community residents with excess levels of alcohol consumption [3]. The bacterium is ubiquitous in the hospital setting (e.g., bedside, bedrail, ventilator, infusion pump, pillow, resuscitation equipment, washbasins) [14, 15]. Furthermore, A. baumannii is found to persist as a contaminant of the hands, gloves and gowns of healthcare workers [16, 17]. This bacterium can survive on inanimate objects for long periods, even after exposure to dry conditions [18] and is also capable of resisting physical and chemical disinfection, often by forming a biofilm [19]. It has a remarkable ability to up-regulate or acquire resistance determinants, making it one of the most important microorganisms threatening the current antibiotic era [11]. Numerous outbreaks of pandrug-resistant A. baumannii have been documented in Asian and Middle East hospitals. Resistance to both tigecycline and polymyxin B (drugs relied on heavily to treat infection with A. baumannii) already exists in these regions [20, 21].
The first genome of an Acinetobacter spp. to be sequenced was that of the highly transformable Acinetobacter spp. strain ADP1 in 2004 [22]. The first completed A. baumannii genome was reported for strain ATCC 17978 in 2007 and included a considerable number of the island-containing genes (16) implicated in virulence, indicating that several of the microorganism’s genes are devoted to pathogenesis. The largest island contains elements homologous to the Legionella/Coxiella Type IV secretion apparatus. Type IV secretion systems have been demonstrated to be important for virulence in other microorganisms and are thus likely to help mediate the pathogenesis of A. baumannii [23]. Several extracellular proteins of A. baumannii, grown in vitro, have been identified by the use of different proteomic approaches [24, 25]. Such approaches have been used for membrane and cytoplasmic proteomic analysis of A. baumannii grown in vitro [26–30]. Comparative proteomic analysis has been performed with cells at three different stages of in vitro growth: exponential, late stationary phase and as biofilms [31]. Comparative proteomic analysis of a multidrug resistant strain with a drug-sensitive strain has also been performed [32]. In vitro culture conditions may stress bacteria (e.g. due to exhaustion of static nutrients, build-up of toxic bacterial by-products and limited physical space). However, simulated in vitro environments do not accurately reflect the protein profile within the lung, because A. baumannii must adjust to the environment inside the host lung (e.g., CO2 concentrations, temperature and immune system) to be able to cause pneumonia.
The goal of this study was to improve our knowledge of the pathogenesis and virulence of A. baumannii by considering how the bacterium adapts to its host. In order to obtain an overview of host-pathogen-interactions during A. baumannii infections, we used, for the first time, a proteomics approach to compare the ex vivo proteomes of A. baumannii grown under different that mimic the physiological conditions that bacteria must face during in vivo host infection.
Results
The objective of this study was to improve our knowledge of the pathogenesis and virulence of A. baumannii and to investigate how this bacterium adapts to its host. For this purpose, A. baumannii cells were cultured in LB (Luria-Bertani) medium with or without BALF (BALF model) and in Dulbecco’s Modified Eagle’s Medium (DMEM) medium with or without RAW 264.7 cells (macrophage model). The proteome composition of the A. baumannii cells in both models was analyzed in parallel, by using iTRAQ labelling and LC-MALDI-TOF/TOF to investigate the response of A. baumannii to exposure to macrophages/BALF (according to the scheme shown in Additional file 1).
Acinetobacter baumannii caused consolidated pneumonia
BALF contains lipids, nucleic acids, secretomes of macrophages, lymphocytes, neutrophils, eosinophils, epithelial cells, pneumocytes, monocytes and basophils. Moreover, infection is capable of causing changes in concentrations, ratios and numbers of proteins detected in BALF. Therefore, to simulate conditions in the respiratory tract in response to A. baumannii infection, rats were inoculated with the bacterium and BALFs were obtained 21 h later. For histological assessment of A. baumannii infection of the airway, lung specimens were stained with haematoxylin and eosin and examined microscopically. Abscess formation, with extensive infiltration of polymorphonuclear leukocytes was noted [see Additional file 2].
Quantitative (iTRAQ) data analysis
In the BALF model, A. baumannii was incubated simultaneously in LB and in LB supplemented with BALF (containing host soluble components present inside the lung 21 h after inoculation with A. baumannii) for 21 h at 37 °C and 5 % CO2 without shaking, in order to study the A. baumannii responses to soluble BALF components at the protein level by iTRAQ labelling and LC-MALDI/TOF analysis. Changes in the proteome of A. baumannii in response to soluble BALF components were examined. The rate of false positive results was estimated to be less than 1 % (with a confidence interval of 95 %) as the search was conducted in parallel with a decoy database using the “PSPEP on” mode, suggesting a high degree of confidence in the reported protein identifications. To evaluate the reproducibility and effectiveness of the iTRAQ experiments, we compared the proteins identified in each iTRAQ set. Overall, 8,310 unique peptides and 896 distinct proteins were detected with more than 95 % confidence in both iTRAQ sets. In biological replicate one, 730 proteins were identified and in biological replicate two, 853 proteins were identified. Only about 15.7 % of the proteins were identified with a single peptide. In total, 687 proteins (76.7 %) were identified in both biological replicates, and only 209 proteins (23.3 %) were unique for a single iTRAQ experiment. These results indicate the reliability of the iTRAQ identification of 687 proteins in all data sets. Compared with the control group, 111 identified proteins displayed significant changes in expression. In total, 50 proteins tended to be over-expressed (fold change >1.5, p <0.05) and 61 proteins tended to be under-expressed (fold change <0.5, p <0.05) relative to the control group (Table 1).
Table 1.
Differentially expressed proteins under ex vivo conditions in BALF model
| Protein description | AbH12O-A2 locus | Sec.a | BALF model/controlb | BALF model/control p-valuec |
|---|---|---|---|---|
| Pathogenesis and virulence | ||||
| Molecular chaperone DnaK | AIS05048.1 | NO | 1.77 (0.65) | 0.027 |
| Clp protease ClpX | AIS05265.1 | NO | 1.51 (0.61) | 0.024 |
| Amino acid ABC transporter substrate-binding protein (NfuA-like) | AIS05727.1 | YES | 2.38 (1.76) | 0.031 |
| Enoyl-CoA hydratase (PaaZ) | AIS06043.1 | NO | 0.30 (0.11) | 0.012 |
| Peptidase M16 | AIS06751.1 | NO | 0.29 (0.16) | 0.007 |
| Aminopeptidase N | AIS06953.1 | NO | 0.39 (0.21) | 0.023 |
| Protein CsuC | AIS07073.1 | YES | 2.19 (0.76) | 0.003 |
| Protein CsuA | AIS07075.1 | YES | 1.87 (0.95) | 0.036 |
| Membrane protein (OmpA) | AIS07737.1 | YES | 1.51 (0.65) | 0.015 |
| Oligopeptidase A | AIS07885.1 | NO | 0.48 (0.11) | 0.001 |
| ABC transporter ATP-binding protein (YjjK) | AIS08062.1 | NO | 1.74 (0.61) | 0.038 |
| Amino acid metabolism and transport | ||||
| Aspartate-semialdehyde dehydrogenase | AIS05215.1 | NO | 0.25 (0.19) | 0.038 |
| 2-Isopropylmalate synthase | AIS05260.1 | NO | 0.31 (0.11) | 0.034 |
| Ketol-acid reductoisomerase | AIS05310.1 | NO | 0.31 (0.05) | 0.027 |
| Ornithine carbamoyltransferase | AIS06867.1 | NO | 0.40 (0.17) | 0.044 |
| Aspartate aminotransferase | AIS07065.1 | NO | 0.08 (0.07) | 0.000 |
| Glutamate synthase | AIS08029.1 | NO | 0.39 (0.20) | 0.025 |
| Carbohydrate metabolism and transport | ||||
| Phosphoglyceromutase | AIS05033.1 | NO | 0.26 (0.14) | 0.020 |
| Phosphoenolpyruvate synthase | AIS07024.1 | NO | 0.19 (0.04) | 0.000 |
| Glucose dehydrogenase | AIS07744.1 | NO | 0.19 (0.13) | 0.009 |
| Cell cycle control and mitosis | ||||
| Cell division inhibitor MinD | AIS05622.1 | NO | 1.63 (1.55) | 0.000 |
| tRNA uridine 5-carboxymethylaminomethyl modification protein | AIS07041.1 | NO | 2.70 (1.89) | 0.025 |
| Cell division protein FtsZ | AIS08195.1 | NO | 4.61 (1.68) | 0.001 |
| Cell wall/membrane/envelope biogenesis | ||||
| Membrane protein(OmpW) | AIS05087.1 | YES | 2.25 (0.89) | 0.032 |
| UDP-N-acetylglucosamine 1-carboxyvinyltransferase (MurA) | AIS05387.1 | NO | 2.27 (1.47) | 0.002 |
| UDP-N-acetylmuramate-alanine ligase (MurC) | AIS08199.1 | NO | 99.08 (0.00) | 0.020 |
| Coenzyme metabolism | ||||
| Coproporphyrinogen III oxidase | AIS07950.1 | NO | 0.31 (0.13) | 0.004 |
| 3-Phosphoglycerate dehydrogenase | AIS07996.1 | NO | 0.38 (0.09) | 0.016 |
| Defense mechanisms (cellular processes and signaling) | ||||
| RND transporter | AIS04803.1 | YES | 0.30 (0.22) | 0.004 |
| Energy production and conversion | ||||
| Malate dehydrogenase | AIS04961.1 | NO | 0.45 (0.23) | 0.038 |
| Inorganic pyrophosphatase | AIS05013.1 | YES | 2.83 (1.43) | 0.030 |
| Aconitate hydratase | AIS05319.1 | NO | 0.21 (0.16) | 0.000 |
| NAD(P) transhydrogenase subunit alpha | AIS05328.1 | NO | 2.49 (1.37) | 0.041 |
| NADH dehydrogenase | AIS05456.1 | NO | 0.44 (0.17) | 0.012 |
| Cytochrome C oxidase subunit II | AIS07026.1 | NO | 0.21 (0.23) | 0.016 |
| Malic enzyme | AIS07199.1 | NO | 0.46 (0.10) | 0.041 |
| Isocitrate dehydrogenase | AIS07371.1 | NO | 0.29 (0.05) | 0.004 |
| Phosphoenolpyruvate carboxykinase | AIS07529.1 | NO | 2.23 (0.66) | 0.000 |
| Succinyl-CoA synthetase subunit beta | AIS07559.1 | NO | 0.41 (0.06) | 0.035 |
| Energy production and conversion | ||||
| 2-Oxoglutarate dehydrogenase | AIS07562.1 | NO | 0.20 (0.04) | 0.000 |
| Succinate dehydrogenase | AIS07563.1 | NO | 1.98 (0.64) | 0.017 |
| Type II citrate synthase | AIS07567.1 | NO | 0.22 (0.05) | 0.002 |
| Oxidoreductase | AIS07724.1 | NO | 0.31 (0.22) | 0.001 |
| Malate dehydrogenase | AIS07865.1 | NO | 0.47 (0.06) | 0.040 |
| Acetyl-CoA hydrolase | AIS08069.1 | NO | 2.05 (0.67) | 0.008 |
| Pyruvate dehydrogenase | AIS08191.1 | NO | 0.36 (0.06) | 0.000 |
| Unknown function | ||||
| Hypothetical protein | AIS05481.1 | YES | 0.07 (0.10) | 0.013 |
| Peptidoglycan-binding protein LysM | AIS05521.1 | YES | 0.05 (0.03) | 0.003 |
| Hypothetical protein | AIS06125.1 | YES | 0.17 (0.07) | 0.027 |
| DNA-binding protein | AIS07086.1 | YES | 0.08 (0.07) | 0.006 |
| Peptidase | AIS07285.1 | YES | 4.57 (3.14) | 0.000 |
| Glyoxalase | AIS07740.1 | YES | 0.39 (0.45) | 0.015 |
| Hypothetical protein | AIS07742.1 | YES | 1.84 (1.06) | 0.048 |
| General functional prediction only (typically, prediction of biochemical activity) | ||||
| Alpha/beta hydrolase | AIS06658.1 | YES | 0.09 (0.07) | 0.001 |
| GTPase obg | AIS07390.1 | NO | 2.58 (1.05) | 0.025 |
| Hypothetical protein | AIS07569.1 | NO | 3.47 (1.83) | 0.027 |
| Inorganic ion transport and metabolism | ||||
| Bacterioferritin | AIS05502.1 | NO | 0.25 (0.18) | 0.013 |
| Hydroperoxidase | AIS06129.1 | YES | 0.26 (0.10) | 0.000 |
| Inorganic ion transport and metabolism | ||||
| Superoxide dismutase | AIS07203.1 | YES | 0.49 (0.14) | 0.013 |
| Sulfurtransferase | AIS07821.1 | NO | 0.37 (0.28) | 0.006 |
| Intracellular trafficking and secretion | ||||
| RNA-binding protein | AIS05494.1 | NO | 2.07 (0.97) | 0.001 |
| Lipid metabolism | ||||
| 3-Ketoacyl-CoA thiolase | AIS05097.1 | NO | 0.35 (0.18) | 0.049 |
| Multifunctional fatty acid oxidation complex subunit alpha | AIS05098.1 | NO | 0.29 (0.08) | 0.000 |
| Acetyl-CoA carboxylase | AIS05368.1 | NO | 2.01 (0.67) | 0.000 |
| 3-Hydroxyacyl-CoA dehydrogenase | AIS06051.1 | NO | 0.32 (0.22) | 0.023 |
| Beta-ketoadipyl CoA thiolase | AIS06052.1 | NO | 0.28 (0.10) | 0.000 |
| 3-Methylcrotonyl-CoA carboxylase | AIS06111.1 | NO | 0.11 (0.04) | 0.002 |
| Short-chain dehydrogenase | AIS06130.1 | YES | 0.04 (0.05) | 0.040 |
| Acetyl-CoA acetyltransferase | AIS06469.1 | YES | 0.09 (0.13) | 0.027 |
| Acetyl-CoA carboxylase | AIS06900.1 | NO | 0.42 (0.17) | 0.001 |
| 3-Hydroxy-2-methylbutyryl-CoA dehydrogenase | AIS07009.1 | NO | 0.25 (0.18) | 0.031 |
| Acetyl-CoA acetyltransferase | AIS07739.1 | YES | 0.24 (0.26) | 0.017 |
| Acyl-CoA dehydrogenase | AIS07780.1 | NO | 0.22 (0.16) | 0.000 |
| Nucleotide metabolism and transport | ||||
| GMP synthase | AIS04952.1 | NO | 0.31 (0.17) | 0.000 |
| Formyltetrahydrofolate deformylase | AIS05238.1 | NO | 1.64 (1.02) | 0.012 |
| Deoxyuridine 5′-triphosphate nucleotidohydrolase | AIS05628.1 | NO | 4.45 (6.37) | 0.039 |
| Dihydroorotase | AIS05812.1 | NO | 1.79 (1.15) | 0.043 |
| Nucleotide metabolism and transport | ||||
| Orotate phosphoribosyltransferase | AIS08207.1 | NO | 2.73 (1.78) | 0.004 |
| Post-translational modification, protein turnover, chaperone functions | ||||
| Peroxidase | AIS07758.1 | YES | 0.25 (0.21) | 0.033 |
| Replication and repair | ||||
| Chromosomal replication initiation protein | AIS04811.1 | NO | 81.66 (31.26) | 0.017 |
| DNA gyrase subunit A | AIS07481.1 | NO | 0.09 (0.16) | 0.004 |
| Transcription | ||||
| Antitermination protein NusG | AIS05075.1 | NO | 2.00 (0.81) | 0.010 |
| DNA-directed RNA polymerase subunit beta | AIS05080.1 | NO | 0.26 (0.05) | 0.000 |
| DNA-directed RNA polymerase subunit beta’ | AIS05081.1 | NO | 0.10 (0.04) | 0.000 |
| RNA polymerase sigma factor RpoD | AIS07572.1 | NO | 1.92 (1.40) | 0.042 |
| Translation | ||||
| Isoleucine-tRNA ligase | AIS04850.1 | NO | 0.28 (0.09) | 0.001 |
| Arginine-tRNA ligase | AIS04960.1 | NO | 0.47 (0.16) | 0.019 |
| 50S ribosomal protein L1 | AIS05077.1 | YES | 3.40 (0.93) | 0.000 |
| 30S ribosomal protein S15 | AIS05149.1 | NO | 15.56 (6.87) | 0.045 |
| 50S ribosomal protein L28 | AIS05235.1 | YES | 1.85 (0.68) | 0.042 |
| Leucyl-tRNA synthetase | AIS05306.1 | NO | 0.39 (0.16) | 0.029 |
| Threonyl-tRNA synthetase | AIS05352.1 | NO | 0.09 (0.16) | 0.000 |
| 30S ribosomal protein S7 | AIS05609.1 | NO | 9.91 (4.58) | 0.000 |
| Elongation factor Tu | AIS05611.1 | NO | 0.34 (0.05) | 0.002 |
| Cysteinyl-tRNA synthetase | AIS05938.1 | NO | 0.18 (0.11) | 0.015 |
| 30S ribosomal protein S6 | AIS07031.1 | NO | 3.56 (0.80) | 0.004 |
| Translation | ||||
| 50S ribosomal protein L9 | AIS07033.1 | NO | 4.70 (2.06) | 0.000 |
| 30S ribosomal protein S21 | AIS07108.1 | YES | 4.33 (3.26) | 0.040 |
| 50S ribosomal protein L21 | AIS07586.1 | NO | 4.45 (2.38) | 0.029 |
| Valyl-tRNA synthetase | AIS07599.1 | NO | 0.17 (0.14) | 0.000 |
| 30S ribosomal protein S9 | AIS07845.1 | YES | 6.19 (2.19) | 0.005 |
| 50S ribosomal protein L17 | AIS07895.1 | YES | 15.14 (7.44) | 0.025 |
| 30S ribosomal protein S11 | AIS07898.1 | YES | 3.40 (1.58) | 0.002 |
| 50S ribosomal protein L15 | AIS07902.1 | NO | 5.06 (1.54) | 0.000 |
| 50S ribosomal protein L6 | AIS07906.1 | YES | 5.35 (2.12) | 0.000 |
| 30S ribosomal protein S8 | AIS07907.1 | YES | 7.52 (1.56) | 0.007 |
| 50S ribosomal protein L5 | AIS07909.1 | NO | 7.18 (2.38) | 0.000 |
| 50S ribosomal protein L14 | AIS07911.1 | NO | 5.20 (1.87) | 0.002 |
| 50S ribosomal protein L22 | AIS07916.1 | NO | 2.03 (0.76) | 0.012 |
| 50S ribosomal protein L23 | AIS07919.1 | YES | 1.77 (0.58) | 0.043 |
| Aminoglycoside phosphotransferase | AIS08113.1 | NO | 2.88 (4.81) | 0.015 |
The protein profiles produced by A. baumannii grown in modified BALF were performed using iTRAQ reagents and LC-MS/MS. Differential expression was defined by a relative abundance ratio >1.5 and <0.5
aSecretion prediction are based on (SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/), Phobius (http://phobius.sbc.su.se/), PrediSi (http://www.predisi.de/), TatP 1.0 (www.cbs.dtu.dk/services/TatP-1.0), Tatfind 1.4 (http://signalfind.org/tatfind.html), SecretomeP 2.0 (www.cbs.dtu.dk/services/SecretomeP), TMHMM (http://www.cbs.dtu.dk/services/TMHMM/), DAS-TMfilter (http://www.enzim.hu/DAS/DAS.html), LipoP 1.0 (www.cbs.dtu.dk/services/LipoP), DOLOP (http://www.mrc-lmb.cam.ac.uk/genomes/dolop/), and LIPO (http://services.cbu.uib.no/tools/lipo))
bAverage relative protein expression level ratio in sample and control, with the standard deviation in parentheses, quantified by Protein Pilot 4.0 software (ABSciex).
cDetermined by Student’s t test. Values of less than 0.05 are considered significant
In the macrophage model, infection of macrophages was induced with A. baumannii (multiplicity of infection 3). Control and infected cells were incubated simultaneously for 21 h at 37 °C and 5 % CO2, without shaking, in order to study the A. baumannii responses to immune system cells at the protein level by iTRAQ labelling and LC-MALDI/TOF analysis. Changes in the proteome of A. baumannii in response to RAW 264.7 cells were also examined. In comparison with the control group, 97 identified proteins displayed significant changes in expression. A total of 76 proteins displayed increased expression levels (fold change >1.5, p <0.05) and 21 proteins displayed decreased expression levels (fold change <0.5, p <0.05) relative to the control group (Table 2).
Table 2.
Differentially expressed proteins under ex vivo conditions in macrophage model
| Protein description | AbH12O-A2 locus | Sec.a | macrophage model/ controlb | macrophage model/ control p-valuec |
|---|---|---|---|---|
| Pathogenesis and virulence | ||||
| Nucleotidyl transferase | AIS04894.1 | NO | 2.83 (0.60) | 0.013 |
| Trigger factor | AIS05263.1 | NO | 3.37 (0.37) | 0.016 |
| ATPase AAA (PaaA) | AIS06044.1 | NO | 5.50 (4.44) | 0.028 |
| Siderophore achromobactin biosynthesis proteína AcsC | AIS06377.1 | NO | 0.44 (0.17) | 0.000 |
| Cyclophilin (PPIase) | AIS06968.1 | NO | 2.83 (0.87) | 0.041 |
| Protein CsuC | AIS07073.1 | YES | 0.16 (0.06) | 0.004 |
| Protein CsuA | AIS07075.1 | YES | 0.31 (0.22) | 0.040 |
| Metallopeptidase | AIS07555.1 | YES | 0.20 (0.16) | 0.028 |
| Membrane protein (OmpA) | AIS07737.1 | YES | 7.80 (2.78) | 0.049 |
| Oligopeptidase A | AIS07885.1 | NO | 4.83 (0.88) | 0.006 |
| ABC transporter ATP-binding protein (YjjK) | AIS08062.1 | NO | 2.33 (0.77) | 0.046 |
| Amino acid metabolism and transport | ||||
| Aspartate aminotransferase | AIS07065.1 | NO | 0.39 (0.18) | 0.010 |
| Serine hydroxymethyltransferase | AIS07167.1 | NO | 2.54 (0.44) | 0.043 |
| Carbamoyl-phosphate synthase | AIS07544.1 | NO | 2.75 (1.74) | 0.035 |
| 4-Hydroxyphenylpyruvate dioxygenase | AIS08279.1 | NO | 4.74 (1.15) | 0.000 |
| Carbohydrate metabolism and transport | ||||
| Phosphoglyceromutase | AIS05033.1 | NO | 3.94 (1.06) | 0.032 |
| Glyceraldehyde-3-phosphate dehydrogenase | AIS07393.1 | NO | 3.08 (0.66) | 0.016 |
| Cell cycle control and mitosis | ||||
| Cell division protein FtsA | AIS08196.1 | NO | 4.74 (7.32) | 0.002 |
| Cell wall/membrane/envelope biogenesis | ||||
| Membrane protein | AIS06856.1 | YES | 8.02 (7.90) | 0.006 |
| Racemase | AIS07059.1 | NO | 0.20 (0.13) | 0.037 |
| UDP-N-acetylmuramate-alanine ligase (MurC) | AIS08199.1 | NO | 8.17 (3.95) | 0.040 |
| Coenzyme metabolism | ||||
| Pantoate-beta-alanine ligase (PanC) | AIS05347.1 | NO | 6.08 (8.59) | 0.010 |
| 3-Phosphoglycerate dehydrogenase | AIS07996.1 | NO | 3.77 (0.88) | 0.014 |
| Defense mechanisms (cellular processes and signaling) | ||||
| Beta-lactamase | AIS07280.1 | YES | 2.09 (0.41) | 0.049 |
| ABC transporter | AIS07290.1 | NO | 0.39 (0.18) | 0.037 |
| Energy production and conversion | ||||
| ATP synthase subunit B | AIS04971.1 | NO | 5.50 (2.54) | 0.044 |
| ATP synthase F0F1 subunit beta | AIS04975.1 | NO | 3.63 (0.48) | 0.002 |
| Bifunctional aconitate hydratase 2/2-methylisocitrate dehydratase | AIS06989.1 | NO | 3.16 (0.31) | 0.002 |
| Isocitrate dehydrogenase | AIS07371.1 | NO | 4.57 (0.55) | 0.002 |
| Electron transfer flavoprotein subunit beta | AIS07482.1 | YES | 5.65 (0.89) | 0.005 |
| Dihydrolipoamide succinyltransferase | AIS07561.1 | NO | 2.44 (0.45) | 0.036 |
| Fumarate reductase | AIS07564.1 | NO | 3.08 (0.59) | 0.010 |
| Acetyl-CoA hydrolase | AIS08069.1 | NO | 4.74 (1.68) | 0.040 |
| NADPH:quinone oxidoreductase | AIS08156.1 | NO | 15.42 (16.20) | 0.034 |
| Dihydrolipoamide acetyltransferase | AIS08190.1 | YES | 3.08 (0.73) | 0.010 |
| Unknown function | ||||
| Hypothetical protein | AIS05430.1 | NO | 19.23 (19.19) | 0.001 |
| Hypothetical protein | AIS05481.1 | YES | 0.17 (0.08) | 0.000 |
| Unknown function | ||||
| Hypothetical protein | AIS05936.1 | YES | 0.23 (0.13) | 0.006 |
| DNA-binding protein | AIS07086.1 | YES | 0.24 (0.17) | 0.037 |
| Hypothetical protein | AIS07091.1 | NO | 0.21 (0.12) | 0.000 |
| DcaP-like protein | AIS07608.1 | YES | 11.07 (6.85) | 0.023 |
| Glyoxalase | AIS07740.1 | YES | 0.30 (0.23) | 0.036 |
| Hypothetical protein | AIS07873.1 | YES | 0.42 (0.16) | 0.011 |
| Inorganic ion transport and metabolism | ||||
| Hydroperoxidase | AIS06129.1 | YES | 0.35 (0.13) | 0.008 |
| ABC transporter permease | AIS07421.1 | YES | 8.17 (3.60) | 0.034 |
| Lipid metabolism | ||||
| 3-Oxoacyl-ACP reductase | AIS05519.1 | YES | 2.68 (0.94) | 0.038 |
| Enoyl-CoA hydratase | AIS06112.1 | NO | 0.17 (0.24) | 0.004 |
| Short-chain dehydrogenase | AIS06130.1 | YES | 0.10 (0.07) | 0.032 |
| Nucleotide metabolism and transport | ||||
| N5-carboxyaminoimidazole ribonucleotide mutase | AIS04832.1 | NO | 4.02 (2.98) | 0.010 |
| Ribose-phosphate pyrophosphokinase | AIS05530.1 | NO | 4.61 (2.76) | 0.000 |
| Deoxyuridine 5′-triphosphate nucleotidohydrolase | AIS05628.1 | NO | 0.28 (0.32) | 0.032 |
| Orotidine 5′-phosphate decarboxylase | AIS06325.1 | NO | 0.19 (0.21) | 0.032 |
| Adenylosuccinate lyase | AIS07333.1 | NO | 6.31 (2.68) | 0.047 |
| Xanthine phosphoribosyltransferase | AIS07874.1 | NO | 2.42 (0.69) | 0.048 |
| Inosine-5-monophosphate dehydrogenase | AIS08184.1 | NO | 3.94 (0.81) | 0.006 |
| Post-translational modification, protein turnover, chaperone functions | ||||
| Molecular chaperone DnaK | AIS04838.1 | NO | 2.13 (0.15) | 0.000 |
| Post-translational modification, protein turnover, chaperone functions | ||||
| Osmotically inducible protein C | AIS04957.1 | NO | 3.16 (1.14) | 0.029 |
| Replication and repair | ||||
| DNA repair protein | AIS04967.1 | YES | 3.87 (1.19) | 0.033 |
| DNA polymerase I | AIS05372.1 | NO | 0.31 (0.34) | 0.038 |
| Secondary metabolites: biosynthesis, transport and catabolism | ||||
| mRNA 3′-end processing factor | AIS06680.1 | NO | 0.10 (0.19) | 0.027 |
| Transcription | ||||
| DNA-directed RNA polymerase subunit beta’ | AIS05081.1 | NO | 2.86 (0.42) | 0.025 |
| Transcription elongation factor NusA | AIS05126.1 | NO | 3.53 (0.94) | 0.036 |
| Transcription termination factor Rho | AIS05365.1 | NO | 3.60 (1.54) | 0.025 |
| DNA-binding protein | AIS07045.1 | NO | 9.73 (6.82) | 0.029 |
| DNA-directed RNA polymerase subunit alpha | AIS07896.1 | NO | 3.28 (0.46) | 0.001 |
| Translation | ||||
| Tyrosyl-tRNA synthetase | AIS04798.1 | NO | 3.37 (0.97) | 0.009 |
| 23S rRNA methyltransferase | AIS05117.1 | NO | 3.37 (4.80) | 0.037 |
| Alanyl-tRNA synthetase | AIS05862.1 | NO | 3.05 (1.16) | 0.004 |
| 30S ribosomal protein S20 | AIS06347.1 | YES | 3.77 (2.31) | 0.026 |
| Ribosome-recycling factor | AIS06861.1 | NO | 3.91 (1.42) | 0.045 |
| Peptide chain release factor 1 | AIS07012.1 | NO | 0.34 (0.27) | 0.021 |
| 30S ribosomal protein S6 | AIS07031.1 | NO | 6.92 (1.66) | 0.000 |
| 30S ribosomal protein S18 | AIS07032.1 | NO | 22.70 (16.56) | 0.028 |
| 50S ribosomal protein L9 | AIS07033.1 | NO | 8.17 (3.02) | 0.002 |
| 30S ribosomal protein S2 | AIS07184.1 | NO | 3.87 (1.07) | 0.040 |
| Translation | ||||
| Tryptophanyl-tRNA synthetase | AIS07557.1 | NO | 8.09 (4.24) | 0.023 |
| 50S ribosomal protein L27 | AIS07585.1 | YES | 10.28 (8.73) | 0.027 |
| Aspartyl-tRNA synthetase | AIS07787.1 | NO | 5.92 (1.79) | 0.038 |
| 30S ribosomal protein S9 | AIS07845.1 | YES | 5.86 (1.38) | 0.009 |
| 50S ribosomal protein L17 | AIS07895.1 | YES | 9.64 (4.81) | 0.003 |
| 30S ribosomal protein S4 | AIS07897.1 | NO | 13.30 (3.19) | 0.010 |
| 50S ribosomal protein L15 | AIS07902.1 | NO | 5.92 (1.17) | 0.000 |
| 30S ribosomal protein S5 | AIS07904.1 | YES | 4.61 (5.00) | 0.003 |
| 50S ribosomal protein L18 | AIS07905.1 | YES | 9.04 (10.58) | 0.033 |
| 50S ribosomal protein L6 | AIS07906.1 | YES | 7.59 (1.58) | 0.037 |
| 30S ribosomal protein S8 | AIS07907.1 | YES | 10.76 (2.98) | 0.011 |
| 50S ribosomal protein L5 | AIS07909.1 | NO | 7.73 (2.20) | 0.000 |
| 50S ribosomal protein L24 | AIS07910.1 | YES | 4.25 (0.70) | 0.038 |
| 50S ribosomal protein L14 | AIS07911.1 | NO | 11.07 (12.57) | 0.002 |
| 30S ribosomal protein S17 | AIS07912.1 | NO | 12.36 (3.91) | 0.048 |
| 50S ribosomal protein L16 | AIS07914.1 | YES | 29.11 (22.95) | 0.038 |
| 30S ribosomal protein S3 | AIS07915.1 | NO | 6.73 (3.82) | 0.026 |
| 50S ribosomal protein L22 | AIS07916.1 | NO | 1.89 (0.85) | 0.035 |
| 50S ribosomal protein L2 | AIS07918.1 | YES | 7.87 (3.98) | 0.038 |
| 50S ribosomal protein L23 | AIS07919.1 | YES | 4.45 (1.04) | 0.018 |
| 50S ribosomal protein L4 | AIS07920.1 | NO | 14.45 (7.02) | 0.002 |
| Translation | ||||
| 50S ribosomal protein L3 | AIS07921.1 | NO | 6.25 (1.29) | 0.042 |
The protein profiles produced by A. baumannii grown in the presence of macrophages were performed using iTRAQ reagents and LC-MS/MS. Differential expression was defined by a relative abundance ratio >1.5 and <0.5
aSecretion prediction are based on (SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/), Phobius (http://phobius.sbc.su.se/), PrediSi (http://www.predisi.de/), TatP 1.0 (www.cbs.dtu.dk/services/TatP-1.0), Tatfind 1.4 (http://signalfind.org/tatfind.html), SecretomeP 2.0 (www.cbs.dtu.dk/services/SecretomeP), TMHMM (http://www.cbs.dtu.dk/services/TMHMM/), DAS-TMfilter (http://www.enzim.hu/DAS/DAS.html), LipoP 1.0 (www.cbs.dtu.dk/services/LipoP), DOLOP (http://www.mrc-lmb.cam.ac.uk/genomes/dolop/), and LIPO (http://services.cbu.uib.no/tools/lipo))
bAverage relative protein expression level ratio in sample and control, with the standard deviation in parentheses, quantified by Protein Pilot 4.0 software (ABSciex).
cDetermined by Student’s t test. Values of less than 0.05 are considered significant
Overall, 179 differentially expressed proteins were identified in both models. 111 proteins were modulated in the BALF model and 97 proteins were modulated in the macrophage model. In the BALF model, 45.0 % of the differentially expressed proteins were over-expressed. In the macrophage model, 78.4 % of the modulated proteins were over-expressed. Of the 179 modulated proteins, 16.2 % (29 proteins) were differentially expressed in both models, 51.7 % (15 proteins) were overexpressed, 20.7 % (6 proteins) were underexpressed and 27.6 % (8 proteins) were opposite (over-expressed in a model and under-expressed in the other model) (Fig. 1, Table 3).
Fig. 1.
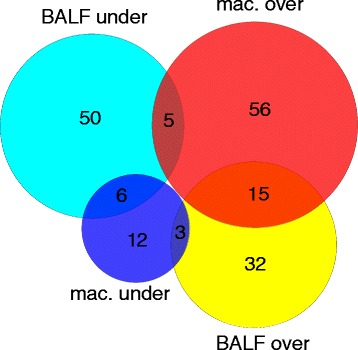
Venn diagram showing the number and relationship between A. baumannii proteins that were differentially expressed in comparison of 2 ex vivo models. Circles represent the set of over-expressed proteins (red) and under-expressed proteins (dark blue) in the macrophage model and the set of over-expressed proteins (yellow) and under-expressed proteins (light blue) in the BALF model. The number of proteins differentially expressed is indicated in each set or subset
Table 3.
Differentially expressed proteins in both ex vivo models
| Protein description | AbH12O-A2 locus | Sec.a | BALF model/controlb | BALF model/control p-valuec | macrophage model/controlb | macrophage model/control p-valuec |
|---|---|---|---|---|---|---|
| Pathogenesis and virulence | ||||||
| Protein CsuC | AIS07073.1 | YES | 2.19 (0.76) | 0.003 | 0.16 (0.06) | 0.004 |
| Protein CsuA | AIS07075.1 | YES | 1.87 (0.95) | 0.036 | 0.31 (0.22) | 0.040 |
| Membrane protein (OmpA) | AIS07737.1 | YES | 1.51 (0.65) | 0.015 | 7.80 (2.78) | 0.049 |
| Oligopeptidase A | AIS07885.1 | NO | 0.48 (0.11) | 0.001 | 4.83 (0.88) | 0.006 |
| ABC transporter ATP-binding protein (YjjK) | AIS08062.1 | NO | 1.74 (0.61) | 0.038 | 2.33 (0.77) | 0.046 |
| Amino acid metabolism and transport | ||||||
| Aspartate aminotransferase | AIS07065.1 | NO | 0.08 (0.07) | 0.000 | 0.39 (0.18) | 0.010 |
| Carbohydrate metabolism and transport | ||||||
| Phosphoglyceromutase | AIS05033.1 | NO | 0.26 (0.14) | 0.020 | 3.94 (1.06) | 0.032 |
| Cell wall/membrane/envelope biogenesis | ||||||
| UDP-N-acetylmuramate-alanine ligase (MurC) | AIS08199.1 | NO | 99.08 (0.00) | 0.020 | 8.17 (3.95) | 0.040 |
| Coenzyme metabolism | ||||||
| 3-Phosphoglycerate dehydrogenase | AIS07996.1 | NO | 0.38 (0.09) | 0.016 | 3.77 (0.88) | 0.014 |
| Energy production and conversion | ||||||
| Isocitrate dehydrogenase | AIS07371.1 | NO | 0.29 (0.05) | 0.004 | 4.57 (0.55) | 0.002 |
| Acetyl-CoA hydrolase | AIS08069.1 | NO | 2.05 (0.67) | 0.008 | 4.74 (1.68) | 0.040 |
| Unknown function | ||||||
| Hypothetical protein | AIS05481.1 | YES | 0.07 (0.10) | 0.013 | 0.17 (0.08) | 0.000 |
| DNA-binding protein | AIS07086.1 | YES | 0.08 (0.07) | 0.006 | 0.24 (0.17) | 0.037 |
| Glyoxalase | AIS07740.1 | YES | 0.39 (0.45) | 0.015 | 0.30 (0.23) | 0.036 |
| Inorganic ion transport and metabolism | ||||||
| Hydroperoxidase | AIS06129.1 | YES | 0.26 (0.10) | 0.000 | 0.35 (0.13) | 0.008 |
| Lipid metabolism | ||||||
| Short-chain dehydrogenase | AIS06130.1 | YES | 0.04 (0.05) | 0.040 | 0.10 (0.07) | 0.032 |
| Nucleotide metabolism and transport | ||||||
| Deoxyuridine 5′-triphosphate nucleotidohydrolase | AIS05628.1 | NO | 4.45 (6.37) | 0.039 | 0.28 (0.32) | 0.032 |
| Transcription | ||||||
| DNA-directed RNA polymerase subunit beta’ | AIS05081.1 | NO | 0.10 (0.04) | 0.000 | 2.86 (0.42) | 0.025 |
| Translation | ||||||
| 30S ribosomal protein S6 | AIS07031.1 | NO | 3.56 (0.80) | 0.004 | 6.92 (1.66) | 0.000 |
| 50S ribosomal protein L9 | AIS07033.1 | NO | 4.70 (2.06) | 0.000 | 8.17 (3.02) | 0.002 |
| 30S ribosomal protein S9 | AIS07845.1 | YES | 6.19 (2.19) | 0.005 | 5.86 (1.38) | 0.009 |
| 50S ribosomal protein L17 | AIS07895.1 | YES | 15.14 (7.44) | 0.025 | 9.64 (4.81) | 0.003 |
| 50S ribosomal protein L15 | AIS07902.1 | NO | 5.06 (1.54) | 0.000 | 5.92 (1.17) | 0.000 |
| 50S ribosomal protein L6 | AIS07906.1 | YES | 5.35 (2.12) | 0.000 | 7.59 (1.58) | 0.037 |
| 30S ribosomal protein S8 | AIS07907.1 | YES | 7.52 (1.56) | 0.007 | 10.76 (2.98) | 0.011 |
| 50S ribosomal protein L5 | AIS07909.1 | NO | 7.18 (2.38) | 0.000 | 7.73 (2.20) | 0.000 |
| 50S ribosomal protein L14 | AIS07911.1 | NO | 5.20 (1.87) | 0.002 | 11.07 (12.57) | 0.002 |
| 50S ribosomal protein L22 | AIS07916.1 | NO | 2.03 (0.76) | 0.012 | 1.89 (0.85) | 0.035 |
| 50S ribosomal protein L23 | AIS07919.1 | YES | 1.77 (0.58) | 0.043 | 4.45 (1.04) | 0.018 |
The protein profiles produced by A. baumannii grown in modified BALF and in the presence of macrophages were performed using iTRAQ reagents and LC-MS/MS. Differential expression was defined by a relative abundance ratio >1.5 and <0.5
aSecretion prediction are based on (SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/), Phobius (http://phobius.sbc.su.se/), PrediSi (http://www.predisi.de/), TatP 1.0 (www.cbs.dtu.dk/services/TatP-1.0), Tatfind 1.4 (http://signalfind.org/tatfind.html), SecretomeP 2.0 (www.cbs.dtu.dk/services/SecretomeP), TMHMM (http://www.cbs.dtu.dk/services/TMHMM/), DAS-TMfilter (http://www.enzim.hu/DAS/DAS.html), LipoP 1.0 (www.cbs.dtu.dk/services/LipoP), DOLOP (http://www.mrc-lmb.cam.ac.uk/genomes/dolop/), and LIPO (http://services.cbu.uib.no/tools/lipo))
bAverage relative protein expression level ratio in sample and control, with the standard deviation in parentheses, quantified by Protein Pilot 4.0 software (ABSciex).
cDetermined by Student’s t test. Values of less than 0.05 are considered significant
Analysis of modulated proteins
According to their predicted biological functions, the differentially expressed proteins were divided into 19 groups (Fig. 2). The largest groups consisted of proteins involved in translation (58), followed by the proteins involved in energy production and conversion (27) and by proteins involved in pathogenesis and virulence (23). Most of the proteins over-expressed in the BALF model were involved in translation (38.0 %), followed by pathogenesis and virulence (14.0 %), and energy production and conversion (10.0 %). In the macrophage model, most were involved in translation (40.8 %), followed by energy production and conversion (13.2 %), and pathogenesis and virulence (10.5 %). Most of the proteins that were under-expressed in the BALF model were involved in energy production and conversion (19.7 %) and in the macrophage model most were proteins of unknown function (28.6 %).
Fig. 2.
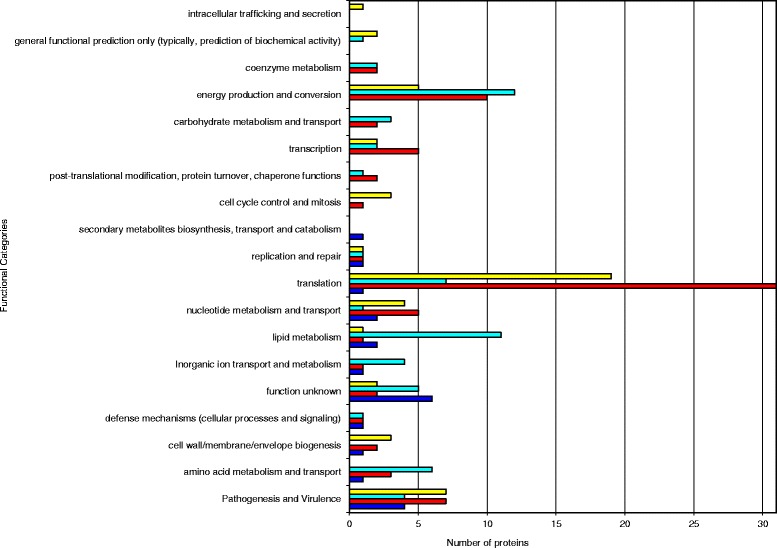
Distribution of differentially expressed proteins in A. baumannii following ex vivo incubation according to functional categories. The stacked bar chart shows the number of over-expressed proteins (red) and under-expressed proteins (dark blue) in the macrophage model and the number of over-expressed proteins (yellow) and under-expressed proteins (light blue) in the BALF model in each functional category
Of the 29 proteins differentially expressed in both models, most were involved in translation (37.9 %), followed by pathogenesis and virulence (17.2 %) and by unknown function (10.3 %). Most of the over-expressed proteins were involved in translation, whereas most of the under-expressed proteins were proteins of unknown function, and 50.0 % of opposite proteins were associated with metabolism (Table 3).
RT-PCR
We selected some proteins for validation of the proteomic data by RT-PCR analysis. The candidate proteins, NfuA-like, YjjK, CsuC, OmpW, OmpA, ClpX, DNA repair, PaaA and PpiA were selected on the basis of their differential expression as representative of pathogenesis and virulence, cell wall/membrane/envelope biogenesis and other functional categories.
BALF model
RT-PCR analysis was applied to 6 genes coding for differentially expressed proteins to confirm the findings of the proteomic analysis. Expression of most, but not all, of these genes paralleled expression of the corresponding proteins revealed by proteomic analysis. Four (NfuA-like, YjjK, CsuC and OmpW) of the six selected genes were over-expressed at the transcriptional level in the sample relative to the control which is consistent with the results of protein expression. However, there was a slight inconsistency between the translational and transcriptional levels of OmpA and ClpX. Expression of OmpA and ClpX mRNA revealed similar patterns in the sample relative to the control, despite the increased levels of the respective mRNAs (Fig. 3, Table 1).
Fig. 3.
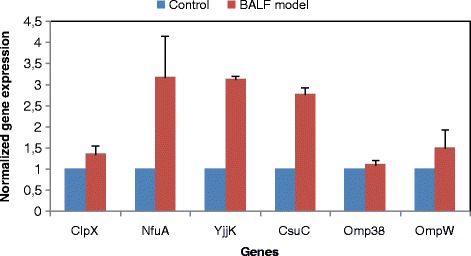
RT-PCR analysis of different genes coding for differentially expressed proteins in controls and samples (BALF model). All expression results were normalized relative to RpoB by the 2−ΔΔCt method. For all genes, relative mRNA expression is presented as a fold-change value relative to the control
Macrophage model
RT-PCR analysis of 7 genes coding for differentially expressed proteins was performed to confirm the results of the proteomic analysis. Expression of most, but not all, of the genes paralleled the expression of the corresponding proteins revealed by proteomic analysis. Five (DNA repair, PaaA, PpiA,YjjK and OmpW) of the seven selected genes were over-expressed at the transcriptional level in the sample, relative to the control, which is consistent with the results of protein expression. However, there was a slight inconsistency between the translational and transcriptional levels of OmpA and an inconsistency between the translational and transcriptional levels of CsuC. The mRNA expression of OmpA revealed similar patterns in the sample, relative to the control, despite the increased levels of the respective mRNAs. The mRNA expression of CsuC was 1.6 times higher and the protein abundance was more than six times lower in the sample than in the control (Fig. 4, Table 2), probably because of post-transcriptional regulation [33].
Fig. 4.
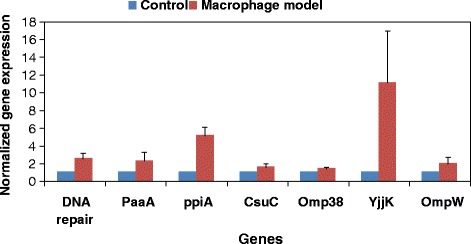
RT-PCR analysis of different genes coding for differentially expressed proteins in controls and samples (macrophage model). All expression results were normalized relative to RpoB by the 2−ΔΔCt method. For all genes, relative mRNA expression is presented as a fold-change value relative to the control
Comparison of the results obtained by iTRAQ labelling-LC-MALDI/TOF and RT-PCR showed that the changes observed in protein abundance between samples of both ex vivo models and controls were also reflected in the mRNA levels.
Discussion
The host activated response to Acinetobacter baumannii is not well understood. Some studies have described changes in A. baumannii gene expression under in vivo-mimicking conditions. However, most of these studies focused on transcriptional changes of one or a few genes of interest, mostly under iron limiting conditions [34–38]. A microarray study defined the expression properties of A. baumannii during growth in human serum and demonstrated significant over-expression of iron acquisition systems, of genes associated with epithelial cell adherence, DNA uptake and of numerous putative drug efflux pumps [39]. At the protein level, comparative analysis of total lysate and outer membrane fractions isolated from A. baumannii cultured under iron-rich and iron-chelated conditions led to identification of 58 modulated proteins [40]. There remains an alarmingly large gap in our knowledge of the pathogenesis and nature of A. baumannii virulence. A better understanding of the adaptation of A. baumannii to the host and its molecular pathogenesis is essential for the development of new therapeutic and diagnostic methods. Research studies that address the A. baumannii proteome in vivo and the proteome that is produced in the presence of fluid or host cells are lacking. In contrast to the numerous proteomic studies of A. baumannii cultivated in vitro laboratory settings, only one microarray study has been carried out ex vivo, and no ex vivo or in vivo proteomic studies have been performed. This is because of technical limitations, which severely limit the number of recovered bacteria, and the potential contamination of sample preparations by host cell nucleic acids or proteins.
To identify putative pathogenesis and virulence factors that mediate A. baumannii pathogenesis, we used 2 ex vivo models and a quantitative proteomics approach. We provide evidence that expression of 179 A. baumannii proteins is affected by the host. Comparison of both models showed that 83 % of the proteins detected were grouped in the same category in both models (under-expressed, over-expressed and un-modulated) and that most of the proteins were unmodulated, which indicates that the host proteins may target and regulate a specific small group of A. baumannii proteins. The modulated proteins possess diverse functions (Fig. 2, Table 1 and Table 2), which suggests a complex interaction between the bacterium and the host. Thus, in the BALF model, 50 of the proteins identified were over-expressed, whereas 61 were under-expressed. Some of these functional groups are clearly over-expressed for functioning in cell wall/membrane/envelope biogenesis, nucleotide metabolism and transport and cell cycle control and mitosis. Other important biological processes, such as pathogenesis and virulence and translation, were mainly over-expressed. However, other metabolic proteins involved in, e.g., amino acid metabolism and lipid metabolism were under-expressed. In the macrophage model, 76 of the proteins identified were over-expressed, whereas 21 were under-expressed. Some of these functional groups are clearly over-expressed for functioning in translation, transcription and energy production and conversion. Other important biological processes, such as pathogenesis and virulence and nucleotide metabolism and transport, were mainly over-expressed. However, other functional groups involved in lipid metabolism were under-expressed. Although both models yielded generally similar results, some differences in the findings are not surprising, for the following reasons. First, different rodents species were used as hosts. Moreover, some of the major influencing factors identified in the BALF model include nutritional differences between samples (LB broth+lipids, nucleic acids, peptides and proteins) and controls (LB) and stress resulting from host inflammatory response (proteins important for immunity and host defence functions, such as surfactant protein A and surfactant protein D). However, in the macrophage model, these factors may compete with macrophages for available nutrients and stress resulting from interaction with a single host cell type.
Proteomic comparison of the levels of protein expression in the models revealed that 82 proteins were only modulated by BALF, 68 proteins were only modulated by macrophages and 29 proteins were modulated by both. Of these 29 proteins, 15 were over-expressed, 6 proteins were under-expressed and 8 proteins were opposite (Fig. 1). Interestingly, 50.0 % of opposite proteins were associated with metabolism (phosphoglyceromutase, 3-phosphoglycerate dehydrogenase, isocitrate dehydrogenase and deoxyuridine 5′-triphosphate nucleotidohydrolase). This is probably due to differences in the concentration of available nutrients in both models and to competition with macrophages for available nutrients in the macrophage model. Over-expressed amino acid catabolism was also observed in the macrophage model and under-expressed amino acid catabolism in the BALF model (Table 3), indicating that amino acids constitute a major source of nitrogen for A. baumannii in the macrophage model. The availability of amino acids is probably also partly due to uptake of oligopeptides. Thus, the oligopeptidase A was over-expressed in the macrophage model and under-expressed in the BALF model. Moreover, CsuC and CsuA were over-expressed in the host cell-free model (the BALF model) and under-expressed in macrophage model, because these proteins are involved in initial attachment to abiotic surfaces but not in the mechanism of adherence to biotic surfaces. Nevertheless, the explanation is not as obvious in the case of the modulation of DNA-directed RNA polymerase subunit beta’.
In the macrophage model, 3 predicted outer membrane proteins were over-expressed (a DcaP-like protein, a membrane protein and OmpA -iTRAQ ratio between 7.80 and 11.07). In the BALF model, 2 predicted outer membrane proteins were over-expressed (OmpW family protein and OmpA -iTRAQ ratio between 1.51 and 2.25). In the macrophage model, 21 predicted secreted proteins were also over-expressed (average iTRAQ ratio 7.88) and 10 were under-expressed (average iTRAQ ratio 0.25). In the BALF model, 17 predicted secreted proteins were over-expressed (average iTRAQ ratio 4.03) and 13 were under-expressed (average iTRAQ ratio 0.20). The modifications that A. baumannii undergoes as a result of host-induced stress include changes in membrane and secretome composition; these changes were greater in the macrophage model than in the BALF model (host cell-free model) probably due to pathogen-host cell interactions. We conclude that the outer membrane and secretome are changed by the host cells and host secretome.
Interestingly, we found that the UDP-N-acetylmuramate-L-alanine ligase (MurC) protein was strongly over-expressed in both ex vivo models (iTRAQ ratio of 99.08 in the BALF model, and iTRAQ ratio of 8.17 in macrophage model). We also observed that UDP-N-acetylglucosamine 1-carboxyvinyl transferase (MurA) was over-expressed in the BALF model, but not significantly over-expressed in the macrophage model (p > 0.05). In peptidoglycan synthesis, uridine-diphospho-N-acetylglucosamine is converted via MurA and MurB to UDP–N-acetylmuramic acid, to which alanine is added by MurC. These are essential steps in cell wall biosynthesis in bacteria [41, 42]. Moreover, MurA is required for full virulence in Listeria monocytogenes [43]. In this context, MurA has previously been suggested as a potential drug target, and effective inhibitors of this protein have been identified [44]. Detection of MurC polypeptides in humans could provide a method for diagnosis and/or prognosis of an infection. Levels of MurC polypeptides could be determined using assay techniques such as include antibody detection, ELISA assays, antibody sandwich assays and Western Blot analysis. Monoclonal antibodies against A. baumannii MurC similar to those generated by Zoeiby et al. could be used to detect MurC polypeptides. Zoeiby et al. used purified P. aeruginosa MurC protein to develop monoclonal antibodies against MurC. Moreover, Zoeiby et al. suggested that a specific inhibitor designed against P. aeruginosa MurC may display universal activity against all bacteria [45]. However, other authors have observed that C-1, a competitive inhibitor of ATP binding to the MurC enzyme, was equally effective at inhibiting MurC from E. coli, Klebsiella pneumoniae and Proteus mirabilis, but that MurC enzymes from more distantly related Gram-negative species such as Acinetobacter baylyi, Pseudomonas aeruginosa and Haemophilus influenzae were not inhibited [46]. More recently, MurC enzymes from Escherichia coli and Pseudomonas aeruginosa have been found to be inhibited by a novel class of pyrazolopyrimidines [47].
Putative virulence-associated factors
Several putative virulence factors were modulated in both ex vivo models. Blast analysis (http://blast.ncbi.nlm.nih.gov/Blast.cgi) [48, 49] revealed significant alignment between the nucleotidyl transferase (AIS04894.1), which was found to be over-expressed in the macrophage model (but not significantly modulated (p > 0.05) in the BALF model) and UTP-glucose-1-phosphate uridylyltransferase (GalU) (query cover 100 % and Evalue = 0.0). The GalU gene encodes for production of UTP-glucose-1-phosphate uridyltransferase, an enzyme that catalyzes the formation of UDP-glucose and is known to have a key role in biosynthesis of cell-envelope-associated carbohydrates (e.g. lipopolysaccharides [LPS] and capsule) in a variety of bacterial species. In many Gram-negative pathogens, mutations in GalU lead to attenuated virulence, mainly because of changes in LPS or capsular structures. Formation of these polysaccharides is critical to bacterial virulence because this enables the bacteria to evade attack by the host immune system. The GalU gene has also been found to be important for pathogenesis of e.g. Pseudomonas aeruginosa and Francisella tularensis [50–54].
Bacteria and their hosts are involved in competition for available iron. In the host, iron is sequestered by ferritin, transferrin, haemoglobin, myoglobin, haptoglobin and haemopexin, thus decreasing the availability of free iron to concentrations below those needed for bacteria to persist in the environment. Therefore, bacteria must survive by competing with the host for iron through the expression of iron acquisition systems. These systems produce high affinity iron chelators known as siderophores, which are secreted into the extracellular milieu [55, 56]. Lipocalin-1 is secreted by the lingual glands, sweat glands, prostate, and secretory glands of the tracheobronchial tract, as well as by the nasal mucosa into BALF, sweat, saliva, tear, sputum and nasal fluids. Lipocalin-2, which is secreted by neutrophils, macrophages, dendritic cells and exocrine glands into trachea, lung, sputum, BAL, stomach, small intestine, pancreas, kidney, prostate, thymus and plasma, interferes with siderophore-mediated iron-uptake. Lipocalin-1 binds to different bacterial siderophores, including bacterial catecholate-type (enterobactin) and hydroxamate-type (ferricrocin) and mixed citrate-hydroxamates (aerobactin). Lipocalin-2 binds to different bacterial siderophores, including enterobactin-type bacterial siderophores and carboxy-mycobactins. To defend themselves against the host, some bacteria use countermeasures to subvert the iron-withholding effects of Lipocalin-2. For example, Salmonella and Klebsiella spp. glycosylate enterobactin, whereas M. tuberculosis modifies its carboxymycobactins. Both mechanisms sterically impair the ability of Lipocalin-2 to bind these siderophores [57–60]. NfuA-like has multiple functional roles related to iron-mediated stress. NfuA-like is involved in intracellular iron metabolism and, as demonstrated in an infection model, it also plays a role in the virulence of A. baumannii by protecting infecting bacteria from oxidative responses rather than providing iron to bacteria grown under iron—limited conditions imposed by cultured human alveolar epithelial cells and G. mellonella larvae [37]. In the present study, we observed that in both models, proteins involved in biosynthesis of the siderophore aerobactin were not significantly modulated (p > 0.05), probably because lipocalin-1 binds to aerobactin. We also observed that one of the under-expressed proteins (not significantly under-expressed (p > 0.05) in the BALF model) is associated with biosynthesis of the siderophore achromobactin (AIS06377.1). This may be because although achromobactin is an efficient hydroxycarboxylate siderophore at low pH (pH < 5), achromobactin could not compete with stronger siderophores such as catecholates at physiological pH [59]. In both models, NfuA-like was also over-expressed (although not significantly (p > 0.05), in the macrophage model). Altogether, these results suggest a third system for iron acquisition in A. baumannii strain AbH12O-A2, but which is not identified by current bioinformatic analysis. In a recent study, the structures of six novel siderophores termed fimsbactin A-F have been elucidated and the biosynthesis gene clusters for fimsbactin production have been identified in A. baylyi ADP1 and A. baumannii strains (ATCC 17978 and 6013150), although BLAST analysis showed that the fimsbactin gene cluster is not present in A. baumannii strain ABH12O-A2 (data not shown) [61].
In the macrophage model, we observed increased expression (not over-expressed in the BALF model: iTRAQ ratio, 1.26) of the Tig factor in the ex vivo samples. De novo protein folding in Escherichia coli is mainly orchestrated by the chaperone trigger factor and by DnaK and GroEL. The ribosome-bound trigger factor is the first chaperone to interact cotranslationally with nascent polypeptides. It has been suggested that a trigger factor homologue in Streptococcus mutans is a key regulator of stress tolerance, genetic competence and biofilm formation, all of which are critical virulence properties of this bacterium. In Listeria monocytogenes, a trigger factor homologue is involved in the stress response and associated pathogenicity [62–64].
We observed increased expression of the DnaK suppressor protein, which was significant (p < 0.05) in the BALF model, but not in the macrophage model. This protein plays an important role in the virulence of Salmonella and Escherichia coli [65].
In the BALF model, expression of the ClpX protein increased (but was not significantly over-expressed (p > 0.05) in the macrophage model). ClpX is required for virulence, biofilm formation and intracellular replication in Staphylococcus aureus [66].
Initial attachment to abiotic surfaces is the first step for colonization and subsequent biofilm formation on e.g. ventilator tubing and catheters. In A. baumannii 19606, the type I pili encoded by CsuA/BABCDE appears to be involved in this process [67]. In our host cell-free model (BALF model), the proteins OmpA (iTRAQ ratio = 1.51), CsuA and CsuC were over-expressed, suggesting biofilm activation by the host secretome that defends A. baumannii against host on medical devices. Another critical step in the pathogenesis of A. baumannii is the ability to adhere to eukaryotic cells, although the mechanisms of adherence are different for abiotic and biotic surfaces. It has been demonstrated that OmpA acts as a virulence factor in A. baumannii and has an important role in cell death through both mitochondrial and nuclear targeting [68]. The OmpA of A. baumannii 19606 also plays a partial role in biofilm formation on plastic, but is essential for attachment to biotic surfaces such as C. albicans and human alveolar epithelial cells. Interestingly, the absence of these cell appendages also favours bacterial attachment and invasion of epithelial cells. This may be due to greater exposure of other unknown bacterial adhesion and biofilm factors in the absence of pili [69]. In our macrophage model, OmpA (iTRAQ ratio = 7.80) was over-expressed and both CsuA and CsuC were under-expressed. Together these results suggest that contact between A. baumannii cells and macrophages may be an important factor determining underexpression of the CsuA/ BABCDE protein. Elucidation of the complex interactions between A. baumanniiand both abiotic and biotic surfaces is important for a better understanding of the pathobiology of A. baumannii and also for identification of novel targets for the development of new antimicrobial strategies.
In the macrophage model, we observed increased expression of the PaaA protein in the ex vivo samples, but we did not observe significant (p > 0.05) repression of the PaaZ protein. In contrast, in the BALF model, we observed repression of PaaZ protein but no modulation (p > 0.05) of the PaaA protein. In the catabolism of phenylacetic acid by some bacteria, PaaA encodes part of the phenylacetic acid—coenzyme A ring hydroxylation system, and opening of the aromatic ring may be performed by PaaZ. In Burkholderia cenocepacia, PaaA insertional mutants were attenuated for virulence and interruption of increased virulence of PaaZ [70].
The success of a bacterial infection greatly depends on the ability of the bacteria to use external nutrients. Therefore, the proteolytic and lipolytic activities of extracellular proteins may play key roles in the establishment of A. baumannii infection [71]. In general, in the macrophage model, proteins with proteolytic activity were over-expressed, and in the BALF model, this type of protein was under-expressed. This may be due to continuous production of host proteins in the macrophage model but not in the BALF model as well as to over-expressed amino acid metabolism in the macrophage model and under-expressed amino acid metabolism in the BALF model.
We identified a cyclophilin (PPIase) (AIS06968.1), which was over-expressed in the macrophage model and not modulated in the BALF model. It has been established that PPIases may play a role during bacterial survival against macrophage attack, and it has been suggested that PPIases may act as virulence factors by interacting with some proteins from the host cell membrane, thus helping render the host cell more susceptible to penetration via conformational changes through cis-trans isomerization of peptidyl-prolyl bonds [72, 73]. These types of proteins have been reported to affect the phagocytosis of Streptococcus pneumoniae by macrophages [74].
In both models, the uncharacterized ABC transporter ATP-binding protein YjjK was over-expressed. The precise role of YjjK, encoding a putative ATP binding protein of an ABC transporter, is not yet known. However, YjjK is required for the entry and survival of Porphyromonas gingivalis in gingival epithelial cells [75]. It has also been suggested that YjjK is involved in Francisella tularensis virulence [76].
Eighteen genes have been identified in A. baumannii as being involved in growth on human ascites and shown to be essential for growth and survival during infection [77]. In the present study, we found 53 % of these essential proteins: 4 of these were over-expressed (OmpF, RpmA, CarA and PyrC), 4 were un-modulated (AroA, AroC, RstA and SecE) and AceE was under-expressed. The present findings are only partly consistent with those of the aforementioned study [77] as SpsC is not contained in the AbH12O-A2 genome and we did not detect8 of the proteins. However, these differences may be due to the undetected proteins occurring in amounts below the detection/identification level or to differences in the host microenvironment.
When A. baumannii infects the host airway, it is present in multiple locations, including in the interstitium between cells, embedded in mucus in the lumen, inside and adhering to macrophages and epithelial cells. Therefore, A. baumannii must adapt to multiple microenvironments and may express different proteins, depending on the microenvironment. We examined 2 ex vivo models of infection: a host cell-free model in which A. baumannii interacts with soluble host components present inside the infected lung, and an immune system cells model, in which A. baumannii interacts with macrophages. One limitation of the study is that we only simulated some conditions in the host airways and A. baumannii is present in many other microenvironments. Caution must therefore be used in extrapolating results of the present study to the human disease because A. baumannii populations that colonize the human airways are probably a mixture of A. baumannii from different microenvironments.
The AbH12O-A2 strain yielded over-expressed ‘hypothetical proteins’. Induced expression of hypothetical proteins strongly indicates that A. baumannii generates host resistance by unknown mechanisms. These genes are obvious candidates for subsequent functional characterization and for research aimed at determining their function within infection processes.
Conclusions
This study is the first comprehensive overview of the ex vivo proteome of the multidrug resistant microbial pathogen A. baumannii and provides some insight into the potential role of putative virulence proteins in vivo. The ex vivo behaviour of A. baumannii was compared, for the first time, under different infection conditions—in BALF and in the presence of macrophages—as well as under in vitro conditions. A rapid enrichment technique was successfully used, together with MS/MS analysis, to characterize the ex vivo proteome of A. baumannii and showed that the proteome is significantly different from that of bacteria cultured in vitro. The changes in the proteome observed for the strain A. baumannii AbH12O-A2 indicate a response to stress resulting from interaction with host and modification of cell wall synthesis. We identified 2 over-expressed virulence-associated proteins with >15 peptides/protein in both ex vivo models (OmpA and an uncharacterized ABC transporter ATP-binding protein YjjK), which appear to be essential for pathogenesis and virulence in the airways. Overall, the data suggest that A. baumannii can use a variety of virulence mechanisms that enable it to adapt to and survive in vastly divergent environments. These data are helpful for elucidating the molecular mechanisms associated with the interaction between A. baumannii and host and represent an important step towards identification of diagnostic biomarkers, novel drug targets and potential vaccine candidates in the fight against pneumonia caused by A. baumannii.
Methods
Bacterial strain
A highly invasive multidrug-resistant (including carbapenems) Acinetobacter baumannii clone (AbH12O-A2), which infected more than 300 patients in the 12 de Octubre Hospital (Madrid, Spain) was used in this study.
The annotated sequence for AbH12O-A2 [GenBank CP009534.1] is available at the National Center for Biotechnology Information (NCBI) [78].
Rat model of pneumonia and BALF (bronchoalveolar lavage fluid)
Male Wistar rats (INIBIC, A Coruña, Spain) of about 350 g weight were used in the experiment. Antibiotic-free pelleted food and water were provided ad libitum during the assay. The experiment was approved by the Ethics Committee for Animal Experimentation of A Coruna Hospital. The rats were anaesthetized intraperitoneally with sodium thiopental and were inoculated intratracheally under direct vision with a bacterial suspension, which was prepared as follows: bacteria were grown at 37 °C in LB until an optical density of 0.7 at 600 nm, washed with sterile saline solution and mixed 1:1 with a saline solution of porcine mucin at 10 % (wt/vol). The final inoculum was about 3 × 108 CFUs/rat. At 21 h after inoculation, animals were euthanized by intraperitoneal injection of an overdose of sodium thiopental. The lungs were washed twice with about 45 ml normal saline (0.9 % NaCl). The concentrations of the original bacterial suspensions and of the BALF were determined by the plate count method. The BALF from three animals was centrifuged (3000 × g, 30 min at 4 °C) to remove cells, and the supernatant was filtered through a 0.22 μm membrane (low protein binding Millex-GP polyethersulfone membrane (Millipore, Bedford, U.S.A.)) to remove residual bacteria. Absence of viable bacteria was confirmed by culture on Mueller Hinton (MH) agar plates. BALF was stored at −70 °C for a maximum of 3 months until use.
Histopathology
Paraformaldehyde-fixed lung tissue was embedded in paraffin, sectioned and stained with hematoxylin and eosin. Two to four sections from each lung of infected rats were examined.
Incubation of A. baumannii in rat BALF
Acinetobacter baumannii strain AbH12O-A2 was streaked on MH nutrient agar. A single colony was then inoculated in 5 mL LB and grown overnight at 37 °C with vigorous shaking. 100 mL of LB (10 g/L tryptone + 5 g/L yeast extract dissolved in saline solution instead of water) and 100 ml of modified LB (40 g/L tryptone + 20 g/L yeast extract dissolved in saline solution instead of water + 75 % (v/v) BALF as a physiologically relevant source of host proteins to simulate conditions in the respiratory tract) were inoculated with a 1:100 dilution of the overnight culture and grown simultaneously for 21 h at 37 °C and 5 % CO2 without shaking. Three independent biological replicates of each culture were prepared but only 2 were used for the ITRAQ. Cells (OD600 nm = 1.0-1.4) were harvested by centrifugation at 2800 × g for 20 minutes at 4 °C, washed twice by suspending in cold 0.9 % NaCl and centrifuged again under the same conditions. The pellets were frozen and stored at −70 °C until needed.
Macrophage infection with A. baumannii
Acinetobacter baumannii strain AbH12O-A2 was streaked on MH nutrient agar. A single colony was then inoculated in 5 mL LB and grown overnight at 37 °C with vigorous shaking. The overnight culture was harvested, washed once and resuspended in tissue culture medium: DMEM with glucose and L-glutamine (4.5 g/L dextrose + 3.7 g/L NaHCO3 + 0.015 g/L phenol red + 0.110 g/L sodium pyruvate + 0.200 g/L CaCl2 + 0.100 mg/L Fe(NO3)3.9H2O + 0.098 g/L MgSO4 + 0.400 g/L KCl + 6.400 g/L NaCl + 0.007 g/L i-Inositol + 0.109 g/L NaH2PO4 + 15 amino acids and vitamins (BioWhittaker Lonza, Verviers, Belgium)) supplemented with 10 % fetal bovine serum and 1 % penicillin/streptomycin. The concentration of bacterial cells was then determined.
To mimic host microbe interactions involving phagocytic immune system cells the RAW 264.7 macrophage-like cell line was plated in cell culture flasks at 3 × 107 cells/flask and incubated overnight at 37 °C with 5 % CO2. Macrophage cells were washed twice with 20 ml/flask of 0.9 % NaCl. The tissue culture medium was then added to each flask, and 1 mL of the A. baumannii in the tissue culture medium was added to each flask with a multiplicity of infection of 3 (i.e. 3 times more bacterial cells than macrophages were used). To increase the uptake of A. baumannii and synchronize infection, flasks were centrifuged at 250 × g for 5 min at room temperature and then incubated at 37 °C with 5 % CO2 for 21 h without shaking. Three independent biological replicates of each culture were prepared but only 2 were used for the ITRAQ. The sample was harvested 21 h post infection. Extracellular bacteria were isolated from infected macrophage cultures. To isolate extracellular bacteria, infected macrophage cultures were washed three times with Hank’s buffered salt solution to remove any bacteria that were not inside adherent cells. The harvested sample and the wash solutions were centrifuged (500 × g for 2 min at 4 °C to pellet any nonadherent macrophage cells) and the supernatants were combined. The supernatant was centrifuged (1500 × g for 20 min at 4 °C) to pellet the bacteria. The pellet was resuspended in phosphate-buffered saline (PBS) and filtered through a Transwell filter membrane system (3.0-μm pore size; BD Falcon, Erembodegem, Belgium). Macrophage cells were very scarce. Bacteria then were washed twice by suspending them in cold PBS and centrifuging (2800 × g for 20 minutes at 4 °C). The pellets were frozen and stored at −70 °C until needed. Control A. baumannii cells were collected, washed and treated in the same way as above but in the absence of macrophages.
Protein extraction
The resultant pellet was resuspended in disintegration buffer (7.8 g/L NaH2PO4, 7.1 g/L Na2HPO4, 0.247 g/L MgSO4.7H2O) + protease inhibitor mix (GE Healthcare, Piscataway, USA) + nuclease mix (GE Healthcare, Piscataway, USA) and sonicated on ice for 3 periods of 5 min. The unbroken cells were separated by centrifugation at 1500 × g at 4 °C. The supernatant was centrifuged for at least 30 min at 4 °C and 4500 × g before being clarified through a 0.22 μm membrane (low protein binding Millex-GP polyethersulfone membrane from Millipore, Bedford, U.S.A.) to remove the cell debris. Finally, the extract was processed with a 2-DE Cleanup Kit (GE Healthcare, Piscataway, USA), following the manufacturer’s instructions. The concentration of protein was measured using the Bio-Rad protein assay (Bio-Rad, Munich, Germany).
iTRAQ labelling and LC-MALDI/TOF analysis
Labelling, 2-D liquid chromatography and MALDI-TOF/TOF MS analysis of the samples were performed as previously described [79], and stage tips were used for peptide desalting. Briefly, 30 μg of protein from each condition was reduced, cysteine was blocked, digested with trypsin and labelled with respective isobaric tags using iTRAQ reagent Multiplex kit (AB Sciex Ltd., Foster City, CA), according to the manufacturer’s protocol. The sample labelling was as follows: iTRAQ tags 114: control BALF model; iTRAQ tags 115: sample BALF model; iTRAQ tags 116: control macrophage model; iTRAQ tags 117: sample macrophage model. In the first dimension, peptides were fractionated by basic reversed-phase chromatography in a 1200 HPLC system (Agilent) and were collected using a Gilson FC203B fraction collector (Gilson, Middleton, WI). The collected peptide fractions were desalted with the aid of home made stage tips and separated by reversed-phase chromatography at acid pH in a nanoLC system (Tempo, ABSciex, Foster City, CA). The peptides were desalted and concentrated in a trapping column (0.5 × 2 mm, Michrom Bioresources, Auburn, CA) at a flow rate of 15 μL/min for 15 min and loaded onto a C18 column (Integrafit C18, Proteopep II, 75 μm i.d., 15 cm, 5 μm, 300 Å; New Objective, Woburn, MA). Peptides were eluted at a flow rate of 0.35 μL/min during a 2 h linear gradient from 2 to 40 % B (mobile phase A: 0.1 % trifluoroacetic acid, 2 % acetonitrile; mobile phase B: 0.1 % trifluoroacetic acid, 95 % acetonitrile), mixed with α-cyano matrix (4 mg/mL at a flow rate of 1.2 μL/ min), automatically deposited on a MALDI plate using a MALDI spotter (SunCollect, Sunchrome, Friedrichsdorf, Germany) and were finally analyzed in a 4800 TOF/TOF system (ABSciex, Foster City, CA). The chromatograms were composed by 480 spots, each comprising a 15 s deposition. Explorer v.4.2 (ABSciex, Foster City, CA) software (series 4000) was used to generate both the spectra and peak list. Plate model and default calibration of the MALDI plate were performed at a laser voltage of 3400 kV and 1000 shots/spectrum. Samples were automatically analyzed in MS mode at a laser voltage of 3800 kV and 1500 shots/spectrum.
Automated precursor selection was carried out by a Jobwide interpretation method, which selects up to 12 precursors per fraction. The lower threshold of signal to noise (50) and trypsin autolytic peptides and other background ions were excluded. The laser voltage was 4800 kV, and 2000 shots/spectrum were acquired using a medium-range CID collision energy.
LC-MALDI-TOF/TOF data were analyzed using Protein Pilot 4.0 software (ABSciex, Foster City, CA) as the search engine for protein identification. Protein Pilot search parameters were as follows: Cys-alkylation, iodoacetamide; iTRAQ 4plex quantitation mode, biological modifications; digestion with trypsin; All searches were performed against the non-redundant NCBI library (http://ncbi.nlm.nih.gov) database comprising annotated proteins of Acinetobacter baumannii AbH12O-A2 and pMMA2 plasmid. The search effort comprised thorough ID and detection protein threshold of unused ProtScore (Conf > 1.3 (95.0 %)). The scoring model was defined by the Paragon algorithm. Bias and background correction were applied to correct experimental differences in total amount of protein used in the experiment and label interference, respectively. Proteins showing sample/control protein expression ratios below 0.5 or above 1.5 (p < 0.05) were considered to be respectively under- or over-expressed and were selected for further analysis.
Purity of the starting material
The MS analysis of A. baumannii in BALF model and in macrophage model did not reveal any of the most abundant BALF proteins (serum albumin, IgG, IgA, transferrin, haptoglobin and antitrypsin) [80], nor the most abundant protein component of alveolar surfactant (surfactant protein A) [81], nor the very abundant enzymes phosphoglycerate kinase (cytosol of macrophages) and metallopeptidase 9 (macrophage membranes) [82]. We therefore, concluded that the starting material for the A. baumannii proteome analysis was not (or only minimally) contaminated with host material. This was not surprising because BALF proteins sediment at much higher centrifugation speeds than bacteria and because we could barely see nonadherent macrophage cells (see section “Macrophage infection with A. baumannii”).
Bioinformatics
Protein function was determined by http://www.ncbi.nlm.nih.gov.
HHomp, http://toolkit.tuebingen.mpg.de/hhomp# was used for the prediction of outer membrane proteins.
The presence of export signals was predicted by use of a combination of algorithms (SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/), Phobius (http://phobius.sbc.su.se/), PrediSi (http://www.predisi.de/), TatP 1.0 (www.cbs.dtu.dk/services/TatP-1.0), Tatfind 1.4 (http://signalfind.org/tatfind.html), SecretomeP 2.0 (www.cbs.dtu.dk/services/SecretomeP), TMHMM (http://www.cbs.dtu.dk/services/TMHMM/), DAS-TMfilter (http://www.enzim.hu/DAS/DAS.html), LipoP 1.0 (www.cbs.dtu.dk/services/LipoP), DOLOP (http://www.mrc-lmb.cam.ac.uk/genomes/dolop/), and LIPO (http://services.cbu.uib.no/tools/lipo)). [see Additional file 3].
RNA extraction and RT-PCR
Total RNA was extracted using the High Pure RNA isolation kit (Roche, Mannheim, Germany), according to the manufacturer’s instructions. PCR without reverse transcriptase confirmed the absence of DNA. Templates of 100 ng of total RNA were used in the target gene studies. Real-time PCR analysis of gene expression was performed in duplicate with specific internal oligonucleotide primers and the TaqMan probe (Universal ProbeLibrary-UPL, Roche, Mannheim, Germany). All primers and UPL probes used in the RT-PCR study are shown in Additional file 4.
Availability of supporting data
The proteomics data sets supporting the results of this article are included within the article and its Additional file 5.
Acknowledgments
This study was funded by grants from the European Community, FP7, ID: 278232 (MagicBullet), and by the Plan Nacional de I+D+I 2008-2011 and Instituto de Salud Carlos III, Subdirección General de Redes y Centros de Investigación Cooperativa, Ministerio de Economía y Competitividad, Spanish Network for Research in Infectious Diseases (REIPI RD12/0015/0014), cofinanced by the European Development Regional Fund (EDRF) “A Way to Achieve Europe.” M. T. and A.B. were financially supported by the Miguel Servet Programme ISCIII-FEDER (CP09/00033 and CP13/00226, respectively). This work was supported by the National Plans for Scientific Research, Development and Technological Innovation 2008-2011 and 2013-2016 and funded by the ISCIII- General Subdirection of Assesment and Promotion of the Research – European Regional Development Fund (ERDF) “A way of making Europe”: PI10/00056 and PI13/02390 to M. T., PI11/01034 to M.P., PI12/00552 to G.B. and PI14/00059 to M.P. and A.B.
We are grateful to Carmen M. Gayoso Babio (Health in Code) for invaluable assistance with use of bioinformatics tools, to Soraya Rumbo Feal (INIBIC), María Merino Carballeira (INIBIC), Patricia Fernández Puente (INIBIC) and Purificación Filgueira Fernández (INIBIC) for expert technical assistance, and to laboratory technician Ma del Carmen Fernández López.
Abbreviations
- BALF
Bronchoalveolar lavage fluid
- LB
Luria-Bertani broth
- LPS
Lipopolysaccharides
- NCBI
National Center for Biotechnology Information
- MH
Mueller Hinton
- DMEM
Dulbecco’s Modified Eagle’s Medium
- PBS
Phosphate-buffered saline
Additional files
Workflow of the proteomic experiment. Strategy used to recover and identify the proteins of A. baumannii in 2 ex vivo models for proteomic analysis.
Histopathology shows signs of consolidated pneumonia in infected animals. A), C) and E) representative low (×4) - and B), D) and F) high (×40)-power histological sections of lungs from three rats infected with A. baumannii for 21 h. A – F haematoxylin and eosin staining.
Prediction of exoproteins. This figure indicates the sequential use of different algorithms into a majority vote decision. Coding sequences were scanned for the presence of signal peptide specific to Sec pathway and Tat pathway. Coding sequences exhibiting no signal peptide were screened as potential nonclassically secreted proteins using SecretomeP 2.0. Proteins predicted as secreted were then asked for the presence of cell-envelope retention domain and erased from the output in positive case. Y, yes; N, no.
RT-PCR analysis of different genes. Primers and Univesal ProbeLibrary (UPL, Roche) probes used in this study.
Protein identification data. Excel workbook containing 2 worksheets with the complete report for the proteins in both models exported from Protein Pilot software.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JAM designed, performed the experiments and wrote the manuscript. JM, AB & ML designed, performed the experiments and critically reviewed the manuscript. MT provided statistical analysis for this project. MP assisted in the identification of proteins and GB conceived the study, participated in all steps of the project as coordinator and critically reviewed the manuscript. All authors read and approved the final manuscript.
Contributor Information
Jose Antonio Méndez, Email: jose.antonio.mendez.arredondo@sergas.es.
Jesús Mateos, Email: Jesus.Mateos.Martin@sergas.es.
Alejandro Beceiro, Email: Alejandro.Beceiro.Casas@sergas.es.
María Lopez, Email: malopedi@gmail.com.
María Tomás, Email: MA.del.Mar.Tomas.Carmona@sergas.es.
Margarita Poza, Email: Margarita.Poza.Dominguez@sergas.es.
Germán Bou, Email: German.Bou.Arevalo@sergas.es.
References
- 1.Howard A, Donoghue MO, Feeney A, Sleator RD. Acinetobacter baumannii an emerging opportunistic pathogen. Virulence. 2012;3:243–250. doi: 10.4161/viru.19700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Visca P, Seifert H, Towner KJ. Acinetobacter infection—an emerging threat to human health. IUBMB Life. 2011;63:1048–1054. doi: 10.1002/iub.534. [DOI] [PubMed] [Google Scholar]
- 3.Anstey NM, Currie BJ, Hassell M, Palmer D, Dwyer B, Seifert H. Community-acquired bacteremic Acinetobacter pneumonia in tropical Australia is caused by diverse strains of Acinetobacterbaumannii with carriage in the throat in at-risk groups. J Clin Microbiol. 2002;40:685–686. doi: 10.1128/JCM.40.2.685-686.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leung WS, Chu CM, Tsang KY, Lo FH, Lo KF, Ho PL. Fulminant community-acquired Acinetobacter baumannii pneumonia as a distinct clinical syndrome. Chest. 2006;129:102–109. doi: 10.1378/chest.129.1.102. [DOI] [PubMed] [Google Scholar]
- 5.Giamarellou H, Antoniadou AK, Kanellakopoulou K. Acinetobacterbaumannii: a universal threat to public health? Int J Antimicrob Agents. 2008;32:106–119. doi: 10.1016/j.ijantimicag.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Theaker C, Azadian B, Soni N. The impact of Acinetobacter baumannii in the intensive care unit. Anaesthesia. 2003;58:271–274. doi: 10.1046/j.1365-2044.2003.30403.x. [DOI] [PubMed] [Google Scholar]
- 7.Chen CH, Lin LC, Chang YJ, Huang CC, Liu CE, Young TG. Analysis of prognostic factors in 95 patients with Acinetobacter baumannii bacteremia. Infection. 2003;31:331–335. doi: 10.1007/s15010-003-3223-1. [DOI] [PubMed] [Google Scholar]
- 8.Rodríguez-Baño J, Cisneros JM, Fernández-Cuenca F, Ribera A, Vila J, Pascual A, Martínez-Martínez L, Bou G, Pachón J. Clinical features and epidemiology of Acinetobacter baumannii colonization and infection in Spanish hospitals. Infect Control Hosp Epidemiol. 2004;25:819–24. doi: 10.1086/502302. [DOI] [PubMed] [Google Scholar]
- 9.Gayoso CM, Mateos J, Méndez JA, Fernández-Puente P, Rumbo C, Tomás M, Martínez de Ilarduya O, Bou G. Molecular mechanisms involved in the response to desiccation stress and persistence in Acinetobacterbaumannii. J Proteome Res. 2014;13:460–76. doi: 10.1021/pr400603f. [DOI] [PubMed] [Google Scholar]
- 10.Eveillard M, Kempf M, Belmonte O, Pailhoriès H, Joly-Guillou ML. Reservoirs of Acinetobacter baumannii outside the hospital and potential involvement in emerging human community-acquired infections. Int J Infect Dis. 2013;17:e802–5. doi: 10.1016/j.ijid.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 11.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seifert H, Dijkshoorn L, Gerner-Smidt P, Pelzer N, Tjernberg I, Vaneechoutte M. Distribution of Acinetobacter species on human skin: comparison of phenotypic and genotypic identification methods. J Clin Microbiol. 1997;35:2819–25. doi: 10.1128/jcm.35.11.2819-2825.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chu YW, Leung CM, Houang ET, Ng KC, Leung CB, Leung HY, Cheng AF. Skin carriage of acinetobacters in Hong Kong. J Clin Microbiol. 1999;35:2962–2967. doi: 10.1128/jcm.37.9.2962-2967.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fournier PE, Richet H. The epidemiology and control of Acinetobacter baumannii in health care facilities. Clin Infect Dis. 2006;42:692–9. doi: 10.1086/500202. [DOI] [PubMed] [Google Scholar]
- 15.Camp C, Tatum OL. A review of Acinetobacter baumannii as a highly successful pathogen in times of war. Lab Medicine. 2010;41:649–657. doi: 10.1309/LM90IJNDDDWRI3RE. [DOI] [Google Scholar]
- 16.Hanlon GW. The emergence of multidrug resistant Acinetobacter species: a major concern in the hospital setting. Lett Appl Microbiol. 2005;41:375–8. doi: 10.1111/j.1472-765X.2005.01791.x. [DOI] [PubMed] [Google Scholar]
- 17.Morgan DJ, Liang SY, Smith CL, Johnson JK, Harris AD, Furuno JP, Thom KA, Snyder GM, Day HR, Perencevich EN. Frequent multidrug-resistant Acinetobacter baumannii contamination of gloves gowns and hands of healthcare workers. Infect Control Hosp Epidemiol. 2010;31:716–721. doi: 10.1086/653201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wendt C, Dietze B, Dietz E, Ruden H. Survival of Acinetobacter baumannii on dry surfaces. J Clin Microbiol. 1997;35:1394–1397. doi: 10.1128/jcm.35.6.1394-1397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rao R, Karthika R, Singh S, Shashikala P, Kanungo R, Jayachandran S, Prashanth K. Correlation between biofilm production and multiple drug resistance in imipenem resistant clinical isolates of Acinetobacter baumannii. Indian J Med Microbiol. 2008;4:333–337. doi: 10.4103/0255-0857.43566. [DOI] [PubMed] [Google Scholar]
- 20.Gales AC, Jones RN, Sader HS. Global assessment of the antimicrobial activity of polymyxin B against 54 731 clinical isolates of Gram-negative bacilli: report from the SENTRY antimicrobial surveillance programme (2001–2004) Clin Microbiol Infect. 2006;12:315–321. doi: 10.1111/j.1469-0691.2005.01351.x. [DOI] [PubMed] [Google Scholar]
- 21.Ko KS, Suh JY, Kwon KT, Jung SI, Park KH, Kang CI, Chung DR, Peck KR, Song JH. High rates of resistance to colistin and polymyxin B in subgroups of Acinetobacter baumannii isolates from Korea. J Antimicrob Chemother. 2007;60:1163–1167. doi: 10.1093/jac/dkm305. [DOI] [PubMed] [Google Scholar]
- 22.Barbe V, Vallenet D, Fonknechten N, Kreimeyer A, Oztas S, Labarre L, Cruveiller S, Robert C, Duprat S, Wincker P, Ornston LN, Weissenbach J, Marlière P, Cohen GN, Médigue C. Unique features revealed by the genome sequence of Acinetobacter sp ADP1 a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 2004;32:5766–5779. doi: 10.1093/nar/gkh910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith MG, Gianoulis TA, Pukatzki S, Mekalanos JJ, Ornston LN, Gerstein M, Snyder M. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 2007;21:601–614. doi: 10.1101/gad.1510307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mendez JA, Soares NC, Mateos J, Gayoso C, Rumbo C, Aranda J, Tomas M, Bou G. Extracellular proteome of a highly invasive multidrug-resistant clinical strain of Acinetobacter baumannii. J Proteome Res. 2012;11:5678–94. doi: 10.1021/pr300496c. [DOI] [PubMed] [Google Scholar]
- 25.Kwon SO, Gho YS, Lee JC, Kim SI. Proteome analysis of outer membrane vesicles from a clinical Acinetobacter baumannii isolate. FEMS Microbiol Lett. 2009;297:150–6. doi: 10.1111/j.1574-6968.2009.01669.x. [DOI] [PubMed] [Google Scholar]
- 26.Tiwari V, Vashistt J, Kapil A, Moganty RR. Comparative proteomics of inner membrane fraction from carbapenem-resistant Acinetobacter baumannii with a reference strain. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0039451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yun SH, Choi CW, Kwon SO, Park GW, Cho K, Kwon KH, Kim JY, Yoo JS, Lee JC, Choi JS, Kim S, Kim SI. Quantitative proteomic analysis of cell wall and plasma membrane fractions from multidrug-resistant Acinetobacter baumannii. J Proteome Res. 2011;10:459–69. doi: 10.1021/pr101012s. [DOI] [PubMed] [Google Scholar]
- 28.Vashist J, Tiwari V, Kapil A, Rajeswari MR. Quantitative profiling and identification of outer membrane proteins of beta-lactam resistant strain of Acinetobacter baumannii. J Proteome Res. 2010;9:1121–8. doi: 10.1021/pr9011188. [DOI] [PubMed] [Google Scholar]
- 29.Yun SH, Choi CW, Park SH, Lee JC, Leem SH, Choi JS, Kim S, Kim SI. Proteomic analysis of outer membrane proteins from Acinetobacter baumannii DU202 in tetracycline stress condition. J Microbiol. 2008;46:720–7. doi: 10.1007/s12275-008-0202-3. [DOI] [PubMed] [Google Scholar]
- 30.Soares NC, Cabral MP, Parreira JR, Gayoso C, Barba MJ, Bou G. 2-DE analysis indicates that Acinetobacter baumannii displays a robust and versatile metabolism. Proteome Sci. 2009;7:37. doi: 10.1186/1477-5956-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cabral MP, Soares NC, Aranda J, Parreira JR, Rumbo C, Poza M, Valle J, Calamia V, Lasa I, Bou G. Proteomic and functional analyses reveal a unique lifestyle for Acinetobacter baumannii biofilms and a key role for histidine metabolism. J Proteome Res. 2011;10:3399–417. doi: 10.1021/pr101299j. [DOI] [PubMed] [Google Scholar]
- 32.Chopra S, Ramkissoon K, Anderson DC. A systematic quantitative proteomic examination of multidrug resistance in Acinetobacter baumannii. J Proteomics. 2013;12:17–39. doi: 10.1016/j.jprot.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 33.Pradet-Balade B, Boulme F, Beug H, Mullner EW, Garcia-Sanz JA. Translation control: bridging the gap between genomics and proteomics? Trends Biochem Sci. 2001;26:225–229. doi: 10.1016/S0968-0004(00)01776-X. [DOI] [PubMed] [Google Scholar]
- 34.Echenique JR, Dorsey CW, Patrito LC, Petroni A, Tolmasky ME, Actis LA. Acinetobacter baumannii has two genes encoding glutathione-dependent formaldehyde dehydrogenase: evidence for differential regulation in response to iron. Microbiology. 2001;147:2805–2815. doi: 10.1099/00221287-147-10-2805. [DOI] [PubMed] [Google Scholar]
- 35.Hood MI, Jacobs AC, Sayood K, Dunman PM, Skaar EP. Acinetobacter baumannii increases tolerance to antibiotics in response to monovalent cations. Antimicrob Agents Chemother. 2010;54:1029–41. doi: 10.1128/AAC.00963-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brossard KA, Campagnari AA. The Acinetobacter baumannii biofilm-associated protein plays a role in adherence to human epithelial cells. Infect Immun. 2012;80:228–33. doi: 10.1128/IAI.05913-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zimbler DL, Park TM, Arivett BA, Penwell WF, Greer SM, Woodruff TM, Tierney DL, Actis LA. Stress response and virulence functions of the Acinetobacter baumannii NfuA Fe-S scaffold protein. J Bacteriol. 2012;194:2884–93. doi: 10.1128/JB.00213-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eijkelkamp BA, Hassan KA, Paulsen IT, Brown MH. Investigation of the human pathogen Acinetobacter baumannii under iron limiting conditions. BMC Genomics. 2011;12:126. doi: 10.1186/1471-2164-12-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jacobs AC, Sayood K, Olmsted SB, Blanchard CE, Hinrichs S, Russell D, Dunman PM. Characterization of the Acinetobacter baumannii growth phase-dependent and serum responsive transcriptomes. FEMS Immunol Med Microbiol. 2012;64:403–12. doi: 10.1111/j.1574-695X.2011.00926.x. [DOI] [PubMed] [Google Scholar]
- 40.Nwugo CC, Gaddy JA, Zimbler DL, Actis LA. Deciphering the iron response in Acinetobacter baumannii: a proteomics approach. J Proteomics. 2011;74:44–58. doi: 10.1016/j.jprot.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X, El-Hajj ZW, Newman E. Deficiency in L-Serine deaminase interferes with one-carbon metabolism and cell wall synthesis in Escherichia coli K-12. J Bacteriol. 2010;192:5515–5525. doi: 10.1128/JB.00748-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emanuele JJ, Jr, Jin H, Jacobson BL, Chang CY, Einspahr HM, Villafranca JJ. Kinetic and crystallographic studies of Escherichia coli UDP-N-acety1muramate:L-alanine ligase. Protein Sci. 1996;5:2566–2574. doi: 10.1002/pro.5560051219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Camejo A, Buchrieser C, Couvé E, Carvalho F, Reis O, Ferreira P, Sousa S, Cossart P, Cabanes D. In vivo transcriptional profiling of listeria monocytogenes and mutagenesis identify new virulence factors involved in infection. PLoS Pathog. 2009;5:e1000449. doi: 10.1371/journal.ppat.1000449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.White TA, Kell DB. Comparative genomic assessment of novel broad-spectrum targets for antibacterial drugs. Comp Funct Genom. 2004;5:304–327. doi: 10.1002/cfg.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.El Zoeiby A, Sanschagrin F, Lamoureux J, Darveau A, Levesque RC. Cloning, over-expression and purification of Pseudomonas aeruginosa murC encoding uridine diphosphate N-acetylmuramate: L-alanine ligase. FEMS Microbiol Lett. 2000;183(2):281–8. doi: 10.1111/j.1574-6968.2000.tb08972.x. [DOI] [PubMed] [Google Scholar]
- 46.Zawadzke LE, Norcia M, Desbonnet CR, Wang H, Freeman-Cook K, Dougherty TJ. Identification of an inhibitor of the MurC enzyme, which catalyzes an essential step in the peptidoglycan precursor synthesis pathway. Assay Drug Dev Technol. 2008;6:95–103. doi: 10.1089/adt.2007.114. [DOI] [PubMed] [Google Scholar]
- 47.Hameed PS, Manjrekar P, Chinnapattu M, Humnabadkar V, Shanbhag G, Kedari C, Mudugal NV, Ambady A, de Jonge BL, Sadler C, Paul B, Sriram S, Kaur P, Guptha S, Raichurkar A, Fleming P, Eyermann CJ, McKinney DC, Sambandamurthy VK, Panda M, Ravishankar S. Pyrazolopyrimidines establish MurC as a vulnerable target in Pseudomonas aeruginosa and Escherichia coli. ACS Chem Biol. 2014;9(10):2274–82. doi: 10.1021/cb500360c. [DOI] [PubMed] [Google Scholar]
- 48.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 49.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deng WL, Lin YC, Lin RH, Wei CF, Huang YC, Peng HL, Huang HC. Effects of galU Mutation on Pseudomonas syringae–plant interactions. Mol Plant-Microbe Interact. 2010;23:1184–1196. doi: 10.1094/MPMI-23-9-1184. [DOI] [PubMed] [Google Scholar]
- 51.Jayakar HR, Parvathareddy J, Fitzpatrick EA, Bina XR, Bina JE, Re F, Emery FD, Miller MA. A galU mutant of francisella tularensis is attenuated for virulence in a murine pulmonary model of tularemia. BMC Microbiol. 2011;179:1471–2180. doi: 10.1186/1471-2180-11-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang SS, Lin TY, Wang WB, Liu MC, Hsueh PR, Liaw SJ. Characterization of UDP-glucose dehydrogenase and UDP-glucose pyrophosphorylase mutants of Proteus mirabilis: defectiveness in polymyxin B resistance swarming and virulence. Antimicrob Agents Chemother. 2010;54:2000–2009. doi: 10.1128/AAC.01384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mollerach M, López R, García E. Characterization of the galU gene of Streptococcus pneumoniae encoding a uridine diphosphoglucose pyrophosphorylase: a gene essential for capsular polysaccharide biosynthesis. J Exp Med. 1998;188:2047–2056. doi: 10.1084/jem.188.11.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Priebe GP, Dean CR, Zaidi T, Meluleni GJ, Coleman FT, Coutinho YS, Noto MJ, Urban TA, Pier GB, Goldberg JB. The galU Gene of Pseudomonas aeruginosa is required for corneal infection and efficient systemic spread following pneumonia but not for infection confined to the lung. Infect Immun. 2004;72:4224–4232. doi: 10.1128/IAI.72.7.4224-4232.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crosa JH, Mey AR, Payne SM. Iron Transport in Bacteria. Washington, DC: ASM Press; 2004. [Google Scholar]
- 56.Skaar EP. The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dittrich AM, Meyer HA, Hamelmann E. The role of lipocalins in airway disease. Clin Exp Allergy. 2013;43:503–511. doi: 10.1111/cea.12025. [DOI] [PubMed] [Google Scholar]
- 58.Välimäki E, Miettinen JJ, Lietzén N, Matikainen S, Nyman TA. Monosodium urate activates Src/Pyk2/PI3 Kinase and Cathepsin dependent unconventional protein secretion from human primary macrophages. Mol Cell Proteomics. 2013;12:749–763. doi: 10.1074/mcp.M112.024661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev. 2007;71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raffatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio SP, Paixao TA. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe. 2009;5:476–486. doi: 10.1016/j.chom.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Proschak A, Lubuta P, Grün P, Löhr F, Wilharm G, Berardinis VD, Bode HB. Structure and Biosynthesis of Fimsbactins A–F, Siderophores from Acinetobacter baumannii and Acinetobacter baylyi. ChemBioChem. 2013;14:633–638. doi: 10.1002/cbic.201200764. [DOI] [PubMed] [Google Scholar]
- 62.Wen ZT, Suntharaligham P, Cvitkovitch DG, Burne RA. Trigger factor in Streptococcus mutans is involved in stress tolerance competence development and biofilm formation. Infect Immun. 2005;73:219–225. doi: 10.1128/IAI.73.1.219-225.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ullers RS, Schwager DAF, Georgopoulos C, Genevaux P. Trigger factor can antagonize both SecB and DnaK/DnaJ chaperone functions in Escherichia coli. PNAS. 2007;104:3101–3106. doi: 10.1073/pnas.0608232104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bigot A, Botton E, Dubail I, Charbit A. A homolog of Bacillus subtilis trigger factor in Listeria monocytogenes is involved in stress tolerance and bacterial virulence. Appl Environ Microbiol. 2006;72:6623–6631. doi: 10.1128/AEM.00624-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Henard CA, Bourret TJ, Song M, Vázquez-Torres A. Control of redox balance by the stringent response regulatory protein promotes antioxidant defenses of Salmonella. J Biol Chem. 2010;285:36785–36793. doi: 10.1074/jbc.M110.160960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Frees D, Chastanet A, Qazi S, Sørensen K, Hill P, Msadek T, Ingmer H. Clp ATPases are required for stress tolerance intracellular replication and biofilm formation in Staphylococcus aureus. Mol Microbiol. 2004;54:1445–1462. doi: 10.1111/j.1365-2958.2004.04368.x. [DOI] [PubMed] [Google Scholar]
- 67.Eijkelkamp BA, Stroeher UH, Hassan KA, Papadimitrious MS, Paulsen IT, Brown MH. Adherence and motility characteristics of clinical Acinetobacter baumannii isolates. FEMS Microbiol Lett. 2011;323:44–51. doi: 10.1111/j.1574-6968.2011.02362.x. [DOI] [PubMed] [Google Scholar]
- 68.Choi CH, Lee EY, Lee YC, Park TI, Kim HJ, Hyun SH, Kim SA, Lee SK, Lee JC. Outer membrane protein 38 of Acinetobacter baumannii localizes to the mitochondria and induces apoptosis of epithelial cells. Cell Microbiol. 2005;7:1127–1138. doi: 10.1111/j.1462-5822.2005.00538.x. [DOI] [PubMed] [Google Scholar]
- 69.Gaddy JA, Tomaras AP, Actis LA. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect Immun. 2009;77:3150–3160. doi: 10.1128/IAI.00096-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Law RJ, Hamlin JN, Sivro A, McCorrister SJ, Cardama GA, Cardona ST. A functional phenylacetic acid catabolic pathway is required for full pathogenicity of Burkholderia cenocepacia in the Caenorhabditis elegans host model. J Bacteriol. 2008;190:7209–7218. doi: 10.1128/JB.00481-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu JW, Chen XL. Extracellular metalloproteases from bacteria. Appl Microbiol Biotechnol. 2011;92:253–262. doi: 10.1007/s00253-011-3532-8. [DOI] [PubMed] [Google Scholar]
- 72.Moro A, Ruiz-Cabello F, Fernandez-Cano A, Stock RP, González A. Secretion by trypanosoma cruzi of a peptidyl-prolpyl cis-trans isomerase involved in cell infection. EMBO J. 1995;14:2483–2490. doi: 10.1002/j.1460-2075.1995.tb07245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moulder JW. Comparative biology of intracellular parasitism. Microbiol Rev. 1985;49:298–337. doi: 10.1128/mr.49.3.298-337.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hermans PWM, Adrian PV, Albert C, Estevao S, Hoogenboezem T, Luijendijk IHT, Kamphausen T, Hammerschmidt S. The Streptococcal lipoprotein rotamase A (SlrA) is a functional peptidyl-prolyl isomerase involved in pneumococcal colonization. J Biol Chem. 2006;281:968–976. doi: 10.1074/jbc.M510014200. [DOI] [PubMed] [Google Scholar]
- 75.Xia Q, Wang T, Taub F, Park Y, Capestany CA, Lamont RJ, Hackett M. Quantitative proteomics of intracellular Porphyromonas gingivalis. Proteomics. 2007;7:4323–4337. doi: 10.1002/pmic.200700543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dieppedale J, Sobral D, Dupuis M, Dubail I, Klimentova J, Stulik J, Postic G, Frapy E, Meibom KL, Barel M. Charbit: a identification of a putative chaperone involved in stress resistance and virulence in Francisella tularensis. Infect Immun. 2011;79:1428–1439. doi: 10.1128/IAI.01012-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Umland TC, Schultz LW, MacDonald U, Beanan JM, Olson R, Russo TA. In vivo-validated essential genes identified in acinetobacter baumanniiby using human ascites overlap poorly with essential genes detected on laboratory media. mBio. 2012;3:e00113–12. doi: 10.1128/mBio.00113-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Merino M, Alvarez-Fraga L, Gómez MJ, Aransay AM, Lavín JL, Chaves F, Bou G, Poza M. Complete genome sequence of the multiresistant Acinetobacter baumannii strain AbH12O-A2, isolated during a large outbreak in Spain. Genome Announc. 2014;2(6):e01182–14. doi: 10.1128/genomeA.01182-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fernández-Puente P, Mateos J, Fernández-Costa C, Oreiro N, Fernández-López C, Ruiz-Romero C, Blanco FJ. Identification of a panel of novel serum osteoarthritis biomarkers. J Proteome Res. 2011;10:5095–5101. doi: 10.1021/pr200695p. [DOI] [PubMed] [Google Scholar]
- 80.Wu J, Kobayashi M, Sousa EA, Liu W, Cai J, Goldman SJ, Dorner AJ, Projan SJ, Kavuru MS, Yongchang Qiu Y, Thomassen MJ. Differential proteomic analysis of bronchoalveolar lavage fluid in asthmatics following segmental antigen challenge. Mol Cell Proteomics. 2005;4:1251–64. doi: 10.1074/mcp.M500041-MCP200. [DOI] [PubMed] [Google Scholar]
- 81.Crowther JE, Kutala VK, Kuppusamy P, Ferguson JS, Beharka AA, Zweier JL, McCormack FX, Schlesinger LS. Pulmonary surfactant protein a inhibits macrophage reactive oxygen intermediate production in response to stimuli by reducing NADPH oxidase activity. J Immunol. 2004;172:6866–74. doi: 10.4049/jimmunol.172.11.6866. [DOI] [PubMed] [Google Scholar]
- 82.Polati R, Castagna A, Bossi A, Campostrini N, Zaninotto F, Timperio AM, Zolla L, Olivieri O, Corrocher R, Girelli D. High resolution preparation of monocyte-derived macrophages (MDM) protein fractions for clinical proteomics. Proteome Sci. 2009;7:4. doi: 10.1186/1477-5956-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]


