Abstract
Background: In western North America, plague epizootics caused by Yersinia pestis appear to sweep across landscapes, primarily infecting and killing rodents, especially ground squirrels and prairie dogs. During these epizootics, the risk of Y. pestis transmission to humans is highest. While empirical models that include climatic conditions and densities of rodent hosts and fleas can predict when epizootics are triggered, bacterial transmission patterns across landscapes, and the scale at which Y. pestis is maintained in nature during inter-epizootic periods, are poorly defined. Elucidating the spatial extent of Y. pestis clones during epizootics can determine whether bacteria are propagated across landscapes or arise independently from local inter-epizootic maintenance reservoirs.
Material and Methods: We used DNA microarray technology to identify single-nucleotide polymorphisms (SNPs) in 34 Y. pestis isolates collected in the western United States from 1980 to 2006, 21 of which were collected during plague epizootics in Colorado. Phylogenetic comparisons were used to elucidate the hypothesized spread of Y. pestis between the mountainous Front Range and the eastern plains of northern Colorado during epizootics. Isolates collected from across the western United States were included for regional comparisons.
Results: By identifying SNPs that mark individual clones, our results strongly suggest that Y. pestis is maintained locally and that widespread epizootic activity is caused by multiple clones arising independently at small geographic scales. This is in contrast to propagation of individual clones being transported widely across landscapes. Regionally, our data are consistent with the notion that Y. pestis diversifies at relatively local scales following long-range translocation events. We recommend that surveillance and prediction by public health and wildlife management professionals focus more on models of local or regional weather patterns and ecological factors that may increase risk of widespread epizootics, rather than predicting or attempting to explain epizootics on the basis of movement of host species that may transport plague.
Key Words: : Yersinia pestis, Plague–Single-nucleotide polymorphisms, Spatial diversity, Western United States, Colorado
Introduction
Yersinia pestis, the causative agent of plague, draws attention because of its documented potential for spillover and devastating human disease outbreaks killing millions (Pollitzer 1954). Today, approximately 3000 human cases occur worldwide annually, mostly in Africa, with smaller numbers occurring in Asia and the Americas. In the United States, the frequency of human disease is low, with an average of eight cases reported annually (range, 1–40) from 1950 to 2008 (World Health Organization 2004, 2010). Nevertheless, case fatality rates are high if appropriate antibiotic therapy is delayed or inadequate (Hull et al. 1986), making plague a disease of high consequence. Currently, Y. pestis is established in hundreds of natural plague foci around the world where it causes sporadic epizootics, primarily in rodents (Link 1955, Barnes 1982, Anisimov et al. 2004, Zhou et al. 2004). During plague epizootics, humans are at the greatest risk of exposure to Y. pestis because mass rodent die offs increase the potential for human exposure to sick or dead animals and infectious fleas (Barnes 1982, Eisen and Gage 2009). Recently, plague also has been identified as a significant concern for the conservation of certain species, including prairie dogs and black-footed ferrets in the western United States where the disease is enzootic in many locations (Matchett et al. 2010).
Despite its historical and public health significance, our understanding of Y. pestis maintenance in nature is limited. Although empirical models have linked plague epizootics and climatic conditions, as well as increased rodent and flea densities (Parmenter et al. 1999, Enscore et al. 2002, Davis et al. 2004, Gage and Kosoy 2005, Gage et al. 2008, Ben-Ari et al. 2011, Savage et al. 2011), how Y. pestis bacteria are maintained during inter-epizootic periods is still not clearly understood. Alternative (but not mutually exclusive) hypotheses include: (1) Maintenance in off-host fleas (Kartman et al. 1962, Bazanova and Maevskii 1996, Gage and Kosoy 2005), (2) via transmission cycles between “enzootic” hosts (Barnes 1982, Gage et al. 1994, Gage and Kosoy 2005, Eisen and Gage 2009), (3) persistence among metapopulations of rodent hosts where epizootics occur in relatively isolated subpopulations (Keeling and Gilligan 2000, Davis et al. 2004, Collinge et al. 2005, George et al. 2013), (4) as a percolation phenomenon highlighting interactions between spatial structure of host populations and abundance (Davis et al. 2008, Salkeld et al. 2010), and (5) bacterial persistence in soil following the death of infected animals (Eisen et al. 2008). Maintenance mechanisms may affect how Y. pestis bacteria spread across landscapes and the scope of epizootic activity. Currently, our understanding of how Y. pestis spreads across expansive land areas is largely based on observations of epizootics in animals and epidemics in humans (Gage and Kosoy 2005). In reality, we do not know whether increased transmission and epizootics arise from geographically isolated bacterial clones or from a chain of infection propagated through sustained transmission and geographic spread across large landscapes.
Using genetic markers to examine the population structure of Y. pestis isolates collected during widespread epizootic activity may elucidate if bacteria are being spread and at what scale, or if they may cause plague locally without transport. Our recent understanding of Y. pestis genetic diversity, evolution, and transmission patterns on several geographic scales has increased dramatically and suggests that plague exists as a clonal pathogen (Klevytska et al. 2001, Achtman et al. 2004, Girard et al. 2004, Lowell et al. 2005, Auerbach et al. 2007, Touchman et al. 2007, Vogler et al. 2007, Zhang et al. 2009, Morelli et al. 2010, Cui et al. 2013).
Much of the diversity detected among North American Y. pestis isolates has been based on analyses of repetitive genetic elements (variable number of tandem repeats, VNTRs) via multilocus VNTR analysis (MLVA), which is useful for molecular epidemiologic studies on geographically local scales. However, the mutation rates within repetitive genomic regions are too great to accurately reconstruct large-scale movements and transmission patterns (Keim et al. 2003, Girard et al. 2004, Lowell et al. 2007). Whole genome comparisons among globally distributed Y. pestis collections have revealed the evolutionary diversification of Y. pestis in its historic range in Asia, its worldwide spread, and among North American isolates (Achtman et al. 2004, Chain et al. 2004, 2006, Auerbach et al. 2007, Morelli et al. 2010, Cui et al. 2013, Wagner et al. 2014). However, we still do not know whether repeated epizootics in North America are caused by large-scale clonal spread of bacteria from an initial focus, or whether near-simultaneous eruptions of plague epizootics over large areas are the result of multiple epidemic clones arising at local scales, perhaps facilitated by a combination of high rodent or flea abundance and favorable environmental conditions (Enscore et al. 2002, Davis et al. 2004, Salkeld et al. 2010, Savage et al. 2011).
We used DNA microarray technology (Hinds et al. 2004) to identify SNPs in 34 Y. pestis isolates collected in the western United States from 1980 to 2006. A subset of 21 isolates from Colorado was collected during plague epizootics between 1999 and 2006. Single-nucleotide polymorphism (SNP) arrays provide an alternative to genome sequencing for standard laboratories, where the costs, labor, and time associated with whole genome library preparation, sequencing, bioinformatic analysis, and data storage are prohibitive. Furthermore, SNP arrays demonstrate concordance rates of up to 99.86% to SNPs discovered using full genome sequences and have yielded identical phylogenies in studies of Bacillus anthracis diversity (Gardner et al. 2013). The objectives of this study were to: (1) test the hypothesis that during widespread epizootic plague activity Y. pestis spreads clonally between the mountainous Front Range of northern Colorado and the eastern plains, and (2) provide additional information on the population structure of Y. pestis in the United States on local and regional scales. Elucidating Y. pestis population structure on local scales may determine how epizootics spread and may provide insight into long-term maintenance of Y. pestis during inter-epizootic periods. An understanding of how Y. pestis bacteria circulate during epizootics, and at what spatial scales they are maintained, will also facilitate development of predictive surveillance and control measures intended to aid public health professionals, wildlife ecologists, and natural resource managers in implementing focused recommendations during and between plague epizootics.
Materials and Methods
For this study, a total of 21 isolates were available for analysis that represented the northern Colorado mountainous Front Range and the plains of the Pawnee National Grasslands (Fig. 1). Isolates were grouped according to collection from mountain or plains locations (Table 1). All were collected from 1992 to 2006 during routine surveillance and human plague case investigations carried out by the Centers for Disease Control and Prevention's (CDC) Division of Vector-Borne Diseases (DVBD) and by researchers from Colorado State University (CSU) carrying out intensive epidemiologic studies of prairie dogs, fleas, and plague in northern Colorado (Tripp et al. 2009). During collection periods, evidence of plague epizootics, such as animal-to-human transmission and rodent die-offs, were apparent. Colorado isolates collected strictly from 1999 to 2006 were used to compare phylogeographic relationships during widespread epizootic activity that occurred during this interval in the northcentral mountains in Colorado and approximately 70–150 km further east on the plains of the Pawnee National Grasslands.
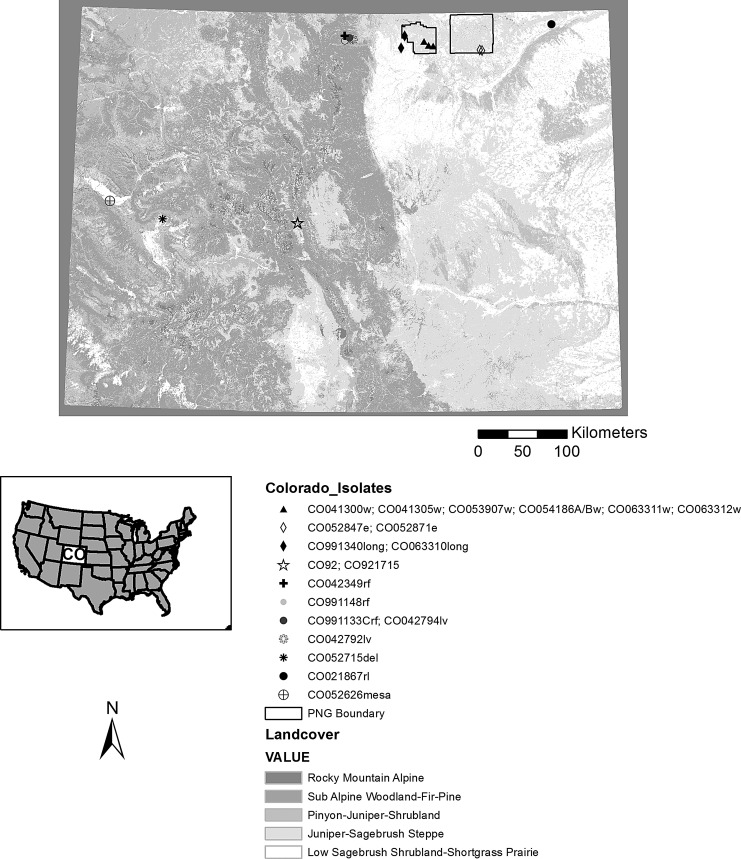
FIG. 1.
Colorado isolate collection locations. Symbols represent isolates with matching SNP profiles. CO054186A/Bw represents two isolates—CO054186Aw and CO054186Bw. Corresponding CDC accession numbers are listed in Table 1.
Table 1.
Origins of Y. pestis Isolates Used for Single-Nucleotide Polymorphism Discovery
| CDC accession numbera | County | Isolate origin | Collection location (general characteristic of area) | CDC collaborator in isolate collection |
|---|---|---|---|---|
| CO92 | Chaffee | human blood | Mountains | Colorado Department of Public Health and Environment |
| CO921715 | Chaffee | Chipmunk | Mountains | Colorado Department of Public Health and Environment |
| CO991133Crf | Larimer | human blood | Mountains | Colorado Department of Public Health and Environment |
| CO991148rf | Larimer | Cediopsylla inaequalis (rabbit flea) | Mountains | Colorado Department of Public Health and Environment |
| CO042349rf | Larimer | Human | Mountains | Colorado Department of Public Health and Environment |
| CO042792liv | Larimer | cat blood | Mountains | Larimer County Health Department, Colorado |
| CO042794liv | Larimer | cat blood | Mountains | Larimer County Health Department, Colorado |
| CO052871e | Weld | Cynomys ludovicianus (black-tailed prairie dog) | Plains | Colorado State University |
| CO052847e | Weld | Cynomys ludovicianus | Plains | Colorado State University |
| CO053907w | Weld | Cynomys ludovicianus | Plains | Colorado State University |
| CO054186Aw | Weld | Pulex sp. (flea) | Plains | Colorado State University |
| CO054186Bw | Weld | Cynomys ludovicianus | Plains | Colorado State University |
| CO041300w | Weld | Cynomys ludovicianus | Plains | Colorado State University |
| CO041305w | Weld | Cynomys ludovicianus | Plains | Colorado State University |
| CO063311w | Weld | Cynomys ludovicianus | Plains | Colorado State University |
| CO063312w | Weld | Cynomys ludovicianus | Plains | Colorado State University |
| CO052715del | Delta | unidentified flea species from coyote | Plains | USDA Wildlife Services |
| CO063310b | Larimer | Cynomys ludovicianus | Plains | Larimer County Health Department, Colorado |
| CO991340b | Larimer | Cynomys ludovicianus | Plains | Larimer County Health Department, Colorado |
| CO052626mes | Mesa | Spermophilus variegatus (rock squirrel) | Mountains | Mesa County Health Department, Colorado |
| CO021867rl | Sedgwick | Oropsylla hirsuta (prairie dog flea) | Plains | Colorado Department of Public Health and Environment |
| 80CA2178 | Los Angeles | Spermophilus beecheyi | Mountains | California Department of Health Services |
| CA812298 | Shasta | Spermophilus lateralis | Mountains | California Department of Health Services |
| AZ921389 | Apache | Human | Mountains | CDC/Indian Health Services |
| AZ962456 | Coconino | Human | Plains | CDC/Indian Health Services |
| NV870978_51 | Clark | Thrassis bacchi (antelope ground squirrel flea) | Mountains | CDC/Environmental Health Department, Clark County, Nevada |
| NM021852 | Santa Fe | Orchopeas sexdentatus (wood rat flea) | Mountains | New Mexico Department of Health |
| NM024479 | Santa Fe | Peromyscopsylla hesperomys (deer mouse flea) | Mountains | New Mexico Department of Health |
| NE054860 | Box Butte | Cynomys ludovicianus | Plains | USDA Wildlife Services |
| WY042773 | Goshen | Sylvilagus audubonii(desert cottontail rabbit) | Plains | Colorado Department of Public Health and Environment |
| WY000345 | Washakie | Human | Plains | Wyoming Department of Health |
| MT922215 | Big Horn | Lynx rufus (bobcat) | Plains | Wyoming Department of Health |
| SD053915_255 | Shannon | Oropsylla hirsuta | Plains | USDA Wildlife Services |
| SD053909 | Shannon | Cynomys ludovicianus | Plains | USDA Wildlife Services |
The CDC accession number consists of the state abbreviation followed by the last two digits of the collection year, except for isolate 80CA2178 in which these two identifiers are reversed. The following digits are unique identifiers. The extension abbreviations on each accession number are as follows: rf, Red Feather; liv, Livermore; e, Pawnee National Grasslands east; w, Pawnee National Grasslands west; rl, Red Lion National Wildlife Refuge; del, Delta County; mes, Mesa County.
Isolates representing temporal comparison.
CDC, Centers for Disease Prevention and Control.
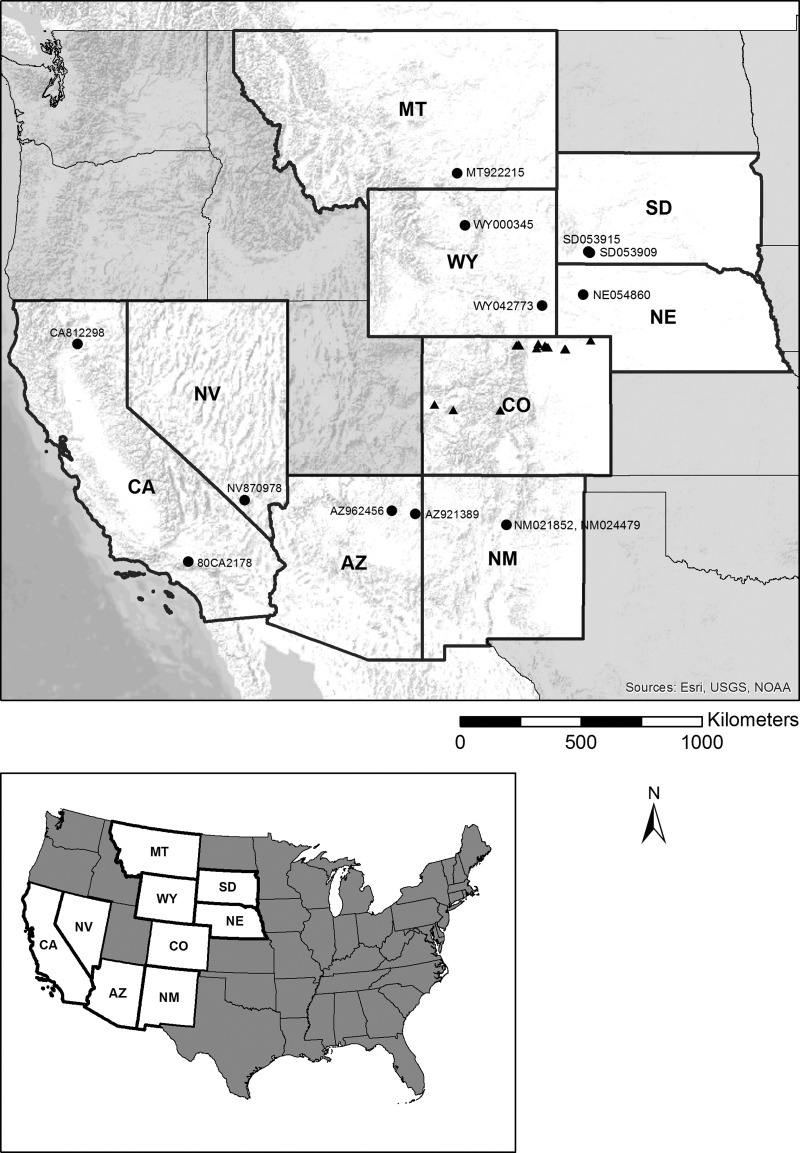
Epizootic activity was especially apparent during 1999 and 2004 in both the mountains (rodent and human plague cases), and plains (prairie dog die-offs), and during 2005 and 2006 on the plains (prairie dog die-offs) (Table 1). One pair of isolates, which were previously collected about 13 km apart in 1999 (CO991340) and 2006 (CO063310), was included for temporal comparison. Another pair, CO92 and CO921715, represented mountain isolates collected ∼1 km apart during the same time period but in a different region of Colorado than the other isolates. These were included for additional geographic comparisons (Table 1). An additional 13 isolates from Arizona, California, Nebraska, New Mexico, Nevada, Montana, South Dakota, and Wyoming, collected as part of routine plague surveillance activities and human plague case investigations carried out by the CDC/DVBD and its partners in state and local health departments (Table 1), were genotyped and included in the analysis to provide a regional ecological scale (Fig. 2). These were selected retrospectively because they were historically well characterized by isolate host and place and time of collection, and they provided a broad geographic and temporal representation of Y. pestis isolates from across the western United States. Isolates were collected from a variety of sources, including humans, rodents, and fleas (Table 1). These samples were collected during the course of human plague case investigations or routine plague surveillance activities for public health purposes rather than as part of research projects. Because each of the isolates made from these samples was obtained as part of a public health response, their use was not subject to the same Institutional Animal Care and Use Committee (IACUC) or Institutional Review Board (IRB) approvals and restrictions required for isolates derived from research activities. All personal identifiers, however, were removed from samples prior to use. An isolate from Kazakhstan (KZ993829), was included as an outgroup for phylogenetic analysis.
FIG. 2.
Regional isolate collection locations. Corresponding CDC accession numbers are listed in Table 1. NV870978 and SD053915 correspond to NV870978_51 and SD053915_255, respectively.
Animal care and welfare protocols were approved by the CSU Animal Care and Use Committee (ACUC) (animal welfare assurance no. A3572-01 and protocol approval no. 03-053A-01). Sample collection permits were issued under the authority of the State of Colorado Department of Natural Resources, Division of Wildlife (permit nos. 06TR983, 07TR983, and 08TR983).
Isolate preparation and DNA extractions
All isolates were grown on Congo Red agar for 48 h at 25°C to screen for the presence of the pgm locus (Burrows and Jackson 1956, Surgalla and Beesley 1969). One inoculation loop full of red colonies (i.e., pgm positive) was selected for each isolate, transferred to 5 mL of Heart Infusion Broth (HIB) (Archibald and Kunitz 1971) (Becton Dickinson, Franklin Lakes, NJ), and shaken overnight (ON) at 25°C and 160 rpm. One milliliter of each 5-mL ON culture was transferred to 25 mL of HIB, and ON culturing was repeated. Cells were pelleted in Oakridge tubes by centrifugation in a Beckman Coulter J series centrifuge (Fullerton, CA) at 8000 rpm for 10 min. Cell pellets were stored at −80°C until DNA extractions were performed. DNA was extracted using the PureGene DNA Purification Kit™ (Qiagen, Valencia, CA) protocol with the following modifications. Cell pellets from 25-mL ON cultures were used rather than 50-mL cultures. Pellets were resuspended in 1.0 mL of 50 mM Tris/50 mM EDTA and transferred to a 50-mL conical tube prior to the cell lysis step. For RNA digestion, RNase A was added to a final concentration of 200 μg/mL to each sample, mixed by inverting the tubes 25 times, and incubated at 37°C for 30 min. Proteinase K (Invitrogen, Grand Island, NY) was then added to a final concentration of 100 μg/mL, and the samples were incubated at 55°C for 1 h. Following DNA precipitation, the pellets were transferred to 1.5-mL microcentrifuge tubes containing 1.0 mL of 70% ethanol, washed, and centrifuged for 3 min at 6000 rpm. The ethanol was removed, and the pellets were air-dried and resuspended in 1.0 mL of kit-supplied DNA Hydration Solution. DNA was shipped overnight to Lawrence Berkeley Laboratory, (Berkeley, CA) where it was transferred to Perlegen Sciences (Mountain View, CA) for microarray analysis.
SNP discovery array
The SNPs were discovered by resequencing and comparison of 147 strains of global origin. For this study, the resequencing microarray was designed by incorporating unique regions of six reference genomes: CO92 (Parkhill et al. 2001), KIM (Deng et al. 2002), Antiqua, Microtus 91001 (Song et al. 2004), Nepal 516 (Chain et al. 2006), and Y. pseudotuberculosis IP32953 (Chain et al. 2004), from which a minimal set of nonredundant contiguous fragments was developed. First, the set of all unique 25-mers present in the source sequences was identified, and the first occurrence of each unique 25-mer by sequence ID and position was chosen. Next, six bases were adjoined to either side of each chosen 25-mer to provide context for SNP discovery. Adjacent sequences were merged into fragments. Unique sequences were tiled. These reference sequences provided 7.85 Mb of potential SNP positions after redundant regions, such as insertion sequence elements and repeat regions, were removed. Bases were called from the resequencing array data using the ABCUS algorithm, a clustering program based on Gaussian mixture models (Cutler et al. 2001). Comparisons were also made between each of the reference sequences to determine base-calling accuracy. For the discovery array, base calling was determined to be 99.8% accurate. Furthermore, similar probe designs have revealed call rates of over 99% when compared to full genome sequencing (Gardner et al. 2013).
Our Y. pestis isolate DNAs were compared to the reference isolates by hybridization of samples to the high-density oligonucleotide array containing the reference oligonucleotides. A total of 20 μg of DNA per sample was fragmented using DNase I and biotinylated. The resulting labeled oligonucleotides were hybridized to the arrays according to Hinds et al. (2004). Hybridization of the labeled sample to the microarray was detected using a confocal laser scanner (Patil et al. 2001).
SNP genotyping
To find SNP locations, positions 9, 13, and 17 of each oligonucleotide were queried for a match or mismatch between the reference and queried sequences. SNPs were determined by measuring the ratios of mean intensity of perfect match features to mismatch features on the microarray (Hinds et al. 2004). Each 25-bp oligonucleotide that contained a SNP at the queried position was compared against the published CO92 Y. pestis genome (GenBank acc. no. NC_003143), using BLAST (www.ncbi.nlm.nih.gov/BLAST/). SNPs were categorized as intergenic or within an open reading frame, synonymous or nonsynonymous, and as a transition or transversion by generating graphical representations of codons using Codon Plot (Stothard 2000).
Phylogenetic analysis
SNPs were used to compare isolates within Colorado and from around the western United States. Data were entered into PAUP* version 4.0b10 (Swofford 2002). The phylogeny was inferred using equally weighted parsimony and 1000 tree–bisection–reconnection (TBR) searches with a maximum of 20 trees held per search. TBR branch swapping was then performed on all of the most parsimonious trees found with a maximum of 100,000 trees held, from which a strict consensus tree was calculated (Schuh and Polhemus 1980). Jackknife (JK) support (Farris et al. 1996) was inferred using 37% deletion and 1000 replicates, each consisting of 10 TBR searches and a maximum of 20 trees held. A Y. pestis isolate (KZ993829) from the Tien Shan region of Kazakhstan, which was included in the microarray analysis, was used as an outgroup (Achtman et al. 2004, Lowell et al. 2007).
Results
SNP discovery
Microarray comparisons of this Y. pestis isolate set yielded 40 previously unidentified, and two previously identified SNPs (Gibbons et al. 2012). Of those, 10 (24%) were intergenic, 23 (55%) were nonsynonymous SNPs, and 9 (21%) were synonymous SNPs (Table 2). Six (23%) of the SNPs found in coding regions were in virulence plasmids. Eighteen SNPs (45%) were present in more than one isolate, whereas the remainder represented single isolates (Table 2). Fifteen SNP (36%) mutations caused transitions and 27 (64%) caused transversions.
Table 2.
Single-Nucleotide Polymorphism Locations and Mutation Outcomes Relative to the CO92 Genome
| CO92 position | CO92 gene | CO92 | Anc | Der | Anc AA | Der AA | Anc codon | Der codon | Mutation | Feature related to sequence |
|---|---|---|---|---|---|---|---|---|---|---|
| 31229 | YPCD1.42 | C | C | A | T | K | aca | aaa | ns | Putative type III secretion protein |
| 38534 | YPO0028 | G | G | A | V | M | gtg | atg | ns | Ribonuclease BN |
| 48123 | YPCD1.09c | G | G | T | M | I | atg | att | ns | Hypothetical protein YPCD1.09c |
| 66130 | YPCD1.91_92 | A | A | T | N | Y | aac | tac | ns | PCD1 |
| 77771 | YPMT1.78 | T | T | C | D | D | gat | gac | s | PMT1_possible pseudogene |
| 78181 | YPO0067 | G | T | T | A | S | gct | tct | ns | Protein-export protein |
| 83402 | YPMT1.83 | A | A | T | N | I | aat | att | ns | caf1A, probable F1 capsule anchoring |
| 89402 | YPMT1.87 | G | G | C | G | R | ggc | cgc | ns | PMT1_possible porphyrin biosynthetic protein |
| 219319 | YPO0203 | T | T | A | D | E | gat | gaa | ns | Elongation factor Tu |
| 268686 | YPO0266 | C | C | T | L | L | cta | tta | s | Putative type III secretion system ATP synthase |
| 519600 | YPO0486_87 | C | C | A | Q | K | caa | aaa | ns | ig |
| 531578 | YPO0497_98 | T | T | A | n/a | n/a | n/a | n/a | ig | ig |
| 617307 | YPO0572 | G | G | A | STOP | STOP | tag | taa | s | Putative exported protein |
| 681456 | YPO0618 | A | A | T | S | C | agt | tgt | ns | Putative exported protein |
| 888395 | YPO0809 | C | C | A | N | K | aac | aaa | ns | General secretion pathway protein K |
| 922389 | YPO0842 | T | T | G | V | G | gtc | ggc | ns | Sulfatase |
| 925344 | YPO0843_44 | C | C | A | n/a | n/a | n/a | n/a | ig | ig |
| 1391154 | YPO1229_31 | C | C | T | n/a | n/a | n/a | n/a | ig | ig |
| 1418458 | YPO1260 | C | C | T | N | N | aac | aat | s | Putative membrane protein |
| 1418459 | YPO1260 | C | C | A | Q | K | cag | aag | ns | Putative membrane protein |
| 1448300 | YPO1288_90 | G | G | A | n/a | n/a | n/a | n/a | ig | ig |
| 1570041 | YPO1390 | C | C | A | A | A | gcc | gca | s | 3-Phosphoshikimate 1-carboxyvinyltransferase |
| 2183336 | YPO1925_26 | C | C | T | n/a | n/a | n/a | n/a | ig | ig |
| 2235130 | YPO1966_67 | G | G | T | n/a | n/a | n/a | n/a | ig | ig |
| 2287501 | YPO2015 | T | T | A | L | Q | ctg | cag | ns | Putative lipoprotein |
| 2348970 | YPO2067_68 | G | G | A | n/a | n/a | n/a | n/a | ig | Putative lipoprotein |
| 2507851 | YPO2232_34 | T | T | C | n/a | n/a | n/a | n/a | ig | ig |
| 2520100 | YPO2243 | C | C | T | H | Y | cat | tat | ns | Putative AraC-family transcriptional regulatory protein |
| 2669972 | YPO2377 | T | T | C | I | T | atc | acc | ns | Putative membrane protein |
| 2893787 | YPO2573 | G | G | T | T | T | acg | act | s | Putative membrane protein |
| 2968425 | YPO2641 | G | G | A | Q | Q | cag | caa | s | Phage family integrase (partial) |
| 3112196 | YPO2777 | C | C | A | H | N | cat | aat | ns | Histidine transport ATP-binding protein HisP |
| 3155657 | YPO2828 | T | T | A | START | K | atg | aag | ns | Phosphoribosylaminoimidazole synthetase |
| 3225580 | YPO2886 | C | C | T | T | I | act | att | ns | Putative autotransporter protein |
| 3457057 | YPO3098 | G | G | T | L | F | ttg | ttt | ns | Probable glycosyltransferase |
| 3464694 | YPO3108 | T | T | C | S | S | agt | agc | s | Putative glycosyltransferase (pseudogene) |
| 4083888 | YPO3663 | G | G | T | G | C | ggt | tgt | ns | Probable zinc-binding dehydrogenase |
| 4108908 | YPO3678 | G | G | A | S | N | agc | aac | ns | Insecticidal toxin complex |
| 4145723 | YPO3711 | G | G | A | A | T | gct | act | ns | Maltoporin |
| 4422198 | YPO3937 | G | G | A | Q | Q | cag | caa | s | Aerobic glycerol-3-phosphate dehydrogenase (partial) |
| 4489139 | YPO3984_85 | C | C | A | n/a | n/a | n/a | n/a | ig | ig |
| 4503423 | YPO3995_96 | T | T | C | n/a | n/a | n/a | n/a | ig | ig |
SNP, single-nucleotide polymorphism; Anc, ancestral genotypes; Der, derived genotypes; s, synonymous SNPs that do not alter protein sequence; ns, nonsynonymous SNPs that alter protein sequence; ig, intergenic SNPs; n/a, not applicable.
Phylogenetic analysis
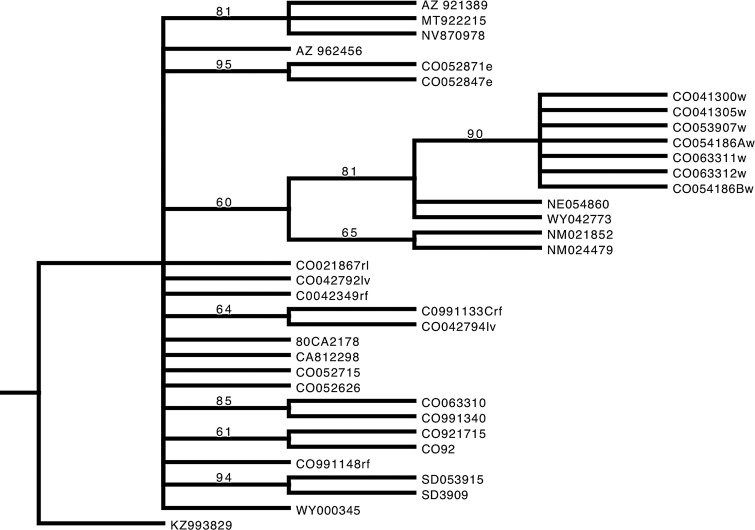
Eighteen parsimony informative SNPs were identified among this isolate collection (Table 3). SNPs that appeared only in single isolates were termed “singletons,” and, although interesting for describing the full diversity within this group of isolates, singletons do not provide information about common ancestry among isolates. Phylogenetic analysis showed that unique genotypes corresponded with local geographic scales, and that regionally, genetic differences approximated geographic distance. First, three unique genotypes identified among the Colorado plains isolates signified near simultaneous emergence of several localized epidemic clones during widespread plague activity. Isolates from the western Pawnee National Grasslands, eastern Pawnee National Grasslands, and the temporal comparison (Table 3, Fig. 3) were highly supported by JK consensus, with values of 90%, 95%, and 85%, respectively. Second, two similar genotypes were detected among Colorado mountain isolates. The JK consensus yielded 64% support among isolates from Red Feather and Livermore (C0991133Crf and CO042794lv), collected in 1999 and 2004, respectively, and 61% between CO92 and CO921715 from Chaffee County. The Colorado isolates collected from mountainous Delta and Mesa counties, and the majority of the remaining plains and mountain isolates yielded singleton SNPs and were unresolved in the tree (Fig. 3). One isolate, C0042349rf, yielded no SNPs when compared to the other North American isolates. Among regionally defined samples, the JK consensus supported a unique genotype among the South Dakota isolates (94%) and the two isolates from New Mexico (65%).
Table 3.
Genomic Diversity in North American Collection
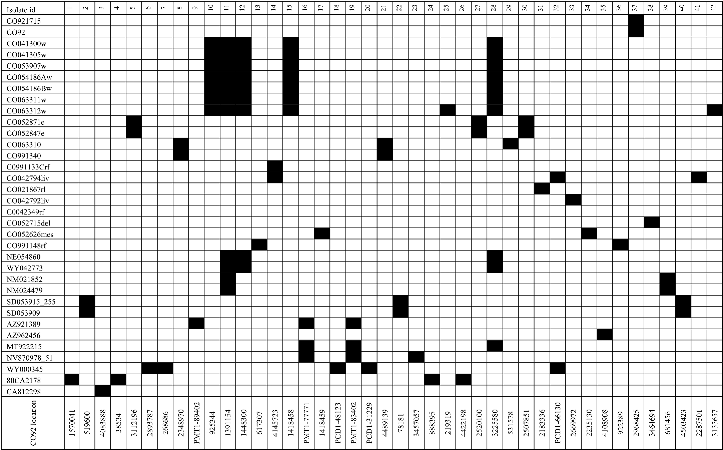 |
The positions of base changes are indicated according to location in the CO92 genome. Black squares indicate SNP presence. The base changes caused by each SNP mutation are shown in Table 2.
SNP, single-nucleotide polymorphism.
FIG. 3.
Maximum parsimony tree of isolates used for SNP discovery. Isolate abbreviations are listed in Table 1 with the following modifications: Extensions del, mes, _51, and _255 were removed from CO052715, CO052626, NV870978, and SD053915, respectively, during phylogentic analysis. Jackknife support is indicated above branches.
When considered as pairs, these comparisons further exemplified localized emergence of unique genotypes. The inferred phylogeny grouped regional isolates collected in 1987 and 1992 from Nevada, Montana, and Arizona (Fig. 3). Whereas this group shared a single common mutation, MT922215 and AZ921389 contained several singleton mutations that may have occurred following regional distribution from a common ancestor (Fig. 3). Interestingly, isolates from Nebraska, Wyoming, and New Mexico fell into a weakly supported group with the geographically nearby isolates from the western Pawnee National Grasslands and shared a previously described SNP from a New Mexico Y. pestis collection (Gibbons et al. 2012). A second common ancestral mutation, also previously described by (Gibbons et al. 2012), was shared by the western Pawnee National Grasslands and New Mexico isolates, lending additional support to a historic Y. pestis radiation event after long-range spread from a plague focus originally located around Santa Fe, NM (Gibbons et al. 2012). The remaining isolates collected regionally were unresolved at the base of the tree. An ensemble consistency index (CI) (Kluge and Farris 1969) of 0.88 and an ensemble retention index (RI) of 0.95 (Farris 1989) suggested that identical mutations that arise independently (homplasies) are unlikely to have occurred and therefore would not have biased the results.
Discussion
We discovered SNPs using high-density oligonucleotide microarray technology for genome-wide comparisons of a North American Y. pestis isolate collection. In the 34 isolates studied, 42 variable SNPs were identified, 18 of which were informative for delineating the geographical extent of Y. pestis clones in the western United States (Table 2). Results from the northern Colorado transect, which spanned sample locations from the Front Range mountains eastward to the Pawnee National Grasslands, suggested that plague epizootics in this region are localized events and that widespread plague activity is the result of several unique epizootic clones emerging simultaneously. The data do not support the notion of large-scale pandemics sweeping across the landscape. Perhaps the most compelling evidence of local maintenance of Y. pestis was that of our temporal comparison, in which unique SNPs supported the relationship between two Colorado isolates (CO991340, CO063310) collected from similar locations during epizootics that occurred 7 years apart (Table 3, Fig. 1). Existence of locally confined genotypes supports the idea that transmission and persistence of Y. pestis is facilitated by metapopulation turnover with localized epizootics.
Previous analyses have also demonstrated limited pathogen dispersal distances and subpopulation structure of Y. pestis during widespread epizootic activity (Girard et al. 2004, Snall et al. 2008, Gibbons et al. 2012). On the basis of the tree topology generated by the Colorado transect isolates, chain-reaction transmission of Y. pestis among many host species may not be necessary for widespread pathogen dispersal during epizootics, as previously suggested (Girard et al. 2004).
Several mechanisms of Y. pestis dispersal that would result in distances greater than prairie dog dispersal have been proposed. They include increased host abundance and contact with putative reservoir hosts (small rodents) (Snall et al. 2008), attraction of rodent-consuming predators that could carry infected fleas between Cynomys spp. colonies (Cully and Williams 2001, Salkeld et al. 2007), and interspecies transmission events facilitating Y. pestis translocation among dispersal-limited reservoir populations (Girard et al. 2004, George et al. 2013). Explanations of bacterial dispersal mechanisms have depended in the past on interpretation of rapidly mutating VNTR markers, suggesting that plague epizootics are initiated by precipitous spread of a single Y. pestis genotype across landscapes and subsequent genotypic differentiation within local, dispersal-limited reservoir populations (Girard et al. 2004). These mechanisms were no doubt important during the eastward expansion of Y. pestis, but our implementation of more slowly evolving SNPs suggests that multiple epidemic clones may have emerged independently (Fig. 1) and that longer-distance plague transmission is not necessary to explain the occurrence of widespread plague epizootics. Furthermore, our mountain isolates, which were subject to higher levels of geographic isolation, did not yield well-supported genetic relationships with each other or with plains isolates. Isolation by geographic barriers would speed differentiation between Y. pestis clones and prevent regular interspecies Y. pestis dispersal at large scales.
Phylogenetic relationships inferred among our samples are consistent with historic translocation across broad regional scales and provide clues to directional spread and establishment of Y. pestis in the United States (Enscore et al. 2002). Two SNPs were shared by isolates collected in 1987 from Nevada and in 1992 from Arizona and Montana. Whereas these isolates shared mutations, several singleton SNPs signify historic spread followed by local divergence (Table 3, Fig. 3). Furthermore, Nebraska and Wyoming isolates shared an ancestral SNP with the western Pawnee National Grasslands isolates, suggesting historic spread of a single clone, followed by local divergence. Additional sampling should reveal whether the singleton mutations mark clones that have spread regionally within mountain valleys.
Conclusion
The Y. pestis collection analyzed here suggested: (1) local-scale emergence of unique Y. pestis epizootic clones during widespread epizootic activity in Colorado, and that widespread interspecies chain reaction transmission events are not the sole mechanism of spread during epizootics; and (2) regional-level relationships among isolates, suggesting that Y. pestis diversifies at relatively local scales following long-range translocation events. Although this study would be strengthened by additional samples, the results strongly evoke a local maintenance mechanism for Y. pestis independent of interspecies and landscape-scale dispersal by rodent hosts or their predators. Future studies should focus on similar analyses using more samples clustered locally and more samples collected through time. A focus on potential local reservoirs, such as off-host fleas (Gage and Kosoy 2005), soil (Eisen et al. 2008), and nematodes (Tan and Darby 2004), should be included. Surveillance and public health recommendations could be improved by focusing more on climate models that may predict widespread epizootic activity based on ecological factors, rather than relying on prediction of directional spread.
Acknowledgments
We thank John Montenieri and Scott Bearden from the Centers for Disease Control and Prevention, Division of Vector Borne Diseases, respectively, for geographic information systems and laboratory support. We also thank Dan Tripp and the field crews from the Antolin Laboratory at Colorado State University, Fort Collins, CO. This work was supported by the ISTC Biotechnology Engagement Program (award K- 584 p) and National Science Foundation–Ecology of Infectious Diseases (NSF-EID) (grant no. 0327052). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Disclosure Statement
Renee Stokowski is employed by Ariosa Diagnostics. Neither Renee Stokowski nor anyone within Ariosa Diagnostics itself has any competing interests with regard to this publication or any of the work associated with it. All other authors have declared that no competing financial interests exist.
References
- Achtman M, Morelli G, Zhu P, Wirth T, et al. Microevolution and history of the plague bacillus, Yersinia pestis. Proc Natl Acad Sci USA 2004; 101:17837–17842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimov AP, Lindler LE, Pier GB. Intraspecific diversity of Yersinia pestis. Clin Microbiol Rev 2004; 17:434–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald WS, Kunitz SJ. Detection of plague by testing serums of dogs on the Navajo Reservation. HSMHA Health Rep 1971; 86:377–380 [PMC free article] [PubMed] [Google Scholar]
- Auerbach RK, Tuanyok A, Probert WS, Kenefic L, et al. Yersinia pestis evolution on a small timescale: Comparison of whole genome sequences from North America. PLoS One 2007; 2:e770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes AM. Surveillance and control of bubonic plague in the United States. Symp of Zool Soc Lond 1982; 50:237–270 [Google Scholar]
- Bazanova LP, Maevskii MP. The duration of the persistence of plague microbe in the body of the flea Citellophilus tesquorum altaicus in Tuva plague focus. Sci J Cent Control Res Nat Infect Dis 1996; 8:67–74 [Google Scholar]
- Ben-Ari T, Neerinckx S, Gage KL, Kreppel K, et al. Plague and climate: Scales matter. PLoS Pathog 2011; 7:e1002160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows TW, Jackson S. The pigmentation of Pasteurella pestis on a defined medium containing haemin. Br J Exp Pathol 1956; 37:570–576 [PMC free article] [PubMed] [Google Scholar]
- Chain PS, Carniel E, Larimer FW, Lamerdin J, et al. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc Natl Acad Sci USA 2004; 101:13826–13831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chain PS, Hu P, Malfatti SA, Radnedge L, et al. Complete genome sequence of Yersinia pestis strains Antiqua and Nepal 516: Evidence of gene reduction in an emerging pathogen. J Bacteriol 2006; 188:4453–4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinge SK, Johnson WC, Ray C, Matchett R, et al. Landscape structure and plague occurrence in black-tailed prairie dogs on grasslands of the western USA. Landscape Ecology 2005; 20:941–955 [Google Scholar]
- Cui Y, Yu C, Yan Y, Li D, et al. Historical variations in mutation rate in an epidemic pathogen, Yersinia pestis. Proc Natl Acad Sci USA 2013; 110:577–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cully JF, Williams ES. Interspecific comparisons of sylvatic plague in prairie dogs. J Mammalogy 2001; 82:894–905 [Google Scholar]
- Cutler DJ, Zwick ME, Carrasquillo MM, Yohn CT, et al. High-throughput variation detection and genotyping using microarrays. Genome Res 2001; 11:1913–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S, Begon M, De Bruyn L, Ageyev VS, et al. Predictive thresholds for plague in Kazakhstan. Science 2004; 304:736–738 [DOI] [PubMed] [Google Scholar]
- Davis S, Trapman P, Leirs H, Begon M, et al. The abundance threshold for plague as a critical percolation phenomenon. Nature 2008; 454:634–637 [DOI] [PubMed] [Google Scholar]
- Deng W, Burland V, Plunkett G, 3rd, Boutin A, et al. Genome sequence of Yersinia pestis KIM. J Bacteriol 2002; 184:4601–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen RJ, Gage KL. Adaptive strategies of Yersinia pestis to persist during inter-epizootic and epizootic periods. Vet Res 2009; 40:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen RJ, Petersen JM, Higgins CL, Wong D, et al. Persistence of Yersinia pestis in soil under natural conditions. Emerg Infect Dis 2008; 14:941–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enscore RE, Biggerstaff BJ, Brown TL, Fulgham RE, et al. Modeling relationships between climate and the frequency of human plague cases in the southwestern United States, 1960–1997. Am J Trop Med Hyg 2002; 66:186–196 [DOI] [PubMed] [Google Scholar]
- Farris JS. The retention index and the rescaled consistency index. Cladistics 1989; 5:417–419 [DOI] [PubMed] [Google Scholar]
- Farris JS, Albert VA, Kallersjo M, Lipscomb D, et al. Parsimony jackknifing outperforms neighbor-joining. Cladistics 1996; 12:99–181 [DOI] [PubMed] [Google Scholar]
- Gage KL, Kosoy MY. Natural history of plague: Perspectives from more than a century of research. Annu Rev Entomol 2005; 50:505–528 [DOI] [PubMed] [Google Scholar]
- Gage KL, Montenieri JA, Thomas RE. The role of predators in the ecology, epidemiology, and surveillance of plague in the United States. Proceedings of the Sixteenth Vertebrate Pest Conference 1994: 200–206 [Google Scholar]
- Gage KL, Burkot TR, Eisen RJ, Hayes EB. Climate and vectorborne diseases. Am J Prev Med 2008; 35:436–450 [DOI] [PubMed] [Google Scholar]
- Gardner SN, Thissen JB, McLoughlin KS, Slezak T, et al. Optimizing SNP microarray probe design for high accuracy microbial genotyping. J Microbiol Methods 2013; 94:303–310 [DOI] [PubMed] [Google Scholar]
- George DB, Webb CT, Pepin KM, Savage LT, et al. Persistence of black-tailed prairie-dog populations affected by plague in northern Colorado, USA. Ecology 2013; 94:1572–1583 [DOI] [PubMed] [Google Scholar]
- Gibbons HS, Krepps MD, Ouellette G, Karavis M, et al. Comparative genomics of 2009 seasonal plague (Yersinia pestis) in New Mexico. PLoS One 2012; 7:e31604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard JM, Wagner DM, Vogler AJ, Keys C, et al. Differential plague-transmission dynamics determine Yersinia pestis population genetic structure on local, regional, and global scales. Proc Natl Acad Sci USA 2004; 101:8408–8413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds DA, Stokowski RP, Patil N, Konvicka K, et al. Matching strategies for genetic association studies in structured populations. Am J Hum Genet 2004; 74:317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull HF, Montes JM, Mann JM. Plague masquerading as gastrointestinal illness. West J Med 1986; 145:485–487 [PMC free article] [PubMed] [Google Scholar]
- Kartman L, Quan SF, Lechleitner RR. Die-off of a Gunnison's prairie dog colony in central Colorado. II. Retrospective determination of plague infection in flea vectors, rodents, and man. Zoonoses Res 1962; 1:201–224 [PubMed] [Google Scholar]
- Keeling MJ, Gilligan CA. Bubonic plague: a metapopulation model of a zoonosis. Proc Biol Sci 2000; 267:2219–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keim ME, Pesik N, Twum-Danso NA. Lack of hospital preparedness for chemical terrorism in a major US city: 1996–2000. Prehosp Disaster Med 2003; 18:193–199 [DOI] [PubMed] [Google Scholar]
- Klevytska AM, Price LB, Schupp JM, Worsham PL, et al. Identification and characterization of variable-number tandem repeats in the Yersinia pestis genome. J Clin Microbiol 2001; 39:3179–3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluge A, Farris JS. Quantitative phyletics and the evolution of Anurans. Systemat Zool 1969; 18:1–32 [Google Scholar]
- Link VB. A History of Plague in the United States of America. Washington, DC: United States Department of Health, Education, and Welfare, 1955:1–2 [Google Scholar]
- Lowell JL, Wagner DM, Atshabar B, Antolin MF, et al. Identifying sources of human exposure to plague. J Clin Microbiol 2005; 43:650–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowell JL, Zhansarina A, Yockey B, Meka-Mechenko T, et al. Phenotypic and molecular characterizations of Yersinia pestis isolates from Kazakhstan and adjacent regions. Microbiology 2007; 153:169–177 [DOI] [PubMed] [Google Scholar]
- Matchett MR, Biggins DE, Carlson V, Powell B, et al. Enzootic plague reduces black-footed ferret (Mustela nigripes) survival in Montana. Vector Borne Zoonotic Dis 2010; 10:27–35 [DOI] [PubMed] [Google Scholar]
- Morelli G, Song Y, Mazzoni CJ, Eppinger M, et al. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat Genet 2010; 42:1140–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J, Wren BW, Thomson NR, Titball RW, et al. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 2001; 413:523–527 [DOI] [PubMed] [Google Scholar]
- Parmenter RR, Yadav EP, Parmenter CA, Ettestad P, et al. Incidence of plague associated with increased winter-spring precipitation in New Mexico. Am J Trop Med Hyg 1999; 61:814–821 [DOI] [PubMed] [Google Scholar]
- Patil N, Berno AJ, Hinds DA, Barrett WA, et al. Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science 2001; 294:1719–1723 [DOI] [PubMed] [Google Scholar]
- Pollitzer R. Plague. World Health Organization Monograph Series. Geneva, Switzerland: World Health Organization, 1954:698 [Google Scholar]
- Salkeld DJ, Eisen RJ, Stapp P, Wilder AP, et al. The potential role of swift foxes (Vulpes velox) and their fleas in plague outbreaks in prairie dogs. J Wildl Dis 2007; 43:425–431 [DOI] [PubMed] [Google Scholar]
- Salkeld DJ, Salathe M, Stapp P, Jones JH. Plague outbreaks in prairie dog populations explained by percolation thresholds of alternate host abundance. Proc Natl Acad Sci USA 2010; 107:14247–14250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage LT, Reich RM, Hartley LM, Stapp P, et al. Climate, soils, and connectivity predict plague epizootics in black-tailed prairie dogs (Cynomys ludovicianus). Ecological Applications 2011; 21:2933–2943 [Google Scholar]
- Schuh RT, Polhemus JT. Analysis of taxonomic congruence among morphological, ecological, and biogeographic data sets for the Leptopodomorpha (Hemiptera). Syst Zool 1980; 29:1–26 [Google Scholar]
- Snall T, O'Hara RB, Ray C, Collinge SK. Climate-driven spatial dynamics of plague among prairie dog colonies. Am Nat 2008; 171:238–248 [DOI] [PubMed] [Google Scholar]
- Song YJ, Tong ZZ, Wang J, Wang L, et al. Complete genome sequence of Yersinia pestis strain 91001, an isolate avirulent to humans. DNA Res 2004; 11:179–197 [DOI] [PubMed] [Google Scholar]
- Stothard P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 2000; 28:1102, 1104 [DOI] [PubMed] [Google Scholar]
- Surgalla MJ, Beesley ED. Congo red-agar plating medium for detecting pigmentation in Pasteurella pestis. Appl Microbiol 1969; 18:834–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (*and other methods). Sunderland, MA: Sinauer Associates, 2002 [Google Scholar]
- Tan L, Darby C. A movable surface: formation of Yersinia sp. biofilms on motile Caenorhabditis elegans. J Bacteriol 2004; 186:5087–5092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchman JW, Wagner DM, Hao J, Mastrian SD, et al. A North American Yersinia pestis draft genome sequence: SNPs and phylogenetic analysis. PLoS One 2007; 2:e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripp DW, Gage KL, Montenieri JA, Antolin MF. Flea abundance on black-tailed prairie dogs (Cynomys ludovicianus) increases during plague epizootics. Vector Borne Zoonotic Dis 2009; 9:313–321 [DOI] [PubMed] [Google Scholar]
- Vogler AJ, Keys CE, Allender C, Bailey I, et al. Mutations, mutation rates, and evolution at the hypervariable VNTR loci of Yersinia pestis. Mutat Res 2007; 616:145–158 [DOI] [PubMed] [Google Scholar]
- Wagner DM, Klunk J, Harbeck M, Devault A, et al. Yersinia pestis and the plague of Justinian 541–543 AD: A genomic analysis. Lancet Infect Dis 2014; 14:319–326 [DOI] [PubMed] [Google Scholar]
- World Health Organization. Weekly epidemiological record. WHO Weekly Epidemiological Record 2004; 33:301–308 [Google Scholar]
- World Health Organization. Human plague: Review of regional morbidity and mortality, 2004–2009. Geneva, Switzerland: World Health Organization; 2010:37–48 [Google Scholar]
- Zhang X, Hai R, Wei J, Cui Z, et al. MLVA distribution characteristics of Yersinia pestis in China and the correlation analysis. BMC Microbiol 2009; 9:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Han Y, Song Y, Huang P, et al. Comparative and evolutionary genomics of Yersinia pestis. Microbes Infect 2004; 6:1226–1234 [DOI] [PubMed] [Google Scholar]