Abstract
The transcription factor atonal homolog 1 (ATOH1) has multiple homologues that are functionally conserved across species and is responsible for the generation of sensory hair cells. To evaluate potential functional differences between homologues, human and mouse ATOH1 (HATH1 and MATH-1, respectively) were nonvirally delivered to human Wharton's jelly cells (hWJCs) for the first time. Delivery of HATH1 to hWJCs demonstrated superior expression of inner ear hair cell markers and characteristics than delivery of MATH-1. Inhibition of HES1 and HES5 signaling further increased the atonal effect. Transfection of hWJCs with HATH1 DNA, HES1 siRNA, and HES5 siRNA displayed positive identification of key hair cell and support cell markers found in the cochlea, as well as a variety of cell shapes, sizes, and features not native to hair cells, suggesting the need for further examination of other cell types induced by HATH1 expression. In the first side-by-side evaluation of HATH1 and MATH-1 in human cells, substantial differences were observed, suggesting that the two atonal homologues may not be interchangeable in human cells, and artificial expression of HATH1 in hWJCs requires further study. In the future, this line of research may lead to engineered systems that would allow for evaluation of drug ototoxicity or potentially even direct therapeutic use.
Introduction
Hair cells located in the cochlea and vestibular organs of the inner ear are responsible for hearing and balance, respectively. Sensorineural hearing loss occurs when the hair cells are irreversibly damaged. Mammalian hair cells in the inner ear do not regenerate and are susceptible to damage from noise-induced trauma, genetic diseases, viral infections, ototoxic antibiotics, and age-related wear and tear.1,2 Hearing aids and cochlear implants are the only available therapies for sensorineural hearing loss. As such, considerable effort has been invested into developing ways to regenerate damaged hair cells through gene delivery or replace hair cells using transplantation through stem cell therapy.3–6
Atonal homolog 1 (ATOH1) is a basic helix–loop–helix (bHLH) transcription factor necessary for hair cell differentiation that is negatively regulated by HES1 and HES5 through the NOTCH signaling pathway.7–12 Several groups have demonstrated that delivery of ATOH1 or MATH-1 (mouse homolog of ATOH1) in vivo to neuroprogenitors and supporting cells has enabled the target cells to transdifferentiate into hair cells.13–16 However, while highly encouraging, most studies have focused on targeting the inner ear epithelium in mouse and rat models that rely on treatment during embryogenesis or shortly after birth. Several research groups have focused on differentiating stem cells into neuroprogenitors or hair cells through gene delivery, coculture, or growth factor exposure using MATH-1 with limited success.17–21
Transdifferentiation has been demonstrated, but not postmitotic cell division and differentiation, which are key barriers that need to be overcome for hair cell regeneration. Transdifferentiation induces one differentiated cell type to change into another differentiated cell type without self-renewal of the original cell.22 Thus, there is still much that is unknown about how hair cells develop and the mechanisms required for regenerating functional hair cells.
The potential to engineer terminal cell phenotypes outside of the body through cellular reprogramming may provide significant insights into the physiology of inner ear sensory and nonsensory epithelia. The ability to engineer a stable inner ear sensory epithelium outside of the body may allow for the screening of ototoxic or therapeutic agents, which may be beneficial in developing new therapies for hearing loss. Thus, we endeavored to explore the possibility of producing hair cells outside of the body by transfecting human Wharton's jelly cells (hWJCs) with two different homologues of ATOH1.
hWJCs are a highly desirable cell population because hWJCs are abundant in supply, not ethically controversial, exhibit no risk of injury to the donor, are highly proliferative, and have demonstrated a differentiation potential similar to human bone marrow stem cells.23–26 As human umbilical cords are a potential source of mesenchymal stem cells, the ability to use human mesenchymal stem cells for hair cell studies could be highly advantageous for avoiding ethical concerns regarding human embryonic stem cells and avoiding performing an invasive procedure on a patient. Furthermore, mesenchymal stem cells have been shown to be capable of differentiating down neural lineages.27–34
We were the first to demonstrate that hWJCs are amenable to inner ear hair cell lineage differentiation when transduced with MATH-1 through adenovirus.35 In addition to viral gene delivery, we are interested in the efficacy and efficiency of nonviral gene delivery for ex vivo tissue engineering experiments. Thus, hWJCs in the current study were transfected with HATH1 (human homolog of ATOH1) for the first time and MATH-1 through Nucleofection™. Nucleofection is a highly effective electroporation method for transfecting primary cells and stem cells, which are known to be notoriously difficult to transfect.36–39 While, electroporation has been known to cause high cell death, cell pretreatment and post-treatment with a Y-27632 ROCK inhibitor can mitigate cell death and low gene expression by preventing apoptosis associated with the RhoA GTP signaling pathways.40
MATH-1 has received more attention in investigations in both mouse and human tissues, but focus on HATH1 has been limited.41–43 The atonal homologues, HATH1 and MATH-1, share 86.04% nucleotide identity and 89.17% amino acid identity.44 More specifically, HATH1 is 1065 base pairs (bp) in length and located on human chromosome 4 (Entrez Gene ID: 474), whereas MATH-1 is 2098 bp in length and located on mouse chromosome 6 (Entrez Gene ID: 11921), yet no side-by-side evaluation of the two atonal homologues in the same tissue exists. Mulvaney et al.45 reported significant differences regarding sequence homology and protein function between ATOH1 homologues, CATH1 (chicken ATOH1 homologue) and HATH1, which underscore the need to further investigate differences between ATOH1 homologues.
While MATH-1 and HATH1 show similar identity, it was hypothesized that the differences in sequences were not interchangeable and that MATH-1 may not interact with human signaling pathways in human tissues as MATH-1 would interact with mouse signaling pathways in mouse tissues. Furthermore, given that HES1 and HES5 are known negative regulators of ATOH1, it was hypothesized that knocking down HES1 and HES5 could enhance the expression of ATOH1 and promote development of hair cell characteristics in hWJCs.
Thus, the objectives of the current study were to evaluate the expression of the atonal effect when HATH1 and MATH-1 were delivered side by side to hWJCs, to evaluate the effect of knocking down HES1 and HES5 concurrently with overexpression of ATOH1 homologues, and also to evaluate hWJCs for hair cell markers after transfection with different combinations of HATH1, MATH-1, and siRNA against HES1 and HES5.
Materials and Methods
Procurement and expansion of hWJCs
hWJCs were isolated from Wharton's jelly of five human umbilical cords with informed consent (KU-IRB #15402) following a modification of our previous protocol.35 Two cords were from males who were born at full term and delivered under normal delivery conditions. Two cords were from females born at 38.3 and 39 weeks under normal delivery conditions. The gender of the last cord used was not available; however, the child was born at full term under normal conditions. Within 24 h of delivery, umbilical cords were soaked in sterile 2% antibiotic–antimycotic (AA, 100×; Life Technologies, Grand Island, NY) in phosphate-buffered saline (PBS) and drained of excess cord blood.
Umbilical cords were cut into ∼3-cm segments, which were filleted open and stripped of blood vessels. The umbilical cord segments were minced finely and suspended in sterile digesting media comprising 0.2% type II collagenase (298 U per mg; Worthington Biochemical, Lakewood, NJ) and 1% penicillin–streptomycin (10,000 U per mL; Life Technologies) in low-glucose Dulbecco's modified Eagle's medium (DMEM; Life Technologies), and then incubated at 37°C in a 5% CO2 environment on an orbital shaker table at 50 rpm for 6 h.
After digestion, the homogenous solution was diluted in sterile 2% AA in PBS at a 1:16 ratio and centrifuged. The supernatant was discarded and cells were combined and plated at a density of 7×103 cells per cm2 in tissue culture-treated T-75 flasks (MidSci, St. Louis, MO). hWJCs were cultured in traditional hWJC medium (10% fetal bovine serum [FBS-MSC qualified] and 1% penicillin–streptomycin in low-glucose DMEM [Life Technologies]). The hWJC medium was changed thrice per week, and hWJCs were maintained at 37°C with 5% CO2 in a cell culture grade incubator. hWJCs were trypsinized with 0.05% Trypsin-EDTA (1×; Life Technologies) at 80% to 90% confluency.
All hWJCs were expanded to passage 1 (P1). Upon reaching 90% confluency, cells were washed with PBS twice, trypsinized, and resuspended at a concentration of 1×106 cells per 1 mL of Recovery™ cell culture freezing medium (Life Technologies) in 2-mL round-bottom cryogenic vials (Corning Incorporated, Acton, MA). Cryogenic vials were immediately placed in a Nalgene® Mr. Frosty container (Sigma-Aldrich, St. Louis, MO) filled with isopropanol and stored at −80°C for 12 h. Cryogenic vials were then transferred and stored in liquid nitrogen.
When ready for use, cells were thawed by transferring cryogenic vials into a 10-cm petri dish filled with PBS warmed to room temperature. Cells were diluted into 50 mL of thawing medium (low-glucose DMEM, 20% FBS-MSC qualified, and 1% penicillin–streptomycin; Life Technologies) and transferred to a T-300 flask (MidSci). Cells were expanded from P2 to P5, then used for experiments. Five umbilical cords (n=5) were used in total for this study. All experiments were performed in triplicate for each cord.
Plasmid and siRNA
Two PrecisionShuttle mammalian vectors with independent turboGFP expression from OriGene (Rockville, MD) were used to deliver target genes to hWJCs. Cloning and verification services were provided by Blue Heron (Blue Heron Biotech LLC, Bothell, WA) to manufacture the vectors. In one vector, an MATH-1 insert (NCBI Reference Sequence: NC_000072.6) was cloned in, and in the other vector, an HATH1 insert (NCBI Reference Sequence: NC_000004.12) was cloned in. The MATH-1 and HATH1 gene inserts were driven by a cytomegalovirus promoter, followed by a Kozak sequence, and the turboGFP gene was driven by a simian virus 40 (SV40) promoter. The PrecisionShuttle vectors contained a kanamycin resistance gene for bacterial selection.
Upon arrival, vectors were reconstituted in 10 mM Tris and 1 mM EDTA (TE) buffer solution and stored at −20°C. Plasmids were verified and sequenced by Blue Heron to ensure no mutations or shifts in reading frame occurred after MATH-1 or HATH1 plasmid generation from a Qiagen Plasmid Plus Giga kit (Qiagen, Valencia, CA).
Based on data from pilot studies (data not shown), custom HES1 siRNA (Hs_HES1_5, gene accession no.: NM_005524, gene ID: 3280) modified with 3′-Alexa Fluor 555 and custom HES5 siRNA (Hs_HES5_5, gene accession no.: NM_001010926, gene ID: 388585) modified with 3′-Alexa Fluor 647 (Qiagen) were selected for experiments. Upon arrival, siRNA was reconstituted with RNase-free water and both HES1 and HES5 siRNA were diluted to 100 nM and stored at −20°C.
Experimental design and transfection
Twenty-four hours before transfection, hWJCs were trypsinized and plated into tissue culture-treated six-well plates (BD Biosciences, San Jose, CA) at a density of 5×105 cells per well. On the day of transfection, media from all wells were removed, and cells were washed with PBS twice. Afterward, cells were incubated for 1 h in a 37°C culture grade incubator supplied with 5% CO2 in 37°C prewarmed traditional hWJC medium (10% FBS-MSC qualified, 1% penicillin–streptomycin, low-glucose DMEM) with 10 μM of Y-27632 ROCK inhibitor (Reagents Direct, Encinitas, CA). After 1 h, hWJCs were washed twice with PBS, trypsinized, and then resuspended in 4D Nucleofector™ P1 primary solution (4DNP1; Lonza, Basel, Switzerland) and one of six treatment solutions.
All cells were suspended at a concentration of 5×105 cells per 100 μL solution at one of the five following ratios: 100 μL 4DNP1 (untreated), 95 μL 4DNP1: 5 μL MATH-1 pDNA (1 μg per μL) (MATH-1), 95 μL 4DNP1: 5 μL HATH1 pDNA (1 μg per μL) (HATH1), 99 μL 4DNP1: 0.5 μL HES1 siRNA (100 nM): 0.5 μL HES5 siRNA (100 nM) (H1/H5), 94 μL 4DNP1: 5 μL MATH-1 pDNA (1 μg per μL): 0.5 μL HES1 siRNA (100 nM): 0.5 μL HES5 siRNA (100 nM) (MATH-1/H1/H5), and 94 μL 4DNP1: 5 μL HATH1 pDNA (1 μg per μL): 0.5 μL HES1 siRNA (100 nM): 0.5 μL HES5 siRNA (100 nM) (HATH1/H1/H5).
The untreated control cells were not nucleofected and were immediately pipetted into six-well plates (BD Biosciences) or Nunc™ Lab-Tek™ eight-well chambered coverglass slides (Thermo Scientific, Waltham, MA) precoated with fibronectin (BD Biosciences) containing 1.5 or 0.5 mL, respectively, of 37°C prewarmed traditional hWJC medium. Cells were placed into a cell culture grade incubator set at a temperature of 37°C and supplied with 5% CO2. hWJC suspensions were transferred to 100-μL 4D nucleofection cuvettes through separate pipettes.
The cuvettes were placed in a 4D Nucleofector (Lonza) and nucleofected with the FF-104 program. Afterward, hWJCs were allowed to incubate at room temperature (ca. 22°C) for 10 min. hWJCs were transferred to a six-well plate (BD Biosciences) or Nunc Lab-Tek eight-well chambered coverglass slides (Thermo Scientific) precoated with fibronectin (BD Biosciences) containing 1.5 mL or 0.5 mL, respectively, of prewarmed 37°C traditional hWJC medium with 10 μM of Y-27632 ROCK inhibitor, and placed into a cell culture grade incubator at 37°C with 5% CO2.
Gene expression
At 1, 3, and 7 days after transfection, cells were collected and harvested for gene expression analysis through real-time quantitative polymerase chain reaction (RT-qPCR). At each time point, RNA was collected from each cell sample according to the manufacturer's instructions of the Qiagen RNeasy Plus Minikit (Qiagen). RNA purity and quality were assessed through NanoDrop 2000 (Thermo Scientific) and Agilent 2200 TapeStation (Agilent Technologies, Santa Clara, CA), respectively. An RNA integrity number of 7–10 was considered acceptable for cDNA conversion.
RNA was converted to cDNA using the High-Capacity cDNA conversion kit (Life Technologies) and the Eppendorf Realplex Mastercycler (Eppendorf, Hamburg, Germany). Converted cDNA purity and quality were assessed quantitatively through the NanoDrop 2000 (Thermo Scientific) and qualitatively through the Agilent 2200 TapeStation (Agilent Technologies), respectively.
cDNA from each sample was loaded into a MicroAmp® Fast Optical 96-well reaction plate (0.1 mL; Life Technologies). Individual wells were loaded sequentially in the following ratios: 1 μL TaqMan assay (Life Technologies), 9 μL sample cDNA, 10 μL TaqMan Fast Universal PCR Master Mix (Life Technologies). The TaqMan assays used are listed in Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/tea).
Plates were sealed with MicroAmp Optical Adhesive Film (Life Technologies) and centrifuged at 1500 rpm (∼500 g) for 5 min at 4°C. Afterward, the samples were loaded into the Eppendorf Realplex Mastercycler and run according to the recommended TaqMan Fast Universal PCR Master Mix protocol (Life Technologies). Cycle threshold (Ct) values were recorded and analyzed through the Delta-Delta Ct method. Values were normalized to day 0 untreated control samples and the endogenous controls. Three biological replicates from each of five umbilical cords (n=5) were taken for gene expression analysis at 1, 3, and 7 days post-transfection.
Live cell fluorescent imaging and flow cytometry
hWJCs were collected for live stain imaging 24 h after transfection. A 0.5-μL aliquot of Hoechst 33342 dye (Life Technologies) was added to each well, and hWJCs were incubated for 10 min at 37°C in 5% CO2. Afterward, hWJCs were imaged using a custom epifluorescent and confocal microscope comprising the following components: an Olympus IX81 inverted spinning disc confocal microscope base (Olympus America, Center Valley, PA), a Prior microscope stage for automated image acquisition (Prior Scientific, Rockland, MA), an Olympus 20× or 40× long working distance air objective (Olympus), and a Hamamatsu electron multiplying charge-coupled device (EMCCD) camera (Hamamatsu, Hamamatsu, Shizuoka Prefecture, Japan).
Images were captured using the acquisition and analysis software, SlideBook (Intelligent Imaging Innovations [3i], Denver, CO). A mercury arc lamp was used with the following excitation filters (excitation/emission) for image collection: Hoechst (387±11 nm/447±60 nm), GFP (494±20 nm/531±22 nm), Alexa Fluor 555 (575±25 nm/624±40 nm), and Alexa Fluor 647 (650±25 nm/684±25 nm). A montage was generated from 49 (seven by seven arrangement) neighboring fields of view that were aligned together to generate one comprehensive composite image of the sample. No bacteria, fungi, or mycoplasma entities were observed in cell cultures that were imaged and stained with Hoechst.
In a pilot study, cell surface areas were measured using the imaging software package, CellProfiler, and quantified using the imaging software package, CellProfiler Analyst (BROAD Institute of MIT and Harvard).46–49
So as to not bias cell viability after imaging, the spent medium containing nonadhering or loose adhering cells was collected from each sample and transferred into a prelabeled 15-mL conical tube (Phenix, Candler, NC). Remaining adherent hWJCs were washed twice with PBS, then trypsinized, and added to corresponding 15-mL conical tubes containing the previous spent medium with the unattached cells so that both live cells and dead cells would be retained for analysis. Then, 4 mL of fresh traditional hWJC medium was added to each conical tube, and all conical tubes were centrifuged at 1500 rpm (∼500 g) for 5 min. The supernatant was discarded, and hWJCs were resuspended in 500 μL of PBS and pipetted through a 70-μm nylon mesh cell strainer (BD Biosciences); 0.5 μL of Sytox Red (Life Technologies) was added just before analysis.
A minimum of 20,000 events were analyzed through flow cytometry on the MoFlo XDP fluorescent-activated cell sorter (FACS; Beckman Coulter, Brea, CA). Flow Cytometry was used to analyze cell viability and transfection efficiency. Live hWJCs were characterized as hWJCs expressing Hoechst at an intensity of 102 relative fluorescent units (RFU) or above, with expression of Sytox Red at an intensity below 100 RFU. Dead hWJCs were characterized as hWJCs that expressed Hoechst at an intensity below 102 RFU and expressed Sytox Red at an intensity above 100 RFU. GFP-positive hWJCs were characterized as live hWJCs that expressed GFP at an intensity of 100 RFU or greater. Transfection efficiency was determined by dividing the number of live GFP-positive cells in a sample by the total population of the sample.
All experiments were performed in triplicate for each umbilical cord 24 h after transfection.
Additional images were taken using an Axio Observer inverted researcher microscope (Zeiss, Peabody, MA) with 20×long working distance objectives using a plastic differential interference contrast (PlasDIC; Zeiss) at 4, 6, and 10 days after transfection/cell seeding.
FM® 1-43 staining
Cells from each cord were stained with FM 1-43 and imaged 7 days after transfection under a confocal microscope. Briefly, cells were washed twice with PBS, and 500 μL of FM 1-43 (5 μg/mL in Hanks' balanced salt solution [HBSS]) was added to the cells while on ice. Cells were fixed with 500 μL of 4% formaldehyde in HBSS on ice for 10 min, then washed thrice with HBSS. Cells were then sealed with ProLong® Gold antifade reagent with DAPI (Life Technologies) and imaged on the same custom confocal microscope as stated previously. A 488-nm solid-state laser was used for excitation with confocal emission of 625±11 nm. No bacteria, fungi, or mycoplasma entities were observed in cell cultures that were imaged and stained with DAPI.
Immunocytochemistry
At 1 and 7 days after transfection, cells were collected for immunocytochemistry. Primary antibodies were preconjugated to quantum dots (Qdot®) using the 565, 605, 655, and 805 Qdot® Antibody Conjugation Kits (Life Technologies). Primary antibodies were conjugated to Qdots according to the manufacturer's instructions and stored at 4°C for immediate use. The following primary antibodies were conjugated to the following Qdots: Anti-human HES1 (Cat. No. AB5702, Millipore) preconjugated to Qdot 565 (1:200; Life Technologies), Anti-human myosin VIIA (MYO7A, Cat. No. NBP1-84266; Novus, Littleton, CO) preconjugated to Qdot 605 (1:500; Life Technologies), Anti-human HES5 (Cat. No. AB5708; Millipore) preconjugated to Qdot 655 (1:200; Life Technologies), and Anti-human glial fibrillary acidic protein (GFAP, Cat. No. AB5541; Millipore) preconjugated to Qdot 800 (1:100; Life Technologies).
Cells were fixed by first washing cells in 37°C PBS, followed by fixation with 4% formaldehyde in PBS for 15 min. Cells were then washed and incubated for 5 min with PBS thrice. Afterward, cells were permeabilized with 0.25% Triton X-100 in PBS for 15 min, then washed thrice in PBS. Cells were blocked with 4% bovine serum albumin in PBS for 60 min. Afterward, cells were incubated with all primary antibodies preconjugated to respective Qdots as listed above for 60 min in blocking buffer. Following three PBS washes, cells were counterstained with Syto 9 (10 nM; Life Technologies) for 30 min, and then dehydrated with graded ethanol, followed by double exposure to 100% toluene.
Cells were then mounted in Qmount™ Qdot® mounting media (Life Technologies). Cells were imaged using confocal microscopy using a 405-nm solid-state laser for Qdot excitation, a 488-nm solid-state laser for Syto 9 excitation, and the following emission filters: Syto 9 (531±22 nm), Qdot 525 (531±22 nm), Qdot 565 (560±25 nm), Qdot 605 (613±20 nm), Qdot 655 (655±15 nm), and Qdot 800 (788±20 nm). Cells were collected from each umbilical cord at 1 and 7 days after transfection and images were taken from individual wells on eight-well chambered glass slides.
Analysis of stem cell characteristics
A subculture of cells from each cord was characterized through cell surface marker identification through flow cytometry. hWJCs were trypsinized and centrifuged. The supernatant was discarded and hWJCs were suspended in 5% FBS-MSC qualified (Life Technologies) in PBS and placed on ice and kept in the dark for 20 min. Aliquots containing 5×105 hWJCs in ∼200 μL were pipetted into 50-mL conical tubes (Phenix). Primary cell surface antibodies and secondary antibodies were added sequentially one at a time per incubation–wash cycle to avoid cross-reaction; however, preconjugated primary antibodies with secondary antibodies were added simultaneously. A single incubation–wash cycle consisted of adding a primary antibody, secondary fluorescent antibody, or primary antibody preconjugated to a specific fluorescent secondary antibody to the cell suspension.
After incubation, 800 μL of 5% FBS in PBS was added to the cell suspension to bring the total volume of the cell suspension up to 1 mL, and the cell suspension was then centrifuged. The supernatant was discarded, and cell pellets were suspended in 5% FBS-MSC qualified (Life Technologies) in PBS, which concluded one incubation–wash cycle.
Cell surface marker antibodies and secondary antibodies were added in the following order at the following ratios: STRO-1 mouse IgM (2.5:200, 1 mg per mL, Cat. No. MAB1038; R&D Systems, Minneapolis, MN); Alexa Fluor 568® rabbit anti-mouse IgG (2:200, 2 mg per mL, Cat. No. A-11061; Life Technologies); CD105 mouse IgG (2.5:200, 1 mg per mL, Cat. No. MAB10971; R&D Systems); Qdot® 525 donkey anti-mouse IgG (2:200, 1 μM, Cat. No. Q22073; Life Technologies); human CD45 preconjugated to Qdot® 800 (2:200, Cat. No. Q10156; Life Technologies); human CD73 preconjugated to FITC (5:200, Cat. No. 561254; BD Biosciences); human CD34 preconjugated to Brilliant Violet (5:200, Cat. No. 562577; BD Biosciences); and human CD90 preconjugated to APC (5:200, Cat. No. 559869; BD Biosciences).
At the end of the last incubation–wash cycle, hWJCs were resuspended in PBS and pipetted through a 70-μm nylon mesh cell strainer (BD Biosciences). hWJCs were analyzed by flow cytometry on a MoFlo XDF FACS (Beckman Coulter). Positive identification of cell markers was defined as fluorescent emission that exceeded the fluorescent threshold of cells stained with corresponding isotype (negative) controls. The isotype controls used in these studies were rabbit IgG Alexa Fluor 568, donkey IgG Qdot 525, IgG2 Qdot 800 (all from Life Technologies), and IgG1 FITC, IgG1 Brilliant Violet, and IgG1 APC (all from BD Biosciences). An example of how cells were gated is displayed in Supplementary Fig. S1.
The cell characterization experiments were repeated thrice for each cord to show expression of mesenchymal stem cell surface markers. Cells were not sorted due to the stress imposed on cells during sorting and the need for a large quantity of viable cells for experiments.
Statistical analyses
All values are reported as statistical means with standard deviations, unless otherwise noted. Cells were isolated from five (n=5) different umbilical cords, and three technical replicates were used for each quantitative analysis, unless otherwise noted. Five samples were considered adequate for discriminating statistical significance based on Mead's resource equation and previous published studies using human umbilical cords. A one-way analysis of variance was performed with a least sum of differences post hoc in conjunction with Dunnett's (multicomparison) test to assess statistical significance with p set at ≤0.05 and power >0.8. Dunnett's test was set up as a one-tailed assessment to examine increased values against control samples. The software SPSS (IBM) version 22 was used to compute all statistical analyses.
Results
Cells transfected with HATH1 showed greater cell density than cells transfected with MATH-1
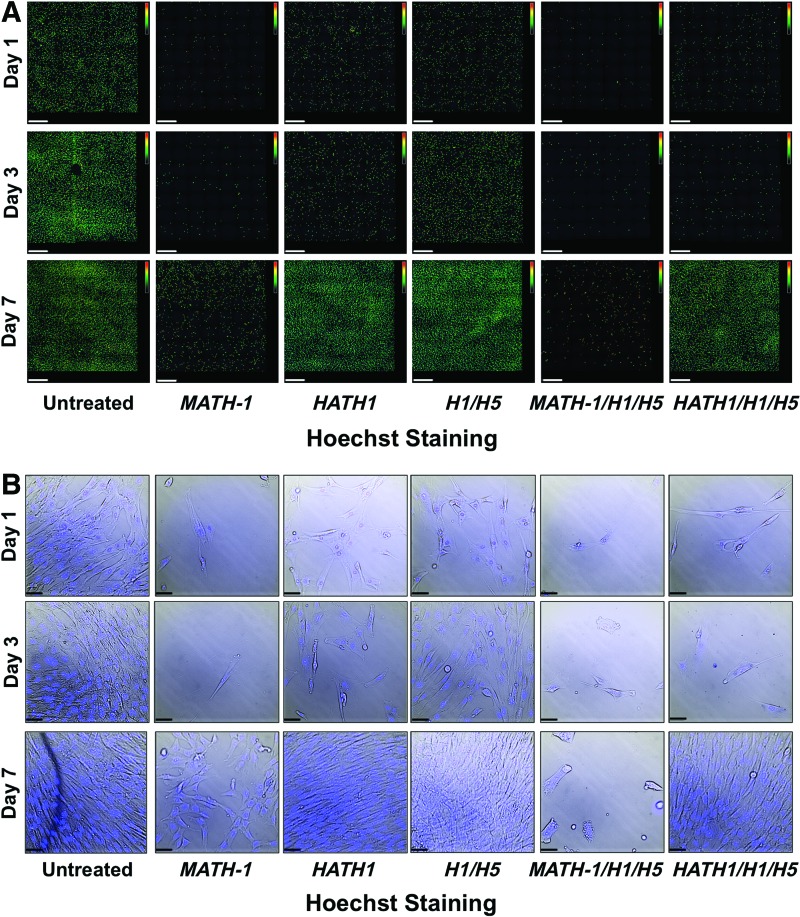
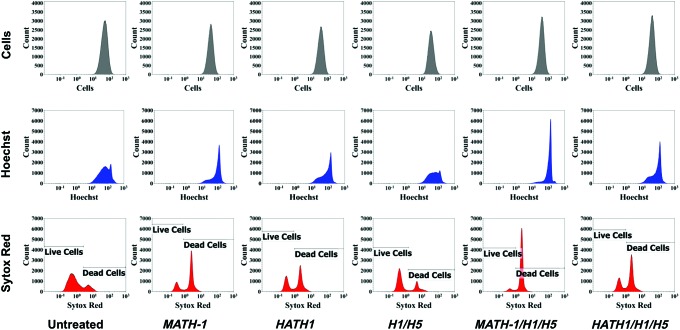
hWJCs were transfected through nucleofection, an electroporative technique (Lonza), with one of five different treatments: MATH-1 pDNA, HATH1 pDNA, siRNA against HES1 and HES5, MATH-1 pDNA and siRNA against HES1 and HES5, or HATH1 pDNA and siRNA against HES1 and HES5. At 24 h post-transfection, there was a noticeable visual difference in cell numbers between cells treated with HATH1 versus cells treated with MATH-1 (Fig. 1). Twenty-four hours after transfection, flow cytometry revealed that there were 1.9 times more viable cells transfected with HATH1, and 2.2 times more viable cells transfected with HATH1 and siRNA against HES1 and HES5, than viable cells transfected with MATH-1 (Fig. 2).
FIG. 1.
Cell density and proliferation. Cells transfected with HATH1 or HATH1 and siRNA against HES1 and HES5 displayed more intact nuclei than MATH-1 at 1, 3, and 7 days after transfection. Cells cotransfected with MATH-1 and siRNA against HES1 and HES5 displayed few intact nuclei at 1, 3, and 7 days after transfection. Cells from each group were stained with Hoechst and imaged through an inverted epifluorescent microscope. (A) Composite image montages comprising 49 neighboring fields of view stitched together into a 7×7 image. Images are shown as intensity maps of fluorescence, where green fluorescence represents low fluorescence and red represents high fluorescence. (B) Bright-field images with the fluorescent overlay of Hoechst taken from the corresponding first image (top left corner) in each montage. Untreated control populations had the greatest cell density at each time point. Cells treated with MATH-1 had the lowest cell densities across all three time points, whereas cells treated with only siRNA against HES1 and HES5 had the greatest cell densities behind untreated control cells. Images are representative of cells from each treatment group across five umbilical cords (n=5) at each time point. MATH-1 represents cells transfected with MATH-1 pDNA. HATH1 represents cells transfected with HATH1 pDNA. H1/H5 represents cells transfected with HES1 and HES5 siRNA. MATH-1/H1/H5 represents cells cotransfected with MATH-1 pDNA, HES1 siRNA, and HES5 siRNA. HATH1/H1/H5 represents cells cotransfected with HATH1 pDNA, HES1 siRNA, and HES5 siRNA. White scale bar=500 μm (A). Black scale bar=50 μm (B). Color images available online at www.liebertpub.com/tea
FIG. 2.
Live/Dead analysis of transfected hWJCs. Transfection with HATH1 led to greater cell viability than transfection with MATH-1. A minimum of 20,000 events were analyzed through flow cytometry to determine the viability of treatments 1 day after transfection. The top row shows the distribution of cells from forward scatter. The middle row displays cells with an intact nucleus positively identified by Hoechst staining. The bottom row displays the distribution of cells identified as live or dead based on Sytox Red (dead cell stain) staining. The histograms shown are an arbitrary selection from one umbilical cord of five tested (n=5). MATH-1 represents cells transfected with MATH-1 pDNA. HATH1 represents cells transfected with HATH1 pDNA. H1/H5 represents cells transfected with HES1 and HES5 siRNA. MATH-1/H1/H5 represents cells cotransfected with MATH-1 pDNA, HES1 siRNA, and HES5 siRNA. HATH1/H1/H5 represents cells cotransfected with HATH1 pDNA, HES1 siRNA, and HES5 siRNA. hWJCs, human Wharton's jelly cells. Color images available online at www.liebertpub.com/tea
In addition, 24 h post-transfection cells cotransfected with HATH1 and siRNA against HES1 and HES5 displayed 3.7 times more viable cells than cells cotransfected with MATH-1 and siRNA against HES1 and HES5. At 24 h post-transfection, cells transfected with HATH1 displayed transfection efficiency that was 0.2 times greater than cells transfected with MATH-1. Moreover, 7 days after transfection, cell counts revealed that there were 2.8 times more viable cells transfected with HATH1, and 3.1 times more viable cells transfected with HATH1 and siRNA against HES1 and HES5, than cells transfected with MATH-1. At 7 days post-transfection, cells cotransfected with HATH1 and siRNA against HES1 and HES5 displayed 2.8 times more viable cells than cells cotransfected with MATH-1 and siRNA against HES1 and HES5.
Only cells transfected with HATH1 revealed significant visual changes in morphology
Visual morphological differences were evident between untreated control cells and cells treated with HATH1 and siRNA against HES1 and HES5 starting at day 3 (Fig. 3). Cells treated with MATH-1 displayed a fibroblastic morphology, consistent with hWJCs. However, cells treated with HATH1, siRNA against HES1 and HES5, or a combination of both showed an elongated cell body with small projections expanding away from the cell body. Cells treated with both HATH1 and siRNA against HES1 and HES5 displayed a bipolar phenotype with cell extensions reaching out from the nucleus and terminating with multiple slender projections, which was uncharacteristic of hWJCs. Additional images displaying changes in morphology are presented in Supplementary Fig. S2.
FIG. 3.
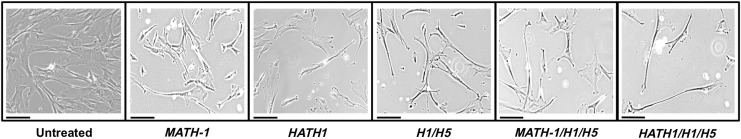
Phase-contrast images of transfected hWJCs. The images were taken at a 10×magnification 4 days after cells were transfected. Untreated control cells displayed a fibroblastic phenotype, while cells treated with both HATH1 and siRNA against HES1 and HES5 displayed an elongated body and bipolar phenotype with projections at the terminal ends of the cell. The images shown are an arbitrary selection from one umbilical cord of five tested (n=5). Additional images are available in Supplementary Figure S2. MATH-1 represents cells transfected with MATH-1 pDNA. HATH1 represents cells transfected with HATH1 pDNA. H1/H5 represents cells transfected with HES1 and HES5 siRNA. MATH-1/H1/H5 represents cells cotransfected with MATH-1 pDNA, HES1 siRNA, and HES5 siRNA. HATH1/H1/H5 represents cells cotransfected with HATH1 pDNA, HES1 siRNA, and HES5 siRNA. Scale bar=50 μm.
In a pilot study, cell surface area for treatment groups from one umbilical cord was analyzed. Cell surface area data are presented in Supplementary Fig. S3. No significant differences were detected in surface area measurements over the course of 7 days; however, cells treated with HATH1 increased in surface area over a period of 7 days.
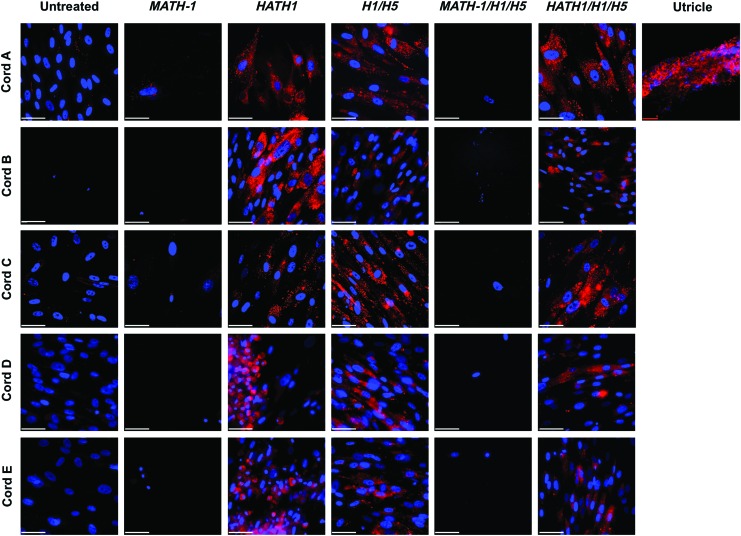
HATH1-transfected cells revealed infiltration of lipophilic dye, FM® 1-43
To further evaluate the development of morphological features of hair cells, controls and treatment groups were stained 7 days after transfection with FM 1-43, a lipophilic dye that is known to enter cells through transduction channels found in hair cells and neurons. Cells treated with HATH1 stained positive for FM 1-43, as did cells treated with only siRNA against HES1 and HES5. FM 1-43 entered HATH1-transfected cells more readily and robustly than MATH-1-transfected cells. Across cells from all five umbilical cords, positive FM 1-43 staining was observed in the greatest quantities in cells treated with HATH1 only or HATH1 and HES1 siRNA and HES5 siRNA.
The amount of positive FM 1-43 staining varied between cells treated only with HATH1 and cells treated with HATH1 and siRNA against HES1 and HES5 (Fig. 4). Limited infiltration of FM 1-43 was observed in some of the samples cotransfected with MATH-1 and siRNA against HES1 and HES5 across cell samples from all five umbilical cords.
FIG. 4.
Transfected hWJCs stained with FM® 1-43. Transfected cells were imaged with an epifluorescent microscope 7 days after transfection. Images were taken from each treatment of cells from each of five umbilical cords (n=5) tested. Cells transfected with HATH1 and HATH1/H1/H5 displayed superior infiltration of FM 1-43 than cells transfected with MATH-1. FM 1-43 (red) intensely stained hWJCs transfected HATH1/H1/H5. Cell nuclei are represented by DAPI staining (blue). A single field of view from a C57BL mouse utricle is shown as a positive control for FM 1-43 staining. MATH-1 represents cells transfected with MATH-1 pDNA. HATH1 represents cells transfected with HATH1 pDNA. H1/H5 represents cells transfected with HES1 and HES5 siRNA. MATH-1/H1/H5 represents cells cotransfected with MATH-1 pDNA, HES1 siRNA, and HES5 siRNA. HATH1/H1/H5 represents cells cotransfected with HATH1 pDNA, HES1 siRNA, and HES5 siRNA. Utricle (red) scale bar=20 μm. White scale bar=50 μm. Color images available online at www.liebertpub.com/tea
HATH1-transfected cells upregulated different genes from MATH-1-transfected cells
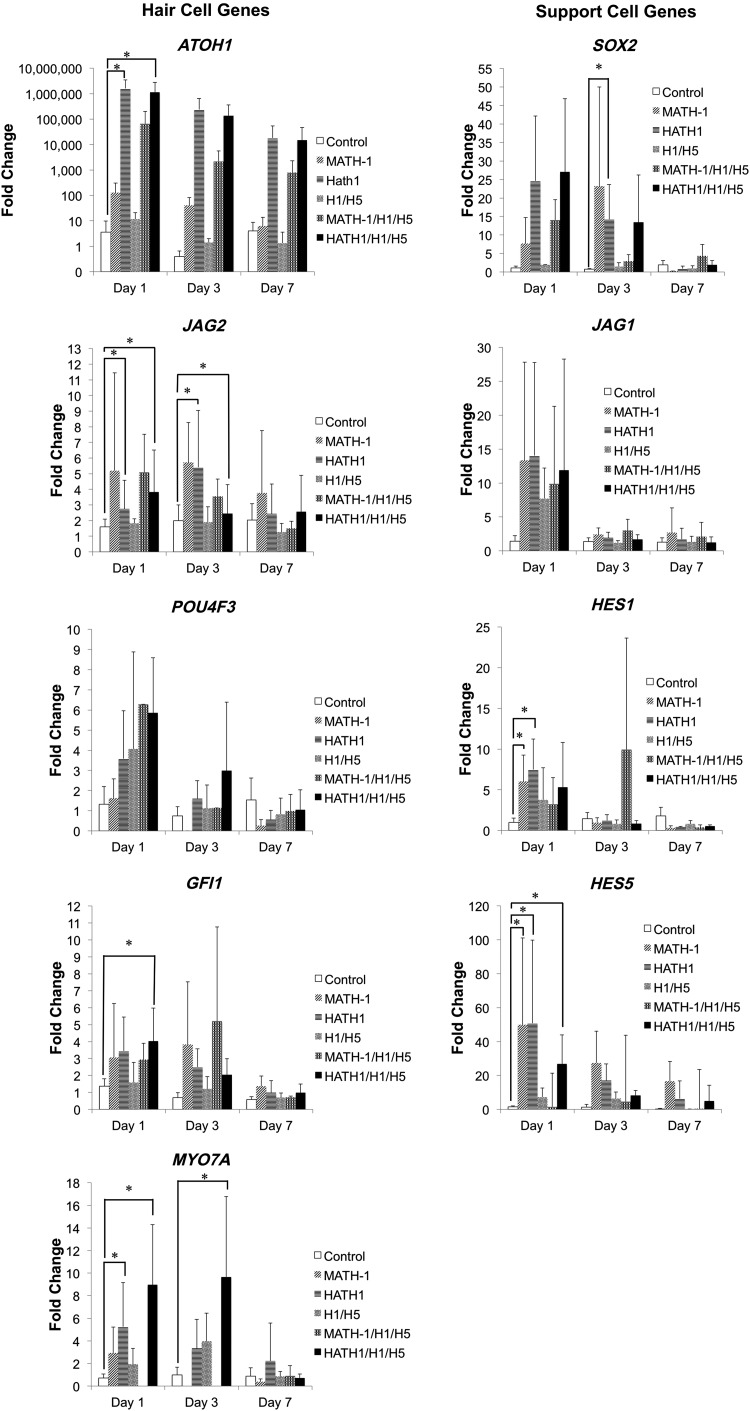
Gene expression was evaluated across all treated cells from all human umbilical cords at 1, 3, and 7 days after transfection. The relationship regarding how all of the analyzed genes are related to each other is illustrated in Supplementary Fig. S4.
The common trend observed across all analyzed genes was an upregulation of gene expression 1 day after transfection, and gene expression levels returned to levels similar to untreated controls 7 days after transfection (Fig. 5). Gene expression in MATH-1-transfected cells did not significantly differ from untreated control cells within the 7-day time period following transfection, except for jagged 2 (JAG2), HES1, and HES5 genes, 1 day after transfection.
FIG. 5.
Gene expression of transfected hWJCs. HATH1/H1/H5-transfected cells showed significant increases in gene expression across most genes compared with untreated control cells 1 day after transfection, whereas MATH-1-transfected cells only showed significant increases in gene expression in JAG2, HES1, and HES5. Treated hWJCs were assessed for gene expression using RT-qPCR at 1, 3, and 7 days after transfection. *Statistically significant difference from untreated hWJCs (p<0.05). ATOH1, HES1, and HES5 are genes pertaining to pDNA and siRNA used for different treatments. SOX2 initiates ATOH1 expression and is then downregulated by ATOH1. No significant differences were detected among groups regarding JAG1 expression. JAG1 and JAG2 are NOTCH ligands that participate in the NOTCH signaling pathway. MYO7A, GFI1, and POU4F3 are key hair cell markers. MYO7A is a critical marker that was significantly expressed in hWJCs treated with HATH1/H1/H5 at 1 and 3 days after transfection. The results are representative of cells collected from five different umbilical cords (n=5) and are reported as statistical means. All experiments were performed in triplicate. Error bars represent standard deviations. H1/H5 represents cells transfected with HES1 and HES5 siRNA. MATH-1/H1/H5 represents cells cotransfected with MATH-1 pDNA, HES1 siRNA, and HES5 siRNA. HATH1/H1/H5 represents cells cotransfected with HATH1 pDNA, HES1 siRNA, and HES5 siRNA. ATOH1, atonal homolog 1.
Cells cotransfected with MATH-1 and siRNA against HES1 and HES5 displayed no significant gene expression differences from untreated control samples within the 7-day time period following transfection. Perhaps, most importantly, MATH-1-transfected cells failed to show any significant increase in gene expression over the 7-day time period, except for JAG2, HES1, and HES5, whereas HATH1-transfected cells showed significant (p<0.05) increases in gene expression 1 day after transfection compared with untreated control cells in ATOH1 (4.5×105-fold change), HES1 (6.8-fold change), HES5 (33.3-fold change), and myosin VIIA (MYO7A; 6.5-fold change).
Cells that were cotransfected with HATH1 and siRNA against HES1 and HES5 displayed significant (p<0.05) increases in gene expression across ATOH1 (3.2×105-fold change), HES5 (17.6-fold change), MYO7A (11.0-fold change), GFI1 (2.9-fold change), and JAG2 (2.4-fold change) 1 day after transfection. hWJCs cotransfected with HATH1 and siRNA against HES1 and HES5 displayed significant (p<0.05) increases in gene expression across MYO7A (9.1-fold change) and JAG2 (1.2-fold change) 3 days after transfection. Expression of BARHL1 was measured; however, it was not detected in any samples throughout the entire study, thus data are not shown. In addition, jagged 1 (JAG1) gene expression was measured, and no significant differences were detected between any treated samples and controls over a period of 7 days.
Myosin VI (MYO6) gene expression was measured; however, no significant difference was detected between controls and treatment groups at any time points.
Only cells transfected with HATH1 displayed increased protein expression of MYO7A
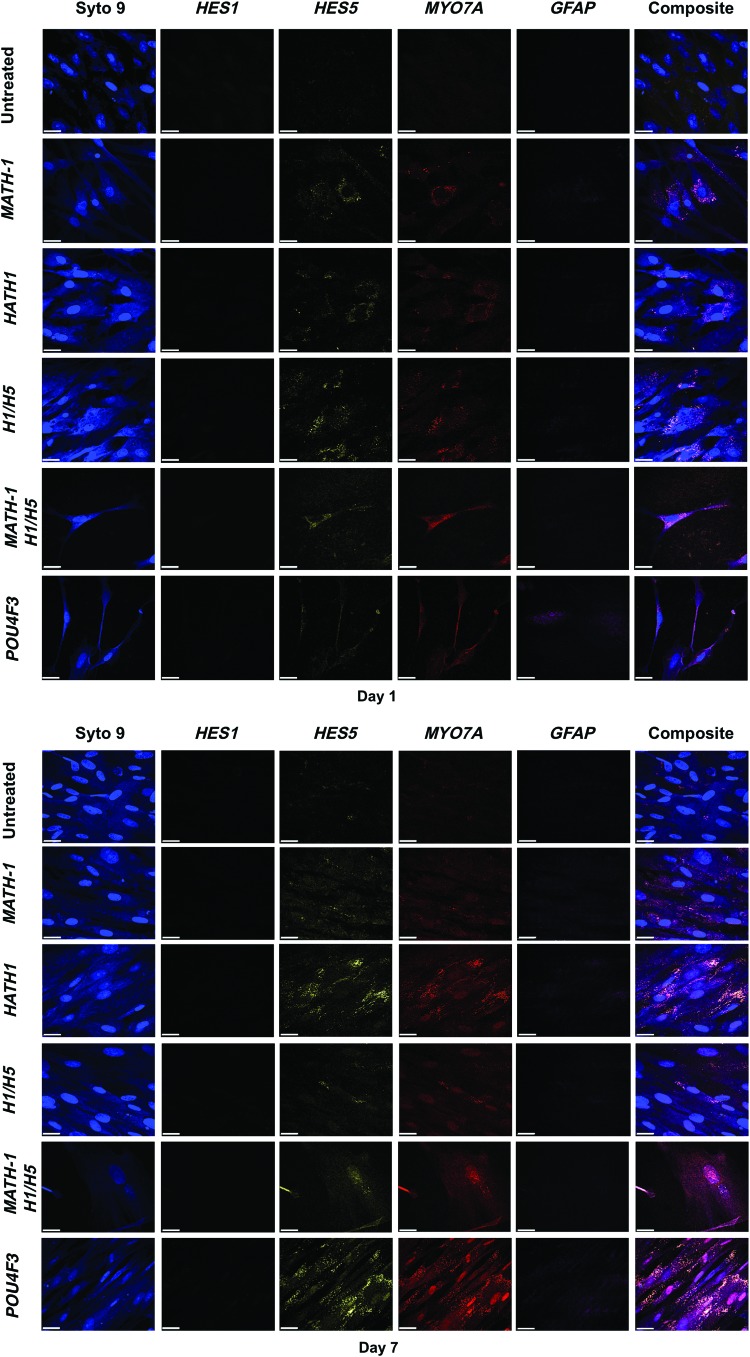
Cells were analyzed for protein expression through immunocytochemistry 1 and 7 days after transfection (Fig. 6). All treated cells displayed positive identification of MYO7A and HES5 1 day after transfection. However, cells cotransfected with HATH1 and siRNA against HES1 and HES5 displayed positive identification of GFAP 1 day after transfection. MATH-1-transfected cells displayed a visual decrease in MYO7A and HES5 expression, whereas HATH1-transfected cells displayed a visual increase in immunostaining for MYO7A and HES5 7 days after transfection. No GFAP expression was detected in any treated group 7 days after transfection. Untreated control cells displayed no presentation of any hair cell marker proteins at 1 or 7 days after culture.
FIG. 6.
Immunostaining of transfected hWJCs. HATH1/H1/H5-transfected cells showed increased expression of MYO7A and HES5 from 1 to 7 days after transfection, whereas MATH-1-transfected cells showed a decrease in MYO7A and HES5 expression. Treated hWJCs were assessed for protein expression by immunostaining at 1 and 7 days after transfection. Primary antibodies were preconjugated to quantum dots. Cell nuclei are represented by Syto 9 staining (blue). HES1 (orange) was not positively identified. HES5 (yellow) and MYO7A (red) were positively identified in all treated groups. Cells transfected with HATH1/H1/H5 displayed positive identification of GFAP (pink) 1 day after transfection. H1/H5 represents cells transfected with HES1 and HES5 siRNA. The images shown are an arbitrary selection from one umbilical cord of five tested (n=5). MATH-1/H1/H5 represents cells cotransfected with MATH-1 pDNA, HES1 siRNA, and HES5 siRNA. HATH1/H1/H5 represents cells cotransfected with HATH1 pDNA, HES1 siRNA, and HES5 siRNA. Scale bar=20 μm. GFAP, glial fibrillary acidic protein. Color images available online at www.liebertpub.com/tea
In a pilot study, 7 days after transfection, treated cells from one cord were stained with a fluorescent probe, developed by Vytla et al.,50 against the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor to identify active calcium-permeable ion channels. MATH-1-transfected cells and cells cotransfected with MATH-1 and siRNA against HES1 and HES5 visually displayed limited presentation of the AMPA receptor, whereas HATH1-transfected cells and cells cotransfected with HATH1 and siRNA against HES1 and HES5 displayed strong presentation of active AMPA receptors (Supplementary Fig. S5).
Only minor changes in cell surface markers were observed between untreated and treated cells
hWJCs were characterized for CD markers associated with stem cells 10 days after transfection, and no significant changes between untreated and treated cells were found. All cell populations were strongly negative for CD34 and CD45, which indicated that cell populations were nonhematopoietic. Additionally, all cell populations displayed presentation of CD73, CD90, and CD105, which are surface markers found on mesenchymal stem cells. Supplementary Table S2 summarizes the flow cytometry data collected from cell characterization.
Discussion
For the first time, HATH1 was delivered to hWJCs, and ATOH1 homologues (HATH1 and MATH-1) were compared side by side. The data have suggested that there may be a functional difference between MATH-1 and HATH1 in human tissues. In our previous study, we transduced hWJCs with MATH-1 and showed that hWJCs were amenable to MATH-1 gene delivery.35 The current study set out to deliver MATH-1 nonvirally to hWJCs. In addition, HATH1 was delivered to hWJCs for the first time to compare differences between ATOH1 homologues as it was hypothesized that HATH1 and MATH-1 were not functionally interchangeable in human tissues. Furthermore, HES1 and HES5 were knocked down in conjunction with overexpression of ATOH1 homologues as it was hypothesized that increasing the atonal effect could enhance the development of hair cells.
Live cell imaging and flow cytometry revealed noticeable differences between the number of viable cells between groups that were transfected with MATH-1 and HATH1. There were substantially fewer viable cells 1 day after transfection in groups that were transfected with MATH-1 than in groups transfected with HATH1 or siRNA against HES1 and HES5. In addition, cells transfected with MATH-1 displayed little to no infiltration of FM 1-43, whereas cells transected with HATH1 or siRNA against HES1 and HES5 showed varying levels of FM 1-43 infiltration 7 days after transfection. While MATH-1 and HATH1 are ATOH1 homologues, there may be a difference in the efficiency of cellular signaling between MATH-1 and HATH1. Furthermore, expression of MATH-1 in hWJCs may be negatively affecting hWJC viability.
The expression of ATOH1 induces the development of Merkel cells (touch sensory cells in the skin), cerebellar granule cells, and intestinal cells (Paneth and goblet cells) in addition to the development of hair cells in the cochlea and vestibular organs.51–54 In the current study, several morphologies, such as elongated cell bodies, bipolar cells, branching, condensed globular, or pear-shaped cells, were observed in cells that were transfected with HATH1 or a combination of HATH1 and siRNA against HES1 and HES5. Hair cells normally have a pear-shaped morphology, while neurons can be branched or bipolar, and nonsensory support cells can be elongated.55–59
In hWJCs, treatment with HATH1 or HATH1 and siRNA against genes, HES1 and HES5, displayed significant immediate increases in mRNA and protein expression of key hair cell markers compared with cells treated with MATH-1, which displayed limited increases in gene expression and protein expression 1 day after transfection. Kiernan et al. have shown that JAG1 is required for the development of sensory epithelium in the cochlea and it maintains barriers between sensory and nonsensory epithelia.60–63 ATOH1 has been shown to produce hair cell phenotypes when activated by SOX2 in conjunction with EYA1 and SIX1; however, if SOX2 activates NEUROG1 and NEUROD1, then ATOH1 expression will be downregulated and a neuronal phenotype may emerge.64,65
However, expression of ATOH1 in the absence of NEUROG1 expression downregulates SOX2 and leads to presentation of JAG2, which binds to the NOTCH1 receptor on adjacent cells and initiates lateral inhibition by enabling the NOTCH intercellular domain to translocate to the nucleus and initiate transcription of HES1 and HES5, which negatively regulate ATOH1.66–71 Neuroprogenitor cells that express ATOH1 develop into hair cells and further express MYO6 and MYO7A motor proteins in the actin fiber bundles located on the apical side of the hair cells, and hair cells express POU4F3, GFI1, and BARHL1, while cells that express HES1 and HES5 and SOX2 develop into nonsensory support cells.72–80
GFAP is an intermediate filament found in astrocytes in the central nervous system.81 The positive identity of GFAP in cells cotransfected with HATH1 and siRNA against HES1 and HES5 taken with the observation of bipolar cell morphology and infiltration of FM 1-43 suggests that some cells are exhibiting an initial differentiation toward a neural-like phenotype. The positive identification of AMPA receptors on cells transfected with both HATH1 and siRNA against HES1 and HES5 in the pilot study (Supplementary Fig. S5) further supports the potential of development of a basic neuron phenotype in some cells as AMPA receptors are not present on hWJCs or hair cells, but are present on motor neurons.82
In addition, all treated cells displayed positive presentation of MYO7A 1 day after transfection through immunocytochemistry; however, presentation of MYO7A decreased in groups transfected with MATH-1. MYO7A expression is expected if cells are differentiating toward a hair cell lineage, but HES5 expression is surprising because HES5 encourages support cell differentiation by negatively regulating ATOH1. The positive expression of HES5 both at the gene and protein levels suggested that hWJCs may be differentiating into both hair cells and supporting cells concurrently.
The significant upregulations of the GFI1 gene expression in cells cotransfected with HATH1 and siRNA against HES1 and HES5 suggested that presentation of a hair cell phenotype had started within at least a subpopulation of treated cells. However, it is peculiar that POU4F3 expression did not parallel GFI1 expression in cells transfected with HATH1 and siRNA against HES1 and HES5 as POU4F3 is upstream of GFI1. The mismatched gene expression between POU4F3 and GFI1 may have indicated that multiple cell types differentiated within the population of cells transfected with HATH1 and siRNA against HES1 and HES5. NEUROG1 is a transcription factor that can inhibit expression of ATOH1 and specify neuronal differentiation, whereas NEUROD1 is another bHLH transcription factor found in hair cells and regulated by ATOH1.83,84
The examination of NEUROG1 and NEUROD1 may be beneficial in revealing sensory and nonsensory differentiation in hWJCs transfected with HATH1 in follow-up studies. Furthermore, the differentiation of multiple cell populations may be supported by the surface area measurements taken in the pilot study. No significant changes were detected in cell surface area measurements collectively between groups; however, the variation in cell surface area was notable. A follow-up study where different cell types can be quantified and modeled based on general cell measurements may be beneficial in providing evidence to categorize different emerging cell types from treatment with HATH1 or MATH-1. In addition, follow-up studies that track the gene expression and protein expression of neural-specific Merkel cell and goblet cell-specific genes in addition to hair cell and support cell genes will be beneficial in illuminating the cell types induced by ATOH1 when expressed in hWJCs and providing further information on the use of hWJCs in hearing regeneration.
Now that measureable and observable differences have been established in hWJCs transfected with HATH1 and hWJCs transfected with MATH-1, there is motivation and a strong rationale for future investigation of functional testing such as full electrophysiological testing and full AMPA receptor staining. Mechanistic analyses such as microarray analysis will help illuminate which biochemical pathways are active after hWJCs are transfected, which may help determine the terminal lineages hWJCs are moving toward (i.e., hair cells, cerebellar neurons, Merkel cells) outside the body.
Additionally, examining the presentation of ATOH1 and the neurofilament through immunocytochemistry would be helpful for identifying hair cells and neurons in a follow-up study. Conversely, transfecting HATH1 and MATH-1 into mouse Wharton's jelly cells should yield interesting results that provide more evidence on whether HATH1 and MATH-1 are truly able to be used interchangeably between species or not. Mulvaney et al.45 reported significant differences in sequence homology and functionality when embryonic cochlear mouse explants were transfected with either the avian ATOH1 homologue (CATH1) or the mammalian ATOH1 homologue (HATH1). It is well known that hair cells in avian species regenerate, whereas hair cells in mammalian species do not regenerate.85–87
Cochlear explants transfected with HATH1 produced a higher percentage of functional hair cells than CATH1-transfected explants. However, when explants were cotransfected with SOX2 and CATH1, Mulvaney et al.45 found that CATH1 more efficiently out-competed SOX2 and produced a significantly higher ratio of hair cells to support cells than explants that were transfected with SOX2 and HATH1. Thus, further investigation and evaluation of ATOH1 homologues is needed for engineering and regenerating sensory epithelium in the inner ear.
Furthermore, culturing treated cells in a three-dimensional environment similar to the native cochlea or coculture of treated cells with a combination of native hair cells and support cells may further enhance the atonal effect and potential display of hair cell characteristics.
In summary, the data revealed that hWJCs transfected with HATH1 displayed far superior expression of key hair cell markers in relation to presentation of mRNA transcripts, proteins, and morphological features in contrast to hWJCs transfected with MATH-1.
The development and presentation of hair cell markers were further enhanced when HATH1-transfected hWJCs were cotransfected with siRNA against HES1 and HES5. The current study demonstrated that hWJCs can be manipulated outside of a target tissue to produce a rare and complex phenotype that may aid in illuminating how hair cells develop in the human body. For the first time, the ATOH1 homologues, HATH1 and MATH-1, were compared in cells from human tissue, and human cells conclusively responded differently to HATH1 and MATH-1, which suggests that the two homologues may not be interchangeable among species, and that further study of HATH1 expression in hWJCs is needed.
Supplementary Material
Author Contribution Statement
The idea to deliver MATH-1 to hWJCs was conceived by K.D., H.S., and M.S.D. The idea to compare HATH1 against MATH-1 and use siRNA against HES1 and HES5 was conceived by A.J.M. The idea to incorporate siRNA into the experiment was inspired by E.T. and S.N. The initial experimental design was developed by A.J.M., M.S.D., M.L.F., H.E.S., and D.S.M. The experiments were executed by A.J.M. and H.E.S. Advice and technical support was provided by D.S.M., Z.T., and H.S., and modifications to experimental design were based on advice from M.L.F. and J.S.L. The manuscript was written by A.J.M. and edited and revised by K.D., H.E.S., D.S.M., J.S.L., M.L.S., S.N., E.T., H.S., and M.S.D.
Acknowledgments
This project was funded by NIH R01 AR056347 and the State of Kansas. The authors would like to acknowledge the efforts of the nursing staff at Lawrence Memorial Hospital (Lawrence, KS) for their assistance in obtaining human umbilical cords. The authors would like to acknowledge the efforts of Peggy Keefe, Jenny Nelson-Brantley, Mary Krause, and Talia Martin for their helpful instructions on hWJC harvest, immunocytochemistry, DNA isolation, and DNA cloning, respectively. The authors would like to acknowledge the efforts of Anthony Frei, Megan Godsey, Austin Smith, and Mackenzie Bloom for their assistance on maintaining cells for this project.
Disclosure Statement
No competing financial interests exist.
References
- 1.Devarajan K., Staecker H., and Detamore M.S. A review of gene delivery and stem cell based therapies for regenerating inner ear hair cells. J Funct Biomater 2, 249, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dror A.A., and Avraham K.B. Hearing loss: mechanisms revealed by genetics and cell biology. Annu Rev Genet 43, 411, 2009. [DOI] [PubMed] [Google Scholar]
- 3.Chonko K.T., et al. Atoh1 directs hair cell differentiation and survival in the late embryonic mouse inner ear. Dev Biol 381, 401, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Izumikawa M., et al. Auditory hair cell replacement and hearing improvement by Atoh1 gene therapy in deaf mammals. Nat Med 11, 271, 2005. [DOI] [PubMed] [Google Scholar]
- 5.Jeon S.J., Oshima K., Heller S., and Edge A.S. Bone marrow mesenchymal stem cells are progenitors in vitro for inner ear hair cells. Mol Cell Neurosci 34, 59, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park Y.H., et al. Conditioning the cochlea to facilitate survival and integration of exogenous cells into the auditory epithelium. Mol Ther 22, 873, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukui H., and Raphael Y. Gene therapy for the inner ear. Hear Res 297, 99, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du X., et al. Regeneration of mammalian cochlear and vestibular hair cells through Hes1/Hes5 modulation with siRNA. Hear Res 304, 91–110, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jung J.Y., et al. siRNA targeting Hes5 augments hair cell regeneration in aminoglycoside-damaged mouse utricle. Mol Ther 21, 834, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kageyama R., and Ohtsuka T. The Notch-Hes pathway in mammalian neural development. Cell Res 9, 179, 1999. [DOI] [PubMed] [Google Scholar]
- 11.Mizutari K., et al. Notch inhibition induces cochlear hair cell regeneration and recovery of hearing after acoustic trauma. Neuron 77, 58, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoon H., Lee D.J., Kim M.H., and Bok J. Identification of genes concordantly expressed with Atoh1 during inner ear development. Anat Cell Biol 44, 69, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Driver E.C., Sillers L., Coate T.M., Rose M.F., and Kelley M.W. The Atoh1-lineage gives rise to hair cells and supporting cells within the mammalian cochlea. Dev Biol 376, 86, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly M.C., Chang Q., Pan A., Lin X, and Chen P. Atoh1 directs the formation of sensory mosaics and induces cell proliferation in the postnatal mammalian cochlea in vivo. J Neurosci 32, 6699, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kraft S., Hsu C., Brough D.E., and Staecker H. Atoh1 induces auditory hair cell recovery in mice after ototoxic injury. Laryngoscope 123, 992, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Z., et al. Age-dependent in vivo conversion of mouse cochlear pillar and Deiters' cells to immature hair cells by Atoh1 ectopic expression. J Neurosci 32, 6600, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boddy S.L., et al. Inner ear progenitor cells can be generated in vitro from human bone marrow mesenchymal stem cells. Regen Med 7, 757, 2012. [DOI] [PubMed] [Google Scholar]
- 18.Cox B.C., et al. Spontaneous hair cell regeneration in the neonatal mouse cochlea in vivo. Development 141, 816, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Z., et al. Reprogramming of single-cell-derived mesenchymal stem cells into hair cell-like cells. Otol Neurotol 33, 1648, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pauley S., Kopecky B., Beisel K., Soukup G., and Fritzsch B. Stem cells and molecular strategies to restore hearing. Panminerva Med 50, 41, 2008. [PMC free article] [PubMed] [Google Scholar]
- 21.Ronaghi M., et al. Inner ear hair cell-like cells from human embryonic stem cells. Stem Cells Dev 23, 1275, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manohar R., and Lagasse E. Transdetermination: a new trend in cellular reprogramming. Mol Ther 17, 936, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bailey M., Wang L., Bode C., Mitchell K., and Detamore M. A comparison of human umbilical cord matrix stem cells and temporomandibular joint condylar chondrocytes for tissue engineering temporomandibular joint condylar cartilage. Tissue Eng 13, 2003, 2007. [DOI] [PubMed] [Google Scholar]
- 24.Detamore M.S. Human umbilical cord mesenchymal stromal cells in regenerative medicine. Stem Cell Res Ther 4, 142, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitchell K., et al. Matrix cells from Wharton's jelly form neurons and glia. Stem Cells 21, 50, 2003. [DOI] [PubMed] [Google Scholar]
- 26.Wang L., Ott L., Seshareddy K., Weiss M., and Detamore M. Musculoskeletal tissue engineering with human umbilical cord mesenchymal stromal cells. Regen Med 6, 95, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J., and Streit A. Induction of the inner ear: stepwise specification of otic fate from multipotent progenitors. Hear Res 297, 3, 2013. [DOI] [PubMed] [Google Scholar]
- 28.Rooney G.E., et al. Gene-modified mesenchymal stem cells express functionally active nerve growth factor on an engineered poly lactic glycolic acid (PLGA) substrate. Tissue Eng Part A 14, 681, 2008. [DOI] [PubMed] [Google Scholar]
- 29.Duan P., et al. miR-29a modulates neuronal differentiation through targeting REST in mesenchymal stem cells. PLoS One 9, e97684, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guan M., Xu Y., Wang W., and Lin S. Differentiation into neurons of rat bone marrow-derived mesenchymal stem cells. Eur Cytokine Netw 25, 58, 2014. [DOI] [PubMed] [Google Scholar]
- 31.Huat T.J., et al. IGF-1 enhances cell proliferation and survival during early differentiation of mesenchymal stem cells to neural progenitor-like cells. BMC Neurosci 15, 91, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nandy S.B., Mohanty S., Singh M., Behari M., and Airan B. Fibroblast Growth Factor-2 alone as an efficient inducer for differentiation of human bone marrow mesenchymal stem cells into dopaminergic neurons. J Biomed Sci 21, 83, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsai H.L., et al. Wnts enhance neurotrophin-induced neuronal differentiation in adult bone-marrow-derived mesenchymal stem cells via canonical and noncanonical signaling pathways. PLoS One 9, e104937, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao H.B., et al. Salidroside induces neuronal differentiation of mouse mesenchymal stem cells through Notch and BMP signaling pathways. Food Chem Toxicol 71, 60, 2014. [DOI] [PubMed] [Google Scholar]
- 35.Devarajan K., Forrest M.L., Detamore M.S., and Staecker H. Adenovector-mediated gene delivery to human umbilical cord mesenchymal stromal cells induces inner ear cell phenotype. Cell Reprogram 15, 43, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aluigi M., et al. Nucleofection is an efficient nonviral transfection technique for human bone marrowñderived mesenchymal stem cells. Stem Cells 24, 454, 2006. [DOI] [PubMed] [Google Scholar]
- 37.Cesnulevicius K., et al. Nucleofection is the most efficient nonviral transfection method for neuronal stem cells derived from ventral mesencephali with no changes in cell composition or dopaminergic fate. Stem Cells 24, 2776, 2006. [DOI] [PubMed] [Google Scholar]
- 38.Gresch O., and Altrogge L. Transfection of difficult-to-transfect primary mammalian cells. Methods Mol Biol 801, 65, 2012. [DOI] [PubMed] [Google Scholar]
- 39.Mellott A.J., Forrest M.L., and Detamore M.S. Physical non-viral gene delivery methods for tissue engineering. Ann Biomed Eng 41, 446, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mellott A.J., et al. Improving viability and transfection efficiency with human umbilical cord Wharton's jelly cells through use of a ROCK inhibitor. Cell Reprogram 16, 91, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kong J., Crissey M.A., Sepulveda A.R., and Lynch J.P. Math1/Atoh1 contributes to intestinalization of esophageal keratinocytes by inducing the expression of Muc2 and Keratin-20. Dig Dis Sci 57, 845, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salsano E., Pollo B., Eoli M., Giordana M.T., and Finocchiaro G. Expression of MATH1, a marker of cerebellar granule cell progenitors, identifies different medulloblastoma sub-types. Neurosci Lett 370, 180, 2004. [DOI] [PubMed] [Google Scholar]
- 43.Yang J., Cong N., Han Z., Huang Y., and Chi F. Ectopic hair cell-like cell induction by Math1 mainly involves direct transdifferentiation in neonatal mammalian cochlea. Neurosci Lett 549, 7, 2013. [DOI] [PubMed] [Google Scholar]
- 44.Blake J.A., et al. The Mouse Genome Database: integration of and access to knowledge about the laboratory mouse. Nucleic Acids Res 42, D810, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mulvaney J.F., Amemiya Y., Freeman S.D., Ladher R.K., and Dabdoub A. Molecular cloning and functional characterisation of chicken Atonal homologue 1: a comparison with human Atoh1. Biol Cell 107, 41, 2015. [DOI] [PubMed] [Google Scholar]
- 46.Carpenter A.E., et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7, R100, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones T.R., et al. Scoring diverse cellular morphologies in image-based screens with iterative feedback and machine learning. Proc Natl Acad Sci U S A 106, 1826, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kamentsky L., et al. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics 27, 1179, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lamprecht M.R., Sabatini D.M, and Carpenter A.E. CellProfiler: free, versatile software for automated biological image analysis. Biotechniques 42, 71, 2007. [DOI] [PubMed] [Google Scholar]
- 50.Vytla D., Combs-Bachmann R.E., Hussey A.M., Hafez I., and Chambers J.J. Silent, fluorescent labeling of native neuronal receptors. Org Biomol Chem 9, 7151, 2011. [DOI] [PubMed] [Google Scholar]
- 51.Gazit R., Krizhanovsky V., and Ben-Arie N. Math1 controls cerebellar granule cell differentiation by regulating multiple components of the Notch signaling pathway. Development 131, 903, 2004. [DOI] [PubMed] [Google Scholar]
- 52.Leonard J.H., et al. Proneural and proneuroendocrine transcription factor expression in cutaneous mechanoreceptor (Merkel) cells and Merkel cell carcinoma. Int J Cancer 101, 103, 2002. [DOI] [PubMed] [Google Scholar]
- 53.Mulvaney J., and Dabdoub A. Atoh1, an essential transcription factor in neurogenesis and intestinal and inner ear development: function, regulation, and context dependency. J Assoc Res Otolaryngol 13, 281, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Q., Bermingham N.A., Finegold M.J., and Zoghbi H.Y. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science 294, 2155, 2001. [DOI] [PubMed] [Google Scholar]
- 55.Davies D. Cell-extracellular matrix versus cell-cell interactions during the development of the cochlear-vestibular ganglion. J Neurosci Res 2011 89, 1375. [DOI] [PubMed] [Google Scholar]
- 56.Driver E.C., and Kelley M.W. Specification of cell fate in the mammalian cochlea. Birth Defects Res C Embryo Today 87, 212, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Forge A., and Wright T. The molecular architecture of the inner ear. Br Med Bull 63, 5, 2002. [DOI] [PubMed] [Google Scholar]
- 58.Gerchman E., Hilfer S.R., and Brown J.W. Involvement of extracellular matrix in the formation of the inner ear. Dev Dyn 202, 421, 1995. [DOI] [PubMed] [Google Scholar]
- 59.Goodyear R.J., and Richardson G.P. Extracellular matrices associated with the apical surfaces of sensory epithelia in the inner ear: molecular and structural diversity. J Neurobiol 53, 212, 2002. [DOI] [PubMed] [Google Scholar]
- 60.Kiernan A.E. Notch signaling during cell fate determination in the inner ear. Semin Cell Dev Biol 24, 470, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kiernan A.E., et al. The Notch ligand Jagged1 is required for inner ear sensory development. Proc Natl Acad Sci U S A 98, 3873, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kiernan A.E., Li R., Hawes N.L., Churchill G.A, and Gridley T. Genetic background modifies inner ear and eye phenotypes of jag1 heterozygous mice. Genetics 177, 307, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kiernan A.E., Xu J, and Gridley T. The Notch ligand JAG1 is required for sensory progenitor development in the mammalian inner ear. PLoS Genet 2, e4, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wong E.Y., Ahmed M., and Xu P.X. EYA1-SIX1 complex in neurosensory cell fate induction in the mammalian inner ear. Hear Res 297, 13, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Evsen L., Sugahara S., Uchikawa M., Kondoh H., and Wu D.K. Progression of neurogenesis in the inner ear requires inhibition of Sox2 transcription by neurogenin1 and neurod1. J Neurosci 33, 3879, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cai T, and Groves A.K. The role of atonal factors in mechanosensory cell specification and function. Mol Neurobiol 2014. [Epub ahead of print]; DOI: 10.1007/s12035-014-8925-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cotanche D.A., and Kaiser C.L. Hair cell fate decisions in cochlear development and regeneration. Hear Res 266, 18, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jarman A.P., and Groves A.K. The role of Atonal transcription factors in the development of mechanosensitive cells. Semin Cell Dev Biol 24, 438, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu Z., et al. In vivo visualization of Notch1 proteolysis reveals the heterogeneity of Notch1 signaling activity in the mouse cochlea. PLoS One 8, e64903, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Petrovic J., Galvez H., Neves J., Abello G., and Giraldez F. Differential regulation of Hes/Hey genes during inner ear development. Dev Neurobiol 2014; DOI: 10.1002/dneu.22243 [DOI] [PubMed] [Google Scholar]
- 71.Yang S.M., et al. Regeneration of stereocilia of hair cells by forced Atoh1 expression in the adult mammalian cochlea. PLoS One 7, e46355, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dabdoub A., et al. Sox2 signaling in prosensory domain specification and subsequent hair cell differentiation in the developing cochlea. Proc Natl Acad Sci U S A 105, 18396, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Frolenkov G.I., Belyantseva I.A., Friedman T.B., and Griffith A.J. Genetic insights into the morphogenesis of inner ear hair cells. Nat Rev Genet 5, 489, 2004. [DOI] [PubMed] [Google Scholar]
- 74.Groves A.K., Zhang K.D., and Fekete D.M. The genetics of hair cell development and regeneration. Annu Rev Neurosci 36, 361, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hongmiao R., Wei L., Bing H., Xiong D.D., and Jihao R. Atoh1: landscape for inner ear cell regeneration. Curr Gene Ther 14, 101, 2014. [DOI] [PubMed] [Google Scholar]
- 76.Ikeda R., Pak K., Chavez E., and Ryan A.F. Transcription factors with conserved binding sites near ATOH1 on the POU4F3 gene enhance the induction of cochlear hair cells. Mol Neurobiol 2014. [Epub ahead of print]; DOI: 10.1007/s12035-014-8801-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lanford P.J., et al. Notch signalling pathway mediates hair cell development in mammalian cochlea. Nat Genet 21, 289, 1999. [DOI] [PubMed] [Google Scholar]
- 78.Masuda M., et al. Regulation of POU4F3 gene expression in hair cells by 5′ DNA in mice. Neuroscience 197, 48, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Neves J., Vachkov I., and Giraldez F. Sox2 regulation of hair cell development: incoherence makes sense. Hear Res 297, 20, 2013. [DOI] [PubMed] [Google Scholar]
- 80.Woods C., Montcouquiol M., and Kelley M.W. Math1 regulates development of the sensory epithelium in the mammalian cochlea. Nat Neurosci 7, 1310, 2004. [DOI] [PubMed] [Google Scholar]
- 81.Placone A.L., et al. Human astrocytes develop physiological morphology and remain quiescent in a novel 3D matrix. Biomaterials 42, 134, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sanchez J.T., Ghelani S., and Otto-Meyer S. From development to disease: diverse functions of NMDA-type glutamate receptors in the lower auditory pathway. Neuroscience 285, 248, 2015. [DOI] [PubMed] [Google Scholar]
- 83.Jahan I., Pan N., Kersigo J., and Fritzsch B. Neurod1 suppresses hair cell differentiation in ear ganglia and regulates hair cell subtype development in the cochlea. PLoS One 5, e11661, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jahan I., Pan N., Kersigo J., and Fritzsch B. Beyond generalized hair cells: molecular cues for hair cell types. Hear Res 297, 30, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Roberson D.W., Alosi J.A., and Cotanche D.A. Direct transdifferentiation gives rise to the earliest new hair cells in regenerating avian auditory epithelium. J Neurosci Res 78, 461, 2004. [DOI] [PubMed] [Google Scholar]
- 86.Rubel E., Oesterle E., and Weisleder P. Hair cell regeneration in avian inner ear. Ciba Foundation Symposium 166, 77, 1991. [DOI] [PubMed] [Google Scholar]
- 87.Weisleder P., and Rubel E. Hair cell regeneration in the avian vestibular epithelium. Exp Neurol 115, 2, 1992. [DOI] [PubMed] [Google Scholar]
- 88.Chellappa R., et al. Barhl1 regulatory sequences required for cell-specific gene expression and autoregulation in the inner ear and central nervous system. Mol Cell Biol 28, 1905, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.