Abstract
Alzheimer’s disease (AD) is associated with the formation of toxic Aβ42 oligomers and recent evidence supports a role for Aβ dimers as building blocks for oligomers. Molecular dynamics (MD) simulation studies have identified clans for the dominant conformations of Aβ42 forming dimers; however, it is unclear if a larger spectrum of dimers is involved and which set(s) of dimers might evolve to oligomers verse fibrils. Therefore, for this study we generated multiple structural conformations of Aβ42, using explicit all-atom MD, and then clustering the different structures based on key conformational similarities. Those matching a selection threshold were then used to model a process of oligomerization. Remarkably, we showed a greater diversity in Aβ dimers than previously described. Depending on the clan family, different types of Aβ dimers were obtained. While some had the tendency to evolve into oligomeric rings, others formed fibrils of diverse characteristics. Then we selected the dimers that would evolve to membranephilic annular oligomers. Nearly one third of the 28 evaluated annular oligomers had the dimer interfaces between the neighboring Aβ42 monomers with possible salt bridges between the residue K28 from one side and either residue E22 or D23 on the other. Based on these results, key amino acids were identified for point mutations that either enhanced or suppressed the formation and toxicity of oligomer rings. Our studies suggest a greater diversity of Aβ dimers. Understanding the structure of Aβ dimers might be important for the rationale design of small molecules that block formation of toxic oligomers.
Keywords: Oligomers, dimers, synaptoxicity, neurodegeneration, molecular dynamics
Introduction
Aggregation of misfolded neuronal proteins and peptides may play a primary role in the pathogenesis of neurodegenerative disorders including Alzheimer’s disease (AD) [1–3]. Amyloid-β (Aβ) is a small (40 to 43 amino acids (aa) long) [4] amyloidogenic peptide generated following the proteolytic cleavage of the amyloid precursor protein (APP). Aβ accumulates extracellularly in AD, forming amyloid fibrils that are the main component of plaques. The physiological and pathological roles of Aβ are areas of active investigation [5], with some authors proposing Aβ*56 as the toxic species [6], while others have proposed arrays involving higher-order species such as Aβ-derived diffusible ligands (ADDLs) [7]. Recently, the evidence had been shown that small oligomers of Aβ42 [8], and even dimers [9], may play a primary role in the formation of neurotoxic annular oligomers that occur in AD. Aβ dimers were identified in cerebral spinal cord fluid in AD patients [10], and were shown to decrease long-term potentiation (LTP) both in slices or when infused into rats [9]. Recently, various groups developed molecular models of Aβ dimers; however, there are a number of challenges to modeling Aβ dimerization. Therefore, in order to develop rationally designed therapeutics for the treatment and prevention of AD, it is crucial to understand the tertiary and quaternary structures of Aβ.
There are several challenges to identifying the tertiary structure of the Aβ; perhaps the most significant is that the peptide is “naturally unfolded,” and as such, it lacks a stable tertiary structure [11, 12]. Moreover, there are experimental limitations, such as Aβ aggregates very quickly in water, Aβ fibrils are insoluble, and the peptide has yet to be crystalized for X-ray crystallography, have hampered this area of research [13–16]. Since Aβ has an unstable tertiary structure, it is likely that there are actually a number of conformations for the Aβ monomer. Therefore, researchers turned to computational experiments to identify the theoretical structure(s) of Aβ. There are a number of reports of using molecular dynamics (MD) to simulate the behavior of Aβ structures in solution. However most were performed with shortened sequences of Aβ (residues 16–22), focusing on the hydrophobic core where, according to these studies, the α-helix unfolds into the β-strand [17–30]. Other studies used an implicit solvent [22, 28, 31–36], or simplified the simulated structure, using coarse-grained approaches [28, 35, 37–41]. These approximations were used to overcome limitations in CPU speeds, which, until recently, prevented researchers from simulating the structure(s) of Aβ using all-atom force field of the full-length peptide in explicit solvent [42].
Molecular dynamics simulation of Aβ dimers is the starting point for elucidating the process of Aβ peptide oligomerization. Initially, molecular models of dimers were constructed using traditional molecular docking methods where the best structural pair of Aβ monomers was identified by the energies of intermolecular interactions [43–45]. Using discrete MD (DMD) with simplified potentials and coarse-grain modeling, another set of Aβ dimers was obtained [40]. In addition, replica exchange MD (REMD) studies of Aβ dimers using coarse-grain and implicit solvents were used to simulate dimers and tetramers based on structures obtained from solid-state NMR experiments [46]. This study found that residues 10–23 account for most of the interpeptide interactions during aggregation [47] and identified the importance of Ile41 and Ala42 to Aβ dimerization. In addition, Asp23-to-Lys28 has a role in forming a salt bridge in a number of the computational models that enhances nucleation and fibrillization [45, 48]. In the recent studies Yu and Zheng [49], using explicit solvent MD and their peptide-packing program, showed that the Aβ monomers and dimers can be expanded to energetically favorable and stable annular oligomers.
Taken together, these studies identified different sets of dimers that can be potentially involved in the development of annular oligomers in AD. For example, while Côté and co-authors described six main clusters for the dominant morphologies of the dimers [48], authors of more recent studies have shown another variety of possible dimers starting with ten different configurations with very diverse secondary structures of different peptide regions [50. 51]. Therefore, it is unclear if a larger spectrum of dimers is involved in AD, and which set(s) of dimers might evolve to toxic oligomers verse fibrils. For this purpose, we utilized a strategy that used a combination of MD simulations, molecular docking, and studies of molecular interactions with the membrane, as well as mutagenesis, biochemical, and electron microscopy studies to characterize the evolution of dimers to oligomers. Our studies suggest that there is a greater diversity of Aβ dimers that previously described and that depending on their conformations some can be nonpropagating—not leading to any further oligomerization, and some lead to oligomer rings or fibrils.
Materials and Methods
Overview of approach
To study the structural diversity of Aβ aggregates, in particular dimers, as well as identifying key aa residues involved in preventing dimerization, we developed a new combined modeling approach (Figure 1) that enabled the characterization of the large variety of Aβ42 conformations. Rather than limiting the molecular modeling to one rigid conformation of Aβ42 (Figure 1A), the approach for this study was to generate different structural conformations of Aβ42 (Figure 1B) using all-atom MD in explicit water environment, and then clustering the different conformers based on key structural similarities (Figure 1C). This enabled the selection of the best centroid conformer from each conformational clan (Figure 1D), which was then used for modeling the homodimers (Figure 1E). The best centroid conformer (referred to as “centroid”) was selected for each clan as the conformer with the best RMSD and Z-scores when compared to each member within its clan (Table S1). The best-energy dimers (Figure 1F) were used as the starting points for modeling the oligomers, using the consecutive docking procedure to evaluate the possibility of quaternary structural formation. As the result we obtained rings, fibrils, or nonpropagating dimers that cannot form any oligomers (Figure 1G). Then we selected the rings having the best propensities to the membrane (Figure 1H) for further analysis (Figure 1I) and studied the dimers that belong to these rings (Figure 1J). We conducted a frequency analysis of the intermolecular interactions between the neighboring Aβ42 monomers within the annular oligomers. Based on these studies, key amino acids were proposed (Figure 1K) for point mutations. The wild type (WT) and mutant peptides have been synthesized (Figure 1L). The WT and two mutant peptides that were predicted to have significantly different properties were then evaluated for peptide aggregation using Western blot (Figure 1M), ring and fibril formation in a cell-free environment using transmitted electron microscopy (TEM; Figure 1N), and for cytotoxicity in primary neuronal cultures (Figure 1O).
Figure 1.
Overview of the scientific approach. Level 1 is focused on (A) the preparation of Aβ rings starting from an initial NMR conformation of Aβ, which lead to (B) the generation of 1000 different conformers of Aβ. (C) These conformers were clustered into 77 groups, and (D) the best centroid conformer was selected from each cluster as a representative structure. (E) Centroid monomers were docked to form dimers, and (F) the dimers with the best energy were selected. (G) Consistent docking of the conformers to the dimers was conducted. It resulted in generation of rings, fibrils, or nonpropagating dimers. Level 2 is focused on the analysis of ring structures. (H) Rings were evaluated for membranephilicity and (I) selected for further analysis. (J) Dimers from the rings with the best energy of interaction with the membrane were examined after which (K) the interpeptide contacts were studied to determine key amino acid residues. Level 3 is focused on the biological effects of mutating key aa residues. The wild type (WT) and two mutant peptides (L) were evaluated for peptide aggregation using Western blot (M), for ring and fibril formation in a cell-free environment using transmitted electron microscopy (N), and for cytotoxicity in primary neuronal cultures (O).
Molecular dynamics simulations of Aβ
Molecular dynamics were conducted as previously described [52, 53] for 100 ns using Aβ1–42 NMR structure (PDB ID 1IYT) [54] as the initial structure, with NAMD MD program (version 2.6) [55]. The CHARMM27 force-field parameters [55] were used for MD simulations of Aβ in water under normal conditions. The temperature was maintained at 310K by means of Langevin dynamics using a collision frequency of 1/ps. A fully flexible cell at constant pressure (1 ATM) was employed using the Nosé-Hoover Langevin piston algorithm [57, 58] as used in the NAMD software package. At least 30 Å separated the Aβ molecule from the box walls of the water box. TIP3P model for water molecules was used. The van der Waals (vdW) interactions were switched smoothly to zero over the 10 Å region and electrostatic interactions were included via the smooth particle-mesh Ewald summation [59]. CMAP correction was used to avoid the known tendency of CHARMM27 to overstabilize π-helices.
The simulation was performed in four steps. Initially the systems were minimized for 10,000 iterations. Then the systems were heated in 0.1° increments and equilibrated for 1 ns; then the molecular dynamics simulation was conducted. Data for analysis were taken every 100 ps throughout the simulation.
Clustering, dimer and oligomer construction
Conformers were clustered by structural similarity using Shindialov’s algorithm [60] with the maximum possible cutoff Z-scores and minimal possible root mean squared difference (RMSD) between selected members of the clan. The conformer having the best Z-score along with the smallest summary RMSDs of C-α atoms to the same atoms of other conformers was selected as the centroid and used in further studies as a representative of its clan. Using the docking program Hex [61], Aβ centroids conformers were docked to the copies of themselves. The Hex program, based on the Fast Fourier Transform (FFT) method, conducts comprehensive docking of two molecules in all possible positions with the further selection of the best docked complexes based on the shape of the complex and electrostatic interactions. Then we consecutively docked to each of the dimers additional copies of the centroid (one at a time) with docking rotation and translation parameters, as were used in the initial docking complex configuration selected by the Hex program, to extend the dimers. This procedure used three-dimensional (3D) transformation matrices that were preliminarily calculated for each of the starting dimers. A similar procedure was used by Yu and Zheng [49]. This stepwise extension of the original dimer led to three different possible scenarios. The first case was a “nonpropagating dimer” where addition of the second conformer was impossible. The second case was a dimer that propagated to a fibril (Tables S1, S2). The third case was a dimer that propagated to an annular structure. As a result of the latter procedure sets of different rings were generated for each clan (Table S1). There were also clans that did not generated any rings.
Elucidation of the membranephilic rings
In order to check the stability of the annular oliogmers, 2 ns MD in the explicit water environment was conducted. Only those oligomers that remained stable after this procedure were selected for further study.
Then the annular oligomers were filtered to select structures with a significant propensity for the membrane surface. During the first step we filtered the generated annular oligomers for general correspondence to the single conformer propensity to the membrane. All centroid conformers that generated rings were tested with the program MAPAS that predicted their best energy position on the membrane surface. Then we compared the position of the conformers in the ring oligomer located on the membrane surface with the previously defined best position of the monomers. The angles between the lines connecting the Cα atoms of N- and C-terminals of the peptide and the plane of the membrane surface were calculated. Similar calculations were done for the peptide within the ring located on the membrane. For further analysis we selected only the rings with less than a 15% difference in the angles of each of the members of the oligomer to the plane of the membrane, compared to the predicted angle for a single centroid conformer estimated by our program MAPAS [62].
The next step was selecting the annular oligomers that would have the maximum membrane propensities or would have a maximum “membranephilicity.” For this purpose we used the Prediction of Proteins in Membranes (PPM) server [63]. This server is considered as one of the best servers devoted to estimation of possible positions of proteins and peptides in membranes. It uses the polarity profile of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) bilayer determined by neutron and X-ray scattering [64], and distribution of water in DOPC bilayer determined by spin-labeling experiments [65]. The server uses the implicit solvent representation of lipid bilayer that takes in to consideration its anisotropic properties. The server calculates the energy of transfer of protein (peptide) from water to the lipid bilayer as a sum of two terms. First—solvent accessible surface area-dependent term that takes in consideration the vdW and hydrogen-bonding interactions along with entropy of solvent molecules; second—electrostatic term including solvation energies and deionization penalties of ionizable groups in nonpolar environment. The details of this method are described explicitly elsewhere [66]. The Cartesian coordinates of the studied annular oligomers of Aβ were submitted to the PPM server and the membrane transfer free energy per one monomer of Aβ for each of them was calculated. The resulting values were used for selection of the best membranephilic annular oligomers for further studies.
Study of intermolecular interactions between the neighboring Aβ peptides in oligomers
The interpeptide contacts between the neighboring monomers of Aβ in the annular oligomers and fibrils were studied. We measured the distances between the centers of the closest heavy atoms of the neighbors. Such distances were measured between each pair of neighbors in the annular structures or fibrils and then the average distances per a single pair of monomers were calculated. Distances less than 4 Å have been considered as contacts and were used in the following frequency analysis. The frequencies of occurrences of the specific residues in the contacts with the neighboring monomers were calculated for all previously selected rings and fibrils.
Frequency analysis of intermolecular interactions, selection of mutations, and molecular dynamics simulations of mutant Aβ
Based on a frequency analysis of interpeptide contacts within the Aβ rings and analysis of contacts of the best membranephilic ring oligomers, two artificial mutants of Aβ were created in order to evaluate the effects the main intermolecular interfaces in Aβ rings. Mutant 1, E3R, D7R, V36S, V40S, was designed to evaluate substitution of negative to positive changes along with two hydrophobic-to-hydrophilic mutations to mitigate the strong energies of hydrophobic interactions. Mutant 2 evaluated the positive-to-negative substitutions, R5D and K16D, along with one hydrophobic-to-hydrophilic mutation, V39S. MD simulations of the mutants were conducted with the same parameters and protocols as WT peptide (see above).
We checked the possibility of prediction of the mutations impact on the properties of the Aβ peptides and their capability to form rings using relatively short MD simulations. Conformers of the mutant Aβ peptides obtained following only 8.5 ns of MD were docked as homodimers. Next, these dimers underwent the consecutive docking procedure as described above. Rings, fibrils, and nonpropagating dimers were generated. Then we analyzed the percentage of propagating to rings dimers as well as nonpropagating dimers that can be generated from the mutant conformers. These results then were compared with the experimental results.
Preparation and aggregation of Aβ1–42 wild type and mutants
Aβ42 (American Peptide Co., Sunnyvale, Calif.) WT and Mutants 1 and 2 (Eton Biosciences Inc., San Diego, Calif.) were aggregated by incubating the peptides for 16 h at 37°C followed by a 6 h incubation at 56°C. Different concentrations of peptides (1, 10, and 50 μg) were loaded into each lane of a 4–12% Bis-Tris gel (Life Technologies (Invitrogen), Carlsbad, Calif.) using MES running buffer (Invitrogen). Gels were either stained with colloidal blue (Invitrogen), silver stain (Invitrogen), or transferred to a 0.2 μm nitrocellulose membrane (Whatman, Piscataway, N.J.), and probed using 4G8 (1:1000; Signet Laboratories, Inc., Dedham, Mass.) followed by incubation with secondary antibodies tagged with horseradish peroxidase (1:5,000 dilution, Santa Cruz Biotechnology, Santa Cruz, Calif.). Immunoreactive signals were visualized by enhanced chemiluminescence and analyzed using a Versadoc XL imaging machine using the Quantity One program (BioRad, Hercules, Calif.).
Electron microscopy
Briefly, WT and mutated Aβ peptides were aggregated, as described above, before 2 μL was added to a formvar/carbon-coated gold grid (200 mesh grid size; Ted Pella Inc., Redding, Calif.) and allowed to dry over night at room temperature [53]. The peptides were then negative stained with uranyl acetate (50% in ethanol; PolySciences Inc., Warrington, Pa.) and Bismuth subnitrate (2% in 2 M sodium hydroxide) and analyzed using a Zeiss EM10 electron microscope, and electron micrographs were obtained at a magnification of 40,000.
Treatment of adult rat hippocampal neural stem cells with WT and mutant Aβ and cell viability
Adult rat hippocampal neural stem cells (Chemicon International Inc., Billerica, Mass.) were grown in DMEM/Ham’s F-12 medium containing B27 supplements without Vitamin A (Gibco®, Life Technologies, Carlsbad, Calif.). Cells were differentiated in DMEM/F12 medium containing N2 supplements (Gibco®) for 4 days. Cells were then treated with aggregated Aβ WT and mutants at 1, 10, and 50 μM concentrations for 24 hours. Media was collected from the cells and used in the lactate dehydrogenase (LDH) assay (Promega, Madison, Wis.). Assessment of membrane permeabilization was carried out in triplicate as previously described [67], using calcein dye as an indicator (Invitrogen).
Statistical analysis
The data are expressed as mean values ± SEM. Statistical analysis was performed using one-way analysis of variance (ANOVA) followed by post hoc Dunnett’s or Tukey-Kramer tests (Prism Graph Pad Software, San Diego, Calif.). Differences were considered significant at p < 0.05.
Results
Nomenclature
A simplified naming convention was used to identify the different Aβ conformers created during MD simulations. The initial 1000 conformations snapshots were taken every 100 ps and numbered in order. The different configurations of homodimers had an additional label (forming the second part of the two-part label) to distinguish different dimers that were formed from the same conformer. Similarly, the fibrils formed from propagating dimers maintained their two-part label as in the dimer, while when the rings were formed these had a third component to their label indicating the number of monomers in the ring.
Molecular dynamics studies show a large variety of Aβ dimer, annular, and fibrillar structures
Since Aβ is “naturally unfolded,” it has a number of different conformations. Therefore, a large variety of monomeric Aβ conformations were generated using unrestrained all-atom MD in explicit water box, starting with the NMR structural conformation of Aβ (Figure 2A). The conformation of Aβ changed over time from a mostly α-helical conformation defined by NMR to conformations having less α-helical content evolving through the π-helices toward turns and coils (Figure 2A and B). Following 7 ns of modeling, the α-helical content decreasing from 44% to less than 10% (Figure 2B and C). At the same time there was increased π-helix, bend, and coil content and decreased percentage of nonhelical turns (Figure 2C). Around 35 ns of MD, the Aβ conformation had less than 5% α-helices and the maximum number of π-helices, which are a transitional state to the unstructured coil/turn structure. The α-helical structure content increased again before 60 ns and then decreased rapidly to less than 10%. It remained unchanged for the duration of the 100 ns MD. The sharp decrease of π-helical content after 40 ns was accompanied by growth of unstructured peptide regions: turns, bends, and coils. The content of these structures grew to around 100% after 60 ns and remain stable to 100 ns (Figures 2B and C). The 1000 conformers underwent structural superposition and were grouped into 77 clans using the smallest possible RMSD and greatest Z-score values relative to other conformers in the clan (Table S1). The most populated clan contained 82 members and included the selected conformations that occurred from around 30 ns to around 47 ns of MD, while smallest clans were composed of two Aβ conformers. There were also degenerated clans that contained only one conformer. A “centroid conformer” was selected for each clan as the conformer with the best RMSD and Z-scores when compared to each member within its clan (Table S1).
Figure 2.
MD simulation of Aβ over 100 ns. (A) Over time, the conformation of Aβ changes. (B) During the 100 ns time course, α-helices (blue rings) were converted to π-helices (red squares) which are a prerequisite to forming an unstructured loop. Then after 50 ns the π-helices disappear and the α-helices appear again with the growing combined percentage of turns, bends, and coils. After 60 ns rapid transformation of all helices to unstructured conformations is evident. (C) The composition of Aβ secondary structures (reported as a percent).
Using the docking program Hex [61], we constructed the dimers from the centroid conformers from each clan, which were docked to their copies resulting in the generation of distinct dimers. These dimers were then expanded by consecutive docking (see Materials and Methods section). Such expansion led to three different scenarios. The first possible outcome was a nonpropagating dimer (Table 1, Figure 3A). In this case, the addition of the third monomer to the homodimer was not possible. The majority of such cases were so-called head-to-tail dimers where the N-terminal of one monomer was close to the C-terminal of the second monomer. The second situation was a dimer that propagated to a possible fibril (Table 1, Figure 3B, Tables S1 and S2). The third outcome was a dimer that propagated to an annular structure (Table 1, Figure 3C, Table S1). In conclusion, dimers from different clans could lead to the formation of various rings and fibrils.
Table 1.
Examples of MD centroid Aβ conformers and generated by docking dimers and oligomers
| Example type | Centroid ID | Centroid monomer | Dimer ID | Centroid dimer | Oligomer ID | Oligomer |
|---|---|---|---|---|---|---|
| non-propagating conformers | 30 |

|
30 |

|
||
| 79 |

|
79 |

|
|||
| 164 |

|
164 |
|
|||
|
| ||||||
| propagating to fibril conformers | 423 |

|
423_86 |
|
423_86 |
|
| 823 |

|
823_1 |

|
823_1 |
|
|
| 924 |

|
924_3 |

|
924_3 |
|
|
|
| ||||||
| propagating to ring conformers | 389 |

|
389_24 |

|
389_24_7 |

|
| 487 |

|
487_75 |

|
487_75_8 |

|
|
| 600 |

|
600_34 |

|
600_34_5 |

|
|
Figure 3.

Wild type Aβ monomers can form different structural conformations, which lead to a variety of different dimers. Examples of conformers that propagate to (A) only one structural formation and (B) different oligomer conformations within the same structural class (i.e., ring). (C) In some instances the same Aβ monomer structure formed a homodimer that could form both ring and fibril oligomers.
Selection and characterization of oligomeric rings that form on the membrane
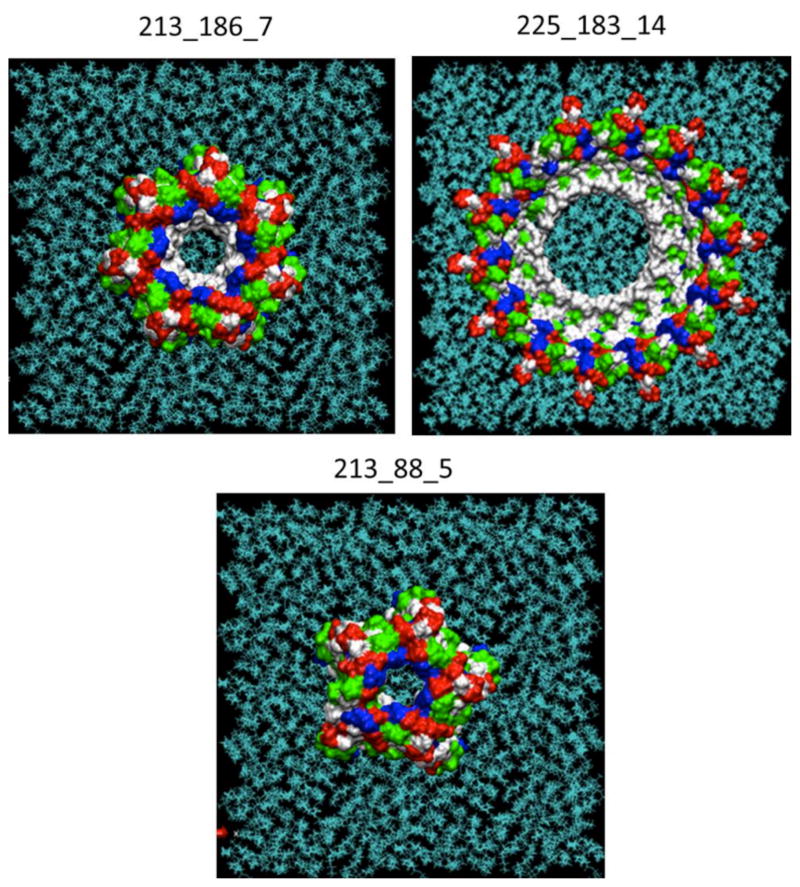
Next we determined which of the oligomer classes were stable in the membrane and therefore prone to form pore-like structures that are relevant to AD. Annular structures were filtered to identify ring structures that had a significant propensity to the membrane surface. For selection of the best membranephilic annular oligomers we used the PPM server [63]. The server calculates the energies of transfer (ΔGtransf) of the oligomers to the DOPS bilayer (Table 2). The accuracy of PPM predictions was tested on the large number of proteins and peptides and compared with the experimental studies [63]. In this method a protein is considered as a rigid body with flexible side chains. We regarded the oligomers having ΔGtransf better than −2 kcal/mol (per a monomer peptide of the ring) as membranephilic rings, which were selected for further analysis. Energy of transfer of the selected rings to the membrane was used as a criterion for selection of the rings that most probably would form and stay in the membrane. These rings can lead to a further pore formation.
Table 2.
Energies of transfer of the rings to the phospholipid membrane
| ID | Number of Aβ monomers in the ring | Energy of the ring transfer to the membrane (per monomer) kcal/mol |
|---|---|---|
| 213_88 | 5 | −4.18 |
| 225_283 | 5 | −4.1- |
| 213_186 | 7 | −3.57 |
| 436_27 | 5 | −3.22 |
| 134_7 | 8 | −3.15 |
| 2_191 | 5 | −2.98 |
| 600_34 | 5 | −2.98 |
| 648_27 | 6 | −2.70 |
| 22_54 | 7 | −2.26 |
| 51_232 | 8 | −2.23 |
| 22_60 | 8 | −2.23 |
| 109_93 | 6 | −2.13 |
| 69_56 | 7 | −2.07 |
| 423_15 | 7 | −1.91 |
| 277_21 | 5 | −1.82 |
| 120_20 | 7 | −1.63 |
| 487_75 | 7 | −1.47 |
| 85_122 | 13 | −1.34 |
| 85_122 | 7 | −1.34 |
| 2_74 | 7 | −0.99 |
| 334_176 | 20 | −0.96 |
| 106_239 | 8 | −0.86 |
| 184_7 | 16 | −0.74 |
| 334_227 | 4 | −0.70 |
| 389_24 | 8 | −0.56 |
| 98_139 | 6 | −0.35 |
| 430_57 | 15 | −0.27 |
| 22_4 | 14 | −0.26 |
It was Interesting to note that the possibility of the rings formation significantly changes during MD. The last conformers that could form annular oligomers were the 60 ns conformers. The time of around 60 ns was the boundary of existence for any helical secondary structure components in the Aβ peptide. After this point only the bends, turns, and coils corresponding to the fully unstructured peptide remained. It appears that such secondary structure components prevented the creation of the ring structures. We want to note that after 60 ns only fibrils could be constructed using the method of consecutive docking (Table S1). These results led to the assumption that ring formation is a property of rather early than later conformers of the Aβ peptide. It is still possible that in the later stages (that we have not explored yet) β-strands would appear that can again increase the probability of the ring-like structures formation. The rings having the best energies of interaction with the membrane (Table 2) are presented on Figure 4. These rings have more negative residues at the top (most distant from the surface of the membrane) and more positive residues at the bottom. This can create the possible profiles of the electrostatic field in their central parts (when embedded to the membrane) that would support the transport of positive ions, for example Ca2+.
Figure 4.
Membranephilic rings on the membrane. Visualization of the Aβ ring structures that have the top favorable energies of interactions with the membrane. The residues colors: white—apolar/hydrophobic, green—polar/hydrophilic, blue— positively charged, and red— negatively charged.
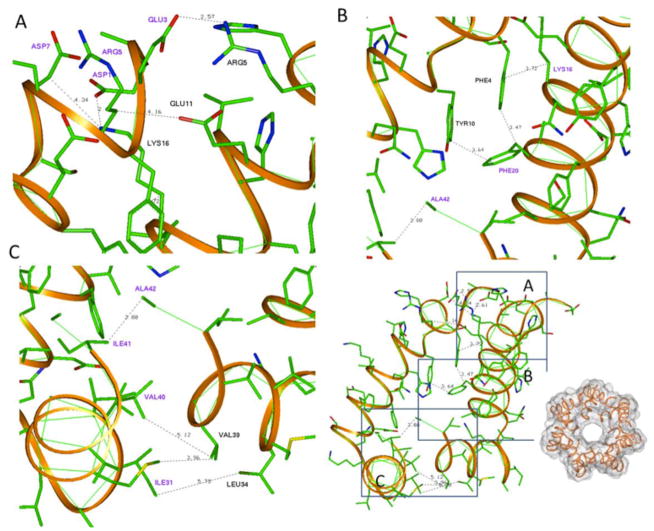
Of the 28 tested rings, one of the best membranephilic annular oligomer 213_186_7 (Figure 5) was further used to elucidate the possible contact residues between neighboring monomers. These data were comparable with data on the frequency of possible interresidue contacts within the Aβ rings. This contact scheme was then used to assign the possible mutation sites to confirm whether the proposed theoretical strategy including MD, docking, and membrane-contact calculations predicted the formation of toxic annular Aβ oligomers.
Figure 5.
Contact residues in neighboring peptides of Aβ participating in a membrane-contacting annular oligomer—representative of the rings having maximum membranephilicity. (A) Contacts of positive and negative residues were found for (B′) the N-terminal part of the dimer 231_186. Hydrophobic residues for the central part (C, C′) and the C-terminal part of the peptide (D, D′).
Identification of key amino acid residues within the dimers related to the formation of annular and fibril structures
Following the filtration steps to select Aβ oligomers that were most likely to form annular rings on the membrane surface and start the membrane penetration process, the frequency of interpeptide contacts in all selected annular formations was determined. We calculated the frequency of the residues involved in the intermolecular contacts between the neighboring monomers in the rings from the set of possible rings generated for the each clan of the conformers (Figure 6A). The residues with the most frequent contacts were divided into three groups, positively and negatively charged, as well as hydrophobic interactions (Figure 6A). The positively charged residues with the most frequent interactions were Arg5, Lys15, and Lys28, and the negatively charged aa residues included Glu3, Asp7, Glu11, Glu22, and Asp23. The hydrophobic residues of the C-terminal were Ile31, Leu34, Met35, Val36, Val39, Val40, and Ile41, while the hydrophobic residues in central part of the Aβ molecule were Val12, Leu17, Val18, Phe19, Phe20, and Val24. The positive residues Arg5 and Lys16 have complementary negative residues (Glu3 and Asp7) on the other side of the interface leading to a decrease in the electrostatic energy of the system and making the formation of the dimer energetically favorable for spontaneous dimerization. In addition, the number of interpeptide contacts for two neighboring Aβ monomers within rings (Figure 6B) and fibrils (Figure 6C) was determined. The regions with the highest contact densities in the ring were between the two neighboring C-terminals (residues 29–42) and the N-terminals (1–16). In fibrils, the highest contact densities were between the central hydrophobic cores (CHC; residues 17–21).
Figure 6.
Identification of key contact residues in neighboring conformers of Aβ participating in the membrane-contacting annular oligomers for selective mutation. (A) Percentage of contacts formed by both right and left peptide residues in the set of all possible membranephilic Aβ rings. Residue colors: negative (red), positive (blue), and hydrophobic (green). Percentage of interpeptide contacts formed in rings (B) and fibrils (C). One can see that the main sets of interaction in the rings are located at the N- and C-terminals and for the fibrils there are the interactions between the N-terminal and central parts of the peptide along with the interactions between its central parts.
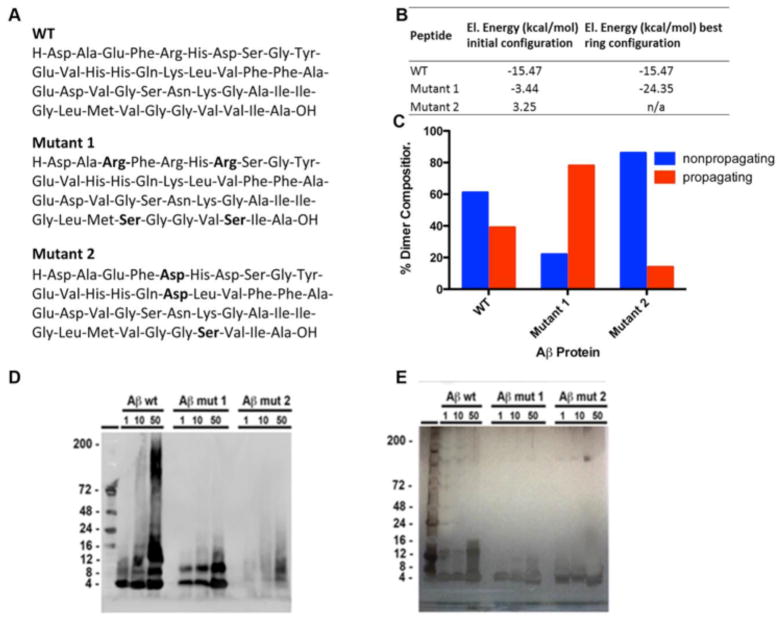
Based on the frequency analysis of the residues participating in the intermolecular contacts within the possible Aβ rings and the configuration of interface between the neighboring peptides in the ring 213_186_7, two Aβ mutants were created with an intention to analyze the possible impact of destroying the complementary contacts of the charged residues of the neighboring peptides in the dimer (Figure 7A). Mutant 1 had substituted negatively charged residues Glu3 and Asp7 for positively charged aa Arg on the “right” conformer (Figure 7A) of the interface between the neighbors of the ring (Figure 5). Additionally, the two hydrophobic residues, Val36 and Val40, were mutated to hydrophilic Ser to diminish the impact of the hydrophobic intermolecular interactions that could mask electrostatic interaction charges. Mutant 2 was designed to examine the “left” member of the interface. In this mutant, the positively charged residues Arg5 and Lys16 of the “left” Aβ molecule were substituted with negatively charged residue—Asp, and the hydrophobic residue Val39 was substituted with a hydrophilic residue—Ser.
Figure 7.
Aβ WT and mutants evaluated. (A) Amino acid sequence for WT and mutant Aβ where aa substitutions are indicated by bold-faced type. (B) Electrostatic energies of interaction between neighboring Aβ molecules in the dimer 213_186 and the mutants. The first column lists the energies for the mutants that were held in the same configuration as the WT with just substituted selected amino acids. Column 2 in the Fig. 7B table shows the energies for the new best energy dimers generated. If for WT dimer this is the same value, because the studied dimer is already in the best energy configuration, for mutants it is quite different. Mutant 1 showed even better energy of interpeptide interaction in the new ring when the peptides found a new configuration for the attractive interpeptide contacts (the positive residues found other complementary negative residues of the neighboring peptide). Mutant 2 in which actually all positively charged amino acids have been replaced to negative could not find any compensatory docking and simply did not form any ring structures.. (C) Percent of dimers formed that were propagating (blue bars) and nonpropagating (red bars) for the WT and mutant Aβ peptides. Compared to WT Aβ, Mutation 1 increased the number of propagating dimers, while Mutation 2 dramatically decreased the formation of propagating dimers indicating that the selected aa residues have key roles in the types of dimer that form. (D) Aβ mutant and WT peptide aggregates (1, 10 and 50 μM) were evaluated using Western blot probed using the 4G8 antibody. (E) While only the WT Aβ peptide formed aggregates that were detected in the Western blot silver stain verified the presence of the mutant peptides on the gel indicating that while the WT peptide forms a variety of aggregates having different molecular weights, key mutations to the primary structure of Aβ prevented the formation of larger aggregates.
We explored the opportunity to predict the impact of the mutations on ring formation and neural toxicity. We calculated the electrostatic energy of interpeptide interactions between the neighboring molecules in the ring. Initially we calculated the energy for the existing WT ring 213_186_7 and Mutants 1 and 2 having the same conformations and configuration as this WT ring. Then we performed other consecutive docking procedures for both mutants selecting the best-energy rings. Figure 7B shows the results of these modeling and calculations. The first column lists the energies for the mutants that were held in the same configuration as the WT with just substituted selected amino acids. It corresponds to the initial naïve suggestion that substitution of charged amino acids to the opposite charge amino acids will significantly decrease the energy of interpeptide interaction. In actuality, the peptides with the substituted amino acids simply do not dock in the same configuration as WT peptides. They try to find the best energy configuration of the docking complexes. Column 2 in the Figure 7B table shows exactly this result. Mutant 1 showed even better energy of interpeptide interaction in the new ring when the peptides found a new configuration for the attractive interpeptide contacts (the positive residues found other complementary negative residues of the neighboring peptide). Mutant 2, in which all positively charged amino acids were replaced with negative amino acids, could not find any compensatory docking and simply did not form any ring structures. In addition, the likelihood of the dimer propagating to the annular oligomers changed because of the introduced mutations. According to the theoretical calculations, compared to the WT dimer, which formed 61% nonpropagating dimers, Mutant 1 formed 22% nonpropagating dimers, and Mutant 2 formed 86% nonpropagating dimers (Figure 7C).
Targeted Aβ point mutations inhibited annular oligomer and fibril formation and modulated cellular toxicity in vitro
In order to confirm our modeling and simulation findings in an in-vitro system, the Aβ WT and two mutants were synthesized and evaluated by immunoblot, TEM, and cytotoxicity. Immunoblot analysis of Aβ aggregates with the 4G8 antibody revealed, as expected, that Aβ WT had the presence of monomers (4 kDa), dimers (8 kDa), and higher order oligomers (> 12 kDa) that were augmented with concentration (Figure 7D). In contrast, Aβ Mutant 1 formed mostly monomers, dimers, and trimers. Aβ Mutant 2 also formed monomers, dimers, and trimers but to a lesser extent than Mutant 1 (Figure 7D). The presence of the WT and mutant Aβ peptides was confirmed by silver staining (Figure 7E). Ultrastructural analysis by TEM showed that while the WT Aβ peptide formed small globular aggregates at 1 μM, at higher concentrations cluster aggregates (10 μM) and fibrils (50 μM) were formed. Interestingly, Mutant 1 formed globular or clustered aggregates at 10 μM, which were still present at 50 μM, but failed to form fibrils (Figure 8A), again suggesting that the selected mutations directly impacted the peptide’s structural conformations. Mutant 2 also formed globular aggregates at 50 μM, but to a much lesser extent than Mutant 1. In vitro, Mutant 1 showed a significant (p-value < 0.01; Figure 8C), dose-dependent decrease in the cellular sequestering of the calcein dye at 50 μM (Figure 8B) indicating greater cytotoxicity. In order to confirm the cytotoxicity, the LDH assay was utilized. We noted that Mutant 1 was cytotoxic in a dose-dependent manner. In contrast to Mutant 1, Mutant 2 was not cytotoxic at any of the tested concentrations (Figure 8C). Consistent with lacking cytotoxicity, Mutant 2 did not affect cellular permeability (calcein assay). Additionally, the targeted Aβ point mutations had a distinct mutation-dependent effect on cell survival (Figure 8B and C). While none of the peptides were cytotoxic at 1 μM both the WT and Mutant 1 significantly reduced the calcein retention and increased the release of LDH at 10 μM compared to vehicle control (p-value < 0.05; Figure 8B and C). In conclusion, substitution of negatively charged residues to positively charged residues in Mutant 1 resulted in greater formation of globular oligomers and toxicity. In contrast, substitution of positively to negatively charged residues in peptide Mutant 2 resulted in the failure to form globular oligomers and was not toxic. This is consistent with the prediction from the modeling data that showed Mutant 1 forming a much higher number of propagating dimers, while Mutant 2 formed much less propagating dimers.
Figure 8.

In-vitro analysis of Aβ WT and mutant peptide aggregates using Western blot, electron microscopy, and cell-based assays. (A) TEM evaluation at 1, 10, and 50 μM further confirmed these findings since only Aβ WT formed protofibrils and fibrils, while the mutants remained as small oligomeric peptides. (B) The toxicity of the Aβ peptides was evaluated using adult rat hippocampal neural stem cells at 1, 10, and 50 μM and indicated that while Mutant 2 was not toxic at any concentration, Mutant 1 had a clear increase in toxicity with increasing dose. WT Aβ was toxic at both 10 and 50 μM concentrations. (C) Changes in intracellular calcium concentrations were evaluated using the calcein assay. Cells were treated with Aβ WT and mutant peptides (1, 10, and 50 μM). WT Aβ altered the endogenous calcium levels, as did Mutant 1, which had a significant dose-dependent effect. On the other hand, Mutant 2 did not affect intracellular calcium concentrations. ANOVA followed by post hoc Dunnett’s or Tukey-Kramer tests. *p < 0.05; #p < 0.01.
Discussion
The present study investigated the structural diversity and fate of Aβ42 dimers utilizing MD simulations to generate a large array of Aβ42 conformers that were then used for modeling in propagating mode the higher order oligomers. Remarkably, we showed a greater diversity in Aβ homodimers compared to previous studies [48]. Depending on the clan family, different types of Aβ42 homodimers were obtained. While some had the tendency to evolve during our propagation modeling into oligomeric rings others formed fibrils of diverse characteristics (Table 1; Figure 3).
Prior to this study, few publications evaluated the structural characteristics of Aβ dimers based on modeling simulation techniques [47, 48, 68, 69]. While the study by Urbanc and colleagues [68] used DMD with a four-bead peptide model to show that unlike Aβ40, Aβ42 had a high propensity to form paranuclei (pentameric or hexameric) structures that could self-associate into higher-order oligomers, the study by Mitternacht and coworkers [69] used implicit solvent all-atom Monte Carlo simulations to show spontaneous dimerization of Aβ42, with 42 residues. They showed that the primary force driving the formation of dimers is hydrophobic attraction and that the conformational differences involve turns centered in the 20–30 region. In addition, that the probability of finding turns centered in the 25–30 region, where there is a loop in Aβ fibrils, was found to increase upon dimerization. In contrast, Takeda and Klimov [47] utilized replica exchange molecular dynamics and all-atom implicit solvent model to show that the sequence region 10–23 in Aβ peptide was found to form most of interpeptide interactions upon aggregation.
In the recent study Côté and co-authors [48] used accurate coarse-grained force field coupled to Hamiltonian-temperature replica exchange molecular dynamics to show that the Aβ42 dimer has a higher propensity than the Aβ40 dimer to form β-strands at the central hydrophobic core (residues 17–21) and at the C-terminal (residues 30–42), which are two segments crucial to the oligomerization of Aβ. This study identified six main cluster centers for Aβ42 dimers. Overall, their analysis pointed to the strong impact of the two aa Ile41–Ala42 and the salt-bridge D23–K28 on the folding of the Aβ dimer. Consistent with this and previous reports [45], we found that nearly one-third of the 28 evaluated membranephilic ring oligomer interfaces between the neighboring Aβ monomers contained possible salt bridges between the residue K28 from one side and either residue E22 or D23 from the other.
Also in agreement with previous simulation studies [47, 48, 68, 69], we found that a significant number of the intermolecular interactions in the homodimers that can evolve to fibrils were hydrophobic involving the central region (CHC) residues 17–28). In our all-atom MD simulations, a higher proportion of α-helical structures were present that gradually transformed to π-helices and then to coil and turn structures that would most probably later evolve to β-strands. The proportions of coils and turns were similar to previous reports [48]. The π-helix is known to be a transitional secondary structure in the evolution of the peptide from helical to nonhelical structures (turns, coils, and β-strands). This transformation of the Aβ secondary structure has been confirmed by combined NMR analysis and MD simulations [70, 71].
Further studies evaluating the assembly of dimers as well as the role of the lipid membrane in dimer and oligomer formation are limited due to the added computational complexities associated with additional elements that are modeled. Oligomer formation studies have identified a stable double-layer ring structure, but not single-layer ring, that formed from the β-strand conformation [43, 44], as well as an Aβ pentamer, which formed via linear propagation of monomers [72]. Interestingly, Aβ monomers modeled in the presence of a self-assembled monolayer (SAM) and bilayer membranes suggested that the peptide prefers to adopt an α-helical or unstructured conformation [73], rather than the β-hairpin structure identified by modeling the monomer in a water box [74, 75]. Self-assembled monolayer also had a strong seeding effect on Aβ, and a trimer was identified as the smallest seeding nucleus [73]. In addition to SAM, Aβ was modeled on a 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) membrane using all-atom traditional MD. Under these conditions, α-helices evolved toward β-strand structures had a higher propensity to aggregate. Finally, the amount of energy required to disconnect an Aβ dimer from the membrane were calculated as a measure of the interactions between the dimers and the membrane. Similar to a previous study, a number of membranephilic dimers were identified, although comparison with the energy to disconnect the Aβ monomer were not reported [76].
Our study differs from other MD reports [48]. We first used explicit MD and then docking to obtain a greater variety of Aβ42 dimers. This enabled the selection of conformational clans, centroids from which were then used for modeling the homodimers and higher oligomers. With this new approach a large variety of Aβ42 dimers was found that, depending on the unique conformation, evolved into oligomer rings or fibrils. This finding supports the notion that the dimers are the starting configurations for the further construction of higher-order Aβ oligomers and that different homodimers can be organized from different conformations of Aβ monomers. Each of the dimer classes and subclasses differ only in conformation but not in a sequence. The precise secondary and tertiary characteristics that differentiate each subclass of dimers forming rings and fibrils are unknown and will be the subject of future more detailed molecular studies. As part of a more detailed analysis of the ring oligomers, we calculated the propensity of the rings to form on the membrane, as well as, which of the rings had the best energies of interaction with the membrane; these rings were selected for further analysis.
To validate the MD studies, two artificial mutants of Aβ were created to modulate the main intermolecular interfaces in Aβ rings that had been found by in-silico modelling. Mutant 1 substituted negatively charged residues for positively charged on the “left” conformer of the interface. Mutant 2 was designed to examine the “right” member of the interface. While Mutant 1 enhanced the formation of ring-like oligomers and toxicity, Mutant 2 blocked the formation of toxic oligomers. A possible explanation as to how these mutants work is that when we mutated the positive residues Arg5 and Lys16 on the “right” monomer this mutation eliminated two of the three most frequently occurring positive residues of the Aβ dimer interface. Since the dimer is formed by a combination of hydrophobic and electrostatic interactions, it is clear that the one remaining positive residue (Lys28) could not maintain the necessary complimentary electrostatic field for a proper configuration. This most likely leads to the warped interface that would not evolve to ring or even fibril structures. On the other hand, eliminating the negative charges (residues Glu3 and Asp7) on the “left” monomer, removes only two of five negative residues that most frequently participate in the interface. Thus just the configuration of this interface would change and lead to a dimer configuration that frequently propagates to more toxic annular oligomers.
Note that in our theoretical studies the annular oligomers have been formed only from the centroids of clans that include the conformers obtained before 60 ns of MD. After this time point we could not generate any ring structures but only fibrils. Such results can lead to a hypothesis that for the formation of annular oligomers the Aβ conformer has to have at least some of α- or π-helical structural components. After the conformer lost all helical structures, no more rings would be formed. Such a hypothesis leads to an assumption that the Aβ rings are formed at the early times after peptide creation. It can explain failures of numerous attempts to stop the rings formation using the molecules that can prevent docking of one Aβ peptide to another. The compounds that can protect Aβ peptides from binding to each other are present in the cell after the ring formation, during the time when the unstructured Aβ already cannot form any rings but only fibrils. In AD, memory and cognitive deficits has been associated with impairments of the synaptic connections among neurons in association areas of the pre-frontal and parietal cortices and limbic system [77, 78]. The mechanisms of synaptic pathology in AD are not completely understood, however several studies point to toxic oligomers including dimers.[9] Aβ dimers were identified in cerebral spinal cord fluid in AD patients [10] and potently disrupt hippocampal LTP, adversely affect dendritic spines in cultured neurons and impair memory consolidation [9, 79]. Although it is unlikely that inhibition of memory consolidation will be associated with a unique Aβ structure or assembly form, the results presented here together with those from prior studies [9, 10, 80] strongly suggest that soluble diffusible dimers and/or prefibrillar intermediates built starting from of Aβ dimers are central stating point in the formation of memory-impairing arrays.
In conclusion, the present study suggests a more complex and dynamical role for Aβ dimers in the process oligomerization and fibril formation. It also suggests that although dimers might be the key starting points for oligomerization, this is a rapidly changing process where diverse conformers and oligomers of various orders are formed and eliminated very quickly. Thus not all dimers behave the same way. Depending on the subclass, some types of dimers do not form oligomers while others do. Overall this, as well as, previous results suggest that targeting selected Aβ dimers may be important in an effort to ameliorate the episodic memory described in MCI and the early stages of AD.
Acknowledgments
This work was supported by NIH grants AG18440, AG031097, AG043384, and AG022074. The authors are grateful for computational support from the San Diego Supercomputer Center at UCSD.
Footnotes
Conflict of Interests
The authors of the manuscript do not have direct financial relations with the commercial identities mentioned in your paper that might lead to a conflict of interest for any of the authors.
References
- 1.Lashuel HA, Hartley DM, Petre BM, Wall JS, Simon MN, Walz T, Lansbury PT., Jr Mixtures of wild-type and a pathogenic (E22G) form of Abeta40 in vitro accumulate protofibrils, including amyloid pores. J Mol Biol. 2003;332:795–808. doi: 10.1016/s0022-2836(03)00927-6. [DOI] [PubMed] [Google Scholar]
- 2.Klein WL. ADDLs & protofibrils--the missing links? Neurobiol Aging. 2002;23:231–235. doi: 10.1016/s0197-4580(01)00312-8. [DOI] [PubMed] [Google Scholar]
- 3.Klein WL, Krafft GA, Finch CE. Targeting small Abeta oligomers: the solution to an Alzheimer’s disease conundrum? Trends Neurosci. 2001;24:219–224. doi: 10.1016/s0166-2236(00)01749-5. [DOI] [PubMed] [Google Scholar]
- 4.Iwatsubo T, Odaka A, Suzuki N, Mizusawa H, Nukina N, Ihara I. Visualization of A beta 42(43) and A beta 40 in senile plaques with end-specific A beta monoclonals: evidence that an initially deposited species in A beta 42(43) Neuron. 1994;13:45–53. doi: 10.1016/0896-6273(94)90458-8. [DOI] [PubMed] [Google Scholar]
- 5.Sakono M, Zako T. Amyloid oligomers: formation and toxicity of Abeta oligomers. FEBS J. 2010;277:1348–1358. doi: 10.1111/j.1742-4658.2010.07568.x. [DOI] [PubMed] [Google Scholar]
- 6.Lesne S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, Gallagher M, Ashe KH. A specific amyloid-beta protein assembly in the brain impairs memory. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- 7.Lambert MP, Barlow AK, Chromy BA, Edwards C, Freed R, Liosatos M, Morgan TE, Rozovsky I, Trommer B, Viola KL, Wals P, Zhang C, Finch CE, Krafft GA, Klein WL. Diffusible, nonfibrillar ligands derived from Abeta1–42 are potent central nervous system neurotoxins. Proc Natl Acad Sci U S A. 1998;95:6448–6453. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 9.Shankar GM, Li S, Mehta TH, Garcia-Munoz A, Shepardson NE, Smith I, Brett FM, Farrell MA, Rowan MJ, Lemere CA, Regan CM, Walsh DM, Sabatini BL, Selkoe DJ. Amyloid-beta protein dimers isolated directly from Alzheimer’s brains impair synaptic plasticity and memory. Nat Med. 2008;14:837–842. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klyubin I, Betts V, Welzel AT, Blennow K, Zetterberg H, Wallin A, Lemere CA, Cullen WK, Peng Y, Wisniewski T, Selkoe DJ, Anwyl R, Walsh DM, Rowan MJ. Amyloid beta protein dimer-containing human CSF disrupts synaptic plasticity: prevention by systemic passive immunization. J Neurosci. 2008;28:4231–4237. doi: 10.1523/JNEUROSCI.5161-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lansbury PT., Jr Evolution of amyloid: what normal protein folding may tell us about fibrillogenesis and disease. Proc Natl Acad Sci U S A. 1999;96:3342–3344. doi: 10.1073/pnas.96.7.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 13.Barrow C, Yasuda A, Kenny P, Zagorski M. Solution conformations and aggregational properties of synthetic amyloid beta-peptides of Alzheimer’s disease. Analysis of circular dichroism spectra. J Mol Biol. 1992;225:1075–1093. doi: 10.1016/0022-2836(92)90106-t. [DOI] [PubMed] [Google Scholar]
- 14.Jarrett JT, Lansbury PT., Jr Seeding “one-dimensional crystallization” of amyloid: a pathogenic mechanism in Alzheimer’s disease and scrapie? Cell. 1993;73:1055–1058. doi: 10.1016/0092-8674(93)90635-4. [DOI] [PubMed] [Google Scholar]
- 15.Bitan G, Kirkitadze MD, Lomakin A, Vollers SS, Benedek GB, Teplow DB. Amyloid beta -protein (Abeta) assembly: Abeta 40 and Abeta 42 oligomerize through distinct pathways. Proc Natl Acad Sci U S A. 2003;100:330–335. doi: 10.1073/pnas.222681699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riek R, Guntert P, Dobeli H, Wipf B, Wuthrich K. NMR studies in aqueous solution fail to identify significant conformational differences between the monomeric forms of two Alzheimer peptides with widely different plaque-competence, A beta(1–40)(ox) and A beta(1–42)(ox) Eur J Biochem. 2001;268:5930–5936. doi: 10.1046/j.0014-2956.2001.02537.x. [DOI] [PubMed] [Google Scholar]
- 17.Baumketner A, Krone MG, Shea JE. Role of the familial Dutch mutation E22Q in the folding and aggregation of the 15–28 fragment of the Alzheimer amyloid-beta protein. Proc Natl Acad Sci U S A. 2008;105:6027–6032. doi: 10.1073/pnas.0708193105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cruz L, Urbanc B, Borreguero JM, Lazo ND, Teplow DB, Stanley HE. Solvent and mutation effects on the nucleation of amyloid beta-protein folding. Proc Natl Acad Sci U S A. 2005;102:18258–18263. doi: 10.1073/pnas.0509276102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fawzi NL, Phillips AH, Ruscio JZ, Doucleff M, Wemmer DE, Head-Gordon T. Structure and dynamics of the Abeta(21–30) peptide from the interplay of NMR experiments and molecular simulations. J Am Chem Soc. 2008;130:6145–6158. doi: 10.1021/ja710366c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gnanakaran S, Nussinov R, Garcia AE. Atomic-level description of amyloid beta-dimer formation. J Am Chem Soc. 2006;128:2158–2159. doi: 10.1021/ja0548337. [DOI] [PubMed] [Google Scholar]
- 21.Han W, Wu YD. A strand-loop-strand structure is a possible intermediate in fibril elongation: long time simulations of amyloid-beta peptide (10–35) J Am Chem Soc. 2005;127:15408–15416. doi: 10.1021/ja051699h. [DOI] [PubMed] [Google Scholar]
- 22.Khandogin J, Brooks CL., 3rd Linking folding with aggregation in Alzheimer’s beta-amyloid peptides. Proc Natl Acad Sci U S A. 2007;104:16880–16885. doi: 10.1073/pnas.0703832104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klimov DK, Straub JE, Thirumalai D. Aqueous urea solution destabilizes Abeta(16–22) oligomers. Proc Natl Acad Sci U S A. 2004;101:14760–14765. doi: 10.1073/pnas.0404570101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krone MG, Hua L, Soto P, Zhou R, Berne BJ, Shea JE. Role of water in mediating the assembly of Alzheimer amyloid-beta Abeta16–22 protofilaments. J Am Chem Soc. 2008;130:11066–11072. doi: 10.1021/ja8017303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lakdawala AS, Morgan DM, Liotta DC, Lynn DG, Snyder JP. Dynamics and fluidity of amyloid fibrils: a model of fibrous protein aggregates. J Am Chem Soc. 2002;124:15150–15151. doi: 10.1021/ja0273290. [DOI] [PubMed] [Google Scholar]
- 26.Ma B, Nussinov R. Stabilities and conformations of Alzheimer’s beta -amyloid peptide oligomers (Abeta 16–22, Abeta 16–35, and Abeta 10–35): Sequence effects. Proc Natl Acad Sci U S A. 2002;99:14126–14131. doi: 10.1073/pnas.212206899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen PH, Li MS, Stock G, Straub JE, Thirumalai D. Monomer adds to preformed structured oligomers of Abeta-peptides by a two-stage dock-lock mechanism. Proc Natl Acad Sci U S A. 2007;104:111–116. doi: 10.1073/pnas.0607440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santini S, Mousseau N, Derreumaux P. In silico assembly of Alzheimer’s Abeta16–22 peptide into beta-sheets. J Am Chem Soc. 2004;126:11509–11516. doi: 10.1021/ja047286i. [DOI] [PubMed] [Google Scholar]
- 29.Tarus B, Straub JE, Thirumalai D. Dynamics of Asp23-Lys28 salt-bridge formation in Abeta10–35 monomers. J Am Chem Soc. 2006;128:16159–16168. doi: 10.1021/ja064872y. [DOI] [PubMed] [Google Scholar]
- 30.Wu C, Wang Z, Lei H, Zhang W, Duan Y. Dual binding modes of Congo red to amyloid protofibril surface observed in molecular dynamics simulations. J Am Chem Soc. 2007;129:1225–1232. doi: 10.1021/ja0662772. [DOI] [PubMed] [Google Scholar]
- 31.Bernstein SL, Wyttenbach T, Baumketner A, Shea JE, Bitan G, Teplow DB, Bowers MT. Amyloid beta-protein: monomer structure and early aggregation states of Abeta42 and its Pro19 alloform. J Am Chem Soc. 2005;127:2075–2084. doi: 10.1021/ja044531p. [DOI] [PubMed] [Google Scholar]
- 32.Dong X, Chen W, Mousseau N, Derreumaux P. Energy landscapes of the monomer and dimer of the Alzheimer’s peptide Abeta(1–28) J Chem Phys. 2008;128:125108. doi: 10.1063/1.2890033. [DOI] [PubMed] [Google Scholar]
- 33.Itoh SG, Okamoto Y. Amyloid-beta(29–42) dimer formations studied by a multicanonical-multioverlap molecular dynamics simulation. J Phys Chem B. 2008;112:2767–2770. doi: 10.1021/jp712170h. [DOI] [PubMed] [Google Scholar]
- 34.Jang S, Shin S. Amyloid beta-peptide oligomerization in silico: dimer and trimer. J Phys Chem B. 2006;110:1955–1958. doi: 10.1021/jp055568e. [DOI] [PubMed] [Google Scholar]
- 35.Lam AR, Teplow DB, Stanley HE, Urbanc B. Effects of the Arctic (E22-->G) mutation on amyloid beta-protein folding: discrete molecular dynamics study. J Am Chem Soc. 2008;130:17413–17422. doi: 10.1021/ja804984h. [DOI] [PubMed] [Google Scholar]
- 36.Urbanc B, Cruz L, Yun S, Buldyrev SV, Bitan G, Teplow DB, Stanley HE. In silico study of amyloid beta-protein folding and oligomerization. Proc Natl Acad Sci U S A. 2004;101:17345–17350. doi: 10.1073/pnas.0408153101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borreguero JM, Urbanc B, Lazo ND, Buldyrev SV, Teplow DB, Stanley HE. Folding events in the 21–30 region of amyloid beta-protein (Abeta) studied in silico. Proc Natl Acad Sci U S A. 2005;102:6015–6020. doi: 10.1073/pnas.0502006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jang H, Hall CK, Zhou Y. Assembly and kinetic folding pathways of a tetrameric beta-sheet complex: molecular dynamics simulations on simplified off-lattice protein models. Biophys J. 2004;86:31–49. doi: 10.1016/S0006-3495(04)74081-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng S, Ding F, Urbanc B, Buldyrev SV, Cruz L, Stanley HE, Dokholyan NV. Discrete molecular dynamics simulations of peptide aggregation. Phys Rev E Stat Nonlin Soft Matter Phys. 2004;69:041908. doi: 10.1103/PhysRevE.69.041908. [DOI] [PubMed] [Google Scholar]
- 40.Urbanc B, Cruz L, Ding F, Sammond D, Khare S, Buldyrev SV, Stanley HE, Dokholyan NV. Molecular dynamics simulation of amyloid beta dimer formation. Biophys J. 2004;87:2310–2321. doi: 10.1529/biophysj.104.040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yun S, Urbanc B, Cruz L, Bitan G, Teplow DB, Stanley HE. Role of electrostatic interactions in amyloid beta-protein (A beta) oligomer formation: a discrete molecular dynamics study. Biophys J. 2007;92:4064–4077. doi: 10.1529/biophysj.106.097766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee C, Ham S. Characterizing amyloid-beta protein misfolding from molecular dynamics simulations with explicit water. J Comput Chem. 2011;32:349–355. doi: 10.1002/jcc.21628. [DOI] [PubMed] [Google Scholar]
- 43.Zheng J, Jang H, Ma B, Nussinov R. Annular structures as intermediates in fibril formation of Alzheimer Abeta17–42. J Phys Chem B. 2008;112:6856–6865. doi: 10.1021/jp711335b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng J, Jang H, Ma B, Tsai CJ, Nussinov R. Modeling the Alzheimer Abeta17–42 fibril architecture: tight intermolecular sheet-sheet association and intramolecular hydrated cavities. Biophys J. 2007;93:3046–3057. doi: 10.1529/biophysj.107.110700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anand P, Nandel FS, Hansmann UH. The Alzheimer’s beta amyloid (Abeta1–39) monomer in an implicit solvent. J Chem Phys. 2008;128:165102. doi: 10.1063/1.2907718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer’s beta-amyloid fibrils. Biochemistry (Mosc) 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takeda T, Klimov DK. Interpeptide interactions induce helix to strand structural transition in Abeta peptides. Proteins. 2009;77:1–13. doi: 10.1002/prot.22406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cote S, Laghaei R, Derreumaux P, Mousseau N. Distinct dimerization for various alloforms of the amyloid-beta protein: Abeta(1–40), Abeta(1–42), and Abeta(1–40)(D23N) J Phys Chem B. 2012;116:4043–4055. doi: 10.1021/jp2126366. [DOI] [PubMed] [Google Scholar]
- 49.Yu X, Zheng J. Polymorphic structures of Alzheimer’s beta-amyloid globulomers. PloS one. 2011;6:e20575. doi: 10.1371/journal.pone.0020575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barz B, Urbanc B. Dimer formation enhances structural differences between amyloid beta-protein (1–40) and (1–42): an explicit-solvent molecular dynamics study. PloS one. 2012;7:e34345. doi: 10.1371/journal.pone.0034345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu X, Bora RP, Barman A, Singh R, Prabhakar R. Dimerization of the full-length Alzheimer amyloid beta-peptide (Abeta42) in explicit aqueous solution: a molecular dynamics study. J Phys Chem B. 2012;116:4405–4416. doi: 10.1021/jp210019h. [DOI] [PubMed] [Google Scholar]
- 52.Tsigelny IF, Bar-On P, Sharikov Y, Crews L, Hashimoto M, Miller MA, Keller SH, Platoshyn O, Yuan JX, Masliah E. Dynamics of alpha-synuclein aggregation and inhibition of pore-like oligomer development by beta-synuclein. FEBS J. 2007;274:1862–1877. doi: 10.1111/j.1742-4658.2007.05733.x. [DOI] [PubMed] [Google Scholar]
- 53.Tsigelny IF, Crews L, Desplats P, Shaked GM, Sharikov Y, Mizuno H, Spencer B, Rockenstein E, Trejo M, Platoshyn O, Yuan JX, Masliah E. Mechanisms of hybrid oligomer formation in the pathogenesis of combined Alzheimer’s and Parkinson’s diseases. PloS one. 2008;3:e3135. doi: 10.1371/journal.pone.0003135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crescenzi O, Tomaselli S, Guerrini R, Salvadori S, D’Ursi AM, Temussi PA, Picone D. Solution structure of the Alzheimer amyloid beta-peptide (1–42) in an apolar microenvironment. Similarity with a virus fusion domain. Eur J Biochem. 2002;269:5642–5648. doi: 10.1046/j.1432-1033.2002.03271.x. [DOI] [PubMed] [Google Scholar]
- 55.Kalé L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K, Schulten K. NAMD2: Greater scalability for parallel molecular dynamics. J Comput Phys. 1999;151:282–312. [Google Scholar]
- 56.Feller S, MacKerell A. An improved empirical potential energy function for molecular simulations of phospholipids. J Phys Chem B. 2000;104:7510–7515. [Google Scholar]
- 57.Tu K, Tobias DJ, Klein ML. Constant pressure and temperature molecular dynamics simulation of a fully hydrated liquid crystal phase dipalmitoylphosphatidylcholine bilayer. Biophys J. 1995;69:2558–2562. doi: 10.1016/S0006-3495(95)80126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feller S, Zhang Y, Pastor RW, Brooks BR. Constant pressure molecular dynamics simulation: The Langevin piston method. J Chem Phys. 1995;103:4613–4621. [Google Scholar]
- 59.Essmann U, Perera L, Berkowitz ML. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 60.Shindyalov IN, Bourne PE. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 61.Macindoe G, Mavridis L, Venkatraman V, Devignes MD, Ritchie DW. HexServer: an FFT-based protein docking server powered by graphics processors. Nucleic Acids Res. 2010;38:W445–449. doi: 10.1093/nar/gkq311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sharikov Y, Walker RC, Greenberg J, Kouznetsova V, Nigam SK, Miller MA, Masliah E, Tsigelny IF. MAPAS: a tool for predicting membrane-contacting protein surfaces. Nat Methods. 2008;5:119. doi: 10.1038/nmeth0208-119. [DOI] [PubMed] [Google Scholar]
- 63.Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 2012;40:D370–376. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kucerka N, Nagle JF, Sachs JN, Feller SE, Pencer J, Jackson A, Katsaras J. Lipid bilayer structure determined by the simultaneous analysis of neutron and X-ray scattering data. Biophys J. 2008;95:2356–2367. doi: 10.1529/biophysj.108.132662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marsh D. Membrane water-penetration profiles from spin labels. Eur Biophys J. 2002;31:559–562. doi: 10.1007/s00249-002-0245-z. [DOI] [PubMed] [Google Scholar]
- 66.Lomize AL, Pogozheva ID, Mosberg HI. Anisotropic solvent model of the lipid bilayer. 2. Energetics of insertion of small molecules, peptides, and proteins in membranes. J Chem Inf Model. 2011;51:930–946. doi: 10.1021/ci200020k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Rooijen BD, Claessens MM, Subramaniam V. Lipid bilayer disruption by oligomeric alpha-synuclein depends on bilayer charge and accessibility of the hydrophobic core. Biochim Biophys Acta. 2009;1788:1271–1278. doi: 10.1016/j.bbamem.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 68.Urbanc B, Betnel M, Cruz L, Bitan G, Teplow DB. Elucidation of amyloid beta-protein oligomerization mechanisms: discrete molecular dynamics study. J Am Chem Soc. 2010;132:4266–4280. doi: 10.1021/ja9096303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mitternacht S, Staneva I, Hard T, Irback A. Monte Carlo study of the formation and conformational properties of dimers of Abeta42 variants. J Mol Biol. 2011;410:357–367. doi: 10.1016/j.jmb.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 70.Armen R, Alonso DO, Daggett V. The role of alpha-, 3(10)-, and pi-helix in helix-->coil transitions. Prot Sci. 2003;12:1145–1157. doi: 10.1110/ps.0240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Espinoza-Fonseca LM, Kast D, Thomas DD. Molecular dynamics simulations reveal a disorder-to-order transition on phosphorylation of smooth muscle myosin. Biophys J. 2007;93:2083–2090. doi: 10.1529/biophysj.106.095802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Masman MF, Eisel UL, Csizmadia IG, Penke B, Enriz RD, Marrink SJ, Luiten PG. In silico study of full-length amyloid beta 1–42 tri- and penta-oligomers in solution. J Phys Chem B. 2009;113:11710–11719. doi: 10.1021/jp901057w. [DOI] [PubMed] [Google Scholar]
- 73.Zhao J, Wang Q, Liang G, Zheng J. Molecular dynamics simulations of low-ordered alzheimer beta-amyloid oligomers from dimer to hexamer on self-assembled monolayers. Langmuir. 2011;27:14876–14887. doi: 10.1021/la2027913. [DOI] [PubMed] [Google Scholar]
- 74.Wang Q, Zhao J, Yu X, Zhao C, Li L, Zheng J. Alzheimer Abeta(1–42) monomer adsorbed on the self-assembled monolayers. Langmuir. 2010;26:12722–12732. doi: 10.1021/la1017906. [DOI] [PubMed] [Google Scholar]
- 75.Wang Q, Zhao C, Zhao J, Wang J, Yang JC, Yu X, Zheng J. Comparative molecular dynamics study of Abeta adsorption on the self-assembled monolayers. Langmuir. 2010;26:3308–3316. doi: 10.1021/la903070y. [DOI] [PubMed] [Google Scholar]
- 76.Davis CH, Berkowitz ML. A molecular dynamics study of the early stages of amyloid-beta(1–42) oligomerization: the role of lipid membranes. Proteins. 2010;78:2533–2545. doi: 10.1002/prot.22763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Terry RD, Masliah E, Salmon DP, Butters N, DeTeresa R, Hill R, Hansen LA, Katzman R. Physical basis of cognitive alterations in Alzheimer’s disease: synapse loss is the major correlate of cognitive impairment. Ann Neurol. 1991;30:572–580. doi: 10.1002/ana.410300410. [DOI] [PubMed] [Google Scholar]
- 78.DeKosky ST, Scheff SW. Synapse loss in frontal cortex biopsies in Alzheimer’s disease: correlation with cognitive severity. Ann Neurol. 1990;27:457–464. doi: 10.1002/ana.410270502. [DOI] [PubMed] [Google Scholar]
- 79.Freir DB, Fedriani R, Scully D, Smith IM, Selkoe DJ, Walsh DM, Regan CM. Abeta oligomers inhibit synapse remodelling necessary for memory consolidation. Neurobiol Aging. 2011;32:2211–2218. doi: 10.1016/j.neurobiolaging.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.O’Nuallain B, Freir DB, Nicoll AJ, Risse E, Ferguson N, Herron CE, Collinge J, Walsh DM. Amyloid beta-protein dimers rapidly form stable synaptotoxic protofibrils. J Neurosci. 2010;30:14411–14419. doi: 10.1523/JNEUROSCI.3537-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]